Abstract
Magnesium transporter 1 (MAGT1) critically mediates magnesium homeostasis in eukaryotes and is highly-conserved across different evolutionary branches. In humans, loss–of–function mutations in the MAGT1 gene cause X-linked magnesium deficiency with Epstein-Barr virus (EBV) infection and neoplasia (XMEN), a disease that has a broad range of clinical and immunological consequences. We have previously shown that EBV susceptibility in XMEN is associated with defective expression of the antiviral natural-killer group 2 member D (NKG2D) protein and abnormal Mg2+ transport. New evidence suggests that MAGT1 is the human homolog of the yeast OST3/OST6 proteins that form an integral part of the N-linked glycosylation complex, although the exact contributions of these perturbations in the glycosylation pathway to disease pathogenesis are still unknown. Using MS-based glycoproteomics, along with CRISPR/Cas9-KO cell lines, natural killer cell-killing assays, and RNA-Seq experiments, we now demonstrate that humans lacking functional MAGT1 have a selective deficiency in both immune and nonimmune glycoproteins, and we identified several critical glycosylation defects in important immune-response proteins and in the expression of genes involved in immunity, particularly CD28. We show that MAGT1 function is partly interchangeable with that of the paralog protein tumor-suppressor candidate 3 (TUSC3) but that each protein has a different tissue distribution in humans. We observed that MAGT1-dependent glycosylation is sensitive to Mg2+ levels and that reduced Mg2+ impairs immune-cell function via the loss of specific glycoproteins. Our findings reveal that defects in protein glycosylation and gene expression underlie immune defects in an inherited disease due to MAGT1 deficiency.
Keywords: transporter, glycoprotein, glycosylation, immunodeficiency, infection, magnesium, immunology, Epstein-Barr virus (EBV) infection, magnesium transporter 1 (MAGT1), N-linked glycosylation (NLG) complex, oligosaccharyltransferase (OST), X-linked magnesium deficiency with Epstein–Barr virus infection and neoplasia (XMEN)
Introduction
MAGT1 is an evolutionally conserved Mg2+-specific ion transport facilitator found in all animals and has been shown to participate in the multienzyme complex responsible for enzymatic coupling of N-glycans onto peptide substrates (1, 2). Null mutations in the MAGT1 gene lead to the rare primary immunodeficiency “X-linked immunodeficiency with Mg2+ defect, Epstein-Barr virus (EBV)7 infection and neoplasia” (XMEN) disease (3, 4). Here, we explore these dual roles by examining cells from both healthy and MAGT1-deficient humans.
Mg2+ is the most abundant divalent cation in eukaryotic cells, with intracellular concentrations ranging from 15 to 20 mm depending on the cell type. Most Mg2+ is tightly bound to cellular substituents, especially nucleic acids, nucleoside triphosphates, and enzymes. The unbound intracellular free Mg2+ is estimated to be 0.4–1.0 mm or ∼1–5% of the total Mg2+ concentration in the cell (5, 6), and because Mg2+ is the biologically active form of Mg, these intracellular concentrations are tightly regulated (5). Despite its critical importance, regulatory functions of Mg2+ remain mostly unknown (3). Our previous work showed that MAGT1 deficiency has two main consequences in T cells: 1) the loss of a T-cell receptor (TCR)–induced Mg2+ flux with resulting suboptimal T-cell activation, and 2) a reduced basal level of intracellular free Mg2+ (3). Recently, Gilmore and co-workers (7) have provided evidence in nonlymphoid tumor lines that a reservoir of MAGT1 is located in the endoplasmic reticulum (ER) and associates with the multisubunit enzymatic complex known as the oligosaccharyltransferase (OST). The OST is the primary mediator of N-linked glycosylation (NLG), and hence perturbations to its subunit composition (e.g. via genetic alterations in MAGT1) may alter glycosylation and/or levels of a subset of N-glycosylation substrates. We wanted to examine whether these functions are important in human nontransformed lymphocytes.
We have shown that chronically low intracellular free Mg2+ is associated with reduced glycosylation and cell-surface expression of NKG2D (also known as killer cell lectin-like receptor K1 or KLRK1), an activating receptor involved in antiviral and antitumor cytotoxicity that is expressed on CD8+ T cells and natural killer (NK) cells (8, 9). NKG2D is especially important for antiviral responses against EBV, an oncogenic virus and ubiquitous human pathogen (9). The dramatically lower level of NKG2D in XMEN is therefore likely the primary cause of the chronic and uncontrolled EBV viral load in these patients, a conspicuous phenotype that frequently progresses to recurrent EBV lymphoma (9). Taking NKG2D as an exemplar, we sought to clarify how NLG is affected by both the presence of MAGT1 in the OST complex as well as the level of intracellular free Mg2+ especially in the context of lymphocytes.
The folding, stability, and function of thousands of secreted and cell-surface proteins depend on NLG, in which a sugar is enzymatically linked to the amino nitrogen atom in an asparagine residue (10, 11). NLG is performed in intimate association with protein translation, as nascent polypeptides are fed into the ER via the translocon machinery (12, 13). The OST catalytically transfers a pre-assembled oligosaccharide from a dolichol-pyrophosphodonor onto asparagine residues of newly-translated polypeptide chains that contain the specific sequence “Asn-Xaa-(Ser/Thr)”, where Xaa (X) is any amino acid except proline (NX(S/T)) (10–12). The glycan transfer process is highly conserved in eukaryotes, as glycosylation is a crucial post-translational modification with significant impact on protein half-lives, localization, and interactions. In mammalian cells, there are two distinct OST complexes distinguished by containing either of two catalytic subunits, STT3A or STT3B. These enzymes are noncovalently complexed with a common set of noncatalytic accessory subunits (MAGT1/TUSC3, OST4, ribophorin I, OST48, and ribophorin II) (Fig. 1A) (13). We wanted to understand the potential impact of MAGT1 deficiency on glycosylation in humans.
Figure 1.
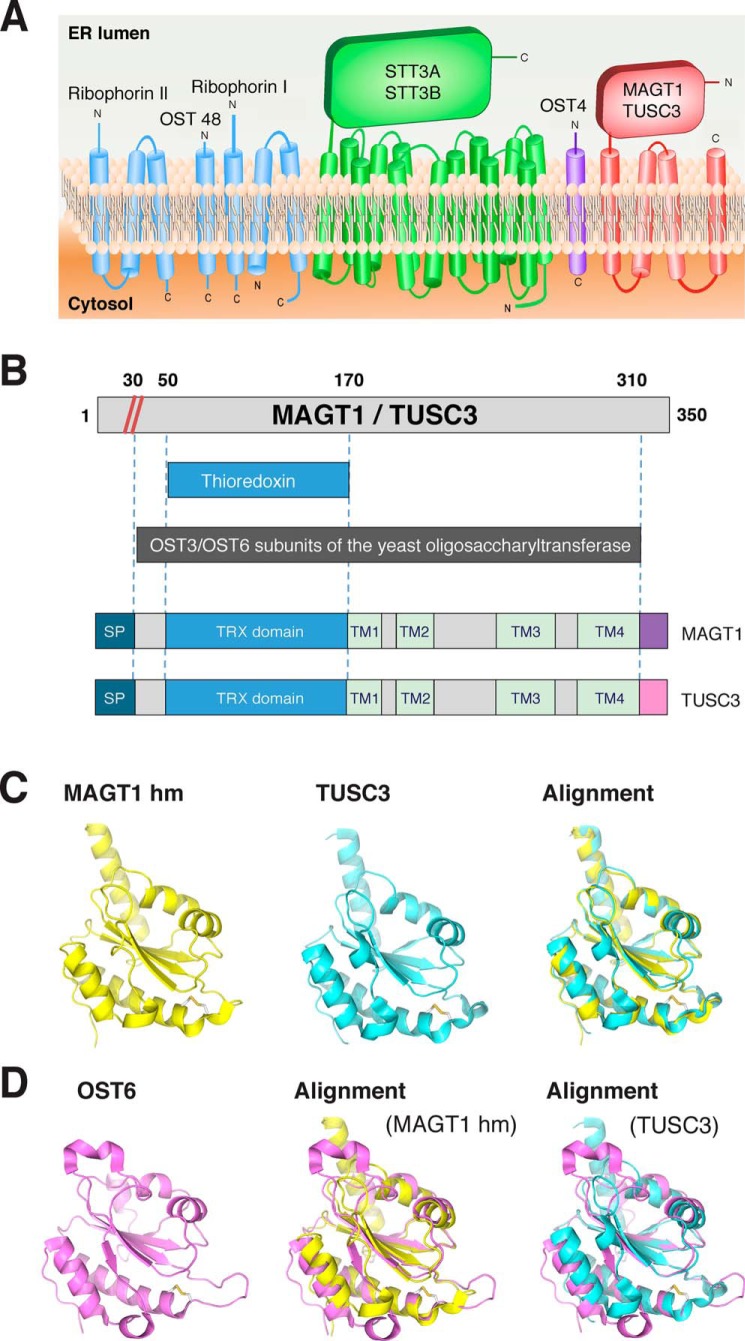
MAGT1 and TUSC3 have conserved structural similarities with OST subunits. A, symbolic representation of OST subunits characterized in the yeast S. cerevisiae. B, domain architecture of MAGT1, TUSC3, and OST3/OST6 subunits. The numeric annotations are for MAGT1, although the analogous numbers for TUSC3 can be approximated by uniformly adding 12 to all numbers or after the signal peptide cleavage site (red diagonal lines). The domains are signal peptide (SP) (teal), thioredoxin domain (blue), TM (green), C termini that are distinct between the two proteins, including two different versions in TUSC3 (purple and pink). C, homology model of Homo sapiens MAGT1 and TUSC3 TRX domain; H. sapiens MAGT1 TRX domain (homology model, left); H. sapiens TUSC3 TRX domain (homology model, middle); alignment of the structures shown in MAGT1 and TUSC3 (right). D, structure of H. sapiens MAGT1 and TUSC3, compared against that predicted from homology modeling and the S. cerevisiae OST6.
In addition to the core components of the OST, several accessory subunits flank the catalytically active STT3 core (14–17). These subunits include ribophorins I and II, OST48 (DDOST), DAD1, and OST4 (Fig. 1A). The composition of these subunits varies depending on whether the central enzyme is either STT3A or STT3B. For instance, in Saccharomyces cerevisiae, the STT3A complex contains keratinocyte-associated protein 2 (KCP2), whereas the STT3B complex contains either of two highly-homologous proteins, OST3 or OST6 (11). Our bioinformatics analysis and other data showed that MAGT1 and tumor suppressor candidate 3 (TUSC3) are the human homologs of OST3/OST6, which appear to function in NLG in an analogous manner (13, 18, 19). The recently reported structure of the entire yeast OST complex showed an intimate association of two transmembrane helices of OST3 with transmembrane segments of the major catalytic STT3 subunits (17). Although this structure has clarified the overall topology of the protein subunits and how the enzyme complex interacts with nascent polypeptides, the precise function of the accessory subunits, including OST3/OST6 or MAGT1/TUSC3, is not fully understood. These recent associations of MAGT1 with the OST complex and its putative role in NLG promise to shed light on the pathophysiology of XMEN disease, which involves the genetic loss of MAGT1.
Recent work has shown that glycoproteomics by lectin capture and dual MS can be used to evaluate cell-associated proteins (i.e. intracellular or membrane-bound) as well as those secreted in plasma or saliva (20, 21). Such studies have been carried out to assess abnormal protein expression as biomarkers in cancer, infections, and neurodegenerative disorders. In contrast, we here employ these technologies to better understand the molecular pathogenesis of an inherited immune disorder (22–24). Our glycoproteome analysis reveals the presence of a highly-selective NLG defect involving a subset of glycoproteins in humans that offers a new understanding of the role of MAGT1 in cellular physiology.
Results
MAGT1 and TUSC3 have conserved structural similarities with OST subunits
More than a decade after MAGT1 was first described as a Mg2+ channel (25), many of its functions and mechanisms of regulation remain poorly understood. MAGT1 was primarily known to play a role in maintaining intracellular Mg2+ homeostasis (2), although its function was noted to partially overlap with that of its homolog, TUSC3 (2). Recent work from nonlymphoid tumor cell lines has suggested that both proteins are localized in the ER and are a subunit of the ER-embedded OST complex (Fig. 1A). MAGT1/TUSC3 share 68% identity and 81% similarity, each possessing three highly-conserved structural features: 1) an N-terminal signal peptide (∼30 amino acids), followed by 2) a thioredoxin (TRX) domain (∼130 amino acids), and 3) a C-terminal domain (160 amino acids) composed of four membrane-spanning segments (Fig. 1B; Fig. S1) (2, 13).
To further elaborate the role of MAGT1, we examined the structural annotation of MAGT1 in several structural databases, which together show it contains both an N-terminal TRX domain and a four-pass transmembrane (TM) region similar to that found in the OST3/OST6 proteins first identified in the S. cerevisiae OST (Fig. 1B and Fig. S2). Detailed sequence comparison revealed that the OST3 and OST6 genes diverge as much from each other as from either MAGT1 or TUSC3 (Fig. 1, C and D, and Fig. S2), indicating that these proteins likely evolved from a single ancestral gene that has duplicated and diverged over yeast and mammalian evolution. The remarkable similarity of the MAGT1/TUSC3 protein domains underscores the potential functional similarity of their parent proteins, despite their minimal sequence conservation. While possessing many of the features of S. cerevisiae OST6, the ligand-bound TUSC3 TRX structures provide further insight into the mode of interaction between cysteine-containing peptides and the bi-cysteine motif of the TRX-active site (14–16). In particular, these structures are consistent with cysteine cross-linking between the MAGT1/TUSC3 TRX domains, and a subset of cysteine-rich substrates could help retain these nascent polypeptides in proximity to the catalytic core of the OST as hypothesized previously (16, 17).
MAGT1 localizes to the ER and Golgi and has a role as a glycosylation accessory protein in human lymphocytes
In previous work, we demonstrated that MAGT1 regulates the basal intracellular Mg2+ concentration in T lymphocytes, which suggested it was operating at the plasma membrane (4). However, recent work in nonimmune tumor cell lines suggests that in nonlymphoid cell lines, MAGT1 preferentially localizes to the ER (Fig. 1A) and co-immunoprecipitates with the STT3B catalytic subunit of the OST (7). These results are consistent with a direct role for MAGT1 in glycosylation (26).
To examine the subcellular distribution of MAGT1 in human T-lymphoid cells, we performed gradient fractionation assays with the Jurkat T-cell line. We found that endogenous MAGT1 was predominantly detected in the fractions containing ER, trans-Golgi, and the first fraction of the cis-Golgi (Fig. 2A). This coincided with the distribution of ribophorin, an essential component of the OST complex, and β-COP1, a membrane-coating protein found in both ER and Golgi complex vesicles. We also detected a modest amount of MAGT1 in the ER–Golgi intermediate compartment (or ERGIC) fraction. Importantly, we observed very little MAGT1 in the plasma membrane (Fig. 2A, lanes 25–27). These results strongly support a general conclusion that the predominant, although not exclusive, localization of MAGT1 is to the ER and Golgi membranes.
Figure 2.
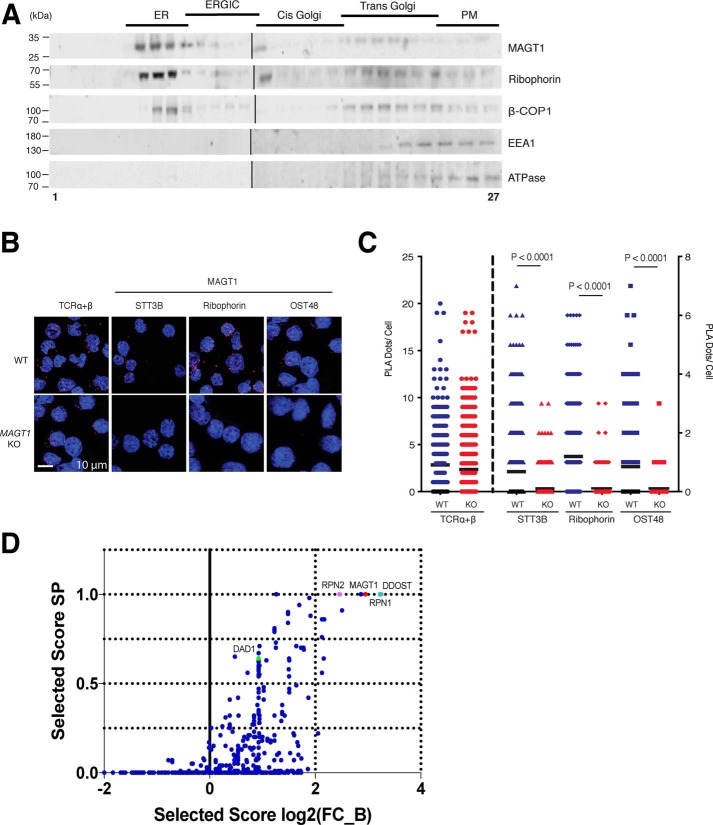
MAGT1 localizes to the ER and Golgi and has a role as a glycosylation accessory protein in human lymphocytes. A, representative Western blot analysis of protein fractions obtained from Jurkat supernatants subjected to density gradient fractionation. Markers for different cell compartments are as follows: ribophorin I for ER, ERGIC, and part of the cis-Golgi; β-COP1 for trans-Golgi; EEA1 and ATPase for plasma membrane (PM). B, PLA confocal photomicrographs. Either WT or MAGT1 KO Jurkat cells were interrogated with the indicated antibodies. Red dots show angstrom proximity. The scale, as indicated, is uniform across all images. C, quantification of the number of PLA dots using >300 cells per condition for images in B. Data in A–C are representative of three independent experiments. Error bars represent the standard error of the mean of X independent experiments, and p values were calculated with a paired t test. D, graph of the SAINT (Significance Analysis of INTeractome) probability scoring versus stringent fold change score (FC_B) for MS for MAGT1 interactors in HEK 293T cells.
We further investigated whether MAGT1 associates with components of the glycosylation complex via a proximity ligation assay (PLA), in which we detected protein interactions using antibodies specific for MAGT1. This method requires that both proteins be close enough to allow the two different antibody–DNA probes to facilitate “rolling circle” replication with labeled nucleotides to generate a bright punctate co-localization signal that can be detected by confocal microscopy (27, 28). We observed a robust association signal of MAGT1 with several members of the OST complex localized in the ER, including ribophorin I, OST48, and STT3B (Fig. 2, B and C). The association signal was as strong as the association between the α- and β-chains of the TCR, implying that these proteins are within ∼40 nm of each other, i.e. they form a protein complex. Moreover, the signal was completely abolished in MAGT1 knockout (KO) Jurkat cell lines (Fig. 2, B and C), whereas the positive control TCRα/β is observed at equivalent levels in WT and MAGT1 KO cells. Interestingly, MAGT1 in situ resides near STT3B, supporting a possible selective role in the STT3B-containing complexes (11, 29).
To further verify the interactors of MAGT1, we performed MS on samples from HEK 293T WT cells and MAGT1 CRISPR KO cells immunoprecipitated with an anti-MAGT1 antibody, similar to what has been previously carried out in HeLa cells (3). We also examined samples from HEK 293T MAGT1 CRISPR KO cells and CRISPR knockins of an HA-tagged MAGT1 immunoprecipitated with an anti-HA antibody. In both cases, MAGT1 was co-immunoprecipitated with both ribophorin 1/2 and DDOST, which are known OST-associated proteins (Fig. 2D and Dataset S1).
Defective glycosylation in XMEN patient cells
We have previously shown that defective antiviral immunity against EBV in XMEN disease is due to the reduced surface expression of NKG2D on patient CD8+ and NK cells (Fig. 3A) (9). This defect is accompanied by the presence of a conspicuously hypoglycosylated form of NKG2D not found in healthy controls (HCs).8 Further glycoproteomic analysis of T cells has additionally revealed a broader, although site-selective, NLG defect in these patients8 (9).
Figure 3.
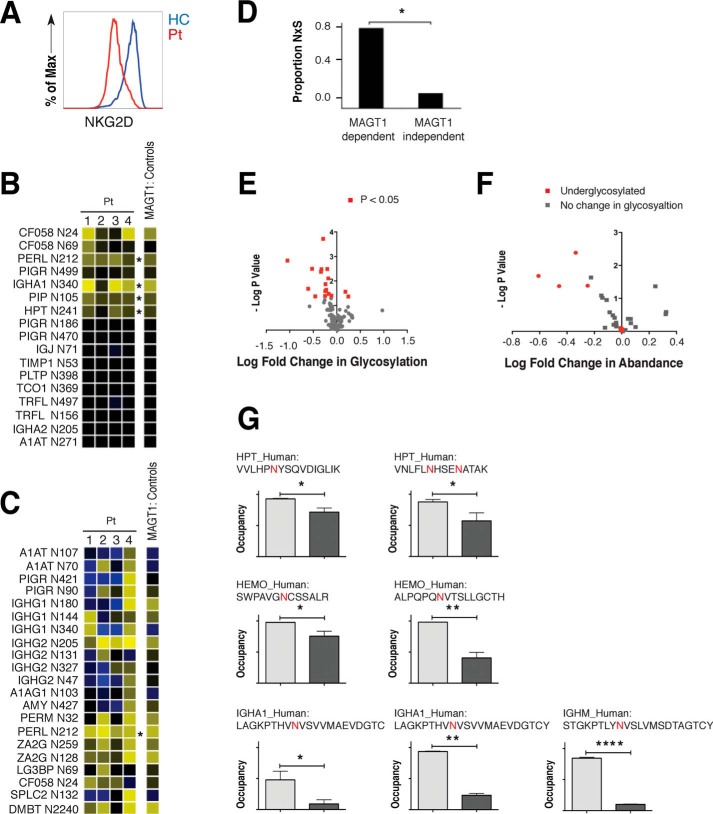
Defective glycosylation in XMEN patient cells. A, flow cytometry analysis of surface expression of NKG2D in cycling T cells from an XMEN patient (Pt) and HC. B, ratio of salivary protein glycosylation site occupancy in XMEN patients after PNGase F and trypsin digestion relative to their HC parents. The N-linked glycosylation site corresponding to each row is indicated at left. C, ratio of salivary protein glycosylation site occupancy in XMEN patients after PNGase F and AspN digestion. B and C; yellow, decreased glycosylation; black, no change in glycosylation; blue, increased glycosylation; *, p < 0.05. D, proportion of the NXS sequons in MAGT1-dependent and -independent glycosylation sites. *, p = 0.02. E, volcano plot of the log fold change in glycosylation versus the negative log of the p value. F, volcano plot of the log fold change in abundance versus the negative log of the p value. G, underglycosylation of glycosites in particular plasma proteins in XMEN patients. *, p < 0.05; **, p < 0.01; ****, p < 0.001.
In addition to characterizing cell-associated proteins, we also examined secreted proteins in saliva and serum from XMEN patients. Although most salivary protein glycosylation sites in XMEN showed comparable occupancy to those found in unaffected individuals, there were four particular glycosylation sites that showed significantly lower occupancy in XMEN patients compared with their unaffected parents: Asn-212 in PERL, Asn-340 in IGHA1, Asn-105 in PIP, and Asn-241 in HPT (Fig. 3, B and C). These proteins are known to contribute to innate and adaptive immune protection (30, 31). For instance, the PERL protein is a lactoperoxidase found in saliva, milk, and airway secretions that catalyzes the generation of the antimicrobial substance hypothiocyanous acid (32). Similarly, insufficient glycosylation of the heavy chain of IgA can lead to decreased intracellular stability and reduce secretion, thereby impairing adaptive immunity (33). We additionally observed that glycosylation sites that were deficient in XMEN and not in their parents (i.e. MAGT1-dependent glycosylation sites) were enriched in NXS over NXT sequons as compared with the comparatively small fraction observed in MAGT1-independent sites (Fig. 3D), as was also the case with peptides with reduced levels identified by an N-linked glycoproteome analysis.8 NXS sequons bind with lower affinity to the peptide acceptor site of the STT3 catalytic subunits of the OST and therefore might be more dependent on facilitation by accessory subunits such as MAGT1 for efficient glycan transfer. Therefore, although the full alignment of N-linked sequons from the N-glycoproteome analysis show the expected NXT bias observed in the human glycoproteome, the peptides with reduced levels and/or stabilities in XMEN cells have a distinct bias toward NXS sequons.8
We subsequently analyzed proteins isolated from XMEN plasma to see whether a similar trend could be detected (Fig. 3E). The bias of the profile toward the left side of the volcano plot is indicative of an underglycosylation phenotype, consistent with the predicted OST-promoting activity of MAGT1. Further analyzing these data as a function of the relative fold changes in abundance demonstrate that the underglycosylated proteins are largely present in less abundance, with only a small subset of these showing no change in abundance (Fig. 3F). These results indicate that the underglycosylated proteins are generally less abundant, fitting with the model that protein glycosylation is often critical for protein folding, secretion, and stability.
In plasma proteins, we found sites in haptoglobin (HPT), hemoglobin (HEMO), and immunoglobulin heavy chain that showed decreased glycosylation (Fig. 3G). HPT binds free HEMO in plasma, allowing subsequent breakdown of HEMO with controlled retention of the iron center of heme (34). Glycosylation changes in both HPT and HEMO could therefore alter the metabolism of iron, as well as the recycling of red blood cells. Similarly, the observation of decreased Ig glycosylation is characteristic of the underglycosylation phenotype of many XMEN proteins and is consistent with observations from saliva. Children with XMEN disease routinely receive injections of Ig to strengthen immune function, which has actually been shown to boost levels of Ig glycosylation in immunocompromised individuals (35, 36). Together, these results suggest that the observed Ig glycosylation deficiency is an inherent phenotype of XMEN disease.
Molecular structure and biochemical characteristics of MAGT1 and TUSC3
The altered glycosylation observed in T cells from XMEN patients led us to examine MAGT1's direct role in the OST complex. Bioinformatic annotation of the MAGT1-coding sequence indeed suggests that the MAGT1 protein is a human homolog of the OST3 and OST6 proteins in S. cerevisiae and is highly conserved across species in eukaryotes (Fig. S1) (2, 11, 13). The conserved structural features across the MAGT1/TUSC3 and OST3/OST6 families, despite a lack of strict sequence identity, indicate a common function (Fig. 4A). The presence of regions with substantial homology to TRX and the proposal that MAGT1 participates in oxidation–reduction reactions during NLG led us first to explore the expression and cellular localization of MAGT1 in more detail (2). Examining protein expression of MAGT1 and TUSC3 across different immune cells types (Fig. 4B), we observed high levels of MAGT1 expression in human T lymphocytes, B lymphocytes, NK cells, monocytes, and the Jurkat T leukemia cell line (Fig. 4B). By contrast, TUSC3 was not detectable in any of the lymphoid or monocytic cells but was present in the Jurkat T leukemia cells, suggesting that although it can appear in transformed cells, it has very little role in normal immune functions. As a negative control, we generated a double-KO Jurkat cell line for both MAGT1 and TUSC3 using CRISPR deletion, and we demonstrated that it was devoid of expression of both proteins (Fig. 4B). We further examined expression in an additional 19 different human tissues (Fig. 4C). We found that liver, spleen, esophagus, and colon express MAGT1, but not TUSC3, whereas brain and prostate preferentially express TUSC3. Lung, stomach, intestine, thymus, testis, ovary, and uterus expressed both proteins, but pancreas, heart, smooth muscle, tongue, and skin have almost undetectable levels of either protein (Fig. 4C). Therefore, the presumptive partially-redundant functions of MAGT1 and TUSC3 in conjunction with their differential expression profiles allow these proteins to additively cover a broad range of tissues for a potentially similar function.
Figure 4.
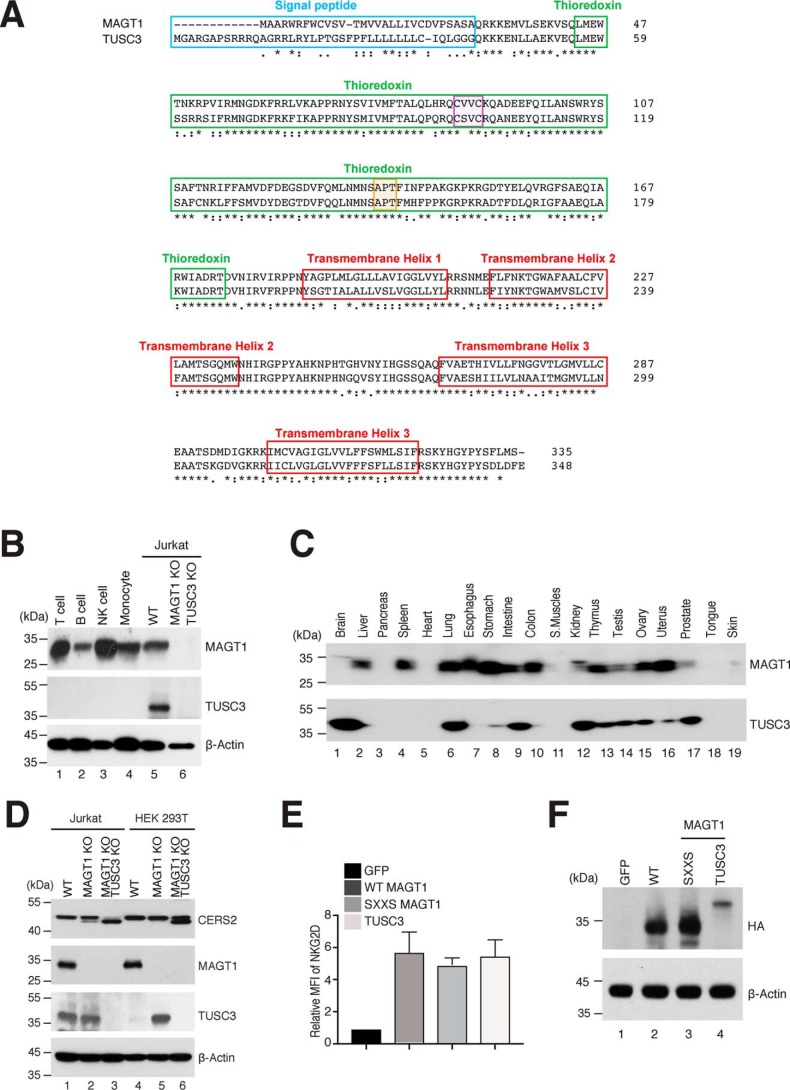
Molecular structure and biochemical characteristic of MAGT1 and TUSC3. A, comparison of the amino acid sequences of MAGT1 and TUSC3. The signal peptide, TRX domain, and transmembrane regions are highlighted in blue, green, and red, respectively. The CXXC and cis-proline motifs are highlighted in purple and orange, respectively. B, representative Western blot analysis showing MAGT1 and TUSC3 expressions in isolated primary human T cells, CD19+ B cells, CD56+ NK cells, CD14+ monocytes as well as Jurkat lines that are WT or harbor CRISPR KOs of both MAGT1 and TUSC3 (MAGT1/TUSC3 KO). C, immunoblotting of MAGT1 and TUSC3 expression in a commercially-prepared human tissue sample membrane. D, representative Western blot analysis of CERS2, MAGT1, TUSC3, and β-actin (loading control) in MAGT1 KO and MAGT1/TUSC3 KO from Jurkat and HEK 293T cells. E, quantification of the mean fluorescent intensity (MFI) of surface NKG2D in MAGT1 KO and MAGT1/TUSC3 KO from Jurkat and HEK 293T cells. Cells were transfected with mRNA encoding HA-tagged versions of GFP, WT MAGT1, the thioredoxin domain mutant of MAGT1 (SXXS), or WT TUSC3 along with NKG2D/DAP10-encoding plasmids. Error bars represent the standard error of the mean of three independent experiments; p values were calculated with a paired t test. F, representative Western blot analysis of MAGT1 and TUSC3 expression in transfected MAGT1/TUSC3 KO HEK 293T cells. The expressions of MAGT1 and TUSC3 were probed by HA antibody with actin as a loading control.
To further examine the degree to which MAGT1 is interchangeable with TUSC3, we queried the glycosylation level of CERS2, a protein whose single N-glycosylation site is impaired in XMEN patients.8 We found that the full-length CERS2 glycosylated species that is observed as a 45-kDa band in both WT Jurkat and HEK 293T cells (Fig. 4D, lanes 1 and 4) shifts to a lower molecular mass band of ∼43 kDa in MAGT1 KO cells (Fig. 4D, lanes 3 and 6), consistent with this being the nonglycosylated form. Moreover, we found TUSC3 expression is up-regulated in MAGT1 KO HEK 293T cells, potentially allowing TUSC3 to substitute for MAGT1 (Fig. 4D, lane 5) (26). Cells that lack both MAGT1 and TUSC3 have more of the 43-kDa form, indicating a greater defect in glycosylation. Surprisingly, there is still some apparently glycosylated CERS2 in the MAGT1/TUSC3 KO cell line, suggesting that the OST complex for this protein does not have an absolute requirement for either accessory factor. These results demonstrate that MAGT1 and TUSC3 indeed may have overlapping and/or compensatory functions, and that their ability to complement each other's activity is heavily influenced by protein expression level in a given tissue.
TRXs and related oxidoreduction proteins possess a bi-cysteine motif separated by two central amino acids (CXXC) that appear to modulate the redox potential of the active site, these motifs are conserved in conserved in both MAGT1 and TUSC3 throughout phylogeny (Fig. 4A and Figs. S1, A and B and S3). Consistent with this model, replacing both active-site cysteines with serines in OST3/OST6 caused a defect in site-specific glycosylation in yeast (37). The active site CXXC motifs in MAGT1/TUSC3 have been proposed to be indispensable for glycosylation of STT3B-dependent acceptor sites by forming a transient mixed disulfide with a free thiol in a glycoprotein substrate to allow access of STT3B to an inaccessible NLG sequon (7). We therefore constructed a MAGT1 mutant by replacing the two active-site cysteines with serines, and we transfected either the WT CVVC mRNA or the SVVS mutant mRNA along with NKG2D/DAP10 expression plasmids (DAP10 is a requisite adaptor protein required for NKG2D stability) (38) into MAGT1/TUSC3 KO HEK 293T cells. Both the WT and SXXS mutant MAGT1 proteins were expressed at comparable levels (Fig. 4F) and restored NKG2D glycosylation as inferred from surface expression with similar efficacy (Fig. 4E). NKG2D surface expression was also fully complemented by expression of WT TUSC3 in MAGT1/TUSC3 KO HEK 293T cells (Fig. 4E). STT3B-dependent NLG of NKG2D was therefore not measurably affected by mutating the CVVC motif in MAGT1, indicating that the ability of this protein to promote glycotransferase activity may not require the disulfide cross-linking at least in the case of NKG2D. Additionally, although TUSC3 can functionally complement MAGT1, its poor expression in lymphocytes likely accounts for the NLG defects in key immune glycoproteins (e.g. NKG2D) in XMEN disease (Figs. 3 and 4).
Mg2+ regulates glycosylation and plays a unique role in T-cell–mediated immunity, especially in immune protection against EBV
Our previous work showed that MAGT1 deficiency causes reduced basal levels of intracellular free Mg2+ in patient cells (3). Because of the presumptive dual role of MAGT1 in facilitating Mg2+ transport and directly participating in the OST protein complex, we used a Mg2+ deprivation assay to examine whether Mg2+ itself is involved in driving glycosylation. This method is based on our previous observation that, unlike Ca2+, which is maintained at an intracellular free concentration of 100 nm by a tightly-controlled pump-leak mechanism (39), the intracellular free Mg2+ varies with the concentration in the extracellular milieu (9). We mimicked the lower intracellular free Mg2+ levels observed in XMEN cells by culturing cycling T cells from HCs in Mg2+-depleted media (dRPMI; <0.1 mm Mg2+) for 72 h. We could then evaluate the effects of low millimolar Mg2+ without changing the level of MAGT1 protein. We tested the expression of putative Mg2+-sensitive glycoproteins, including NKG2D, CD70, and HLA-DR, and found that Mg2+ deprivation caused decreased glycosylation and surface expression of these proteins similar to what we found in XMEN cells (Fig. 5, A–C). Adding back cRPMI with 0.5 mm Mg2+ to the deprived cells caused a partial restoration of surface expression (Fig. 5, A and B, light blue) and total protein abundance (Fig. 5C). Importantly, neither the glycosylated CD5 nor the nonglycosylated actin controls were affected by Mg2+ removal and/or add-back, indicating that the deficient glycosylation is not simply due to poor health of the deprived cells. These data support a model in which the glycosylation defect is selective and can be caused by decreased free basal Mg2+ levels. Moreover, this defect is dynamic and reversible, allowing for therapeutic intervention.
Figure 5.
Mg2+ regulates glycosylation and plays a unique role in T-cell–mediated immunity, especially in immune protection against EBV. A, flow cytometry histograms of surface expression of NKG2D, CD70, HLA-DR, and CD5 in cycling T cells from HCs cultured either in cRPMI for 5 days, dRPMI for 5 days, or cells that were cultured in dRPMI for 3 days followed by the addition of Mg2+ (0.5 mm) back into the dRPMI for 2 days. B, quantification of the MFI in A. Error bars represent the standard error of the mean of eight independent experiments, and p values were calculated with a paired t test. C, representative Western blot analysis of NKG2D, CD70, TCR-β, and β-actin in cycling T cells from a HC as described in A. Numbers at left indicate kDa standards. Glycosylation patterns are shown at right: fully-glycosylated (2); partially-glycosylated (1); and unglycosylated (0). Data are mean of eight (A and B) or are representative of three (C) independent replicates. D, schematic diagram for killing pathway with NK cells (top) and EBV-721.221(target; bottom) cells. E, flow cytometry analysis of surface expression of NKG2D in EBV-specific NK cells from a HC with cRPMI and dRPMI. F, quantification of the MFI in F. G, percent lysis of autologous EBV-LCLs by EBV-specific NK cells from HC with a different dose of the Mg2+ with significance determined by one-way ANOVA. n indicates the number of independent samples. H, quantification from G. Data are representative of three independent biological replicates. NS, nonsignificant.
The N-linked glycopeptides that were observed at decreased levels in XMEN as compared with HCs in a previous study8 were therefore re-examined in the context of Mg2+ supplementation. In agreement with the interplay of Mg2+ levels and glycosylation, a select subset of glycosites demonstrate increases in glycosylation and/or peptide levels, likely both, given that N-glycosylation frequently enhances the half-life of cognate peptides. The peptides corresponding to NKG2D and CD70 were notably present in this dataset, further reinforcing their significance and robustness in responding to augmentation in Mg2+ levels. The Mg2+ deprivation data suggest that Mg2+ levels may control the function of key glycosylated immune molecules. We therefore tested the consequences of reduced NKG2D glycosylation using an in vitro NK killing assay (40). EBV-specific NK cells from HCs, expanded by culturing with irradiated autologous EBV-transformed B-lymphoblastic cell lines (EBV-LCLs) in dRPMI (<0.1 mm Mg2+), exhibited decreased expression of NKG2D (Fig. 5, E and F) and defective cytotoxic activity against EBV-LCL (Fig. 5, G and H) compared with those expanded in Mg2+ complete media (cRPMI). These results support the idea that chronic deprivation of Mg2+ leads to down-regulation or enhanced instability of NKG2D caused by impaired T- and NK-cell cytotoxicity, thereby potentially explaining the XMEN patient's pronounced susceptibility to viral infections (Fig. 5D).
Transcriptome analysis reveals immune gene expression defects in XMEN T cells
To further assess the biological effects of MAGT1 deficiency in XMEN patients on cytotoxic T-cell activation and growth, we performed RNA-Seq of the complete transcriptome of CD8+ cells from four XMEN patients and HCs. After CD8+ purification, cells were activated with anti-CD3 and anti-CD28 antibodies in the presence of IL-2 and cultured for 3 or 12 days. A defect in TCR-mediated T-cell activation has previously been reported in XMEN patients (4). To characterize the extent of the defect over the course of activation and to identify genes or pathways that may play a role in the defect, we compared XMEN patient cells to time-matched HC cells to determine differentially expressed genes (DEGs). Interestingly, the number of significant (p < 0.05, log2 fold change >1) DEGs is much greater at baseline compared with after activation. For instance, we identified 1050 DEGs before activation (day 0) versus 131 and 192 DEGS at days 3 and 12, respectively (Dataset S2), with the majority of the genes at day 0 being down-regulated (Fig. 6A). Many of the genes we find differentially expressed only on day 0 do trend in the same direction of expression on day 3 and even some on day 12, while not passing our threshold for statistical significance (Fig. 6, B and C).
Figure 6.
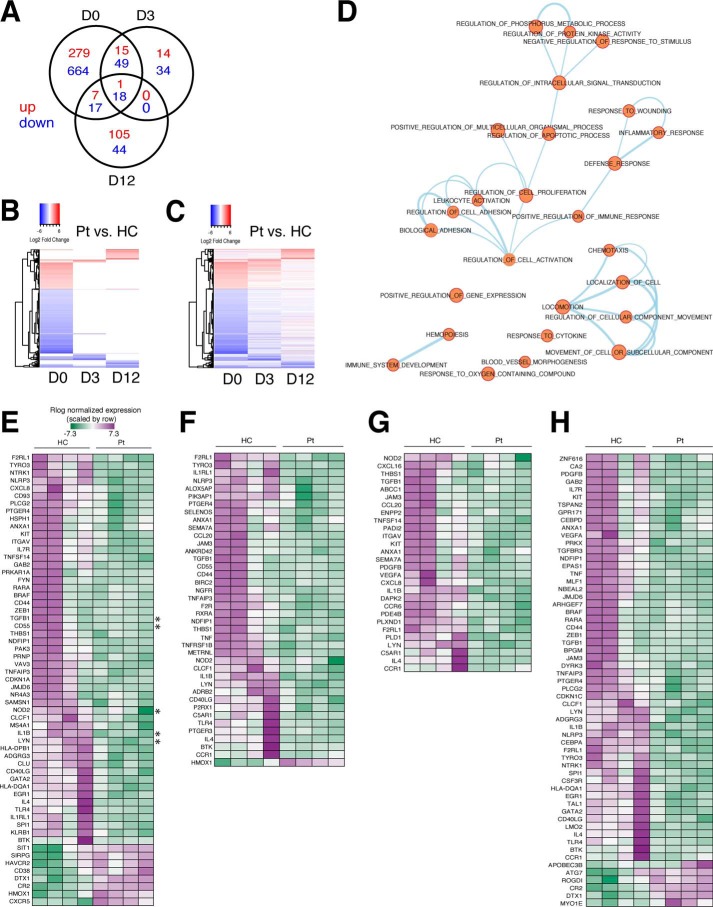
DEGs and functional enrichment in CD8+ T cells from XMEN and healthy controls before and after activation. A, Venn diagram of the number of DEGs found in CD8+ T cells from XMEN patients (Pts) versus HCs on day 0 (D0) before activation, and day 3 (D3) or day 12 (D12) after activation. DEGs are separated into up-regulated (red) and down-regulated (blue) expression status for each time point. B and C, heatmap of log2 fold change for patients versus HCs for DEGs found in at least one time point (n = 1238 genes), with up-regulated expression as red and down-regulated expression as blue. Genes are represented on the y axis, organized by unsupervised hierarchical clustering. B, heatmap showing only data passing significance threshold of adjusted p value <0.05 and log2 ratio >1. C, heatmap corresponding to B showing all data points (including nonsignificant values) to illustrate trends in direction of expression of genes across time points. D, enrichment map of biological processes enriched in DEGs arranged as a network, with connections between nodes signifying at least 50% overlap among the genes assigned to each node. E–H, representative heatmaps of Rlog normalized counts for genes common to all nodes in various subgroupings within the enrichment Map in D (nodes selected for each heatmap are indicated above each table), including the following themes: E, leukocyte activation; F, defense response; G, cell locomotion; and H, hematopoiesis. The color key in E is used across all heatmaps; up-regulated expression is purple and down-regulated expression is green. Asterisks in E mark genes specifically mentioned under “Results.”
Focusing on day 0 DEGs, we identified significantly enriched (p < 0.000002) functional categories affecting multiple biological processes, most of which are related to immune function (Dataset S3). Because many of these processes involve shared sets of genes, we have displayed them as a network where each biological process is a colored circle, with a line connecting two circles representing an overlap of at least 50% for the sets of genes associated with each process (Fig. 6D). Some of the processes and functions affected in XMEN cells include chemotaxis, leukocyte activation, cellular adhesion, immune system development, regulation of immune responses (defense response, inflammatory response, and response to cytokines), regulation of cell death and proliferation, and regulation of protein phosphorylation and signal transduction (Fig. 6D).
Inspection of normalized values for genes common to various clusters shows some variability in gene expression between HC samples (Fig. 6, E–H). For example, for genes associated with leukocyte activation (Fig. 6E), TGFB1 and CD55 are up-regulated only in a subset of HC samples, whereas other genes such as NOD2, IL1B, and LYN are up-regulated broadly in all HC samples (Fig. 6E, asterisks). By contrast, transcript levels of these genes are remarkably consistent between XMEN samples, suggesting a common basal transcriptional and functional defect, regardless of the particular MAGT1 mutation a patient has.
We determined the direction of effect that MAGT1 deficiency has on these processes by performing a Core Analysis of the DEGs using Ingenuity Pathway Analysis (IPA) software (Qiagen). As expected, the predicted effect for these immune-related processes enriched in day 0 DEGs is down-regulation in XMEN compared with HC samples (Fig. S4A). Consistent with a reduced number of DEGs at days 3 and 12, the effect on these processes is greatly diminished after T-cell activation (Fig. S4, B and C). To illustrate this, we examined the Z-scores associated with each subcategory in the treemap plots. According to the Downstream Effects Analysis convention within IPA, functions with a Z-score of < −2 are considered significantly “decreased,” and likewise functions with a Z-score of >2 are considered significantly “increased.” Using this convention, at day 0 prior to activation, there are 40 decreased functions, although in each post-activation time point there are only two decreased functions (Fig. S4 and Dataset S4). The labeled categories that remain decreased at day 3 are “activation of leukocytes” and “activation of blood cells,” and at day 12 are “quantity of mononuclear leukocytes” and “quantity of T lymphocytes” (Dataset S4). Altogether, it appears that T-cell activation minimizes the differences between XMEN and HC cells, which may therefore obscure some of the signature distinctions between patient and normal control cells at later time points.
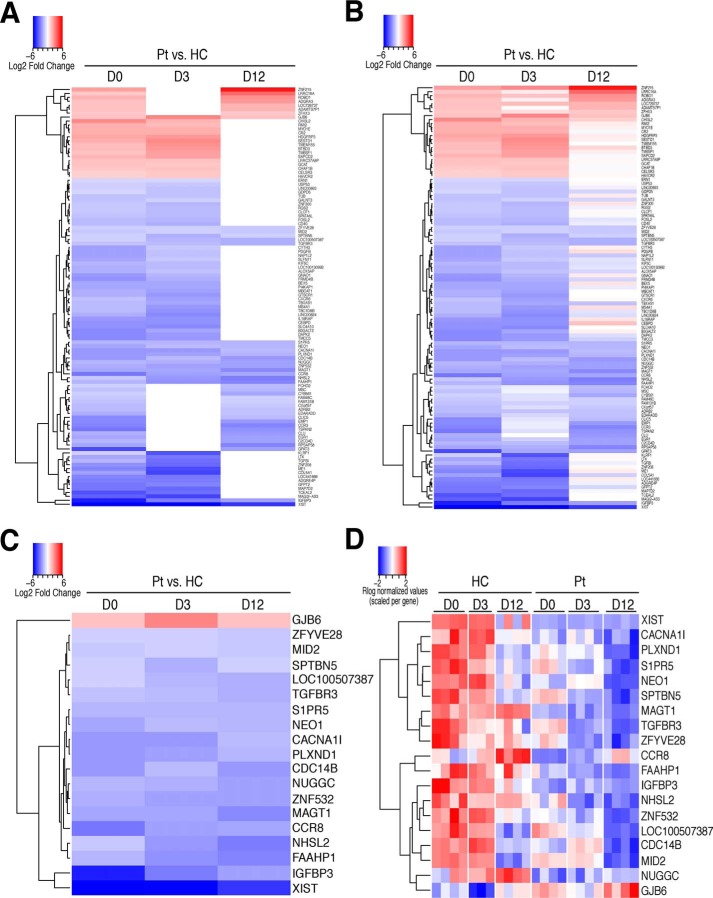
To identify genes responsible for persistent cellular defects in T cells from XMEN patients, and thereby better understand the connection between MAGT1 and these defects, we determined the subset of genes differentially expressed in the same direction in at least two time points (and not differentially expressed in the opposite direction in the third time point) (Fig. 7, A and B) or all three time points (Fig. 7, C and D). Of the 106 genes differentially expressed in at least two time points, 23 are up-regulated and 83 are down-regulated in XMEN. Of these, we found that 19 genes are differentially expressed in all three time points: 18 of these genes are down-regulated, including MAGT1, PLXND1, IGFBP3, CCR8, and TGFBR3, and GJB6 is the single gene that is up-regulated. Note that XIST was also apparently down-regulated in patients, although this is an artifact attributable to the fact that all of the XMEN patients are male but females were included in the controls. Persistent down-regulation of MAGT1 in XMEN patients is consistent with the fact that they all have a genetic defect in the ORF of this gene, leading to nonsense-mediated decay of the transcript. The mouse homolog of PLXND1 has been reported to be involved in chemotaxis of double-positive thymocytes into the thymic medulla (41) as well as T-cell–dependent B-cell activation (42). IGFBP3 expression level has additionally been shown to be correlated with maintenance of naïve CD8+ T-cell populations (43). GJB6, a gap junction protein, has been shown to be involved in neutrophil activity (44), but so far it has no known role in T cells. Despite the significance of several of these genes to T-cell function, the mechanism by which these systematic variations in a subset of genes contribute to the pathogenesis of XMEN disease certainly will require further investigation for future therapies or treatments.
Figure 7.
Differentially expressed genes in CD8+ T cells from XMEN and healthy controls present in multiple time points. A and B, heatmap of log2 fold change for XMEN patients versus HCs for DEGs found in at least two time points (n = 106 genes). A, heatmap showing only data passing significance threshold of adjusted p value <0.05 and log2 ratio >1. B, heatmap corresponding to A showing all data points (including nonsignificant values) to illustrate trends in direction of expression of genes across time points. C and D, heatmap of log2 fold change (C) and Rlog normalized counts for DEGs (D) found in all three time points (n = 19). For all heatmaps, up-regulated expression is red, and down-regulated expression is blue.
Transcriptome analysis links glycosylation defects in XMEN T cells with immunodeficiency
Our transcriptome analysis of CD8+ T cells from XMEN and HC donors established global gene expression pattern differences and helped elucidate functional defects during T-cell activation. To determine whether defective glycosylation might itself contribute to the overall gene expression patterns we observe in cells from XMEN patients, we analyzed the RNA-Seq data of CD8+ cells prior to T-cell activation using IPA “Upstream Analysis” to determine upstream regulators and then intersected them with the 73 proteins found to be differentially glycosylated in XMEN versus HC. Although there are many upstream regulators and master regulators predicted in IPA, one differentially glycosylated factor, CD28, was predicted to be a strongly inhibited (Z-score −6.3) master regulator, based on the predicted activation status of ∼55 regulators upstream of the day 0 DEGs in XMEN cells (Fig. S5). This predicted inhibition is confirmed by our finding that reduced glycosylation in XMEN cells causes reduced stability and surface expression of CD28. CD28 inhibition, through its downstream regulators, can explain the differential expression of up to 231 (22%) of the previously identified DEGs (Dataset S5), and these genes are enriched for most of the same functions that we previously saw enriched in the full DEG list, including leukocyte activation, locomotion, regulation of apoptosis, cellular adhesion, and adaptive immune response. Together, these results provide a link between the glycosylation defect in XMEN and their T-cell activation defect, further unifying our understanding of their immunodeficiency.
Discussion
This study sheds new light on the biomedical importance and functional association of MAGT1 with the canonical subunits of the cellular glycosylation machinery. We additionally show that XMEN disease has certain selective features of congenital disorders of glycosylation. The enzyme STT3 is the catalytic core of the glycosylation machinery, and it has been intensively studied because of its prominent role in the N-glycosylation pathway. STT3 and other OST proteins contain mostly TM helices that have an essential scaffolding function and are thus critical for stabilizing the complex as it recognizes and processes a diverse range of acceptor proteins (Fig. 1A). Mutations in some of the core subunits of the OST therefore produce distinct deficiencies in NLG (10). Previous work has clearly highlighted the important role that MAGT1 can play in regulating intracellular Mg2+ (1, 2). Our current observations raise the possibility that MAGT1 plays a unique dual role in glycosylation and ion regulation, which together may contribute to the specific phenotype observed in XMEN syndrome.
We previously detected down-regulation or increased turnover of a number of N-glycoproteins, including CD70, TCR-β, CD28, and HLA-DR, that are associated with anti-EBV responses and general immune function (Fig. 3D).8 Moreover, aberrant glycosylation of TCR-β and CD28 caused their accelerated degradation, which may explain why these patients have T-cell activation defects. Underglycosylation of CD28 in MAGT1-deficient cell defects could therefore be the cause for much of the gene expression patterns and the functional T-cell defect that we see in XMEN patient samples. We similarly surmise that loss of HLA class II molecules may contribute to patients' susceptibility to viral infections. Furthermore, the glycosylation defect observed for NKG2D appears to result both from loss of MAGT1 in the OST complex, as well as a reliance on normal physiological concentrations of Mg2+ for stability (9). These observations together highlight a select subset of proteins whose perturbed glycosylation and decreased expression contribute to XMEN immunodeficiency.
Beside the lack of TUSC3 expression in immune cells (Fig. 4C), we also demonstrated the tissue specificity of MAGT1 and TUSC3 expression. For example, TUSC3 is expressed predominantly in the brain, consistent with studies showing TUSC3 is involved in autosomal recessive mental retardation (19, 45, 46). Similarly, the abundance of MAGT1 in immune cells and in the liver may help to rationalize why XMEN patients have functional defects in both immune cells and liver function. Of particular interest to XMEN patients' preponderance of extraordinarily high EBV titers, NKG2D glycosylation was not decreased by “rescue” of MAGT1 KO cells using the expression of mutant MAGT1 lacking CXXC motifs. These results suggest that disulfide-mediated cross-linking to peptide substrates is not strictly required for glycosylation by the MAGT1–STT3B complex, as proposed previously (7, 8). Instead, the peptide-binding groove presented on the surface of MAGT1/TUSC3 may be a more general way of explaining the ability of these accessory OST proteins to enhance N-glycosylation of nascent polypeptide substrates. These results help define a broader role for MAGT1 in N-glycosylation, as opposed to prior work strongly implicating disulfide cross-linking to peptide substrates as a primary mode of MAGT1/TUSC3-assisted N-glycosylation (10).
The fact that NKG2D demonstrates a glycosyltransfer defect likely from the loss of MAGT1 in the OST complex, and depends on normal physiological concentrations of Mg2+ for its stability, may explain why this protein, in particular, was so dramatically perturbed in XMEN. NKG2D is particularly suited to curbing EBV infection, which thereby links both a glycosylation defect and Mg2+ deficiency to the XMEN patients' conspicuous susceptibility to chronically active EBV infection. This connection is particularly important because EBV infection often leads to lymphoma, which can cause the premature death of XMEN patients.
Our studies clearly show that Mg2+ deprivation lowers the glycosylation level of a specific subset of N-glycoproteins and reduces the killing function of cytotoxic immune cells. It remains unknown what step Mg2+ is required for in the expression or maintenance of these molecules. Nevertheless, we have demonstrated that NLG's range of substrates is expanded through MAGT1/TUSC3's participation as accessory subunits in the OST complex. MAGT1/TUSC3-containing STT3B complexes may potentially require Mg2+ for their catalytic function, consistent with the role of Mg2+ in many enzymatic processes and with the intimate link between OST function and intracellular Mg2+ levels.
Experimental procedures
Patients
All subjects provided written informed consent for the National Institutes of Health Institutional Review Board (IRB)–approved protocols in accordance with the Declaration of Helsinki.
Cell culture and T-cell expansion
The human epithelial kidney cell line HEK 293T was purchased from the American Type Culture Collection (ATCC). HEK 293T cells were cultured in DMEM with 10% FBS, 2 mm glutamine, 10 units ml−1 penicillin (Sigma), 100 μg ml−1 streptomycin (Sigma), and 55 μm β-mercaptoethanol. Jurkat cells (clone E6.1) were purchased from the ATCC. Jurkat cells were cultured in RPMI 1640 medium (Sigma) with 10% FBS, 2 mm glutamine, 10 units ml−1 penicillin (Sigma), 100 μg ml−1 streptomycin (Sigma), and 55 μm β-mercaptoethanol (cRPMI). In the case of patient cells, freshly isolated or frozen PBMCs were stimulated at 1 × 106 ml−1 in cRPMI with 1 μg ml−1 of anti-CD3 (Biolegend) and 1 μg ml−1 anti-CD28 (Biolegend) for 48 h. The cells were washed twice with cRPMI and resuspended in media at 1 × 106 ml−1 with 100 units ml−1 recombinant human IL-2 (IL-2), and cultured for up to 3 weeks with fresh IL-2 and medium supplemented every 2 days. Mg2+ supplementation was performed using sterile cell-culture grade MgSO4 (Sigma).
CRISPR KO cell lines
We designed four guide RNAs (gRNAs) to target the N-terminal region of human MAGT1 and TUSC3 using the online gRNA Guide Design package. Each gRNA was cloned into the pGuide plasmid (Addgene, catalog no. 64711), and CRISPR/Cas9-mediated cleavage efficiency was evaluated using T7 endonuclease I assay (New England Biolabs, catalog no. M0302S). To generate MAGT1 KO and MAGT1/TUSC3 KOs, HEK 293T cells were transfected and electroporated using 4D Amaxa nucleofector (Lonza), respectively, with pCas9-GFP (Addgene, catalog no. 44719) and gRNA at a 1:1 ratio. 48 h after transfection or nucleofection, GFP-positive clones were selected by FACS and plated as single cells. KO clones were confirmed by Western blotting and DNA sequencing.
Cytotoxic NK cell-killing assay
NK cells were isolated from PBMCs using CD56+-positive selection (Miltenyi Biotec). Cells were cultured in RPMI 1640 medium (Sigma) with 10% FBS, 2 mm glutamine, 10 units ml−1 penicillin (Sigma), 100 μg ml−1 streptomycin (Sigma), and 55 μm β-mercaptoethanol. Cells were cultured with 100 units ml−1 recombinant human IL-2 (IL-2), with fresh IL-2 and medium supplemented every 2 days. NK cells cultured in cRPM (with 0.5 mm Mg2+) or deprived RPMI (with < 0.1 mm Mg2+) for 3 days were used in cytotoxicity assays against the 721.221 cell lines. NK cell cytotoxicity was measured using the GranToxilux Plus kit (OncoImmunin, Inc.) according to the manufacturer's instructions. Target cells were loaded with 1:5000 TFL4 dye for 20 min at 37 °C, washed twice in PBS, and plated with effector cells. NK cells were plated at an effector/target ratio from 2:1 to 4:1, in the presence of the granzyme B substrate. After 60 min of incubation, cytotoxicity was measured using an LSR II flow cytometer (BD Biosciences).
Magnesium deprivation in vitro
For Mg2+ deprivation experiments, FBS was first deprived of Mg2+ and Ca2+ by incubating with Chelex 100 resin (Bio-Rad) twice before the pH was adjusted to 7.4. dRPMI was prepared by addition of 10% resin-treated FBS and 1.2 mm CaCl2 (Sigma) to Mg2+- and Ca2+-free RPMI 1640 medium. Cells were cultured in deprived medium or deprived medium plus 0.5 mm Mg2+ for 3 days before flow cytometric assessment cytotoxicity and NKG2D surface levels were carried out.
Proximity ligation assay
WT or MAGT1 KO Jurkat cells were seeded on 0.01% poly-l-lysine–coated coverslips for 10 min. The cells were washed with PBS and then were fixed in 4% paraformaldehyde for 10 min at room temperature. Cells were then permeabilized with ice-cold methanol at −20 °C for 30 min. After washing twice with PBS, PLA was performed using the Duolink PLA kit (Sigma). Briefly, cells were blocked with Duolink blocking solution for 1 h at 37 °C and then probed with 50 μl of a pair of primary antibodies at 37 °C for another 1 h. The unbound primary antibodies were removed by washing with Duolink buffer A twice for 5 min. Samples were then incubated with Duolink probes for 1 h at 37 °C, followed by Duolink buffer A wash for 5 min. Then 40 μl of diluted ligase were added to each coverslip and incubated at 37 °C for 30 min. Duolink buffer A was applied to wash away the ligase, and 40 μl of diluted polymerase were added to each sample to amplify the PLA signal for 100 min. Finally, cells were washed with Duolink buffer B twice at room temperature and mounted using antifade mountant with 4′,6-diamidino-2-phenylindole on glass slides for further analysis by confocal microscopy.
HEK 293T cell lines and mRNA transfection
The MAGT1 SXXS mutant was generated by replacing both Cys with serine by following the instructions of the QuikChange II site-directed mutagenesis kit (200523, Agilent). mRNAs of MAGT1, MAGT1 SXXS mutant, and TUSC3 were synthesized in vitro using T7 mScript Standard mRNA Production System (C-MSC11610, CellScript). Briefly, DNA templates (pCI-neo-MAGT1, pCI-neo-MagT1-SXXS, and pCI-neo-TUSC3) with T7 promoters were linearized by NotI digestion for 1 h at 37 °C. Linearized DNAs were incubated with NTP, DTT, RNase inhibitor, and mScript T7 enzyme at 37 °C for 30 min to allow in vitro transcription. DNA templates were removed by treatment with DNase I for 15 min at 37 °C, and ammonium acetate precipitation was used to purify mRNA. Purified mRNA was incubated with GTP, S-adenosylmethionine, RNase inhibitor, and ScriptCap 2′-O-methyltransferase at 37 °C for 30 min to add 5′-cap, followed by adding the 3′-poly(A) tail through mScript poly(A) polymerase. The MAGT1/TUSC3 KO HEK 293T cells were co-transfected with 1 μg of NKG2D DNA, 1 μg of DAP10 DNA, and either 3.5 μg of MagT1 mRNA, or MagT1 SXXS mRNA, or TUSC3 mRNA, or GFP mRNA (TriLink) by Lipofectamine 3000 (Thermo Fisher Scientific). After 12 h, surface NKG2D was stained and analyzed by flow cytometry. The expression levels of MAGT1, MAGT1 SXXS, and TUSC3 were analyzed by Western blotting.
Flow cytometry
For surface staining, cells were washed once in PBS and stained with anti-CD3 (clone HIT3a), anti-CD4 (clone RPA-T4), anti-CD8 (clone SK1), and anti-NKG2D (Clone 1D11) (all from BioLegend) in 100 μl of FACS buffer (2% FBS, 1 mm EDTA, 0.01% NaN3) for 30 min on ice. Cells were washed in FACS buffer before acquisition on either an LSR II or LSR Fortessa flow cytometer (BD Biosciences). Data were analyzed using FlowJo versions 9.9.5 and 10.2 (Treestar).
Deglycosylation
5 × 106 cells were lysed in 100 μl of RIPA lysis buffer (1% Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl, pH 7.4) with protease inhibitor (Roche Applied Science). The lysates were then centrifuged at 14,000 rpm at 4 °C for 10 min to remove insoluble material. Supernatants were transferred to a separate tube and used for subsequent experimentation. The samples were then treated with PNGase F (New England BioLabs) according to the manufacturer's instructions. Briefly, a stock of 10× denaturation buffer (5% SDS and 400 mm DTT) was added to the samples to a final concentration of 1×. The samples were denatured by heating reaction at 100 °C for 10 min and then chilled on ice. Denatured samples were mixed with 2 μl of 10× Glyco buffer 2 and 2 μl of 10% Nonidet P-40 and then diluted with 20 μl of ddH2O. 20 μl of the mixture was then transferred to 2 separate tubes (i.e. 40 μl total). One tube was treated with 1 μl of PNGase F (500,000 units ml−1, New England Biolabs) or ddH2O vehicle at 37 °C for 1 h.
Immunoprecipitation
1 × 106 cells were washed in PBS prior to detachment with 0.05% trypsin-EDTA (Invitrogen, catalog no. 25300-054) and trypsin neutralization with complete DMEM growth medium. Cells were washed with ice-cold PBS, and the pellets were subjected to five rounds of flash freezing in liquid nitrogen and thawing on ice. 50 mg of cell pellets were resuspended with 500 μl of ice-cold lysis buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, and 5% glycerol) supplemented with 1× protease inhibitor (Halt Protease Inhibitor Mixture, Thermo Fisher Scientific, catalog no. 87786) (10:1 v/w). Lysates were homogenized by five passages through a 25–5/8-gauge needle and then incubated on ice for 10 min. Samples were centrifuged at 14,000 × g for 10 min at 4 °C to clear the lysate. 2 μg of either anti-MAGT1, rabbit isotype control (Cell Signaling, catalog no. 3900S), or anti-HA (Biolegend, catalog no. 901501) antibody were added to the cell lysates and incubated overnight. 25 μl of pre-washed protein A magnetic beads (Pierce, catalog no. 88846) were then added to the antibody/lysate mixture, and samples were incubated on a tube rotator at room temperature for 45 min. Samples were placed in a magnetic rack, and the supernatant was removed. Protein A magnetic beads were then subject to four washes in 25 mm Tris, 0.15 m NaCl, 0.05% Tween 20, pH 7.5 (TBST). After the final wash, the immunoprecipitated proteins were eluted by incubation in low-pH elution buffer (Pierce IgG elution buffer, pH 2.0, catalog no. 21028).
Immunoblotting
Protein lysates were isolated from cell pellets via incubation in lysis buffer (1% Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl, pH 7.4) with protease inhibitor for 20 min, which were then clarified via centrifugation at 14,000 rpm at 4 °C for 20 min. Total protein levels were measured by BCA assay (Pierce). 100 μg of total protein was denatured by adding 2× SDS loading solution (Quality Biological) supplemented with 5% β-mercaptoethanol and heated at 95 °C for 10 min. Equal amounts of total protein were subjected to 4–20% NuPAGE Tris-glycine gels (Thermo Fisher Scientific) at 150 V for 2 h. Then the gel was transferred to a positively charged nitrocellulose membrane at 100 V for 1.5 h. The membrane was blocked with 5% (w/v) nonfat milk in TBST (25 mm Tris, 150 mm NaCl, 0.05% Tween 20, pH 7.2) and then probed with specific primary antibodies at 4 °C overnight, followed by washing with TBST buffer and further incubated with anti-rabbit or anti-mouse IgG antibody (1:3000) at room temperature for 1 h. Proteins were visualized by chemiluminescent substrate (Luminata Classico/Forte Western horseradish peroxidase substrate or Thermo Fisher Scientific Super Signal West Femto Maximum Sensitivity Substrate).
Antibodies against human antigens
The following antibodies were used: MAGT1 (Mer catalog no. 17-1, 1:50 dilution, produced by Merck); TUSC3 (Mer 85 catalog no. 38-1, affinity-purified, produced by Merck) (Fig. S6)8; β-actin (ab20272, Abcam); NKG2D (D-20, sc-9621, Santa Cruz Biotechnology); tubulin (AA2, 05-661, Merck Millipore); TCR-β (H-197, sc-9101, Santa Cruz Biotechnology); CERS2 (A303-193A, Bethyl Laboratories, Inc.); ribophorin I (C-15, sc-12164, Santa Cruz Biotechnology); COP1 (PA1-061, Thermo Fisher Scientific); EEA1 (C45B10, 3288 Cell Signaling); STT3B (HPA036646, Sigma); OST 48 (Santa Cruz Biotechnology); Na,K-ATPase α1 (D4Y7E, 23565, Cell Signaling).
Density gradient fractionation
500 million Jurkat cells were harvested by conventional tissue culture methods, and all subsequent steps were performed at 4 °C. Cells were washed twice in ice-cold PBS, resuspended in 5 ml of homogenization medium (HM) (0.25 m sucrose, 1 mm EDTA, 10 mm HEPES-NaOH, pH 7.4) with 1× Halt Protease inhibitor (Thermo Fisher Scientific), and disrupted by Dounce homogenization followed by repeat passages through a 25-gauge syringe needle. The homogenate was then centrifuged at 2000 × g for 10 min. A diluent solution (DS) was prepared (0.25 m sucrose, 6 mm EDTA, 60 mm HEPES-NaOH, pH 7.4). 2 and 25% (w/v) iodixanol solutions were prepared by diluting OptiPrep Density Gradient Medium (Sigma) in HM and DS at 24:1 and 1:1 ratios (v/v), respectively. 12–13-ml gradients made with equal volumes of the 2 and 25% iodixanol solutions were prepared, and the vesicle suspension (cell homogenate) was layered on top of the gradient. The tubes were then centrifuged in a swinging-bucket rotor (Beckman SW 41) at 200,000 × g for 3 h. Gradients were collected by tube puncture in 0.5-ml fractions. Protein quantification for each fraction was determined by bicinchoninic acid (BCA) assay prior to immunoblotting.
Human tissue Western blotting
Human tissue Western blotting was purchased from Zyagen (HW-MT-1). The membrane was blotted with anti-MAGT1 antibody and re-probed with anti-TUSC3 antibody.
Salivary and serum protein analysis
Salivary proteins were deglycosylated with PNGase F and then digested with either trypsin (which cleaves polypeptides just C-terminal to arginine and lysine) or AspN (which cleaves polypeptides just N-terminal to aspartic acid). The resulting peptide digests were subsequently analyzed by LC-MS/MS, and the occupancy of glycosylation sites was assessed using the ratio of the deamidated (deglycosylated) and nondeamidated (nonglycosylated) peaks. Changes in occupancy for glycopeptides after AspN digestion were detected using the ratio of the deglycosylated peptide to the sum of all peptides from the corresponding protein. Each value is expressed as a ratio of peptide levels detected in a particular patient versus peptide levels from the respective parent.
Protein structure modeling and analysis
Homology models were generated with I-TASSER and refined using ModRefiner. Images were generated with PyMOL.
RNA-Seq of XMEN and HC cells
CD8+ T cells from XMEN (four patients) or HCs (four donors) were isolated from PBMCs using CD8-positive selection magnetic beads (Miltenyi Biotec). On day 0, the cells were divided into two groups: half of which were immediately harvested prior to activation (see below), while the other half were activated. T cells were stimulated with anti-CD3 (Biolegend) and anti-CD28 (Biolegend) antibodies in the presence of 100 units ml−1 IL-2 in cRPMI. On day 3, aliquots of the activated cells were harvested. The remaining cells were washed twice with cRPMI and resuspended at 1 × 106 ml−1 with 100 units ml−1 IL-2. The cultures were then replenished with fresh IL-2 and medium every 2 days, until final harvesting on day 12. For each time point, harvested cells were pelleted and washed twice with ice-cold PBS. The pellets were suspended in 350 μl of lysis buffer (Qiagen RLT) and homogenized by pipetting. Samples were then transferred to barcoded tubes, flash-frozen in liquid nitrogen, and stored at −80 °C.
For RNA-Seq, poly(A)-selected RNA was isolated, and sequencing libraries were prepared using the Illumina TruSeq RNA Library preparation kit. For each sample replicate, we obtained ∼10 million paired 101-bp reads using the Illumina platform. Sequencing reads were mapped to human hg38 genome using Omicsoft Sequence Aligner (OSA) version 7.2 (47). We measured differential gene expression using DESeq2 version 1.22.1 (48), including log fold shrinkage using an adaptive normal distribution. Differentially expressed genes were those with an adjusted p value (Benjamin and Hochberg (BH) false-discovery rate (FDR)) <0.05 and log2 ratio >1. Genes on chromosome Y were removed from analysis because of the gender bias in patients (all male) versus controls (all female).
We performed gene ontology (GO) enrichment using ToppGene (49), and after filtering terms for BH FDR <0.000002, redundant GO terms were removed using REViGO (http://revigo.irb.hr/index.jsp)9 (50) with allowed similarity of 0.5 and using SimRel semantic similarity measure. Remaining GO terms were displayed as an Enrichment Map version 3.1.0 (51) network in Cytoscape version 3.7.0 (52) using a Jaccard index of 0.5 to draw edges between overlapping nodes/functions. To determine significantly enriched pathways, we performed a Core Analysis in IPA (Qiagen, Redwood City, CA) comparing XMEN to HC samples for each time point (53).
We performed a further Core Analysis in IPA to determine significantly activated (p < 0.05, Z-score >2) or inhibited (p < 0.05, Z-score < −2) upstream regulators and causal network master regulators from each time point, which were then intersected with the list of 73 differentially glycosylated proteins identified in our MS analysis of XMEN patient samples.8
Statistical analyses
Data were analyzed using GraphPad Prism 7.0. Depending on experimental design, statistical significance was tested via either two-tailed unpaired or paired Student's t test. p values ≤0.05 were considered significant, and p values >0.05 were considered nonsignificant (NS). Flow cytometry data were analyzed via Flowjo 9.9.5 and 10.2 (Treestar). No statistical methods were used to pre-determine sample size.
Author contributions
M. M.-L., M. B., and M. J. L. project conceptualization; M. M.-L., M. B., and M. J. L.; identifying resources; M. M.-L., M. B., J. Z., J. C. R., L. Z., C. K., P. J., G. N., H. J., E. M., Y. S., A. J. O., L. R. O., B. L. S., and M. J. L. data curation; M. M.-L., M. B., A. J. O., Y.S., and L. R. O. software analysis; M. M.-L., M. B., C. K., B. L. S., and M. J. L. scientific investigation; M. M.-L., M. B., J. Z., J. C. R., L. Z., C. K., P. J., G. N., H. J., E. M., A. J. O., L. R. O., B. L. S., and M. J. L. experimental design and methodology; M. M.-L. and M. B. writing of original draft; M. M.-L., M. B., and M. J. L. project administration; M. M.-L., M. B., J. Z., L. Z., C. K., P. J., G. N., H. J., E. M., Y. S., A. J. O., L. R. O., B. L. S., and M. J. L. writing, reviewing, and editing manuscript; M. B., J. Z., J. C. R., L. Z., P. J., G. N., H. J., E. M., Y.S., A. J. O., L. R. O., and B. L. S. data analysis; M. J. L. supervision.
Supplementary Material
Acknowledgments
We thank Drs. Reid Gilmore and Natalia Cherepanova for sharing constructs, antibodies, and HEK 293T KO cells. We thank Ryan Kissinger at the Visual and Medical Arts Department, NIAID, for help with illustrations. We thank Dr. Suk See DeRavin and Harry Malech for advice and help in experiments involving mRNA transfection into cells and confocal imaging. We thank Merck, Inc., for antibody preparation and general support of this work through a Nat Cooperative Research and Development Agreement. We thank Ke Huang, Sandhya Xirasagar, Darrell Hurt, and other members of the Bioinformatics and Computational Biosciences Branch (BCBB), NIAID, National Institutes of Health, and Yu Zhang for variant assessment and bioinformatics support.
This work was supported in part by grants from the Uehara Research Fellowship (201330032) from Uehara Memorial Foundation, the Japan Society for the Promotion of Science (JSPS) and NIH grant (71403), and by co-funding through the NIH Office of Disease Prevention, and by the Division of Intramural Research, NIAID, National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S6 and Data Sets S1–S5.
J. Ravell and M. Lenardo, unpublished observations.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- EBV
- Epstein-Barr virus
- XMEN
- X-linked magnesium deficiency with neoplasia infection and neoplasia
- NLG
- N-linked glycosylation complex
- OST
- oligosaccharyltransferase
- TCR
- T-cell receptor
- ER
- endoplasmic reticulum
- TRX
- thioredoxin
- TM
- transmembrane domain
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- NK
- natural killer
- HPT
- haptoglobin
- HEMO
- hemoglobin
- HC
- healthy control
- DEG
- differentially expressed gene
- gRNA
- guide RNA
- PLA
- proximity ligation assay
- ddH2O
- double-distilled H2O
- PNGase F
- peptide:N-glycosidase F
- ERGIC
- ER–Golgi intermediate compartment
- cRPMI
- complete RPMI
- dRPMI
- Mg2+-depleted RPMI
- MFI
- mean fluorescent intensity
- PBMC
- peripheral blood mononuclear cell
- IPA
- Ingenuity Pathway Analysis
- RNA-Seq
- RNA sequencing
- GO
- gene ontology
- BH
- Benjamin and Hochberg
- FDR
- false-discovery rate.
References
- 1. Goytain A., and Quamme G. A. (2005) Identification and characterization of a novel mammalian Mg2+ transporter with channel-like properties. BMC Genomics 6, 48 10.1186/1471-2164-6-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou H., and Clapham D. E. (2009) Mammalian MagT1 and TUSC3 are required for cellular magnesium uptake and vertebrate embryonic development. Proc. Natl. Acad. Sci. U.S.A. 106, 15750–15755 10.1073/pnas.0908332106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li F., Chaigne-Delalande B., Su H., Uzel G., Matthews H., and Lenardo M. J. (2014) XMEN disease: a new primary immunodeficiency affecting Mg2+ regulation of immunity against Epstein-Barr virus. Blood 123, 2148–2152 10.1182/blood-2013-11-538686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li F.-Y., Chaigne-Delalande B., Kanellopoulou C., Davis J. C., Matthews H. F., Douek D. C., Cohen J. I., Uzel G., Su H. C., and Lenardo M. J. (2011) Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature 475, 471–476 10.1038/nature10246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grubbs R. D., and Maguire M. E. (1987) Magnesium as a regulatory cation: criteria and evaluation. Magnesium 6, 113–127 [PubMed] [Google Scholar]
- 6. Romani A. M. (2011) Cellular magnesium homeostasis. Arch. Biochem. Biophys. 512, 1–23 10.1016/j.abb.2011.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cherepanova N. A., Shrimal S., and Gilmore R. (2014) Oxidoreductase activity is necessary for N-glycosylation of cysteine-proximal acceptor sites in glycoproteins. J. Cell Biol. 206, 525–539 10.1083/jcb.201404083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lanier L. L. (2009) DAP10- and DAP12-associated receptors in innate immunity. Immunol. Rev. 227, 150–160 10.1111/j.1600-065X.2008.00720.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaigne-Delalande B., Li F.-Y., O'Connor G. M., Lukacs M. J., Jiang P., Zheng L., Shatzer A., Biancalana M., Pittaluga S., Matthews H. F., Jancel T. J., Bleesing J. J., Marsh R. A., Kuijpers T. W., Nichols K. E., et al. (2013) Mg2+ regulates cytotoxic functions of NK and CD8 T cells in chronic EBV infection through NKG2D. Science 341, 186–191 10.1126/science.1240094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shrimal S., Cherepanova N. A., and Gilmore R. (2015) Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Semin. Cell Dev. Biol. 41, 71–78 10.1016/j.semcdb.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cherepanova N., Shrimal S., and Gilmore R. (2016) N-Linked glycosylation and homeostasis of the endoplasmic reticulum. Curr. Opin. Cell Biol. 41, 57–65 10.1016/j.ceb.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aebi M. (2013) N-Linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437 10.1016/j.bbamcr.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 13. Kelleher D. J., Karaoglu D., Mandon E. C., and Gilmore R. (2003) Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol. Cell 12, 101–111 10.1016/S1097-2765(03)00243-0 [DOI] [PubMed] [Google Scholar]
- 14. Mohorko E., Glockshuber R., and Aebi M. (2011) Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation. J. Inherit. Metab. Dis. 34, 869–878 10.1007/s10545-011-9337-1 [DOI] [PubMed] [Google Scholar]
- 15. Mohorko E., Owen R. L., Malojčić G., Brozzo M. S., Aebi M., and Glockshuber R. (2014) Structural basis of substrate specificity of human oligosaccharyl transferase subunit N33/Tusc3 and its role in regulating protein N-glycosylation. Structure 22, 590–601 10.1016/j.str.2014.02.013 [DOI] [PubMed] [Google Scholar]
- 16. Kelleher D. J., and Gilmore R. (2006) An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R 10.1093/glycob/cwj066 [DOI] [PubMed] [Google Scholar]
- 17. Wild R., Kowal J., Eyring J., Ngwa E. M., Aebi M., and Locher K. P. (2018) Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science 359, 545–550 10.1126/science.aar5140 [DOI] [PubMed] [Google Scholar]
- 18. MacGrogan D., Levy A., Bova G. S., Isaacs W. B., and Bookstein R. (1996) Structure and methylation-associated silencing of a gene within a homozygously deleted region of human chromosome band 8p22. Genomics 35, 55–65 10.1006/geno.1996.0322 [DOI] [PubMed] [Google Scholar]
- 19. Molinari F., Foulquier F., Tarpey P. S., Morelle W., Boissel S., Teague J., Edkins S., Futreal P. A., Stratton M. R., Turner G., Matthijs G., Gecz J., Munnich A., and Colleaux L. (2008) Oligosaccharyltransferase-subunit mutations in nonsyndromic mental retardation. Am. J. Hum. Genet. 82, 1150–1157 10.1016/j.ajhg.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zielinska D. F., Gnad F., Wiśniewski J. R., and Mann M. (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 10.1016/j.cell.2010.04.012 [DOI] [PubMed] [Google Scholar]
- 21. Caragata M., Shah A. K., Schulz B. L., Hill M. M., and Punyadeera C. (2016) Enrichment and identification of glycoproteins in human saliva using lectin magnetic bead arrays. Anal. Biochem. 497, 76–82 10.1016/j.ab.2015.11.024 [DOI] [PubMed] [Google Scholar]
- 22. Hwang H., Zhang J., Chung K. A., Leverenz J. B., Zabetian C. P., Peskind E. R., Jankovic J., Su Z., Hancock A. M., Pan C., Montine T. J., Pan S., Nutt J., Albin R., Gearing M., et al. (2010) Glycoproteomics in neurodegenerative diseases. Mass Spectrom. Rev. 29, 79–125 10.1002/mas.20221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang W., Laeyendecker O., Wendel S. K., Zhang B., Sun S., Zhou J.-Y., Ao M., Moore R. D., Jackson J. B., and Zhang H. (2014) Glycoproteomic study reveals altered plasma proteins associated with HIV elite suppressors. Theranostics 4, 1153–1163 10.7150/thno.9510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang S., Chen L., Chan D. W., Li Q. K., and Zhang H. (2017) Protein signatures of molecular pathways in nonsmall cell lung carcinoma (NSCLC): comparison of glycoproteomics and global proteomics. Clin. Proteomics 14, 31 10.1186/s12014-017-9166-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Quamme G. A. (2010) Molecular identification of ancient and modern mammalian magnesium transporters. Am. J. Physiol. Cell Physiol. 298, C407–C429 10.1152/ajpcell.00124.2009 [DOI] [PubMed] [Google Scholar]
- 26. Cherepanova N. A., and Gilmore R. (2016) Mammalian cells lacking either the cotranslational or posttranslocational oligosaccharyltransferase complex display substrate-dependent defects in asparagine linked glycosylation. Sci. Rep. 6, 20946 10.1038/srep20946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Söderberg O., Leuchowius K.-J., Gullberg M., Jarvius M., Weibrecht I., Larsson L.-G., and Landegren U. (2008) Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 45, 227–232 10.1016/j.ymeth.2008.06.014 [DOI] [PubMed] [Google Scholar]
- 28. Trifilieff P., Rives M.-L., Urizar E., Piskorowski R. A., Vishwasrao H. D., Castrillon J., Schmauss C., Slättman M., Gullberg M., and Javitch J. A. (2011) Detection of antigen interactions ex vivo by proximity ligation assay: endogenous dopamine D2-adenosine A2A receptor complexes in the striatum. BioTechniques 51, 111–118 10.2144/000113719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.-G., and Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- 30. Huntoon K. M., Wang Y., Eppolito C. A., Barbour K. W., Berger F. G., Shrikant P. A., and Baumann H. (2008) The acute phase protein haptoglobin regulates host immunity. J. Leukoc. Biol. 84, 170–181 10.1189/jlb.0208100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Urbaniak A., Jablonska K., Podhorska-Okolow M., Ugorski M., and Dziegiel P. (2018) Prolactin-induced protein (PIP)–characterization and role in breast cancer progression. Am. J. Cancer Res. 8, 2150–2164 [PMC free article] [PubMed] [Google Scholar]
- 32. Shin K., Tomita M., and Lönnerdal B. (2000) Identification of lactoperoxidase in mature human milk. J. Nutr. Biochem. 11, 94–102 10.1016/S0955-2863(99)00082-0 [DOI] [PubMed] [Google Scholar]
- 33. Taylor A. K., and Wall R. (1988) Selective removal of α heavy-chain glycosylation sites causes immunoglobulin A degradation and reduced secretion. Mol. Cell. Biol. 8, 4197–4203 10.1128/MCB.8.10.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andersen C. B., Torvund-Jensen M., Nielsen M. J., de Oliveira C. L., Hersleth H.-P., Andersen N. H., Pedersen J. S., Andersen G. R., and Moestrup S. K. (2012) Structure of the haptoglobin-haemoglobin complex. Nature 489, 456–459 10.1038/nature11369 [DOI] [PubMed] [Google Scholar]
- 35. Ravell J., Chaigne-Delalande B., and Lenardo M. (2014) X-linked immunodeficiency with magnesium defect, Epstein-Barr virus infection, and neoplasia disease: a combined immune deficiency with magnesium defect. Curr. Opin. Pediatr. 26, 713–719 10.1097/MOP.0000000000000156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fokkink W.-J., Selman M. H., Dortland J. R., Durmuş B., Kuitwaard K., Huizinga R., van Rijs W., Tio-Gillen A. P., van Doorn P. A., Deelder A. M., Wuhrer M., and Jacobs B. C. (2014) IgG Fc N-glycosylation in Guillain-Barré syndrome treated with immunoglobulins. J. Proteome Res. 13, 1722–1730 10.1021/pr401213z [DOI] [PubMed] [Google Scholar]
- 37. Schulz B. L., Stirnimann C. U., Grimshaw J. P., Brozzo M. S., Fritsch F., Mohorko E., Capitani G., Glockshuber R., Grütter M. G., and Aebi M. (2009) Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc. Natl. Acad. Sci. U.S.A. 106, 11061–11066 10.1073/pnas.0812515106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garrity D., Call M. E., Feng J., and Wucherpfennig K. W. (2005) The activating NKG2D receptor assembles in the membrane with two signaling dimers into a hexameric structure. Proc. Natl. Acad. Sci. U.S.A. 102, 7641–7646 10.1073/pnas.0502439102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tao J., and Haynes D. H. (1992) Actions of thapsigargin on the Ca2+-handling systems of the human platelet. Incomplete inhibition of the dense tubular Ca2+ uptake, partial inhibition of the Ca2+ extrusion pump, increase in plasma membrane Ca2+ permeability, and consequent elevation of resting cytoplasmic Ca2+. J. Biol. Chem. 267, 24972–24982 [PubMed] [Google Scholar]
- 40. Abolhassani H., Edwards E. S., Ikinciogullari A., Jing H., Borte S., Buggert M., Du L., Matsuda-Lennikov M., Romano R., Caridha R., Bade S., Zhang Y., Frederiksen J., Fang M., Bal S. K., et al. (2017) Combined immunodeficiency and Epstein-Barr virus-induced B cell malignancy in humans with inherited CD70 deficiency. J. Exp. Med. 214, 91–106 10.1084/jem.20160849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi Y. I., Duke-Cohan J. S., Ahmed W. B., Handley M. A., Mann F., Epstein J. A., Clayton L. K., and Reinherz E. L. (2008) PlexinD1 glycoprotein controls migration of positively selected thymocytes into the medulla. Immunity 29, 888–898 10.1016/j.immuni.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Holl E. K., O'Connor B. P., Holl T. M., Roney K. E., Zimmermann A. G., Jha S., Kelsoe G., and Ting J. P. (2011) Plexin-D1 is a novel regulator of germinal centers and humoral immune responses. J. Immunol. 186, 5603–5611 10.4049/jimmunol.1003464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen J., Li J., Lim F. C., Wu Q., Douek D. C., Scott D. K., Ravussin E., Hsu H.-C., Jazwinski S. M., Mountz J. D., and Louisiana Healthy Aging Study. (2010) Maintenance of naïve CD8 T cells in nonagenarians by leptin, IGFBP3 and T3. Mech. Ageing Dev. 131, 29–37 10.1016/j.mad.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pietrzak A., Grywalska E., Gerkowicz A., Krasowska D., Chodorowska G., Michalska-Jakubus M., Roliński J., Wawrzycki B., Radej S., Dybiec E., Wroński J., Sobczyńska-Tomaszewska A., Rudzki M., and Hadj-Rabia S. (2016) Immune system disturbances in Clouston syndrome. Int. J. Dermatol. 55, e241–e249 10.1111/ijd.13152 [DOI] [PubMed] [Google Scholar]
- 45. Garshasbi M., Hadavi V., Habibi H., Kahrizi K., Kariminejad R., Behjati F., Tzschach A., Najmabadi H., Ropers H. H., and Kuss A. W. (2008) A defect in the TUSC3 gene is associated with autosomal recessive mental retardation. Am. J. Hum. Genet. 82, 1158–1164 10.1016/j.ajhg.2008.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garshasbi M., Kahrizi K., Hosseini M., Nouri Vahid L., Falah M., Hemmati S., Hu H., Tzschach A., Ropers H. H., Najmabadi H., and Kuss A. W. (2011) A novel nonsense mutation in TUSC3 is responsible for nonsyndromic autosomal recessive mental retardation in a consanguineous Iranian family. Am. J. Med. Genet. A 155A, 1976–1980 10.1002/ajmg.a.34077 [DOI] [PubMed] [Google Scholar]
- 47. Hu J., Ge H., Newman M., and Liu K. (2012) OSA: a fast and accurate alignment tool for RNA-Seq. Bioinformatics 28, 1933–1934 10.1093/bioinformatics/bts294 [DOI] [PubMed] [Google Scholar]
- 48. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen J., Bardes E. E., Aronow B. J., and Jegga A. G. (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311 10.1093/nar/gkp427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Supek F., Bošnjak M., Škunca N., and Šmuc T. (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Merico D., Isserlin R., Stueker O., Emili A., and Bader G. D. (2010) Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5, e13984 10.1371/journal.pone.0013984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krämer A., Green J., Pollard J. Jr., and Tugendreich S. (2014) Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.