Acute myeloid leukemia (AML) patients with partial tandem duplications (PTDs) in the Mixed Lineage Leukemia (MLL) officially known as the Lysine (K)-specific Methyltransferase 2A (KMT2A) gene, generally have adverse outcomes. Previous mouse studies have shown that Kmt2a-ptd is insufficient to cause AML, indicating additional mutations are required for leukemogenesis. Herein, we evaluated the mutational landscape, gene expression signatures and prognosis of KMT2A-PTD adult AML in comparison to a well-characterized adult AML cohort without KMT2A-PTD. Our study demonstrates that KMT2A-PTD AML has a distinct gene expression signature and that concomitant DNMT3A and NRAS mutations were associated with adverse clinical outcome in this subset of AML.
AML patients with KMT2A-PTD is characterized by an internal duplication spanning exon 3 to 9, exon 3 to 10, or exon 3 to 11 (Fig. S1A, Supplemental Digital Content 1).1KMT2A-PTDs occur in 3.2 to 11% of adult de novo AML and are more frequently present in AML with normal cytogenetics and AML with trisomy of chromosome 11 as a sole cytogenetic aberration.2 The presence of a KMT2A-PTD has been shown to associate with adverse outcome in AML.2,3
The partial-tandem duplications within KMT2A result in in-frame additions of extra N-terminal amino acids and maintain functional proteins, which contribute to leukemogenesis.4 Under normal physiological conditions, the KMT2A gene encodes a SET domain-containing protein, which mediates methylation of histone 3 lysine 4 (H3K4).5 The effect of the PTD on normal KMT2A function and its role in leukemogenesis are currently unknown. 11q23-rearrangements involving KMT2A gene (3–4% of adult AML) result in abrogation of KMT2A transactivation and histone methyltransferase function.6Kmt2a-PTD alone appeared insufficient to cause AML.7 These findings support the notion that additional genetic hits are required for the development of KMT2A-PTD leukemia.7,8
We aimed to evaluate the mutational landscape, gene expression signatures and prognosis of KMT2A-PTD adult AML in comparison to a well-characterized adult AML cohort without KMT2A-PTD (hereafter referred as reference cohort) treated according to the international multicenter HOVON-SAKK AML clinical trials (www.hovon.nl).
cDNA from 1998 AML patients was screened for KMT2A-PTD mutations using RT-PCR and confirmed by Sanger sequencing (Fig. S1B and Supplementary method, Supplemental Digital Content 1 and 2). KMT2A-PTDs were present in 5.5% (109 out of 1998) of all AML cases. The KMT2A-PTD was examined in the context of a number of clinical parameters (summarized in Table S1, Supplemental Digital Content 3 and detailed in Table S4, Supplemental Digital Content 4). The median age of AML patients with or without a KMT2A-PTD was 56 and 51 years, respectively (p = 0.0016). The majority of KMT2A-PTD AML patients had a normal karyotype (57.3%). However, this did not significantly differ from AML patients without a KMT2A-PTD (48.8%) (p = 0.158). In line with previous studies, the presence of KMT2A-PTDs was significantly associated with a concurrent trisomy 11 as compared to the KMT2A wild-type AML reference cohort (7.3% vs 1.1%; respectively, p = 0.002).
Genomic DNA (gDNA) was available for 85 out of the 109 KMT2A-PTD AML cases. We performed next-generation sequencing (NGS) on the 85 KMT2A-PTD AML cases to determine the concurrent driver mutations, with as reference the KMT2A wild-type AML cohort (n = 561). All AML cases were sequenced using the Illumina TruSight Myeloid Sequencing Panel (Table S6, Supplemental Digital Content 4) on the HiSeq 2500 in Rapid mode following the manufacturer's recommendations (Illumina, San Diego, CA). FLT3 internal tandem duplications (ITD) were determined with fragment length analyses as previously described.9 The number of mutations detected in the KMT2A-PTD AML cohort ranged from 0 to 6 mutations, with an average of 2.7 mutations per KMT2A-PTD AML patient, whereas, the AML reference cohort carried 0 to 9 mutations with an average of 3.0 mutations per AML patient. The average number of mutations was not significantly different between both cohorts (p = 0.1).
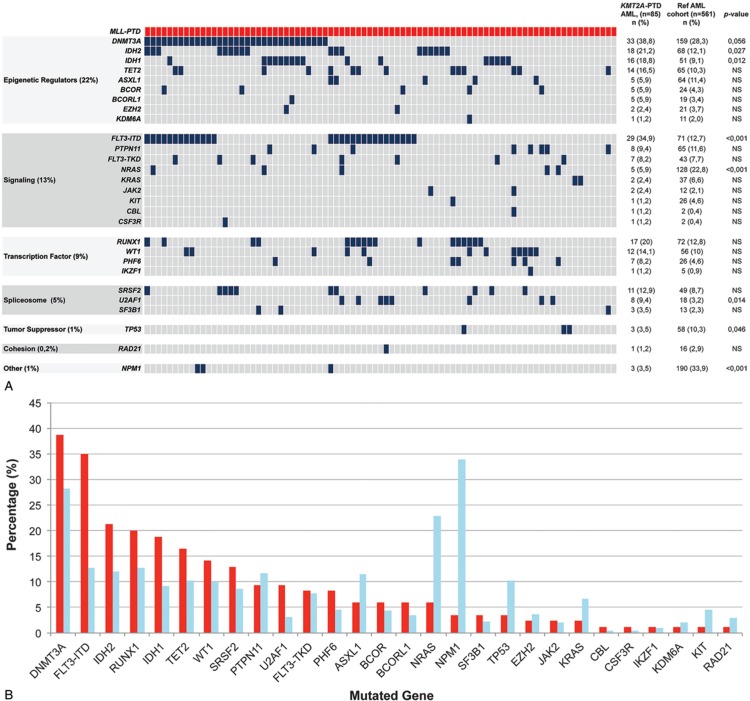
The most frequently mutated genes in KMT2A-PTD AML were DNMT3A (38.8%), FLT3-ITD (34.9%), IDH2 (21.2%), RUNX1 (20%), IDH1 (18.8%), TET2 (16.5%), WT1 (14.1%), and SRSF2 (12.9%) (Fig. 1). In contrast to what has been reported, we found three cases with a concurrent mutation in the NPM1 gene (Fig. 1).10,11
Figure 1.
Mutational landscape and gene expression changes of KMT2A-PTD AML. A) KMT2A-PTD mutation landscape. Each column represents an individual AML patient. The gene mutations are categorized according to gene function and family. The number and the percentage of the gene mutations in KMT2A-PTD AML and the AML reference cohort and p-value are indicated. B) The frequency of concurrent mutations of KMT2A-PTD AML (red) and the reference cohort (blue).
We next examined whether the concurrently mutated genes were significantly associated with KMT2A-PTD AML compared to the KMT2A wild-type AML reference cohort. KMT2A-PTD concurrent mutations were present in proteins involved in epigenetic regulation, signaling, transcription, splicing, chromosome segregation, and tumor suppression (Fig. 1A). None of these mutational categories was significantly associated with KMT2A-PTD AML. However, a number of mutated genes were significantly more frequent in KMT2A-PTD AML compared to other types of AML, i.e., FLT3-ITD (34.9% vs. 23%; p = 0.028) as well as mutations in IDH1 (18.8% vs 9.1%; p = 0.012), U2AF1 (9.4% vs 3.2%; p = 0.014) and IDH2 (21.2% vs 12.1%; p = 0.027) [Fig. 1B and Fig. S2, Supplemental Digital Content 1]. Similar associations with IDH2 and U2AF1 mutations were demonstrated by Papaemmanuil et al12 however, DNMT3A, RUNX1, and STAG2 mutations were not significantly associated with KMT2A-PTD AML in our cohort. Sun et al. demonstrated that KMT2A-PTD AML carried more frequently FLT3, DNMT3A, RUNX1, IDH1 and IDH2 mutations,10 whereas Kao et al showed this correlation for FLT3, U2AF1, RUNX1, STAG2, PTPN11, WT1 and EZH2 mutations.11 However, in the latter 2 studies results were not compared to an internal KMT2A wild-type AML cohort, which could potentially result in misinterpretation as a result of selection biases regarding these positive associations. In our AML cohort, we did see trends for associations, for example, mutations in DNMT3A, RUNX1, PTPN11, and WT1, but these did not reach statistical significance. In contrast, mutations in NPM1 (3.5% vs 33.9%; p < 0.001), TP53 (3.5% vs 10.3%; p = 0.046) and NRAS (5.9% vs 22.8%; p < 0.001) were significantly less frequent in KMT2A-PTD AML in our cohort (Fig. 1 and Fig. S2, Supplemental Digital Content 1). Mutual exclusivity between NPM1 mutations and KMT2A-PTDs have been shown before in AML.10–12
We next investigated which genes were differentially expressed between KMT2A-PTD AML and all other AMLs, in particular, AML with t(11q23), using our previously published gene expression profile (GEP) dataset (n = 513 AML).13 Interestingly, multiple homeobox-related gene family members were consistently overexpressed in KMT2A-PTD AML. The top-35 differentially expressed genes included HOX- and TALE-related genes, such as HOXB5, HOXB6, HOXB7, HOXB8, HOXB9, and NKX2.3, whereas KMT2A itself appeared to be the most consistently overexpressed gene (Table S2, Supplemental Digital Content 3). Since mutations in NPM1 are also associated with dysregulation of HOX gene expression, we next used an association model to see if those differentially expressed HOX genes were also dysregulated in NPM1 mutant AML.14,15 In this association model, which takes a number of relevant clinically and genetically defined subsets of AML into account, HOX-related genes, such as HOXA7, HOXA9, and HOXA5, seemed to be more significantly differentially expressed in mutant NPM1 AML (Table S2, Supplemental Digital Content 3). In contrast, these specific gene expression changes were absent when a similar analysis was performed in AML with t(11q23) involving KMT2A (Table S2, Supplemental Digital Content 3). Thus, KMT2A-PTD may induce overexpression of HOX-related genes in different ways than t(11q23)-related fusion proteins, suggesting that the KMT2A-PTD induces leukemogenesis by mechanisms distinct from t(11q23) abnormalities involving KMT2A.
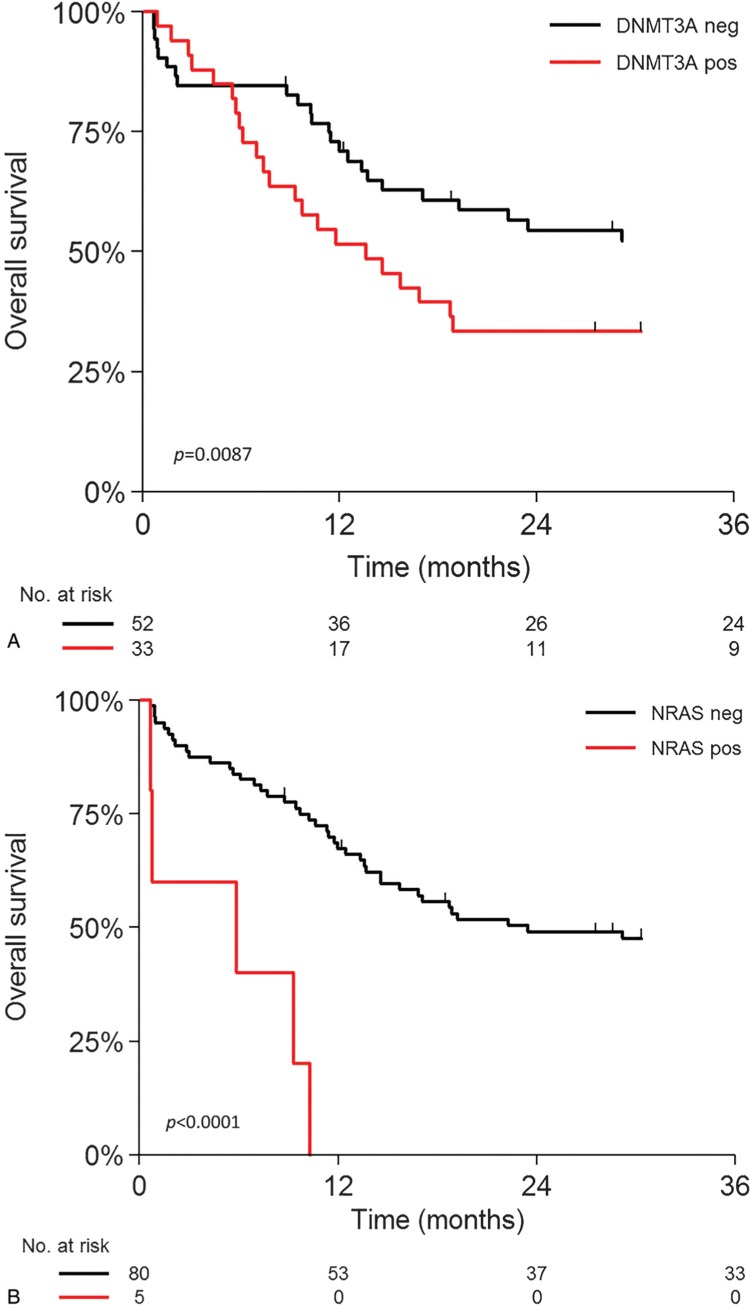
The prognostic impact of KMT2A-PTD on overall survival (OS) and event-free survival (EFS) of this AML cohort appeared to be not significantly different to wild-type KMT2A AML (p = 0.44) (Figure S3, Supplemental Digital Content 1). This is in contrast to what has been shown before but in line with more recent publications.2,3 In normal karyotype AML, this association was also absent (p = 0.7, data not shown). We next addressed the question whether the concurrent mutations might carry prognostic value within KMT2A-PTD AML. Interestingly, in KMT2A-PTD AML, coexisting DNMT3A mutations were significantly associated with inferior overall survival (HR: 2.06; 95%CI: 1.19–3.58; p = 0.010) (Fig. 2A), as was suggested before.11 Moreover, KMT2A-PTD AML patients that harbor NRAS mutations also have an inferior outcome (HR: 6.54; 95%CI: 2.45–17.49; p < 0.001) (Fig. 2B). RAS-related mutations such as FLT3 TKD mutations recently were shown to confer an inferior outcome to patients with KMT2A-PTD AML. Survival analysis of DNMT3A and NRAS mutations in KMT2A wild-type AML patients revealed that mutations in DNMT3A were not associated with treatment outcome (p = 0.99) (Fig. S4, Supplemental Digital Content 1), whereas mutations in NRAS only showed a borderline association with positive outcome (p = 0.044) (Fig. S5, Supplemental Digital Content 1). In multivariable analysis, including white blood cell count (WBC) and cytogenetics, DNMT3A and NRAS mutations remained significantly associated with adverse outcome (Table S3, Supplemental Digital Content 3). Thus, although KMT2A-PTD did not associate with outcome in AML in general, specific mutational subtypes within KMT2A-PTD AML appear to carry poor prognostic value.
Figure 2.
Overall survival analysis KMT2A-PTD AML and concurrent mutations. A) KMT2A-PTD with and without DNMT3A mutations (P = 0.0087). B) KMT2A-PTD with and without NRAS mutations (P < 0.0001).
To validate our findings in KMT2A-PTD AML, we investigated an independent KMT2A-PTD AML cohort of patients included in the treatment protocols of the CETLAM cooperative group. The validation cohort contained 27 KMT2A-PTD AML patients with a median age of 56 years (Table S5, Supplemental Digital Content 4), who were sequenced using Qiagen Human Myeloid Neoplasms GeneRead DNAseq Targeted Panel V2 (Table S6, Supplemental Digital Content 4) (Qiagen, Hilden, Germany) on the MiSeq (Illumina, San Diego, CA) as per manufacturer instructions. The most frequently mutated genes in KMT2A-PTD AML validation cohort were similar to our 85 AML patients cohort (DNMT3A (48%), FLT3-ITD (37%), IDH2 (22.2%), WT1 (14.8%), IDH1 (14.8%) and RUNX1 (11.1%)) (Fig. S2, Supplemental Digital Content 1). In the validation cohort, we confirmed that KMT2A-PTD AML patients with concurrent mutant DNMT3A have inferior outcome (13 out of 27; p = 0.0017; Fig. S6, Supplemental Digital Content 1). Unfortunately, the low numbers in the validation cohort precluded survival analyses of coexisting NRAS mutations, which should be confirmed in a larger KMT2A-PTD AML cohort.
In summary, we revealed within the molecular landscape of KMT2A-PTD AML, which carry specific HOX gene expression signatures, that concurrent DNMT3A mutations and NRAS mutations are associated with an adverse outcome.
Acknowledgments
We thank all the participating centers of the Dutch–Belgian Cooperative Trial Group for Hematology–Oncology (HOVON) and Swiss Group for Clinical Cancer Research (SAKK), where the clinical trials that formed the basis for this study were conducted; H. Berna Beverloo for performing cytogenetic analyses, Eric Bindels for performing next-generation sequencing, Remco Hoogenboezem for assisting with bioinformatics.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Citation: Hinai ASAA, Pratcorona M, Grob T, Kavelaars FG, Bussaglia E, Sanders MA, Nomdedeu J, Valk PJM. The landscape of KMT2A-PTD AML: Concurrent mutations, gene expression signatures, and clinical outcome. HemaSphere, 2019;00:00 http://dx.doi.org/10.1097/HS9.0000000000000181.
Supported by a grant from the Netherlands Organization for Health Research and Development (846002002). ASAH is a recipient of a Ph.D. scholarship from the Ministry of Health, Oman and MAS is supported by a Rubicon fellowship from the Netherlands Organization for Scientific Research (019.153LW.038).
ASAH: performed research, analyzed data and wrote the paper; MP, TG, FGK, EB, MAS and JFN: Performed research, and analyzed data; and PJMV: Designed and performed research, analyzed data and wrote the paper.
The authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Steudel C, Wermke M, Schaich M, et al. Comparative analysis of MLL partial tandem duplication and FLT3 internal tandem duplication mutations in 956 adult patients with acute myeloid leukemia. Genes Chromosomes Cancer. 2003;37:237–251. [DOI] [PubMed] [Google Scholar]
- 2.Basecke J, Whelan JT, Griesinger F, et al. The MLL partial tandem duplication in acute myeloid leukaemia. Br J Haematol. 2006;135:438–449. [DOI] [PubMed] [Google Scholar]
- 3.Shimada A, Taki T, Tabuchi K, et al. Tandem duplications of MLL and FLT3 are correlated with poor prognoses in pediatric acute myeloid leukemia: a study of the Japanese childhood AML Cooperative Study Group. Pediatr Blood Cancer. 2008;50:264–269. [DOI] [PubMed] [Google Scholar]
- 4.Whitman SP, Liu S, Vukosavljevic T, et al. The MLL partial tandem duplication: evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular-targeted therapy. Blood. 2005;106:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. [DOI] [PubMed] [Google Scholar]
- 6.Visani G, Piccaluga PP, Martinelli G, et al. Sustained molecular remission in advanced acute promyelocytic leukemia with combined pulsed retinoic acid and arsenic trioxide. Clinical evidence of synergistic effect and real-time quantification of minimal residual disease. Haematologica. 2003;88:ELT15. [PubMed] [Google Scholar]
- 7.Zhang Y, Yan X, Sashida G, et al. Stress hematopoiesis reveals abnormal control of self-renewal, lineage bias, and myeloid differentiation in Mll partial tandem duplication (Mll-PTD) hematopoietic stem/progenitor cells. Blood. 2012;120:1118–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilliland DG. Hematologic malignancies. Curr Opin Hematol. 2001;8:189–191. [DOI] [PubMed] [Google Scholar]
- 9.Versluis J, In ’t Hout FE, Devillier R, et al. Comparative value of post-remission treatment in cytogenetically normal AML subclassified by NPM1 and FLT3-ITD allelic ratio. Leukemia. 2017;31:26–33. [DOI] [PubMed] [Google Scholar]
- 10.Sun QY, Ding LW, Tan KT, et al. Ordering of mutations in acute myeloid leukemia with partial tandem duplication of MLL (MLL-PTD). Leukemia. 2017;31:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kao HW, Liang DC, Kuo MC, et al. High frequency of additional gene mutations in acute myeloid leukemia with MLL partial tandem duplication: DNMT3A mutation is associated with poor prognosis. Oncotarget. 2015;6:33217–33225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verhaak RG, Wouters BJ, Erpelinck CA, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94:131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mullighan CG, Kennedy A, Zhou X, et al. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007;21:2000–2009. [DOI] [PubMed] [Google Scholar]
- 15.Alcalay M, Tiacci E, Bergomas R, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106:899–902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.