Abstract
Although EGFR mutant-selective TKIs are clinically effective, acquired resistance can occur by reactivating ERK. We show using in vitro models of acquired EGFR TKI resistance with a mesenchymal phenotype that CXCR7, an atypical GPCR, activates the MAPK-ERK pathway via β-arrestin. Depletion of CXCR7 inhibited the MAPK pathway, significantly attenuated EGFR TKI resistance and resulted in mesenchymal to epithelial transition. CXCR7 overexpression was essential in reactivation of ERK1/2 for the generation of EGFR TKI resistant persister cells. Many NSCLC patients harboring an EGFR kinase domain mutation, who progressed on EGFR inhibitors, demonstrated increased CXCR7 expression. These data suggest that CXCR7 inhibition could considerably delay and prevent the emergence of acquired EGFR TKI resistance in EGFR mutant NSCLC.
Keywords: EGFR, Resistance, EMT, CXCR7, NSCLC
INTRODUCTION:
NSCLCs with acquired EGFR TKI resistance that present with histological transformations including Epithelial to Mesenchymal Transition (EMT) have been identified in recent clinical studies (1-4) and remain as a significant clinical problem. In vitro, NSCLC cells with an EMT phenotype are de novo resistant to EGFR TKIs despite the overexpression of mutant EGFR (5-11). EMT is a physiological program required in development, and is implicated in tumor progression and metastasis (12). Cancer cells presenting with a canonical EMT phenotype are poorly differentiated and often refractory to chemotherapy (13,14). For this subset of EGFR TKI resistant tumors, there are no specific targeted therapies and patients are limited to the conventional chemotherapeutic agents, radiation, epigenetic modifiers or the mixing of targeted agents (15-17).
We reported that acquired resistance to first generation EGFR TKIs with an EMT phenotype is a TGFβ-mediated process in HCC4006 EGFR mutant cells that can be blocked with combined inhibition of EGFR and the TGFβ receptor (18). However, the co-treatment failed to prevent acquired EGFR TKI resistance due to an increased emergence of the EGFR T790M allele compared to cells treated with TKI alone (18). Our finding underscores the difficulty in suppressing the EGFR TKI acquired resistance in NSCLC cells lines harboring EGFR kinase domain mutations as intratumoral heterogeneity gives rise to divergent resistance mechanisms in response to treatment. Furthermore, the clinical availability of third generation EGFR TKIs including osimertinib (AZD9291) that overcome the T790M mutation in NSCLC patients increases the prevalence of resistance cases with histological transformation, acquired KRAS mutation, gene fusions or an EGFR C797S mutation (19).
To date, little is known about the oncogenic drivers in EGFR mutant cells with acquired resistance with EMT. Understanding the mechanisms of resistance underlying EMT may help in developing treatment strategies for this subset of resistant NSCLC. Prior in vitro studies have identified that the receptor tyrosine kinase AXL is frequently overexpressed in EGFR TKI-resistant NSCLC cell lines with an EMT phenotype (10,17). AXL inhibition has been shown to sensitize this population to antimitotic agents but not to EGFR TKIs (17). This result suggests that sensitizing the resistant cells with EMT could potentially be difficult, and that a more thorough understanding of the molecular mechanisms by which the inhibition of mutant EGFR in NSCLC cells promotes EMT is required. Consequently, we decided to explore therapeutic targets beyond traditional TKIs and TGFβR in this subset of resistant cells with a hope to sensitize the resistant tumor to EGFR TKIs. We postulated that identifying and inhibiting an EMT-selective therapeutic target would prevent or reverse EMT and resistance to TKIs in EGFR mutant cells.
In this study, we have employed an unbiased approach to find a molecular target that could compensate for the loss of EGFR signaling in NSCLC cell lines with acquired resistance to EGFR TKIs with an EMT phenotype. We have utilized NSCLC patient samples and mouse models of acquired EGFR TKI resistance to test if our approach using these cell lines is instructive. Our studies identify a previously-unrealized molecule that can be targeted to treat or prevent the emergence of EGFR TKI resistant cancers with an EMT phenotype.
MATERIALS AND METHODS:
NSCLC cell lines and STR assays
HCC827, HCC4006, and NCI-H1975 NSCLC cells were obtained from the ATCC and maintained as specified. To generate cell lines resistant to EGFR TKIs, cells were exposed to increasing concentrations of EGFR TKIs over 6 months in a manner similar to previously described (18); however, resulting resistant cell lines are polyclonal and not clones. For EGFR TKI-resistant HCC827 cells, clones were used as described previously (18). All resistant cells are able to proliferate normally in the presence of 10 μmol/L EGFR TKIs. Upon confirming resistance, cells were cultured without drugs and their resistance to TKI was examined periodically. All the cell lines including the drug resistant and engineered cells were tested for the presence of mycoplasma and the cell authenticity using the STR assay. Results for the STR assay are listed in the Supplementary Materials and Methods.
Cell viability assays and cell counting
Live cells were counted using Countess (Thermo Fisher Scientific; heretofore TFS) and an equal number of live cells were seeded in each assay to compare cell growth kinetics using the Cell Counting Kit-8 (CCK-8) colorimetric assay (Dojindo) as previously described (18). The results were analyzed and graphed using Prism (GraphPad Software). For the crystal violet cell viability assays, cells were seeded in plates and grown to 70% confluence followed by drug treatments for the indicated times. Supernatant was removed and replaced with fixing/staining solution. Fixing/staining solution was removed, and plates were washed in dH2O, allowed to dry, and scanned for imaging.
Western blot analysis
Lysate preparation and Western blotting were performed as described previously (18). A list of antibodies is listed in the Supplementary Materials and Methods.
cDNA/shRNA/siRNA/sgRNA constructs and lentiviral infection
cDNAs were cloned into pLX304 lentiviral expression vectors (Addgene) or pBabe (Addgene). pLKO.1 shRNA lentiviral vector construct designed by the RNAi Consortium were used as described previously (18). Lentivirus coding for cDNA or shRNA were packaged in 293LTV cells (Cell Biolabs) and transduced to the target cells as previously described (18). cDNA/shRNA/siRNA/sgRNA source, sequences and RNA depletion procedures are provided in the Supplementary Materials and Methods.
RNA/DNA extraction
RNA from cell lines and fresh frozen tumor or normal lung tissues was extracted and purified using QIAshredder and RNeasy Plus (Qiagen) according to the manufacturer’s instructions. DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen). Samples were quantified using NanoDrop ND-2000 Spectrophotometer followed by Qubit (TFS).
TaqMan real-time PCR
Total RNA was converted to cDNA using a High-Capacity RNA-to-cDNA Kit (TFS). Assays were conducted using TaqMan Gene Expression Master Mix and Gene Expression Assays (TFS) for the given targets with 50ng of cDNA template. Human Beta-D-Glucuronidase (GUSB) was used as an endogenous control. Samples were run in triplicate using a QuantStudio 6 Flex Real-Time PCR System and analyzed via comparative ΔΔCT method. Probes used are listed in the Supplementary Materials and Methods.
Analysis of microarray gene expression data
Gene expression profiling methods are detailed in the Supplementary Materials and Methods.
Flow cytometry
Adherent cells were removed from plates using Accutase (TFS) and pooled. Apoptosis was assessed using an Annexin V-FLUOS Staining Kit (Sigma) according to the manufacturer’s instructions. For the detection of the cell surface expression of CXCR7 and CXCR4, pelleted cells were incubated with fluorochrome-conjugated antibodies (See Supplementary Materials and Methods), and/or DAPI. Following washing, cells were resuspended in buffer and samples were run using either an Amnis ImagestreamX Imaging Flow Cytometer or a BD LSRFortessa. Appropriate unstained and single-stained CXCR7, CXCR4, or DAPI-only samples were used for controls and compensation. Imagestream data was analyzed using IDEAS software (Amnis/EMD Millipore), while BD LSRFortessa data was analyzed with FlowJo software.
Luminex Assay
Details for Luminex-based multibead assays are available in the Supplementary Materials and Methods. Concentration of cytokines and chemokines (pg/ml) were log2 transformed and relative amount for each chemokine and cytokines was displayed using a heat map.
Immunofluorescence
Cells were grown on Nunc Lab-Tek II Chamber Slides (TFS) to 70% confluence. Following treatments, the cells were fixed in 4% Formaldehyde, permeabilized in 0.1% Triton X-100, and washed in PBS. The cells were incubated with Image-iT FX (TFS) and blocked prior to addition of anti-CXCR7 (Genetex) followed by incubation with goat anti-rabbit Alexa Fluor 546-conjugated secondary antibody and Prolong Gold Antifade Reagent with DAPI (TFS). Samples were imaged using an Olympus BX53 Scope with Hamamatsu ORCA-Flash 4.0 LT Digital Camera and analyzed with CellSens Software.
Murine drug treatment studies
All animal treatment studies were reviewed and approved by the Institutional Animal Care and Use Committee at the Dana-Farber Cancer Institute. Human EGFRDel E746_A750/T790M (TD) -inducible bitransgenic mice were generated and have been previously characterized (20). After continuous exposure to doxycycline diets for more than 8 weeks, bitransgenic mice were subjected to MRI to document the lung tumor burden. After initial imaging, mice were treated either with vehicle (10% 1-methyl-2-pyrrolidinone:90% PEG-300) alone or WZ4002, the mutant-selective EGFR TKI with activity against T790M, at 25 mg/kg gavage daily.
Biopsies of Human NSCLC
This study was carried out in accordance with the Declaration of Helsinki including all amendments, and participating hospitals required Institutional Research Ethics Board approval. All patients provided their written informed consent prior to being included in the study and before study procedures, sampling, and analyses. Following routine clinical practice, patients’ data were retrospectively obtained from patients’ medical records. All 29 EGFR mutant NSCLC patients who progressed on EGFR TKI therapy undergoing second biopsy of their tumor at Hospital Clinico Universitario and Hospital Universitario La Fe in Valencia, Spain were included in the study cohort. Samples used for analyses had to have both pre- and post-treatment tumor specimens available. 7 samples were excluded from the analysis due to the lack of sufficient materials for the IHC. The medical records were reviewed retrospectively by board-certified physicians to obtain clinical information under an IRB-approved protocol. All relevant ethical regulations were followed. The mutational status of EGFR (both primary and secondary mutations) was determined by the institutions as part of their diagnostic routine. Detailed procedures are available in Supplementary Materials and Methods.
Histology and immunohistochemistry
In mice studies, the left lung of each mouse was dissected and snap-frozen for biochemical analysis after sacrifice. The right lung was inflated with buffered 10% formalin for 10 minutes and fixed in 10% formalin overnight at room temperature. The specimen was washed once in PBS, placed in 70% ethanol, and embedded in paraffin, from which 5-μm sections were generated. All biopsy specimens from NSCLC patients were stained with H&E, Nkx2.1, and CXCR7 and reviewed by the board-certified pathologists in the Department of Pathology at Hospital Quirónsalud (Valencia, Spain). Detailed procedures and the list of antibodies used are available in the Supplementary Materials and Methods.
Statistical analysis
Unless otherwise stated, comparisons of statistical significance were performed using ANOVA & Tukey’s multiple comparison test or t-test where applicable. P < 0.05 was considered statistically significant.
RESULTS:
CXCR7 expression associates with an EMT phenotype.
Studies show that the EMT phenotype is associated with a subset of EGFR TKI resistant tumors (2,10,21). We hypothesized that an EMT-specific signaling molecule promotes persistent activation of downstream signaling and blocks the apoptosis or impaired cellular proliferation that is normally mediated by EGFR inhibition.
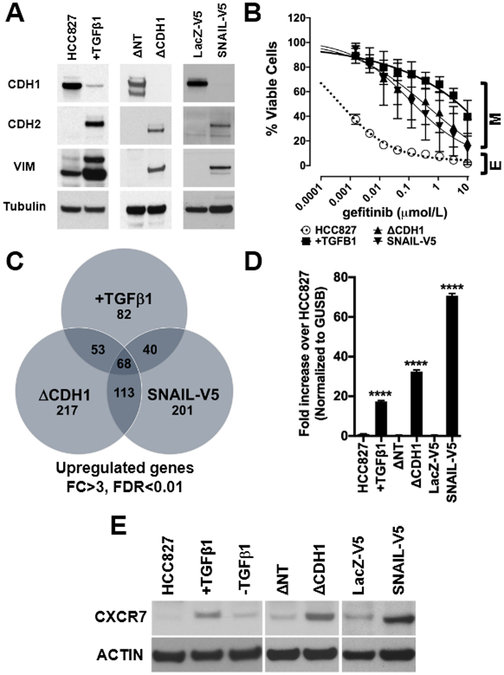
To test this hypothesis, we undertook an unbiased approach by inducing EMT in EGFR-mutated HCC827 cells by TGFβ1 stimulation (18), depleting E-cadherin by shRNA, or ectopically expressing C-terminus V5-tagged SNAIL (SNAIL-V5). The induction of EMT was confirmed by immunoblot with canonical EMT markers, demonstrating the depletion of an epithelial marker, E-cadherin (CDH1), and the increase of mesenchymal markers, N-cadherin (CDH2) and vimentin (VIM, Fig.1A). Gene set enrichment analyses (GSEA) confirmed the induction of an EMT phenotype and predicted gefitinib resistance (Fig.S1A). As we reported previously (18), the EMT induction was sufficient to promote gefitinib resistance (Fig.1B and Fig.S1B) while EGFR and ERBB3 expressions were decreased (Fig.S1C). As oncogenic drivers used to bypass EGFR signaling are often overexpressed, we identified 68 genes significantly upregulated at the mRNA level (FDR<0.01, Fold Change >3) in all HCC827 cells with the EMT induction (Fig.1C, Supplementary Table1). Among the mRNAs, typical G-Protein Coupled Receptor (GPCR), C-X-C chemokine receptor 7 (CXCR7) was significantly overexpressed. (Supplementary Table1). Consistent with this result, TaqMan RT-PCR (Fig.1D) and Western blot (Fig.1E) confirmed the CXCR7 overexpression which was TGF-β1 dependent as previously reported (22). ImageStream revealed that CXCR7 is expressed on the surface and in the cytosol of mesenchymal HCC827 cells (Fig.S1D).
Figure 1. Induction of a mesenchymal phenotype in HCC827 cells promotes overexpression of CXCR7.
A. Chronic exposure to TGFβ1 (+TGFβ1), shRNA-mediated E-cadherin depletion (ΔCDH1), and ectopic expression of SNAIL-V5, induce canonical EMT markers (CDH1: E-cadherin, CDH2: N-cadherin, VIM: vimentin) in EGFR mutant HCC827 cells. Non-target shRNA (ΔNT) and LacZ-V5 are controls. B. The induction of a mesenchymal phenotype (M) renders in HCC827 epithelial cells (E) resistant to gefitinib. (n=5, Error bars: S.D.) C. Gene expression profiling identifies commonly upregulated genes in cells with EMT (fold change >3, FDR<0.01) and identifies CXCR7 as one of 68 mutually overexpressed genes. D. Taqman qRT-PCR using CXCR7 probe in HCC827 with EMT (+TGFβ, ΔCDH1, and SNAIL-V5). (n=3, Error bars: S.D.) (****p≤0.0001). E. Immunoblot showing CXCR7 expression in EGFR mutant NSCLC cells with mesenchymal phenotype (ΔCDH1 and SNAIL-V5). ACTIN as loading control. The image is representative of three independent experiments.
CXCR7 is overexpressed in a subset of tumors from patients with EGFR mutant NSCLC.
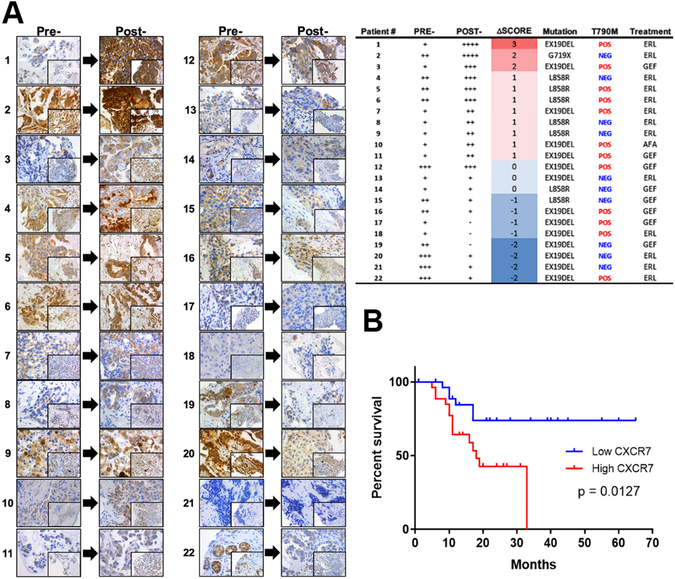
The results in NSCLC cell lines prompted us to analyze consecutive Formalin-Fixed Paraffin-Embedded (FFPE) tissue samples from 29 advanced stage EGFR mutant lung adenocarcinoma patients who radiographically progressed on EGFR TKI treatment for the presence of a T790M secondary mutation and the overexpression of CXCR7. Overall, 12 of the 22 post-treatment specimens (54.5%) harbored a T790M mutation. Of those who had sufficient and evaluable FFPE tissues (Fig.2A), board-certified pathologists scored immunohistochemical (IHC) samples for the expression of CXCR7. 11 of the 22 (50%) patients showed increased CXCR7 protein expression at the time of disease progression; of these 11 patients, 7 also harbored a T790M secondary mutation. However, we were unable to find correlations between CXCR7 expression and T790M status or progression free survival (PFS). Interestingly, 8 of the 22 post-treatment specimens (36.4%) showed decreased CXCR7 expression at the time of disease progression.
Figure 2. CXCR7 is overexpressed in EGFR mutant tumors from NSCLC patients and is an indicator of poor prognosis.
A. Immunohistochemical analyses of tumors from 22 advanced-stage NSCLC patients harboring EGFR mutation using anti-CXCR7 antibody before EGFR TKI therapies (Pre-) and at the time of clinical progression on EGFR TKI therapies (Post-). 40X and 20X (inset). Table: clinicopathological features. B. CXCR7 expression significantly (log rank p value = 0.0127) correlates with poor progression free survival (PFS) in a cohort of 55 early-stage, surgically resected lung adenocarcinoma patients.
We found significant correlation between high CXCR7 expression and poor overall survival in very few GEO datasets, most of them composed of resectable stages; GSE3141 (p=0.0027, n=111), GSE13213 (p=0.016, n=117), and GSE30219 (p=0.0001, n=307, Fig.S2A) using PROGgeneV2 (23,24). In contrast, no significant correlation was found in the TCGA lung cancer dataset. To extend these results to an unbiased and more homogeneous manner, we also assessed the impact of CXCR7 overexpression on PFS in our own cohort of surgically resected stage I/II lung adenocarcinoma patients, comparing normal adjacent tissue vs tumor in each patient by Taqman RT-PCR. All patients (n=55) underwent surgery prior to any therapies and had thorough clinical annotations. The result indicates that high CXCR7 expression levels significantly (p=0.0127) predict poorer PFS in patients with lung adenocarcinoma (Fig.2B).
Genetically engineered mouse models (GEMMs) of NSCLC harboring EGFRL858R/T790M (TL) or EGFRDel E746_A750/T790M (TD) mutation have been used extensively to develop therapeutics to overcome EGFR TKI resistance (18,20,25-29). WZ4002, an irreversible EGFR TKI, is effective in TL and TD GEMMs over a 2 to 6-week treatment; however, prolonged treatment resulted in the development of acquired resistance with an increase in tumor volume due to ERK reactivation (25,26). To investigate if the WZ4002 resistant tumors overexpress CXCR7, we treated TD tumors for 12 weeks with WZ4002 (Fig.S2B). We took serial MRIs and measured tumor volumes (30) (Fig.S2C&D). We used a threshold of 30% decrease in tumor volume to define partial response and a 20% increase in tumor size as a progressive disease based on our previous studies (30,31). At the time of resistance (12 weeks), the expression of CXCR7 was increased compared to tumors from untreated or 2-week treatment mice (Fig.S2E). While no change in epithelial marker, E-cadherin, mesenchymal markers, both vimentin and N-cadherin, increased in WZ4002-resistant tumors compared to untreated or 2-week treated tumors (Fig.S2F).
The CXCR7 signaling axis is activated in EGFR TKI resistant HCC4006Ge-R cells.
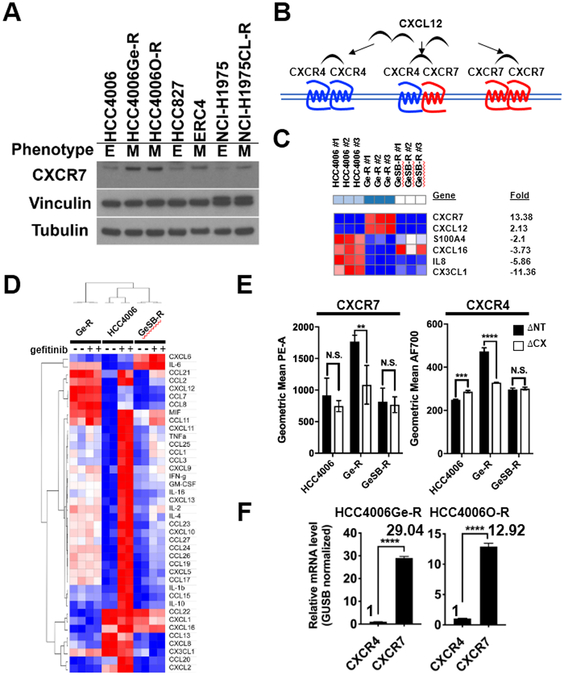
To investigate EGFR TKI-induced EMT, several EGFR mutant cell lines were grown in increasing concentrations of EGFR TKIs (Fig.3A, Fig.S3A, B and C) (18,27). CXCR7 was overexpressed in EGFR TKI-resistant mesenchymal HCC4006Ge-R or HCC4006O-R cells (heretofore Ge-R and O-R, respectively) derived by exposing EGFR TKI-sensitive, EGFR-mutated, epithelial HCC4006 cells to increasing concentrations of gefitinib or osimertinib, respectively (Fig.3A). CXCR7 was also overexpressed in a mesenchymal subset of HCC827 cells grown resistant to erlotinib (ERC4) and a mesenchymal subset of NCI-H1975 grown resistant to an irreversible EGFR TKI, CL-387,785 (NCI-H1975CL-R).
Figure 3. CXCR7 and CXCL12 are overexpressed in gefitinib-resistant HCC4006Ge-R cells with a mesenchymal phenotype.
A. Immunoblot showing CXCR7 expression with vinculin and β-tubulin as loading controls. B. Schematic diagram showing CXCL12-mediated activation of CXCR4 or CXCR7. C. A heat map for the overexpressed chemokine and cytokine genes (FDR<0.05, FC>3) in mesenchymal HCC4006Ge-R cells and epithelial HCC4006GeSB-R cells compared to parental HCC4006 cells. D. A heatmap showing relative concentration of cytokine and chemokines in HCC4006, HCC4006 Ge-R, or HCC4006GeSB-R cells treated with DMSO (−) or 500nmol/L gefitinib (+). E. Cell surface expression of CXCR7 or CXCR4 in cells expressing shRNAs against Non-Target (ΔNT) or CXCR7 (ΔCX). Average of three independent assays, (**p≤0.01, ***p≤0.001, and p<0.0001, N.S., not significant). F. The estimated expression ratio between CXCR4 and CXCR7 mRNAs in HCC4006Ge-R cells or HCC4006O-R cells calculated using GUSB as an internal control. Average of three independent assays, (****p≤0.0001).
CXCR7 could homodimerize or heterodimerize with CXCR4 upon CXCL12 stimulation (32) to activate PI3K and MAPK pathways (33); however, the CXCR7 signaling mechanisms in EGFR mutant NSCLC remain elusive (Fig.3B). EGFR mutant cell lines are instructive in delineating mechanisms of EGFR TKI acquired resistance that have been confirmed to exist in clinical samples (9,34-39). We sought to analyze the acquired EGFR TKI resistant cells with an EMT phenotype for the presence of canonical CXCR7 signaling pathways.
We reported that HCC4006 cells consist of a heterogeneous population that can be grown to develop resistance to gefitinib with an EMT phenotype (Ge-R) or with a T790M secondary mutation if concurrently grown in 1μM TGFβ receptor inhibitor, SB431542 (HCC4006GeSB-R, heretofore GeSB-R) (18). First, we used gene expression profiling followed by GSEA (40,41). The GSEA revealed significant enrichment of gefitinib-resistant, mesenchymal and TGFβ gene signatures in Ge-R cells but not in GeSB-R cells (Fig.S3D). Comparative marker selection analysis identified a marked increase in the expression of CXCR7 and its ligand CXCL12 (Fig.3C). In contrast, β-arrestin and CXCR4 expression did not meet the criteria for a significant increase in mesenchymal Ge-R cells (FDR<0.05, FC>2).
Chemokine or cytokine binding to GPCRs is a prerequisite for receptor activation and it is known that CXCL12 binds to CXCR7 with increased affinity compared to CXCR4, and CXCR7 binds CXCL12 with higher affinity than CXCL11 (33,42-44). To identify the chemokines and cytokines in the supernatants from gefitinib-resistant HCC4006 cells, we used a Luminex-based multiplex assay (Fig.3D). CXCL12, CCL7, and CCL8 were uniquely found in the supernatants from mesenchymal Ge-R but not in epithelial HCC4006 and the T790M-enriched GeSB-R. Of note, little CXCL11 was detected in the supernatant from the mesenchymal Ge-R cells.
As the CXCR7 activation by ligand occurs on the cell surface, we quantified surface expression of CXCR7 and CXCR4 in HCC4006, Ge-R and GeSB-R cells with or without the depletion of CXCR7 using validated shRNA (Fig.S3E) using fluorescently labelled anti-CXCR7 or anti-CXCR4 antibodies in FACS analyses (Fig.3E). CXCR7 was markedly overexpressed on the surface of mesenchymal Ge-R cells and lentiviral shRNA significantly depleted the expression. Modest surface CXCR7 expression was found in HCC4006 or GeSB-R thus CXCR7 shRNA did not substantially change the surface expression. Of note, the depletion of CXCR7 promoted the decrease in CXCR4 expression at protein (Fig.3E, S3F) and mRNA (Fig.S3G) levels.
Since relative quantity between CXCR7 and CXCR4 cannot be assessed by the cell surface staining using antibodies with dissimilar epitope reactivity, the quantity of CXCR7 in Ge-R cells was calculated using TaqMan RT-PCR, with expression being significantly (p<0.0001) higher than CXCR4 when normalized with endogenous control GUSB (Fig.3F).
In summary, the mesenchymal Ge-R overexpresses CXCR7 at a considerably higher level than CXCR4, suggesting that the CXCR7 homodimer might be functionally more relevant in TKI-resistant cells. The overexpression of CXCL12 that binds tightly to CXCR7 homodimers is also unique in the mesenchymal Ge-R cells.
CXCR7 maintains MAPK signaling via β-arrestins in HCC4006Ge-R cells.
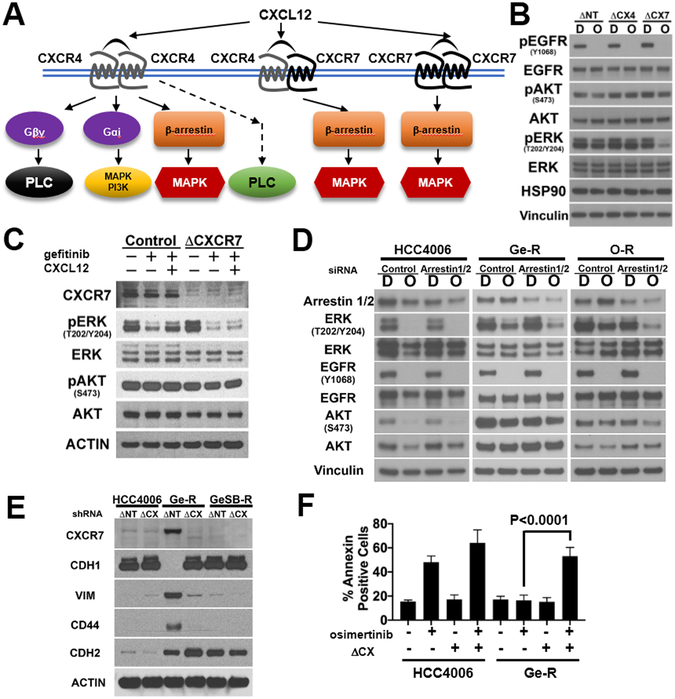
Unlike CXCR4 that predominantly transduces signals through the conventional heterotrimeric G-protein, Gαi, CXCR7 is an atypical GPCR that transduces signals through β-arrestin upon activation by CXCL12 (Fig.4A) (32,33,43). While CXCR7 homodimers are likely more prevalent than CXCR4-CXCR7 heterodimers in Ge-R cells (Fig.3F), it is unclear if the CXCR7 homodimers compensate for the loss of the EGFR signal. Lentiviral shRNAs against non-target, CXCR4, or CXCR7 were transduced to Ge-R cells and the decrease in the surface expression of CXCR4 or CXCR7 was confirmed (Fig.S4A). Ge-R cells constitutively overexpressing the shRNAs were incubated with DMSO or osimertinib (Fig.4B). As expected, CXCR4 depletion did not attenuate the phosphorylation of AKT (pAKTS437) and ERK (pERKT202/Y204), surrogate markers for PI3K and MAPK pathways, respectively. Interestingly, the phosphorylation of ERK but not AKT was markedly decreased by the depletion of CXCR7 following 500nM osimertinib exposure (Fig.4B&S4B). Similarly, combined CXCR7 depletion and EGFR inhibition in mesenchymal HCC827 cells ectopically expressing SNAIL-V5 markedly reduced ERK phosphorylation and the expression of mesenchymal markers, vimentin and N-cadherin (Fig.S4C lane 4).
Figure 4. CXCR7 activates ERK through β-arrestins to confer resistance to EGFR TKIs.
A. Schematic diagram summarizing known CXCL12-mediated activation of CXCR4 or CXCR7 signaling axis. B. HCC4006Ge-R transduced with shRNA targeting non-target (ΔNT), CXCR4 (ΔCX4), and CXCR7 (ΔCX7) were treated with DMSO (D) or 500nmol/L osimertinib (O) for 4 hours. Lysates were subject to immunoblots with antibodies indicated (HSP90 and vinculin: loading controls). A representative of three independent experiments. C. HCC4006Ge-R cells were transduced with control shRNA (ΔNT) or shRNA targeting CXCR7 (ΔCX). Cells were starved in RPMI supplemented with BSA fraction V overnight then treated with 100nmol/L gefitinib or DMSO in starvation media for 30 minutes followed by exposure to 200ng/mL SDF-1α or vehicle for an additional 30 minutes prior to harvest. Lysates were subject to immunoblot with antibodies indicated. A representative of three independent experiments. D. HCC4006, HCC4006Ge-R, and HCC4006O-R cells were transfected with scrambled siRNA (Control) or with siRNA targeting both β-arrestin1 and β-arrestin2 followed by the treatment with DMSO (D) or 500nmol/L osimertinib (O) for 4 hours. The lysates were made for the representative immunoblot from four independent experiments with antibodies indicated. E. HCC4006, HCC4006 Ge-R, or HCC4006GeSB-R cells were transduced with lentivirus coding for shRNA against non-target (ΔNT) or CXCR7 (ΔCX). The cells were passaged in media supplemented with 5μg/mL puromycin for 4 months. Lysates were subject to immunoblots with antibodies indicated. Immunoblot representative of three independent experiments. F. HCC4006 cells and HCC4006Ge-R cells transduced with lentivirus coding for shRNA against Non-Target (−) or CXCR7 (+) were treated with DMSO (−) or 500nmol/L osimertinib (+) for 48 hours and Annexin V assay was performed. Results are average of three independent assays. Error bars: S.D. *** p≤0.001.
To investigate if CXCL12 overexpressed in Ge-R cells (Fig.3D) activates CXCR7 establishing an autocrine loop, Ge-R cells constitutively overexpressing lentiviral shRNA against non-target control or CXCR7 were stimulated with CXCL12 (SDF1α) in the presence of gefitinib. In the control Ge-R cells that overexpress CXCR7 (ΔNT), gefitinib treatment reduced but maintained, phosphorylation of ERK (Fig.4C, lane 2). In the presence of SDF1α, phosphorylation of ERK increased upon gefitinib treatment in the Ge-R control cells (Fig.4C, lane 3). In contrast, ERK phosphorylation in Ge-R cells lacking CXCR7 was not maintained to the same level nor increased upon SDF1α addition (Fig.4C, lanes 5 and 6).
While CXCR7 could heterodimerize with CXCR4 or other GPCRs, or even with EGFR to activate PI3K and MAPK pathways (32,33,45), CXCR7 itself is an atypical GPCR that associates with β-arrestin 2 upon activation by either CXCL11 or CXCL12 (33,43,46) (Fig.4A). To delineate if CXCR7 regulates PI3K and MAPK pathways, we tested if the depletion of β-arrestins 1/2 would inhibit either signaling pathway. While β-arrestin 2 is known to preferentially associate with CXCR7, β-arrestin 1 also binds CXCR7 upon CXCL12 stimulation (33). Therefore, we transfected HCC4006 parental cells, Ge-R cells, or O-R cells with siRNA for a scrambled sequence or the combination of siRNAs against β-arrestins 1 and 2 (Fig.4D). Osimertinib alone was sufficient to dephosphorylate AKT and ERK in parental HCC4006 with or without depleting β-arrestins 1/2; however, osimertinib failed to dephosphorylate AKT and ERK in Ge-R and O-R cells if β-arrestins 1/2 are expressed (Fig.4D, Ge-R and O-R, lane 2). In contrast, osimertinib-mediated EGFR inhibition dephosphorylated ERK but not much AKT in the Ge-R and O-R cells with depleted β-arrestins 1/2, even though the siRNAs only decreased β-arrestins 1/2 protein levels by roughly half (Fig.4D, Ge-R and O-R lane 4). In IHC samples from the GEMM model of WZ4002 resistance, β-arrestins 1/2 were ubiquitously expressed in the course of treatment (Fig.S4D).
Interestingly, a long-term depletion of CXCR7 in the mesenchymal Ge-R cells diminished the expression of canonical mesenchymal markers, vimentin and CD44 but not N-cadherin, while increasing the expression of an epithelial marker, E-cadherin (Fig.4E, lane 4). While EGFR inhibition with gefitinib failed to suppress ERK phosphorylation in Ge-R cells transduced with lentivirus coding for shRNA targeting non-target (ΔNT) sequence (Fig.S4E, lane 6), CXCR7 depletion in Ge-R cells, followed by gefitinib treatment resulted in profound dephosphorylation of EGFR and ERK (Fig.S4E, lane 8). Furthermore, the CXCR7 depletion resulted in a marked increase in apoptosis following exposure to osimertinib 48 hrs (Fig.4F and Fig.S4F).
In summary, EGFR inhibition combined with CXCR7 depletion in the mesenchymal EGFR mutant cells attenuated β-arrestins 1/2-mediated ERK phosphorylation in the HCC4006Ge-R and O-R cells. Exposure of HCC4006Ge-R cells to CXCR7 ligand, CXCL12 (SDF1α), resulted in increased ERK phosphorylation.
CXCR7-driven MAPK signaling is necessary for the EGFR TKI resistant phenotype.
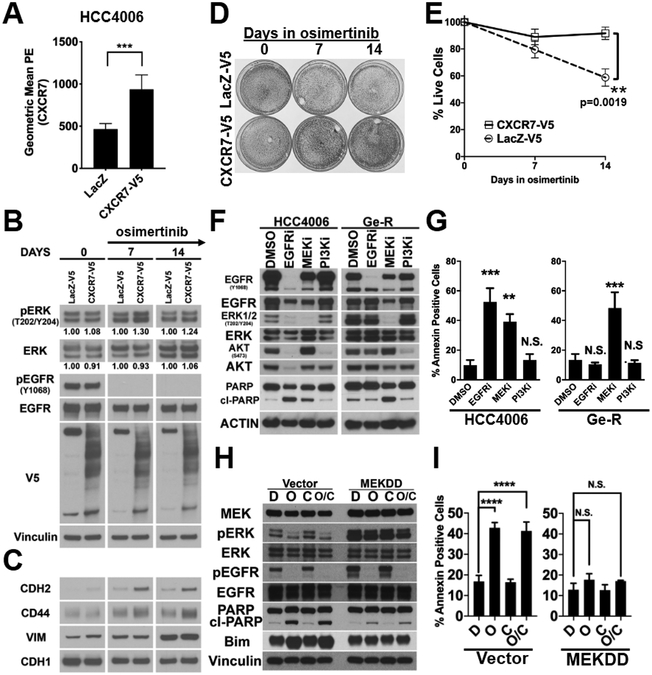
We next investigated if ectopic expression of CXCR7 in EGFR mutant cells promotes phosphorylation of ERK and EGFR TKI resistance. Lentivirus coding for V5-tagged CXCR7 (CXCR7-V5) or LacZ-V5 were transduced to HCC4006 cells and surface expression of the receptor was confirmed (Fig.5A&S5A) and by immunoblot with anti-V5 antibody (Fig.5B). The transduced cells were treated with 1 μmol/L osimertinib for the days indicated. The appearance of heavier bands that represent post-translationally modified CXCR7-V5 suggests but not proves internalization and activation of the CXCR7-V5 (Fig.5B, Lanes 2, 4 & 6). The phosphorylation of ERK was maintained in the osimertinib-treated cells ectopically overexpressing CXCR7-V5 at days 7 and 14 (Fig.5B, Lanes 4&6). Interestingly, increased expression of mesenchymal markers, N-cadherin, CD44, and vimentin was found in the cells ectopically expressing CXCR7-V5 chronically exposed to osimertinib but not in the control cells expressing LacZ-V5 (Fig.5C). Nonetheless, the increased ERK activity and the induction of mesenchymal markers in the cells ectopically expressing CXCR7-V5 coincided with more surviving cells under 7 and 14-days osimertinib treatment (Fig.5D&Fig.5E).
Figure 5. HCC4006 Ge-R cells with mesenchymal phenotype are sensitive to MEK inhibition.
A. Cell surface expression of CXCR7 in HCC4006 cells ectopically expressing LacZ-V5 or CXCR7-V5. Average of five independent assays with duplicate runs, ****p≤0.0001, ***p≤0.001. B. Lysates made from HCC4006 cells ectopically expressing LacZ-V5 or CXCR7-V5, treated with DMSO (DAY0) or 1 μmol/L osimertinib for 7 days or 14 days, were subject to immunoblots with antibodies indicted. Image is representative of three independent experiments. C. Lysates used in Fig.5B was subject to immunoblots with indicated antibodies. Image is representative of three independent experiments. D. HCC4006 cells ectopically expressing LacZ-V5 or CXCR7-V5 were treated with DMSO or 1 μmol/L osimertinib for the days indicated and surviving cells were stained with crystal violet. Image is representative of three independent assays. E. Additional set of cells were prepared and treated as described in Fig.5D. The surviving cells were counted. Average of three independent assays, p=0.0019. F. HCC4006 and Ge-R cells were treated with DMSO, 500nmol/L of gefitinib (EGFRi) or 500nmol/L trametinib (MEKi) or 500nmol/L buparlisib (PI3Ki) for 72 hours. Lysates were subjected to immunoblot with antibodies indicated. Image is representative of three independent experiments. G. HCC4006 and Ge-R cells were treated as in Fig.5F and Annexin V apoptosis assay was performed. Average of three independent assays. Bars, S.D. (***p≤0.001, **p≤0.01, *p≤0.05). H. HCC827 cells ectopically overexpressing constitutively active MEK (MEKDD) or pBabe control vector (Vector) were treated with DMSO (D), osimertinib (O), crizotinib (C) or combination (O/C) and phosphorylation of ERK1/2 for 48hours and EGFR and apoptotic markers were assessed. Image is representative of three independent experiments. I. Annexin V apoptosis assay was performed for the experiment described in Fig.5H. Results are average of three independent assays. (N.S. not significant, ****p≤0.0001). Average of three independent assays. Error Bars, S.D.
In the WZ4002-resistant EGFR TD GEMM tumors with increased CXCR7 expression (Fig.S2E), both phosho-AKT (pAKTS437) and phosho-ERK (pERKT202/Y204) are significantly increased (Fig.S5B). Since the depletion of CXCR7 or β-arrestins 1/2 suppressed MAPK signaling in our cell line models (Fig.4), we hypothesized that CXCR7-activated MAPK signaling is necessary for the survival of mesenchymal Ge-R cells. To test the hypothesis, we first challenged the HCC4006 and Ge-R cells with EGFR, MEK, or PI3-K inhibitors for 72 hours (Fig.5F). The treatment of HCC4006 with EGFR TKI induced cleaved PARP while the same treatment in Ge-R cells did not. The treatment of HCC4006 cells with MEK inhibitor promoted PARP cleavage; however, the resulting apoptosis was less than that induced by EGFR TKI (Fig.5G&S5C). Of note, the inhibition of PI3K alone was not sufficient to induce cleaved-PARP or cell death in both HCC4006 and Ge-R cells. Cell viability assays confirmed these results (Fig.S5D). Exposure of mesenchymal Ge-R or O-R cells to MEK inhibitor was the only treatment that completely eliminated the cells in the long-term cell viability assay (Fig.S5E). Similarly, HCC827 cells ectopically expressing SNAIL-V5 with CXCR7 overexpression (Fig.1A) remained sensitive to MEK inhibitor (Fig.S5F).
It has not been demonstrated if ectopic activation of the MAPK pathway is sufficient for survival. On the basis of our results that CXCR7 drives the MAPK pathway but not PI3K pathway (Fig.4B&5B), we wanted to investigate if the CXCR7-activated MAPK pathway is sufficient to confer resistance in EGFR mutant NSCLC. Constitutively active MEK, MEKDD, was ectopically expressed in EGFR TKI sensitive HCC827. Since HCC827 cells have a propensity to develop EGFR TKI resistance MET amplification (39), we performed apoptosis assays with or without the MET inhibitor crizotinib. In HCC827 cells constitutively overexpressing vector control, osimertinib treatment, regardless of the use of crizotinib, resulted in dephosphorylation of ERK and induction of Bim and cleaved-PARP (Fig.5H, Lane4). In contrast, osimertinib treatment of HCC827 cells constitutively overexpressing MEKDD did not diminish phosphorylation of ERK and induced little Bim and cleaved-PARP (Fig.5H, Lane8). This result was also confirmed with Annexin V apoptosis assays (Fig.5I, Fig.S5G).
In summary, ectopic overexpression of CXCR7-V5 in EGFR mutant cells promoted survival under the inhibition of EGFR by activating the MAPK-ERK signaling that is directly linked to the survival of the EGFR TKI resistant cells overexpressing CXCR7.
CXCR7 inhibition potentiates EGFR inhibition in EGFR mutant NSCLC cells by limiting ERK reactivation.
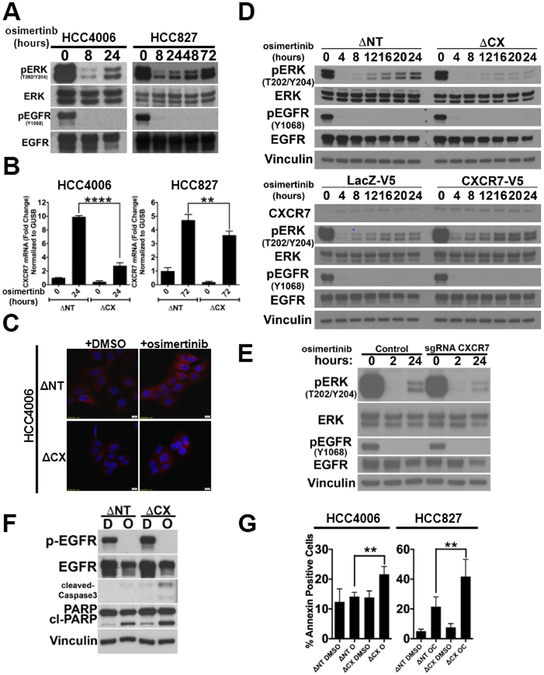
EGFR TKI-resistant cells with a secondary T790M EGFR mutation can both pre-exist and/or emerge from drug-tolerant cells called persisters (47). We and others reported that exposure of EGFR mutant NSCLC cells (epithelial) to an EGFR TKI results in the emergence of persisters in the short term (16,18,47-49). Previous studies have shown that ERK is reactivated in the persisters and EGFR-MEK dual inhibition significantly delays their emergence (25,26). We have reproduced ERK reactivation in HCC4006 and HCC827 cells (Fig.6A) and realized that the ERK reactivation occurs in parallel with the increase in CXCR7 expression increase in the persister cells (Fig.6B). The overexpression of shRNA targeting CXCR7 in HCC4006 and HCC827 cells, while incomplete, significantly impaired the induction of CXCR7 following 24hr (HCC4006) or 72hr (HCC827) osimertinib treatment (Fig.6C). Based on our results that CXCR7 activated the MAPK pathway (Fig.4) in long-term resistance, we hypothesized that CXCR7 reactivates the MAPK pathway in persister EGFR mutant NSCLC cells.
Figure 6. CXCR7 depletion in EGFR mutant cells attenuates ERK reactivation following EGFR TKI treatment.
A. HCC4006 or HCC827 cells were treated with 100nM gefitinib for the indicated time and phosphorylation of ERK1/2 and EGFR was assessed by immunoblot. A representative of three independent experiments. B. HCC4006 or HCC827 cells were transduced with shRNA targeting non-target (ΔNT) or shRNA against CXCR7 (ΔCX). The cells were treated with 100nmol/L osimertinib and TaqMan RT-PCR assay with CXCR7 probe was performed. GUSB as the reference control. Average of three independent assays with duplicate runs, ****p≤0.0001, **p≤0.01. C. HCC4006 cells stably expressing shRNA targeting non-target (ΔNT) or shRNA against CXCR7 (ΔCX) were treated with DMSO or 100nmol/L gefitinib for 8 hours. CXCR7 expression was measured using immunofluorescence. A representative image from three independent experiments. Red: CXCR7, Blue: DAPI. D. HCC4006 cells (Top) stably expressing shRNA targeting non-target (ΔNT) or shRNA against CXCR7 (ΔCX) or (Bottom) ectopically expressing LacZ-V5 or CXCR7-V5 were treated with DMSO or 100nmol/L osimertinib for the indicated hours. Lysates were made and subject to immunoblot with antibodies indicated. A representative image from three independent experiments. E. HCC4006 cells were stably transfected with vectors coding for the indicated single-guided RNA (sgRNA). The cells were treated with 100nmol/L osimertinib for the indicated time. Lysates were subject to immunoblots with antibodies indicated. A representative of three independent experiments. F. HCC4006 cells stably expressing shRNA targeting non-target (ΔNT) or shRNA against CXCR7 (ΔCX) were treated with DMSO (D) or 100nmol/L osimertinib (O) for 24 hours and apoptosis was assessed using cleaved PARP and cleaved-caspase3 in Western blot. Vinculin was used as loading control. A representative image from three independent experiments. G. (Left) HCC4006 cells or (right) HCC827 cells stably overexpressing shRNA against non-target (ΔNT) or CXCR7 (ΔCX) were treated with DMSO control or combination of 100 nmol/L osimertinib for 24 hours (HCC4006) or a combination of 100 nmol/L osimertinib and 10 nmol/L crizotinib (AC) for 72 hours (HCC827). Annexin V assay was performed and % change in annexin positive cells was calculated relative to ΔNT overexpressing parental cells treated with DMSO. (average of three independent experiments, mean ± SD, **p≤0.01).
We then treated EGFR TKI sensitive HCC4006 cells with or without impaired CXCR7 expression with osimertinib to monitor ERK phosphorylation (Fig.6D top). While the reactivation of ERK was evident in the control cells with CXCR7 expression, it was significantly attenuated in the cells with CXCR7 depletion. We have also developed CXCR7-depleted mass culture of HCC4006 cell lines using CRISPR/Cas9-mediated biallelic disruption of the CXCR7 locus to obtain identical results (Fig.6E&S6A). Similar results were obtained in HCC827 cells (Fig.S6B), PC9 cells (Fig.S6C), NCI-H1975 cells (Fig.S6D). In HCC4006 cells ectopically expressing CXCR7-V5, the ERK reactivation in drug tolerant cells was more profound than that in the control HCC4006 cells expressing LacZ-V5 (Fig.6D bottom). Similarly, HCC827 cells ectopically overexpressing CXCR7-V5 (Fig.S6E) resulted in the increased maintenance of phosphorylated ERK upon EGFR inhibition even after 24 hours of osimertinib/crizotinib combination treatment.
Motivated by these findings, we sought to investigate if CXCR7 depletion potentiates efficient cell death in EGFR TKI treated NSCLC cells. HCC4006 cells constitutively overexpressing shRNA against non-target or CXCR7 were treated with osimertinib and apoptotic markers were assessed (Fig.6F). Annexin V assay demonstrated significantly (p<0.01) increased apoptosis by osimertinib treatment in HCC4006 cells (Fig.6G left and S6F) or HCC827 cells (Fig.6G right and S6G) where normal EGFR TKI-induced CXCR7 production is attenuated by shRNA depletion.
In summary, our results demonstrate that CXCR7 expression is an early event following EGFR inhibition and depletion of CXCR7 hampers the efficient reactivation of ERK resulting in increased apoptosis. The expression of CXCR7 following EGFR inhibition was a ubiquitous event among the EGFR mutant NSCLC cells that we tested. Ectopic expression of CXCR7 in HCC4006 cells potentiated the reactivation of ERK following initial EGFR inhibition.
DISCUSSION:
While the G-protein coupled receptor (GPCR) family represents the largest family of membrane receptors, their therapeutic values have not been extensively explored in NSCLC. The up-regulation of tri-heteromeric GPCR, adenosine A2a receptor, in gefitinib resistant NSCLC cell models has been reported (50); however, its signaling mechanisms and therapeutic potential remain unknown. Our analyses of the EMT-promoted EGFR mutant NSCLC cell line models revealed that (GPCR), C-X-C chemokine receptor 7 (CXCR7), is ubiquitously overexpressed in acquired EGFR TKI resistant cell line models with an EMT phenotype (Fig.1E and 3A). Coincidentally, 50% of specimens from EGFR mutation-positive NSCLC patients who progressed on EGFR TKI treatment exhibit increased CXCR7 expression (Fig.2A). Furthermore, CXCR7 expression correlates with poor progression-free survival in treatment-naïve, early-stage NSCLC patients regardless of their EGFR or any other oncogene (i.e., KRAS) statuses, suggesting CXCR7 activity is important in tumor progression and relapse (Fig.2B and Fig.S2A). However, the correlation between CXCR7 expression and T790M status or PFS was not evident in our EGFR-mutant patient cohort (Fig.2A). This discrepancy is due to the differences in disease stages and sample size of the specimens used for the IHC studies in Fig.2A compared to survival analyses displayed in Fig.2B and Fig.S2A. Nonetheless, the overexpression of CXCR7 was also evident in the tumors from transgenic murine models with EGFR TKI acquired resistance (Fig.S2) supporting its implications in the acquired resistance.
These clinical samples indicate CXCR7 overexpression occurs with or without T790M secondary mutations in tumors from NSCLC patients who progressed on EGFR TKI treatment samples (Fig.2A). From the limited amount of patient specimens, it was unclear if the CXCR7 overexpression and T790M emerge in the same subpopulation or in distinct subsets. However, our EGFR TKI acquired resistance study using HCC4006 cells revealed that the cells with T790M or a mesenchymal phenotype exist as different populations and the treatments dictate the emergence of dominant resistance mechanisms (18). Thus, the types of drug treatment or environmental cues from the tumor microenvironment might impact the dominant EGFR TKI resistance mechanisms. Further studies are necessary in a larger cohort of patient samples to correlate basal CXCR7 expression with the TKI response, or to understand why tumors with high expression of CXCR7 prior to EGFR TKI therapy showed a decrease in expression following the treatment.
CXCR7 is an atypical GPCR and the CXCR7 homodimer associates with β-arrestin2 instead of the conventional heterotrimeric G-protein, Gαi, upon activation by either CXCL11 or CXCL12 (43). Accumulating evidence suggests that CXCR7 plays pivotal roles in metastasis (22,51,52), angiogenesis (53,54), proliferation of cancer cells (33,55,56) and de novo drug resistance (57,58). CXCR7 overexpression has recently been linked to acquired enzalutamide resistant prostate cancer (59) by activating MAPK signaling pathway in a ligand-independent fashion (60). While CXCR7 is linked to TGFβ1-induced EMT in lung cancer cell line models (61), its roles in acquired drug resistance, and in particular, its therapeutic value in lung cancer has not been fully elucidated.
Our results demonstrate that the CXCL12-CXCR7 axis is necessary for the activation of the MAPK signaling (Fig.4D, Fig.4E and S4C) and ectopic expression of CXCR7 is sufficient to increase the MAPK signaling when the CXCR7 overexpressing cells were exposed to osimertinib (Fig.5A and Fig.5B). The ectopic CXCR7 overexpression also increased the expression of mesenchymal markers (Fig.5C) and promoted resistance to osimertinib (Fig.5D and Fig.5E). Ge-R and O-R cells with CXCR7 overexpression (Fig.3A) are exquisitely sensitive to MEK inhibitors (Fig.5F and G) and aberrant activation of MEK in otherwise EGFR TKI-sensitive HCC4006 is sufficient for conferring resistance to EGFR TKI (Fig.5H and I). These results cumulatively indicate CXCR7 activates the MAPK signaling to promote EMT and EGFR TKI resistance. CXCR7 could heterodimerize with other GPCRs, or even bind to EGFR (32,45) to activate MAPK pathways (33); however, the activation of the MAPK signaling by CXCR7 occurs under the EGFR inhibition with TKIs. Accordingly, this resistance mechanism resembles the EGFR TKI acquired resistance with MET amplification (39,62), in which activation of MET sustains downstream signaling despite inhibition of oncogenic mutant EGFR.
Several questions remain unanswered regarding the regulation of EMT by CXCR7 (Fig.5C). While CXCR4 is known to promote EMT (63-65), the depletion of CXCR7 alone was sufficient to reverse the EMT phenotype in our model (Fig.4E). While CXCR4 expression was significantly lower than CXCR7 (Fig.3F), the depletion of CXCR7 itself attenuated the expression of CXCR4 (Fig.3E, right). It is likely that the decreased expression of CXCR4 is due to the mesenchymal to epithelial transition following CXCR7 depletion; however, additional studies are required to dissect the CXCR7-CXCR4 interaction in this model. Additionally, further study is required to investigate if CXCR7-driven MAPK signaling exclusively activates EMT promoting transcription factors or other pathways are involved in the activation. We have shown that exposure of CXCR7-overexpressing Ge-R cells to CXCL12 increases ERK phosphorylation (Fig.4C) and that these cells also express several other chemokines and cytokines known to activate CXCR7 (Fig.3D). Additional experiments are required to elucidate if specific chemokines preferentially promote EMT by activating CXCR7.
With the introduction of third generation EGFR TKIs that overcome the T790M mutation in NSCLC patients (50%), the prevalence of resistance cases with pathological transformation including EMT may rise. This underscores the need for new biomakers and improved therapeutic strategies for this subset of patients. Here, we demonstrated that co-targeting EGFR and CXCR7 may be an effective combination strategy against EGFR TKI resistant cells with a mesenchymal phenotype. The depletion of CXCR7 not only re-sensitized the TKI resistant EGFR mutant NSCLC with a mesenchymal phenotype to EGFR TKI (Fig.4F), but also promoted partial mesenchymal to epithelial transition (Fig.4E). These results support the unique concept that the combination of osimertinib and CXCR7 inhibitor might be particularly effective in NSCLC patients who developed resistance to first line EGFR TKIs, as a significant number of patients who received the treatment developed resistant tumors with both T790M and CXCR7 overexpression (Fig.2). CXCR7 and EGFR dual inhibition can also be applied for the prevention of drug resistance, which offers broad clinical applicability, as our results demonstrate that the EGFR inhibition combined with CXCR7 depletion diminishes ERK reactivation (Fig.6). We have previously reported that a surviving population of TKI-naïve EGFR mutant NSCLC cells treated with EGFR TKIs, so-called persister cells, secrete TGFβ upon EGFR TKI treatment priming the surviving cells for EMT(18). Surprisingly, the overexpression of CXCR7 following EGFR TKI treatment was observed in persisting populations from 4 different EGFR mutant NSCLC cell lines (Fig.6). The depletion of CXCR7 in the persister cells suppressed ERK reactivation following EGFR inhibition and the EGFR-CXCR7 dual inhibition promoted a significant increase in cell death. Taken together, our study gives credence to the future development of potent CXCR7 inhibitors and provides guidance as to where CXCR7-inhibition should be evaluated, specifically either as initial therapy in EGFR TKI–näive patients or in those who have developed EGFR TKI resistance with a mesenchymal phenotype. The proposed concurrent inhibition of EGFR and CXCR7 may offer significant advantages over co-targeting EGFR and MEK, as CXCR7 overexpression seems to be specific to a subset of NSCLC cells compared to the more ubiquitously expressed MEK allowing for more selective targeting while attenuating toxicity.
Supplementary Material
SIGNIFICANCE.
Increased expression of the chemokine receptor CXCR7 constitutes a mechanism of resistance to EGFR tyrosine kinase inhibition in patients with non-small cell lung cancer through reactivation of ERK signaling.
ACKNOWLEDGEMENTS:
We would like to acknowledge the INCLIVA Biobank (PT13/0010/0004) integrated in the Valencian Biobanking Network and the Spanish National Biobanks Network for its collaboration. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
GRANT SUPPORT:
This work is supported by American Cancer Society Illinois Division Basic Science Grant #254563 (T.S.), the American Cancer Society Research Scholar Grant, 126638-RSG-14-229-01-TBG (T.S.), MINECO (SAF2013-49056, SAF2017-85352, J.C.), and Domingo Martinez Foundation (J.C.), National Institutes of Health grants CA140594, CA122794, CA163896, CA166480 (K.K.W), CA114465, CA135257, and CA154303 (P.A.J.).
Footnotes
CONFLICT OF INTEREST DISCLOSURE STATEMENT
On behalf of all authors on this manuscript, we disclose the following conflicts of interest. All authors have completed separate conflict of interest forms, and we have no additional conflicts of interest to report.
Takeshi Shimamura
Consultant/Advisory Board/Ownership
K-K.Wong is a founder and equity holder of G1 therapeutics. K-K.Wong reports receiving commercial research grants from MedImmune, Takeda, TargImmune, and BMS. K-K.Wong also reports consulting & Sponsored Research Agreements with AstraZeneca, Janssen, Pfizer, Novartis, Merck, Ono, and Array.
P.A. Jänne reports receiving commercial research grants from AstraZeneca, Boehringer Ingelheim, Daiichi Sankyo, Astellas Pharmaceuticals, Eli Lilly, Takeda Oncology and PUMA; holds ownership interest (including patents) in Gatekeeper Pharmaceuticals and LOXO Oncology; and is a consultant/advisory board member for AstraZeneca, Boehringer-Ingelheim, Pfizer, Roche/Genentech, Merrimack Pharmaceuticals, Chugai, Ariad, Ignyta, LOXO Oncology, SFJ Pharmaceuticals, Voronoi, Takeda Oncology, Daiichi Sankyo, Novartis, Eli-Lilly Biocartis, and Mirati Therapeutics. Dr. Jänne receives post-marketing royalties from DFCI owned intellectual property on EGFR mutations licensed to Lab Corp
Amelia Insa reports receiving honoraria from or advisory roles in Pfizer, Roche, Bristol-Myers Squibb, Boeringher Ingelheim.
Paloma Martin reports receiving honoraria from or advisory roles in Boehringer Ingelheim, AstraZeneca, Roche, Bristol-Myers Squibb.
Oscar Juan reports receiving honoraria from or advisory roles in Boehringer Ingelheim, Bristol-Myers Squibb, Merck Sharp & Dohme, Roche/Genetech, AstraZeneca, Pfizer, Eli Lilly, Abbvie.
Research Funding: Bristol-Myers Squibb, AstraZeneca.
Javier Alcacer reports receiving research Funding from Imegen.
No potential conflicts of interest were disclosed by the other authors.
REFERENCES
- 1.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Science translational medicine 2011;3:75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uramoto H, Iwata T, Onitsuka T, Shimokawa H, Hanagiri T, Oyama T. Epithelial-mesenchymal transition in EGFR-TKI acquired resistant lung adenocarcinoma. Anticancer Res 2010;30:2513–7 [PubMed] [Google Scholar]
- 4.Chung JH, Rho JK, Xu X, Lee JS, Yoon HI, Lee CT, et al. Clinical and molecular evidences of epithelial to mesenchymal transition in acquired resistance to EGFR-TKIs. Lung Cancer 2011;73:176–82 [DOI] [PubMed] [Google Scholar]
- 5.Coldren CD, Helfrich BA, Witta SE, Sugita M, Lapadat R, Zeng C, et al. Baseline gene expression predicts sensitivity to gefitinib in non-small cell lung cancer cell lines. Molecular cancer research : MCR 2006;4:521–8 [DOI] [PubMed] [Google Scholar]
- 6.Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer research 2005;65:9455–62 [DOI] [PubMed] [Google Scholar]
- 7.Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, et al. Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer research 2006;66:944–50 [DOI] [PubMed] [Google Scholar]
- 8.Yauch RL, Januario T, Eberhard DA, Cavet G, Zhu W, Fu L, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research 2005;11:8686–98 [DOI] [PubMed] [Google Scholar]
- 9.Suda K, Tomizawa K, Fujii M, Murakami H, Osada H, Maehara Y, et al. Epithelial to mesenchymal transition in an epidermal growth factor receptor-mutant lung cancer cell line with acquired resistance to erlotinib. J Thorac Oncol 2011;6:1152–61 [DOI] [PubMed] [Google Scholar]
- 10.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clinical cancer research : an official journal of the American Association for Cancer Research 2013;19:279–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nature genetics 2012;44:852–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation 2009;119:1420–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010;141:69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochimica et biophysica acta 2009;1796:75–90 [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Daemen A, Hatzivassiliou G, Arnott D, Wilson C, Zhuang G, et al. Metabolic and transcriptional profiling reveals pyruvate dehydrogenase kinase 4 as a mediator of epithelial-mesenchymal transition and drug resistance in tumor cells. Cancer Metab 2014;2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson C, Nicholes K, Bustos D, Lin E, Song Q, Stephan JP, et al. Overcoming EMT-associated resistance to anti-cancer drugs via Src/FAK pathway inhibition. Oncotarget 2014;5:7328–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson C, Ye X, Pham T, Lin E, Chan S, McNamara E, et al. AXL Inhibition Sensitizes Mesenchymal Cancer Cells to Antimitotic Drugs. Cancer research 2014;74:5878–90 [DOI] [PubMed] [Google Scholar]
- 18.Soucheray M, Capelletti M, Pulido I, Kuang Y, Paweletz CP, Becker JH, et al. Intratumoral Heterogeneity in EGFR-Mutant NSCLC Results in Divergent Resistance Mechanisms in Response to EGFR Tyrosine Kinase Inhibition. Cancer research 2015;75:4372–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 2009;462:1070–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song KA, Niederst MJ, Lochmann TL, Hata AN, Kitai H, Ham J, et al. Epithelial-to-Mesenchymal Transition Antagonizes Response to Targeted Therapies in Lung Cancer by Suppressing BIM. Clinical cancer research : an official journal of the American Association for Cancer Research 2018;24:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu PF, Huang Y, Xu CL, Lin LY, Han YY, Sun WH, et al. Downregulation of CXCL12 in mesenchymal stromal cells by TGFbeta promotes breast cancer metastasis. Oncogene 2017;36:840–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goswami CP, Nakshatri H. PROGgene: gene expression based survival analysis web application for multiple cancers. J Clin Bioinforma 2013;3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer 2014;14:970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tricker EM, Xu C, Uddin S, Capelletti M, Ercan D, Ogino A, et al. Combined EGFR/MEK Inhibition Prevents the Emergence of Resistance in EGFR-Mutant Lung Cancer. Cancer discovery 2015;5:960–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ercan D, Xu C, Yanagita M, Monast CS, Pratilas CA, Montero J, et al. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer discovery 2012;2:934–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimamura T, Li D, Ji H, Haringsma HJ, Liniker E, Borgman CL, et al. Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer research 2008;68:5827–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Shimamura T, Ji H, Chen L, Haringsma HJ, McNamara K, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell 2007;12:81–93 [DOI] [PubMed] [Google Scholar]
- 29.Ji H, Li D, Chen L, Shimamura T, Kobayashi S, McNamara K, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell 2006;9:485–95 [DOI] [PubMed] [Google Scholar]
- 30.Xu L, Kikuchi E, Xu C, Ebi H, Ercan D, Cheng KA, et al. Combined EGFR/MET or EGFR/HSP90 inhibition is effective in the treatment of lung cancers codriven by mutant EGFR containing T790M and MET. Cancer research 2012;72:3302–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z, Sasaki T, Tan X, Carretero J, Shimamura T, Li D, et al. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer research 2010;70:9827–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Decaillot FM, Kazmi MA, Lin Y, Ray-Saha S, Sakmar TP, Sachdev P. CXCR7/CXCR4 heterodimer constitutively recruits beta-arrestin to enhance cell migration. J Biol Chem 2011;286:32188–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Martin L, Sanchez-Mateos P, Cabanas C. CXCR7 impact on CXCL12 biology and disease. Trends in molecular medicine 2013;19:12–22 [DOI] [PubMed] [Google Scholar]
- 34.Engelman JA. The role of phosphoinositide 3-kinase pathway inhibitors in the treatment of lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2007;13:s4637–40 [DOI] [PubMed] [Google Scholar]
- 35.Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proceedings of the National Academy of Sciences of the United States of America 2005;102:3788–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. The Journal of clinical investigation 2006;116:2695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ercan D, Zejnullahu K, Yonesaka K, Xiao Y, Capelletti M, Rogers A, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene 2010;29:2346–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786–92 [DOI] [PubMed] [Google Scholar]
- 39.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039–43 [DOI] [PubMed] [Google Scholar]
- 40.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics 2003;34:267–73 [DOI] [PubMed] [Google Scholar]
- 41.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balabanian K, Lagane B, Infantino S, Chow KY, Harriague J, Moepps B, et al. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem 2005;280:35760–6 [DOI] [PubMed] [Google Scholar]
- 43.Burns JM, Summers BC, Wang Y, Melikian A, Berahovich R, Miao Z, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med 2006;203:2201–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cruz-Orengo L, Holman DW, Dorsey D, Zhou L, Zhang P, Wright M, et al. CXCR7 influences leukocyte entry into the CNS parenchyma by controlling abluminal CXCL12 abundance during autoimmunity. J Exp Med 2011;208:327–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh RK, Lokeshwar BL. The IL-8-regulated chemokine receptor CXCR7 stimulates EGFR signaling to promote prostate cancer growth. Cancer research 2011;71:3268–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coggins NL, Trakimas D, Chang SL, Ehrlich A, Ray P, Luker KE, et al. CXCR7 controls competition for recruitment of beta-arrestin 2 in cells expressing both CXCR4 and CXCR7. PLoS One 2014;9:e98328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 2010;29:4741–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhang HE, Ruddy DA, Krishnamurthy Radhakrishna V, Caushi JX, Zhao R, Hims MM, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med 2015;21:440–8 [DOI] [PubMed] [Google Scholar]
- 50.Kuzumaki N, Suzuki A, Narita M, Hosoya T, Nagasawa A, Imai S, et al. Multiple analyses of G-protein coupled receptor (GPCR) expression in the development of gefitinib-resistance in transforming non-small-cell lung cancer. PLoS One 2012;7:e44368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian T, Liu Y, Dong Y, Zhang L, Dong Y, Sun Y, et al. CXCR7 regulates breast tumor metastasis and angiogenesis in vivo and in vitro. Mol Med Rep 2018;17:3633–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stacer AC, Fenner J, Cavnar SP, Xiao A, Zhao S, Chang SL, et al. Endothelial CXCR7 regulates breast cancer metastasis. Oncogene 2016;35:1716–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miao Z, Luker KE, Summers BC, Berahovich R, Bhojani MS, Rehemtulla A, et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proceedings of the National Academy of Sciences 2007;104:15735–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang M, Qiu L, Zhang Y, Xu D, Zheng JC, Jiang L. CXCL12 enhances angiogenesis through CXCR7 activation in human umbilical vein endothelial cells. Sci Rep 2017;7:8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kallifatidis G, Munoz D, Singh RK, Salazar N, Hoy JJ, Lokeshwar BL. beta-Arrestin-2 Counters CXCR7-Mediated EGFR Transactivation and Proliferation. Mol Cancer Res 2016;14:493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saha A, Ahn S, Blando J, Su F, Kolonin MG, DiGiovanni J. Proinflammatory CXCL12-CXCR4/CXCR7 Signaling Axis Drives Myc-Induced Prostate Cancer in Obese Mice. Cancer research 2017;77:5158–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hattermann K, Held-Feindt J, Lucius R, Muerkoster SS, Penfold ME, Schall TJ, et al. The chemokine receptor CXCR7 is highly expressed in human glioma cells and mediates antiapoptotic effects. Cancer research 2010;70:3299–308 [DOI] [PubMed] [Google Scholar]
- 58.Waldschmidt JM, Simon A, Wider D, Muller SJ, Follo M, Ihorst G, et al. CXCL12 and CXCR7 are relevant targets to reverse cell adhesion-mediated drug resistance in multiple myeloma. Br J Haematol 2017;179:36–49 [DOI] [PubMed] [Google Scholar]
- 59.Luo Y, Azad AK, Karanika S, Basourakos SP, Zuo X, Wang J, et al. Enzalutamide and CXCR7 inhibitor combination treatment suppresses cell growth and angiogenic signaling in castration-resistant prostate cancer models. International journal of cancer Journal international du cancer 2018;142:2163–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li S, Fong KW, Gritsina G, Zhang A, Zhao JC, Kim J, et al. Activation of MAPK signaling by CXCR7 leads to enzalutamide resistance in prostate cancer. Cancer research 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu YC, Tang SJ, Sun GH, Sun KH. CXCR7 mediates TGFbeta1-promoted EMT and tumor-initiating features in lung cancer. Oncogene 2015 [DOI] [PubMed] [Google Scholar]
- 62.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proceedings of the National Academy of Sciences of the United States of America 2007;104:20932–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cives M, Quaresmini D, Rizzo FM, Felici C, D’Oronzo S, Simone V, et al. Osteotropism of neuroendocrine tumors: role of the CXCL12/ CXCR4 pathway in promoting EMT in vitro. Oncotarget 2017;8:22534–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tu Z, Xie S, Xiong M, Liu Y, Yang X, Tembo KM, et al. CXCR4 is involved in CD133-induced EMT in non-small cell lung cancer. Int J Oncol 2017;50:505–14 [DOI] [PubMed] [Google Scholar]
- 65.Yin H, Wang Y, Chen W, Zhong S, Liu Z, Zhao J. Drug-resistant CXCR4-positive cells have the molecular characteristics of EMT in NSCLC. Gene 2016;594:23–9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.