Abstract
Treatment of locally advanced rectal cancer includes chemotherapy, radiation, and surgery but patient responses to neoadjuvant treatment are variable. We have shown that rectal tumors are comprised of multiple genetically distinct sub-clones. Unique sub-clones within tumors may harbor mutations which contribute to inter-patient variation in response to neoadjuvant chemoradiotherapy (nCRT). Analysis of the influence of nCRT on the extent and nature of intra-tumoral genetic heterogeneity in rectal cancer may provide insights into mechanisms of resistance.
Locally advanced rectal cancer patients underwent pre-treatment biopsies. At the time of surgery, tissue from the treated tumor was obtained and analyzed. Pre- and post-treatment specimens were subjected to whole exome and confirmatory deep sequencing for somatic mutations. Copy number variation was assessed using OncoScan SNP arrays. Genomic data were analyzed using PyClone to identify sub-clonal tumor population following nCRT. Alterations that persisted or were enriched in the post-treatment tumor specimen following nCRT were defined for each patient.
Thirty-two samples were obtained from ten patients. PyClone identified 2 to 10 genetic sub-clones per tumor. Substantial changes in the proportions of individual sub-clones in pre- versus post-treatment tumor material were found in all patients. Resistant sub-clones recurrently contained mutations in TP53, APC, ABCA13, MUC16, and THSD4. Recurrent copy number variation was observed across multiple chromosome regions after nCRT. Pathway analysis including variant alleles and copy number changes associated with resistant sub-clones revealed significantly altered pathways, especially those linked to the APC and TP53 genes, which were the two most frequently mutated genes.
Intra-tumoral heterogeneity is evident in pre-treatment rectal cancer. Following treatment, sub-clonal populations are selectively modified and enrichment of a subset of pre-treatment sub-clones is seen. Further studies are needed to define recurrent alterations at diagnosis that may contribute to resistance to nCRT.
Abbreviations: CRC, colorectal cancer; SNP, single nucleotide polymorphism; nCRT, neoadjuvant chemoradiotherapy; cCR, clinical complete response
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth most common cause of cancer death globally [1]. Rectal cancer accounts for approximately one third of all CRC's [2]. Due to an increased risk of local recurrence, treatment of locally advanced rectal cancer requires a multimodal approach which includes neoadjuvant chemoradiotherapy (nCRT), followed by surgery including a total mesorectal excision (TME) with partial or complete proctectomy and often a temporary or permanent ostomy [3]. Approximately 20% of rectal cancer patients who undergo nCRT will have a complete response (CR), or no detectable tumor during preoperative evaluation or at the time or surgery [4], [5]. However, to date there have been no identified biomarkers that accurately predict the degree of response in a given patient.
Being able to predict who will and will not respond to cancer therapies has substantial clinical significance for cancer patients in general and rectal cancer patients specifically. Nonetheless, this continues to be an elusive challenge due in part to intra-tumoral heterogeneity, where single tumors are composed of multiple sub-clones that develop through complex evolutionary trajectories [6]. These sub-clones may have differential growth potential, metastatic potential, and resistance to cancer therapies [6]. We and others have shown that CRC exhibits intra-tumor genetic heterogeneity [7], [8], where multiple sub-clones exist in the primary tumor harbor shared somatic alterations along with diverse and sub-clone-preferential or -specific somatic point mutations and/or copy number changes [7], [9]. We hypothesized that intra-tumoral heterogeneity may impact on the effectiveness of nCRT because some sub-clones are sensitive to treatment and thus eliminated and others are resistant and therefore, persist. Analysis of the nature and extent of intra-tumoral heterogeneity in the primary tumors of individual rectal cancer patients before and after nCRT might provide insights into possible factors and mechanisms in therapeutic resistance and might also help to highlight opportunities for more effective treatment and improved outcomes in patients with rectal cancer.
In the work presented here, we studied multiple primary rectal tumor regions pre- and post-nCRT from 10 rectal cancer patients, with the goal of understanding treatment effects on genetic sub-clonal populations. We found that the genetic composition of each patient's tumor was changed substantially by treatment, and we identified multiple apparent sensitive and resistant sub-clones in each patient as well as the somatic mutations and copy number variance (CNV) associated with resistant sub-clones. The composition of the tumor sub-clones after treatment varied substantially among the patient's tumors, highlighting the potentially important role of heterogeneity in treatment resistance for rectal cancer.
Materials and Methods
Sample Collection and DNA Extraction
The Michigan Medicine Institutional Review Board approved this study. Ten consented patients with newly diagnosed clinical stage 2 or 3 rectal cancer patients underwent 1 to 4 pre-treatment biopsies of their tumors and adjacent normal tissue during routine proctoscopy to clinically assess tumor location. Although we did take each pre-treatment tumor biopsies from distinct areas of the tumor, samples were not spatially oriented in any particular way. Patients then underwent standard of care treatment for their rectal cancers with combination 5-flourouracil (or the oral equivalent, xeloda) and radiation 5 days per week for 6 weeks for a total of 50.4 Gray (Gy) followed by a 6- to10-week waiting period and then proctectomy. Post-treatment samples were provided by the pathologist focusing on the area containing the most residual tumor. Biopsies and post-treatment tumor samples were assessed by a gastrointestinal pathologist to confirm the presence of adenocarcinoma and to grade response to treatment [10]. Samples which were not found to contain adenocarcinoma or that yielded poor quality DNA were excluded and this resulted in some included samples not being sequentially numbered such that T2 or R1 was not included in the analysis. In general, grade ranges from 0, which indicates no residual cancer, to grade 3, indicating minimal to no response to treatment. Patients with grade 0 were excluded from our study due to our interest in studying treatment resistant tumors. Tissue was isolated from the relevant areas of each frozen or FFPE tumor block and manually dissected from the blocks. DNA was extracted, using the Allprep mini kit (QIAGEN) according to the manufacturer's instructions. Clinical data on each patient was abstracted from the medical records.
Whole-Exome Sequencing
Genomic DNA samples were fragmented to a target size of 125–300 bp using a Covaris S2 fragmentation system. The samples were end-repaired, a-tailed, and custom adapters were ligated using the NEBNext DNA Library Prep kit according to the manufacturer's recommended protocols. The custom adapters included 6-bp barcodes designed using BARCRAWL software [11] and synthesized by Integrated DNA Technologies. After ligation, the samples were size selected to 300 bp on a 2% agarose gel and 1-mm gel slices were retained. Samples were isolated from the gel using the QIAGEN QIAquick gel extraction system. Either 10 mL or 15 mL of each ligation product was enriched using the Phusion master mix kit and custom polymerase chain reaction (PCR) primers for a total of 14 cycles of PCR amplification. The PCR products were purified using AmpureXP beads. Library QC was performed using the Agilent Bioanalyzer and qPCR. Each pool was captured using the Roche/Nimblegen SeqCap EZ V3 Exome Enrichment Kit. The captured pools were combined and sequenced on an lllumina HiSeq 4000 platform with paired-end 150 bp reads using v4 reagents at a targeted depth of 75×. Paired reads were adapter-trimmed and mapped to the hg19 reference genome using the Burrows-Wheeler Aligner (BWA mem7 v0.7.8) [12]. Duplicates were removed using Picard v2.7.1 and indel realignment and base quality recalibration performed using GATK v3.8–0. Tumor/normal pairs of the analysis-ready alignment files were analyzed using three somatic variant callers: MuTect v1.1.7, VarScan somatic v2.3.7, and Strelka v1.0.14 using default parameters, with the exception of VarScan for which the minimum variant allele frequency was reduced to 0.05. VarScan variants were filtered using the fpfilter tool. The resulting variant calls were compiled using a custom tool called Jacquard (GitHub repository: https://github.com/umich-brcf-bioinf/Jacquard). High-confidence somatic variants identified by any one of the three callers were retained as targets for deep-sequencing and validation. Variants were annotated using VarSeq (Golden Helix, v.1.4.7), classifying them by region (e.g., exon, intron, UTR) and amino acid impact (e.g., synonymous, nonsynonymous).
Deep Targeted Sequencing
To validate mutations identified from exome sequencing and to facilitate sub-clone profiling, we designed custom capture panels targeting somatic loci detected in the primary tumors using the Agilent SureSelect XT platform [13]. Samples were sequenced on an Illumina HiSeq 4000 using paired-end 150 bp reads to a targeted depth of 500×. Deep sequencing data were processed via a workflow identical to exome sequencing described above except that VarScan alternate allele frequency variants were permitted down to 0.01. Variants were retained from the deep sequencing data if at least two callers reported the mutation as somatic and the coverage depth was at least 75x in at least one tumor sample of the patient. In addition to variant validation, these data were used as input into the PyClone tool for assessing sub-clonality.
OncoScan SNP Arrays and Copy Number Variation Analysis
ThermoFischer (Affymetrix) OncoScan v3 arrays were run on all samples. The assay detects copy number change by generating data at 50–100-kb resolution across a set of 891 cancer genes and 300–400 kb resolution across the rest of the genome. Raw array florescence intensity data generated on the Affymetrix scanners in the form of CEL files were loaded into the OncoScan Console software v.1.1.0 (ThermoFisher) and processed using the standard Affymetrix reference control files, based on sample type (normal or FFPE). Copy number data were processed using the Nexus Copy Number software v7.5 (BioDiscovery) as previously described [9]. For individual samples where the median probe set value was incorrectly assigned to unbalanced, non-diploid regions, regions of balanced heterozygosity were manually identified as most likely diploid by visual inspection of log-ratio and BAF plots, permitting the Nexus software to re-center the entire probe set. CNV events were mapped to genes and resistant CNV was defined as an alteration that was present both before and after treatment.
Purity Estimation
The tumor purity estimates generated by the TuScan algorithm implemented in the Affymetrix Oncoscan Console software were used for samples in which a solution was available. For the remaining (typically lower-purity) samples, tumor purity was estimated manually from the log2R and BAF plots based on the equations 1 and 2 in the ASCAT publication [14]. Purity estimates for each tumor are utilized as input into the PyClone algorithm.
PyClone Analysis
PyClone datasets were assembled for each patient including variant and reference read depth as well as major and minor copy number values for the region in which each variant resides. The reference and alternate read depths were extracted from the alignment (bam) files at identified mutation sites for each deep sequencing sample using a custom software tool called Zither (https://github.com/umich-brcf-bioinf/Zither), permitting data at each variant loci across all samples rather than only samples in which the variant is present. Copy number data processed via Nexus as described above were compiled for each variant in a semi-manual manner using the TAPS tool in the Patchwork software library [14]. Copy number analysis results were exported from the Nexus software as text and imported into TAPS, and chromosomal plots were generated representing the log-ratio vs. allelic imbalance of every copy number segment. Chromosomal plots were manually inspected to assign copy number states to clustered regions on the plots. TAPS then used this information to assign a copy number state and allelic ratio to each copy number segment. The segmental results from TAPS were mapped to detected somatic variants based on each variant's genomic coordinate using a custom Python script.
PyClone is a hierarchical Bayesian model that infers the cellular prevalence of each variant (the proportion of tumor cells in a sample that contain the variant), clustering variants based on covariance of those prevalence estimates across multiple samples of the same patient [15], [16], [17], [18] to identify possible sub-clones. The PyClone v0.13 Markov Chain Monte Carlo model was run for 10,000 iterations for each patient, discarding the first 1000 as burn-in. PyClone clusters were retained for subsequent analysis and reporting if they contained at least one non-silent mutation.
PyClone cluster prevalences across patient pre-treatment and post-treatment tumor samples were manually inspected and flagged as resistant if cluster cellular prevalence was increased or maintained above 5% in post-treatment samples. A gene level frequency of resistant variants was obtained by filtering to resistant clusters and counting the number of patients with resistant variants in each gene. Patient missense variants were mapped to OncoKB gain-of-function/loss-of-function annotations downloaded using the OncoKB web API [19].
Pathway Analysis
The Hotnet2 algorithm was used to identify regions of the human protein interaction network that were enriched for resistant mutation or copy number events [20]. Hotnet2 uses an insulated heat diffusion algorithm to model network co-locality of aberrations and assigns a p-value using network permutations to describe the significance of the size and number of subnetworks identified. We used resistant cluster variants to quantify mutation frequencies per gene and resistant copy number gain, loss, or loss of heterozygosity (LOH) events to quantify copy number aberration frequencies per gene. Hotnet2 identified consensus subnetworks enriched for resistant mutation and copy number events. Hotnet2 v1.2.1 was used with 100 network permutations, 1000 heat permutations, and 100 consensus permutations. Our analysis focused on sub-networks with P-values <.05 and sub-networks with at least 10 nodes. DAVID web services were used to identify Gene Ontology functional and KEGG pathway gene set enrichment for each sub-network [21].
Data Sharing
The called mutation data can be found at the Figshare website: https://figshare.com/articles/rectal_tumor_targeted_sequencing/9770444. The OncoScan array data are available from the NCBI's Gene Expression Omnibus [22] at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136872 accession number: GSE136872.
Results
Tumor Characteristics
The patient and tumor characteristics, including sex, age at diagnosis, clinical stage, chemotherapy regimen, pathologic stage, and grade of therapy response, of the 10 rectal cancer patients included in this study are summarized in Table 1. All of the rectal tumors analyzed were clinical stage 2 or 3. In total, 24 pre-treatment and 10 post-treatment samples were collected from ten rectal cancers (one to five pre-treatment and one post-treatment sample per patient). Grade of pathologic response achieved following nCRT ranged from 1–3. Patients with CR - grade 0 - were excluded from our study.
Table 1.
Clinical characteristics of patients and tumors.
| Patient ID | Sex | Age at diagnosis | Clinical stage | Neoadjuvant treatment regimen | Surgical procedure | Pathology stage | Grade of response to treatment |
|---|---|---|---|---|---|---|---|
| NP01 | M | 54 | uT3N1 | Xeloda+50.4 Gy radiation | APR | ypT3N1b | 2 |
| NP04 | M | 61 | uT3N1 | Xeloda+50.4 Gy radiation | APR | ypT2N0 | 1 |
| NP10 | M | 60 | uT4N1 | Xeloda+50.4 Gy radiation | Exenteration | ypT3N1b | 2 |
| NP11 | F | 30 | uT4N2 | Xeloda+50.4 Gy radiation | LAR with Vaginectomy | ypT2N0 | 2 |
| NP12 | M | 54 | uT3N0 | Xeloda+50.4 Gy radiation | LAR | ypT2N0 | 2 |
| NP14 | M | 68 | mT2N1 | Xeloda+50.4 Gy radiation | APR | ypT3N0 | 1 |
| NP16 | F | 45 | mT4N1 | Xeloda+50.4 Gy radiation | APR | ypT4N1 | 1 |
| NP17 | F | 33 | uT3N1 | Xeloda+50.4 Gy radiation | LAR | ypT2N0 | 2 |
| NP18 | F | 53 | uT3N1 | Xeloda+50.4 Gy radiation | LAR | ypT2N0 | 2 |
| NP19 | F | 54 | mT4N0 | Xeloda+50.4 Gy radiation | LAR | ypT3N0 | 2 |
Clinical Stage is determined by endorectal ultrasound (u) or MRI (m) and is according to AJCC, 8th Edition; Gy = Gray; APR = Abdominoperineal resection of the rectum; LAR = Low anterior resection of the rectum; yp indicates pathology stage after neo-adjuvant treatment.
Pre-Treatment Tumor Alterations and Genetic Sub-Clones
The nature of genetic alterations in tumor samples can be described in a number of ways, and we emphasized somatic mutations, copy number variance, and tumor sub-clones. Between 13 and 758 somatic mutations were identified per sample. Mutations varied between tumor regions of any individual patient's tumor patient before treatment, similar to our previous studies [7]. The number of pre-treatment of mutations did not correlate with response. Inactivating mutations in the APC and TP53 genes are common in colorectal cancer and were present in 7 out of the 10 patients studied. APC somatic mutations of presumptive pathogenic significance (nonsense or frameshift mutations) were found in pre-treatment tumor samples NP01, NP04, NP10, NP11, NP12, NP14, and NP19. TP53 somatic mutations of presumptive pathogenic significant (missense mutations in the DNA binding domain or truncating mutations in any region of the gene) were found in NP01, NP04, NP10, NP11, NP16, NP17, and NP19.
Each patient's primary tumor also had multiple examples of copy number heterogeneity among the different regions sampled (plots in Supplemental Figure 1). In CRC's, it is well known that the copy number of multiple different genomic regions can differ from the diploid state [23], [24]. Differences in copy number variation (CNV) between samples for the same patient's tumor pre- and post-treatment provide orthogonal information about genomic alteration events. Neoplastic cell purity and the overall fraction of each tumor sample altered by a copy number event was calculated using TuScan/ASCAT (Supplemental Table 1). These pre-treatment tumors showed substantial overall copy number changes across the genome, with averages of 39.3% of the genome modified by a copy number event, respectively. An average of 9.6% of the genome of pre-treatment samples exhibited copy number neutral LOH as well. Intra-tumor heterogeneity was observable between the pre- therapy tumor samples as has been seen previously [7], [9]. For example, in NP10, each tumor has LOH in Chr 4 and gain in Chr 8 but alterations in Chr 3, 7, 9, 11, 12, and 18 are only seen in some of the samples.
PyClone was then utilized to integrate deep sequencing data and copy number data from SNP arrays to profile the sub-clonality of each tumor (Figure 1, Supplemental Table 2). As discussed above, PyClone uses tumor purity as well as copy number data to estimate the cellular prevalence of each variant based on its variant allele frequency (VAF), and clusters the variants into distinct groups of different VAF within each tumor. All tumor samples showed evidence of sub-clonal structure, with the number of mutation clusters detected by PyClone varying between two and 10 individual sub-clonal populations per tumor. Additionally, heterogeneity among spatially distinct tumor samples from an individual patient's primary tumor was evident, as reflected by the differing prevalence measurements of clusters across samples even prior to treatment.
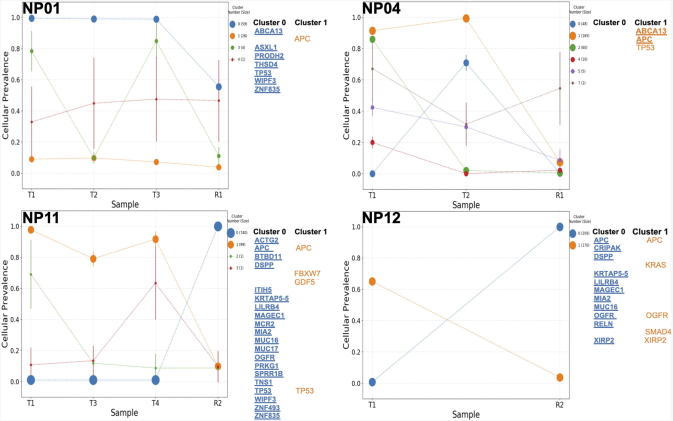
Figure 1.
Genetic sub-clones across tumors. Tumor samples were taken before treatment (T) and after treatment (R). Each tumor contained multiple sub-clones and the composition changed after treatment. Genes that previously identified as a commonly mutated genes in colorectal cancer as identified by Giannakis et al. and the most commonly resistant genes identified in our patients were used to label genes present in each sub-clone cluster. Bold and underlined genes are those genes classified as resistant in our patients.
Alterations in Tumor Sub-clones in Response to Therapy
The cellular prevalence plots provide a profile of all sub-clones and indicate the changing sub-clonal compositions among samples from each patient's tumor (Figure 1, Supplemental Figure 2). Note, that the cellular prevalence plots are not temporal in order, but instead represent the composition of a sample at a single time point. After analysis, if post-treatment sub-clones did not contain non-silent somatic mutations that could be functionally clinically relevant, these sub-clones were manually removed from the PyClone figures.
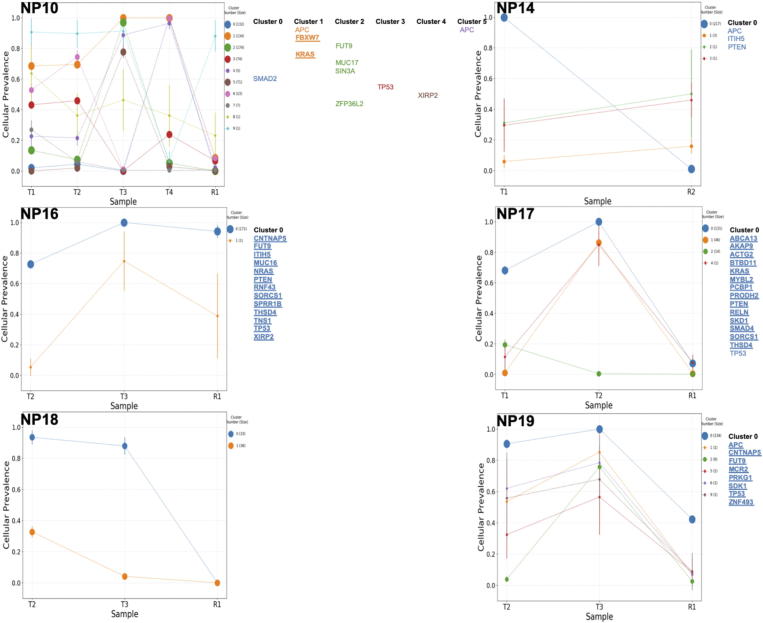
Supplemental Figure 2.
Genetic sub-clones across all remaining patient tumors. Tumor samples were taken before treatment (T) and after treatment (R). Each tumor contained multiple sub-clones and the composition changed after treatment. Genes that previously identified as a commonly mutated genes in colorectal cancer as identified by Giannakis et al and the most commonly resistant genes identified in our patients, were used to label genes present in each sub-clone cluster. Bold and underlined genes are those genes classified as resistant in our patients.
We found that nCRT often resulted in substantial changes in the relative proportions of individual sub-clones or clusters within the primary tumor. For example, in NP11, clusters 1–3 were nearly gone after treatment but cluster 1, which was very low prevalence before treatment, was the most prevalent after treatment. Another example is NP12, which had 2 sub-clones in the T1 sample: cluster 1 that had high cellular prevalence and cluster 0 that had low cellular prevalence. After nCRT, the prevalence of these 2 clusters were changed relative to the pre-treatment sample (T1), with the cellular prevalence of Cluster 0 going from near zero to near 1.0 and the cellular prevalence of the apparently sensitive sub-clone going from 0.65 to near zero. Similarly, in NP04, a dominant sub-clone containing the APC and TP53 mutations shows a marked reduction in cellular prevalence from roughly 0.9 VAF to 0.09 post-nCRT. Likewise, NP14, NP17 and NP18 also show a very large decrease in the cellular prevalence of the dominant pre-treatment sub-clone. In other tumors, such as NP01, the pattern of clusters was not altered much by nCRT. These changes indicate that sub-clonal populations are selectively modified following nCRT. Somatic mutations in the APC and TP53 genes were present in different sub-clones of the same primary tumors but responded differently to neoadjuvant therapy depending upon the tumor and sub-clone in which they were present. To further describe this phenomenon, the APC gene is mutated in both sub-clone 0 and 1 in NP12. However, in Cluster 0, the APC gene has a missense mutation that is present before and after therapy, while in Cluster 1, the APC gene has a loss of function (LOF) mutation that is found in a cluster that was eliminated following nCRT. In NP11, mutations in the TP53 gene were found both in Clusters 0 and 1. In Cluster 0, a TP53 missense mutation was identified and the cluster was resistant to therapy while there was a TP53 LOF mutation in Cluster 1 and this cluster was sensitive to nCRT (Figure 1).
Characterization of Genetic Alterations Associated with Resistance
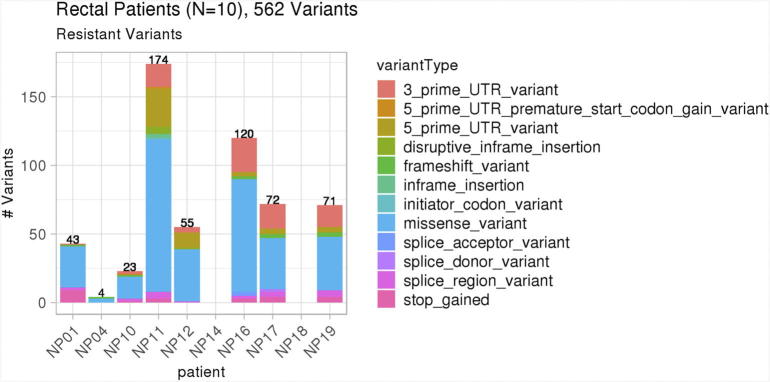
The sub-clonal composition following nCRT changed for each tumor assessed, with some sub-clones being eliminated after treatment and others persisted after treatment. We assessed the genetic composition of each sub-clone and its response to treatment and looked for genetic alterations found in resistant sub-clones across different patient's tumors. Across all 10 rectal patients, there were 562 variants in resistant sub-clones in multiple variant types, as shown in Figure 2. The number of resistant sub-clones varied from 1 to 5 sub-clones per patient.
Figure 2.
Variants in resistant tumor sub-clones across all patients. There were 562 non-silent, variants in resistant tumor sub-clones across all 10 patient tumors.
To better understand the roles of genetic subclones in treatment resistance, we combined the functionally significant mutation and copy number data into gene-level annotations for each of our 10 patient tumors (Table 2). Genes with mutations found in sub-clones that persisted after therapy, here defined as resistant to treatment, and found in more than 1 patient are listed in the left side of Table 2. In Table 2, the mutations found in resistant sub-clones are in bold print. Resistant mutations in TP53 were found in 4 patients and resistant CNV (loss of TP53) in 3. For APC, resistant mutations were found in 4 patients and resistant CNV (loss of APC) was found in 1 patient. Oncoprints of each tumor showing mutations across samples from each tumor are shown in Supplemental Figure 3, again revealing intratumor heterogeneity and that some alterations persist after treatment while others do not. Oncoprint cell color represents Pyclone cellular prevalence and patient samples with cellular prevalence <0.05 are colored black. We did not find evidence of a single mechanism for therapeutic resistance across tumors based on our data but instead our findings suggested multiple alterations potentially associated with resistance. Resistant sub-clones contained mutations in APC (NP04, NP11, NP12, and NP19), TP53 (NP01, NP11, NP16, and NP19), ABCA13 (NP01, NP04, and NP17), MUC16 (NP11, NP12, and NP16), and THSD4 (NP01, NP16, and NP17).
Table 2.
Summary of genes which were mutated in resistant sub-clones across multiple patients and CNV affecting those genes.
| Gene | Resistant alterations in Gene |
Mutations in All Sub-clone (bold = resistant) |
All CNV |
||||
|---|---|---|---|---|---|---|---|
| # Patients: Mutation | # Patients: CNV | # Pts | CN Loss | CN Gain | LOH | ||
| APC | 4 | 1 | 7 | NP01 (missense) NP04 (LoF) NP10(LoF and missense) NP11 (LoF and other) NP12 (missense and LoF) NP14 (LoF) NP19 (LoF) |
NP16 NP17 NP18 NP19 |
- | NP04 |
| TP53 | 4 | 3 | 7 |
NP01 (missense) NP04 (missense) NP10 (missense) NP11 (missense and LoF) NP16 (missense) NP17 (missense) NP19 (LoF) |
NP04 NP11 NP17 NP19 |
- | - |
| ABCA13 | 3 | 5 | 3 |
NP01 (iSRV) NP04 (missense) NP17 (missense) |
- | NP01 NP04 NP10 NP11 NP12 NP16 NP18 NP19 |
- |
| MUC16 | 3 | 0 | 3 |
NP11 (missense) NP12 (iSRV and missense) NP16 (missense) |
NP16 | NP01 NP04 |
NP16 |
| THSD4 | 3 | 1 | 3 |
NP01 (missense) NP16 (LoF) NP17 (LoF) |
NP18 | - | NP17 |
| ACTG2 | 2 | 0 | 2 |
NP11 (iSRV) NP17 (missense) |
- | NP01 NP04 NP16 |
- |
| BTBD11 | 2 | 0 | 2 |
NP11 (missense) NP17 (UTR5) |
- | - | - |
| CNTNAP5 | 2 | 0 | 2 |
NP16 (missense) NP19 (missense) |
- | - | NP11 |
| DSPP | 2 | 0 | 2 |
NP11 (missense) NP12 (missense) |
- | - | - |
| FUT9 | 2 | 0 | 3 | NP10 (UTR3) NP16 (UTR3) NP19 (UTR3) |
- | NP01 NP04 NP17 |
- |
| ITIH5 | 2 | 0 | 3 |
NP11 (missense) NP14 (missense) NP16 (missense) |
- | - | - |
| KRAS | 2 | 0 | 3 |
NP10 (missense) NP12 (missense) NP17 (missense) |
- | NP04 NP10 |
NP16 |
| KRTAP5–5 | 2 | 1 | 2 |
NP11 (missense and UTR3) NP12 (missense) |
- | NP01 NP12 NP17 |
- |
| LILRB4 | 2 | 0 | 2 |
NP11 (missense) NP12 (missense) |
- | - | NP16 |
| MAGEC1 | 2 | 2 | 2 |
NP11 (missense) NP12 (missense) |
- | NP18 NP19 |
- |
| MC2R | 2 | 1 | 2 |
NP11 (UTR3) NP19 (missense) |
NP04 NP17 NP18 NP19 |
- | - |
| MIA2 | 2 | 3 | 2 |
NP11 (missense) NP12 (missense) |
NP19 | NP18 NP19 |
- |
| OGFR | 2 | 5 | 2 |
NP11 (missense) NP12 (missense and missense) |
- |
NP01 NP11 NP14 NP16 NP17 NP18 NP19 |
- |
| PRKG1 | 2 | 0 | 2 |
NP11 (UTR5) NP19 (UTR3) |
- | - | - |
| PRODH2 | 2 | 1 | 2 |
NP01 (LoF) NP17 (missenses) |
NP19 | - | NP16 |
| PTEN | 2 | 1 | 3 | NP14 (missense) NP16 (missense) NP17 (LoF) |
NP01 NP10 NP11 NP16 |
- | NP17 |
| RELN | 2 | 3 | 2 |
NP12 (UTR5) NP17 (missense) |
- | NP01 NP04 NP10 NP11 NP18 NP19 |
- |
| SDK1 | 2 | 4 | 2 |
NP17 (missense) NP19 (missense) |
- | NP01 NP04 NP10 NP11 NP12 NP19 |
- |
| SORCS1 | 2 | 0 | 2 |
NP16 (UTR3) NP17 (missense) |
NP17 | - | NP04 NP16 |
| SPRR1B | 2 | 0 | 2 |
NP11 (missense and UTR3) NP16 (UTR3) |
NP11 | NP01 NP04 |
NP11 |
| TNS1 | 2 | 1 | 2 |
NP11 (missense) NP16 (missense) |
- | NP04 NP11 NP19 |
- |
| WIPF3 | 2 | 4 | 2 |
NP01 (missense) NP11 (missense) |
- |
NP10 NP11 NP12 NP19 |
- |
| XIRP2 | 2 | 1 | 3 | NP10 (missense) NP12 (missense and LoF) NP16 (missense) |
- | NP01 NP04 NP19 |
NP04 NP19 |
| ZNF493 | 2 | 0 | 2 |
NP11 (missense and UTR3) NP19 (UTR3) |
- | NP01 NP04 NP17 |
NP16 NP19 |
| ZNF835 | 2 | 0 | 2 |
NP01 (missense) NP11 (missense) |
- | - | NP16 |
Bold, Resistant alteration.
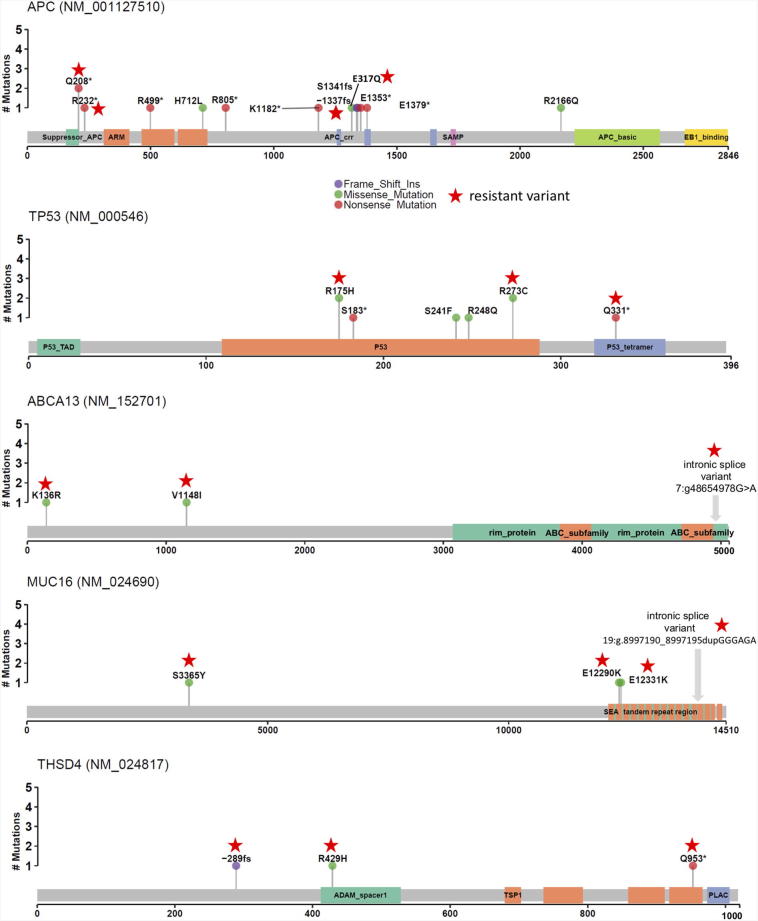
Lollipop plots of the mutations found in the APC, TP53, ABCA13, MUC16, and THSD genes to assess mutational clustering around gene regions (Figure 3). No particular pattern was apparent with mutations scattered across each gene. Interestingly, one recurrent APC variant (Q208*) and two recurrent TP53 variants (R175H, R273C) were observed in multiple patients. The role of APC and TP53 as likely driver genes in CRC has been previously well described [25], [26], but their role in resistance to treatment is unclear. We also identified ABCA13, MUC16, and THSD4 as potentially important in therapeutic resistance in rectal cancer. Unlike APC and TP53, which had mutations in 7 distinct patients but only 4 of these patients were classified as resistant mutations, ABCA13 and THSD4 are both classified as resistant in all three patients where alterations are present. An additional 30 genes contained mutations that were in resistant sub-clones in more than 1 patient, including many potential drivers of resistance that haven't been previously been described (Supplemental Table 2).
Figure 3.
Mutation lollipop visualizations highlight location of resistant variants (stars) across gene products. Mutations in APC, PT53, ABCA13, MUC16, and THSD4 are demonstrated.
Furthermore, CNV that persisted after nCRT was frequently observed and when it included genes identified as altered in resistant sub-clones, this is annotated in Table 2. CN loss, CN gain, and LOH (copy neutral) were all present. In 4 out of the 5 most commonly mutated genes (APC, TP53, ABCA13, MUC16 and THSD4), resistant CNV were also observed involving these genes (Table 2). Additionally, resistant gene variants and resistant copy number changes existed simultaneously in 3 (APC, TP53, and THSD4) out of the 5 most commonly mutated genes. NP19 had a resistant LOF variant of the APC gene as well as a CN loss. NP11 had a resistant missense variant mutation and NP19 had a LOF variant mutation in the TP53 gene and both also contained resistant CN loss.
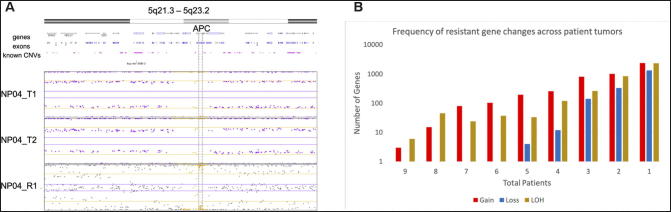
When characterizing resistant copy number variants across tumors, the most common form of resistant alteration was LOH. An example of resistant LOH is shown in Figure 4A, where the alteration is found in both the pre- and post-treatment samples. Nine patients (NP01, NP10, NP11, NP12, NP14, NP16, NP17, NP18, and NP19) had 11 genes (abParts, TCF19, HLA-B, IGVK-A2, IGKV, PSORS1C3, HLA-C, AK128525, POU5F1, IG kappa, and HCG27) with resistant LOH and 6 genes (JH8, PSCA, AX747544, LOC101928087, JRK, and ARC) with resistant CN gain. As illustrated by Figure 4B, as the number of total patients decreases, the number of resistant genes increases. Thus, most resistant copy number changes were unique to individual tumors and were not shared across patients. Resistant CN loss first appears in 5 total patients and contains the following 4 altered genes, ARHGEF10L, RCC2, CRK, and WRAP53. To note, the most common genes effected by resistant CNV across tumors does not overlap with the most common resistant somatic mutations.
Figure 4.
Panel A: Resistant loss of heterozygosity (LOH) in APC. LOH in APC was found in all samples (pre- and post-treatment), illustrating resistant LOH. Panel B: Resistant copy number variants (CNV) in rectal cancer.
Resistant Pathways
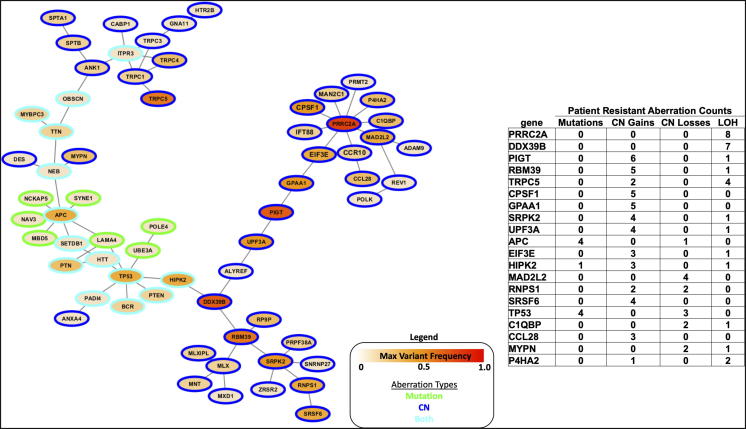
Hotnet 2 was used to perform a network analysis to identify regions of the human interactome enriched for resistant mutations and copy number changes including CN gain, CN loss, and LOH. Eleven sub-networks were identified as significant. The number of altered networks found again supports the hypothesis that no single gene or pathway is fully responsible for resistance. The largest sub-network also contained genes with the highest frequencies of resistant variants including alterations in APC and TP53 (Figure 5, Supplemental Table 3A). This sub-network was enriched for genes involved in negative regulation of cell cycle process (GO:0010948) with LOF aberrations in APC, DDX39B, MAD2L2, and TP53. Over-expression of DDX39, an RNA helicase, was recently shown to promote cell migration, invasion, growth, and metastasis in hepatocellular carcinoma [27]. MAD2L2 is a spindle assembly checkpoint protein shown to inhibit colorectal cancer growth and is furthermore a favorable prognostic in colorectal patients demonstrating a strong potential resistance mechanism in this patient cohort [28].
Figure 5.
Hotnet2 pathway analysis including resistant CNV and resistant mutations (P < .01). Sub-network consists of 65 genes connected by 68 curated protein interactions constructed from consensus of mutation and CN gain, loss, and LOH. Table shows 20 sub-network genes with the highest frequency of resistant variants across patients.
Figure 5 annotates the top 20 sub-network genes most frequently aberrant in resistant patient samples and shows their network. Sub-network genes were also enriched for cell differentiation (GO:0030154), regulation of apoptotic process (GO:0042981), and calcium ion homeostasis (GO:0055074) gene sets providing additional mechanistic hypotheses for pathways involved in resistance. Interestingly two amplified genes in the Figure 5 table, TRCP5 and CCL28, have been shown to be involved in calcium ion homeostasis. Calcium signaling remodeling has been linked to cancer cell proliferation, metastasis, and has been proposed as a therapeutic target [29], [30], [31]. A complete list of sub-network genes is available as Supplemental Table 3B.
Discussion
We have previously highlighted the potential clinical significance of intra-tumor genetic heterogeneity in rectal cancer [7]; however, the role of intra-tumoral genetic heterogeneity and sub-clonal architecture in resistance to nCRT in rectal cancer patients is unknown. This is the first study using comprehensive genomic analysis of intra-tumor heterogeneity in treatment response in rectal cancer patients. Using current informatic tools for inferring sub-clonal populations from bulk tumor samples, we found that intra-tumoral heterogeneity is evident in pre-treatment rectal cancer. Following nCRT, the prevalence of sub-clonal populations was selectively modified in each patient, suggesting that some sub-clones are likely resistant to treatment. It is well understood that gene alterations do not act in isolation and systems biology analyses leveraging current understanding of the human interactome may be used to effectively search for and recognize canonical cancer pathway events. We also performed a systems biology network analysis, Hotnet2, to identify resistant signaling pathways observed across multiple patients, including both somatic mutation and CNV data. Such system-level strategies provide statistically rigorous framework to integrate disparate data types and generate hypotheses for future studies. We highlighted a sub-network enriched for resistant variants with genes that have been shown to be involved in cell cycle regulation, apoptosis, and calcium signaling. We did not identify any individual gene alteration that appears to be responsible for resistance, but instead found that there are potentially several genetic alterations associated with therapeutic resistance in rectal cancer.
In our study, resistant sub-clones demonstrate high frequency of mutation in genes previously known to be frequently altered in CRC including TP53 and APC [32], along with multiple additional potential drivers of resistance. Because APC and TP53 are frequently altered in rectal cancer before treatment, it is difficult to assess whether their presence in sub-clones after treatment contributes to resistance or not. Prior investigators have found evidence that APC and TP53 may contribute to resistance. Sakai et al. assessed 9 post-treatment rectal cancer patients with targeted sequencing of commonly mutated regions across 50 genes and found TP53 mutations in 8 patients and APC mutations in 3 patients, in addition to increased TP53 expression pre vs post treatment [25]. Additionally, Emons et al. found that in the cell line SW1463, radiation resistance was mediation by activation of the Wnt/ß-catenin pathway [33]. KRAS alone or concurrent with TP53 mutation has also previously been shown to be enriched patients who did not achieve a complete response to neo-adjuvant therapy [26]. We found mutated KRAS in 2 resistant sub-clones. One drawback of prior studies in that they focus only on selected genes and there have not been prior agnostic studies of gene alterations that are resistant.
Our data suggest that there may likely be complex relationships between molecular alterations and the sub-clones that persist in rectal cancers treated with nCRT. We identified recurrently altered genes that persisted after treatment and are therefore potentially associated in some functional manner with resistance. Candidate drivers of resistance from our work include, ABCA13, MUC16, and THSD4. A search of TCGA reveals mutation or amplification in ABCA13 in 15% of colorectal cancers, mutation THSD4 in 3%, and mutation or deletion in MUC16 in 28% of cases. However, because these are surgical specimens in untreated patients, this only tells us that these mutations do exist in colorectal cancer, not what their behavior would be following treatment. The ABCA13 (ATP-Binding Cassette, Sub-Family A, Member 13) gene, which has not been studied in depth in rectal cancer, is a member of the ABC transporter family. Other members of this family of transporters have been shown to play a crucial role in the development of resistance through efflux of anticancer agents outside of cancer cells including 5-FU [34]. We found both mutations and amplifications in this gene associated with resistance, which could be consistent with up-regulation of a protein causing efflux of the radiation sensitizer 5-FU. Additionally, these transporters have been shown to be upregulated in colorectal cancer compared to control tissues [34]. Furthermore, ABCA13 may play additional roles in cancer progression as amplification has been shown to increase the risk of lymph node metastasis in gastric adenocarcinoma [35] and it is a marker of shorter overall survival in ovarian serous carcinoma [36]. The MUC16 gene product is CA-125 which is a membrane protein mucin whose serum level has been shown to negatively correlate with survival in colorectal cancer [37].THSD4 (Thrombospondin, Type I Domain Containing 4) is a member of the extracellular calcium-binding protein family involved in cell adhesion and migration and is being investigated as a potential tumorigenesis gene in hemangioblastomas [38], as well as being evaluated as a prognosticating marker in glioblastomas [39].
Our study has multiple limitations. One limitation is our small sample size, which impacts on our ability to definitively highlight potential contributing molecular lesions associated with resistance to nCRT among rectal cancers and patients. Nonetheless, we have shown that nCRT affects the genetic sub-clonal populations in a rectal cancer in complex ways, and presumptive sensitive and resistant sub-clones, somatic mutations, and CNV likely vary from one patient to another. Additionally, our work highlighted some alterations previously associated with therapy resistance, along with suggesting some other candidates. The design of our study only allows us to observe the behavior of sub-clones and test our hypothesis that they are altered by therapy and potentially important in resistance. Another limitation of our study and all broad assessments of genetic alterations in cancer is our inability to definitively identify whether particular alterations are responsible for resistance or whether they are simply carried forward in the residual tumor sub-clones following treatment. We acknowledge that other biologic variables such as the presence of other mutations in that sub-clone or the immune environment in the patient may have contributing roles in shaping the sub-clonal landscape and the molecular alterations found. We did not perform separate validation studies of the genetic alterations identified in the sub-clones that persisted after treatment due to lack of tissue for these studies. Lastly, there are limitations to the PyClone algorithm used to identify tumor sub-clones. The number of sub-clones identified by PyClone is dependent upon having multiple samples and the algorithm generally only identifies groups of sensitive and resistant mutations if there are only 2 samples (NP12 for example, Supplemental 2 and 3). This means, we would likely have identified more sub-clones if we had been able to analyze more pre- and post-treatment samples and that our results may have been altered by sampling error.
Conclusions
In summary, our analyses reveal the complex and dynamic genomic architecture of pre- and post-treatment rectal cancers, with individual tumors having multiple examples of mutational and copy number heterogeneity among samples before treatment as well as after treatment. Furthermore, multiple known and previously understudied genetic alterations that may be contributing to resistance were identified. Assessment of a larger collection of tumor samples and rectal cancer patients treated with nCRT may help to highlight potentially significant recurrent driver alterations associated with resistance to nCRT. Additional future studies could also include assessment of interactions of sub-clones with immune cells as well as the potential contributions of other tumor microenvironment cell populations to response or resistance.
The following are the supplementary data related to this article.
Purity for each specimen as determined by TuScan/ASCAT, which utilizes copy number variation data from the OncoScan SNP arrays. The percent of genome changed is also provided which is calculated by Nexus. T samples are pre-treatment samples and R samples are post-treatment samples.
Complete list of all mutations in sub-clones across all tumors.
Excel file A: Eleven sub-networks identified using Hotnet2 network analysis. Sub-network 3 is highlighted as excel file B annotates the complete list of sub-network genes in sub-network 3.
CNV plots for all samples. There is heterogeneity in CNV between samples from individual tumors before treatment and some CNV persists after treatment and some is lost.
Oncoprints of each sample from each tumor: Oncoprints show the cellular prevalence (as calculated by PyClone) of each genetic alteration in each specimen before (T) and after treatment (R).
Funding
Grant Support: LMF: National Institutes of Health T32 HL007517. KMH: American Surgical Association Foundation Fellowship 5P50CA130810; John S. and Suzanne C. Munn Cancer Research Fund 5P30A046592; National Institutes of Health K08CA190645. This work was also supported by the National Institutes of Health, National Cancer Institute Comprehensive Cancer Center Core Grant [5 P30 CA 46592].
Statement of Author Contributions
LMF wrote the first draft of the manuscript. LMF, PU, and AB created the figs. PU, AB, and CS completed the computational analyses. JG was the gastrointestinal pathologist. LM assisted with manuscript review and project design. RI performed DNA isolation. EF mentored KMH and assisted with project design. KMH was the supervisor of these studies and, as such, had full access to all of the data and takes responsibility for the integrity of the data and the accuracy of the data analyses.
Footnotes
Conflicts of Interest: None
Contributor Information
Lynn M Frydrych, Email: lfrydryc@med.umich.edu.
Peter Ulintz, Email: pulintz@umich.edu.
Armand Bankhead, Email: bankhead@med.umich.edu.
Christopher Sifuentes, Email: cjsifuen@med.umich.edu.
Joel Greenson, Email: facjkgmd@med.umich.edu.
Lillias Maguire, Email: maguirel@med.umich.edu.
Regina Irwin, Email: irwinre@med.umich.edu.
Eric R. Fearon, Email: fearon@med.umich.edu.
Karin M Hardiman, Email: khardiman@uabmc.edu.
References
- 1.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.van Gijn W, Marijnen CA, Nagtegaal ID, Kranenbarg EM, Putter H, Wiggers T, Rutten HJ, Pahlman L, Glimelius B, van de Velde CJ. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet Oncol. 2011;12:575–582. doi: 10.1016/S1470-2045(11)70097-3. [DOI] [PubMed] [Google Scholar]
- 4.Habr-Gama A, de Souza PM, Ribeiro U, Jr., Nadalin W, Gansl R, Sousa AH, Jr., Campos FG, Gama-Rodrigues J. Low rectal cancer: impact of radiation and chemotherapy on surgical treatment. Dis Colon Rectum. 1998;41:1087–1096. doi: 10.1007/BF02239429. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Aguilar J, Hernandez de Anda E, Sirivongs P, Lee SH, Madoff RD, Rothenberger DA. A pathologic complete response to preoperative chemoradiation is associated with lower local recurrence and improved survival in rectal cancer patients treated by mesorectal excision. Dis Colon Rectum. 2003;46:298–304. doi: 10.1007/s10350-004-6545-x. [DOI] [PubMed] [Google Scholar]
- 6.Kreso A, O'Brien CA, van Galen P, Gan OI, Notta F, Brown AM, Ng K, Ma J, Wienholds E, Dunant C. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardiman KM, Ulintz PJ, Kuick RD, Hovelson DH, Gates CM, Bhasi A, Rodrigues Grant A, Liu J, Cani AK, Greenson JK. Intra-tumor genetic heterogeneity in rectal cancer. Lab Investig. 2016;96:4–15. doi: 10.1038/labinvest.2015.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, Marjoram P, Siegmund K, Press MF, Shibata D. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulintz PJ, Greenson JK, Wu R, Fearon ER, Hardiman KM. Lymph Node Metastases in Colon Cancer Are Polyclonal. Clin Cancer Res. 2018;24:2214–2224. doi: 10.1158/1078-0432.CCR-17-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler JM, Dodds E, Warren BF, Cunningham C, George BD, Jones AC, Mortensen NJ. Preoperative chemoradiotherapy and total mesorectal excision surgery for locally advanced rectal cancer: correlation with rectal cancer regression grade. Dis Colon Rectum. 2004;47:2025–2031. doi: 10.1007/s10350-004-0713-x. [DOI] [PubMed] [Google Scholar]
- 11.Frank DN. BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinformatics. 2009;10:362. doi: 10.1186/1471-2105-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagemann IS, Devarakonda S, Lockwood CM, Spencer DH, Guebert K, Bredemeyer AJ, Al-Kateb H, Nguyen TT, Duncavage EJ, Cottrell CE. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer. 2015;121:631–639. doi: 10.1002/cncr.29089. [DOI] [PubMed] [Google Scholar]
- 14.Mayrhofer M, DiLorenzo S, Isaksson A. Patchwork: allele-specific copy number analysis of whole-genome sequenced tumor tissue. Genome Biol. 2013;14:R24. doi: 10.1186/gb-2013-14-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McPherson A, Roth A, Laks E, Masud T, Bashashati A, Zhang AW, Ha G, Biele J, Yap D, Wan A. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat Genet. 2016;48:758–767. doi: 10.1038/ng.3573. [DOI] [PubMed] [Google Scholar]
- 16.Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, Ha G, Aparicio S, Bouchard-Cote A, Shah SP. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11:396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamy P, Nordentoft I, Birkenkamp-Demtroder K, Thomsen MB, Villesen P, Vang S, Hedegaard J, Borre M, Jensen JB, Hoyer S. Paired Exome Analysis Reveals Clonal Evolution and Potential Therapeutic Targets in Urothelial Carcinoma. Cancer Res. 2016;76:5894–5906. doi: 10.1158/0008-5472.CAN-16-0436. [DOI] [PubMed] [Google Scholar]
- 18.Findlay JM, Castro-Giner F, Makino S, Rayner E, Kartsonaki C, Cross W, Kovac M, Ulahannan D, Palles C, Gillies RS. Differential clonal evolution in oesophageal cancers in response to neo-adjuvant chemotherapy. Nat Commun. 2016;7:11111. doi: 10.1038/ncomms11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leiserson MD, Vandin F, Wu HT, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2015;47:106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao X, Sherman BT, Huang da W, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie T, G DA, Lamb JR, Martin E, Wang K, Tejpar S, Delorenzi M, Bosman FT, Roth AD, Yan P. A comprehensive characterization of genome-wide copy number aberrations in colorectal cancer reveals novel oncogenes and patterns of alterations. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai K, Kazama S, Nagai Y, Murono K, Tanaka T, Ishihara S, Sunami E, Tomida S, Nishio K, Watanabe T. Chemoradiation provides a physiological selective pressure that increases the expansion of aberrant TP53 tumor variants in residual rectal cancerous regions. Oncotarget. 2014;5:9641–9649. doi: 10.18632/oncotarget.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chow OS, Kuk D, Keskin M, Smith JJ, Camacho N, Pelossof R, Chen CT, Chen Z, Avila K, Weiser MR. KRAS and Combined KRAS/TP53 Mutations in Locally Advanced Rectal Cancer are Independently Associated with Decreased Response to Neoadjuvant Therapy. Ann Surg Oncol. 2016;23:2548–2555. doi: 10.1245/s10434-016-5205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang T, Ma Z, Liu L, Sun J, Tang H, Zhang B, Zou Y, Li H. DDX39 promotes hepatocellular carcinoma growth and metastasis through activating Wnt/beta-catenin pathway. Cell Death Dis. 2018;9:675. doi: 10.1038/s41419-018-0591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Li L, Chen M, Yu X, Gu Z, Qiu H, Qin G, Long Q, Fu X, Liu T. MAD2L2 inhibits colorectal cancer growth by promoting NCOA3 ubiquitination and degradation. Mol Oncol. 2018;12:391–405. doi: 10.1002/1878-0261.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roderick HL, Cook SJ. Ca2+ signalling checkpoints in cancer: remodelling Ca2+ for cancer cell proliferation and survival. Nat Rev Cancer. 2008;8:361–375. doi: 10.1038/nrc2374. [DOI] [PubMed] [Google Scholar]
- 30.Prevarskaya N, Skryma R, Shuba Y. Calcium in tumour metastasis: new roles for known actors. Nat Rev Cancer. 2011;11:609–618. doi: 10.1038/nrc3105. [DOI] [PubMed] [Google Scholar]
- 31.Fiorio Pla A, Avanzato D, Munaron L, Ambudkar IS. Ion channels and transporters in cancer. 6. Vascularizing the tumor: TRP channels as molecular targets. Am J Physiol Cell Physiol. 2012;302:C9–15. doi: 10.1152/ajpcell.00280.2011. [DOI] [PubMed] [Google Scholar]
- 32.Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016;17:1206. doi: 10.1016/j.celrep.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emons G, Spitzner M, Reineke S, Moller J, Auslander N, Kramer F, Hu Y, Beissbarth T, Wolff HA, Rave-Frank M. Chemoradiotherapy Resistance in Colorectal Cancer Cells is Mediated by Wnt/beta-catenin Signaling. Mol Cancer Res. 2017;15:1481–1490. doi: 10.1158/1541-7786.MCR-17-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hlavata I, Mohelnikova-Duchonova B, Vaclavikova R, Liska V, Pitule P, Novak P, Bruha J, Vycital O, Holubec L, Treska V. The role of ABC transporters in progression and clinical outcome of colorectal cancer. Mutagenesis. 2012;27:187–196. doi: 10.1093/mutage/ger075. [DOI] [PubMed] [Google Scholar]
- 35.Araujo TM, Seabra AD, Lima EM, Assumpcao PP, Montenegro RC, Demachki S, Burbano RM, Khayat AS. Recurrent amplification of RTEL1 and ABCA13 and its synergistic effect associated with clinicopathological data of gastric adenocarcinoma. Mol Cytogenet. 2016;9:52. doi: 10.1186/s13039-016-0260-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nymoen DA, Holth A, Hetland Falkenthal TE, Trope CG, Davidson B. CIAPIN1 and ABCA13 are markers of poor survival in metastatic ovarian serous carcinoma. Mol Cancer. 2015;14:44. doi: 10.1186/s12943-015-0317-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang SJ, Ge XS, Xu ZY, Qi XW, Chen XP. Lower serum CA125 level, negative vascular invasion, and wild BRAF were strongly associated with better 2-year disease-free survival in patients with stage III colorectal cancer who received adjuvant chemotherapy. Cancer Biomark. 2018;22:161–168. doi: 10.3233/CBM-181179. [DOI] [PubMed] [Google Scholar]
- 38.Ma D, Yang J, Wang Y, Huang X, Du G, Zhou L. Whole exome sequencing identified genetic variations in Chinese hemangioblastoma patients. Am J Med Genet A. 2017;173:2605–2613. doi: 10.1002/ajmg.a.38350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J, Hou X, Li M, Ren H, Fang S, Wang X, He C. Genome-wide methylation profiling reveals new biomarkers for prognosis prediction of glioblastoma. J Cancer Res Ther. 2015;11(Suppl 2):C212–C215. doi: 10.4103/0973-1482.168188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purity for each specimen as determined by TuScan/ASCAT, which utilizes copy number variation data from the OncoScan SNP arrays. The percent of genome changed is also provided which is calculated by Nexus. T samples are pre-treatment samples and R samples are post-treatment samples.
Complete list of all mutations in sub-clones across all tumors.
Excel file A: Eleven sub-networks identified using Hotnet2 network analysis. Sub-network 3 is highlighted as excel file B annotates the complete list of sub-network genes in sub-network 3.
CNV plots for all samples. There is heterogeneity in CNV between samples from individual tumors before treatment and some CNV persists after treatment and some is lost.
Oncoprints of each sample from each tumor: Oncoprints show the cellular prevalence (as calculated by PyClone) of each genetic alteration in each specimen before (T) and after treatment (R).