Abstract
Protein tyrosine phosphorylation is a crucial signaling mechanism that plays a role in epithelial carcinogenesis. Protein tyrosine kinases (PTKs) control various cellular processes including growth, differentiation, metabolism, and motility by activating major signaling pathways including STAT3, AKT, and MAPK. Genetic mutation of PTKs and/or prolonged activation of PTKs and their downstream pathways can lead to the development of epithelial cancer. Therefore, PTKs became an attractive target for cancer prevention. PTK inhibitors are continuously being developed, and they are currently used for the treatment of cancers that show a high expression of PTKs. Protein tyrosine phosphatases (PTPs), the homeostatic counterpart of PTKs, negatively regulate the rate and duration of phosphotyrosine signaling. PTPs initially were considered to be only housekeeping enzymes with low specificity. However, recent studies have demonstrated that PTPs can function as either tumor suppressors or tumor promoters, depending on their target substrates. Together, both PTK and PTP signal transduction pathways are potential therapeutic targets for cancer prevention and treatment.
Keywords: Carcinogenesis, tyrosine phosphorylation, PTK, PTP, EGFR, IGF-1R, STAT3, AKT
INTRODUCTION
Protein phosphorylation is a post-translational modification that regulates protein function in response to exogenous factors. Phosphorylation of substrate proteins on serine, threonine, or tyrosine residues by protein kinases modulates diverse physiological functions. While more than 95% of protein phosphorylation occurs on serine or threonine residues, tyrosine phosphorylation only makes up less than 1% of total cellular phosphorylation. Even though its contribution to total phosphorylation is very small compared with serine or threonine phosphorylation, tyrosine phosphorylation is still a critical signaling mechanism needed to maintain cellular function and homeostasis, and abnormal phosphotyrosine signaling can facilitate the development of cancer [1,2].
Approximately 35 years ago, protein tyrosine kinases (PTKs), enzymes that catalyze tyrosine phosphorylation of specific target proteins, were identified. Since that time 90 genes that encode PTKs have been discovered in the human genome. Among them, 58 PTKs are receptor type kinases and 32 PTKs are non-receptor type kinases, located in the cytoplasm [1–3]. Both the activation of receptor type PTKs through the binding of ligands such as growth factor receptors, and the activation of non-receptor type PTKs modulate major cellular processes including cell proliferation, apoptosis, angiogenesis, and cell motility via signal transduction. Aberrant PTK signaling has been implicated in various types of cancers, with the observation that 51 of 90 PTKs are associated with cancer through mutation or overexpression [4–6]. As a result, PTKs have been studied extensively in order to develop PTK inhibitors for the prevention and treatment of cancer. Subsequently, several PTK inhibitors have been developed by pharmaceutical companies and approved for medical use by the FDA [7–9].
Besides exogenous regulation by inhibitors, PTK activity is endogenously regulated by negative feedback mechanisms. Protein tyrosine phosphatases (PTPs) can be activated to directly dephosphorylate target proteins and thereby negatively regulate phosphotyrosine signaling [10–12]. PTPs were first identified in the late 1980s by Nicholas Tonks and colleagues, approximately 10 years after the discovery of PTKs [13]. Since then, studies using the conserved catalytic domain of PTPs to search the human genome database have identified at least 107 PTPs encoded in the human genome [14,15]. PTPs are classified into four groups based on the amino acid sequences of their catalytic domains. The largest group, the class I cysteine-based PTPs, consists of 99 PTPs, including 38 well-known classical PTPs that have strict specificity for phosphotyrosine. These classical PTPs are further categorized as either receptor-like PTPs or nonreceptor-like PTPs [16–19]. Importantly, functional studies have demonstrated that PTPs are associated with carcinogenesis similar to PTKs. Of the 38 classical PTPs, 22 have been shown to play a tumor suppressive role in different types of human cancer. These tumor suppressive PTPs include eleven members of the receptor-like PTP subfamily (PTPRA, PTPRD, PTPRF, PTPRG, PTPRH, PTPRJ, PTPRK, PTPRM, PTPRO, PTPRS, and PTPRT) and eleven members of the nonreceptor-like PTP subfamily (PTP1B, TC-PTP, PTPH1, STEP, SHP1, HePTP, PTP-PEST, PTPBAS, PTP36, BDP, and PTPD1). Given this tumor suppressive capability, it is reasonable to hypothesize that cancer cells may bear somatic mutations of PTPs and/or underexpress PTPs [15]. On the other hand, other PTPs have been shown to act as oncogenes by stimulating cell proliferation and survival. Studies have identified 11 of the 38 classical PTPs act as potential oncogenes [15,20,21]. These oncogenic PTPs include five members of the receptor-like PTP subfamily (PTPRA, PTPRB, PTPRF, PTPRG, and PTPRH) and six members of the nonreceptor-like PTP subfamily (PTP1B, PTPH1, SHP1, HePTP, SHP2, and PTPD1). In order to combat the oncogenic functions of some PTPs, inhibitors were developed to use as potent anti-tumor drugs.
PTK IN CARCINOGENESIS
The roles of PTKs in cell signaling
Protein tyrosine kinases (PTKs) are essential enzymes which can transfer a phosphate to the amino acid tyrosine within a protein [22]. Phosphorylation of tyrosine residues by PTKs changes the properties of a protein so that it becomes active and transmits the cellular signal downstream in a cascading manner [23]. The activation of PTKs is involved in various major cellular processes including cell growth, differentiation, cytoskeletal rearrangement, cell migration, and apoptosis [4]. These PTK–mediated signaling pathways are regulated by protein tyrosine phosphatase (PTPs). Until now, approximately 20 receptor tyrosine kinase (RTK) families and at least 10 distinct groups of non-receptor tyrosine kinases have been identified in humans and represent about 2% of all human genes [3]. Most RTKs are single-pass, transmembrane proteins. Upon binding of a ligand like epithelial growth factor (EGF), RTKs promote their dimerization and the subsequent autophosphorylation of receptor tyrosine residues transmits extracellular signals to the cytoplasm or to the nucleus (Figure 1). Non-receptor tyrosine kinase is localized in the cytosol or nuclear matrix [24]. Interestingly, those kinases found in the nuclear envelope may be involved in DNA stability, mitosis, or DNA repair [25]. The signals transmitted by RTKs to the nucleus may modify gene expression by activating transcriptional factors or by regulating cell cycle-associated proteins. Therefore, mutation of PTKs which permit aberrant expression and constitutive activation of PTKs, initiates and promotes carcinogenesis [26].
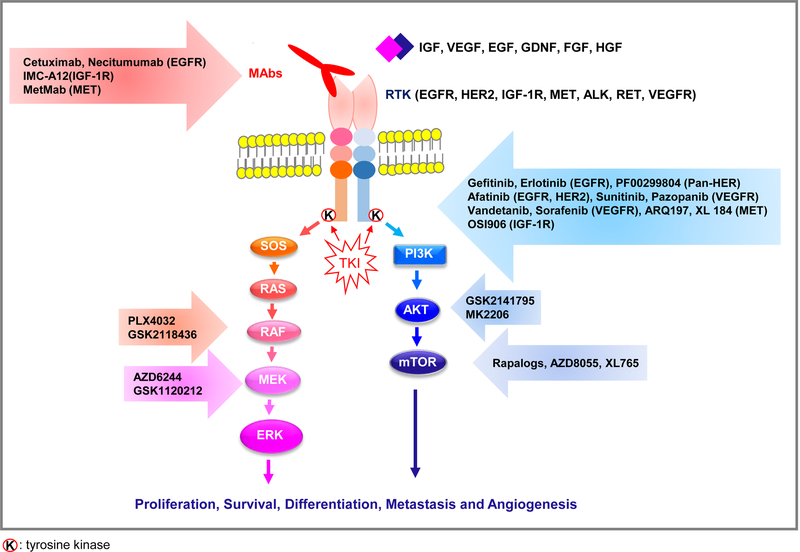
Figure 1. Schematic representation of the RTK signaling network and nodes of therapeutic blockade.
Activation of RTKs can result in signaling via two pathways: PI3/AKT and RAS/RAF/MEK/ERK. PI3K/AKT signaling induces cell survival, increases protein synthesis, activates glucose metabolism, and decreases apoptosis. RAS/RAF/MEK/ERK increases cellular proliferation, angiogenesis, migration, and differentiation by activating transcriptional factors in a cascade. Each ligand binds to RTK and transfers the extracellular signal to the cytosol and then, ultimately, to the nucleus. Targeted therapy using monoclonal antibody (-MAB or -IB) or TKI can block the extracellular signals which enter the cell through the RTK pathway.
PTK in skin carcinogenesis
Since the discovery of the association between multistage carcinogens and Harvey-ras gene in 1983 [27], the mouse skin model of multistage carcinogenesis has been a relatively simple, well-designed, and useful tool, providing evidence on how accumulated genetic change can lead to tumorigenesis and providing clues towards the development of therapeutic methods to prevent carcinogenesis [28]. In this model for mouse skin carcinogenesis, skin tumor development occurs via three major stages - initiation, promotion and progression [29]. Tumor initiation by a single topical subcarcinogenic dose of the genotoxic carcinogen, such as 7, 12-dimethylbenz[α]anthrancene (DMBA), induces a mutation in a critical gene (Ha-ras) or genes. Typically, this initiation stage does not produce morphological changes in mouse skin. Following initiation, tumor promotion occurs by repeatedly applying the non-mutagenic tumor promoter, such as 12-O-tetradecanoylphorbol-13-acetate (TPA), triggering a variety of tumor-related gene expression which affects epidermal cell proliferation and hyperplasia. Cells undergo clonal expansion, resulting in the development of premalignant papilloma. During this process, many kinases and phosphatases, such as RTKs and PTPs, are activated by posttranslational modification. For example, investigation of the potential regulatory role of epidermal growth factor receptor (EGFR) in the activity of signal transducer and activator of transcription 3 (STAT3) during tumor promotion revealed that addition of exogenous EGF in primary cultures of mouse keratinocytes led to activation of STAT3 as evidenced by an elevation in tyrosine phosphorylation and nuclear translocation. Also, in the epidermis of transgenic mice expressing TGFα under control of the keratin 14 promoter, STAT3 was constitutively activated. However, abrogation of EGFR function in mouse epidermis using an EGFR kinase inhibitor or by overexpressing a dominant negative form of EGFR led to a reduction in STAT3 activation in response to TPA treatment which activates STAT1, 3 and 5 under normal conditions. Immunoprecipitation analyses using lysates from TPA-treated epidermis and skin papillomas showed enhanced interaction between EGFR and STAT3. Furthermore, STAT3 deficiency in mouse epidermis significantly reduced the proliferative response after TPA treatment. These results revealed STAT3 activation by EGFR in tumor promoter-treated epidermis and in skin papillomas may be a critical event during mouse skin tumor promotion, possibly through regulation of keratinocyte proliferation [30,31]. In the final stage of multistage mouse skin carcinogenesis, progression, papillomas convert to squamous carcinoma in correlation with additional epigenetic modulation such as gene addition, deletion or chromosomal switch [29]. Research of skin carcinogenesis has contributed a great deal to understanding how PTKs can play a role in cancer.
PTK as a therapeutic target for chemotherapy
Over half of the 90 PTK genes that have been identified have been shown to be involved in human cancer either as tumor promoters through gain of function mutations (Bcr-Abl); gene amplification (EGFR); or overexpression (c-Src) or as tumor suppressors (Syk, c-Fes, Csk, EphB2, 3 or 4). Since the mid-2000s when gene target therapy began, PTKs have emerged as validated targets for novel anti-cancer drugs (Figure 2). Recently, inhibitors for EGFR (afatinib [32], gefitinib [33], erlotinib [34,35], cetuximab [36,37], panitumumab [38], necitumumab [39], and osimertinib [40]), inhibitors for anaplastic lymphoma kinase (ALK) (alectinib [41,42], crizotinib [43], brigatinib [44], and ceritinib [45,46]), inhibitors for RET (cabozantinib [47,48] or and vandetanib [49]) (Table 1), inhibitors for insulin-like growth factor 1 receptor (IGF-1R) (Cixutumumab [50,51], figitumumab [52], Dalotuzumab [53,54], Ganitumab [55,56], R1507 [57], Robatumumab [58], AVE1642 [59,60], MEDI-573 [61,62], Linsitinib [63], BMS-754807 [64,65], and BVP-51004 [66]) (Table 2 and 3), and inhibitors for Src (Dasatinib [67–72], Saracatinib [73], and Bosutinib [74–78]) (Table 4) have been in clinical trials for the treatment of a variety of cancers. In this current review, we summarize the signaling mechanisms and target therapy for representative PTKs including EGFR, IGF-1R and proto-oncogene c-Src (Src).
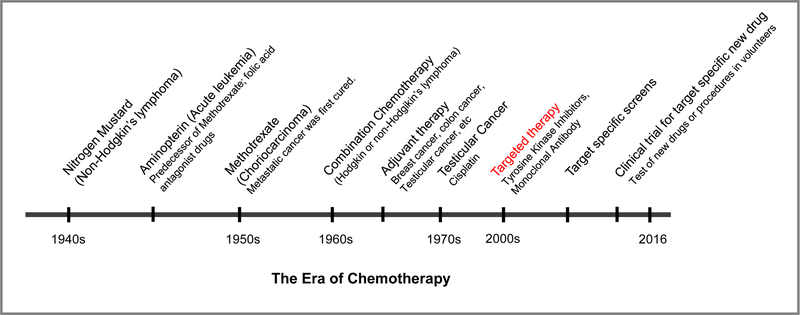
Figure 2. The era of chemotherapy.
Since the discovery of nitrogen mustards and folic acid antagonist drugs in the 1940s, the history of chemotherapy has begun. In the mid-2000s, when PTKs were discovered and revealed to be involved in carcinogenesis, the approach toward chemotherapy evolved to target specific cancer-associated molecules like PTKs. However, due to chemoresistance, targeted therapy has still been challenged by combination chemotherapy in clinical trials.
Table 1.
Targeted therapy for EGFR, ALK, and RET
| Inhibitor | Target | Types of cancer |
|---|---|---|
| Gilotrif(Afatinib) [32] | EGFR | NSCLC, pancreatic cancer, colon cancer, head and neck cancer |
| Iressa (Gefitinib) [33] | ||
| Tarceva (Erlotinib) [34, 35] | ||
| Erbitux (Cetuximab) [36, 37] | ||
| Vectibix (Panitumumab) [38] | ||
| Portrazza (Necitumumab) [39] | Squamous cell lung cancer | |
| Tagrisso (Osimertinib) [40] | EGFR (T790M) | Cancers failed with Gilotrif, Iressa or Tarceva |
| Xalkori (Crizotinib) [43] | ALK | Lung cancer, metastatic NSCLC with ROS1 mutation |
| AP26113 (Brigatinib) [44] | ALK, EGFR (T790M) | NSCLC |
| Alecensa (Alectinib) [41, 42] | ALK | Metastatic tumor in the brain, central nervous system |
| Zykadia (Ceritinib) [45, 46] | ALK | Metastatic tumor in the brain, central nervous system, NSCLC |
| Cometriq (Cabozantinib) [47, 48] | RET | Thyroid cancer, prostate cancer, melanoma |
| Caprelsa (Vandetanib) [49] |
Table 2.
Targeted therapy for IGF-1R
| Inhibitor | Target | Types of cancer |
|---|---|---|
| IMC-A12 (Cixutumumab) [50, 51] | IGF-1R | Advanced nonsquamous NSCLC, metastatic docetaxel-pretreated castration-resistant prostate cancer |
| CP-751, 871 (Figitumumab) [52] | IGF-1R | Advanced solid tumor |
| MK-0646 (Dalotuzumab) [53, 54] | IGF-1R | Advanced solid tumor, KRAS wild-type, metastatic colorectal cancer |
| AMG 479 (Ganitumab) [55, 56] | IGF-1R | Mutant KRAS metastatic colorectal cancer, metastatic adenocarcinoma of the pancreas |
| R1507 [57] | IGF-1R | Recurrent or refractory rhabdomyosarcoma, osteosarcoma, dynovial sarcoma, other soft tissue sarcoma |
| SCH-717454 (Robatumumab) [58] | IGF-1R | Relapsed osteosarcoma and Ewing sarcoma |
| AVE1642 [59, 60] | IGF-1R | Advanced solid tumor |
| MEDI-573 [61, 62] | IGF-1R and IGF-2 | Advanced solid tumor |
Table 3.
Small molecular inhibitors targeting IGF-1R
Table 4.
Targeted therapy for Src
| Inhibitor | Target | Types of cancer |
|---|---|---|
| BMS-354825 (Dasatinib) [67–72] | Src Bcr-Abl PDGFR c-Kit EphA2 |
HR/HER2-positive breast cancer, triple-negative breast cancer, castration-resistant prostate cancer, NSCLC, colon cancer, HNSCC metastatic breast cancer, myeloid leukemia |
| AZD0530 (Saracatinib) [73] | Src Bcr-Abl |
castration-resistant prostate cancer |
| SKI-606 (Bosutinib) [74–78] | Src Bcr-Abl |
Myeloid leukemia, metastatic breast cancer |
EGFR
The epidermal growth factor receptor (EGFR; ErbB-1; HER1 in humans) was the first structure discovered as a receptor tyrosine kinase of the EGF family and it is a member of the ErbB family of receptors which includes EGFR (ErbB-1), HER2/c-neu (ErbB-2), Her 3 (ErbB-3) and Her 4 (ErbB-4) [79]. Upon binding with its ligands, such as EGF or TGFα, EGFR can transform to an active homodimer and autophosphorylate [80,81]. In addition, EGFR may create an active heterodimer with another member of the ErbB receptor family, such as ErbB2/Her2/neu. Dimerization of EGFR subsequently leads to the autophosphorylation of several tyrosine (Tyr) residues in the C-terminal domain including Tyr992, Tyr1045, Tyr1068, Tyr1148 and Tyr1173 [82]. Consequently, autophosphorylated EGFR can transmit extracellular signals to downstream signaling proteins through several signal transduction cascades, like the Mitogen-activated protein kinase (MAPK), AKT, Src, and Janus kinase (JAK) pathways, which leads to DNA synthesis and cell proliferation, cell survival, cell migration and focal adhesion (Figure 3). Thus, overexpression or aberrant activity of EGFR can contribute to the development of several cancers, including lung cancer, head and neck carcinoma, and glioblastoma [83–85]. For example, in skin cancer arising from exposure to ultraviolet B radiation (UVB), UVB has been known to activate EGFR and induce EGFR-mediated pathways, such as AKT-, PKC-, and protein kinase A-dependent signal transduction pathways [86,87]. Additionally, low dose UVB irradiation of cancer cells overexpressing EGFR prior to adding EGF halted the EGFR signaling pathway due to conformational change of EGFR by UVB [88,89].
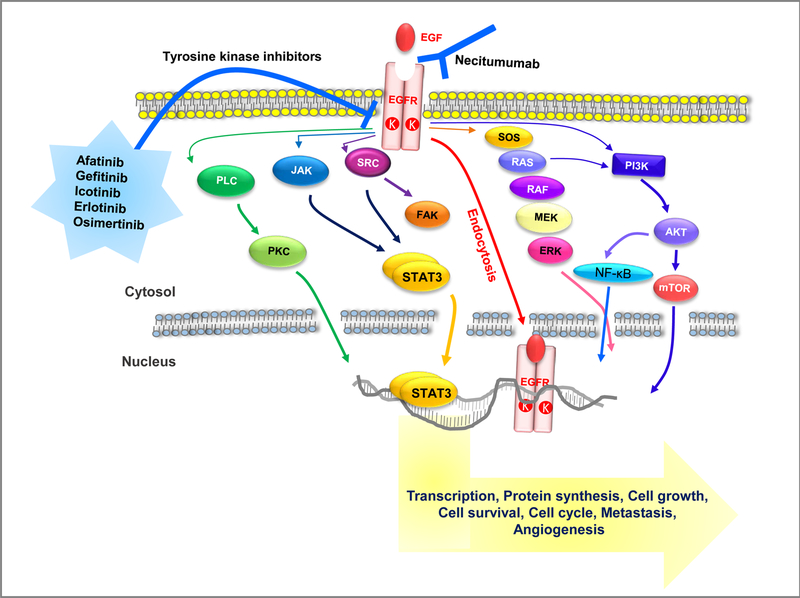
Figure 3. EGF/EGFR signal transduction and its targeted therapy.
The binding of EGF to its receptor initiates a variety of signaling cascade via five main pathways; 1) RAS/RAF/MAPK, 2) PI3K/AKT, 3) JAK/STAT, 4) PLCγ/PKC/Ca2+-dependent, and 5) Src/FAK/MMP. Furthermore, the EGF/EGFR dimer can directly regulate the expression of specific genes through an endocytosis mechanism. Constitutive activation of EGFR as a result of mutation of the EGFR gene can promote cancer by facilitating DNA synthesis, cell proliferation, angiogenesis, invasion, and metastasis. Ongoing clinical trials of anticancer drugs, like monoclonal antibodies of the EFG binding site or small molecules against the EGF catalytic domain, may prove to inhibit EGF/EGFR signaling by blocking its autophosphorylation.
Since the identification of EGFR as an oncogene, a variety of therapeutic approaches have been developed and clinical trials investigating cancer treatments have been performed with EGFR inhibitors, including gefitinib, erlotinib, and, afatinib for lung cancer, and cetuximab for lung cancer or colon cancer as shown in Table 1. Generally, monoclonal antibody inhibitors are divided into two groups by the route, or method of treatment: –MAB is for intravenous injection (IV) and –IB is for oral drug. With the binding site blocked by a monoclonal antibody, extracellular signaling molecules can no longer attach and activate the tyrosine kinase as shown in Figure 1. Another therapeutic approach that targets EGFR is using small molecules called tyrosine kinase inhibitors, such as gefitinib, brigatinib and lapatinib, to inhibit the EGFR tyrosine kinase, which is on the cytoplasmic side of the receptor [90].
In addition, given the issue of drug resistance, other new approaches aimed at EGFR have been developed. Two sources of resistance, T790M and MET oncogene have been found. While in clinical trial phase II, brigatinib received breakthrough therapy designation status by the FDA in October 2014 and is on track to file for approval in the U.S. in the third quarter of 2016 [44,91].
IGF-1R
Insulin-like growth factor 1 (IGF-1) is secreted primarily by the liver upon stimulation with human growth hormone (HGH), whereas IGF-2 is non-HGH dependent and is expressed in a variety of tissues [92,93]. At least six well-characterized IGF-binding proteins (IGFBP-1 through-6) bind IGFs and prevent their action on the receptors. In serum, only approximately 2% of IGF ligands exist in the unbound form. At the tissue level, bioavailability of IGF-1 and IGF-2 is modulated by IGFBP protease and the presence of the non-signaling, IGF-2-binding IGF-2R [94,95]. IGF-1 binds to at least two receptor tyrosine kinases, including the IGF-1 receptor (IGF-1R) and the insulin receptor, which transmits cellular signals by causing the addition of a phosphate molecule on tyrosine residues.
IGF-1R is a transmembrane receptor that is activated by the IGF-1 hormone and by the related hormone IGF-2. It belongs to the large class of tyrosine kinase receptors and it is a member of a family which consists of the insulin receptor (60% homology with IGF-1R) and the IGF-2R. IGF-1 receptor is comprised of two alpha subunits and two beta subunits. Both α and β subunits are synthesized from a single mRNA precursor. The precursor is then glycosylated, cleaved, and crosslinked by cysteine bonds to form a functional transmembrane αβ chain. The α chains are located at the extracellular membrane, while the β subunit spans the membrane and is responsible for intracellular signal transduction upon ligand stimulation. Like EGFR, IGF-1 binding to IGF-1R induces autophosphorylation on tyrosine residues 1165 and 1166 of the receptor and triggers IR substrates (IRS-1 through −4) and the Src-homology collagen protein (Shc)-mediated cascade signaling pathway [96,97]. In turn, the Shc-mediated cascade leads to activation of the MAPK pathway and the phosphatidylinositol-4,5-biphosphate 3-kinase (PI3K)-AKT pathway. These events promote cell survival and cell proliferation in mitosis-competent cells and in tissues such as skeletal muscle and cardiac muscle [98,99]. In a recent study using a mouse model lacking the IGF-1 receptor, the loss of IGF-1 resulted in the failure of late development and a dramatic reduction in body mass [100,101]. Furthermore, aberrant IGF signaling has been shown to be associated with numerous cancers, including colon cancer [102], prostate cancer [103], pancreatic cancer [104], melanoma [105], and osteosarcoma [106], as well as childhood malignancies [107]. For example, increased levels of the IGF-IR are expressed in the majority of primary and metastatic prostate cancer patient tumors and required for survival and growth when prostate cancer cells progress to androgen independence [103]. However, unlike other growth factor receptors such as EGFR and HER-2, activating mutations of the IGF-1R gene have not been reported.
Based on the previous successful approach to inhibitors directed against the EGFR family members, more than 10 IGF/IGF-1R inhibitors have entered clinical trials to test the effect in the treatment of cancer patients, and these divided into three group: (1) monoclonal antibodies against IGF-1R, (2) monoclonal antibodies against IGF-1R ligands (IGF-1 and IGF-2), and (3) IGF-1R tyrosine kinase inhibitors. As shown in Table 2, among IGFR monoclonal antibodies [50–62], MEDI-573 is the only monoclonal antibody in clinical development that targets the ligands IGF-1 and IGF-2 [61,62]. MEDI-573 inhibits IGF-induced IGF-1R and IR-activation without inhibiting insulin signaling. Several small molecule inhibitors against IGF-1R are under clinical investigation [63–66] (Table 3). Among them, OSI-906 is the most specific, whereas others also inhibit receptor tyrosine kinases beyond the IGF-1R and IR family because of the high degree of homology between IGF-1R and IR. Currently, OSI-906 is being tested in combination with erlotinib on patients with non-small-cell lung carcinoma (NSCLC) possessing EGFR activating mutation; in combination with standard of care for pancreatic cancer; and combination with cetuximab for head and neck cancer (https://clinicaltrials.gov). However, thus far, none have shown a significant benefit. The reason for this failure seems to be the complexity of the IGF-1R signaling pathway, such as the crosstalk among EGFR, IGF-1R, the estrogen receptor, and HER-2 which may induce resistance to a drug, as well as a lack of tumor selection markers. The development of chemoresistance is also an issue which seems to be supported by the recent data – trastuzumab-resistant ovarian cancer by IGF-1R and ErbB3/HER3 [108] or centuximab-resistant metastatic colorectal cancer by IGF-1R and c-met [109]. Consequently, understanding the complexity of the IGF-1R system may be a breakthrough to develop new therapeutic strategies and approaches including combinational target therapy.
Src
Proto-oncogene tyrosine-protein kinase Src, known as cellular Src kinase (c-Src), is a non-receptor tyrosine kinase protein in normal mammalian cells discovered in 1979 [110]. Src belongs to the Src family of kinases (SFKs) including blk, c-fgr, fyn, hck, lck, lyn, c-Src, c-yes, and yrk [111,112]. The constitutive activation of c-Src tyrosine kinase caused by genetic mutations is implicated in cancer progression given that c-Src activity promotes signaling by RTKs like EGFR, HER2, platelet-derived growth factor receptor, IGF-1R and c-Met/hepatocyte growth factor receptor and c-Src directly transduces survival signals to the downstream PI3K-AKT and STAT3 pathways [113,114]. Src activation also stabilizes the focal adhesion complex, which is composed of FAK, paxillin, RhoA and other components and which stimulates cell adhesion to the extracellular matrix [115,116]. The structure of c-Src consists of six functional regions including an SH4 domain, a unique region, an SH3 domain, an SH2 domain, a tyrosine kinase domain (catalytic domain) and a short regulatory tail. When Src is inactive, the phosphorylated tyrosine group at the 527 position interacts with the SH2 domain which helps the SH3 domain interact with the flexible linker domain and thereby keeps the inactive unit tightly bound. The activation of c-Src causes the dephosphorylation of the Tyr527 residue and autophosphorylation of the residue Tyr416 [117]. c-Src can be activated by a variety of transmembrane proteins including adhesion receptors, receptor tyrosine kinases (EGFR), G-protein coupled receptors and cytokine receptors. Src activation promotes survival, angiogenesis, proliferation and motility. Its kinase activity can be inhibited via phosphorylation by C-terminal Src kinase which phosphorylates tyrosine residues located in the C-terminal end of SFKs [118]. The expression of SFKs depends on tissue and cell types [119–121]. Thus, the aberrant expression or activation of c-Src in tissue indicates that it is involved in cancer progression. For example, in colon cancer, increased c-Src expression promotes tumor progression including metastasis [122,123]. In breast cancer, EGFR not only activates c-Src but also increases the activity of c-Src. In addition, overexpression of c-Src increases the response of EGFR-mediated processes, and EGFR and c-Src enhance the effects of one another in metastatic breast cancer [124,125].
A number of tyrosine kinase inhibitors of c-Src tyrosine kinase have been developed and utilized for targeted anti-cancer therapy as shown in Table 4 [67–78]. Among them, dasatinib (BMS-354825, Sprycel) is a multi-targeted inhibitor of RTKs, including BCR-ABL fusion protein, stem cell factor receptor, and platelet-derived growth factor receptor, which led to its approval for the treatment of chronic myeloid leukemia and Philadelphia chromosome-positive acute lymphocytic leukemia. Dasatinib is also in clinical trials for use in treating gastrointestinal stromal tumors [126], malignant pleural mesothelioma [126], sarcomas [127], NSCLC [128], colorectal cancer [129,130], glioblastoma [131], multiple myeloma [126], melanoma [132,133], head and neck cancer [72], metastatic breast cancer [134], and prostate cancer [126]. However, recent studies of ongoing combination therapy with drugs such as erlotinib, FOLFOX and cetuximab reveal the various, complicated roles of Src signaling.
IMPORTANT FACTORS INVOLVED IN SIGNALING PATHWAYS REGULATED BY PTK ACTIVATION
STAT3
Signal transducer and activator of transcription (STAT) proteins were characterized as a family of cytoplasmic transcription factors that mediate normal cellular responses to cytokines, growth factors, and other polypeptide ligands [135,136]. The activation of STATs is an important event for the regulation of cytokine and growth factor-induced cellular and biological processes, including proliferation, differentiation, survival, apoptosis and inflammation. The STAT family of proteins is comprised of seven structurally and functionally related proteins: STAT1, 2, 3, 4, 5a, 5b, and 6. All of the family members share six distinct domains: the N-terminal domain, the coiled-coil domain, the DNA-binding domain, the linker domain, the Src homology 2 (SH2) domain, and the transactivation domain which contains a critical tyrosine residue (Tyr705 in STAT3) at the C-terminus that is phosphorylated during activation. STAT activation by phosphorylation is mediated by growth factor receptor tyrosine kinases and cytoplasmic kinases, such as cytokine receptor-associated JAKs and Src family kinases. Phosphorylation induces STAT-STAT homodimer complex formation via the interaction of the phosphorylated tyrosine of one monomer and the SH2 domain of another [135,136]. From the cytoplasm, STATs translocate to the nucleus where they regulate gene transcription by binding to specific DNA response elements [135]. Nuclear accumulation of STATs as monomers or dimers remains to be fully defined and may involve several mechanisms [137], including active shuttling between the cytoplasm and nucleus [138].
In contrast to the transient STAT activation in normal cells, approximately 70% of many human solid and hematological tumors display overexpression of STAT3 or constitutive activation of STAT3. These results strongly implicate a major role for aberrantly active STAT3 in tumor formation. Accumulating data show that constant STAT3 activation is required for aberrant cell proliferation in carcinogenesis. In gastric cancer, STAT3 activation by IL-26 mediates up-regulated expression of Bcl-2, Bcl-xL and c-Myc, which in turn facilitates cell proliferation [139]. Activated STAT3 is involved in cell proliferation of endometrial, bladder, colon and renal cancers [140–143]. STAT3 promotes cell survival in esophageal, colon, gastric and other types of cancer [139,144,145]. On the contrary, inhibition of STAT3 results in decreased proliferation and increased apoptosis in cancer cells. The microRNA miRNA130b, which targets STAT3, inhibits proliferation in pancreatic cancer cell [146] and STAT3 inhibitors also decrease cell proliferation and promote apoptosis in breast cancer, colorectal cancer, gastric cancer and lung cancer [147–152].
STAT3 interacts with several factors involved in angiogenesis which includes degradation of the vascular basement membrane, vascular epithelial cell proliferation, migration and new vessel formation [153]. STAT3 inhibition causes decreased angiogenesis by down-regulation of metalloproteinase 2 (MMP-2), which has a role in degradation of vascular basement membrane. Conversely, STAT3 activation induces elevated MMP-2 expression [154]. STAT3 also induces VEGF expression, which plays a crucial role in invasion and metastasis of human cancers such as ovarian carcinoma [155]. Thus, inhibition of angiogenesis by blocking STAT3 signaling would be an attractive strategy in preventing or delaying tumor formation.
Studies using STAT3-deficient mice showed that STAT3 activation is engaged in both the initiation and promotion stages in skin carcinogenesis [156]. Loss of STAT3 in K5.Cre × STAT3flox/flox mice resulted in a significant reduction of epidermal hyperproliferation compared to control mice following TPA treatment [31,156]. Mechanistic studies showed that recovery of cell cycle regulatory proteins cyclin D1 and cyclin E was delayed and c-myc expression was constantly downregulated in the epidermis of K5.Cre × STAT3flox/flox mice after treatment with TPA in comparison to control mice. Thus, deletion of STAT3 in the basal layer of epidermis inhibited TPA-induced epidermal hyperproliferation during tumor promotion. Using the temporally regulated epidermis-specific STAT3-deficient mouse model, temporal disruption of STAT3 at the stage of carcinogenesis initiation resulted in an increased number of apoptotic cells following treatment with carcinogen 7,12-dimethylbenz[a]anthracene (DMBA, initiation chemical of skin carcinogenesis) [157]. Inducible deletion of STAT3 in epidermis prior to DMBA treatment delayed tumor onset and reduced the number of papillomas. Similarly, inducible deletion of STAT3 prior to TPA treatment during the tumor promotion stage delayed tumor onset and tumor growth [157]. The epidermis-specific STAT3-deficient mouse model facilitated further study of this important molecule in UVB-mediated skin carcinogenesis. UVB radiation is the prime risk factor for nonmelanoma skin cancer in humans [158]. Following exposure to UVB, the level of phosphorylated STAT3 (p-STAT3) is initially decreased, followed by a significant increase at later time points in the mouse epidermis. The levels of STAT3 target genes, such as cyclin D1, Bcl-xL, and c-Myc, followed the changes in activated STAT3 in response to UVB irradiation [159]. Epidermal-specific STAT3-deficient mice were very sensitive to UVB radiation as revealed by a higher number of sunburned mice and a high number of apoptotic cells following UVB irradiation [160]. On the other hand, the epidermis of K5.STAT3C (constitutive active STAT3 form) mice was resistant to UVB-induced apoptosis [161]. These results demonstrate that STAT3 plays an important role in the development of UVB-induced skin tumors through its effects on both proliferation and survival of keratinocytes [162].
As mentioned, STAT3 regulates gene expression involved in proliferation, apoptosis, and angiogenesis. Also, STAT3 activation is required for both the initiation and promotion stages during skin carcinogenesis. Therefore, targeting STAT3 using a specific inhibitor may be an attractive cancer treatment approach. A number of small molecule compounds directly inhibit the activity and function of STAT3 which have been developed for use in cancer treatment and prevention. The three domains of STAT3 – NH-2 terminal domain, DNA-binding domain and SH2 domain – were selective targets for the development of STAT3 inhibitors, which block STAT3 function(s) and signaling by preventing phosphorylation, dimerization, nuclear translocation and DNA binding [163,164]. Table 5 summarizes the small molecular inhibitors targeting STAT3.
Table 5.
Small molecular inhibitors targeting STAT3
| Inhibitor | Target | Types of cancer/transformed cell lines |
|---|---|---|
| PY*LKTK [165] | SH2 domain | Transformed fibroblasts |
| STA-21 [166] | SH2 domain | Breast cancer |
| LLL-3 [167] | SH2 domain | Breast cancer |
| Stattic [168] | SH2 domain | Breast cancer |
| S3I-201 [169] | SH2 domain | Breast cancer, hepatocellular carcinoma |
| S3I-M2001 [170] | SH2 domain | Breast cancer, pancreatic cancer |
| BP-1–102 [171] | SH2 domain | Breast cancer, lung cancer |
| HIC 1 [172] | DNA binding domain | Breast cancer |
| IS3–295 [173] | DNA binding domain | Colon cancer |
| DBD-1 [174] | DNA binding domain | Melanoma |
| InS3–54 [175] | DNA binding domain | Breast cancer, lung cancer |
| ST3-H2A2 [178] | N-terminal domain | Prostate cancer |
| G-quartet ODN [184, 185] | SH2 domain | Head and neck cancer, breast cancer, prostate cancer |
Inhibitors targeting the SH2 Domain of STAT3.
The SH2 domain of STAT3 plays a pivotal role in STAT3 activation by mediating the interaction of STAT3 with phosphorylated tyrosine residues on the cytoplasmic region of any activated receptors. Inhibition of this target can block the formation of STAT3 dimer and consequently inhibit nuclear translocation and STAT3-dependent gene regulation. A peptide composed of PY*LKTK (Y* is the phosphorylated tyrosine) was derived from the STAT SH2 domain-binding peptide sequence. It can directly form a complex with STAT3 monomer and inhibit STAT3 activity by disrupting STAT3 dimerization [165]. STA-21 is a natural compound that specifically binds to SH2 domain and inhibits STAT3 dimerization and nuclear translocation [166]. LLL-3 is STA-21 derivative that possesses comparable anti-proliferative activity to STA-21 but exhibits increased cell permeability [167]. Stattic selectively inhibits dimerization and prevents STAT3 translocation to the nucleus [168]. As a result, stattic induces apoptosis in breast cancer apoptosis. S3I-201, a salicylic acid derivative, blocks the formation of STAT3 homodimers through SH2 domain binding and inhibits proliferation of breast and hepatocellular cancer cells in mice [169]. BP-1–102, a S3I-201 analog, inhibits STAT3 via the same mechanism and selectively suppresses malignant cell growth, transformation, survival and migration [170]. Additionally, this compound displays oral bioavailability. S3I-M2001, an oxazole-based peptidomimetic, selectively blocks STAT3 dimerization and inhibits STAT3 dependent transcription, transformation, survival and migration [171].
Inhibitors targeting the DNA binding domain of STAT3.
To regulate gene expression, it is essential for the STAT3 DNA-binding domain (DBD) to physically interact with the consensus DNA-binding sequence in the target gene’s promoter. Thus, STAT3 activity can be inhibited by targeting the STAT3 DBD to prevent interaction with the target gene’s promoter and thus block its tumor-promoting functions. Hypermethylated in cancer 1 (HIC1) gene naturally forms a complex with STAT3 protein via direct binding between the C-terminal domain of HIC1 and the STAT3 DBD [172]. This interaction prevents STAT3 binding to the promoters of its target genes, such as VEGF and c-myc. Platinum compounds, such as CPA-1 and CPA-7 inhibit STAT3 DNA binding, hence suppressing cell growth and increasing cell death in several human cancers. More recently, another platinum compound, IS3–295, was shown to inhibit STAT3 DNA binding capability although its mechanism remains unclear [173]. DBD-1, a small peptide aptamer, also blocks STAT3 DNA binding and induces significant apoptosis in murine melanoma cells [174]. More recently, the compound InS3–54 was identified; it inhibits STAT3 activity and it is capable of inducing apoptosis in breast and lung cancer cell lines [175].
Inhibitors targeting the STAT3 N-terminal domain.
The N-terminal domain of STAT3 comprises ~130 amino acids and contains eight helices, which have multiple biological activities, including dimer formation, binding to promoter and assembly of transcriptional machinery [176,177]. Compounds targeting the N-terminal domain of STAT3 may therefore inhibit tumorigenesis. ST3-H2A2, a synthetic compound, binds to the STAT3 N-terminal domain and activates expression of proapoptotic genes, thereby initiating apoptosis in cancer cells [178].
Oligonucleotide approaches to inhibit STAT3 signaling.
Approaches targeting gene expression based on oligonucleotide technology include antisense RNA, small interfering RNA (siRNA), and decoy oligodeoxynucleotide (ODN). The knockdown of the STAT3 protein by antisense RNA or siRNA approaches has been demonstrated in studies which showed the induction of tumor cell apoptosis and tumor regression following loss of STAT3 expression [179–181]. A STAT3-decoy oligonucleotide (ODN) can trap an activated STAT3 dimer in the cytoplasm by inhibiting interaction between active STAT3 and importin, which can result in increased apoptosis in colorectal cancer cells [182]. Also, ODN is a competitive inhibitor of STAT3, and thereby can suppress STAT3-dependent transcription of genes such as cyclin D, c-Myc, Survivin, and Bcl-xL [183]. Decreased expression of these genes inhibits proliferation and increases apoptosis in tumor cell lines [184,185].
PI3K/AKT
The PI3K/AKT pathway is activated by various different cellular factors, including binding of ligands to RTKs and G-protein coupled receptors and GTP binding of RAS proteins. These signals activate the catalytic activity of PI3K, which consists of the regulatory (p85) subunit, harboring two SH2 domains, and the catalytic (p110) subunit. PI3K phosphorylates the 3’ position of the inositol ring of lipids in the cytosolic membrane, resulting in the production of phosphatidylinositol-(3,4)-P2 (PIP2) and phosphatidylinositol-(3,4,5)-P3 (PIP3). PIP2 and PIP3 interact with pleckstrin homology (PH) domains of intracellular proteins, resulting in the localization of PH domain containing proteins to the inner surface of the plasma membrane. One such protein is AKT (also known as protein kinase B, or PKB), which was initially identified as the oncogene in a transforming murine retrovirus [186]. AKT, which has three homologous isoforms (AKT1, AKT2, AKT3), is a serine-threonine kinase that normally exists in the cytoplasm in an inactive state [187]. All three isoform possess a similar structure: an N-terminal PH domain, a central S/T catalytic domain and a C-terminal regulatory domain. After activation of PI3K, AKT is translocated to the cell membrane where it is phosphorylated at two regulatory sites, Thr308 and Ser473. The Thr308 residue, which is in the catalytic domain of AKT, is phosphorylated by phosphoinositide-dependent kinase 1, which is another PH domain containing kinase that is recruited to the plasma membrane [188,189]. Members of the PI3K-related kinase family, including DNA-PK also phosphorylate AKT at Ser473. The Ser473 residue is in the regulatory domain and is phosphorylated in response to growth factor stimulation by the mechanistic target of rapamycin complex 2 (mTORC2) complex, which contains one of the PI3K-AKT pathways’ important downstream effectors, mTOR [190]. The activity of the PI3K-AKT pathway is regulated by the lipid phosphatase PTEN. PTEN dephosphorylates the 3’ position of PIP2 and PIP3 and thus directly antagonizes the activity of PI3K. Loss of PTEN results in constitutive activation of AKT.
AKT phosphorylates proteins that contain the R-X-R-X-X-S/T-B motif. The first AKT substrate identified was glycogen synthase kinase 3, which is an important metabolic enzyme and also a key node of other signaling cascades [191]. AKT modulates cellular proliferation, survival and cell cycle progression by phosphorylating numerous substrates, such as BAD, CDK inhibitors p21 and p27, MDM2, IKK-alpha and caspase 9 [192]. AKT regulates a variety of processes by inhibiting the function of the FOXO transcription factors, which are localized to the cytoplasm following phosphorylation by AKT [193]. Also, AKT regulates cellular metabolism, proliferation and survival through its effects on mTOR signaling, which is activated by AKT’s inhibitory phosphorylation of TSC2 [194,195]. AKT activity has also been shown to play a critical role in the regulation of other important cellular behaviors, including motility, invasion, and angiogenesis [187].
As previously mentioned, activation of the PI3K/AKT signaling pathway contributes to cell proliferation, survival and motility as well as angiogenesis, and thereby contributes to all the important aspects of tumorigenesis and tumor metastasis. Evidence has shown that AKT is overexpressed or activated in a variety of human cancers, including lung, breast, ovarian, gastric and pancreatic carcinomas [196]. Therefore, PI3K/AKT is considered to be an attractive target for cancer therapy and many specific inhibitors with acceptable pharmaceutical properties have been identified and developed (Table 6).
Table 6.
Inhibitors targeting PI3K/AKT in clinical trials for cancer treatment
| Inhibitor | Target | Types of cancer |
|---|---|---|
| BEZ235 (Dactolisib) [199, 200] | ATP competitive PI3K α/β/γ/δ | Glioblastoma multiforme, advanced breast cancer |
| BGT226 [201] | ATP competitive PI3K α/β/γ | Solid tumors, advanced breast cancer |
| BKM-120 (Buparlisib) [202] | ATP competitive PI3K α/β/γ/δ | Breast cancer, glioblastoma multiforme |
| GDC-0941 (Pictilisib) [203] | ATP competitive PI3K α/δ | Advanced breast cancer, NSCLC, melanoma, pancreatic cancer |
| SF1126 [204] | ATP competitive PI3K α/β/δ | Advanced solid tumors |
| PX-866 [205] | ATP competitive PI3K α/γ/δ | Ovarian cancer, prostate cancer, glioblastoma multiforme |
| CAL-101 (Idelalisib) [206] | ATP competitive PI3K δ | Chronic lymphocytic leukemia, acute myeloid leukemia, non-Hodgkin’s lymphoma |
| GSK690693 [211] | ATP competitive Akt 1/2/3 | Acute lymphoblastic leukemia |
| AZD5363 [212] | ATP competitive Akt 1/2/3 | Breast cancer, gastric cancer, prostate cancer |
| GSK2110183 (Afuresertib) [213] | ATP competitive Akt 1/2/3 | Multiple myeloma |
| GSK2141795 (Uprosertib) [214] | ATP competitive Akt 1/2/3 | Multiple myeloma |
| GDC-0068 (Ipatasertib) [215] | ATP competitive Akt 1/2/3 | Triple-negative breast cancer |
| MK2206 [217] | Allosteric Akt 1/2/3 | Advanced solid tumor |
| NSC-154020 (Triciribine) [218] | Allosteric Akt 1/2/3 | Hematologic malignancies, NSCLC |
Inhibitors targeting PI3K.
Two well-known PI3K inhibitors are the fungal metabolite wortmannin and LY294002. Wortmannin binds irreversibly to PI3K enzymes through covalent modification of a lysine essential for catalytic activity, whereas LY294002 is a classical reversible, ATP-competitive PI3K modulator in micromolar concentration. However, both wortmannin and LY294004 have little or no selectivity for individual PI3K isoforms and show substantial toxicity in animals [197,198]. In spite of the crossover inhibition of other lipid and protein kinases and their pharmaceutical properties, the preclinical studies of these PI3K inhibitors have greatly contributed to understanding the biological importance of PI3K signaling in the signal transduction network of human cancers and provided a platform for the discovery of novel PI3K inhibitors. At present, numerous PI3K-targeted compounds have been developed and introduced to clinical trials (Table 6). BEZ235 (dactolisib) is an imidazoquinazoline derivative that inhibits multiple class I PI3K isoforms and mTOR kinase activity via binding to the ATP-binding site of these enzymes [199]. BEZ235 showed strong anti-proliferative activity against tumor xenografts showing abnormal PI3K signaling including loss of PTEN function or PI3K gain of function mutations [200]. BGT226 is another potent pan-PI3K/mTOR inhibitor similar to BEZ235 [201]. Unlike BEZ235 and BGT226, BKM120 (buparlisib) is selective for class I PI3K enzymes with no mTOR inhibitory activity and it is capable of inducing apoptotic cell death in multiple myeloma cells [202]. GDC0941 (pictilisib), a thienopyrimidine derivative, inhibits all isoforms of class I PI3Ks in a nanomolar concentration [203]. It displayed potent antitumor activity against a panel of mouse xenograft models of human glioblastoma, breast cancer, small bowel gastrointestinal stromal tumor, follicular cell lymphoma and it is the first PI3K inhibitor to enter clinical trials in patients with advanced solid tumors or lymphoma. SF1126 is a covalent conjugate of LY294002 with an RGD (arg-gly-asp) peptide designed for increased solubility and enhanced delivery of the active PI3K inhibitor to the tumor resulting in significant antitumor activity in xenograft models [204]. In addition to its direct activity on cancer cells, SF1126 also had significant antiangiogenic activity in vivo with lowered toxicity compared to the LY294002. A number of compounds that preferentially target selected isoforms of class I PI3Ks are also under development. For example, PX-866 targets p110α, p110δ and p110γ with nanomolar half-maximal inhibitory concentration (IC50) values, while CAL-101 (idelalisib) is a p110 γ-selective inhibitor [205,206].
Inhibitors targeting AKT.
As the most well-known downstream effector of the RTK/PI3K complex, AKT is another attractive therapeutic target. Several AKT inhibitors have been developed, which can be grouped into a number of classes including lipid-based phosphatidylinositol analogs, ATP competitive inhibitors, and allosteric inhibitors. KRX 0401 (perifosine), the most clinically advanced inhibitor, is a lipid-based PI analog that targets the PH domain of AKT, thereby preventing binding to PIP3 and its membrane translocation [207]. In several preclinical models, such as the murine neuroblastoma model, KRX 0401 demonstrated substantial activity. Other AKT PH domain inhibitors, including PX316 and PIAs, showed inhibitory effects on the growth of tumor cells exhibiting increased PI3K/ AKT activity [208–210]. Most ATP-competitive AKT inhibitors are non-selective, targeting all AKT isoforms. GSK690693 is an ATP-competitive AKT kinase inhibitor, which targets all three AKT isoforms at nanomolar concentrations and is also active against additional kinases from the cAMP-dependent protein kinase C family [211]. In xenograft models, administration of GSK690693 led to significant growth inhibition in mice bearing SKOV-3 ovarian cancer cells, BT474 breast cancer cells and LNCap prostate cancer cells. AZD5363, another ATP-competitive inhibitor, inhibited all AKT isoforms [212]. Treatment with AZD5363 inhibited proliferation of 41 out of 182 solid and hematological tumor cell lines with the highest frequency of sensitivity occurring in breast cancer cells. Several clinical trials in phase I and II are being undertaken for breast, prostate and gastric cancers. GSK2110183 (afuresertib) is a highly potent inhibitor of AKT and showed the most sensitivity within hematological cell lines, such as acute lymphoblastic leukemia amd chronic lymphocytic leukemia [213]. GSK2141795 (uprosertib) is an analog of GSK2110183, the difference being the substitution of a bioisostere furan ring for a thiophene core [214]. This compound showed a similar capacity for AKT inhibition and subsequently, had a similar anti-proliferative effect as GSK2110183 but it demonstrated outstanding off-target kinase inhibition. ATP-competitive inhibitors are non-selective against AKT isozymes and are inadequately selective against similar kinases. GDC-0068 (ipatasertib) is an orally bioavailable inhibitor capable of inhibiting all three AKT isoforms. Treatment with GDC-0068 blocked cell cycle progression and decreased viability of cancer cell lines [215]. To address a major issue regarding the potential benefits of isoform specificity, efforts to identify AKT-specific and isoform-selective inhibitors have resulted in the discovery of allosteric inhibitors. These allosteric AKT inhibitors have exhibited isoform selectivity, reduced side-effects and lower toxicity [216]. AKTi-1/2, a naphthyridinone allosteric dual inhibitor of AKT1 and AKT2, showed potent antitumor activity in tumor xenograft models, and its analogue MK2206 led to around 60 % growth inhibition in ovarian cancer cell line. In preclinical studies, MK-2206 showed significant synergistic effect when combined with other chemotherapeutic drugs [217]. NSC-154020 (triciribine), a tricyclic purine nucleoside derivative, strongly inhibited cell growth and induced apoptosis in human cancer cell [218].
MAPK/ERK
MAPKs/extracellular signal-regulated kinases (ERKs) are serine/threonine kinases that mediate extracellular stimuli into a wide range of cellular responses including cell proliferation, differentiation, survival, death and transformation [219,220]. MAPK pathways incorporate a three-step kinase series in which MAPK is activated upon phosphorylation by MAPK kinase (MAPKK, MEK), which in turn is activated when it phosphorylated by MAPKK kinase (MAPKKK, MEKK, Raf). The ERK1 and ERK2 MAPKs are activated by mitogens and were found to be upregulated in human tumors. Components of the ERK signaling cascade are frequently mutated in cancer, with mutations occurring in approximately one-third of human tumors [219]. The mechanism(s) whereby growth factors and mitogens activate ERK signaling is of particular relevance to human cancer. Consequently, inhibitors targeting components of the ERK signaling pathway have been developed to be used as cancer therapeutics [220]. Two other major MAPK pathways, the Jun N-terminal kinase (JNK) and p38 MAPK pathways, which are referred to as the stress activated protein kinase pathways, are also often deregulated in cancers. JNKs and p38 MAPKs are activated by environmental and genotoxic stresses and have key roles in inflammation, as well as in tissue homeostasis given that they control cell proliferation, differentiation, survival and the migration of specific cell types [221–225]. The expression or activity of JNK and p38 MAPK pathway components is often altered in human tumors and cancer cell lines. Given the many tumorigenesis-related functions that these kinases can control, both in the cancer cell and in the tumor microenvironment, it is important to carefully consider the type of tumor before attempting to modulate these pathways for cancer therapy. Here, we focused on the basic and progressing research on MEK-ERK MAPK signaling inhibitors and its implications.
Inhibitors targeting ERK signaling.
In the ERK/MAPK module, ERK is activated upon phosphorylation by MEK, thus MEK also has been a target for anticancer drug development for almost 15 years [226] (Table 7). Trametinib (GSK1120212; GSK) became the first MEK inhibitor to be approved by the US Food and Drug Administration (FDA) for the treatment of metastatic melanoma with the BRAF(V600E/K) mutation [227]. It is a potent inhibitor of both MEK1/2 that, unlike the MEK inhibitors discussed above, preferentially binds to dephosphorylated MEK1/2 and prevents RAF-dependent MEK phosphorylation and activation. PD0325901, with a IC50 of 1 nM against purified MEK1 and MEK2, showed significant antitumor activity in several in vitro and in vivo models [228]. The anticancer drug activity also has been demonstrated in a variety of human tumor xenografts. AZD6244 (selumetinib) is an oral potent second generation inhibitor [229]. This is another allosteric MEK1 and MEK2 inhibitor that is highly selective for MEK1/2 with an IC50 of 14nM against purified MEK1/2. GDC-0973 (cobimetinib) is an oral active inhibitor of MEK1/2 [230]. In vitro studies demonstrated it was able to inhibit ERK1/2 phosphorylation at nanomolar range and it demonstrated antiproliferative effects on multiple tumor cell lines. RO5126766 is a potent and selective dual RAF/MEK inhibitor and it was more effective at reducing colony formation than other MEK inhibitors [231]. In addition, this compound suppressed tumor growth in a SK-MEL-2 xenograft model.
Table 7.
Inhibitors targeting MEK/ERK signaling
| Inhibitor | Target | Types of cancer |
|---|---|---|
| Trametinib (GSK) [227] | Allosteric MEK | Melanoma, colorectal cancer, neuroblastoma, lung cancer |
| PD0325901[228] | Allosteric MEK | Colorectal cancer, solid tumor, |
| Selumetinib [229] | Allosteric MEK | Triple-negative breast cancer, melanoma, lung cancer, head and neck carcinoma |
| Cobimetinib [230] | Allosteric MEK | Melanoma, |
| RO5126766 [231] | Allosteric MEK | Solid tumor, multiple myeloma |
| SCH772984 [235] | ATP competitive ERK | KRAS-mutant lung cancer |
| VX11e [237] | ATP competitive ERK | BRAF-inhibitor progressed melanoma |
Activated ERK regulates a number of cellular events, including cell proliferation and survival [232,233]. In contrast to the advanced development and evaluation of RAF and MEK inhibitors, there has been limited progress in the development of ERK1- and ERK2-selective inhibitors. This is partly due to the earlier assumption that, as ERK is the only known downstream target of MEK, no additive benefit would result from an ERK inhibitor compared to a MEK inhibitor. Thus, development of ERK inhibitors lagged behind RAF/MEK inhibitors. However, interest in the discovery and development of ERK inhibitors has recently intensified, for several reasons. First, after the experience with RAF and MEK inhibitors, there has been an increasing appreciation of the complexity and diversity of the biochemical effects of different small-molecule inhibitors targeting components of the same pathway. Second, the negative feedback loops that are promoted by small-molecule inhibitors of different components of the ERK signaling cascade may show important differences depending on the molecule that is targeted. Finally, resistance to RAF and MEK inhibitors frequently involves the recovery of ERK signaling, suggesting the potential use of an ERK inhibitor [234]. To date, a few potent and selective cell-active preclinical ERK inhibitors have been described in the patent literature. SCH772984 is an ATP-competitive ERK1 and ERK2 inhibitor that was derived from an affinity-based high throughput screen for small molecules that bind to the dephosphorylated, or inactive, form of ERK2 [235]. Binding of SCH772984 to ERK results in a dual mechanism of inhibition: inhibition of ERK1 and ERK2 intrinsic kinase activity and the prevention of phosphorylation of ERK1 and ERK2 by MEK. This latter activity is thought to occur as a result of a large conformational change induced by SCH772984 binding, which opens up a new side pocket on ERK [235,236]. SCH772984 inhibited cellular proliferation and induced apoptosis selectively in tumor cell lines that carry RAS or BRAF mutations, and it induced significant tumor regression in mice with BRAF- or RAS-mutant xenografts. SCH772984 also demonstrated activity in cells that were resistant to either BRAF or MEK inhibitors and in cells that became resistant to the dual combination of these inhibitors. SCH900353, a clinical grade analogue of SCH772984, is currently being tested in Phase I clinical trials. VTX-11e is a potent, selective, and orally bioavailable ERK2 inhibitor with Ki of < 2 nM [237].
PTPS IN CARCINOGENESIS AND ITS POTENTIAL APPLICATION
PTPs with potential tumor suppressive function
As previously mentioned, PTPs can antagonize oncogenic PTK signaling by catalyzing the reverse function of PTKs. In this regard, the function of PTPs as tumor suppressors have been studied in various cancers. A mutational analysis of the PTP gene superfamily in human colorectal cancer through systematic sequencing revealed 83 somatic mutations in six PTP genes, including three members of the receptor-like PTP subfamily (PTPRF, PTPRG, and PTPRT) and three members of the nonreceptor-like PTP subfamily (PTPN3, PTPN13, and PTPN14) [238]. Fifteen mutations of the 83 somatic mutations detected were nonsense, frameshift, or splice-site alterations that were predicted to result in truncated proteins lacking phosphatase activity. In this study, the most frequently mutated PTP gene was PTPRT. Biochemical analysis of five missense mutations of PTPRT indicated that these mutations reduced phosphatase activity of PTPRT. Consistent with this observation, exogenous expression of wild-type PTPRT in HCT116 and DLD1 colorectal cancer cells significantly inhibited cell growth, whereas mutant PTPRT expression did not have an effect on cancer cell growth [238], suggesting that PTPRT functions as a tumor suppressor in colorectal cancer. High-frequency microsatellite instability (MSI-H), which is characterized by length alterations within simple repeated sequences, is induced by defective DNA mismatch repair [239]. An analysis of 54 MSI-H colorectal cancers identified frameshift mutations in six PTP genes, including three members of the receptor-like PTP subfamily (PTPRA, PTPRS, and PTPRE) and three members of the nonreceptor-like PTP subfamily (PTPN21, PTPN5, and PTPN23) [240]. These studies showed that about 32% of MSI-H tumors had frameshift mutations in at least one of the six PTP genes identified, with the highest mutation frequency occurring in PTPN21. Recent studies using whole-exome sequencing to identify novel risk factors for early-onset of colorectal cancers revealed that PTPN12 is a potential candidate that can contribute to the heterogeneous susceptibility to colorectal cancer [241].
Genetic alterations of PTPs were also found in other human cancers [15,242]. For example, loss of function mutations in receptor-like PTPs, such as PTPRT, PTPRC, PTPRD, and PTPRM, were found in head and neck squamous cell carcinoma (HNSCC) [243]. In particular, PTPRT has been found to be the most frequently mutated PTP gene in human cancers [244,245]. According to the Catalogue Of Somatic Mutations In Cancer (COSMIC) (http://cancer/sanger.ac.uk/cancergenome/projects/cosmic/), PTPRT mutations have been identified in a variety of human cancers including colon (11%), bladder (6%), endometrium (8%), esophagus (11%), head and neck (6%), lung (10%), and stomach (9%) cancers [238,243,244]. One oncogenic substrate of PTPRT that has been identified is STAT3 and overexpression of PTPRT in colorectal cancer cells reduced the expression of STAT3 target genes [246]. In accordance with this observation, HNSCC tumors harboring PTPRT mutations exhibited a significantly higher level of phosphorylated, or activated, STAT3 compared with HNSCC tumors without mutation. Overexpression of wild-type PTPRT in HNSCC cells reduced the level of phosphorylated STAT3 expression, whereas expression of mutant PTPRT increased the level of phosphorylated STAT3 expression [243]. In addition to somatic mutations of PTPRT gene, the analysis of the Cancer Genome Atlas (TCGA) showed that the PTPRT promoter is frequently hypermethylated in HNSCC tumors which was associated with the downregulation of PTPRT mRNA expression and upregulation of phosphorylated STAT3 expression. Further, mouse xenograft study using HNSCC cells with PTPRT methylation demonstrated that increased PTPRT promoter methylation is associated with increased sensitivity to STAT3 inhibition [247].
Functional studies including work with PTP-specific transgenic mouse models have provided in vivo evidence that PTPs can function tumor suppressors. Generation of PTPRT knockout mice showed that PTPRT deficiency increased levels of colonic paxillin phosphorylation at residue Y88 and increased the susceptibility to carcinogen azoxymethane-induced colon tumor development [248]. PTPRD, also known as PTPR-delta, is a receptor-like PTP that has been shown to be involved in the regulation of cell growth, migration, and angiogenesis [15,242]. PTPRD inactivation was found in different cancers including glioblastoma multiforme (GBM) [249–251]. Studies showed that STAT3 is one of the substrates of PTPRD and mutations of PTPRD abrogate the ability of this phosphatase to dephosphorylate STAT3 [252]. Increased levels of phosphorylated STAT3 have frequently been found in solid tumors including GBM [253,254]. Recent studies using PTPRD-knockout mice showed that PTPRD deficiency promotes gliomagenesis corresponding with the accumulation of phosphorylated STAT3 and STAT3 hyperactivation [255].
T-cell protein tyrosine phosphatase (TC-PTP; encoded by PTPN2) is one of 17 intracellular and non-receptor PTPs and was originally cloned from a human T-cell cDNA library [256,257]. It has been shown that TC-PTP is involved in the regulation of various physiological functions including cell cycle regulation and apoptosis through dephosphorylation of its target substrates, such as JAK1, JAK3, STAT1, STAT3 and STAT5 [258,259]. Recent studies revealed that focal deletion of PTPN2 was detected in human T-cell acute lymphoblastic leukemia, suggesting TC-PTP has the potential to act as a tumor suppressor [260]. In accordance with this observation, studies have shown that TC-PTP has a tumor suppressive function in breast and colorectal cancers mainly through its regulation of STAT3 signaling [261,262]. Decreased levels of TC-PTP expression was detected in a subset of breast cancer cell lines and a large proportion of triple-negative primary human breast cancers. In addition, TC-PTP overexpression in human breast cancer cell lines suppressed cell proliferation and anchorage-independent growth with reduced tyrosine phosphorylation of STAT3 and SRC family kinase [261]. GdX (X-linked gene in the G6PD cluster at Xq28) is known to act as a chaperon in protein processing in the endoplasmic reticulum [263,264]. Studies showed that GdX stabilizes the steady-state association of phosphorylated STAT3 with TC45, a nuclear form of TC-PTP, and promotes STAT3 dephosphorylation. Deletion of GdX in mice significantly accelerated colitis-associated colorectal tumorigenesis that corresponded with an increased level of phosphorylated STAT3 [262].
The Src homology 2 domain-containing PTP-1 (SHP-1; encoded by PTPN6) is a non-receptor PTP that is expressed most abundantly in hematopoietic cells [265]. Loss of SHP-1 expression was frequently found in anaplastic lymphoma kinase (ALK)-positive anaplastic large cell lymphoma (ALK+ ALCL), a type of non-Hodgkin lymphoma of T/null-cell immunophenotype [266]. Transfection and expression of exogenous SHP-1 in SHP-1-negative ALK+ ALCL cell lines significantly reduced phosphorylated JAK3 and phosphorylated STAT3 and, consequently, the downregulation of STAT3 downstream targets. In contrast, knockdown of SHP-1 with siRNA in SHP-1-positive ALK+ ALCL cell lines increased the levels of phosphorylated JAK3 and phosphorylated STAT3 [267]. SHP-1 was also involved in the induction of apoptosis in epithelial cancer cells by downregulating phosphorylated STAT3. Increased SHP-1 activity induced by sorafenib or its derivatives inhibited STAT3 phosphorylation, which contributed to the increase in apoptosis in breast cancer and hepatocellular carcinoma cell lines [268,269]. Regorafenib, an inhibitor of multiple protein kinases that has anti-tumor and anti-metastatic capabilities in metastatic colorectal cancer, triggered apoptotic cell death by decreasing STAT3 phosphorylation through enhanced SHP-1 activity [270].
SHP-2, which is encoded by PTPN11, is a ubiquitously expressed PTP that plays an important role in developmental process and its germline mutations are known to cause both Noonan syndrome and LEOPARD syndrome, two clinically similar autosomal dominant developmental disorders [271]. Generation of hepatocyte-specific SHP-2 knockout mice showed that SHP-2 deficiency significantly promoted diethylnitrosamine-induced hepatocellular carcinoma development with increased STAT3 signaling [272].
Studies indicate that constitutive activation of STAT3, a common substrate of PTPs, is found in human tumors and cancer cell lines, and its inhibition can suppress the growth of cancer cells, implying that it possesses a critical role in cancer cell proliferation. In this regard, STAT3 signaling was downregulated by PTPs in different models of carcinogenesis (Table 8). These results suggest that elucidation of the cellular signaling mechanism(s) that regulate STAT3 phosphorylation/dephosphorylating will be important for cancer prevention and the development of more effective cancer treatments.
Table 8.
PTPs involved in the regulation of STAT3 signaling in carcinogenesis.
| PTP | Target | Types of cancer |
|---|---|---|
| PTPRT [243, 246] | STAT3 | Colon cancer, head and neck cancer |
| TC-PTP [261, 284] | EGFR, STAT3, JAK1, JAK3, Src | Breast cancer, skin cancer |
| PTPRD [252] | STAT3 | Glioblastoma |
| SHP1 [267–270] | EGFR, JAK2, STAT3 | Lymphoma, liver cancer, breast cancer, colon cancer |
| SHP2 [272] | EGFR, JAK2, STAT3 | Liver cancer |
While the frequent mutation/inactivation of PTPs in human cancers which result in tumor suppressive roles for PTPs that has been observed in in vitro cell culture systems was confirmed by in vivo transgenic mouse models in most cases, other recent studies showed that there is a discrepancy between the potential tumor suppressive role of PTPs and their actual effects on in vivo tumorigenesis. As already mentioned, HNSCC tumors can express somatic mutations of the PTPRT gene or hypermethylation of the PTPRT promoter and theses alterations resulted in an increased expression of phosphorylated STAT3 [243,247]. However, PTPRT-knockout mice were not more susceptible to 4-nitroquinoline 1-oxide (4-NQO)-induced HNSCC carcinogenesis compared to wild-type mice [273]. Stattic is a nonpeptidic small molecule that selectively inhibits the function of the STAT3 SH2 domain and prevents its activation, dimerization, and nuclear translocation [168]. Even though targeting of STAT3 with stattic resulted in a chemopreventive effect against 4-NQO-induced oral carcinogenesis, both PTPRT-knockout and wild type mice responded similarly to stattic-mediated chemoprevention [273]. These results indicate that functional loss of PTPRT in mice does not support its tumor suppressive role with the phenotype of mutation or promoter methylation in this model of carcinogenesis. There are several possible explanations for this discrepancy. First, it is possible that the C57BL/6J mice used in this study were sensitive to 4-NQO, whereas this genotype of mice were resistant to azoxymethane because PTPRT-knockout mice showed increased susceptibility to azoxymethane-induced colon carcinogenesis [248]. Second, it is possible that PTPRT may have divergent or pleiotropic roles in oral and colonic epithelium during carcinogenesis [273]. Third, it may be that the redundant regulation of STAT3 signaling by other PTPs can compensate for the loss of PTPRT in oral carcinogenesis induced by 4-NQO. Finally, 4-NQO treatment or cell signaling pathways activated by 4-NQO treatment may cause the inhibition of PTPRT activity.
Collectively, genetic mutation analysis in human cancers and subsequent functional studies using transgenic mice have suggested that PTPs play a critical role in attenuating carcinogenesis by inhibiting oncogenic signaling pathways, such as STAT3 signaling. It further suggests that development of small molecule PTP activators will be another efficient strategy for targeted cancer therapy, in addition to PTK inhibitors.
Tumor suppressive PTPs in skin carcinogenesis
As previously mentioned, PTKs have been shown to play a prominent role in skin carcinogenesis. Consequently, PTPs also can function in skin cancer though their exact role is less well-characterized. For example, PTP expression is known to be induced during the proliferation and maturation of keratinocytes but paradoxically their expression levels remain unchanged within epidermal tissue [274]. Still, microarray analysis of human melanoma tissues revealed that expression of PTPs, such as PTPκ and PTPλ, decreases in human melanoma when compared with analogous noncancerous tissue [275,276], indicating a tumor suppressor role for PTPs. Also, exposure to acute UV radiation increases the ligand-independent activation of PTKs [277,278]. One possible explanation for this result would be that UV irradiation induces PTP inactivation in order to allow for the activation PTKs. In fact, biochemical studies revealed that reactive oxygen species (such as H2O2) produced by UV irradiation caused the inactivation of PTPs by oxidizing the cysteine residue within the conserved active-site of the PTP catalytic domain [279–281]. Furthermore, studies from different groups have demonstrated that acute UV irradiation resulted in the inactivation of PTPs, such as PTPκ, in keratinocytes as well [282,283].
Recent studies have shown that STAT3 plays an important role in UVB-mediated skin carcinogenesis. In this regard, STAT3-overexpressing keratinocytes were resistant to UVB-induced epidermal apoptosis, whereas STAT3-deficient keratinocytes were sensitive to UVB-induced apoptosis compared with control keratinocytes [160]. Further studies using transgenic mice either deficient in STAT3 or expressing constitutively active STAT3 in keratinocytes demonstrated a critical role for STAT3 in UVB-mediated skin carcinogenesis [161]. While STAT3 has a role in skin cancer formation, UVB irradiation initially caused rapid STAT3 dephosphorylation in keratinocytes and pretreatment of sodium vanadate, a pan PTP inhibitor, desensitized keratinocytes to UVB-induced apoptosis corresponding with the recovery of phosphorylated STAT3 expression, suggesting the involvement of PTPs in this mechanism [160]. Further studies showed that three PTPs, TC-PTP, SHP-1, and SHP-2, can cooperate in the dephosphorylation of STAT3 in response to UVB irradiation. Following irradiation of mouse skin with UVB, the protein expression level of phosphorylated STAT3 in the epidermis was reduced, though the level recovered at later time points [159]. It suggests that PTP-mediated signaling may serve as part of a protective mechanism against skin carcinogenesis. Knockdown of each of these three PTPs using siRNA revealed that only TC-PTP had a major effect on STAT3 regulation in skin keratinocytes. The level of phosphorylated STAT3 in TC-PTP knockdown keratinocytes was higher relative to SHP-1 knockdown or SHP-2 knockdown keratinocytes, implying that TC-PTP has a greater effect on STAT3 dephosphorylation than the other two PTPs [284]. In this regard, TC-PTP deficiency in keratinocytes significantly reduced UVB-induced apoptosis with increased cell proliferation. TC-PTP activity was increased in response to UVB irradiation, and overexpression of TC-PTP in keratinocytes showed greater increased activity in the presence of UVB [284]. These studies suggest that TC-PTP may be a novel therapeutic target for the prevention of UVB-induced skin cancer. Identification of the mechanism of UVB-induced PTP activation using a TC-PTP specific transgenic mouse model will be helpful to understand the PTP-mediated protective mechanism in skin carcinogenesis and so that it can be applied to the prevention of skin cancer.
PTPs with potential oncogenic function
While PTPs initially were thought to be potential tumor suppressors as they appear to be in skin cancer, studies have shown that PTPs can also promote tumorigenesis by triggering negative-feedback mechanisms that terminate activation signals or by dephosphorylating the inhibitory factors of oncogenic PTK signaling pathways. In this section, we discuss two oncogenic PTPs, SHP-2 and PTP1B and the recent development of their inhibitors.
Despite its tumor suppressive role in hepatocellular carcinogenesis [272], SHP-2 primarily has been identified as an oncogenic PTP for the following reasons: a) it can mediate the activation of the Ras-ERK pathways by growth factors, cytokines, and hormones; b) several types of leukemia possess mutations that activate SHP-2 [285–287]. In this regard, inhibitors targeting SHP-2 have been identified and/or developed. Chen et al. identified Fumosorinone as a potent SHP-2 inhibitor [288]. Fumosorinone, which originates from entomogenous fungi, exhibited selective inhibition of SHP-2 over other PTPs tested. Fumosorinone effectively inhibited SHP-2-dependent activation of the Ras-ERK signaling pathway downstream of EGFR, while it had little effect on SHP-2-independent ERK activation induced by TPA [288]. Recently, the highly potent, selective and orally bioavailable small molecule inhibitor SHP099 was also developed [289]. SHP099 inhibited SHP-2 activity through an allosteric mechanism by binding to the interface of the N-terminal SH2, C-terminal SH2, and PTP activation domain. SHP099 inhibited the proliferation of RTK-driven human cancer cells by suppressing Ras-ERK signaling pathway [289].
PTP1B, which is encoded by PTPN1, is an intracellular and non-receptor PTP that has a critical role in diabetes and obesity and which also has been shown to function as a tumor promoter [290]. PTP1B overexpression was found in human breast cancers. Its overexpression was observed in more than 70% of mammary tumor sections compared with normal counterparts [291]. PTP1B increased c-Src activity by dephosphorylating it negative regulatory site, tyrosine-530 in human breast cancer cell lines [292]. PTP1B has been implicated in gastric carcinogenesis as a potential oncogenic PTP as well. PTP1B increased gastric cancer cell proliferation and survival by regulating Src-mediated signaling pathways. Further clinicopathological examination of gastric cancer patients indicated that PTP1B amplification is associated with poor survival of gastric cancer patients [293]. In this regard, the small molecule inhibitor MSI-1436, which can inhibit PTP1B by targeting its disordered C terminal noncatalytic domain, has been identified as a potential anti-cancer drug. MSI-1436 inhibited tumorigenesis in xenografts and abrogated metastasis in the NDL2 mouse model of breast cancer [294]. However, like SHP-2, PTP1B has demonstrated some tumor suppressive capabilities. PTP1B was underexpressed in ovarian carcinoma-derived cell lines and its expression decreased proliferation, migration, and invasion of ovarian cancer cell lines through the dephosphorylation of the IGF-1R β-subunit and BRK/PTK6, a Src-like PTK that physically and functionally interacts with the IGF-1R β-subunit [295].
Taken together, studies have shown that PTPs also have a potential in promoting carcinogenesis by activating oncogenic signaling pathways, such as Src signaling. It suggests that development of small molecule PTP inhibitors to block PTP oncogenic function could be a potential approach to prevent carcinogenesis dependent on the type of cancer that is being targeting.
CONCLUDING REMARKS
Tyrosine phosphorylation signaling is one major therapeutic target in carcinogenesis. PTKs and their downstream signal transduction pathways are aberrantly activated in various cancers. Specific inhibitors targeting PTKs and their downstream pathways have been developed and used to kill cancer cells. However, most PTK inhibitors have not been able to completely block cancer cell growth, even though they showed significant effects in abrogating carcinogenesis in both in vitro cell lines and in vivo mouse models. This reduction in efficacy may be due to more complex cancer signaling in humans, cancer heterogeneity, and/or the development of drug resistance following long term treatment. PTPs are involved in carcinogenesis as both tumor suppressors and tumor promoters. Like PTK inhibitors, PTP inhibitors targeting oncogenic function have recently been developed. However, the development of activators to target the tumor suppressive function(s) of PTPs and the consequences of their application have not been investigated given the difficulty of designing and creating specific activators of PTPs. In order to develop better therapeutic methods for preventing and treating cancer, we must better understand the function of tyrosine phosphorylation signaling during carcinogenesis, create novel, more effective and specific inhibitors of PTKs or PTPs, create tumor suppressive PTP activators, and investigate the efficacy of the combinatorial use of these inhibitors and activators.
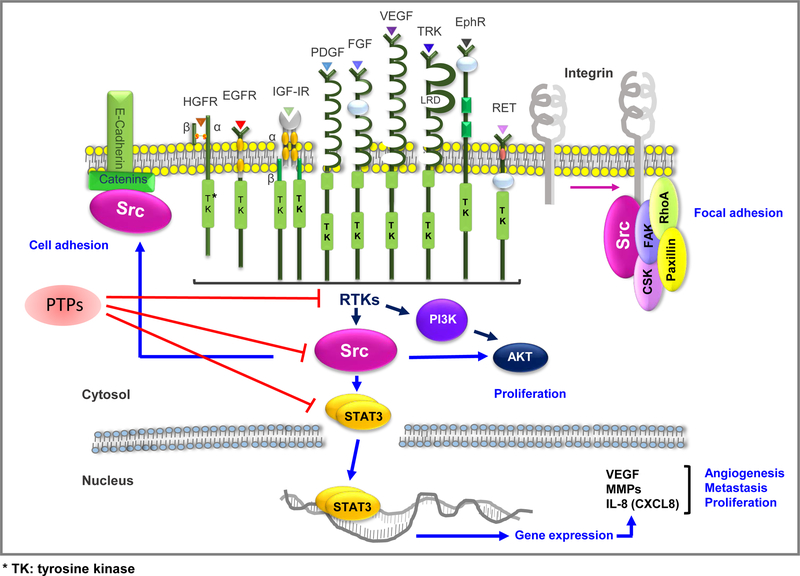
Figure 4. The role of Src in cells.
Src may interact with a number of kinases which can regulate cell proliferation, migration, adhesion, and angiogenesis. RTKs can trigger the phosphorylation of the Src Tyr416 residue and promote activation of the transcription factor STAT3, which can regulate gene expression to stimulate cell migration, angiogenesis, and cell survival. PI3K activation following loss of PTEN may induce Src/AKT cascade signaling to enhance cell growth. Moreover, Src can mediate cell adhesion and migration by interacting with catenin or integrin/focal adhesion proteins, such as FAK, CSK, Paxillion, and RhoA.
ACKNOWLEDGMENTS:
This work was supported by National Institutes of Health, NIEHS Grant ES022250 (to D.J. Kim).
LIST OF ABBREVIATIONS
- 4-NQO
4-nitroquinoline 1-oxide
- ALK
anaplastic lymphoma kinase
- ALCL
anaplastic large cell lymphoma
- DBD
DNA-binding domain
- DMBA
7,12-dimethylbenz[a]anthracene
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- ERK
extracellular signal-regulated kinases
- GBM
glioblastoma multiforme
- GdX
X-linked gene in the G6PD cluster at Xq28
- HGH
human growth hormone
- HIC1
hypermethylated in cancer 1
- HNSCC
head and neck squamous cell carcinoma
- IGF-1
insulin-like growth factor 1
- IGFBP
insulin-like growth factor binding protein
- IGF-1R
insulin-like growth factor 1 receptor
- JAK
Janus kinase
- JNK
Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- MMP-2
metalloproteinase 2
- MSI-H
high-frequency microsatellite instability
- NSCLC
non-small-cell lung carcinoma
- mTOR
mechanistic target of rapamycin
- ODN
oligodeoxynucleotide
- PH
pleckstrin homology
- PKC
protein kinase C
- PTK
protein tyrosine kinase
- PTP
protein tyrosine phosphatase
- RTK
receptor tyrosine kinase
- SFK
Src family of kinase
- SH2
Src homology 2
- SHP-1
Src homology 2 domain-containing PTP-1
- siRNA
small interfering RNA
- STAT3
signal transducer and activator of transcription 3
- TC-PTP
T-cell protein tyrosine phosphatase
- TGFα
transforming growth factor α
- TPA
12-O-tetradecanoylphorbol-13-acetate
- Tyr
tyrosine
- UVB
ultraviolet B
REFERENCE
- 1.Lim WA, Pawson T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell 2010;142:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter T Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol 2009;21:140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene 2000;19:5548–5557. [DOI] [PubMed] [Google Scholar]
- 4.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell 2010;141:1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casaletto JB, McClatchey AI. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat Rev Cancer 2012;12:387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature 2001;411:355–365. [DOI] [PubMed] [Google Scholar]
- 7.Levitzki A, Mishani E. Tyrphostins and other tyrosine kinase inhibitors. Annu Rev Biochem 2006;75:93–109. [DOI] [PubMed] [Google Scholar]
- 8.Shawver LK, Slamon D, Ullrich A. Smart drugs: tyrosine kinase inhibitors in cancer therapy. Cancer Cell 2002;1:117–123. [DOI] [PubMed] [Google Scholar]
- 9.Chow LQ, Eckhardt SG. Sunitinib: from rational design to clinical efficacy. J Clin Oncol 2007;25:884–896. [DOI] [PubMed] [Google Scholar]
- 10.Tonks NK, Neel BG. From form to function: signaling by protein tyrosine phosphatases. Cell 1996;87:365–368. [DOI] [PubMed] [Google Scholar]
- 11.Stoker AW. Protein tyrosine phosphatases and signalling. J Endocrinol 2005;185:19–33. [DOI] [PubMed] [Google Scholar]
- 12.Hendriks WJ, Elson A, Harroch S, Pulido R, Stoker A, den Hertog J. Protein tyrosine phosphatases in health and disease. FEBS J 2013;280:708–730. [DOI] [PubMed] [Google Scholar]
- 13.Tonks NK, Diltz CD, Fischer EH. Purification of the major protein-tyrosine-phosphatases of human placenta. J Biol Chem 1988;263:6722–6730. [PubMed] [Google Scholar]
- 14.Alonso A, Sasin J, Bottini N, et al. Protein tyrosine phosphatases in the human genome. Cell 2004;117:699–711. [DOI] [PubMed] [Google Scholar]
- 15.Julien SG, Dube N, Hardy S, Tremblay ML. Inside the human cancer tyrosine phosphatome. Nat Rev Cancer 2011;11:35–49. [DOI] [PubMed] [Google Scholar]
- 16.Tiganis T, Bennett AM. Protein tyrosine phosphatase function: the substrate perspective. Biochem J 2007;402:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol 2006;7:833–846. [DOI] [PubMed] [Google Scholar]
- 18.Andersen JN, Jansen PG, Echwald SM, et al. A genomic perspective on protein tyrosine phosphatases: gene structure, pseudogenes, and genetic disease linkage. FASEB J 2004;18:8–30. [DOI] [PubMed] [Google Scholar]
- 19.Tabernero L, Aricescu AR, Jones EY, Szedlacsek SE. Protein tyrosine phosphatases: structure-function relationships. FEBS J 2008;275:867–882. [DOI] [PubMed] [Google Scholar]
- 20.Hendriks WJ, Elson A, Harroch S, Stoker AW. Protein tyrosine phosphatases: functional inferences from mouse models and human diseases. FEBS J 2008;275:816–830. [DOI] [PubMed] [Google Scholar]
- 21.Labbe DP, Hardy S, Tremblay ML. Protein tyrosine phosphatases in cancer: friends and foes! Prog Mol Biol Transl Sci 2012;106:253–306. [DOI] [PubMed] [Google Scholar]
- 22.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 1988;241:42–52. [DOI] [PubMed] [Google Scholar]
- 23.Ptasznik A, Prossnitz ER, Yoshikawa D, Smrcka A, Traynor-Kaplan AE, Bokoch GM. A tyrosine kinase signaling pathway accounts for the majority of phosphatidylinositol 3,4,5-trisphosphate formation in chemoattractant-stimulated human neutrophils. J Biol Chem 1996;271:25204–25207. [DOI] [PubMed] [Google Scholar]
- 24.Taagepera S, McDonald D, Loeb JE, et al. Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc Natl Acad Sci USA 1998;95:7457–7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahajan K, Mahajan NP. Cross talk of tyrosine kinases with the DNA damage signaling pathways. Nucleic Acids Res 2015;43:10588–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia Y, Yun CH, Park E, et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 2016;534:129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balmain A, Pragnell IB. Mouse skin carcinomas induced in vivo by chemical carcinogens have a transforming Harvey-ras oncogene. Nature 1983;303:72–74. [DOI] [PubMed] [Google Scholar]
- 28.DiGiovanni J Modification of multistage skin carcinogenesis in mice. Prog Exp Tumor Res 1991;33:192–229. [DOI] [PubMed] [Google Scholar]
- 29.DiGiovanni J Multistage carcinogenesis in mouse skin. Pharmacol Ther 1992;54:63–128. [DOI] [PubMed] [Google Scholar]
- 30.Macias E, Rao D, Digiovanni J. Role of stat3 in skin carcinogenesis: insights gained from relevant mouse models. J Skin Cancer 2013;2013:684050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan KS, Carbajal S, Kiguchi K, Clifford J, Sano S, DiGiovanni J. Epidermal growth factor receptor-mediated activation of Stat3 during multistage skin carcinogenesis. Cancer Res 2004;64:2382–2389. [DOI] [PubMed] [Google Scholar]
- 32.Park K, Tan EH, O’Byrne K, et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol 2016;17:577–589. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Y, Murakami H, Yang PC, et al. Randomized phase II trial of gefitinib with and without pemetrexed as first-line therapy in patients with advanced nonsquamous non-small-cell lung cancer with activating epidermal growth factor receptor mutations. J Clin Oncol 2016;34:3258–3266. [DOI] [PubMed] [Google Scholar]
- 34.Brower JV, Robins HI. Erlotinib for the treatment of brain metastases in non-small cell lung cancer. Expert Opin Pharmacother 2016;17:1013–1021. [DOI] [PubMed] [Google Scholar]
- 35.De Greve J, Van Meerbeeck J, Vansteenkiste JF, et al. Prospective evaluation of first-line erlotinib in advanced non-small cell lung cancer (NSCLC) carrying an activating EGFR mutation: a multicenter academic phase II study in caucasian patients (FIELT). PLoS One 2016;11:e0147599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walraven I, van den Heuvel M, van Diessen J, et al. Long-term follow-up of patients with locally advanced non-small cell lung cancer receiving concurrent hypofractionated chemoradiotherapy with or without cetuximab. Radiother Oncol 2016;118:442–446. [DOI] [PubMed] [Google Scholar]
- 37.Deming DA, Cavalcante LL, Lubner SJ, et al. A phase I study of selumetinib (AZD6244/ARRY-142866), a MEK1/2 inhibitor, in combination with cetuximab in refractory solid tumors and KRAS mutant colorectal cancer. Invest New Drugs 2016;34:168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tebbutt NC, Price TJ, Ferraro DA, et al. Panitumumab added to docetaxel, cisplatin and fluoropyrimidine in oesophagogastric cancer: ATTAX3 phase II trial. Br J Cancer 2016;114:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elez E, Hendlisz A, Delaunoit T, et al. Phase II study of necitumumab plus modified FOLFOX6 as first-line treatment in patients with locally advanced or metastatic colorectal cancer. Br J Cancer 2016;114:372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sequist LV, Piotrowska Z, Niederst MJ, et al. Osimertinib responses after disease progression in patients who had been receiving rociletinib. JAMA Oncol 2016;2:541–543. [DOI] [PubMed] [Google Scholar]
- 41.Shaw AT, Gandhi L, Gadgeel S, et al. Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol 2016;17:234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gadgeel SM, Gandhi L, Riely GJ, et al. Safety and activity of alectinib against systemic disease and brain metastases in patients with crizotinib-resistant ALK-rearranged non-small-cell lung cancer (AF-002JG): results from the dose-finding portion of a phase 1/2 study. Lancet Oncol 2014;15:1119–1128. [DOI] [PubMed] [Google Scholar]
- 43.Krytska K, Ryles HT, Sano R, et al. Crizotinib synergizes with chemotherapy in preclinical models of neuroblastoma. Clin Cancer Res 2016;22:948–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosell R, Gettinger SN, Bazhenova LA, et al. 1330: Brigatinib efficacy and safety in patients (Pts) with anaplastic lymphoma kinase (ALK)-positive (ALK+) non-small cell lung cancer (NSCLC) in a phase 1/2 trial. J Thorac Oncol 2016;11:S114. [Google Scholar]
- 45.Kim DW, Mehra R, Tan DS, et al. Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): updated results from the multicentre, open-label, phase 1 trial. Lancet Oncol 2016;17:452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S, Qi X, Huang Y, Liu D, Zhou F, Zhou C. Ceritinib (LDK378): a potent alternative to crizotinib for ALK-rearranged non-small-cell lung cancer. Clin Lung Cancer 2015;16:86–91. [DOI] [PubMed] [Google Scholar]
- 47.Miles DR, Wada DR, Jumbe NL, Lacy SA, Nguyen LT. Population pharmacokinetic/pharmacodynamic modeling of tumor growth kinetics in medullary thyroid cancer patients receiving cabozantinib. Anticancer Drugs 2016;27:328–341. [DOI] [PubMed] [Google Scholar]
- 48.Leibowitz-Amit R, Pintilie M, Khoja L, et al. Changes in plasma biomarkers following treatment with cabozantinib in metastatic castration-resistant prostate cancer: a post hoc analysis of an extension cohort of a phase II trial. J Transl Med 2016;14:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta A, Roberts C, Tysoe F, et al. RADVAN: a randomised phase 2 trial of WBRT plus vandetanib for melanoma brain metastases - results and lessons learnt. Br J Cancer 2016;115:1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novello S, Scagliotti G, de Castro G Jr., et al. An open-label, multicenter, randomized, phase II study of cisplatin and pemetrexed with or without cixutumumab (IMC-A12) as a first-line therapy in patients with advanced nonsquamous non-small cell lung cancer. J Thorac Oncol 2016; In press Available at ( 10.1016/j.jtho.2016.07.013). [DOI] [PubMed]
- 51.Hussain M, Rathkopf D, Liu G, et al. A randomised non-comparative phase II trial of cixutumumab (IMC-A12) or ramucirumab (IMC-1121B) plus mitoxantrone and prednisone in men with metastatic docetaxel-pretreated castration-resistant prostate cancer. Eur J Cancer 2015;51:1714–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calvo E, Soria JC, Ma WW, et al. A phase I clinical trial and independent patient-derived xenograft study of combined targeted treatment with dacomitinib and figitumumab in advanced solid tumors. Clin Cancer Res 2016; In press Available at ( 10.1158/1078-0432.CCR-15-2301). [DOI] [PubMed]
- 53.Frappaz D, Federico SM, Pearson AD, et al. Phase 1 study of dalotuzumab monotherapy and ridaforolimus-dalotuzumab combination therapy in paediatric patients with advanced solid tumours. Eur J Cancer 2016;62:9–17. [DOI] [PubMed] [Google Scholar]
- 54.Sclafani F, Kim TY, Cunningham D, et al. A randomized phase II/III study of dalotuzumab in combination with cetuximab and irinotecan in chemorefractory, KRAS wild-type, metastatic colorectal cancer. J Natl Cancer Inst 2015;107:djv258. [DOI] [PubMed] [Google Scholar]
- 55.Cohn AL, Tabernero J, Maurel J, et al. A randomized, placebo-controlled phase 2 study of ganitumab or conatumumab in combination with FOLFIRI for second-line treatment of mutant KRAS metastatic colorectal cancer. Ann Oncol 2013;24:1777–1785. [DOI] [PubMed] [Google Scholar]
- 56.Fuchs CS, Azevedo S, Okusaka T, et al. A phase 3 randomized, double-blind, placebo-controlled trial of ganitumab or placebo in combination with gemcitabine as first-line therapy for metastatic adenocarcinoma of the pancreas: the GAMMA trial. Ann Oncol 2015;26:921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pappo AS, Vassal G, Crowley JJ, et al. A phase 2 trial of R1507, a monoclonal antibody to the insulin-like growth factor-1 receptor (IGF-1R), in patients with recurrent or refractory rhabdomyosarcoma, osteosarcoma, synovial sarcoma, and other soft tissue sarcomas: results of a Sarcoma Alliance for Research Through Collaboration study. Cancer 2014;120:2448–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson PM, Bielack SS, Gorlick RG, et al. A phase II study of clinical activity of SCH 717454 (robatumumab) in patients with relapsed osteosarcoma and Ewing sarcoma. Pediatr Blood Cancer 2016;63:1761–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soria JC, Massard C, Lazar V, et al. A dose finding, safety and pharmacokinetic study of AVE1642, an anti-insulin-like growth factor-1 receptor (IGF-1R/CD221) monoclonal antibody, administered as a single agent and in combination with docetaxel in patients with advanced solid tumours. Eur J Cancer 2013;49:1799–1807. [DOI] [PubMed] [Google Scholar]
- 60.Macaulay VM, Middleton MR, Protheroe AS, et al. Phase I study of humanized monoclonal antibody AVE1642 directed against the type 1 insulin-like growth factor receptor (IGF-1R), administered in combination with anticancer therapies to patients with advanced solid tumors. Ann Oncol 2013;24:784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iguchi H, Nishina T, Nogami N, Kozuki T, Yamagiwa Y, Yagawa K. Phase I dose-escalation study evaluating safety, tolerability and pharmacokinetics of MEDI-573, a dual IGF-I/II neutralizing antibody, in Japanese patients with advanced solid tumours. Invest New Drugs 2015;33:194–200. [DOI] [PubMed] [Google Scholar]
- 62.Haluska P, Menefee M, Plimack ER, et al. Phase I dose-escalation study of MEDI-573, a bispecific, antiligand monoclonal antibody against IGFI and IGFII, in patients with advanced solid tumors. Clin Cancer Res 2014;20:4747–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leighl NB, Rizvi NA, de Lima LG Jr., et al. Phase 2 study of erlotinib in combination with linsitinib (OSI-906) or placebo in chemotherapy-naive patients with non-small-cell lung cancer and activating epidermal growth factor receptor mutations. Clin Lung Cancer 2016; In press Available at ( 10.1016/j.cllc.2016.07.007). [DOI] [PMC free article] [PubMed]
- 64.Kolb EA, Gorlick R, Lock R, et al. Initial testing (stage 1) of the IGF-1 receptor inhibitor BMS-754807 by the pediatric preclinical testing program. Pediatr Blood Cancer 2011;56:595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee J, Jain A, Kim P, et al. Activated cMET and IGF1R-driven PI3K signaling predicts poor survival in colorectal cancers independent of KRAS mutational status. PLoS One 2014;9:e103551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stromberg T, Ekman S, Girnita L, et al. IGF-1 receptor tyrosine kinase inhibition by the cyclolignan PPP induces G2/M-phase accumulation and apoptosis in multiple myeloma cells. Blood 2006;107:669–678. [DOI] [PubMed] [Google Scholar]
- 67.Ishida Y, Murai K, Yamaguchi K, et al. Pharmacokinetics and pharmacodynamics of dasatinib in the chronic phase of newly diagnosed chronic myeloid leukemia. Eur J Clin Pharmacol 2016;72:185–193. [DOI] [PubMed] [Google Scholar]
- 68.Mitri Z, Nanda R, Blackwell K, et al. TBCRC-010: phase I/II study of dasatinib in combination with zoledronic acid for the treatment of breast cancer bone metastasis. Clin Cancer Res 2016;22:5706–5712.. [DOI] [PubMed] [Google Scholar]
- 69.Schott AF, Barlow WE, Van Poznak CH, et al. Phase II studies of two different schedules of dasatinib in bone metastasis predominant metastatic breast cancer: SWOG S0622. Breast Cancer Res Treat 2016;159:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schuetze SM, Wathen JK, Lucas DR, et al. SARC009: Phase 2 study of dasatinib in patients with previously treated, high-grade, advanced sarcoma. Cancer 2016;122:868–874. [DOI] [PubMed] [Google Scholar]
- 71.Sueda T, Kudo T, Sakai D, et al. Safety and pharmacokinetics of S-1 in a recurrent colon cancer patient with chronic myeloid leukemia treated with dasatinib: a case report. Cancer Chemother Pharmacol 2014;74:1321–1324. [DOI] [PubMed] [Google Scholar]
- 72.Brooks HD, Glisson BS, Bekele BN, et al. Phase 2 study of dasatinib in the treatment of head and neck squamous cell carcinoma. Cancer 2011;117:2112–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Posadas EM, Ahmed RS, Karrison T, et al. Saracatinib as a metastasis inhibitor in metastatic castration-resistant prostate cancer: A University of Chicago Phase 2 Consortium and DOD/PCF Prostate Cancer Clinical Trials Consortium Study. Prostate 2016;76:286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whiteley J, Reisman A, Shapiro M, Cortes J, Cella D. Health-related quality of life during bosutinib (SKI-606) therapy in patients with advanced chronic myeloid leukemia after imatinib failure. Curr Med Res Opin 2016;32:1325–1334. [DOI] [PubMed] [Google Scholar]
- 75.Brummendorf TH, Cortes JE, Khoury HJ, et al. Factors influencing long-term efficacy and tolerability of bosutinib in chronic phase chronic myeloid leukaemia resistant or intolerant to imatinib. Br J Haematol 2016;172:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Isakoff SJ, Wang D, Campone M, et al. Bosutinib plus capecitabine for selected advanced solid tumours: results of a phase 1 dose-escalation study. Br J Cancer 2014;111:2058–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moy B, Neven P, Lebrun F, et al. Bosutinib in combination with the aromatase inhibitor letrozole: a phase II trial in postmenopausal women evaluating first-line endocrine therapy in locally advanced or metastatic hormone receptor-positive/HER2-negative breast cancer. Oncologist 2014;19:348–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campone M, Bondarenko I, Brincat S, et al. Phase II study of single-agent bosutinib, a Src/Abl tyrosine kinase inhibitor, in patients with locally advanced or metastatic breast cancer pretreated with chemotherapy. Ann Oncol 2012;23:610–617. [DOI] [PubMed] [Google Scholar]
- 79.Zhang H, Berezov A, Wang Q, et al. ErbB receptors: from oncogenes to targeted cancer therapies. J Clin Invest 2007;117:2051–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bergeron JJ, Di Guglielmo GM, Dahan S, Dominguez M, Posner BI. Spatial and Temporal Regulation of Receptor Tyrosine Kinase Activation and Intracellular Signal Transduction. Annu Rev Biochem 2016;85:573–597. [DOI] [PubMed] [Google Scholar]
- 81.Koschut D, Richert L, Pace G, Niemann HH, Mely Y, Orian-Rousseau V. Live cell imaging shows hepatocyte growth factor-induced Met dimerization. Biochim Biophys Acta 2016;1863:1552–1558. [DOI] [PubMed] [Google Scholar]
- 82.Guo L, Kozlosky CJ, Ericsson LH, Daniel TO, Cerretti DP, Johnson RS. Studies of ligand-induced site-specific phosphorylation of epidermal growth factor receptor. J Am Soc Mass Spectrom 2003;14:1022–1031. [DOI] [PubMed] [Google Scholar]
- 83.Lin EH, Kao YR, Lin CA, et al. Hedgehog pathway maintains cell survival under stress conditions, and drives drug resistance in lung adenocarcinoma. Oncotarget 2016;7:24179–24193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stauber RH, Knauer SK, Habtemichael N, et al. A combination of a ribonucleotide reductase inhibitor and histone deacetylase inhibitors downregulates EGFR and triggers BIM-dependent apoptosis in head and neck cancer. Oncotarget 2012;3:31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Feng H, Hu B, Jarzynka MJ, et al. Phosphorylation of dedicator of cytokinesis 1 (Dock180) at tyrosine residue Y722 by Src family kinases mediates EGFRvIII-driven glioblastoma tumorigenesis. Proc Natl Acad Sci USA 2012;109:3018–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tu Y, Ji C, Yang B, et al. DNA-dependent protein kinase catalytic subunit (DNA-PKcs)-SIN1 association mediates ultraviolet B (UVB)-induced Akt Ser-473 phosphorylation and skin cell survival. Mol Cancer 2013;12:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu Y, Voorhees JJ, Fisher GJ. Epidermal growth factor receptor is a critical mediator of ultraviolet B irradiation-induced signal transduction in immortalized human keratinocyte HaCaT cells. Am J Pathol 2006;169:823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Correia M, Thiagarajan V, Coutinho I, Gajula GP, Petersen SB, Neves-Petersen MT. Modulating the structure of EGFR with UV light: new possibilities in cancer therapy. PLoS One 2014;9:e111617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kostyuk V, Potapovich A, Stancato A, et al. Photo-oxidation products of skin surface squalene mediate metabolic and inflammatory responses to solar UV in human keratinocytes. PLoS One 2012;7:e44472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dempke WC. Targeted therapy for NSCLC--a double-edged sword? Anticancer Res 2015;35:2503–2512. [PubMed] [Google Scholar]
- 91.Caccese M, Ferrara R, Pilotto S, et al. Current and developing therapies for the treatment of non-small cell lung cancer with ALK abnormalities: update and perspectives for clinical practice. Expert Opin Pharmacother 2016;17:2253–2266. [DOI] [PubMed] [Google Scholar]
- 92.Berisha B, Schams D, Rodler D, Pfaffl MW. Angiogenesis in the ovary - the most important regulatory event for follicle and corpus luteum development and function in cow - an overview. Anat Histol Embryol 2016;45:124–130. [DOI] [PubMed] [Google Scholar]
- 93.Martin-Estal I, de la Garza RG, Castilla-Cortazar I. Intrauterine growth retardation (IUGR) as a novel condition of insulin-like growth factor-1 (IGF-1) deficiency. Rev Physiol Biochem Pharmacol 2016;170:1–35. [DOI] [PubMed] [Google Scholar]
- 94.Brandt K, Grunler J, Brismar K, Wang J. Effects of IGFBP-1 and IGFBP-2 and their fragments on migration and IGF-induced proliferation of human dermal fibroblasts. Growth Horm IGF Res 2015;25:34–40. [DOI] [PubMed] [Google Scholar]
- 95.Thomsen J, Hjortebjerg R, Espelund U, et al. PAPP-A proteolytic activity enhances IGF bioactivity in ascites from women with ovarian carcinoma. Oncotarget 2015;6:32266–32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen WF, Wong MS. Genistein enhances insulin-like growth factor signaling pathway in human breast cancer (MCF-7) cells. J Clin Endocrinol Metab 2004;89:2351–2359. [DOI] [PubMed] [Google Scholar]
- 97.Guvakova MA, Surmacz E. Tamoxifen interferes with the insulin-like growth factor I receptor (IGF-IR) signaling pathway in breast cancer cells. Cancer Res 1997;57:2606–2610. [PubMed] [Google Scholar]
- 98.Glass DJ. Molecular mechanisms modulating muscle mass. Trends Mol Med 2003;9:344–350. [DOI] [PubMed] [Google Scholar]
- 99.Glass DJ. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol 2003;5:87–90. [DOI] [PubMed] [Google Scholar]
- 100.Zhang W, Liu K, Liu S, Ji B, Wang Y, Liu Y. MicroRNA-133a functions as a tumor suppressor by targeting IGF-1R in hepatocellular carcinoma. Tumour Biol 2015;36:9779–9788. [DOI] [PubMed] [Google Scholar]
- 101.O’Neill BT, Lauritzen HP, Hirshman MF, Smyth G, Goodyear LJ, Kahn CR. Differential role of insulin/IGF-1 receptor signaling in muscle growth and glucose homeostasis. Cell Rep 2015;11:1220–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park JY, Murakami T, Lee JY, Zhang Y, Hoffman RM, Bouvet M. Fluorescent-antibody targeting of insulin-like growth factor-1 receptor visualizes metastatic human colon cancer in orthotopic mouse models. PLoS One 2016;11:e0146504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chien SW, Kuo DY, Liao JM, Wang PS, Yu CH. Growth Modulation of Diabetic Factors and Antidiabetic Drugs on Prostate Cancer Cell Lines. Chin J Physiol 2016;59:109–118. [DOI] [PubMed] [Google Scholar]
- 104.Subramani R, Lopez-Valdez R, Arumugam A, Nandy S, Boopalan T, Lakshmanaswamy R. Targeting insulin-like growth factor 1 receptor inhibits pancreatic cancer growth and metastasis. PLoS One 2014;9:e97016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ramcharan R, Aleksic T, Kamdoum WP, et al. IGF-1R inhibition induces schedule-dependent sensitization of human melanoma to temozolomide. Oncotarget 2015;6:39877–39890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen G, Fang T, Huang Z, et al. MicroRNA-133a inhibits osteosarcoma cells proliferation and invasion via targeting IGF-1R. Cell Physiol Biochem 2016;38:598–608. [DOI] [PubMed] [Google Scholar]
- 107.Kurmasheva RT, Dudkin L, Billups C, Debelenko LV, Morton CL, Houghton PJ. The insulin-like growth factor-1 receptor-targeting antibody, CP-751,871, suppresses tumor-derived VEGF and synergizes with rapamycin in models of childhood sarcoma. Cancer Res 2009;69:7662–7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jia Y, Zhang Y, Qiao C, et al. IGF-1R and ErbB3/HER3 contribute to enhanced proliferation and carcinogenesis in trastuzumab-resistant ovarian cancer model. Biochem Biophys Res Commun 2013;436:740–745. [DOI] [PubMed] [Google Scholar]
- 109.Hu S, Dai H, Li T, et al. Broad RTK-targeted therapy overcomes molecular heterogeneity-driven resistance to cetuximab via vectored immunoprophylaxis in colorectal cancer. Cancer Lett 2016;382:32–43. [DOI] [PubMed] [Google Scholar]
- 110.Oppermann H, Levinson AD, Varmus HE, Levintow L, Bishop JM. Uninfected vertebrate cells contain a protein that is closely related to the product of the avian sarcoma virus transforming gene (src). Proc Natl Acad Sci USA 1979;76:1804–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kefalas P, Brown TR, Brickell PM. Signalling by the p60c-src family of protein-tyrosine kinases. Int J Biochem Cell Biol 1995;27:551–563. [DOI] [PubMed] [Google Scholar]
- 112.Jackson EK, Gillespie DG, Jackson TC. Phospholipase C and Src modulate angiotensin II-induced cyclic AMP production in preglomerular microvascular smooth-muscle cells from spontaneously hypertensive rats. J Cardiovasc Pharmacol 2007;49:106–110. [DOI] [PubMed] [Google Scholar]
- 113.Ma J, Lyu H, Huang J, Liu B. Targeting of erbB3 receptor to overcome resistance in cancer treatment. Mol Cancer 2014;13:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.You L, Wang Z, Li H et al. The role of STAT3 in autophagy. Autophagy 2015;11:729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Digiacomo G, Tusa I, Bacci M, Cipolleschi MG, Dello Sbarba P, Rovida E. Fibronectin induces macrophage migration through a SFK-FAK/CSF-1R pathway. Cell Adh Migr 2016:1–11. [DOI] [PMC free article] [PubMed]
- 116.Vultur A, Villanueva J, Krepler C, et al. MEK inhibition affects STAT3 signaling and invasion in human melanoma cell lines. Oncogene 2014;33:1850–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Piwnica-Worms H, Saunders KB, Roberts TM, Smith AE, Cheng SH. Tyrosine phosphorylation regulates the biochemical and biological properties of pp60c-src. Cell 1987;49:75–82. [DOI] [PubMed] [Google Scholar]
- 118.Levinson NM, Visperas PR, Kuriyan J. The tyrosine kinase Csk dimerizes through Its SH3 domain. PLoS One 2009;4:e7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sadasivam G, Willmann R, Lin S, et al. Src-family kinases stabilize the neuromuscular synapse in vivo via protein interactions, phosphorylation, and cytoskeletal linkage of acetylcholine receptors. J Neurosci 2005;25:10479–10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hermiston ML, Zikherman J, Zhu JW. CD45, CD148, and Lyp/Pep: critical phosphatases regulating Src family kinase signaling networks in immune cells. Immunol Rev 2009;228:288–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Adam AP, Lowery AM, Martino N, Alsaffar H, Vincent PA. Src family kinases modulate the loss of endothelial barrier function in response to TNF-alpha: crosstalk with p38 signaling. PLoS One 2016;11:e0161975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rengifo-Cam W, Konishi A, Morishita N, et al. Csk defines the ability of integrin-mediated cell adhesion and migration in human colon cancer cells: implication for a potential role in cancer metastasis. Oncogene 2004;23:289–297. [DOI] [PubMed] [Google Scholar]
- 123.Sirvent A, Benistant C, Pannequin J, et al. Src family tyrosine kinases-driven colon cancer cell invasion is induced by Csk membrane delocalization. Oncogene 2010;29:1303–1315. [DOI] [PubMed] [Google Scholar]
- 124.Matalkah F, Martin E, Zhao H, Agazie YM. SHP2 acts both upstream and downstream of multiple receptor tyrosine kinases to promote basal-like and triple-negative breast cancer. Breast Cancer Res 2016;18:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tai YL, Chu PY, Lai IR, et al. An EGFR/Src-dependent beta4 integrin/FAK complex contributes to malignancy of breast cancer. Sci Rep 2015;5:16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gnoni A, Marech I, Silvestris N, Vacca A, Lorusso V. Dasatinib: an anti-tumour agent via Src inhibition. Curr Drug Targets 2011;12:563–578. [DOI] [PubMed] [Google Scholar]
- 127.Kantarjian H, le Coutre P, Cortes J, et al. Phase 1 study of INNO-406, a dual Abl/Lyn kinase inhibitor, in Philadelphia chromosome-positive leukemias after imatinib resistance or intolerance. Cancer 2010;116:2665–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gold KA, Lee JJ, Harun N, et al. A phase I/II study combining erlotinib and dasatinib for non-small cell lung cancer. Oncologist 2014;19:1040–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Strickler JH, McCall S, Nixon AB, et al. Phase I study of dasatinib in combination with capecitabine, oxaliplatin and bevacizumab followed by an expanded cohort in previously untreated metastatic colorectal cancer. Invest New Drugs 2014;32:330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sharma MR, Wroblewski K, Polite BN, et al. Dasatinib in previously treated metastatic colorectal cancer: a phase II trial of the University of Chicago Phase II Consortium. Invest New Drugs 2012;30:1211–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lassman AB, Pugh SL, Gilbert MR, et al. Phase 2 trial of dasatinib in target-selected patients with recurrent glioblastoma (RTOG 0627). Neuro Oncol 2015;17:992–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Algazi AP, Weber JS, Andrews SC, et al. Phase I clinical trial of the Src inhibitor dasatinib with dacarbazine in metastatic melanoma. Br J Cancer 2012;106:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kluger HM, Dudek AZ, McCann C, et al. A phase 2 trial of dasatinib in advanced melanoma. Cancer 2011;117:2202–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pusztai L, Moulder S, Altan M, et al. Gene signature-guided dasatinib therapy in metastatic breast cancer. Clin Cancer Res 2014;20:5265–5271. [DOI] [PubMed] [Google Scholar]
- 135.Darnell JE, Jr. STATs and gene regulation. Science 1997;277:1630–1635. [DOI] [PubMed] [Google Scholar]
- 136.Turkson J STAT proteins as novel targets for cancer drug discovery. Expert Opin Ther Targets 2004;8:409–422. [DOI] [PubMed] [Google Scholar]
- 137.Sehgal PB. Paradigm shifts in the cell biology of STAT signaling. Semin Cell Dev Biol 2008;19:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc Natl Acad Sci USA 2005;102:8150–8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.You W, Tang Q, Zhang C, et al. IL-26 promotes the proliferation and survival of human gastric cancer cells by regulating the balance of STAT1 and STAT3 activation. PLoS One 2013;8:e63588. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Liu Y, Lv L, Xiao W, et al. Leptin activates STAT3 and ERK1/2 pathways and induces endometrial cancer cell proliferation. J Huazhong Univ Sci Technolog Med Sci 2011;31:365–370. [DOI] [PubMed] [Google Scholar]
- 141.Chen RJ, Ho YS, Guo HR, Wang YJ. Rapid activation of Stat3 and ERK1/2 by nicotine modulates cell proliferation in human bladder cancer cells. Toxicol Sci 2008;104:283–293. [DOI] [PubMed] [Google Scholar]
- 142.Corvinus FM, Orth C, Moriggl R, et al. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia 2005;7:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Horiguchi A, Oya M, Marumo K, Murai M. STAT3, but not ERKs, mediates the IL-6-induced proliferation of renal cancer cells, ACHN and 769P. Kidney Int 2002;61:926–938. [DOI] [PubMed] [Google Scholar]
- 144.Zhang Y, Du XL, Wang CJ, et al. Reciprocal activation between PLK1 and Stat3 contributes to survival and proliferation of esophageal cancer cells. Gastroenterology 2012;142:521–530 e523. [DOI] [PubMed] [Google Scholar]
- 145.Lin L, Liu A, Peng Z, et al. STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res 2011;71:7226–7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao G, Zhang JG, Shi Y, et al. MiR-130b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting STAT3. PLoS One 2013;8:e73803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen H, Yang Z, Ding C, et al. Discovery of O-alkylamino tethered niclosamide derivatives as potent and orally bioavailable anticancer agents. ACS Med Chem Lett 2013;4:180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen H, Yang Z, Ding C, et al. Fragment-based drug design and identification of HJC0123, a novel orally bioavailable STAT3 inhibitor for cancer therapy. Eur J Med Chem 2013;62:498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lin W, Zheng L, Zhuang Q, et al. Spica prunellae promotes cancer cell apoptosis, inhibits cell proliferation and tumor angiogenesis in a mouse model of colorectal cancer via suppression of stat3 pathway. BMC Complement Altern Med 2013;13:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhuang Q, Hong F, Shen A, et al. Pien Tze Huang inhibits tumor cell proliferation and promotes apoptosis via suppressing the STAT3 pathway in a colorectal cancer mouse model. Int J Oncol 2012;40:1569–1574. [DOI] [PubMed] [Google Scholar]
- 151.Kanai M, Konda Y, Nakajima T, et al. Differentiation-inducing factor-1 (DIF-1) inhibits STAT3 activity involved in gastric cancer cell proliferation via MEK-ERK-dependent pathway. Oncogene 2003;22:548–554. [DOI] [PubMed] [Google Scholar]
- 152.Pancotti F, Roncuzzi L, Maggiolini M, Gasperi-Campani A. Caveolin-1 silencing arrests the proliferation of metastatic lung cancer cells through the inhibition of STAT3 signaling. Cell Signal 2012;24:1390–1397. [DOI] [PubMed] [Google Scholar]
- 153.Kalluri R Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer 2003;3:422–433. [DOI] [PubMed] [Google Scholar]
- 154.Kang SH, Yu MO, Park KJ, Chi SG, Park DH, Chung YG. Activated STAT3 regulates hypoxia-induced angiogenesis and cell migration in human glioblastoma. Neurosurgery 2010;67:1386–1395. [DOI] [PubMed] [Google Scholar]
- 155.Chen H, Ye D, Xie X, Chen B, Lu W. VEGF, VEGFRs expressions and activated STATs in ovarian epithelial carcinoma. Gynecol Oncol 2004;94:630–635. [DOI] [PubMed] [Google Scholar]
- 156.Chan KS, Sano S, Kiguchi K, et al. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J Clin Invest 2004;114:720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kataoka K, Kim DJ, Carbajal S, Clifford JL, DiGiovanni J. Stage-specific disruption of Stat3 demonstrates a direct requirement during both the initiation and promotion stages of mouse skin tumorigenesis. Carcinogenesis 2008;29:1108–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Preston DS, Stern RS. Nonmelanoma cancers of the skin. N Engl J Med 1992;327:1649–1662. [DOI] [PubMed] [Google Scholar]
- 159.Kim DJ, Tremblay ML, Digiovanni J. Protein tyrosine phosphatases, TC-PTP, SHP1, and SHP2, cooperate in rapid dephosphorylation of Stat3 in keratinocytes following UVB irradiation. PLoS One 2010;5:e10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sano S, Chan KS, Kira M, et al. Signal transducer and activator of transcription 3 is a key regulator of keratinocyte survival and proliferation following UV irradiation. Cancer Res 2005;65:5720–5729. [DOI] [PubMed] [Google Scholar]
- 161.Kim DJ, Angel JM, Sano S, DiGiovanni J. Constitutive activation and targeted disruption of signal transducer and activator of transcription 3 (Stat3) in mouse epidermis reveal its critical role in UVB-induced skin carcinogenesis. Oncogene 2009;28:950–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rho O, Kim DJ, Kiguchi K, Digiovanni J. Growth factor signaling pathways as targets for prevention of epithelial carcinogenesis. Mol Carcinog 2011;50:264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Debnath B, Xu S, Neamati N. Small molecule inhibitors of signal transducer and activator of transcription 3 (Stat3) protein. J Med Chem 2012;55:6645–6668. [DOI] [PubMed] [Google Scholar]
- 164.Deng J, Grande F, Neamati N. Small molecule inhibitors of Stat3 signaling pathway. Curr Cancer Drug Targets 2007;7:91–107. [DOI] [PubMed] [Google Scholar]
- 165.Turkson J, Ryan D, Kim JS, et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem 2001;276:45443–45455. [DOI] [PubMed] [Google Scholar]
- 166.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci USA 2005;102:4700–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bhasin D, Cisek K, Pandharkar T, et al. Design, synthesis, and studies of small molecule STAT3 inhibitors. Bioorg Med Chem Lett 2008;18:391–395. [DOI] [PubMed] [Google Scholar]
- 168.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol 2006;13:1235–1242. [DOI] [PubMed] [Google Scholar]
- 169.Fletcher S, Page BD, Zhang X, et al. Antagonism of the Stat3-Stat3 protein dimer with salicylic acid based small molecules. ChemMedChem 2011;6:1459–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Siddiquee KA, Gunning PT, Glenn M, et al. An oxazole-based small-molecule Stat3 inhibitor modulates Stat3 stability and processing and induces antitumor cell effects. ACS Chem Biol 2007;2:787–798. [DOI] [PubMed] [Google Scholar]
- 171.Zhang X, Yue P, Page BD, et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc Natl Acad Sci USA 2012;109:9623–9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lin YM, Wang CM, Jeng JC, Leprince D, Shih HM. HIC1 interacts with and modulates the activity of STAT3. Cell Cycle 2013;12:2266–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Turkson J, Zhang S, Palmer J, et al. Inhibition of constitutive signal transducer and activator of transcription 3 activation by novel platinum complexes with potent antitumor activity. Mol Cancer Ther 2004;3:1533–1542. [PubMed] [Google Scholar]
- 174.Nagel-Wolfrum K, Buerger C, Wittig I, Butz K, Hoppe-Seyler F, Groner B. The interaction of specific peptide aptamers with the DNA binding domain and the dimerization domain of the transcription factor Stat3 inhibits transactivation and induces apoptosis in tumor cells. Mol Cancer Res 2004;2:170–182. [PubMed] [Google Scholar]
- 175.Huang W, Dong Z, Wang F, Peng H, Liu JY, Zhang JT. A small molecule compound targeting STAT3 DNA-binding domain inhibits cancer cell proliferation, migration, and invasion. ACS Chem Biol 2014;9:1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature 1998;394:145–151. [DOI] [PubMed] [Google Scholar]
- 177.Timofeeva OA, Gaponenko V, Lockett SJ, et al. Rationally designed inhibitors identify STAT3 N-domain as a promising anticancer drug target. ACS Chem Biol 2007;2:799–809. [DOI] [PubMed] [Google Scholar]
- 178.Timofeeva OA, Tarasova NI, Zhang X, et al. STAT3 suppresses transcription of proapoptotic genes in cancer cells with the involvement of its N-terminal domain. Proc Natl Acad Sci USA 2013;110:1267–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gao L, Zhang L, Hu J, et al. Down-regulation of signal transducer and activator of transcription 3 expression using vector-based small interfering RNAs suppresses growth of human prostate tumor in vivo. Clin Cancer Res 2005;11:6333–6341. [DOI] [PubMed] [Google Scholar]
- 180.Konnikova L, Kotecki M, Kruger MM, Cochran BH. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer 2003;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ling X, Arlinghaus RB. Knockdown of STAT3 expression by RNA interference inhibits the induction of breast tumors in immunocompetent mice. Cancer Res 2005;65:2532–2536. [DOI] [PubMed] [Google Scholar]
- 182.Souissi I, Najjar I, Ah-Koon L, et al. A STAT3-decoy oligonucleotide induces cell death in a human colorectal carcinoma cell line by blocking nuclear transfer of STAT3 and STAT3-bound NF-kappaB. BMC Cell Biol 2011;12:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Weerasinghe P, Garcia GE, Zhu Q, et al. Inhibition of Stat3 activation and tumor growth suppression of non-small cell lung cancer by G-quartet oligonucleotides. Int J Oncol 2007;31:129–136. [PubMed] [Google Scholar]
- 184.Jing N, Li Y, Xu X, et al. Targeting Stat3 with G-quartet oligodeoxynucleotides in human cancer cells. DNA Cell Biol 2003;22:685–696. [DOI] [PubMed] [Google Scholar]
- 185.Jing N, Li Y, Xiong W, Sha W, Jing L, Tweardy DJ. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res 2004;64:6603–6609. [DOI] [PubMed] [Google Scholar]
- 186.Staal SP, Hartley JW, Rowe WP. Isolation of transforming murine leukemia viruses from mice with a high incidence of spontaneous lymphoma. Proc Natl Acad Sci USA 1977;74:3065–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2002;2:489–501. [DOI] [PubMed] [Google Scholar]
- 188.Alessi DR, Deak M, Casamayor A, et al. 3-Phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr Biol 1997;7:776–789. [DOI] [PubMed] [Google Scholar]
- 189.Stephens L, Anderson K, Stokoe D, et al. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science 1998;279:710–714. [DOI] [PubMed] [Google Scholar]
- 190.Jacinto E, Facchinetti V, Liu D, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 2006;127:125–137. [DOI] [PubMed] [Google Scholar]
- 191.Grimes CA, Jope RS. The multifaceted roles of glycogen synthase kinase 3beta in cellular signaling. Prog Neurobiol 2001;65:391–426. [DOI] [PubMed] [Google Scholar]
- 192.Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, Gonzalez-Baron M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev 2004;30:193–204. [DOI] [PubMed] [Google Scholar]
- 193.Altomare DA, Khaled AR. Homeostasis and the importance for a balance between AKT/mTOR activity and intracellular signaling. Curr Med Chem 2012;19:3748–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol 2005;17:596–603. [DOI] [PubMed] [Google Scholar]
- 195.Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med (Berl) 2011;89:221–228. [DOI] [PubMed] [Google Scholar]
- 196.Shi Y, Liu X, Han EK, et al. Optimal classes of chemotherapeutic agents sensitized by specific small-molecule inhibitors of akt in vitro and in vivo. Neoplasia 2005;7:992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Knight ZA, Shokat KM. Chemically targeting the PI3K family. Biochem Soc Trans 2007;35:245–249. [DOI] [PubMed] [Google Scholar]
- 198.Marone R, Cmiljanovic V, Giese B, Wymann MP. Targeting phosphoinositide 3-kinase: moving towards therapy. Biochim Biophys Acta 2008;1784:159–185. [DOI] [PubMed] [Google Scholar]
- 199.Maira SM, Stauffer F, Brueggen J, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther 2008;7:1851–1863. [DOI] [PubMed] [Google Scholar]
- 200.Serra V, Markman B, Scaltriti M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res 2008;68:8022–8030. [DOI] [PubMed] [Google Scholar]
- 201.Chang KY, Tsai SY, Wu CM, Yen CJ, Chuang BF, Chang JY. Novel phosphoinositide 3-kinase/mTOR dual inhibitor, NVP-BGT226, displays potent growth-inhibitory activity against human head and neck cancer cells in vitro and in vivo. Clin Cancer Res 2011;17:7116–7126. [DOI] [PubMed] [Google Scholar]
- 202.Burger MT, Pecchi S, Wagman A, et al. Identification of NVP-BKM120 as a potent, selective, orally bioavailable class I PI3 kinase inhibitor for treating cancer. ACS Med Chem Lett 2011;2:774–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Folkes AJ, Ahmadi K, Alderton WK, et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-t hieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J Med Chem 2008;51:5522–5532. [DOI] [PubMed] [Google Scholar]
- 204.Garlich JR, De P, Dey N, et al. A vascular targeted pan phosphoinositide 3-kinase inhibitor prodrug, SF1126, with antitumor and antiangiogenic activity. Cancer Res 2008;68:206–215. [DOI] [PubMed] [Google Scholar]
- 205.Ihle NT, Paine-Murrieta G, Berggren MI, et al. The phosphatidylinositol-3-kinase inhibitor PX-866 overcomes resistance to the epidermal growth factor receptor inhibitor gefitinib in A-549 human non-small cell lung cancer xenografts. Mol Cancer Ther 2005;4:1349–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Herman SE, Gordon AL, Wagner AJ, et al. Phosphatidylinositol 3-kinase-delta inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood 2010;116:2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hilgard P, Klenner T, Stekar J, Nossner G, Kutscher B, Engel J. D-21266, a new heterocyclic alkylphospholipid with antitumour activity. Eur J Cancer 1997;33:442–446. [DOI] [PubMed] [Google Scholar]
- 208.Meuillet EJ, Ihle N, Baker AF, et al. In vivo molecular pharmacology and antitumor activity of the targeted Akt inhibitor PX-316. Oncol Res 2004;14:513–527. [DOI] [PubMed] [Google Scholar]
- 209.Gills JJ, Dennis PA. The development of phosphatidylinositol ether lipid analogues as inhibitors of the serine/threonine kinase, Akt. Expert Opin Investig Drugs 2004;13:787–797. [DOI] [PubMed] [Google Scholar]
- 210.Gills JJ, Holbeck S, Hollingshead M, Hewitt SM, Kozikowski AP, Dennis PA. Spectrum of activity and molecular correlates of response to phosphatidylinositol ether lipid analogues, novel lipid-based inhibitors of Akt. Mol Cancer Ther 2006;5:713–722. [DOI] [PubMed] [Google Scholar]
- 211.Rhodes N, Heerding DA, Duckett DR, et al. Characterization of an Akt kinase inhibitor with potent pharmacodynamic and antitumor activity. Cancer Res 2008;68:2366–2374. [DOI] [PubMed] [Google Scholar]
- 212.Davies BR, Greenwood H, Dudley P, et al. Preclinical pharmacology of AZD5363, an inhibitor of AKT: pharmacodynamics, antitumor activity, and correlation of monotherapy activity with genetic background. Mol Cancer Ther 2012;11:873–887. [DOI] [PubMed] [Google Scholar]
- 213.Spencer A, Yoon SS, Harrison SJ, et al. The novel AKT inhibitor afuresertib shows favorable safety, pharmacokinetics, and clinical activity in multiple myeloma. Blood 2014;124:2190–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Dumble M, Crouthamel MC, Zhang SY, et al. Discovery of novel AKT inhibitors with enhanced anti-tumor effects in combination with the MEK inhibitor. PLoS One 2014;9:e100880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Blake JF, Xu R, Bencsik JR, et al. Discovery and preclinical pharmacology of a selective ATP-competitive Akt inhibitor (GDC-0068) for the treatment of human tumors. J Med Chem 2012;55:8110–8127. [DOI] [PubMed] [Google Scholar]
- 216.Lindsley CW, Barnett SF, Yaroschak M, Bilodeau MT, Layton ME. Recent progress in the development of ATP-competitive and allosteric Akt kinase inhibitors. Curr Top Med Chem 2007;7:1349–1363. [DOI] [PubMed] [Google Scholar]
- 217.Hirai H, Sootome H, Nakatsuru Y, et al. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol Cancer Ther 2010;9:1956–1967. [DOI] [PubMed] [Google Scholar]
- 218.Sampath D, Malik A, Plunkett W, et al. Phase I clinical, pharmacokinetic, and pharmacodynamic study of the Akt-inhibitor triciribine phosphate monohydrate in patients with advanced hematologic malignancies. Leukemia Res 2013;37:1461–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.McCubrey JA, Lahair MM, Franklin RA. Reactive oxygen species-induced activation of the MAP kinase signaling pathways. Antioxid Redox Signal 2006;8:1775–1789. [DOI] [PubMed] [Google Scholar]
- 220.Kholodenko BN, Birtwistle MR. Four-dimensional dynamics of MAPK information processing systems. Wiley Interdiscip Rev Syst Biol Med 2009;1:28–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nebreda AR, Porras A. p38 MAP kinases: beyond the stress response. Trends Biochem Sci 2000;25:257–260. [DOI] [PubMed] [Google Scholar]
- 222.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev 2001;81:807–869. [DOI] [PubMed] [Google Scholar]
- 223.Karin M, Gallagher E. From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life 2005;57:283–295. [DOI] [PubMed] [Google Scholar]
- 224.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol 2007;19:142–149. [DOI] [PubMed] [Google Scholar]
- 225.Rincon M, Davis RJ. Regulation of the immune response by stress-activated protein kinases. Immunol Rev 2009;228:212–224. [DOI] [PubMed] [Google Scholar]
- 226.Kohno M, Pouyssegur J. Targeting the ERK signaling pathway in cancer therapy. Ann Med 2006;38:200–211. [DOI] [PubMed] [Google Scholar]
- 227.Gilmartin AG, Bleam MR, Groy A, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res 2011;17:989–1000. [DOI] [PubMed] [Google Scholar]
- 228.Sebolt-Leopold JS, Dudley DT, Herrera R, et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat Med 1999;5:810–816. [DOI] [PubMed] [Google Scholar]
- 229.Davies BR, Logie A, McKay JS, et al. AZD6244 (ARRY-142886), a potent inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1/2 kinases: mechanism of action in vivo, pharmacokinetic/pharmacodynamic relationship, and potential for combination in preclinical models. Mol Cancer Ther 2007;6:2209–2219. [DOI] [PubMed] [Google Scholar]
- 230.Choo EF, Belvin M, Boggs J, et al. Preclinical disposition of GDC-0973 and prospective and retrospective analysis of human dose and efficacy predictions. Drug Metab Dispos 2012;40:919–927. [DOI] [PubMed] [Google Scholar]
- 231.Martinez-Garcia M, Banerji U, Albanell J, et al. First-in-human, phase I dose-escalation study of the safety, pharmacokinetics, and pharmacodynamics of RO5126766, a first-in-class dual MEK/RAF inhibitor in patients with solid tumors. Clin Cancer Res 2012;18:4806–4819. [DOI] [PubMed] [Google Scholar]
- 232.Boulton TG, Nye SH, Robbins DJ, et al. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 1991;65:663–675. [DOI] [PubMed] [Google Scholar]
- 233.Haystead TA, Dent P, Wu J, Haystead CM, Sturgill TW. Ordered phosphorylation of p42mapk by MAP kinase kinase. FEBS Lett 1992;306:17–22. [DOI] [PubMed] [Google Scholar]
- 234.Morris EJ, Jha S, Restaino CR, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov 2013;3:742–750. [DOI] [PubMed] [Google Scholar]
- 235.Deng Y, Shipps GW Jr., Cooper A, et al. Discovery of novel, dual mechanism ERK inhibitors by affinity selection screening of an inactive kinase. J Med Chem 2014;57:8817–8826. [DOI] [PubMed] [Google Scholar]
- 236.Chaikuad A, Tacconi EM, Zimmer J, et al. A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics. Nat Chem Biol 2014;10:853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Aronov AM, Tang Q, Martinez-Botella G, et al. Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control. J Med Chem 2009;52:6362–6368. [DOI] [PubMed] [Google Scholar]
- 238.Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 2004;304:1164–1166. [DOI] [PubMed] [Google Scholar]
- 239.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis 2008;29:673–680. [DOI] [PubMed] [Google Scholar]
- 240.Korff S, Woerner SM, Yuan YP, Bork P, von Knebel Doeberitz M, Gebert J. Frameshift mutations in coding repeats of protein tyrosine phosphatase genes in colorectal tumors with microsatellite instability. BMC Cancer 2008;8:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.de Voer RM, Hahn MM, Weren RD, et al. Identification of novel candidate genes for early-onset colorectal cancer susceptibility. PLoS Genet 2016;12:e1005880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer 2006;6:307–320. [DOI] [PubMed] [Google Scholar]
- 243.Lui VW, Peyser ND, Ng PK, et al. Frequent mutation of receptor protein tyrosine phosphatases provides a mechanism for STAT3 hyperactivation in head and neck cancer. Proc Natl Acad Sci USA 2014;111:1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhao S, Sedwick D, Wang Z. Genetic alterations of protein tyrosine phosphatases in human cancers. Oncogene 2015;34:3885–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Scott A, Wang Z. Tumour suppressor function of protein tyrosine phosphatase receptor-T. Biosci Rep 2011;31:303–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Zhang X, Guo A, Yu J, et al. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc Natl Acad Sci USA 2007;104:4060–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Peyser ND, Freilino M, Wang L, et al. Frequent promoter hypermethylation of PTPRT increases STAT3 activation and sensitivity to STAT3 inhibition in head and neck cancer. Oncogene 2016;35:1163–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Zhao Y, Zhang X, Guda K, et al. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proc Natl Acad Sci USA 2010;107:2592–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Solomon DA, Kim JS, Cronin JC, et al. Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer Res 2008;68:10300–10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Meehan M, Parthasarathi L, Moran N, et al. Protein tyrosine phosphatase receptor delta acts as a neuroblastoma tumor suppressor by destabilizing the aurora kinase A oncogene. Mol Cancer 2012;11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Du Y, Su T, Tan X, et al. Polymorphism in protein tyrosine phosphatase receptor delta is associated with the risk of clear cell renal cell carcinoma. Gene 2013;512:64–69. [DOI] [PubMed] [Google Scholar]
- 252.Veeriah S, Brennan C, Meng S, et al. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci USA 2009;106:9435–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Brantley EC, Benveniste EN. Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res 2008;6:675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010;463:318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Ortiz B, Fabius AW, Wu WH, et al. Loss of the tyrosine phosphatase PTPRD leads to aberrant STAT3 activation and promotes gliomagenesis. Proc Natl Acad Sci USA 2014;111:8149–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cool DE, Tonks NK, Charbonneau H, Walsh KA, Fischer EH, Krebs EG. cDNA isolated from a human T-cell library encodes a member of the protein-tyrosine-phosphatase family. Proc Natl Acad Sci USA 1989;86:5257–5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Mosinger B Jr., Tillmann U, Westphal H, Tremblay ML. Cloning and characterization of a mouse cDNA encoding a cytoplasmic protein-tyrosine-phosphatase. Proc Natl Acad Sci USA 1992;89:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Dube N, Tremblay ML. Involvement of the small protein tyrosine phosphatases TC-PTP and PTP1B in signal transduction and diseases: from diabetes, obesity to cell cycle, and cancer. Biochim Biophys Acta 2005;1754:108–117. [DOI] [PubMed] [Google Scholar]
- 259.Xu D, Qu CK. Protein tyrosine phosphatases in the JAK/STAT pathway. Front Biosci 2008;13:4925–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kleppe M, Lahortiga I, El Chaar T, et al. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat Genet 2010;42:530–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Shields BJ, Wiede F, Gurzov EN, et al. TCPTP regulates SFK and STAT3 signaling and is lost in triple-negative breast cancers. Mol Cell Biol 2013;33:557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wang Y, Ning H, Ren F, et al. GdX/UBL4A specifically stabilizes the TC45/STAT3 association and promotes dephosphorylation of STAT3 to repress tumorigenesis. Mol Cell 2014;53:752–765. [DOI] [PubMed] [Google Scholar]
- 263.Mariappan M, Li X, Stefanovic S, et al. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 2010;466:1120–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wang Q, Liu Y, Soetandyo N, Baek K, Hegde R, Ye Y. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell 2011;42:758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Yi TL, Cleveland JL, Ihle JN. Protein tyrosine phosphatase containing SH2 domains: characterization, preferential expression in hematopoietic cells, and localization to human chromosome 12p12-p13. Mol Cell Biol 1992;12:836–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Khoury JD, Rassidakis GZ, Medeiros LJ, Amin HM, Lai R. Methylation of SHP1 gene and loss of SHP1 protein expression are frequent in systemic anaplastic large cell lymphoma. Blood 2004;104:1580–1581. [DOI] [PubMed] [Google Scholar]
- 267.Han Y, Amin HM, Franko B, Frantz C, Shi X, Lai R. Loss of SHP1 enhances JAK3/STAT3 signaling and decreases proteosome degradation of JAK3 and NPM-ALK in ALK+ anaplastic large-cell lymphoma. Blood 2006;108:2796–2803. [DOI] [PubMed] [Google Scholar]
- 268.Tai WT, Cheng AL, Shiau CW, et al. Signal transducer and activator of transcription 3 is a major kinase-independent target of sorafenib in hepatocellular carcinoma. J Hepatol 2011;55:1041–1048. [DOI] [PubMed] [Google Scholar]
- 269.Liu CY, Tseng LM, Su JC, et al. Novel sorafenib analogues induce apoptosis through SHP-1 dependent STAT3 inactivation in human breast cancer cells. Breast Cancer Res 2013;15:R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Fan LC, Teng HW, Shiau CW, et al. SHP-1 is a target of regorafenib in colorectal cancer. Oncotarget 2014;5:6243–6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Yu ZH, Zhang RY, Walls CD, et al. Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations. Biochemistry 2014;53:4136–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Bard-Chapeau EA, Li S, Ding J, et al. Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular carcinogenesis. Cancer Cell 2011;19:629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Peyser ND, Wang L, Zeng Y, et al. STAT3 as a Chemoprevention Target in Carcinogen-Induced Head and Neck Squamous Cell Carcinoma. Cancer Prev Res 2016;9:657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Hendriks W, Brugman C, Richter KH, et al. Protein-tyrosine phosphatases expressed in mouse epidermal keratinocytes. J Invest Dermatol 1996;106:972–976. [DOI] [PubMed] [Google Scholar]
- 275.McArdle L, Rafferty M, Maelandsmo GM, et al. Protein tyrosine phosphatase genes downregulated in melanoma. J Invest Dermatol 2001;117:1255–1260. [DOI] [PubMed] [Google Scholar]
- 276.McArdle L, Rafferty MM, Satyamoorthy K, et al. Microarray analysis of phosphatase gene expression in human melanoma. Br J Dermatol 2005;152:925–930. [DOI] [PubMed] [Google Scholar]
- 277.Sachsenmaier C, Radler-Pohl A, Zinck R, Nordheim A, Herrlich P, Rahmsdorf HJ. Involvement of growth factor receptors in the mammalian UVC response. Cell 1994;78:963–972. [DOI] [PubMed] [Google Scholar]
- 278.Coffer PJ, Burgering BM, Peppelenbosch MP, Bos JL, Kruijer W. UV activation of receptor tyrosine kinase activity. Oncogene 1995;11:561–569. [PubMed] [Google Scholar]
- 279.Caselli A, Marzocchini R, Camici G, et al. The inactivation mechanism of low molecular weight phosphotyrosine-protein phosphatase by H2O2. J Biol Chem 1998;273:32554–32560. [DOI] [PubMed] [Google Scholar]
- 280.Denu JM, Tanner KG. Redox regulation of protein tyrosine phosphatases by hydrogen peroxide: detecting sulfenic acid intermediates and examining reversible inactivation. Methods Enzymol 2002;348:297–305. [DOI] [PubMed] [Google Scholar]
- 281.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell 2005;121:667–670. [DOI] [PubMed] [Google Scholar]
- 282.Gulati P, Markova B, Gottlicher M, Bohmer FD, Herrlich PA. UVA inactivates protein tyrosine phosphatases by calpain-mediated degradation. EMBO Rep 2004;5:812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Xu Y, Shao Y, Voorhees JJ, Fisher GJ. Oxidative inhibition of receptor-type protein-tyrosine phosphatase kappa by ultraviolet irradiation activates epidermal growth factor receptor in human keratinocytes. J Biol Chem 2006;281:27389–27397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Lee H, Morales LD, Slaga TJ, Kim DJ. Activation of T-cell protein-tyrosine phosphatase suppresses keratinocyte survival and proliferation following UVB irradiation. J Biol Chemy 2015;290:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Chan RJ, Feng GS. PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood 2007;109:862–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Tartaglia M, Mehler EL, Goldberg R, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet 2001;29(4):465–468. [DOI] [PubMed] [Google Scholar]
- 287.Tartaglia M, Gelb BD. Germ-line and somatic PTPN11 mutations in human disease. Eur J Med Genet 2005;48:81–96. [DOI] [PubMed] [Google Scholar]
- 288.Chen C, Cao M, Zhu S, et al. Discovery of a Novel Inhibitor of the Protein Tyrosine Phosphatase Shp2. Sci Rep 2015;5:17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Chen YN, LaMarche MJ, Chan HM, et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 2016;535:148–152. [DOI] [PubMed] [Google Scholar]
- 290.Yip SC, Saha S, Chernoff J. PTP1B: a double agent in metabolism and oncogenesis. Trends Biochem Sci 2010;35:442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Wiener JR, Kerns BJ, Harvey EL, et al. Overexpression of the protein tyrosine phosphatase PTP1B in human breast cancer: association with p185c-erbB-2 protein expression. J Natl Cancer Inst 1994;86:372–378. [DOI] [PubMed] [Google Scholar]
- 292.Bjorge JD, Pang A, Fujita DJ. Identification of protein-tyrosine phosphatase 1B as the major tyrosine phosphatase activity capable of dephosphorylating and activating c-Src in several human breast cancer cell lines. J Biol Chem 2000;275:41439–41446. [DOI] [PubMed] [Google Scholar]
- 293.Wang N, She J, Liu W, et al. Frequent amplification of PTP1B is associated with poor survival of gastric cancer patients. Cell Cycle 2015;14:732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Krishnan N, Koveal D, Miller DH, et al. Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nat Chem Biol 2014;10:558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Fan G, Lin G, Lucito R, Tonks NK. Protein-tyrosine phosphatase 1B antagonized signaling by insulin-like growth factor-1 receptor and kinase BRK/PTK6 in ovarian cancer cells. J Biol Chem 2013;288:24923–24934. [DOI] [PMC free article] [PubMed] [Google Scholar]