SUMMARY
Chronic or persistent stimulation of the programmed cell death-1 (PD-1) pathway prevents T cells from mounting anti-tumor and anti-viral immune responses. Blockade of this inhibitory checkpoint pathway has shown therapeutic importance by rescuing T cells from their exhausted state. Cognate ligands of the PD-1 receptor include the tissue-specific PD-L1 and PD-L2 proteins. Engineering a human PD-1 interface specific for PD-L1 or PD-L2 can provide a specific reagent and therapeutic advantage for tissue-specific disruption of the PD-1 pathway. We utilized ProtLID, a computational framework, which constitutes a residue-based pharmacophore approach, to custom-design a human PD-1 interface specific to human PD-L1 without any significant affinity to PD-L2. In subsequent cell assay experiments, half of all single-point mutant designs proved to introduce a statistically significant selectivity, with nine of these maintaining a close to wild-type affinity to PD-L1. This proof-of-concept study suggests a general approach to re-engineer protein interfaces for specificity.
Graphical Abstract
In Brief
Shrestha et al. present a computational approach that employs a residue-based pharmacophore approach to design mutations for the interface of PD-1, specific to one of its cognate ligands only, PD-L1 without any significant affinity to PD-L2. In subsequent cell assay experiments half of all single-point mutant designs proved to introduce a statistically significant selectivity.
INTRODUCTION
Lymphocyte activation requires two signals mediated by protein-protein interactions: the T cell receptor interaction with antigenic peptide/MHC complex that provides the specificity of the immune response and an antigen-independent co-modulatory signal that modulates T cell clonal expansion and acquisition of effector function (Lafferty and Cunningham, 1975). Engagement of co-stimulatory receptors such as CD28 and ICOS with their cognate B7 family ligands expressed on antigen-presenting cells results in T cell activation and proliferation. In contrast, engagement of cytotoxic T lymphocyte associate antigen, programmed cell death protein-1 (PD-1), and B and T lymphocyte attenuator with their corresponding ligands negatively regulate T cell activation (Chen and Flies, 2013).
PD-1 is a type I transmembrane protein, composed of an extracellular immunoglobulin variable (IgV) domain, a transmem-brane domain, and an intracellular domain with tyrosine-based signaling motifs (Ishida et al., 1992). The PD-1 ligands, PD-L1 (Dong et al., 1999) and PD-L2 (Latchman et al., 2001), are also type I transmembrane proteins, and possess ectodomains composed of tandem IgV and immunoglobulin constant domains, a single-pass helical transmembrane region and a cytoplasmic domain (Keir et al., 2008; Chattopadhyay et al., 2009). T cells, natural killer T cells, B cells, and some myeloid cells, express PD-1 receptor (Keir et al., 2008). PD-L1 is constitutively expressed on immune cells such as T cells, B cells, and dendritic cells (DCs) (Yamazaki et al., 2002). Nonhematopoietic cells can also express PD-1, with expression levels depending on a variety of other stimuli (Keir et al., 2008). Expression of PD-L2 is limited to DCs, macrophages, and bone marrow-derived mast cells (Latchman et al., 2001).
Engagement of PD-1 with either of its two known ligands, PD-L1 (B7-H1) or PD-L2 (B7-DC) from the B7 family (Chemnitz et al., 2004), provides inhibitory signals that control autoimmune responses by maintaining immune tolerance to self-anti-gens (Riella et al., 2012). PD-1 signaling also makes important contributions to responses against microbial pathogens. Persistent PD-1 expression can cause T cell exhaustion, which reduces antiviral and anti-tum or immune responses, and leads to unfavorable disease progression by inhibiting T cell proliferation and cytotoxicity, as well as cytokine production (Wherry, 2011). Blockade of the PD-1 pathway can reverse T cell exhaustion and enhance anti-tum or responses (Sakuishi et al., 2010). Malignant cells utilize a range of mechanisms to evade host immune surveillance, including the overexpression of PD-L1 on their surface, which can result in a highly immune-suppressive milieu in the tum or microenvironment (Iwai et al., 2002; Dong et al., 2002). Therapeutic agents, especially monoclonal antibodies (mAbs), have been developed to block the PD-1 signaling pathways (Nguyen and Ohashi, 2015) and have shown activity against several cancers, with US Food and Drug Administration approval for multiple indications (Topalian et al., 2012; Brahmer et al., 2012). Among these, nivolumab and pembrolizumab have shown clinical efficacy for the treatment of metastatic melanoma, non-small-cell lung cancer and other malignancies (Postow et al., 2015). Similarly, re-invigorating T cells through PD-1 blockade can elicit beneficial immune responses in chronic viral infections (Lazar-Molnar et al., 2010; Barber et al., 2006; Day et al., 2006). To further increase potency, engineered T cells have been combined with other therapeutics targeting the PD-1 signaling pathway (Hoos, 2016; Fesnak et al., 2016). However, mAbs have inherent limitations such as antigenicity, poor tissue penetrance due to their large size (~150 kDa), and detrimental Fc-effector functions that deplete immune cells (Lee and Tannock, 2010). As a complementary approach to mAbs, PD-1 ectodomains and their engineered variants could be used to interrupt the PD-1 pathway by directly binding to the PD ligands. Directed evolution via yeast-surface display has been used to engineer a PD-1 variant that specifically antagonizes PD-L1. The resulting PD-1 construct had 10 residues mutated compared with wild-type (WT) PD-1, resulting in a 15-to 40,000-fold increase affinity to PD-L1, while not binding to PD-L2 (Maute et al., 2015). While possessing remarkable affinity and selectivity, the large number of altered residues could result in undesirable antigenic properties, making variants with the smallest number of mutations desirable. In a recent study, a cross-reactive single mutant (A132L) in PD-1 was reported to enhance binding activity to PD-L1 and PD-L2 by 45- and 30-fold, respectively, compared with WT PD-1 (Lazar-Molnar et al., 2017).
Computational protein design methods generate and assess large numbers of sequence variants at predetermined binding surfaces. These approaches dramatically reduce the number of designs for subsequent experimental evaluation (Mandell and Kortemme, 2009; Lippow et al., 2007). Computational design algorithms have been used to design new folds (Kuhlman et al., 2003), enzymatic functions (Rothlisberger et al., 2008), and novel binding functions (Looger et al., 2003). Computational approaches for optimizing selectivity often require both positive design considerations to stabilize the desired interactions and negative design considerations to distinguish among a number of sequence and structurally similar competitor molecules (Havranek and Harbury, 2003; Bolon et al., 2005).
In the context of PD-1 pathway modulation, our goal is to computationally redesign the ligand recognition surface of PD-1 to antagonize the PD-1:PD-L2 interaction, while maintaining or enhancing the affinity of the PD-1:PD-L1 interaction. We utilized our recently developed ProtLID (protein-ligand interface design) method to predict residue-based pharmacophore (rs-pharmacophore) signatures over the binding surface for PD-L1 and PD-L2 (Yap and Fiser, 2016). We then compared these rs-pharmacophores with the known binding interfaces of PD-1 and predicted positions and residue types to alter specificities. In subsequent cell-based assays, we vali-dated a number of these designs. Half of all predicted single-mutant PD-1 designs exhibited statistically significantly reduced interaction with PD-L2, nine of which maintained close to WT interaction with PD-L1, and among these, three designs showed no detectable affinity to PD-L2. These new constructs are both reagents and possible drug leads for modulation of the PD-1 pathway.
RESULTS
Residue-Specific Pharmacophore Generation
Understanding the intermolecular interactions between PD-1 and its two cognate ligands (PD-L1 and PD-L2) can help selectively block the PD-1 signaling pathway for mechanistic analysis and potentially provide therapeutic leads. We focused on rede-signing the human PD-1 (hPD-1) interface to selectively bind to hPD-L1 (positive design), but not to hPD-L2 (negative design), to obtain a selective reagent.
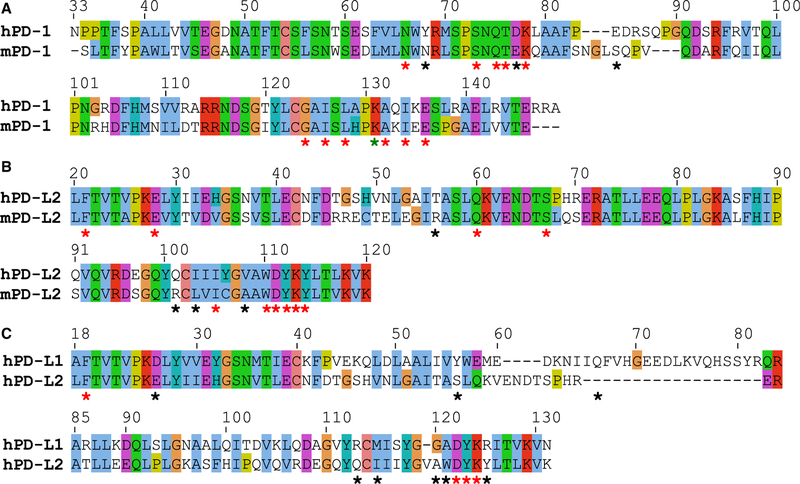
hPD-1 and hPD-L1 contribute 15 and 12 residues, respectively, to the recognition interface (4ZQK) (Zak et al., 2015) (Figure 1; Table S1). As there is no existing structure of the human hPD-1:hPD-L2 complex, the murine mPD-1:mPD-L2 complex (3BP5) (Lazar-Molnar et al., 2008) was used as a proxy. Out of the 14 interface residues that are structurally superposable between hPD-1 and mPD-1, 11 residues (N66, S73, Q75, T76, K78, G124, I126, K131, A132, I134, and E136) are identical. In the case of PD-L2, 10 out of 14 superposable interface residues (F21, E28, Q60, S67, I105, W110, D111, Y112, K113, and Y114) are identical between hPD-L2 and mPD-L2 orthologs (Figure 1; Table S1). A computational homology model (Sali and Blundell, 1993) was built for hPD-1:hPD-L2 using hPD-1 and mPD-1:mPD-L2 as a template; these orthologs share 64% and 70% overall sequence identity for the entire length of the proteins, respectively. Superposition of the hPD-1 and mPD-1 structures with DALI (Holm and Sander, 1995) resulted in Cα root-mean-square deviation = 1.8 Å over 107 out of 113 residues. Interface residues were identified by the CSU program (Sobolev et al., 1999): by this definition, K98 of mPD-1 is not part of the interface of the mPD-1:mPD-L2 complex, although K98 of mPD-1 aligns with K131 of hPD-1.
Figure 1. Sequence Alignments Derived from DALI Structural Superpositions.
(A and B) Pairwise alignments between orthologs: (A) human and mouse PD-1 (hPD-1 and mPD-1), and (B) human and mouse PD-L2 (hPD-L2 and mPD-L2). (C) Alignment between Ig-like variable-type domain of hPD-L1 and hPD-L2. Stars indicate interface residues, red and black refer to conserved and non-conserved ones, respectively. The green star indicates a conserved residue that was not found to be part of mPD-1 by the CSU program. All structures and homology models were energetically relaxed in a 25-ns-long molecular dynamics (MD) simulation prior to superposition.
To design a PD-L1-specific interface on hPD-1, we used our recently developed computational algorithm, ProtLID, to generate rs-pharmacophores for the hPD-L1 and mPD-L2 interfaces (Figure 2). Rs-pharmacophores are descriptions of idealized complementary interacting surface patches to these ligands, which are obtained through the analysis of single-amino acid binding preferences after an extensive molecular dynamics simulation. When the calculated rs-pharmacophore for hPD-L1 is compared with the actual binding residues of hPD-1, 10 out of 15 WT residues (67%) were correctly recapitulated (Table S1). Likewise, 11 out of 19 (58%) WT binding residues of mPD-1 were recapitulated, of which six (N66, Q75, T76, K78, I126, and E136) were identical to hPD-1 interface residues. Four residues are conserved between the interfaces of hPD-L1 and hPD-L2 (the PD-L2 interface residues were obtained through the comparative model built using the mPD-1:mPD-L2 experimental structure) (Table S1).
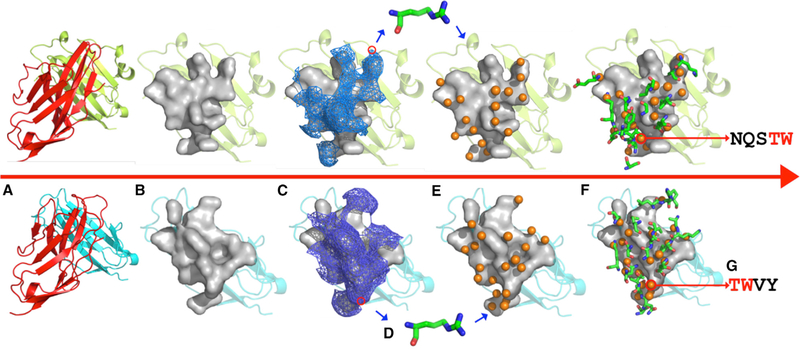
Figure 2. Schema of ProtLID Algorithm.
Major steps of the algorithm are shown: (A) input protein complexes(upper PD-1 [red]:PD-L1 [green], lower PD-1 [red]:PD-L2 [blue]), (B) identification of interface residues in target protein (in gray surface representation), (C) generation of mesh over interacting atoms of the interface residues of target proteins, (D) amino acid probes are placed in each mesh point and undergo extensive MD simulation to determine their preferencesover the surface, (E) prediction of rs-pharmacophores (orange spheres) from the analysis of MD simulation, consisting of the position and functional atom types (FAs), and (F) comparison of suggested FAs after com paring alternative rs-pharmacophores for different cognate ligands, (G) identifying differences of residue preferences between the two pharmacophores.
Selecting Single-Mutant Designs
Differences between the rs-pharmacophores generated for each ligand and the observed interface of WT hPD-1 suggested residue types and positions to modify for enhanced PD-L1 selectivity (Table S1). One set of design targets consisted of positions where the two ligands (PD-L1 and PD-L2) have different residues interacting with the receptor (PD-1), and there were also differences between the rs-pharmacophores designed for these ligand residues. These differences can be utilized to suggest mutations that selectively prefer only one ligand. For instance, in the case of K131 in hPD-1, the interacting residue in hPD-L1 is Q66, while in both hPD-L2 and mPD-L2 it is S67. The differences between the calculated rs-pharmacophores suggest that hPD-L1 uniquely preferred H and P as interacting partners. After visual inspection of the local structural environment, the K131H variant was selected for testing. Other selections involved cases where both ligands had the same residue type interacting with the receptor, but the calculated rs-pharmacophores, influenced by other residues in the environment, suggested differences in preferences between the two ligands. An example is F19 in hPD-L1 (and the equivalent F21 in mPD-L2), interacting with K78 in hPD-1. The rs-pharmacophore for hPD-L1 had two unique residue preferences, R and T, which are not preferred by PD-L2; however, K78T was previously studied (Maute et al., 2015). After visual inspection, we tested K78R, which in a subsequent cell assay showed selective binding to hPD-L1, relative to hPD-L2 (Figure 3).
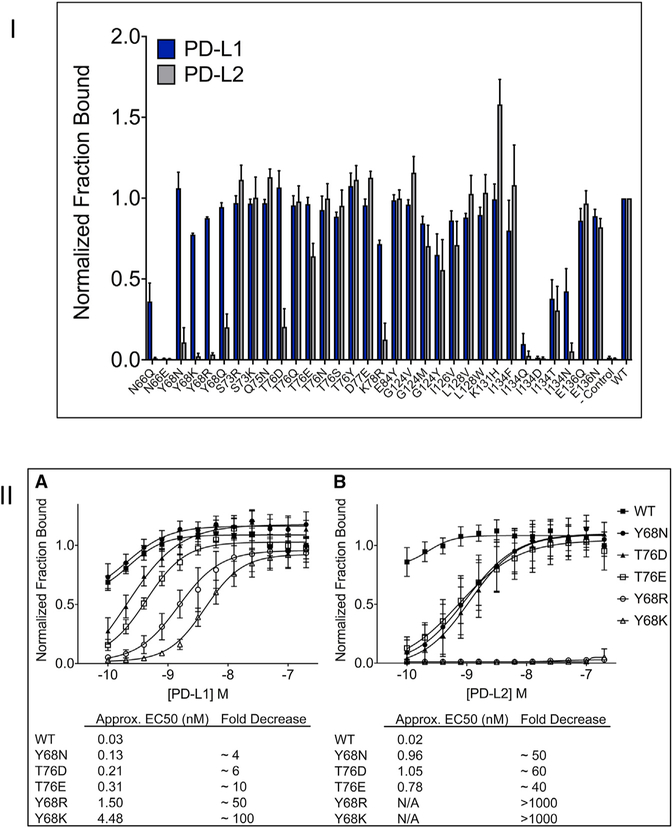
Figure 3. Experimental Testing of Selectivity of PD-1 Mutants.
(I) Flow cytometric analysis of hPD-1 mutants binding to PD-L1 and PD-L2. The data represent the average of three independent experiments with error bars showing the standard deviation. (II) Titration of hPD-1 mutants show s selective binding to PD-L1.
(II) (A) HEK293 suspension cells transiently expressing WT or mutant PD-1 were challenged with increasing concentrations of hPD-L1 hIgG1 (0.1–200 nM) and fluorescence-activated cell sorting analysis was used to determine the percent of PD-1 expressing cells (mCherry) bound to PD-L1. For each experiment, the titration data were normalized to WT PD-1 binding to the highest concentration of PD-L1. Data shown represents four independent experiments with SD. (B) The same analysis as shown in (A) except titrating hPD-L2 hIgG1 protein. Half maximal effective concentration (EC50) values could not be estimated for Y68R and Y68K (N/A) as no detectable binding was observed within the range of concentrations analyzed.
Other design elements required more elaboration due to the complex network of interactions between interface residues, which are not readily deconvolved into simple pairwise contacts. These positions often exhibit “promiscuity,” as they can accommodate a wider range of amino acid substitutions. Once these positions are identified from rs-pharmacophore preferences, they provide a more flexible target environment for exploration. For instance, one of the conserved interface residues is Y123 of hPD-L1, the equivalent of which is Y112 in mPD-L2 (and is also identical in hPD-L2). The calculated rs-pharmacophores for the two ligands suggested similar complementary interacting patches, to accommodate the two “functional atos” of Tyr (aromatic ring center and hydroxyl group, see the STAR Methods). The rs-pharmacophore contains residues that are hydrogen bond donors or acceptors (DEPNQRHT), hydrophobic (LM), and aromatic (FYW) (Table S1). The wide spectrum of tolerated residues in the rs-pharmacophore is explained by the interacting region on the hPD-1 receptor side, where six residues with diverse properties are found in spatial proximity to Y123 of hPD-L1, including hydrogen bond acceptors or donors (E136, G124, and T76), hydrophobic (I126 and I134), and aromatic (Y68) residues. The corresponding residue in mPD-L2, Y112, interacts with hydrogen bond acceptors or donors (N35, E103, and T43) and a hydrophobic residue (I101) in the WT interface of mPD-1. Interestingly, T76 (T43) and E136 (E103) are conserved residues between mPD-1 and hPD-1. Once the rs-pharmacophore was rationalized in this local context, a total of 13 mutations were explored in three positions of hPD-1. Two of these variants, T76D and T76E, achieved high selectivity for PD-L1 (Figure S1; Table S1).
Another promising location that was revealed by the rs-pharmacophore analysis was position Y68 in hPD-1, of which the equivalent is N35 in mPD-1; however, despite being different residue types, both interact with a Tyr in hPD-L1 (Y123) and mPD-L2 (Y112). Y68 is part of a cluster of interacting residues in hPD-1, whose members include E136, G124, I126, I134, and T76, making the resulting rs-pharmacophores relatively accommodating and suggesting a highly tolerant position ripe for exploration. We explored six mutants for Y68, four of which induced selectivity for PD-L1 (Figure 3).
Experimental Validation
After excluding all the previously studied mutations (Lazar-Molnar et al., 2017; Maute et al., 2015), we prioritized 34 mutants covering 14 residues in hPD-1 for experimental validation. Previously studied mutations were excluded from the current study to focus on novelty. Only four of the mutations defined in Maute et al. (2015) overlapped with our interface definition, and three out of these four, and the one defined in Lazar-Molnar et al. (2017), are part of the suggested preferences of rs-pharmacophores (Table S1). Site-directed mutagenesis was performed, resulting in 32 single-point mutations, which were all sequence validated and expressed as GFP fusions presented on the surface of suspension-adapted HEK293 cells. Analysis of the hPD-1 mutants binding to hPD-L1 and hPD-L2 was performed by high-throughput flow cytometry. The percent of hPD-1-expressing cells bound to either hPD-L1 or hPD-L2 was determined and the data normalized to the highest ligand concentration for WT hPD-1 binding. Out of the 32 hPD-1 mutants, 16 (N66Q, Y68N, Y68K, Y68R, Y68Q, S73R, Q75N, T76D, T76E, D77E, K78R, G124V, L128V, K131H, I134N, and I134F) showed statistically significant (p < 0.05, two-tailed t test) increases in selectivity toward PD-L1 (Table S2). Of these, six maintained close to WT binding interaction to PD-L1. We selected five of these designs for titration experiments, where HEK293 cells expressing WT or mutant hPD-1 were challenged with hPD-L1- or hPD-L2-expressing cells (Figure 3). Two mutants, Y68R and Y68K, showed undetectable PD-L2 binding, while half maximal effective concentration values for PD-L1 were 1.50 and 4.48 nM, respectively. The other three mutants (Y68N, T76D, and T76E) showed a 4- to 12-fold selectivity for PD-L1. All the successful mutants achieved selectivity by diminishing PD-1 binding to PD-L2 (negative design), but none of the selectivity was achieved by increasing PD-1 binding to PD-L1 (positive design) in a statistically significant manner.
Structural Insights
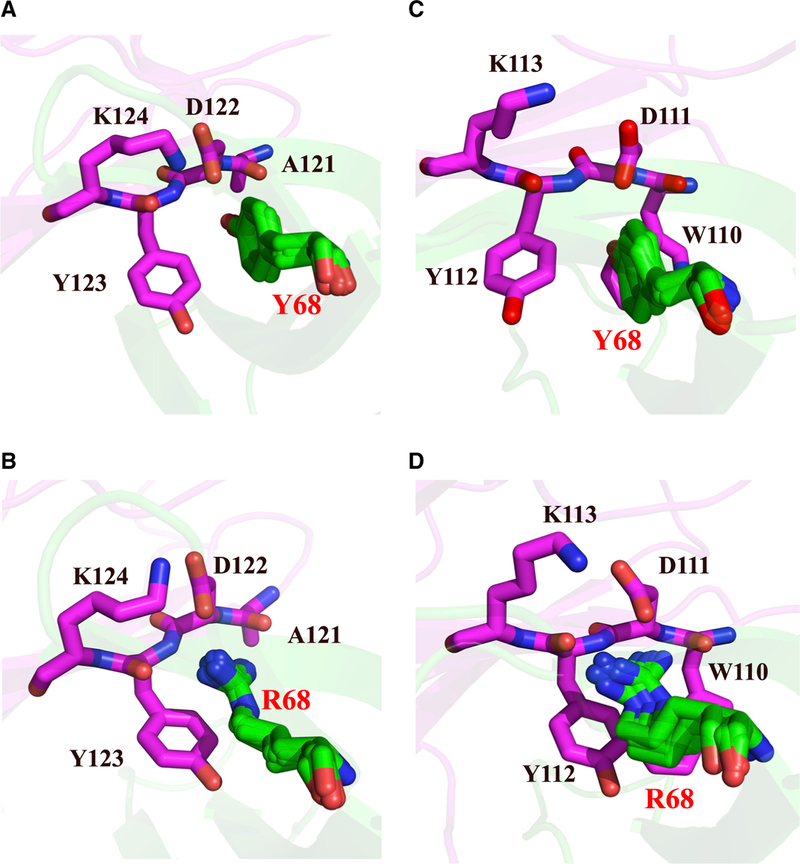
We generated comparative protein structure models to gain further insight about mutations that achieved high selectivity between the two ligands. The models of the complexes were subject to 25-ns molecular dynamics simulations using GROMACS (Lundborg and Lindahl, 2015) to accommodate the rearrangement of local contacts. Interestingly, the Y68 mutant interacts with a highly conserved cluster of residues on hPD-L1 and hPD-L2, which includes D122 (D111 for hPD-L2), Y123 (Y112), and K124 (K113). However, the modeling suggests that residue A121 in hPD-L1 (W110 in hPD-L2) is the most relevant for recognition of Y68 of PD-1. When mutated to long, polar side chains (i.e., Y68R and Y68K), a possible steric conflict emerged with W110 of hPD-L2 despite the otherwise favorable complementary charge interactions (Figure 4). When a shorter side chain is introduced, Y68N, selectivity is still achieved, but to a lesser extent (Figure 4).
Figure 4. Structural Models of Mutant PD-1:L1:L2 Interfaces.
(A-D) (A) Wild-type PD-1 (green):PD-L1 (magenta) complex, (B) mutant (Y68R) PD-1 (green):PD-L1 (magenta) complex, (C) W T PD-1 (green):PD-L2 (magenta) complex, and (D) mutant (Y68R) PD-1 (green):PD-L2 (magenta) complex. In all panels, for PD-1 five conformations taken every 4 ns during the MD simulation are shown.
When exploring the other most selective site for mutation, hPD-1 T76D or T76E, it appears that a more favorable charged or hydrogen bond interaction is established with R125 and K124 of PD-L1, while the Y114 side chain of hPD-L2 is not suitable to support this mutant (Figure S1).
Correlation with Predicted Free Energy Changes
Once experimental data were obtained we also attempted to retrospectively correlate the results with methods that predict the energetic effect of point mutations. We ran three different programs, FoldX (Schymkowitz et al., 2005), Mutabind (Li et al., 2016), and BeAtMuSiC (Dehouck et al., 2013), all of which returned random predictions (calculated correlations between predicted and measured binding affinity changes are 0.31, −0.1 2, and 0.03, respectively, Figure S2). These approaches were either unable to distinguish the differential effect of mutations on the two ligands (BeAtMuSiC), or, if differences were detected, these turned out not to correlate with the observations (FoldX and Mutabind). These results highlight the difficulty of predicting specificity-inducing mutations correctly and support the utility of the rs-pharmacophore-based approach described in this work in efficiently capturing these designs.
DISCUSSION
The PD-1 signaling pathway is one of the inhibitory checkpoints that shapes T cell activity for anti-cancer (Sznol and Chen, 2013) and anti-viral (Gardiner et al., 2013) immune responses (Sznol and Chen, 2013; Nguyen and Ohashi, 2015). Highly effective mAbs have been developed to disrupt both sides of the PD-1:PD-L1 interaction for the treatment of cancer (Topalian et al., 2012; Brahmer et al., 2012). As an alternative approach, a re-en-gineered PD-1 with picomolar affinity to hPD-L1 was developed to block the WT hPD-1:hPD-L1 interaction, with some possible advantages over conventional mAbs (Maute et al., 2015).
In this proof-of-concept study, we utilized the ProtLID computational method (Yap and Fiser, 2016) to re-engineer the protein binding interface of PD-1 for selective recognition of PD-L1, with the goal of introducing as few mutations as possible. ProtLID reduces the theoretical number of possible mutations to an experimentally manageable set using the concept of pharmacophore elaboration (Xu et al., 2012). The construction of a high-specificity interface to discriminate among multiple proteins with similar structure from the same superfamily, as in the current work, is highly challenging (Schreiber and Keating, 2011), in this case because PD-1 and its ligands share the same immunoglobulin fold. Interestingly, the most effective mutant designs of PD-1 to induce selectivity involved two residues (Y68 and T76), which interact with a highly conserved cluster of residues in PD-L1 and PD-L2. Retrospective structural analysis can explain the effect of these mutations, but these are hard to predict a priori. A parallel aim to introduce selectivity was to increase affinity of PD-1 to PD-L1, which was not successful (positive design). A possible synergistic construct design with results from previous works may achieve that. In this study we focused on the proof-of-concept of using the ProtLID program alone to locate possible spatial locations and residue types for introducing specificity of binding, and we intentionally avoided previously tested constructs. In Maute et al. (2015), ten mutations were identified that contributed to an increase in affinity of PD-L1 binding. Out of these ten, only four are part of the strictly defined interface, while the others were either mutations in the core of the protein or just outside of the periphery of the interface. Of the four positions that we explored with ProtLID, we identified the same mutations in three cases (Y68H, K78T, and A132I), but we intentionally did not pursue them further. Meanwhile, the mutation described in Lazar-Molnar et al. (2017), A132L, was also identified in our study (Table S1), and intentionally not explored further. The failure to achieve improved binding affinity to PD-L1 can be explained either by the fact that we steered clear of these already proven mutations, and no or very few additional combinations are left that can achieve the same effect, or that ProtLID is more suitable to identify incompatible sites and residue types for a given interface than compatible ones.
In this work we provided a proof-of-concept study of how residue-based pharmacophore design can aid mutational studies to achieve desired binding selectivity. The resulting variants of PD-1 should prove to be useful reagents for mechanistic studies. Meanwhile, understanding the possible biomedical impact of these PD-1 variants will require a better insight about the role PD-L2 plays in modulating the immune response (Yearley et al., 2017).
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for further information or resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andras Fiser (andras.fiser@einstein.yu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The HEK 293 Freestyle suspension adapted cells sold by Thermo Fisher (Invitrogen) were used in this study. We obtained this cell line directly from the company, we did not authenticate the line ourselves. The sex of these cells is reported as female. We culture these cells in Freestyle 293 media (Thermo Fisher Cat# 12338018). The HEK293 cells are maintained in a 150mL culture volume in a 500 mL baffled tissue culture flask. Cultures are routinely split to 0.5×10^6 cells/mL and not allowed to grow above 3×10^6 cells/mL. Cells are routinely checked for mycoplasma about every 4 – 6 months.
Site-Directed Mutagenesis of Human PD-1 Variants
The coding sequence for the full-length ectodomain of human PD-1 (Leu 25 – Thr 168) was cloned by ligation-independent cloning into a vector that adds the leader sequence from erythropoietin (EPO) and the transmembrane domain from mouse PD-L1 followed by mCherry (hPD-1 Type I mCherry). Site-specific mutagenesis was performed as described previously using high fidelity KOD polymerase (Ramagopal et al., 2017). After two rounds of primer design 32 of the 34 predicted mutations were successfully cloned and sequence validated (94% success rate). These mutants were tested for expression by transient transfection of 1 mL suspension HEK 293 cells. Prior to utilization in downstream binding experiments, all of the hPD-1 mutants were shown to express at levels com parable to the parental hPD-1 construct as analyzed by FACS and showed correct membrane localization as observed by fluorescence microscopy (Figure S3).
Analysis of PD-1 Variants Binding to PD-L1 and PD-L2 by High-Throughput Flow Cytometry
Wild-type and mutant hPD-1 constructs were transiently transfected into 1 mL suspension of HEK 293 cells at a density of 1×106 cells/mL in 24-well plates using 0.5 μg plasmid DNA and 2 μg linear PEI. Two days post transfection, cells were counted and diluted to 1×106 cells/mL with 1× PBS with 2% BSA. In 96-well V-bottom plates, 100,000 cells were challenged with 0.1 μg of hPD-L1 or hPD-L2 Fc-fusion protein (R&D Systems) for 1 hour at room temperature while shaking in a 96-well plate shaker at 900rpm. Cells were subsequently pelleted by centrifugation at 500×g and washed with 1×PBS with 2% BSA two times. Goat anti-human Alexa 488 secondary antibody (0.25 μg) was added to the cells and they were incubated at 4°C for 45 min. After washing three times, antibody binding was assessed by FACS analysis on a BD Accuri cytometer connected to an Intellicyt Hypercyte auto sampler. Flow data were gated for mCherry positive events (hPD-1 expression) and then sub-gated for hPD-L1/L2 binding (Alexa 488 channel). The experiment was performed in triplicate and the data from each experiment were normalized to wild type PD-1 binding.
For titration experiments, WT and selected PD-1 mutants were transiently transfected as described above. Two days post transfection cells were diluted to 1×106 cells/mL with 1× PBS with 2% BSA and 100,000 cells were challenged with increasing concentrations of hPD-L1 hIgG1 or hPD-L2 hIgG1 (R&D Systems) from 0.1 – 200 nM final concentrations. After binding for 1 hour at room temperature, the cells were washed once with 1× PBS with 2% BSA and goat anti-human Alexa 488 secondary antibody (0.25 μg) was added. After incubation with secondary antibody, the cells were washed twice with 1× PBS with 2% BSA and analyzed by FACS as described above. The percent of PD-1 expressing cells bound to either PD-L1 or PD-L2 was determined and the data normalized to the highest ligand concentration for wild-type PD-1 binding. The EC50s were estimated by plotting the normalized titration data in Graphpad Prism software and fitting the data using the equation for a three-parameter dose response non-linear regression analysis Y = Bmin + (Bmax – Bmin/(1 + 10(LogEC50 - X)).
METHOD DETAILS
PD-1, PD-L1, and PD-L2 Proteins and Their Complexes
The crystallographic structures of the human ectodomains of PD-1 and PD-L1 complex (4ZQK) were recently published (Zak et al., 2015). The hPD-1 and hPD-L2 complex is not available, but the structure of the orthologous mouse mPD-1:mPDL2 (PDB ID: 3BP5) complex has been determined (Lazar-Molnar et al., 2008). Since hPD-1 and mPD-1 proteins share 64% sequence identity, a computational homology model for the hPD-1:hPD-L2 complex was generated for this study (Rai et al., 2007; Fernandez-Fuentes et al., 2007; Rai and Fiser, 2006), using the mPD-1:mPD-L2 complex and hPD1 (5GGS.Y) as the templates.
ProtLID, Residue-Specific Pharmacophore Approach for Interface Design
We recently developed a residue-based pharmacophore (rs-pharmacophore) approach for interface design (ProtLID, Protein Ligand Interface Design) that was used to identify cognate ligand binding partners for given target proteins (Yap and Fiser, 2016). In this current application, we used ProtLID to generate rs-pharmacophores for the interfaces of ligand proteins (PD-L1 and PD-L2) in order to identify the ideal matching three-dimensional residue pattern signatures (rs-pharmacophore). These rs-pharmacophores were compared with each other and with the wild type receptor interface to identify differences that could be utilized to design ligand-specific single mutant variants of hPD-1. The rs-pharmacophore generation consists the following steps (Figure 2):
Identify Interface Residues
Interface residues for hPD-L1 (4ZQK.A) and mPD-L2 (3BP5.B) were identified with the CSU program (Sobolev et al., 1999): residues interacting between the receptor and ligand were identified if these satisfied CSU classification for legitimate interactions and the nearest atomic distance fell within 4.0 Å (Sobolev et al., 1999). Interface residues were required to have at least 1 Å2 accessible surface area.
Mesh Generation over Interface Residues
A hypothetical mesh was constructed over the interface of the ligands, using a 1 Å distance and probe radius over the solvent accessible region of the interface residues of hPD-L1 and mPD-L2 (Xu and Zhang, 2009). These mesh points served as starting points for subsequent MD simulations.
Single-Residue Probe Simulation Using Molecular Dynamics
Extensive molecular dynamics (MD) simulations were performed for hPD-L1 and mPD-L2 from each mesh point constructed over their interfaces using AMBER (Case et al., 2005) with seven replicas, each with different starting orientations using the 20 amino acid residues as probes. The system was minimized from 5.0 kcal/mol to 0 with 5000 steps using harmonic restraints on heavy atoms and simulated to 30 ps using the Generalized Born implicit solvation model with no periodic boundary condition at 300 K, using Andersen thermal coupling. The system contains either hPD-L1 or mPD-L2, with single-residue probes (uncapped N-terminal N-H and C-terminal C=O). Each probe residue is uniquely defined by one or more side-chain atoms that represent the most characteristic chemical functional group -these were defined as functional atoms (FA). There are 26 Fas to define 18 amino acids (non-specific mainchain interactions were not considered, and consequently neither Gly nor Ala). For example, the amino acid Trp is represented by aromatic ring center (RC_W) and hydrogen-bond donor (NE1_W) (Yap and Fiser, 2016).
Functional Atom Preference over the Mesh Point
The propensity of an FA in the proximity of the mesh point determines residue preferences in various spatial locations. Actual preferences are estimated using the actual to expected (A/E) ratio. The A/E ratio compares the actual FA propensity to the expected FA propensities observed from the similar interaction of all snapshots nearest to the mesh point, and as such implicitly accounts for geometrical artifacts on the molecular surface. In addition, only legitimate molecular interactions were considered such as hydrogen bond acceptor-donor or hydrophobic contacts according to CSU definitions (Yap and Fiser, 2016).
Match the Predicted rs-Pharmacophores with the Interface Residues of PD-1
The calculated rs-pharmacophore for hPD-L1 (4ZQK.A) is then matched with the known residues from the interaction surface of hPD-1 (4ZQK.B) and similarly, the pharmacophores calculated for mPD-L2 (3BP5.B) were matched to mPD-1 (3BP5.A) using CSU (Sobolev et al., 1999). When multiple residues are in close proximity, the union of their suggested variants was considered. The rs-pharmacophores generated using hPD-L1 and mPD-L2 (equivalent residues with hPD-L2) were compared with one another and with the observed interface of hPD-1 to select hPD-1 mutants with selectivity for hPD-L1.
Molecular Dynamics Simulations for Selected Mutant Complexes
Input models for hPD1:PD-L1 and hPD1:PD-L2 complexes were prepared in the following way: The complex of hPD1:PD-L1 was modeled with Modeller (Sali and Blundell, 1993) using the combination of templates of hPD1:PD-L1 (4ZQK) and hPD1 (5GGS.Y) (Lee et al., 2016) in order to obtain a full atom starting model without missing residues. In the absence of an experimentally known hPD1:PD-L2 complex, a homology model was created using the mouse ortholog (3BP5) (Lazar-Molnar et al., 2008) and human PD1 (5GGS.Y) with Modeller. The mutant variants (Y68R and T76D) of hPD1:PD-L1 and hPD1:PD-L2 complexes were generated in a similar way.
Wild type and mutant complexes were simulated using Gromacs v5.0.6 (Abraham et al., 2015) with the AMBER99sb force field (Hornak et al., 2006) and the TIP3P water model (Jorgensen et al., 1983) in a dodecahedron box. Each system was neutralized by adding sodium or chloride ions and was further relaxed by energy minimization with force threshold of 1000.0 kJ/mol/nm. Subsequently, the system was equilibrated by a 5ns MD simulation under an NVT ensemble followed by an NPT ensemble. After completion of the two equilibrations, the simulation was continued using the NPT ensemble for a production run for 25 ns. The V-rescale thermostat (Bussi et al., 2007) and Parrinello-Rahman pressure (Parrinello, 1981) coupling were utilized throughout the simulation. Periodic boundary conditions were applied. The thresholds for electrostatic and van der Waals interactions were set to 1 nm whereas long range electrostatic interactions were treated using particle-mesh Ewald method (Darden et al., 1993). The bonds were constrained using LINCS algorithm (Hess et al., 1997).
To explore dominant conformations of wild type PD1 and its mutants (Y68R and T76D), we collected 5 snapshots from intermediate conformations separated by 4ns. The first 5 ns of production were ignored from each simulation.
QUANTIFICATION AND STATISTICAL ANALYSIS
For statistical analysis of data on Figures 3A and 3B, we used Graphpad Prism (https://www.graphpad.com/scientific-software/prism/). Statistical t-tests in Table S2 were performed by scipy (https://docs.scipy.org/doc/scipy/reference/stats.htm l).
DATA AND SOFTWARE AVAILABILITY
N/A.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human (H+L) Alexa 488 | Thermofisher | Cat# A-11013 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Human PD-L1 hIgG1 | R&D Systems | Cat# 156-B7–100 |
| Human PD-L2 hIgG1 | R&D Systems | Cat# 1224-PL-100 |
| Deposited Data | ||
| mouse PD-1 and PD-L2 complex | (Lazar-Molnar et al., 2008) | PDB: 3BP5 |
| human programmed death-1 (PD-1) and its ligand PD-L1 | (Zak et al., 2015) | PDB: 4ZQK |
| Experimental Models: Cell Lines | ||
| HEK 293-F Freestyle | Thermofisher | Cat# R79007 |
| Oligonucleotides | ||
| DNA primers | see Table S3 for all primers | |
| Recombinant DNA | ||
| human PD1 cDNA | GeneCopoeia | Cat# EX-B0169-M98 |
| mChery N1 | ClonTech | Cat# 632523 |
| Software and Algorithms | ||
| Gromacs | Gromacs v5.0.6 | http://www.gromacs.org |
| Pymol | Pymol v1.7 | https://sourceforge.net/projects/pymol/files/pymol/1.7/ |
| Modeller | (Sali and Blundell, 1993) | https://salilab.org/modeller/ |
| BeAtMusic | (Dehouck et al., 2013), | http://babylone.ulb.ac.be/beatmusic/ |
| FoldX | (Schymkowitz et al., 2005) | http://foldxsuite.crg.eu/ |
| MutaBind | (Li et al., 2016) | https://www.ncbi.nlm.nih.gov/research/mutabind/ |
| ProtLID | (Yap and Fiser, 2016). | N/A |
| matplotlib | Python version 2.7.10 | https://matplotlib.org |
| pandas | Python version 2.7.10 | https://pandas.pydata.org/index.html |
| numpy | Python version 2.7.10 | http://www.numpy.org/reference/stats.html |
| scipy | Python version 2.7.10 | https://docs.scipy.org/doc/scipy |
Highlights.
A residue-based pharmacophore approach is used to design specific protein interfaces
Designed point mutations of PD-1 interface made it specific to PD-L1 only
Cell-based assays confirmed selectivity of mutants
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 GM118709, R01 HG008325, R01 AI141816, and the Extreme Science and Engineering Discovery Environment (XSEDE) project (NSF grant ACI-1053575).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016Zj.str.2019.03.006.
DECLARATION OF INTERESTS
A provisional patent application has been fled. US Patent Application no. 62/736,477.
REFERENCES
- Abraham MJ, Teemu M, Schulz R, Pall S, Smith JC, Hess B, and Lindahl E (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25. [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, and Ahmed R (2006). Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439, 682–687. [DOI] [PubMed] [Google Scholar]
- Bolon DN, Grant RA, Baker TA, and Sauer RT (2005). Specificity versus stability in computational protein design. Proc. Natl. Acad. Sci. U S A 102, 12724–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. (2012). Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med 366, 2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G, Donadio D, and Parrinello M (2007). Canonical sampling through velocity rescaling. J. Chem. Phys 126, 014101. [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE 3rd, Darden T, Gohlke H, Luo R, Merz KMJR, Onufriev A, Simmerling C, Wang B, and Woods RJ (2005). The Amber biomolecular simulation programs. J. Comput. Chem 26, 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay K, Lazar-Molnar E, Yan Q, Rubinstein R, Zhan C, Vigdorovich V, Ramagopal UA, Bonanno J, Nathenson SG, and Almo SC (2009). Sequence, structure, function, immunity: structural genomics of costimulation. Immunol. Rev 229, 356–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemnitz JM, Parry RV, Nichols KE, June CH, and Riley JL (2004). SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of program med death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J. Immunol 173, 945–954. [DOI] [PubMed] [Google Scholar]
- Chen L, and Flies DB (2013). Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol 13, 227–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T, York D, and Pedersen L (1993). Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys 98, 5. [Google Scholar]
- Day CL, Kaufmann DE, Kizepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, Depierres C, et al. (2006). PD-1 expression on HIV-specific T cells isassociated with T-cell exhaustion and disease progression. Nature 443, 350–354. [DOI] [PubMed] [Google Scholar]
- Dehouck Y, Kwasigroch JM, Rooman M, and Gilis D (2013). BeAtMuSiC: prediction of changes in protein-protein binding affinity on mutations. Nucleic Acids Res. 41, W 333–W 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhu G, Tamada K, and Chen L (1999). B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med 5, 1365–1369. [DOI] [PubMed] [Google Scholar]
- Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al. (2002). Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med 8, 793–800. [DOI] [PubMed] [Google Scholar]
- Fernandez-Fuentes N, Madrid-Aliste CJ, Rai BK, Fajardo JE, and Fiser A (2007). M4T: a comparative protein structure modeling server. Nucleic Acids Res. 35, W 363–W 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesnak AD, June CH, and Levine BL (2016). Engineered T cells: the promise and challenges of cancer immunotherapy. Nat. Rev. Cancer 16, 566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner D, Lalezari J, Lawitz E, Dimicco M, Ghalib R, Reddy KR, Chang KM, Sulkowski M, Marro SO,Anderson J, et al. (2013).Arandomized, double-blind, placebo-controlled assessment of BMS-936558, a fully human monoclonal antibody to programmed death-1 (PD-1), in patients with chronic hepatitis C virus infection. PLoS One 8, e63818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havranek JJ, and Harbury PB (2003). Automated design of specificity in molecular recognition. Nat. Struct. Biol 10, 45–52. [DOI] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, and Fraaije JGEM (1997). LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem 18, 1463–1472. [Google Scholar]
- Holm L, and Sander C (1995). Dali: a network tool for protein structure comparison. Trends Biochem. Sci 20, 478. [DOI] [PubMed] [Google Scholar]
- Hoos A (2016). Development of immuno-oncology drugs - from CTLA4 to PD1 to the next generations. Nat. Rev. Drug Discov 15, 235–247. [DOI] [PubMed] [Google Scholar]
- Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, and Simmerling C (2006). Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Agata Y, Shibahara K, and Honjo T (1992). Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 11, 3887–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, and Minato N (2002). Involvement of PD-L1 on tum or cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. U S A 99, 12293–12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, and Madura JD (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 79, 10. [Google Scholar]
- Keir ME, Butte MJ, Freeman GJ, and Sharpe AH (2008). PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol 26, 677–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, and Baker D (2003). Design of a novel globular protein fold with atomic-level accuracy. Science 302, 1364. [DOI] [PubMed] [Google Scholar]
- Lafferty KJ, and Cunningham AJ (1975). A new analysis of allogeneic interactions. Aust. J. Exp. Biol. Med. Sci 53, 27–42. [DOI] [PubMed] [Google Scholar]
- Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, Iwai Y, Long AJ, Brown JA, Nunes R, et al. (2001). PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol 2, 261–268. [DOI] [PubMed] [Google Scholar]
- Lazar-Molnar E, Yan QR, Cao E, Ramagopal U, Nathenson SG, and Almo SC (2008). Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2. Proc. Natl. Acad. Sci. U S A 105, 10483–10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar-Molnar E, Chen B, Sweeney KA, Wang EJ, Liu W, Lin J, Porcelli SA, Almo SC, Nathenson SG, and Jacobs WR Jr. (2010). Programmed death-1 (PD-1)-deficient mice are extraordinarily sensitive to tuberculosis. Proc. Natl. Acad. Sci. U S A 107, 13402–13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar-Molnar E, Scandiuzzi L, Basu I, Quinn T, Sylvestre E, Palmieri E, Ramagopal UA, Nathenson SG, Guha C, and Almo SC (2017). Structure-guided development of a high-affinity human Programmed Cell Death-1: implications for tumor immunotherapy. EBioMedicine 17, 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, and Tannock IF (2010). The distribution of the therapeutic monoclonal antibodies cetuximab and trastuzumab within solid tumors. BMC Cancer 10, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Lee HT, Shin W, Chae J, Choi J, Kim SH, Lim H, Won Heo T, Park KY, Lee YJ, et al. (2016). Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy. Nat. Commun 7,13354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Simonetti FL, Goncearenco A, and Panchenko AR (2016). MutaBind estimates and interprets the effects of sequence variants on protein-protein interactions. Nucleic Acids Res. 44, W 494–W 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippow SM, Wittrup KD, and Tidor B (2007). Computational design of antibody-affinity improvement beyond in vivo maturation. Nat. Biotechnol 25, 1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looger LL, Dwyer MA, Smith JJ, and Hellinga HW (2003). Computational design of receptor and sensor proteins with novel functions. Nature 423, 185. [DOI] [PubMed] [Google Scholar]
- Lundborg M, and Lindahl E (2015). Automatic GROMACS topology generation and comparisons of force fields for solvation free energy calculations. J. Phys. Chem. B 119, 810–823. [DOI] [PubMed] [Google Scholar]
- Mandell DJ, and Kortemme T (2009). Computer-aided design of functional protein interactions. Nat. Chem. Biol 5, 797–807. [DOI] [PubMed] [Google Scholar]
- Maute RL, Gordon SR, Mayer AT, Mccracken MN, Natarajan A, Ring NG, Kimura R, Tsai JM, Manglik A, Kruse AC, et al. (2015). Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc. Natl. Acad. Sci. U S A 112, E6506–E6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, and Ohashi PS (2015). Clinical blockade of PD1 and LAG3-potential mechanisms of action. Nat. Rev. Immunol 15, 45–56. [DOI] [PubMed] [Google Scholar]
- Parrinello M (1981). Polymorphic transitions in single crystals: a new molecular dynamics method. J. Chem. Phys 52, 9. [Google Scholar]
- Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, Mcdermott D, Linette GP, Meyer N, Giguere JK, Agarwala SS, et al. (2015). Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med 372, 2006–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai BK, and Fiser A (2006). Multiple mapping method: a novel approach to the sequence-to-structure alignment problem in comparative protein structure modeling. Proteins 63, 644–661. [DOI] [PubMed] [Google Scholar]
- Rai BK, Madrid-Aliste CJ, Fajardo JE, and Fiser A (2007). MMM: a sequence-to-structure alignment protocol. Bioinformatics 22, 2691–2692. [DOI] [PubMed] [Google Scholar]
- Ramagopal UA, Liu W, Garrett-Thomson SC, Bonanno JB, Yan Q, Srinivasan M, Wong SC, Bell A, Mankikar S, Rangan VS, et al. (2017). Structural basis for cancer immunotherapy by the first-in-class check-point inhibitor ipilimumab. Proc. Natl. Acad. Sci. U S A 114, E4223–E4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riella LV, Paterson AM, Sharpe AH, and Chandraker A (2012). Role of the PD-1 pathway in the immune response. Am. J. Transplant 12, 2575–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, Dechancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, et al. (2008). Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195. [DOI] [PubMed] [Google Scholar]
- Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, and Anderson AC (2010). Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tum or immunity. J. Exp. Med 207, 2187–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, and Blundell TL (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Schreiber G, and Keating AE (2011). Protein binding specificity versus promiscuity. Curr. Opin. Struct. Biol 21, 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, and Serrano L (2005). The FoldX web server: an online force field. Nucleic Acids Res. 33, W 382–W 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev V, Sorokine A, Prilusky J, Abola EE, and Edelman M (1999). Automated analysis of interatomic contacts in proteins. Bioinformatics 15, 327–332. [DOI] [PubMed] [Google Scholar]
- Sznol M, and Chen L (2013). Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer-response. Clin. Cancer Res. 19, 5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, Mcdermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. (2012). Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med 366, 2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ (2011). T cell exhaustion. Nat. Immunol 12, 492–499. [DOI] [PubMed] [Google Scholar]
- Xu D, and Zhang Y (2009). Generating triangulated macromolecular surfaces by Euclidean distance transform. PLoS One 4, e8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Juan V, Ivanov A, Ma Z, Polakoff D, Powers DB, Dubridge RB, Wilson K, and Akamatsu Y (2012). Affinity and cross-reactivity engineering of CTLA4-Ig to modulate T cell costimulation. J. Immunol 189, 4470–4477. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Akiba H, Iwai H, Matsuda H, Aoki M, Tanno Y, Shin T, Tsuchiya H, Pardoll DM, Okumura K, et al. (2002). Expression of programmed death 1 ligands by murine T cells and APC. J. Immunol 169, 5538–5545. [DOI] [PubMed] [Google Scholar]
- Yap EH, and Fiser A (2016). ProtLID, a residue-based pharmacophore approach to identify cognate protein ligands in the immunoglobulin superfamily. Structure 24, 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yearley JH, Gibson C, Yu N, Moon C, Murphy E, Juco J, Lunceford J, Cheng J, Chow LQM, Seiwert TY, et al. (2017). PD-L2 expression in human tumors: relevance to Anti-PD-1 therapy in cancer. Clin. Cancer Res 23, 3158–3167. [DOI] [PubMed] [Google Scholar]
- Zak KM, Kitel R, Przetocka S, Golik P, Guzik K, Musielak B, Domling A, Dubin G, and Holak TA (2015). Structure of the complex of human programmed death 1, PD-1, and its ligand PD-L1. Structure 23, 2341–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
N/A.