Abstract
ZEB1 and ZEB2 play pivotal roles in solid cancer metastasis by allowing cancer cells to invade and disseminate through the transcriptional regulation of epithelial-to-mesenchymal transition. ZEB expression is also associated with the acquisition of cancer stem cell properties and therapy resistance. Consequently, expression levels of ZEB1/2 and of their direct target genes are widely seen as reliable prognostic markers for solid tumor aggressiveness and cancer patient outcome.
Recent loss-of-function mouse models demonstrated that both ZEBs are also essential hematopoietic transcription factors governing blood lineage commitment and fidelity. Interestingly, both gain- and loss-of-function mutations have been reported in multiple hematological malignancies. Combined with emerging functional studies, these data suggest that ZEB1 and ZEB2 can act as tumor suppressors and/or oncogenes in blood borne malignancies, depending on the cellular context. Here, we review these novel insights and discuss how balanced expression of ZEB proteins may be essential to safeguard the functionality of the immune system and prevent leukemia.
ZEB1 and ZEB2: 2 structurally related E-box-binding transcription factors
The ZEB (Zinc finger E-box-binding homeobox) protein family of transcription factors was discovered in Drosophila melanogaster and consists of 2 structurally conserved multidomain proteins, ZEB1 (originally called ZFHX1A, TCF8, or δEF1) and ZEB2 (originally called ZFHX1B or SIP1).1,2 Both ZEBs contain an amino-terminal (NZF) and carboxy-terminal zinc finger cluster (CZF), which enables them to bind regulatory DNA sequences in their target promoters. Each zinc finger cluster binds an independent 5′-CANNTG-3′ motif, which overlaps with the core of an E2-box sequence,3–5 a binding site for transcription factors of the basic helix-loop-helix (bHLH) family. ZEBs can repress target gene transcription through the recruitment of the CtBP corepressor complex via their CtBP interaction domain (CID).6 Additional conserved domains were documented by which ZEBs can recruit other transcriptional complexes to their target promoters. For instance, the amino-terminus of both ZEBs can bind the Nucleosome Remodeling and Deacetylase complex (NuRD, also known as the Mi-2 complex).7,8 Also, a SMAD Binding Domain (SBD) is located between the NZF and the central homeobox domain.4 Although this SBD is highly conserved and both ZEBs can directly interact with SMADs, they may have an opposite downstream effect9: ZEB1 synergizes with SMAD proteins to activate SMAD-mediated transcription, while ZEB2 seems to inhibit SMAD-mediated transcription. It is believed that this opposite effect is regulated by the differential recruitment of ZEB1/2-specific coactivators/repressors, which may be tissue-specifically expressed. Initially, the N-terminal region of ZEB1, but not ZEB2, was documented to interact with p300 and P/CAF. This differential recruitment of P/CAF may switch ZEB1 from a repressor to an activator through the displacement of CtBP1 from the CID.9,10 Later, others demonstrated that, depending on the experimental context, ZEB2 and ZEB1 are equally potent to bind p300 and P/CAF.11 This suggests that other mechanisms/cofactors are contributing to the differential ZEB1/2-specific effects on SMAD-mediated transcription.
Based on the extensive and continuously growing list of interaction partners and putative downstream targets, one can expect that ZEBs have very pleiotropic functions, which largely depend on the cellular context. Indeed, the chromatin status and the presence/absence of tissue-specific interaction partners strongly influence their role as a transcriptional repressor/activator. This can, to some extent, explain why in some cell types ZEBs play complementary or synergistic roles while in others seemingly opposite roles.
ZEBs are master regulators of (cancer) cell plasticity
ZEBs during EMT
ZEB proteins are primarily known as inducers of epithelial-mesenchymal transition (EMT), a reversible multistep process during which polarized epithelial cells undergo a morphological switch to become motile mesenchymal cells that have lost their polarity.12 ZEBs orchestrate EMT by the direct repression of epithelial genes involved in cellular adhesion and cytoskeleton (re)organization,3,13,14 and subsequent direct or indirect upregulation of mesenchymal genes.15,16 This epithelial-to-mesenchymal cell plasticity is essential at various stages of embryonic development, but also often seen aberrantly activated in cancer cells.12 Induced expression of ZEB1 or ZEB2 in xenotransplanted epithelial cancer cells endows them to invade and disseminate from the primary tumor site. These circulating tumor cells will then need to undergo a reversed process called mesenchymal-epithelial transition or MET to colonize a distant tissue and grow out as a metastasis.
In addition, EMT-inducing transcription factors (EMT-TF), like ZEB1 and ZEB2, are more than “only” regulators of cancer cell invasion and they dictate multiple other steps during cancer initiation and progression.17 Increased ZEB expression has been correlated with the acquisition and/or function of cancer stem cell (CSC) properties.18–20 CSCs or tumor-initiating cells have the potential to self-renew and form secondary tumors when transplanted into immune deficient or syngeneic mice. The molecular mechanisms controlled by EMT-TF that induce these CSC properties remain elusive until today.17,19 In addition, a large body of literature suggests that CSC subpopulations are responsible for emerging residual subpopulations following radiotherapy, chemotherapy, or other targeted therapies. Also EMT-TF-driven cancer cell plasticity facilitates drug adaptation and protects the cancer cells against genotoxic insults and therapy-induced stress via enhanced cell survival, induced DNA damage repair,21,22 antiproliferative mechanisms,23 and increased expression of genes encoding drug-metabolizing enzymes and drug-transporters.24 As such, ZEBs can act as important regulators of therapy resistance.
Next to ZEB proteins, also SNAI and TWIST transcription factors have been demonstrated to catalyze similar EMT phenotypes in the context of solid tumors.12,17 Consistent results in cancer cell lines suggest cooperativity between the different EMT inducers with complex regulatory feedback mechanisms involving Notch signaling and the miR-200 family.25
To conclude, the pivotal roles of the EMT modulators in solid tumor progression have been extensively documented. It is therefore no surprise that ZEB expression levels and the expression of their targets are used in the clinic as reliable prognostic markers for solid tumor aggressiveness and poor patient outcome.
Emerging roles of ZEB proteins in hematopoiesis
Gain- and loss-of-function mouse models revealed that ZEB proteins also play pivotal roles in cellular plasticity of other cell lineages, including the hematopoietic lineage.
Next to the full Zeb1 knockout, which is perinatal lethal due to skeletal defects,26 2 other Zeb1 mutant mouse models expressing truncated C-terminal ZEB1 deletions lacking the CZF domain have been generated.26–28 The first one, Zeb1ΔC-fin mice have a profound thymic atrophy with spontaneous development of CD4+ T-cell lymphoma at older age.29 Their hypocellular thymi show an increased proportion of CD4/CD8 double negative cells and a drastic reduction in double positive cells, suggestive of an early partial block in T-cell development.27 The few Zeb1ΔC-fin thymocytes that can circumvent this early block and differentiate into mature T-cells are skewed toward the CD4+ T-cell lineage. In line with this, previous work has shown that ZEB1 can directly regulate CD4 expression by competing with transcriptional activators E12 and HEB for the binding to an E-box-containing enhancer of the CD4 promoter.30 The Zeb1cellophane mice, the second Zeb1 mutant model with a C-terminal truncation, are also characterized by hypocellular thymi that display an early T-cell differentiation block. Detailed analysis of other lineages suggested B-cell and NK-cell maturation defects in the spleen of these Zeb1 mutant mice.28 Until now, it is unclear whether these defects are cell autonomous or due to aberrant paracrine signaling. Similarly, it remains to be established whether these truncated ZEB1 versions are true loss-of-function mutations and/or can act in a dominant negative manner. A recently generated conditional Zeb1fl/fl knockout mouse line31 will allow further investigation of the role of Zeb1 in specific hematopoietic lineages.
The role of Zeb2 in hematopoiesis has been studied with conditional loss- and gain-of-function mouse models.32,33 Inactivation of Zeb2 in the hematopoietic lineage resulted in vast multilineage differentiation defects, associated with an accumulation of stem and progenitor cells and a significant drop in fully matured functional blood cells.34,35 Differentiation defects appeared at early and later stages of hematopoiesis affecting most lineages, except the granulocytes. These studies were extended by using more lineage-restricted Cre-lines allowing to demonstrate essential roles for ZEB2 for NK-cell maturation,36 terminal differentiation of CD8+ cytotoxic T-cells,37,38 and dendritic cell (DC) development.39 In addition, strong evidence suggests that ZEB2 is also important for Langerhans cell40 and mast cell maturation/activation,41 although the latter is purely based on in vitro work and the relevance of these observations will have to be confirmed in vivo. Altogether, these studies suggest that ZEB2 is a transcription factor that controls lineage commitment and fidelity at various stages of hematopoiesis. In addition, ZEB2 is crucial for proper immune functioning and loss of Zeb2 can lead to defective response to pathogens.
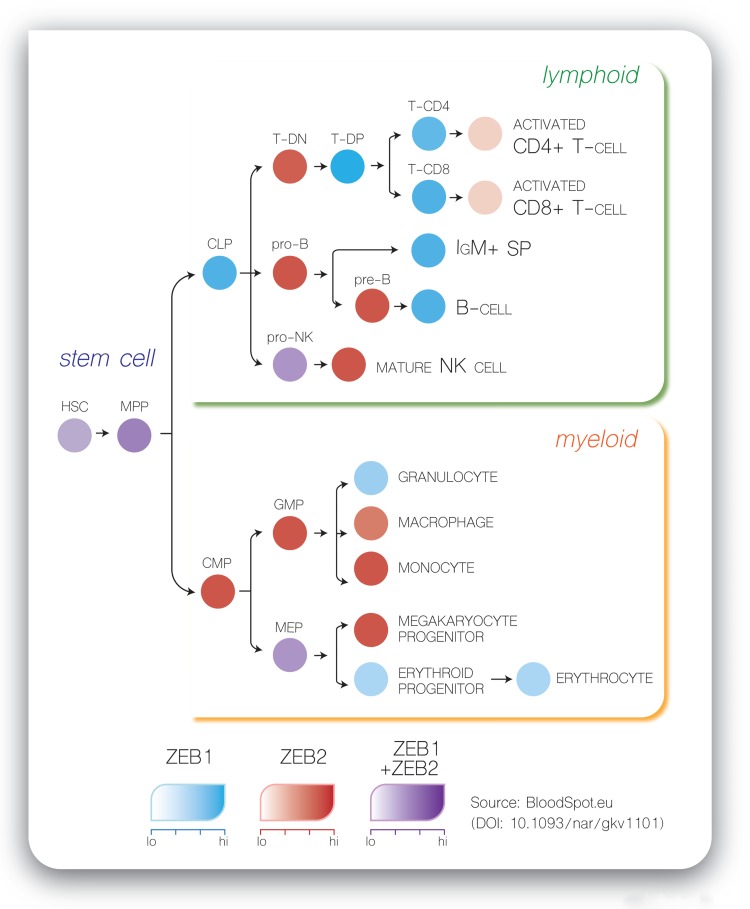
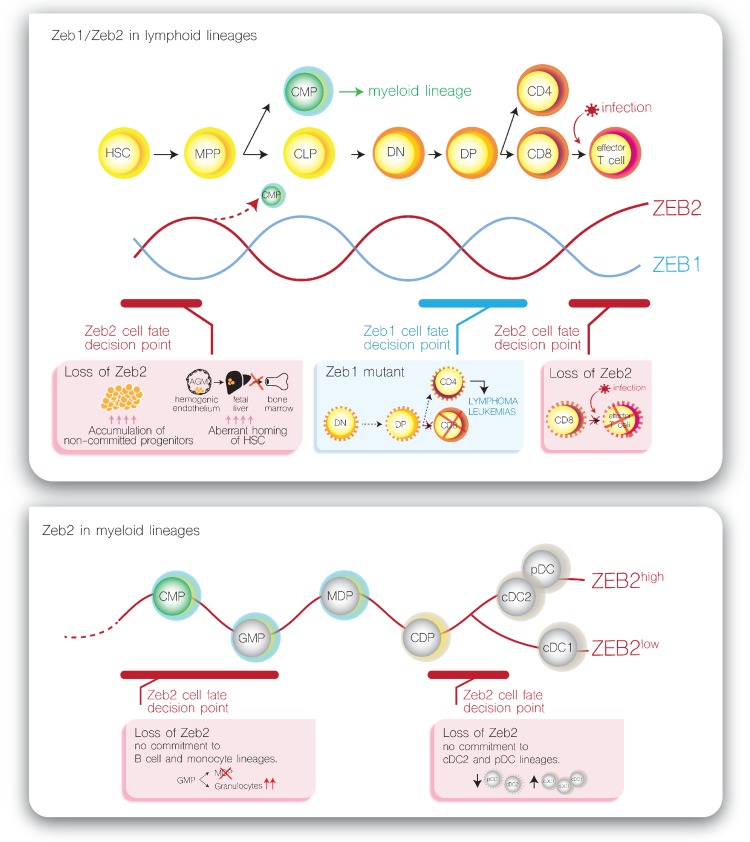
Based on the observed complementary expression patterns of Zeb1 and Zeb2 mRNAs during embryonic development and aggravated phenotypes in compound Zeb1/Zeb2 loss-of-function mice, it was suggested that ZEB transcription factors may have partly overlapping, compensatory functions.42 Remarkably, we observed that Zeb1 and Zeb2 mRNA expression is also mostly mutually exclusive during hematopoietic differentiation, except in the hematopoietic stem and multipotent progenitor cell (HSPC) compartment where Zeb1 and Zeb2 are co-expressed at moderate levels (Fig. 1). Once lineage commitment is initiated, counter-oscillating levels of both mRNAs are observed with fast Zeb1/2 switching at distinct cell fate decision points. In addition, most of the hematopoietic differentiation defects seen in the Zeb1/2 loss- or gain-of-function mouse models occur exactly at these decision checkpoints (Fig. 2, examples are shown for the T-cell lineage [upper panel] and dendritic cell lineage [lower panel]). Based on these observations, one could hypothesize that oscillations of ZEB levels may control hematopoietic differentiation. At this point, the molecular mechanisms that control such a Zeb mRNA switching remain largely unexplored. However, negative feedback mechanisms via the miRNA-200 family might partially explain these Zeb1/2 oscillations.25,43 Indeed, miRNA-200 family members are able to inhibit expression of ZEB1/2 at the post-transcriptional level by binding to highly conserved target sites in their 3′-untranslated region. In addition, ZEBs are also able to transcriptionally repress the miRNA-200 family, suggesting a negative feedback loop that can fuel this ZEB1/2 switching. Interestingly, also in nonhematopoietic cell lineages, including the melanocytes, a similar oscillation between Zeb1/2 mRNAs has been described.44,45 Of note, this concept has also been suggested for other protein families, including the GATA2/GATA1 switch as an important driver of molecular development.46 However, this ZEB switching hypothesis does not exclude that ZEB1 and ZEB2 could also have family member-specific functions, which may be dependent on the cellular context.10
Figure 1.
Zeb1 and Zeb2 are mostly mutually exclusive expressed during hematopoietic differentiation. The figure is based on expression data available via http://servers.binf.ku.dk/bloodspot/. CLP = common lymphoid progenitor, GMP = granulocyte monocyte progenitor, HSC = hematopoietic stem cell, IgM+ SP = IgM positive side population, MEP = megakaryocytic erythroid progenitor, MPP = multipotent progenitor, PreB = Pre-B cell, ProB = Pro-B cell, pro-NK = natural killer cell progenitor, T-CD4 = T-cell CD4+ single positive, T-CD8 = T-cell CD8+ single positive, T-DN = T-cell CD4−CD8− double negatives, T-DP = T-cell CD4+CD8+ double positives.
Figure 2.
Examples of ZEB expression switching as molecular drivers of important cell fate decisions during hematopoietic differentiation. (A) Counter oscillating expression levels of Zeb1 and Zeb2 during T-cell differentiation with indications of known differentiation defects in Zeb1 and Zeb2 loss-of-function mouse models. (B) Oscillating Zeb2 expression levels during dendritic cell (DC) differentiation with indications of known differentiation defects in Zeb2 loss-of-function mouse models. CD4 = T-cell CD4+ single positive, CD8 = T-cell CD8+ single positive, cDC1 = conventional DC subtype 1, cDC2 = conventional DC subtype 2, CDP = common dendritic cell progenitor, CLP = common lymphoid progenitor, CMP = common myeloid progenitor, DN = T-cell CD4−CD8− double negatives, DP = T-cell CD4+CD8+ double positives, GMP = granulocyte monocyte progenitor, HSC = hematopoietic stem cell, MDP macrophage/DC progenitor, MPP = multipotent progenitor, pDC = plasmacytoid DC.
Genetic alterations of ZEBs in leukemia and lymphoma
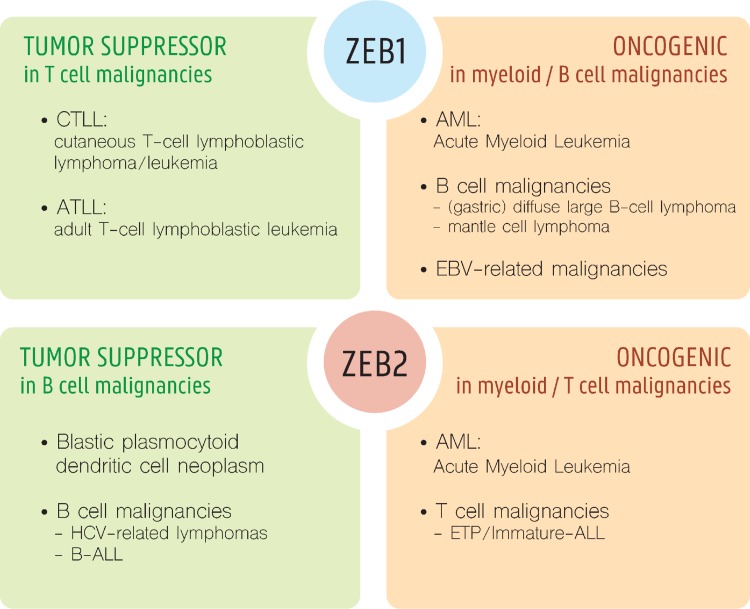
Based on the importance of ZEB expression during normal hematopoiesis, and their pleiotropic roles in progression of solid cancers, it is expected that ZEB1 and ZEB2 could also play pivotal roles in hematologic malignancies. Indeed, both genetic and in vivo functional studies indicate that ZEBs can act, depending on the lineage, as oncogenic drivers and/or tumor suppressors (Fig. 3).
Figure 3.
ZEBs as oncogene or tumor suppressors of leukemia. Schematic representation of known or suggestive oncogenic and/or tumor suppressive roles for ZEB1 and ZEB2 in various leukemia subtypes.
ZEBs in T-cell malignancies
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological cancer of thymic T-cell progenitors that gradually accumulate epigenetic and genetic changes, leading to a block in differentiation, increased survival and proliferative expansion of a malignant clone.47,48 Over the last decade, the prognosis of T-ALL has gradually improved with the introduction of intensified chemotherapy. However, the outcome of T-ALL patients with primary resistant or relapsed disease remains poor.49
A rare (0.2%; 2/1084 patients), but recurrent, t(2;14)(q22;q32) translocation involving the ZEB2 and BCL11B loci has been identified in early T-cell progenitor ALL (ETP-ALL),18 a heterogeneous subgroup of T-ALL with a unique gene expression profile similar to that of the most immature ETPs.50,51 It was hypothesized that this translocation retains ZEB2 expression during T-cell commitment leading to a differentiation block and leukemic transformation.18 Hematopoietic-specific Zeb2 overexpression in the mouse was sufficient to spontaneously develop T-ALL with an immature Lyl1+ expression profile, with profound similarities to the human disease.18 Overexpression of Zeb2 resulted in increased expression of the interleukin-7 receptor (IL7R) and aberrant activation of the IL7R-JAK/STAT signaling pathway. In addition, ZEB2 overexpression was associated with the acquisition of enhanced leukemia stem cell properties. Activating IL7R mutations are recurrently found in ETP-ALL patients.50 In addition, a similar immature ETP-ALL leukemia initiation with increased self-renewal was observed after overexpression of a gain-of-function mutant variant of IL7R in p19Arf−/− mouse hematopoietic progenitors.52
In contrast to ZEB2, ZEB1 seems to act as a tumor suppressor in T-cell derived hematological neoplasms. ZEB1 has been identified as an essential downstream mediator of the LMO2 oncogene in T-ALL.53 In line with this, Zeb1 mutant loss-of-function mice showed drastic defects in early T-cell development and spontaneously develop T-cell lymphoma/leukemia with a median onset of 6 months,27,54 similar to what has been observed in LMO255,56 and ZEB2 overexpressing mice.18 Combined, these data suggest opposing roles for ZEB1 and ZEB2 in T-ALL initiation/progression.
Furthermore, expression analysis of genes mapped within a common (34.6% of patients) breakpoint cluster in the 10p11.2 region of adult T-cell leukemia/lymphoma patients (ATLL), suggested that ZEB1 may act also as a tumor suppressor in these patients.29 ATLL is a peripheral T-cell malignancy with a mature CD4+ immunophenotype.57 Most ATLL cell lines and primary cells display low mRNA expression levels of ZEB1, as a consequence of either chromosomal translocations with heterozygous deletion, intragenic mutations, or epigenetic dysregulation.29 Mechanistically, low ZEB1 levels may result in resistance to TGFβ1-mediated growth arrest.58 Binding of the TGFβ1 ligand to its receptor activates its kinase activity, leading to phosphorylation of receptor-associated SMAD proteins. These phospho-SMADs accumulate in the nucleus as dimers, and in conjunction with other transcription factors like ZEB1 and ZEB2, they bind regulatory elements within their target gene promoters. As an example, TGFβ1 stimulation induces cell cycle arrest in various tumor cell types59 via the direct upregulation of the cyclin-dependent kinase inhibitor p21. ATLL cells with low ZEB1 levels appear to be resistant to these antiproliferative effects of TGFβ1. Overexpression of ZEB1 restored the TGFβ1-mediated growth suppression in these cells, associated with increased p21 expression. Mechanistically, ZEB1 expression, and its direct binding to the phospho-SMAD3 complex, was demonstrated to be essential for the recruitment of this complex to the SMAD-Response Element within the p21 promoter.58
Aberrant ZEB1 expression has also been linked to 2 other lymphoproliferative T-cell disorders: mycosis fungoides and Sézary syndrome. Mycosis fungoides is a cutaneous T-cell lymphoma (CTCL) developing from clonally expanded skin-homing CD4+ T-cells. Sézary syndrome can arise de novo, but mostly occurs in patients with mycosis fungoides and can be considered as the leukemic variant of this disease.60,61 Several genetic alterations targeting ZEB1 have been reported in both forms of CTCL (up to 65%), including translocations, mutations, and both heterozygous and homozygous deletions.62–66 Loss-of-ZEB1 expression or function has been associated with the pathogenic role of IL15 signaling in CTCL. Either loss-of-ZEB1 expression, mutation or hypermethylation of the ZEB1 binding sites in the IL15 promoter resulted in a vast upregulation and activation of oncogenic signals.67 Altogether, these loss-of-function alterations of ZEB1 suggest that the transcription factor acts as a key tumor suppressor in peripheral T-cell lymphoma.
ZEBs in B-cell malignancies
Altered ZEB1 expression and mutations have been associated with 2 types of B-cell malignancies, namely mantle cell lymphoma (MCL) and diffuse large B-cell lymphoma (DLBCL). MCL is an infrequent subtype of non-Hodgkin B-cell lymphoma (B-cell NHL) with a poor response to chemotherapy.68,69 The molecular hallmark of this disease is overexpression of cyclin D1 due to the chromosomal t(11;14)(q13;q32) translocation. Half of MCLs display constitutive active Wnt signaling with nuclear localization of beta-catenin and concomitant high expression of ZEB1. Downregulation of ZEB1 expression in MCL cell lines reduced their tumor growth capacity in mouse xenograft models. In addition, the cell lines with reduced ZEB1 expression were more sensitive to chemotherapeutics, associated with a differential expression of drug influx/efflux transporters, and genes involved in cell survival/apoptosis.70 Therefore, this study suggested that ZEB1 could serve as a potential predictive biomarker and putative therapeutic target in MCL.
DLBCL is the most frequent form of adult NHL.61 ZEB1 can be considered as an oncogene in DLBCL for several reasons. First, strong nuclear immunohistochemical staining for ZEB1 was associated with an adverse 3-year overall survival of DLBCL patients compared to those with no or weak nuclear ZEB1 staining.71 Next to this, higher levels of the miR-200 family, a known negative regulator of ZEB1 mRNA expression levels, results in a less aggressive behavior of this disease.72Helicobacter pylori positive gastric DLBCLs, which typically show lower ZEB1 expression, have less lymph node metastasis, better response to chemotherapy, and are less aggressive. This last subgroup is also characterized by higher expression levels of BCL6, a known predictor of better prognosis in DLBCL73 and a direct target of ZEB1.74 Combined, these expression data suggest that ZEB1 may act as an oncogene in these 2 B-cell NHL forms.
Interestingly, loss and not gain of 2q22.3, the genomic region spanning the ZEB2 locus, has been recurrently (18.75%) observed in B-cell lymphoma patients.75 In addition, rare but recurrent (7.2%) somatic ZEB2 point mutations were found in B-cell precursor acute lymphoblastic leukemia (B-ALL). B-ALL is the most common childhood malignancy that initiates in the bone marrow (BM) with oncogenic transformation of B-cell progenitor cells.76 In all 5 reported B-ALL patients with mutant ZEB2 locus, a single AA mutation specifically affected the carboxy-terminal Cys2His2 Zinc fingers,8 which have proven to be essential for the DNA binding and E-box recognition capacity of ZEB2.3 Notably, all of these patients had a very similar expression profile, associated with deregulated expression of ERG8,77 (https://pecan.stjude.org/proteinpaint/ZEB2). The fact that these mutations are uncommon in other B-ALL subtypes, suggests that loss of the ZEB2 DNA binding capacity may be involved in the progression of this disease, which comprises up to 7% of all B-ALLs. The same laboratory also reported a ZEB2-PDGFRB translocation in 1 B-ALL patient. In this translocation event, the carboxy-terminal zinc finger domain of ZEB2 is missing and in frame fused to the catalytic domain of the Platelet-derived Growth Factor Receptor-B77,78 (https://pecan.stjude.org/proteinpaint/ZEB2). However, no additional data are available whether this chimeric protein is expressed, functional and whether it contributed to the disease progression. The notion that ZEB2, or a mutant version, may play an important role in the initiation and/or progression of B-ALL is further supported by the observed high occurrence of viral insertions at the Zeb2 locus in 2 independent retroviral mutagenesis screens using mouse models that are predisposed to develop spontaneously B-ALL, the CALM-AF10 transgenic and heterozygote Pax5−/+ mice.79,80 However, no information is available whether these viral integrations result in loss- or gain-of-Zeb2 functions.
Combining the mutation data of the human patients and the mutagenesis screenings in the B-ALL mouse models, we hypothesize that ZEB2 acts as a tumor suppressor in B-cell malignancies, in contrast to ZEB1, which seems to act as an oncogene. More research will be necessary with conditional Zeb1/2 gain/loss-of-function mouse models using a B-cell restricted Cre line to further test this hypothesis.
ZEBs in myeloid malignancies
Acute myeloid leukemia (AML) is a clinically and genetically heterogeneous malignancy, characterized by uncontrolled accumulation of immature myeloid cells mostly in the blood and the BM of the patient. This accumulation of blast cells in the BM interferes with normal hematopoiesis and leads to a general deficiency of hematopoietic cells. AML is understood as the product of a rather limited number of genetic alterations including balanced chromosomal translocations. The MLL1 histone 3 lysine 4 (H3K4) methyltransferase (aka KMT2A, ALL1, TRX) has been found involved in over 100 leukemia-associated rearrangements of which the MLL-AF4, MLL-ENL, and MLL-AF9 are among the most prevalent.81 Several MLL fusions have been shown to be potent oncogenes in vitro and in vivo in murine as well as in human cells.82,83 Recently, a conditional mouse model was used to address the role of the cellular origin of MLL-AF9+ AML in the biology and clinical outcome of the disease. Activation of MLL-AF9 in long-term hematopoietic stem cells (LT-HSC) induces a particularly invasive and chemoresistant disease.84 Strikingly, about 10% to 20% of human AMLs express a very similar gene signatures like LT-HSC-derived MLL-AF9 AMLs in mice, which is characterized by expression of high Zeb1 mRNA levels. Chromatin immunoprecipitation (ChIP) experiments suggested that Zeb1 is a direct target of the MLL-AF9 fusion. Interestingly, knockdown of Zeb1 expression compromised the invasive behavior of MLL-AF9 AML cells in vitro and in vivo. Notably, LT-HSC-derived MLL-AF9 AML cells were also characterized by increased expression of many genes regulating migration and invasion, and thus showing at least at the transcriptional level some similarities to that of solid cancer cells undergoing EMT.
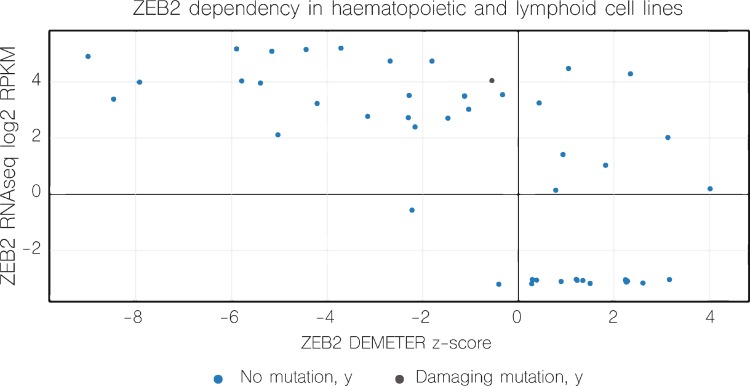
In addition to ZEB1, increasing evidence also suggests an important role for ZEB2 in AML development. An AML patient was found with tumor cells carrying a similar t(2;14)(q22;q32) translocation, as earlier identified in immature T-ALL, involving the ZEB2 and BCL11B loci.85 In the resulting ZEB2-BCL11B fusion transcript, the first 19AA of BCL11B are replaced by the first 24AA of ZEB2. As all functional domains of BCL11B are retained in this fusion product and the inverse BCL11B-ZEB2 transcript could not be detected, the authors concluded that the principal result of this translocation is aberrant expression of BCL11B controlled by the ZEB2 promoter/enhancer. Interestingly, other genetic events driving BCL11B overexpression have been reported in AML, further reinforcing the putative oncogenic role of BCL11B in the myeloid lineage.86,87 These observations are in sharp contrast with the proposed role of BCL11B in T-ALL as a tumor suppressor. These data therefore suggest that a similar genetic aberration, t(2;14)(q22;q32), can drive leukemic transformation both in the myeloid as well as the lymphoid lineage. In the case of AML, BCL11B overexpression most probably drives malignant transformation whereas retained ZEB2 expression during T-cell commitment is most probably the oncogenic driver in the case of immature T-ALL.18 Although the authors mainly focused on the role of BCL11B, it should be noted that ZEB2 mRNA levels in this t(2;14)(q22;q23) positive AML patient were normal, and most likely compensated by the unaffected allele via downregulation of the miR-200 family.88 More recent experiments convincingly showed that ZEB2 expression is essential for maintenance of leukemic growth of AML.88,89 Using an in vitro genome-wide shRNA screening method, followed by an in vivo secondary screen using a murine AML model driven by retroviral expression of an MLL-AF9 fusion, the authors identified ZEB2 as an essential gene for AML progression. Further molecular analysis demonstrated that ZEB2 represses transcription of genes important in myeloid differentiation. Consequently, ZEB2 depletion in AML cells will force differentiation of the leukemic cells. Interestingly, the notion that AML cells depend on sustained ZEB2 expression was recently confirmed by a large-scale deep RNAi screen that unraveled cancer dependencies in an extensive series of human tumor cell lines, including AML90 (Fig. 4).
Figure 4.
ZEB2 is cancer dependency factor in various hematopoietic and leukemia/lymphoma cell lines. Scatter plot Dependency versus Expression. This figure is based on data available via https://depmap.org/rnai/genedeps?gene=ZEB2,90 filtered on “hematopoietic and lymphoid.” Dependency for ZEB2 was determined in a genome-scale loss-of-function study in diverse cancer cell lines. DEMETER is an analytical that segregates on- from off-target effects of RNAi. This figure clearly depicts that large proportion of the analyzed leukemia cell lines do express ZEB2 and are dependent on it, with negative ZEB2 DEMETER z-score. RPKM = reads per kilobase per million mapped reads.
In a recent study, recurrent focal deletions of ZEB2 were found in pediatric AML patients (6.6%; 13/197 patients).91 Unfortunately, no expression data were presented in this study to conclude whether these single-allele mutations indeed resulted in loss-of-ZEB2 expression and/or functionality.
Similarly, whole genome sequencing identified ZEB2 putative loss-of-function mutations in 16% of blastic plasmacytoid dendritic cell neoplasm (BPDCN) patients.92 BPDCN is a very rare and aggressive myeloid neoplasm originating from precursors of a specialized subset of DCs,93–95 with no defined standard of care.
ZEBs in pathogen-induced hematologic malignancies
ZEB2 expression has been shown to be essential for differentiation, maturation, and/or function of CD8+ cytotoxic T cells (CTLs) and NK cells, 2 types of immune cells involved in antiviral host defense.36,38 Interestingly, recurrent deletions of the ZEB2 locus are significantly enriched in the hepatitis C virus (HCV)-related NHL patients.75,96 HCV is a known risk factor to develop B-cell lymphomas. The most convincing evidence is the observation of B-lymphoma regression after HCV eradication by antiviral therapy.
This was further supported by the fact that viral integrations at the Zeb2 locus are sufficient to induce leukemia in mice haplosufficient for Pax5.79,80 This Pax5−/+ mouse model spontaneously develops B-ALL only in a conventional animal house and not under specific pathogen-free conditions suggesting that exposure to infectious agents can act as a trigger for the development of B-ALL.97 The expression of ZEB2 may be essential for the functionality of the innate and adaptive immune system to efficiently eradicate these pathogens, but also infected and (partially) transformed cells from the body, before they can develop into an overt lymphoma/leukemia. We recently demonstrated that ZEB2 also plays a pivotal role in the immunosurveillance and clearance of melanoma cells after transplantation in syngeneic mice,36 suggesting it could serve as a more common mechanism also outside of hematologic malignancies.
Also, altered expression of ZEB1 has been seen in a few pathogen-induced leukemia/lymphoma subtypes, like H pylori positive gastric DLBCLs71 and ATLL, which is associated with HTLV-1 infection.29 Infections typically occur around birth and HTLV-1 carriers have a cumulative risk of 2.5% to develop ATLL with an average latency of 55 years, suggesting the need for extra tumorigenic events. However, no studies have been reported yet that specifically investigated the role of ZEB1 in the defense system of the body against infectious agents.
It has been estimated that over 90% of the world's population is carrying the Epstein-Barr herpes virus (EBV) usually acquired by an infection, often asymptomatic, during childhood. EBV has been linked to many types of malignancies including several epithelial cancers and some B-cell malignancies, such as Burkitt and Hodgkin lymphoma. EBV causes either a latent infection which is maintained stably in the host or a lytic phase with active production of viral particles and killing of the host cell.98 This latent-lytic switch can be initiated by expression of the viral immediate-early BZLF1 gene, a transactivator of viral genes for lytic replication. Interestingly, ZEB proteins are directly involved in the transcriptional regulation of BZLF1.99–101 For example, in B-cell lymphocytes, ZEB1 binds 2 consensus ZEB binding sites in the proximal promoter of the BZLF1 gene to actively repress its transcription. Therefore, inhibition of ZEB1 could be a potential mechanism to break through latency, activate the lytic phase of infection, leading to dead of the virally infected malignant cells. Therefore, ZEB expression may have a direct effect on the viral infectious cycle independent of the immune system and, as such, affect the initiation of EBV-related B-cell lymphomas.
ZEB downstream signaling in epithelial and hematopoietic cells
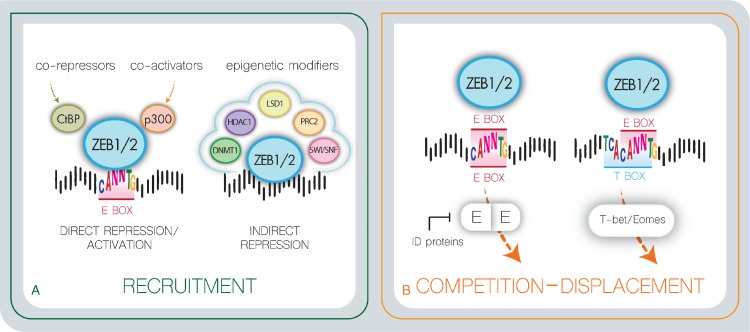
ZEB downstream signaling events have mostly been studied in epithelial cells. Differential gene expression analysis upon drug-inducible ZEB1/2 expression resulted in an extensive list of putative ZEB1/2 target genes in the context of EMT.13,16 Most of these genes are involved in cellular adhesion, cytoskeletal reorganization, cell polarity, and extracellular matrix composition. Subsequent promoter reporter studies in combination with ChIP identified a common ZEB recognition site consisting of a double E/Z-box in promoters of epithelial marker genes repressed by ZEB1/2.3,14 How ZEBs can activate mesenchymal promoters is less understood. Recent work suggested that ZEB1 might act as a direct activator in a complex with others transcription factors, like YAP1102 (Fig. 5A). Based on in silico predictions, in combination with in vitro studies using epithelial cancer cell lines, the spectrum of downstream targets of ZEB1/2, at least in the context of EMT, is relatively well established.
Figure 5.
Various modes of ZEBs transcriptional activity. (A) ZEBs recognize via their zinc finger clusters E-box sequence, which partially overlap T-box recognition sites. ZEBs may therefore compete or affect the function of other tissue-specific transcription factors that recognizes these similar regulatory regions, like E-proteins, Id proteins, and the T-box recognition proteins Eomes and T-bet. (B) ZEBs directly repress/activate target promoters via the recruitment of co-activator/repressor complexes or indirectly via altering chromatin landscape and promoter accessibility. E = E-protein.
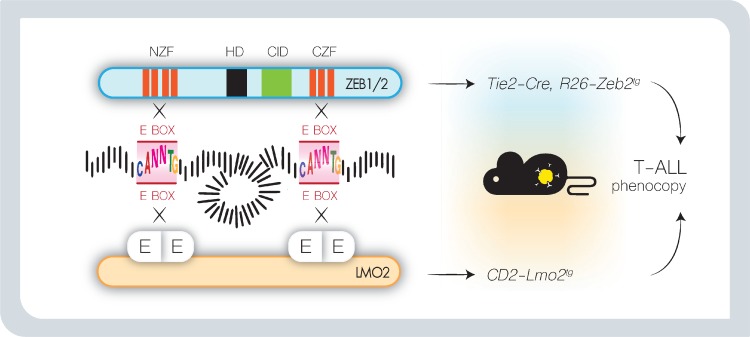
In contrast to solid tumor cells, the ZEB transcriptional targets in hematopoietic cells only start to emerge. Gene expression profiling after conditional loss of ZEB2 identified putative effector genes involved in cellular adhesion/homing and chemotaxis (Cxcr3, Cxcr4, Cxcr5, S1PR5, Cxcr3r1, Itgb2, Ccr7, Epcam, α4-integrin)35–39 as well as lineage-specific cytokines (IL2, IL7R, IL15, IL6, G-CSF).6,18,34,38,48,103 Interestingly, a large proportion of these genes are known targets of E2A.38 E-proteins are widely expressed bHLH transcription factors that cooperate with tissue/lineage-specific bHLH proteins by forming heterodimers that recognize a single E-box in their target promoter.104 Id (Inhibitors of DNA binding) proteins counteract E-protein function. They lack a basic DNA-binding domain and tether E-proteins and other bHLH proteins away from E-boxes by forming heterodimers thereby preventing transcriptional E2A activation. Both E-proteins and Id proteins have been shown to play essential roles in hematopoiesis,105 lymphocyte development, and lymphoid disease,106 and have been identified as master regulators of CSC and cancer aggressiveness.107 Therefore, competition between ZEBs and tissue-specific E-protein heterodimers, that bind similar or overlapping DNA recognition sites, could partially explain the ZEB lineage-specific downstream effects30,108–110 (Fig. 5B). Interesting to note in this context is the similarity in function between ZEBs and LMOs. Indeed, while ZEB proteins directly bind bipartite E-boxes via their amino and carboxyterminal zinc finger domains, LMOs are part of a multiprotein transcription complex that can also bridge 2 distant E-protein heterodimers. Interestingly, these similarities might explain the observed phenotypic similarities between ZEB2 overexpression and LMO2 overexpression in the context of T-ALL, spontaneous immature T-ALL formation associated with gain of self-renewal and stem cell properties18,55,56,111,112 (Fig. 6).
Figure 6.
Hypothetic model in which ZEB2 and LMO2 use a similar oncogenic mechanism of action in T-cell acute lymphoblastic leukemia. LMO proteins do not bind DNA themselves, but nucleate a core multiprotein complex by acting as a bridge between 2 bHLH protein dimers that each recognizes a single E-box. As ZEB2 binds a similar double E-box DNA motif in its target promoters, we speculate that both LMO2 and ZEB2 activate a similar downstream signaling cascade and may explain the observed phenotypic similarities in Zeb2 overexpressing mouse and CD2-LMO2 transgenic animals. bHLH = basic helix-loop-helix, CID = CtBP interaction domain, CZF = carboxyterminal zinc finger domain, E = E-protein, HD = homeobox, NZF = aminoterminal zinc finger domain.
We noted that multiple ZEB binding sites in promoters of differentially expressed genes upon ZEB2 knockout in NK cells and CD8+ CTLs,36,38 partially overlap with T-box recognition sites, suggesting that also ZEBs and T-box binding proteins, such as T-bet/Eomes, could influence each other's DNA binding capacity and transcriptional activity. Such a cross-competition may explain some of the overlapping or synergistic functions in different hematopoietic lineages36–38 (Fig. 5B). More research will be necessary to fully understand the synergistic and competitive interactions between these E- and T-box binding transcription factor families. Also competitive or synergistic functions of ZEB proteins with other transcription factor families needs further investigation. As example, Zeb1 indirectly regulates the expression of α4-integrin by inhibiting the transcriptional activity of c-Myb and Ets individually. However, synergy between c-Myb and Ets can overcome this repression, and highlights the fine balance and complex interactions between ZEB proteins and other essential hematopoietic transcription factors.113
The combination of the above-described oscillating ZEB1/2 expression, the cell-state specific chromatin accessibility, the competition with multiple other transcription factors’ families for DNA binding, and the presence or absence of coactivators and repressors, suggests that the transcriptional regulation of ZEB target genes is highly dynamic and cell type-specific resulting in different phenotypes in the loss- and gain-of-function mouse models affecting various stages of normal and malignant hematopoiesis.
Interestingly, EMT transcription factors of the SNAI family are also expressed during hematopoiesis114 and evidence is accumulating that they are also involved in malignant transformation toward leukemia.115 Further research will be necessary to determine whether they use similar converging/complementary pathways in the context of leukemia, as was previously observed during carcinoma progression.12,17
Conclusion
Until recently, ZEB proteins have mainly been studied as EMT inducing transcription factors allowing dissemination of epithelial cancers from the primary tumor site, and gaining stem cell properties and features for therapy resistance. Although EMT is not a hallmark of hematologic malignancies, we here enlisted extensive evidence that ZEBs do also play an important role in initiation and progression of different subtypes of lymphomas and leukemias.
Depending on the lineage of origin of these malignancies, ZEB1/2 can both act as oncogenes or tumor suppressors. In certain T-cell malignancies, ZEB1 can be considered as a tumor suppressor and ZEB2 as an oncogene, whereas this seems to be opposite for B-cell malignancies. Finally, in AML, both TFs can act as oncogenes and/or dependency factors. These often contradictory, synergistic, and/or complementary functions of ZEBs on lymphoma/leukemia initiation, progression, or maintenance may be, at least in part, explained by their very pleiotropic functions (cellular adhesion and mobilization/homing, stem cell properties and therapy resistance, immune regulation) at various stages during hematopoiesis; but may also be influenced by their complex oscillating expression profile, in combination with the presence/absence of cell-context dependent cofactors and downstream targets.
This raises the question if it would be feasible to therapeutically target ZEBs in the context of leukemia. Nevertheless, next to the inherent difficulty of targeting transcription factors, the main concern for therapeutic inhibition or reactivation of ZEB proteins might reside in the possible side effects. Indeed, ZEB1/2 have physiological functions throughout the body, including control of the immune system, where they safeguard development and functioning of different immune cell types. Interfering with this could not only disturb response to pathogens, but could also interrupt tumor immunosurveillance. To prevent these important side effects, specific pathways acting downstream of ZEBs, specific interaction with leukemia specific cofactors or their competition for DNA binding with other hematopoietic transcription factors, could be targeted. In that respect, it is interesting to mention that pleiotropic functions of ZEB proteins seem to utilize different protein domains,116 and as such the oncogenic roles may be uncoupled from the tumor suppressor roles. More research will be required to dissect the mode of action of ZEB1/2 both in leukemic cells and nontransformed immune cells, which may open the avenue to less toxic and more specific therapies for hematologic malignancies.
Footnotes
Citation: Soen B, Vandamme N, Berx G, Schwaller J, Van Vlierberghe P, Goossens S. ZEB Proteins in Leukemia: Friends, Foes, or Friendly Foes? HemaSphere, 2018;2:3. http://dx.doi.org/10.1097/HS9.0000000000000043
Funding: Our work is supported by the Research Foundation Flanders (FWO, 1504317N), Special Research Fund Ghent University (BOF24Y2017002001), the Worldwide Cancer Research (16-1157), Stand Up To Cancer (Kom op tegen Kanker, Flanders), the Swiss Bridge Foundation, the Swiss National Science Foundation (31003A_149714/1), and the Swiss Cancer League (KFS-3019-08-2012).
Disclosure: The authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Fortini ME, Lai ZC, Rubin GM. The Drosophila zfh-1 and zfh-2 genes encode novel proteins containing both zinc-finger and homeodomain motifs. Mech Dev 1991; 34:113–122. [DOI] [PubMed] [Google Scholar]
- 2.Lai ZC, Fortini ME, Rubin GM. The embryonic expression patterns of zfh-1 and zfh-2, two Drosophila genes encoding novel zinc-finger homeodomain proteins. Mech Dev 1991; 34:123–134. [DOI] [PubMed] [Google Scholar]
- 3.Comijn J, Berx G, Vermassen P, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell 2001; 7:1267–1278. [DOI] [PubMed] [Google Scholar]
- 4.Verschueren K, Remacle JE, Collart C, et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J Biol Chem 1999; 274:20489–20498. [DOI] [PubMed] [Google Scholar]
- 5.Remacle JE, Kraft H, Lerchner W, et al. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. EMBO J 1999; 18:5073–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Lee S, Teh CE, et al. The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Int Immunol 2009; 21:227–235. [DOI] [PubMed] [Google Scholar]
- 7.Verstappen G, van Grunsven LA, Michiels C, et al. Atypical Mowat-Wilson patient confirms the importance of the novel association between ZFHX1B/SIP1 and NuRD corepressor complex. Hum Mol Genet 2008; 17:1175–1183. [DOI] [PubMed] [Google Scholar]
- 8.Zhang JH, McCastlain K, Yoshihara H, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet 2016; 48:1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J 2003; 22:2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Postigo AA, Depp JL, Taylor JJ, et al. Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J 2003; 22:2453–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Grunsven LA, Taelman V, Michiels C, et al. deltaEF1 and SIP1 are differentially expressed and have overlapping activities during Xenopus embryogenesis. Dev Dyn 2006; 235:1491–1500. [DOI] [PubMed] [Google Scholar]
- 12.Nieto MA, Huang RY, Jackson RA, et al. Emt. Cell 2016; 166:21–45. [DOI] [PubMed] [Google Scholar]
- 13.Vandewalle C, Comijn J, De Craene B, et al. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res 2005; 33:6566–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eger A, Aigner K, Sonderegger S, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 2005; 24:2375–2385. [DOI] [PubMed] [Google Scholar]
- 15.Bindels S, Mestdagt M, Vandewalle C, et al. Regulation of vimentin by SIP1 in human epithelial breast tumor cells. Oncogene 2006; 25:4975–4985. [DOI] [PubMed] [Google Scholar]
- 16.Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol Biol Cell 2007; 18:3533–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goossens S, Vandamme N, Van Vlierberghe P, et al. EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim Biophys Acta Rev Cancer 2017; 1868:584–591. [DOI] [PubMed] [Google Scholar]
- 18.Goossens S, Radaelli E, Blanchet O, et al. ZEB2 drives immature T-cell lymphoblastic leukaemia development via enhanced tumour-initiating potential and IL-7 receptor signalling. Nat Commun 2015; 6:5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell 2012; 22:699–701. [DOI] [PubMed] [Google Scholar]
- 20.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009; 11:1487–1495. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P, Wei Y, Wang L, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol 2014; 16:864–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayan AE, Griffiths TR, Pal R, et al. SIP1 protein protects cells from DNA damage-induced apoptosis and has independent prognostic value in bladder cancer. Proc Natl Acad Sci USA 2009; 106:14884–14889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugo HJ, Pereira L, Suryadinata R, et al. Direct repression of MYB by ZEB1 suppresses proliferation and epithelial gene expression during epithelial-to-mesenchymal transition of breast cancer cells. Breast Cancer Res 2013; 15:R113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu CL, Chung FH, Chen CH, et al. Genotypes of cancer stem cells characterized by epithelial-to-mesenchymal transition and proliferation related functions. Sci Rep 2016; 6:32523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop-a motor of cellular plasticity in development and cancer? EMBO Rep 2010; 11:670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takagi T, Moribe H, Kondoh H, et al. DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development 1998; 125:21–31. [DOI] [PubMed] [Google Scholar]
- 27.Higashi Y, Moribe H, Takagi T, et al. Impairment of T cell development in deltaEF1 mutant mice. J Exp Med 1997; 185:1467–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnold CN, Pirie E, Dosenovic P, et al. A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity. Proc Natl Acad Sci USA 2012; 109:12286–12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hidaka T, Nakahata S, Hatakeyama K, et al. Down-regulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood 2008; 112:383–393. [DOI] [PubMed] [Google Scholar]
- 30.Brabletz T, Jung A, Hlubek F, et al. Negative regulation of CD4 expression in T cells by the transcriptional repressor ZEB. Int Immunol 1999; 11:1701–1708. [DOI] [PubMed] [Google Scholar]
- 31.Brabletz S, Lasierra Losada M, Schmalhofer O, et al. Generation and characterization of mice for conditional inactivation of Zeb1. Genesis 2017; 55:e23024. [DOI] [PubMed] [Google Scholar]
- 32.Tatari MN, De Craene B, Soen B, et al. ZEB2-transgene expression in the epidermis compromises the integrity of the epidermal barrier through the repression of different tight junction proteins. Cell Mol Life Sci 2014; 71:3599–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higashi Y, Maruhashi M, Nelles L, et al. Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis 2002; 32:82–84. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Riedt T, Goossens S, et al. The EMT transcription factor Zeb2 controls adult murine hematopoietic differentiation by regulating cytokine signaling. Blood 2017; 129:460–472. [DOI] [PubMed] [Google Scholar]
- 35.Goossens S, Janzen V, Bartunkova S, et al. The EMT regulator Zeb2/Sip1 is essential for murine embryonic hematopoietic stem/progenitor cell differentiation and mobilization. Blood 2011; 117:5620–5630. [DOI] [PubMed] [Google Scholar]
- 36.van Helden MJ, Goossens S, Daussy C, et al. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J Exp Med 2015; 212:2015–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dominguez CX, Amezquita RA, Guan T, et al. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J Exp Med 2015; 212:2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omilusik KD, Best JA, Yu B, et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J Exp Med 2015; 212:2027–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott CL, Soen B, Martens L, et al. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J Exp Med 2016; 213:897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konradi S, Yasmin N, Haslwanter D, et al. Langerhans cell maturation is accompanied by induction of N-cadherin and the transcriptional regulators of epithelial-mesenchymal transition ZEB1/2. Eur J Immunol 2014; 44:553–560. [DOI] [PubMed] [Google Scholar]
- 41.Barbu EA, Zhang J, Berenstein EH, et al. The transcription factor Zeb2 regulates signaling in mast cells. J Immunol 2012; 188:6278–6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyoshi T, Maruhashi M, Van De Putte T, et al. Complementary expression pattern of Zfhx1 genes Sip1 and deltaEF1 in the mouse embryo and their genetic interaction revealed by compound mutants. Dev Dyn 2006; 235:1941–1952. [DOI] [PubMed] [Google Scholar]
- 43.Hill L, Browne G, Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int J Cancer 2013; 132:745–754. [DOI] [PubMed] [Google Scholar]
- 44.Vandamme N, Berx G. Melanoma cells revive an embryonic transcriptional network to dictate phenotypic heterogeneity. Front Oncol 2014; 4:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denecker G, Vandamme N, Akay O, et al. Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ 2014; 21:1250–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bresnick EH, Lee HY, Fujiwara T, et al. GATA switches as developmental drivers. J Biol Chem 2010; 285:31087–31093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest 2012; 122:3398–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durinck K, Goossens S, Peirs S, et al. Novel biological insights in T-cell acute lymphoblastic leukemia. Exp Hematol 2015; 43:625–639. [DOI] [PubMed] [Google Scholar]
- 49.Chiaretti S, Gianfelici V, O’Brien SM, et al. Advances in the genetics and therapy of acute lymphoblastic leukemia. Am Soc Clin Oncol Educ Book 2016; 35:e314–e322. [DOI] [PubMed] [Google Scholar]
- 50.Coustan-Smith E, Mullighan CG, Onciu M, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol 2009; 10:147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meijerink JP. Genetic rearrangements in relation to immunophenotype and outcome in T-cell acute lymphoblastic leukaemia. Best Pract Res Clin Haematol 2010; 23:307–318. [DOI] [PubMed] [Google Scholar]
- 52.Treanor LM, Zhou S, Janke L, et al. Interleukin-7 receptor mutants initiate early T cell precursor leukemia in murine thymocyte progenitors with multipotent potential. J Exp Med 2014; 211:701–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun W, Yang S, Shen W, et al. Identification of DeltaEF1 as a novel target that is negatively regulated by LMO2 in T-cell leukemia. Eur J Haematol 2010; 85:508–519. [DOI] [PubMed] [Google Scholar]
- 54.Larson RC, Fisch P, Larson TA, et al. T cell tumours of disparate phenotype in mice transgenic for Rbtn-2. Oncogene 1994; 9:3675–3681. [PubMed] [Google Scholar]
- 55.Cleveland SM, Smith S, Tripathi R, et al. Lmo2 induces hematopoietic stem cell-like features in T-cell progenitor cells prior to leukemia. Stem Cells 2013; 31:882–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cleveland SM, Goodings C, Tripathi RM, et al. LMO2 induces T-cell leukemia with epigenetic deregulation of CD4. Exp Hematol 2014; 42:581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuoka M. Human T-cell leukemia virus type I and adult T-cell leukemia. Oncogene 2003; 22:5131–5140. [DOI] [PubMed] [Google Scholar]
- 58.Nakahata S, Yamazaki S, Nakauchi H, et al. Downregulation of ZEB1 and overexpression of Smad7 contribute to resistance to TGF-beta1-mediated growth suppression in adult T-cell leukemia/lymphoma. Oncogene 2010; 29:4157–4169. [DOI] [PubMed] [Google Scholar]
- 59.Jahn SC, Law ME, Corsino PE, et al. TGF-beta antiproliferative effects in tumor suppression. Front Biosci (Schol Ed) 2012; 4:749–766. [DOI] [PubMed] [Google Scholar]
- 60.Olsen E, Vonderheid E, Pimpinelli N, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 2007; 110:1713–1722. [DOI] [PubMed] [Google Scholar]
- 61.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016; 127:2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiel MJ, Sahasrabuddhe AA, Rolland DC, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat Commun 2015; 6:8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prasad A, Rabionet R, Espinet B, et al. Identification of gene mutations and fusion genes in patients with Sezary syndrome. J Invest Dermatol 2016; 136:1490–1499. [DOI] [PubMed] [Google Scholar]
- 64.Wang L, Ni X, Covington KR, et al. Genomic profiling of Sezary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet 2015; 47:1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi J, Goh G, Walradt T, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet 2015; 47:1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGirt LY, Jia P, Baerenwald DA, et al. Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides. Blood 2015; 126:508–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mishra A, La Perle K, Kwiatkowski S, et al. Mechanism, consequences, and therapeutic targeting of abnormal IL15 signaling in cutaneous T-cell lymphoma. Cancer Discov 2016; 6:986–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jares P, Colomer D, Campo E. Molecular pathogenesis of mantle cell lymphoma. J Clin Invest 2012; 122:3416–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kahl BS, Dreyling M, Gordon LI, et al. Recent advances and future directions in mantle cell lymphoma research: report of the 2016 mantle cell lymphoma consortium workshop. Leuk Lymphoma 2017; 58:1561–1569. [DOI] [PubMed] [Google Scholar]
- 70.Sanchez-Tillo E, Fanlo L, Siles L, et al. The EMT activator ZEB1 promotes tumor growth and determines differential response to chemotherapy in mantle cell lymphoma. Cell Death Differ 2014; 21:247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lemma S, Karihtala P, Haapasaari KM, et al. Biological roles and prognostic values of the epithelial-mesenchymal transition-mediating transcription factors Twist, ZEB1 and Slug in diffuse large B-cell lymphoma. Histopathology 2013; 62:326–333. [DOI] [PubMed] [Google Scholar]
- 72.Huang WT, Kuo SH, Cheng AL, et al. Inhibition of ZEB1 by miR-200 characterizes Helicobacter pylori-positive gastric diffuse large B-cell lymphoma with a less aggressive behavior. Mod Pathol 2014; 27:1116–1125. [DOI] [PubMed] [Google Scholar]
- 73.Chen YW, Hu XT, Liang AC, et al. High BCL6 expression predicts better prognosis, independent of BCL6 translocation status, translocation partner, or BCL6-deregulating mutations, in gastric lymphoma. Blood 2006; 108:2373–2383. [DOI] [PubMed] [Google Scholar]
- 74.Papadopoulou V, Postigo A, Sanchez-Tillo E, et al. ZEB1 and CtBP form a repressive complex at a distal promoter element of the BCL6 locus. Biochem J 2010; 427:541–550. [DOI] [PubMed] [Google Scholar]
- 75.Matteucci C, Bracci M, Barba G, et al. Different genomic imbalances in low- and high-grade HCV-related lymphomas. Leukemia 2008; 22:219–222. [DOI] [PubMed] [Google Scholar]
- 76.Woo JS, Alberti MO, Tirado CA. Childhood B-acute lymphoblastic leukemia: a genetic update. Exp Hematol Oncol 2014; 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou X, Edmonson MN, Wilkinson MR, et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat Genet 2016; 48:4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 2014; 371:1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caudell D, Zhang Z, Chung YJ, et al. Expression of a CALM-AF10 fusion gene leads to Hoxa cluster overexpression and acute leukemia in transgenic mice. Cancer Res 2007; 67:8022–8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dang J, Wei L, de Ridder J, et al. PAX5 is a tumor suppressor in mouse mutagenesis models of acute lymphoblastic leukemia. Blood 2015; 125:3609–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meyer C, Burmeister T, Groger D, et al. The MLL recombinome of acute leukemias in 2017. Leukemia 2018; 32:273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Slany RK. The molecular mechanics of mixed lineage leukemia. Oncogene 2016; 35:5215–5223. [DOI] [PubMed] [Google Scholar]
- 83.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol 2012; 7:283–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stavropoulou V, Kaspar S, Brault L, et al. MLL-AF9 expression in hematopoietic stem cells drives a highly invasive AML expressing EMT-related genes linked to poor outcome. Cancer Cell 2016; 30:43–58. [DOI] [PubMed] [Google Scholar]
- 85.Torkildsen S, Gorunova L, Beiske K, et al. Novel ZEB2-BCL11B fusion gene identified by RNA-sequencing in acute myeloid leukemia with t(2;14)(q22;q32). PLoS ONE 2015; 10:e0132736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abbas S, Sanders MA, Zeilemaker A, et al. Integrated genome-wide genotyping and gene expression profiling reveals BCL11B as a putative oncogene in acute myeloid leukemia with 14q32 aberrations. Haematologica 2014; 99:848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gutierrez A, Kentsis A, Sanda T, et al. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood 2011; 118:4169–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li H, Mar BG, Zhang H, et al. The EMT regulator ZEB2 is a novel dependency of human and murine acute myeloid leukemia. Blood 2017; 129:497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meyer SE. From EMT to HSC to AML: ZEB2 is a cell fate switch. Blood 2017; 129:400–401. [DOI] [PubMed] [Google Scholar]
- 90.Tsherniak A, Vazquez F, Montgomery PG, et al. Defining a cancer dependency map. Cell 2017; 170:564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bolouri H, Farrar JE, Triche T, Jr, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med 2018; 24:103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Menezes J, Acquadro F, Wiseman M, et al. Exome sequencing reveals novel and recurrent mutations with clinical impact in blastic plasmacytoid dendritic cell neoplasm. Leukemia 2014; 28:823–829. [DOI] [PubMed] [Google Scholar]
- 93.Chaperot L, Bendriss N, Manches O, et al. Identification of a leukemic counterpart of the plasmacytoid dendritic cells. Blood 2001; 97:3210–3217. [DOI] [PubMed] [Google Scholar]
- 94.Pilichowska ME, Fleming MD, Pinkus JL, et al. CD4+/CD56+ hematodermic neoplasm (“blastic natural killer cell lymphoma”): neoplastic cells express the immature dendritic cell marker BDCA-2 and produce interferon. Am J Clin Pathol 2007; 128:445–453. [DOI] [PubMed] [Google Scholar]
- 95.Petrella T, Comeau MR, Maynadie M, et al. “A granular CD4+ CD56+ hematodermic neoplasm” (blastic NK-cell lymphoma) originates from a population of CD56+ precursor cells related to plasmacytoid monocytes. Am J Surg Pathol 2002; 26:852–862. [DOI] [PubMed] [Google Scholar]
- 96.Visentini M, Conti V, Cristofoletti C, et al. Clonal expansion and functional exhaustion of monoclonal marginal zone B cells in mixed cryoglobulinemia: the yin and yang of HCV-driven lymphoproliferation and autoimmunity. Autoimmun Rev 2013; 12:430–435. [DOI] [PubMed] [Google Scholar]
- 97.Martin-Lorenzo A, Hauer J, Vicente-Duenas C, et al. Infection exposure is a causal factor in B-cell precursor acute lymphoblastic leukemia as a result of Pax5-inherited susceptibility. Cancer Discov 2015; 5:1328–1343. [DOI] [PubMed] [Google Scholar]
- 98.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpes virus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA 1985; 82:4085–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yu X, Wang Z, Mertz JE. ZEB1 regulates the latent-lytic switch in infection by Epstein-Barr virus. PLoS Pathog 2007; 3:e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kraus RJ, Perrigoue JG, Mertz JE. ZEB negatively regulates the lytic-switch BZLF1 gene promoter of Epstein-Barr virus. J Virol 2003; 77:199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ellis AL, Wang Z, Yu X, et al. Either ZEB1 or ZEB2/SIP1 can play a central role in regulating the Epstein-Barr virus latent-lytic switch in a cell-type-specific manner. J Virol 2010; 84:6139–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lehmann W, Mossmann D, Kleemann J, et al. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat Commun 2016; 7:10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Williams TM, Moolten D, Burlein J, et al. Identification of a zinc finger protein that inhibits IL-2 gene-expression. Science 1991; 254:1791–1794. [DOI] [PubMed] [Google Scholar]
- 104.Wang LH, Baker NE. E proteins and ID proteins: helix-loop-helix partners in development and disease. Dev Cell 2015; 35:269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cochrane SW, Zhao Y, Welner RS, et al. Balance between Id and E proteins regulates myeloid-versus-lymphoid lineage decisions. Blood 2009; 113:1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Belle I, Zhuang Y. E proteins in lymphocyte development and lymphoid diseases. Curr Top Dev Biol 2014; 110:153–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer 2014; 14:77–91. [DOI] [PubMed] [Google Scholar]
- 108.Postigo AA, Dean DC. ZEB, a vertebrate homolog of Drosophila Zfh-1, is a negative regulator of muscle differentiation. EMBO J 1997; 16:3935–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol 1994; 14:6153–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sekido R, Murai K, Funahashi J, et al. The delta-crystallin enhancer-binding protein delta EF1 is a repressor of E2-box-mediated gene activation. Mol Cell Biol 1994; 14:5692–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Goodings C, Tripathi R, Cleveland SM, et al. Enforced expression of E47 has differential effects on LMO2-induced T-cell leukemias. Leuk Res 2015; 39:100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grutz GG, Bucher K, Lavenir I, et al. The oncogenic T cell LIM-protein LMO2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J 1998; 17:4594–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Postigo AA, Sheppard AM, Mucenski ML, et al. c-Myb and Ets proteins synergize to overcome transcriptional repression by ZEB. EMBO J 1997; 16:3924–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carmichael CL, Haigh JJ. The snail family in normal and malignant haematopoiesis. Cells Tissues Organs 2017; 203:82–98. [DOI] [PubMed] [Google Scholar]
- 115.Carmichael C, Goossens S, Kile B, et al. A novel role for the Emt regulator, snail in hematopoiesis and leukemia. Exp Hematol 2015; 43:S56–S156. [Google Scholar]
- 116.Postigo AA, Dean DC. Independent repressor domains in ZEB regulate muscle and T-cell differentiation. Mol Cell Biol 1999; 19:7961–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]