Abstract
Interferon-stimulated gene product 15 (ISG15) is a key component of host responses to microbial infection. Despite having been known for four decades, grasping the functions and features of ISG15 has been a slow and elusive process. Substantial work over the past two decades has greatly enhanced this understanding, revealing the complex and variable nature of this protein. This has unveiled multiple mechanisms of action that are only now beginning to be understood. In addition, it has uncovered diversity not only between how ISG15 affects different pathogens but also between the function and structure of ISG15 itself between different host species. Here we review the complexity of ISG15 within the context of viral infection, focusing primarily on its antiviral function and the mechanisms viruses employ to thwart its effects. We highlight what is known regarding the impact of ISG15 sequence and structural diversity on these interactions and discuss the aspects presenting the next frontier toward elucidating a more complete picture of ISG15 function.
Keywords: ISGylation, interferon, USP18, DUB, RIG-I
Abbreviations: IFN, interferon; ISG15, interferon-stimulated gene product 15; Ub, ubiquitin; LFA-1, leukocyte function-associated antigen-1; CCHFV, Crimean-Congo hemorrhagic fever virus; EAV, equine arteritis virus; SARS-CoV, severe acute respiratory syndrome-related coronavirus; MERS-CoV, Middle East respiratory syndrome-related coronavirus; DUB, deubiquitinating protein; OTU, ovarian tumor domain protease; PLP, papain-like protease; ISRE, IFN response stimulated elements
Graphical abstract
Introduction
Interferon (IFN)-stimulated gene product 15 (ISG15) is one of the most highly induced genes in response to viral infection. ISG15 is part of a class of proteins sharing structural homology to ubiquitin (Ub), known as Ub-like (proteins), that also includes SUMO, Nedd8, and FAT10 [1], [2], [3], [4], [5], [6], [7], [8], [9]. Both Ub and Ubl proteins are key mediators and regulators of numerous cellular processes, and ISG15 is no exception. Despite being the first Ubl protein discovered, efforts to attain a cohesive characterization of ISG15 lagged behind relative to most of its sister Ubl proteins. While its immunological function, particularly as an antiviral protein, was inferred based on its upregulated levels upon IFN treatment, a detailed understanding of its roles was lacking. Recent studies have begun to unveil the more specific mechanisms of ISG15 action, revealing varied and apparently contrasting functions for ISG15. These have introduced a complexity to our understanding of ISG15, showing it to not only to be intrinsically multifunctional but also to display functional and structural diversity between different species. Understanding these differences will be key to ascertaining the impact on the microbe-host interface, as well as the potential connections this could have to host immune responses, microbe species tropism, and pathogenesis. In this review, we focus on ISG15 within the context of its antiviral function and especially how various viruses have evolved strategies to counter the effects of ISG15. We highlight the emerging picture of how ISG15 species diversity influences its immune function and the ability of viral proteins to thwart its effects.
Discovery of ISG15
ISG15 was first identified in 1979 in IFN-treated cells [10], although its Ub-like nature was not reported until 1987, when it was found to cross-react with Ub antibodies [11]. ISG15 was renamed in 1987 from general references to a “15-kDa protein” to its current name after the discovery that its transcription was driven by IFN-β, and the term “interferon-stimulated gene” was coined [12], [13]. This IFN-dependent expression prompted studies investigating the contribution of ISG15 to antiviral responses. The innate immune response is the first line of defense against invading pathogens, which are sensed by host pattern-recognition receptors. For example, viral RNA is detected by cytoplasmic sensors such as RIG-I and MDA5 (Fig. 1a). This triggers various downstream signaling pathways resulting in the expression of type I IFNs and proinflammatory cytokines. Type I IFNs are released from infected cells and subsequently bind to IFN α/β receptors of the infected cell as well as neighboring cells, thus exerting both autocrine and paracrine effects. Binding of secreted IFN to its receptor activates downstream signaling that ultimately results in the expression of hundreds of IFN-stimulated genes (ISGs), whose promoters contain IFN response stimulated elements (ISRE) (Fig. 1a). ISG-encoded proteins, including ISG15, are critical orchestrators of the host cell defense arsenal to viral infection. The role of ISG15 in antiviral immunity has been investigated extensively in mouse models, and several viruses were found to be more pathogenic in ISG15 KO mice [14].
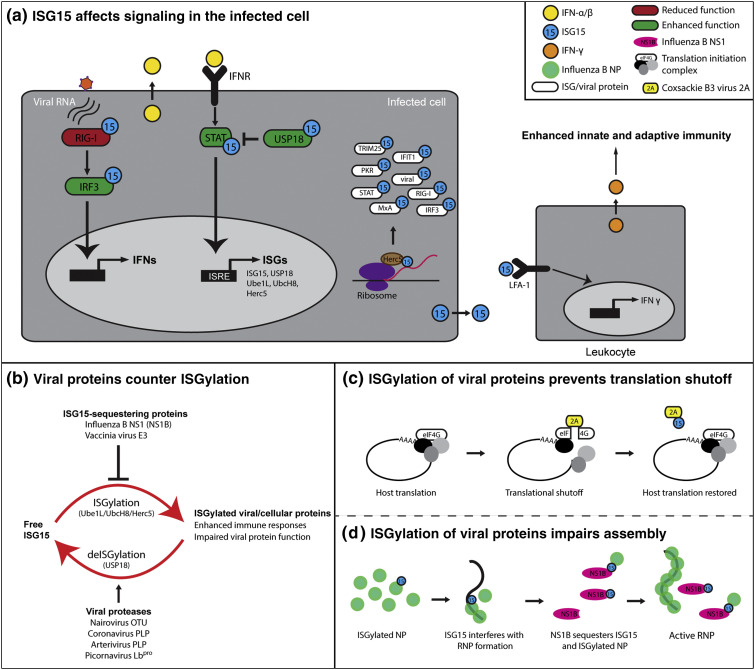
Fig. 1.
ISG15 is conjugated to a wide range of viral and cellular proteins, influencing immune responses. (a) The infected host cell senses viral RNA via RIG-I, which induces signaling leading to the expression and secretion of type I IFN. Type I interferon binding to IFN receptor (IFNR) result in the expression of IFN-stimulated genes (ISGs), including ISG15 and its conjugating enzymes (Ube1L, UbcH8, and Herc5). Herc5 association to ribosomes results in the ISGylation of newly synthesized host and viral proteins including several ISGs (white). ISG15 conjugation can inhibit (red) or enhance (green) the activation state of innate immune signaling proteins. USP18 is a negative regulator of IFNR signaling by limiting STAT activation. USP18 protected from degradation by ISG15 binding, thereby enhancing the inhibition of IFNR signaling. Extracellular ISG15 binds to LFA-1 receptors of leukocytes and induces the expression and secretion of IFN-ɣ. (b) Viral proteins can counter ISGylation by sequestering ISG15/ISGylated proteins, or by deconjugating ISG15. (c) The 2A protease of Coxsackie B3 virus induces host translational shutoff by cleaving eIF4G. ISGylation of 2A restores host translation by preventing eIF4G cleavage. (d) ISGylation of viral proteins interferes with virus replication. ISGylation of Influenza B virus NP reduces the oligomerization and formation of viral ribonucleoprotein complexes (RNPs). NS1B counters ISGylation by sequestering ISG15 and ISGylated NP.
The Intracellular Role of ISG15
In keeping with its similarity to Ub, our current knowledge about ISG15 suggests its role to be predominantly intracellular through its conjugation to lysine (K) residues in a process called ISGylation, although unconjugated (free) ISG15 also has various functions. ISGylation requires a three-step enzymatic cascade involving an E1 activating enzyme (Ube1L), an E2 conjugating enzyme (UbcH8), and an E3 ligase (Herc5 or TRIM25/EFP). ISGylation is reversed by Ub-specific protease USP18 [15]. Like ISG15, the expression of its conjugating enzymes and USP18 is upregulated by IFN (Fig. 1a). In contrast to the hundreds of E1–E2–E3 enzymes available for formation of complex poly-Ub chains, ISG15 is conjugated to target proteins as a monomer by a very limited set of E1–E2–E3 enzymes. ISGylation appears to take place predominantly at the ribosomes, due to the localization of the dominant ISG15 E3 ligase (Herc5), which largely limits ISGylation to newly synthesized proteins [16]. Therefore, during viral infection, actively translated proteins such as ISGs and viral proteins are preferentially ISGylated (Fig. 1a) [16], [17], [18], [19], [20], [21], [22].
The effects of free ISG15 and ISGylation on cell biology are diverse and can be stimulatory as well as inhibitory. ISGylation can enhance antiviral signaling pathways by prolonging the activation state of signaling proteins (e.g., IRF3, STAT1), resulting in a higher production of type I IFN and ISGs [23], [24]. Although ISG15 plays an important role activating antiviral immunity, it also provides negative feedback suppression of antiviral signaling pathways. Type I IFN activation is tightly regulated to prevent excessive immune responses. ISG15 negatively regulates type I IFN signaling at multiple levels. For example, ISGylation of RIG-I results in reduced levels of IFN promoter activity, and ISG15 binding targets RIG-I for autophagic degradation [25], [26]. USP18 regulates antiviral responses by removing ISG15 conjugates and can directly inhibit type I IFN receptor signaling by binding the subunit 2 of the receptor via STAT-2 [27], [28]. This interaction prevents the dimerization of the IFNAR subunits and recruitment of JAK1 necessary for the phosphorylation and activation of STAT1, which induces the transcription activation of ISGs. Therefore, USP18 dampens the immune responses by reversing protein ISGylation and directly inhibiting IFN signaling (Fig. 1a,b). In human cells, binding of ISG15 to USP18 increases USP18 levels by preventing its degradation [29], further reinforcing the inhibition of IFN receptor signaling. Therefore, USP18 and/or ISG15 depletion results in prolonged type I IFN signaling and enhanced levels of ISGs [30]. Similar to human cells, murine USP18 dampens IFN signaling by binding to the IFN receptor, but mouse USP18 is not stabilized by mouse or human ISG15 [30]. This greatly limits the extrapolation of conclusions obtained in murine systems to humans and emphasizes evolutionary divergence of ISG15 between species and potential different roles in regulating the antiviral responses. ISG15 KO mice are more susceptible to various viruses, and in most cases, the antiviral activity of ISG15 appeared to require conjugation, as preventing conjugation by knocking out Ube1L recapitulated the increased viral replication observed in ISG15 KO mice [14]. Unlike mice, ISG15-deficient patients are not more susceptible for viral infection; in fact, they appear to be more resistant due to higher basal levels of ISG expression [29].
ISG15 as an extracellular cytokine
Given its structural similarity to Ub, a somewhat surprising feature of ISG15 is its ability to function as an extracellular signaling molecule. ISG15 is secreted or released by various cell types, including fibroblasts, neutrophils, monocytes, and lymphocytes [31], [32], [33], [34]. The receptor for extracellular ISG15 was identified recently as the leukocyte function-associated antigen-1 (LFA-1), an adhesion molecule of the integrin family composed of an ɑL and β2 subunit [35]. LFA-1 binding to the intercellular adhesion molecule 1 is critical in the homing of leukocytes to sites of inflammation. ISG15 does not compete with intercellular adhesion molecule 1 binding to LFA-1. Instead, secreted ISG15 acts on natural killer cell and T lymphocytes to enhance the secretion of IFN-ɣ (type II IFN) secretion, which is important for the activation of innate and adaptive immune responses [34], [36]. ISG15-deficient patients are highly susceptible to mycobacterial disease because of their IFN-ɣ deficiency [31]. In addition, ISG15 induces natural killer cell proliferation and enhanced lytic capabilities of lymphokine-activated killer-like cells [36].
ISG15 and viral proteins
Beyond ISG15 conjugation to host proteins, various viral proteins have also been identified as ISG15 targets. ISGylation of viral proteins can affect their function by interfering with their localization, protease activity, or ability to interact with host proteins or other viral proteins. For example, ISGylation of Coxsackie B3 virus (CVB3) 2A protease prevents the induction of cellular host shut-off by preventing cleavage of eIF4G (Fig. 1c) [37]. ISGylation of viral proteins that oligomerize, required for the formation of viral replication complexes and/or particle assembly, is especially efficient, as only a small fraction of viral proteins has to be modified in order to cause a dominant inhibitory effect. This has been described for NP of Influenza B virus (Fig. 1d) and the human papillomavirus L1 capsid protein [16], [38]. A particularly well-studied ISGylated viral protein is NS1 of Influenza A virus, a major virulence factor. ISGylation of Influenza A virus NS1 prevents its ability to form homodimers and interaction with PKR, which is required for suppression of host antiviral responses. Mutating the lysine residues predominantly targeted for ISGylation in NS1 produces viruses that demonstrate increased viral growth in infected cells, and enhanced virulence in mice [22].
Considering the potential negative impact of ISG15 on viral processes, it is not surprising that viruses have evolved various mechanisms to inhibit or reverse ISGylation. The main viral strategies to counter ISGylation fall into two general categories: the use of viral proteins to sequester ISG15/ISGylated proteins (e.g., Vaccinia virus E3 and NS1 of Influenza B (Fig. 1c)), or the use of virally encoded proteases to remove ISG15 conjugates from their target proteins (Fig. 1b). Viruses that encode viral proteases able to reverse ISG15 conjugation include nairoviruses [e.g., Crimean-Congo hemorrhagic fever virus (CCHFV), Nairobi sheep disease virus, and Erve virus) [39], arteriviruses [e.g., equine arteritis virus (EAV) and porcine reproductive and respiratory syndrome virus] [39], picornaviruses (e.g., foot and mouth disease virus) [40], and coronaviruses [e.g., severe acute respiratory syndrome-related coronavirus (SARS-CoV), Middle East respiratory syndrome-related coronavirus (MERS-CoV), and mouse hepatitis virus] [41]. These cysteine proteases typically also possess substantial activity in reversing Ub conjugation and thus are often referred to as viral deubiquitinating proteins (DUBs). These DUBs are generally classified as ovarian tumor domain proteases (OTUs), papain-like proteases (PLPs), or leader proteases (Lbpro) with some overlap in the descriptions. Although viral proteins are a primary target of ISGylation and thus a natural target for deISGylating activity, it is equally likely that viral DUBs counter ISGylation of host proteins to disrupt immune regulation. Nairovirus OTUs are one of the better characterized viral DUB families with a total of 12 x-ray crystal structures solved, including three OTU–ISG15 complex structures. OTU activity has been associated with immune suppression and reduction in ISG15 conjugates [42], [43], [44]. In addition, a recent study has tentatively associated the deISGylase activity of nairovirus OTUs with higher levels of the L protein in CCHFV [43]. Coronavirus PLPs have also been studied relatively extensively at the molecular and cellular level. This includes five PLP–ISG15 complex structures. The SARS-CoV PLP was among the first viral DUBs to be recognized as possessing deISGylase activity and has been associated with the reduction of ISG15 conjugates and enhanced immune suppression [41], [44], [45]. Similar effects were observed in the more recently emergent MERS-CoV, suggesting an association between coronavirus pathogenicity and the presence of strong deISGylating activity [46]. Currently, the precise role for viral deISGylase activity is unknown, as no specific cellular targets have been firmly established. However, the fact that human pathogens like CCHFV, SARS-CoV, and MERS-CoV all possess robust deISGylase activity, which in the case of SARS-CoV exceeds its deubiquitinating activity, suggests that it may have an important function [45], [46], [47]. In addition to the more extensively studied nairovirus OTUs and coronavirus PLPs, deISGylating activity has also been observed in arterivirus OTU-like PLPs, EAV PLP2, and the porcine reproductive and respiratory syndrome virus PLP2 [42], [48], [49], [50], [51], [52]. Picornavirus Lbpro's also possess deISGylating activity, and for foot and mouth disease virus, this is the most prominent deconjugating activity [40]. Overall, the widespread occurrence of viral proteins targeting ISG15 conjugation suggests that the ISG15–virus interface is a major factor impacting the balance between host responses and viral countermeasures.
ISG15 Possesses Interspecies Diversity that Translates to Variation in Protein Structure
In comparison to Ub and other Ubl proteins, the sequence diversity of ISG15 has long stood out as an intriguing feature. ISG15 consists of two Ubl domains bearing the characteristic β-grasp fold containing four β-sheets and single α-helix per domain. These two domains are connected by a polypeptide sequence described as a “hinge” [53] (Fig. 2 ). ISG15 ultimately terminates with the familiar LRLRGG sequence, also found in Ub, that is critical for conjugation to proteins as a posttranslational modification. Beyond the C-terminus and other select regions of the protein, however, ISG15 shows substantial sequence variation (Fig. 2). In the most extreme cases, such as between some mammalian and fish species, ISG15s can share sequence identities of just 30%–35%. Even between two different mammals, the sequence identities can be less than 60%. Although substantial, this marked variance is not entirely unexpected, as it would be in keeping with ISG15's function as an immune molecule [55]. Related to this, it has been suggested that only certain features of ISG15, such as those driving the Ubl folds and interactions with proteins in the conjugation system, need to be conserved in order to serve its functions [56]. The cross-species compatibility of ISG15 enzymes is generally consistent with this idea [57], [58] and may account for the higher degree of sequence variability in ISG15 compared to its interactive partners in the conjugation system. Other observations, however, suggest a more nuanced picture and that sequence diversity could have functional implications. Pattyn and coworkers [57] reported that ISG15 from Old World monkeys demonstrated a higher degree of ISGylation in human and mouse cells compared to the native ISG15s. Mapping of the differences within the predicted interface with the E1 enzyme UbE1L revealed the causal residues mediating this effect. Interestingly, some of the positions with the most influence possessed highly similar residues, such as an asparagine versus aspartate at position 89, demonstrating that even subtle differences may contribute to species–species differences. Whether the differences in ISGylation efficiency are meaningful in the context of a viral infection remains to be determined. It conceptually lends possible credence, however, to the suggested mechanism for the differences in the effects of ISG15 on USP18 in humans versus mice [30]. Human ISG15 was observed to bind more strongly to USP18 compared to mouse ISG15, accounting for the protection of USP18 from degradation in human cells. The sequence divergence of ISG15 and USP18 between humans and mice led the authors to suggest that this may lie at the root of this difference in function. While a crystal structure of mouse USP18–ISG15 has been utilized to account for the specificity of ISG15 over Ub, analyses have yet to be performed that clearly determine the cause for the differences in USP18–ISG15 interactions between humans and mice [59], [60].
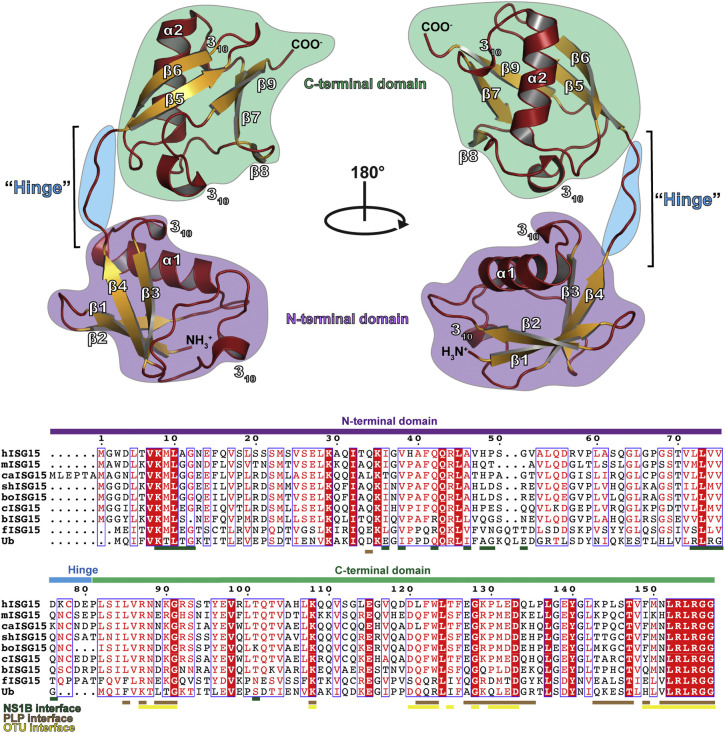
Fig. 2.
Structure of human ISG15 (PDB entry 1Z2M). The secondary structure is denoted with helices and loops shown in red and sheets shown in gold. The C-terminal domain is denoted by a green background, hinge region by a blue background, and the N-terminal domain by a purple background. A sequence alignment of human ISG15 (hISG15), mouse ISG15 (mISG15), canine ISG15 (caISG15), sheep ISG15 (shISG15), bovine ISG15 (boISG15), camel ISG15 (cISG15), vesper bat ISG15 (bISG15), fish ISG15 (fISG15), and Ub is shown with the domain architecture indicated with colored bars. The residues of ISG15 known to interact with the influenza B NS1 protein, coronavirus PLPs, and nairovirus OTUs are indicated. The sequence alignment graphic was generated using the ESPript server [54].
Beyond the effect of species variance within potential protein–protein interfaces, evidence has been accumulating suggesting that ISG15 sequence diversity impacts the tertiary structure, particularly the orientation of the two Ubl domains relative to each other (Fig. 3a). Prior to 2017, the only full-length structure of ISG15 available was human. This included crystal structures of the ISG15 by itself and in complex with viral proteins, including the NS1 protein from Influenza B and the OTU from CCHFV [53], [61], [62], [63]. Although present in different biochemical and crystallographic environments, the ISG15 molecules in these structures showed a remarkable degree of consistency in the orientation of the Ubl domains. From this alone, it would be natural to assume that all ISG15s possess similar structural characteristics. Structures of mouse ISG15 crystallized alone and in complex with USP18, however, revealed this to not be the case [59], [64]. These structures revealed a wide variability in the interdomain arrangement of both individual mouse ISG15 molecules and in comparison to the human ISG15 structures. Examination of the interface between the two domains uncovered specific points of divergence in mouse versus human ISG15 that may account for the differences observed in the structures. In particular, at residue position 39 human ISG15 possesses a histidine, in contrast to proline for most other ISG15s, that may contribute steric and electrostatic factors limiting the conformations it is able to adopt [64]. Mouse ISG15, on the other hand, possesses a wider hydrophobic interface that is less constricting of the interdomain arrangement. This theme was further bolstered with the recently reported structure of vesper bat ISG15 that showed an even wider range of structural modes than previously observed [66]. It also revealed the central role of a highly conserved phenylalanine (Phe40/41) in the N-terminal domain in forming the interdomain contact regardless of the overall structure. Mutation of this residue diminished binding with the SARS-CoV PLP, which is known to interact with both Ubl domains of ISG15, showing this structure-stabilizing effect of the interdomain interface to be of biochemical importance. Beyond this direct domain–domain interaction, the vesper bat ISG15 structure also unveiled an underappreciated role of the hinge region in influencing tertiary structure. Specifically, the sequence composition of the hinge region in bat ISG15 allows it to form a type I reverse turn that is absent from human and mouse ISG15, possibly stabilizing the structural conformation. While this particular role of the hinge region is a novel observation, it is not the only example of species diversity in the hinge region having structural impact. Sorensen and colleagues [72] demonstrated that the lack of three residues within the hinge region of bovine ISG15 compared to sheep ISG15 contributed to the lower stability of bovine ISG15 in solution, potentially as a result of different spatial arrangement of the two Ubl domains. The recently reported crystal structure of bovine ISG15 bound to the Influenza B NS1 protein tentatively supports this conclusion, as the two Ubl domains are twisted in a completely different orientation compared to what has been observed in other ISG15s [65]. Solving the structure of the highly similar sheep ISG15 would be valuable in validating the contribution of these hinge region residues to the tertiary structure.
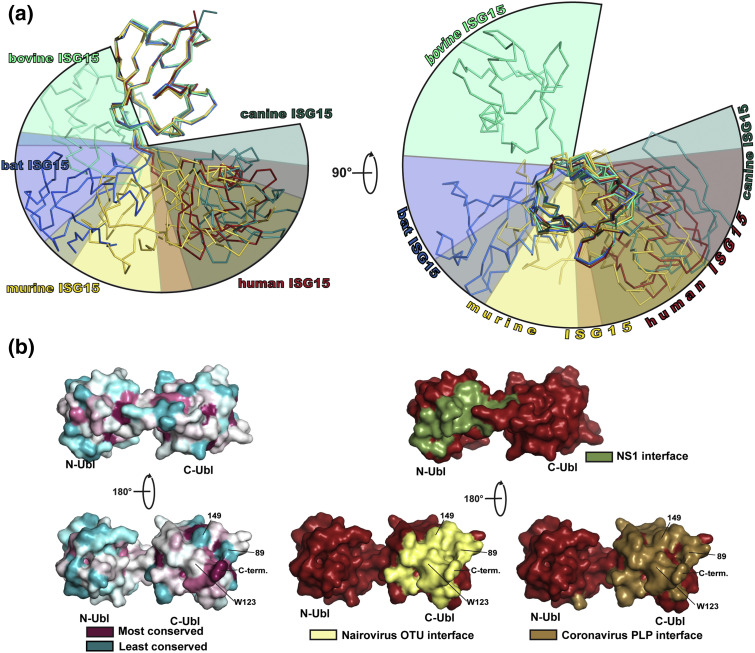
Fig. 3.
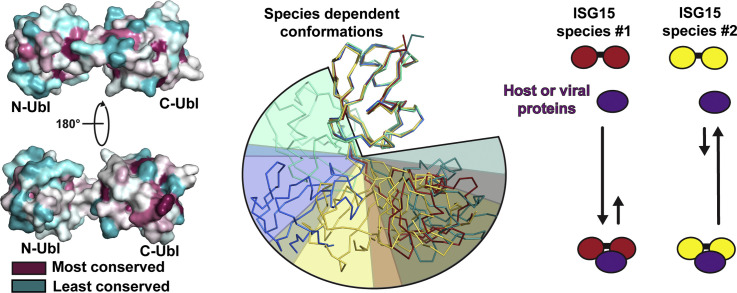
ISG15 structural diversity. (a) The C-terminal domains of ISG15 chains from human ISG15 (red, [53], [61], [62], [63] and PDB entries 3R66 and 6BI8), murine ISG15 (gold, [59], [64], [65] and PDB entries 5CHF and 5CHW), bat ISG15 (blue, [66]), bovine ISG15 (cyan green, [65]), and canine ISG15 (dark teal, [65]) were overlaid to identify the trends in domain–domain orientations between species. Representative chains were selected for each species. For murine ISG15, two structures were included to show the wider range of conformations that have been observed. Colored wedges show the approximate spatial range occupied by the N-terminal domain of ISG15 from each species. (b) Surface rendering of ISG15 showing the relative conservation among > 100 species (left). Calculations and renderings were performed using the ConSurf server and PyMOL [67], [68], [69], [70], [71]. Surface of ISG15 known to interact with viral proteins based on x-ray crystal complex structures (right), with the interface with the influenza B NS1 protein shown in green, nairovirus OTU interface in yellow, and coronavirus PLP in brown. The template ISG15 structure for surface renderings was human ISG15 (PDB entry 1Z2M).
Overall, the current structural evidence suggests that ISG15 species diversity influences tertiary structure characteristics. The primary drivers center around conserved residues forming a hydrophobic interface between the domains that permits an array of possible conformations, with other interdomain interactions and characteristics of the hinge region limiting the range of motion. Regrettably, limited information exists to determine the full structural dynamics of different ISG15s as the only full-length structures available were determined by x-ray crystallography. Although NMR assignments have been made for human ISG15, a complete structure has not been determined [73]. Further structural characterization would bring greater clarity. In addition, it remains to be seen what effects this could have within the ISGylation system of different species, and what magnitude of biochemical effects is needed to observe cellular ones. While the N-terminal Ubl domain of ISG15 has been shown to influence ISG15 conjugation, it is not clear how it contributes to this and whether variability in preferred ISG15 interdomain arrangements would influence it [57], [74]. Furthermore, it is unclear what degree of difference in the efficiency of ISGylation would be necessary to have a biological impact. Additional work will be needed in order to address these questions.
ISG15 Sequence and Structural Diversity Impacts Virus Protein–Protein Interactions
Although it appears that ISG15 diversity may affect cellular proteins involved in the ISGylation/deISGylation system, the impact of ISG15 interspecies differences on endogenous proteins and processes still remains ambiguous. In contrast, ISG15 sequence diversity has been shown to unequivocally affect the countermeasures some viruses have mounted against it. The most well-studied case remains the first characterized example of viral protein ISG15 species specificity with the Influenza B NS1 protein. Influenza B has a narrow host tropism, with humans serving as one of the primary hosts. The observation that the NS1B protein was able to more efficiently bind human and non-human primate ISG15 compared to other species' ISG15, such as mouse, canine, and bovine, suggested that it could be a major factor in determining host susceptibility [58], [75], [76]. Studies utilizing mutagenesis and structural approaches were able to map the determinant interactions for strong binding to the N-terminal Ubl domain and hinge region of ISG15 (Figs. 2, 3b) [58], [62], [63], [65], [76]. Mutations to key residues within the hinge region alone are sufficient to drastically impact the binding [65], [76]. Exchanging two residues in human ISG15 with the corresponding ones in murine, canine, and bovine ISG15 reduced binding to levels comparable with each of those species. Conversely, the opposite exchange increased the binding of murine, canine, and bovine ISG15 to similar levels as human ISG15. This provides a molecular mechanism that can account for the different cellular phenotypes observed in response to influenza B infection in different species and indicates that the hinge may be the primary driver of species specificity for NS1B.
Although Influenza B via the NS1 protein is the best-characterized example of a virus affected by ISG15 interspecies diversity, recent studies indicate that the impact may extend to other viruses. Nairovirus OTUs have been observed to interact with ISG15s from different species to different degrees in vitro [77], [78]. Comparison between crystal structures of the Erve virus OTU bound to the C-terminal Ubl domain of mouse ISG15 and the OTU of CCHFV bound to human ISG15 provided insights into the possible contributing factors to these differences. This highlighted four residues (residue positions 89 and 149–151 in human ISG15) that show a high variability among ISG15s [78]. Interestingly, position 150 is not within the direct binding interface. Instead, the difference in this position between human and mouse ISG15, methionine and lysine, respectively, influences the orientation of the residue present in position 89, illustrating that factors beneath the protein surface can impact the binding interface with viral OTUs. In a similar vein, coronavirus PLPs were demonstrated to possess substantial variation in their ability to interact with different ISG15s [64]. Importantly, it also reflected differences in the known breadth of host ranges between different coronaviruses. The PLP2 from mouse hepatitis virus, which primarily infects mice, showed essentially no activity for some ISG15s (including human) while possessing robust activity for mouse ISG15. In contrast, the PLPs from SARS-CoV and MERS-CoV, which are known to have a wider host range, were somewhat promiscuous deISGylases. In addition to these substrate specificity trends, crystal structures of the SARS-CoV PLP bound to the C-terminal Ubl domains of human and mouse ISG15 revealed novel insights into how interspecies sequence diversity influences the nature of enzyme substrate binding. Surprisingly, mouse ISG15 bound in an orientation shifted 27° compared to human ISG15, with corresponding shifts in the protease structure to accommodate it. Coupling the structural analysis with isothermal titration calorimetry data revealed that the C-terminal Ubl domain of mouse ISG15 contained less optimal elements for protease binding, and that the N-terminal Ubl domain contributed substantially more to binding when compared to human ISG15. While the regions of SARS-CoV PLP that interact with both domains of ISG15 are known, it remains to be determined what portion(s) of the N-terminal Ubl domain of ISG15s is involved in this interaction and how potential differences in the domain–domain orientation impact the interface [45], [64], [79]. Interestingly, similar analyses with the MERS-CoV PLP revealed it to interact solely with the C-terminal Ubl domain, indicating that the driving factors for interaction may not be universal even within a family of viruses [80].
Altogether, a picture is emerging of ISG15 sequence and structural diversity impacting the ability of viruses to counteract its effects. In the case of Influenza B, these differences could be highly influential in determining host tropism. For other viruses affected by ISG15 species diversity, including nairoviruses and coronaviruses, it remains to be determined what the threshold of difference may be in order to observe host-specific effects. The fact that various viruses interact with different portions of the ISG15 molecule opens the possibility that most, or all, of the ISG15 surface could be targeted by viral proteins. Thus, in addition to there being host-specific effects due to ISG15 diversity, the potential also exists for diverse and virus-specific mechanisms to disrupting ISG15 function. Further complicating these is the potential that, as in the case of humans compared to mice, ISG15 from different species may not necessarily play analogous roles. Differences in the relative importance of ISG15 between species could raise the importance of other IFN antagonists and produce unique relationships at each virus–host interface.
Current Outlook and Challenges to Evaluating the Specific Effects of ISG15
While the effects and function of ISG15 are slowly emerging, many details remain to be determined regarding the mechanism of its action. Although ISG15 and ISGylation knockouts have been key to establishing the basic modes of action against several viruses, more nuanced methods may be necessary to fully grasp ISG15 function and viral countermeasures. One approach consists in specifically attenuating the action of ISG15-interacting viral proteins and assessing the activity using mutant viruses. Beyond the technical hurdles associated with developing reverse genetics systems required to modify the viral genomes, another challenging aspect to achieving this lies in the fact that viruses often encode multiple protein antagonists targeting the innate immune responses at multiple levels. This complicates the study to identify individual contribution of each protein, especially when they have redundant or overlapping functions. Therefore, the function of an individual viral protein, or domain thereof, is often studied in biochemical assays using purified forms, allowing insight into mechanistic details that can complement studies using mutant recombinant viruses and their pathogenic role in vivo. In addition to the challenges of distinguishing between the individual contributions of different immune antagonists, the multifunctional nature of many viral proteases can make it difficult even within a single protein to selectively influence one interaction it displays over others. For example, viral deISGylating enzymes such as OTUs and PLPs often possess substantial deubiquitinating activity and in some cases play a critical role in virus polyprotein processing required for polymerase maturation. Due to such critical roles in the viral life cycle, elucidating the function cannot rely on simply deleting the protease from the genome or removing its baseline catalytic activity, and must instead rely on specific reduction in the ability to interact with Ub and/or ISG15. While this presents a challenging task, several encouraging advancements have been made recently that indicate this to be a feasible approach. A reverse genetics system for EAV was successfully developed with structure-guided OTU mutants that reduced deubiquitinating activity while leaving the proteolytic function unperturbed [42]. This study was able to show the impact of the loss on the ability of EAV to block the expression of IFN-β in cellular systems. Further work was able to successfully apply these mutants to in vivo assessment in shetland mares as a possible vaccine candidate [81]. This provided protection against EAV infection and demonstrated the feasibility of creating viable DUB-deficient mutant recombinant viruses as vaccine candidates, though under the conditions studied it could not be established that this provided greater benefit compared to wild-type virus. Another recent achievement demonstrated a similar decoupling of activity with PLP mutants in a SARS-CoV reverse genetics system, also providing key insights into virus immune suppression and pathogenesis [44]. Regrettably, these studies focused solely on the effect of these mutants on ubiquitination. It would be interesting to revisit these mutants to assess whether they have any impairment in deISGylase activity, or possibly pursue additional ones that might provide insight into the impact of ISGylation for these viruses. Currently, the only reverse genetics system that has been utilized to investigate the impact on ISGylation is for CCHFV [43], [82]. This was able to substantiate different effects that were mediated by ubiquitination versus ISGylation in the context of a nairovirus infection by comparing a mutant lacking both Ub and ISG15 activity with one deficient in only Ub activity. Unfortunately, current insights are limited by the lack of a deISGylase-specific OTU mutant, as the most promising candidates to date have also yielded an impairment in deubiquitinase activity [43], [61], [83]. Similar difficulty has been encountered with the SARS-CoV PLP, with mutants typically exhibiting a moderate effect or substantially impacting both Ub and ISG15 activities [64], [79]. Recently, however, Daczkowski and co-workers [80] were able to perform structure-guided mutagenesis to selectively reduce in vitro deISGylase activity of the MERS-CoV PLP without appreciably impacting the other enzymatic functions. Application of these mutants to cellular and in vivo assays would provide novel insights into the relative importance of ISGylation in combatting coronavirus infection.
The nonconserved nature of ISG15 remains another challenge. Apart from specific examples of how this impacts innate immunity and virus–host interactions, we still have a poor understanding of the full influence of this diversity. Much work remains to ascertain the functional drivers of ISG15 variability and how this impacts its role between species. Until there is a good grasp of these aspects, each ISG15 species will have to be tested separately to evaluate the nuances in function. This may ultimately reveal whether there are lineage-specific characteristics or motifs in ISG15 that serve as the primary drivers in dictating its function.
Potential Applications to Leverage ISG15 Interspecies Diversity
Although ISG15's genetic diversity has created challenges to elucidating its function, this same feature may also present the opportunity to leverage it for biomedical purposes. Classical biosurveillance methods rely on a laborious process of sample acquisition and testing that can be time-consuming and expensive. Knowledge of how ISG15 interspecies diversity impacts viral protein interactions could expedite this process. In the case of emerging or re-emerging viruses that possess ISG15-interacting proteins, in vitro assays could be performed in a medium- to high-throughput manner to determine the relative preference for different species' ISG15s. This would guide a more targeted approach in the field for those species most likely to be susceptible to the virus. In an age of increasingly available genomic data, the utility of such a methodology will only grow over time. As more becomes known on the features of ISG15 that are most influential for particular families of viruses, this also lends itself to computational approaches to predict interactions in silico.
In addition to the identification of competent hosts in the present, ISG15 genetic diversity could provide insight into long-term trends in virus evolution. Although cell entry is the first step to virus infection and a predominant factor governing virus tropism, intracellular factors also play a role, and over time a virus will naturally become adapted to these components of the host immune system. If a virus has moved to a different host, it is possible it may retain some of the features for optimization with the prior host. Thus, although ISG15 may not be a primary determining factor for all viruses, it could serve as a tracer for the natural history of the virus. Adding to the value of ISG15 in such a probe is the fact that viruses in the same family possess the same repertoire of potential ISG15-interacting proteins, likely creating a common interface for comparison. While viruses in the same family sometimes utilize different cell-surface receptors, ISG15 may present a consistent point of interaction that makes it ideal for tracing a virus's evolutionary past. Alternatively, interspecies ISG15 diversity may have predictive value regarding the potential hosts to which the virus could most easily adapt. For example, MERS-CoV is unable to infect mouse cells due to an inability to interact with the mouse DPP4 receptor. Exogenous or transgenic expression of the human DPP4 receptor results in successful infection that after serially passaging can produce mouse-adapted pathogenic strains [84], [85], [86]. Interestingly, non-adapted MERS-CoV PLP is already able to interact with mouse ISG15, and while some of the mutations generated in the mouse-adapted strains are present in the nsp3 protein, none of them map to the PLP domain [64], [85], [86], [87]. Looking at the residues of ISG15 that PLPs are known to engage reveals that there is a mix of conserved and variable features with which the PLP interacts (Fig. 3b). This includes, for example, Trp123 that is highly conserved among mammals, and the almost perfectly conserved C-terminal tail. However, there are also notable areas of variation, particularly at positions 89 and 149, that the PLP must be able to accommodate. While the conserved regions may provide a platform for interaction with mammalian ISG15s in general, these other less conserved regions can markedly influence how well the PLP engages specific species' ISG15 [64]. This is reflected in the relatively broad, but not universal activity of the PLP for different ISG15 species. For example, although bat, camel, and human ISG15 only share 62%–66% sequence identity to each other, they all show susceptibility to the MERS-CoV PLP as would be consistent with the virus's known and predicted host range. For other species, including sheep, shrews, and fish that are not predicted to be hosts for the virus, the activity is noticeably lower suggesting the need to accommodate specific sequence variations. Considering this, the combination of the PLP's ability to interact with mouse ISG15 and the lack of mutations in the PLP of mouse-adapted strains suggests that the MERS-CoV, in a sense, may have been partially “pre-adapted” to mice as potential hosts, and that overcoming the threshold of cell entry was all that was required to replicate and eventually fully adapt. It would be interesting to apply this reasoning to other systems to ascertain its predictive value. Such knowledge could aid in understanding the risk posed by a particular virus during the course of genetic drift, as well as inform the design of appropriate model systems of disease.
Beyond surveillance and research applications, the features of ISG15 diversity also raise questions regarding its potential for direct therapeutic use. Could a modified ISG15, or one from a different species, be used to combat viral infection? Given the central place of ISGylation in antiviral mechanisms, such a strategy would rely on providing the conjugation machinery or assuming cross-species compatibility of the conjugation system enzymes. Though far from comprehensive, the current data suggest the latter to possibly be the case [57], [58]. From this standpoint, it could be envisioned how some agriculturally important animals could be genetically engineered for ISG15 to enhance resistance to particular virus threats. Alternatively, perhaps ISG15, or an ISG15 gene, could be administered to transiently introduce a modified or different species version of ISG15 to combat acute viral infection. Further work will be needed to assess the potential of these intriguing possibilities.
Conclusion
ISG15 is an influential component in mediating and regulating host responses to viral infection. Study of its form and function has yielded several surprises, and many aspects remain to be fully comprehended. Grasping its roles has proven elusive due to diverse mechanisms of action that can vary in importance between different pathogens, and possibly even between different host species. The sequence and structural diversity of ISG15 adds an additional layer to this complexity, particularly in how it impacts virus–host interfaces. Although much remains to be known, the tools and knowledge base needed to probe these questions have begun to come together, presenting exciting and unprecedented opportunities to understand and leverage ISG15 function.
Acknowledgments
This work was supported by the National Institutes of Health (application number 1R01AI109008, to S.D.P. and E.B.). The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. The authors thank Xinquan Wang and Yinan Jiang for sharing the structure coordinates for the NS1B-bound murine ISG15, canine ISG15, and bovine ISG15 structures in advance of PDB deposition.
Edited by Dr Carolyn Coyne
References
- 1.Liu M., Reimschuessel R., Hassel B.A. Molecular cloning of the fish interferon stimulated gene, 15 kDa (ISG15) orthologue: a ubiquitin-like gene induced by nephrotoxic damage. Gene. 2002;298:129–139. doi: 10.1016/s0378-1119(02)00932-0. https://www.ncbi.nlm.nih.gov/pubmed/12426101 [DOI] [PubMed] [Google Scholar]
- 2.Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/S0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 3.Matunis M.J., Coutavas E., Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. http://www.ncbi.nlm.nih.gov/pubmed/8978815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamitani T., Kito K., Nguyen H.P., Yeh E.T. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. http://www.ncbi.nlm.nih.gov/pubmed/9353319 [DOI] [PubMed] [Google Scholar]
- 5.Kumar S., Tomooka Y., Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 1992;185:1155–1161. doi: 10.1016/0006-291X(92)91747-E. [DOI] [PubMed] [Google Scholar]
- 6.Bates E.E.M., Ravel O., Dieu M.-C., Ho S., Guret C., Bridon J.-M., Ait-Yahia S., Brière F., Caux C., Banchereau J., Lebecque S. Identification and analysis of a novel member of the ubiquitin family expressed in dendritic cells and mature B cells. Eur. J. Immunol. 1997;27:2471–2477. doi: 10.1002/eji.1830271002. [DOI] [PubMed] [Google Scholar]
- 7.Gruen J.R., Nalabolu S.R., Chu T.W., Bowlus C., Fan W.F., Goei V.L., Wei H., Sivakamasundari R., Liu Y., Xu H.X., Parimoo S., Nallur G., Ajioka R., Shukla H., Bray-Ward P., Pan J., Weissman S.M. A transcription map of the major histocompatibility complex (MHC) class I region. Genomics. 1996;36:70–85. doi: 10.1006/geno.1996.0427. http://www.ncbi.nlm.nih.gov/pubmed/8812418 [DOI] [PubMed] [Google Scholar]
- 8.Fan W., Cai W., Parimoo S., Schwarz D.C., Lennon G.G., Weissman S.M. Identification of seven new human MHC class I region genes around the HLA-F locus. Immunogenetics. 1996;44:97–103. doi: 10.1007/BF02660056. http://www.ncbi.nlm.nih.gov/pubmed/8662070 [DOI] [PubMed] [Google Scholar]
- 9.Liu Y.C., Pan J., Zhang C., Fan W., Collinge M., Bender J.R., Weissman S.M. A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4313–4318. doi: 10.1073/pnas.96.8.4313. http://www.ncbi.nlm.nih.gov/pubmed/10200259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrell P.J., Broeze R.J., Lengyel P., Farrell P., Broeze Paul J., Lengyel Robert J. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature. 1979;279:523–525. doi: 10.1038/279523a0. [DOI] [PubMed] [Google Scholar]
- 11.Haas A.L., Ahrens P., Bright P.M., Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 1987;262:11315–11323. https://www.ncbi.nlm.nih.gov/pubmed/2440890 [PubMed] [Google Scholar]
- 12.Kessler D.S., Levy D.E., Darnell J.E. Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc. Natl. Acad. Sci. U. S. A. 1988;85:8521–8525. doi: 10.1073/pnas.85.22.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reich N., Evans B., Levy D., Fahey D., Knight E., Jr., Darnell J.E., Jr. Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc. Natl. Acad. Sci. U. S. A. 1987;84:6394–6398. doi: 10.1073/pnas.84.18.6394. https://www.ncbi.nlm.nih.gov/pubmed/3476954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morales D.J., Lenschow D.J. The antiviral activities of ISG15. J. Mol. Biol. 2013:1–14. doi: 10.1016/j.jmb.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malakhov M.P., Malakhova O.A., Kim K.I., Ritchie K.J., Zhang D.E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 16.Durfee L.A., Lyon N., Seo K., Huibregtse J.M. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol. Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malakhov M.P., Kim K.I., Malakhova O.A., Jacobs B.S., Borden E.C., Zhang D.E. High-throughput immunoblotting. Ubiquitiin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 2003;278:16608–16613. doi: 10.1074/jbc.M208435200. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi T., Inoue S., Yokosawa H. Identification and Herc5-mediated ISGylation of novel target proteins. Biochem. Biophys. Res. Commun. 2006;348:473–477. doi: 10.1016/j.bbrc.2006.07.076. [DOI] [PubMed] [Google Scholar]
- 19.Zhao C., Denison C., Huibregtse J.M., Gygi S., Krug R.M. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giannakopoulos N.V., Luo J.-K., Papov V., Zou W., Lenschow D.J., Jacobs B.S., Borden E.C., Li J., Virgin H.W., Zhang D.-E. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem. Biophys. Res. Commun. 2005;336:496–506. doi: 10.1016/j.bbrc.2005.08.132. [DOI] [PubMed] [Google Scholar]
- 21.Wong J.J., Pung Y.F., Sze N.S., Chin K.C. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10735–10740. doi: 10.1073/pnas.0600397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y., Zhong G., Zhu L., Liu X., Shan Y., Feng H., Bu Z., Chen H., Wang C. Herc5 attenuates influenza a virus by catalyzing ISGylation of viral NS1 protein. J. Immunol. 2010;184:5777–5790. doi: 10.4049/jimmunol.0903588. [DOI] [PubMed] [Google Scholar]
- 23.Ganesan M., Poluektova L.Y., Tuma D.J., Kharbanda K.K., Osna N.A. Acetaldehyde disrupts interferon alpha signaling in hepatitis C virus-infected liver cells by up-regulating USP18. Alcohol. Clin. Exp. Res. 2016;40:2329–2338. doi: 10.1111/acer.13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi H.X., Yang K., Liu X., Liu X.Y., Wei B., Shan Y.F., Zhu L.H., Wang C. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell. Biol. 2010;30:2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim M.-J., Hwang S.-Y., Imaizumi T., Yoo J.-Y. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J. Virol. 2008;82:1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du Y., Duan T., Feng Y., Liu Q., Lin M., Cui J., Wang R. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J. 2017:e96781. doi: 10.15252/embj.201796781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malakhova O.A., Yan M., Malakhov M.P., Yuan Y., Ritchie K.J., Il Kim K., Peterson L.F., Shuai K., Zhang D.E. Protein ISGylation modulates the JAK–STAT signaling pathway. Genes Dev. 2003;17:455–460. doi: 10.1101/gad.1056303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arimoto K.I., Löchte S., Stoner S.A., Burkart C., Zhang Y., Miyauchi S., Wilmes S., Fan J.-B.B., Heinisch J.J., Li Z., Yan M., Pellegrini S., Colland F., Piehler J., Zhang D.-E.E. STAT2 is an essential adaptor in USP18-mediated suppression of type i interferon signaling. Nat. Struct. Mol. Biol. 2017;24:279–289. doi: 10.1038/nsmb.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Bogunovic D., Payelle-Brogard B., Francois-Newton V., Speer S.D., Yuan C., Volpi S., Li Z., Sanal O., Mansouri D., Tezcan I., Rice G.I., Chen C., Mansouri N., Mahdaviani S.A., Itan Y., Boisson B., Okada S., Zeng L., Wang X., Jiang H., Liu W., Han T., Liu D., Ma T., Wang B., Liu M., Liu J.-Y., Wang Q.K., Yalnizoglu D., Radoshevich L., Uzé G., Gros P., Rozenberg F., Zhang S.-Y., Jouanguy E., Bustamante J., García-Sastre A., Abel L., Lebon P., Notarangelo L.D., Crow Y.J., Boisson-Dupuis S., Casanova J.-L., Pellegrini S. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature. 2015;517:89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speer S.D., Li Z., Buta S., Payelle-Brogard B., Qian L., Vigant F., Rubino E., Gardner T.J., Wedeking T., Hermann M., Duehr J., Sanal O., Tezcan I., Mansouri N., Tabarsi P., Mansouri D., Francois-Newton V., Daussy C.F., Rodriguez M.R., Lenschow D.J., Freiberg A.N., Tortorella D., Piehler J., Lee B., Garcia-Sastre A., Pellegrini S., Bogunovic D. ISG15 deficiency and increased viral resistance in humans but not mice. Nat. Commun. 2016;7:11496. doi: 10.1038/ncomms11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogunovic D., Byun M., Durfee L.A., Abhyankar A., Sanal O., Mansouri D., Salem S., Radovanovic I., Grant A.V., Adimi P., Mansouri N., Okada S., Bryant V.L., Kong X., Kreins A., Velez M.M., Boisson B., Khalilzadeh S., Ozcelik U., Darazam I.A., Schoggins J.W., Rice C.M., Al-muhsen S., Behr M., Vogt G., Puel A., Bustamante J., Gros P., Huibregtse J.M. Mycobacterial disease and impaired IFN-ɣ immunity in humans with inherited ISG15 deficiency. Science. 2012;2(80):1684–1689. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D'Cunha J., Ramanujam S., Wagner R.J., Witt P.L., Knight E., Jr., Borden E.C. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J. Immunol. 1996;157:4100–4108. https://www.ncbi.nlm.nih.gov/pubmed/8892645 [PubMed] [Google Scholar]
- 33.Knight E., Jr., Cordova B. IFN-induced 15-kDa protein is released from human lymphocytes and monocytes. J. Immunol. 1991;146:2280–2284. https://www.ncbi.nlm.nih.gov/pubmed/2005397 [PubMed] [Google Scholar]
- 34.Recht M., Borden E.C., Knight E., Jr. A human 15-kDa IFN-induced protein induces the secretion of IFN-gamma. J. Immunol. 1991;147:2617–2623. https://www.ncbi.nlm.nih.gov/pubmed/1717569 [PubMed] [Google Scholar]
- 35.Swaim C.D., Scott A.F., Canadeo L.A., Huibregtse J.M. Extracellular ISG15 signals cytokine secretion through the LFA-1 integrin receptor. Mol. Cell. 2017;68:581–590.e5. doi: 10.1016/j.molcel.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D'Cunha J., Knight E., Jr., Haas A.L., Truitt R.L., Borden E.C. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. U. S. A. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. https://www.ncbi.nlm.nih.gov/pubmed/8552607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahnefeld A., Klingel K., Schuermann A., Diny N.L., Althof N., Lindner A., Bleienheuft P., Savvatis K., Respondek D., Opitz E., Ketscher L., Sauter M., Seifert U., Tschöpe C., Poller W., Knobeloch K.-P., Voigt A. Ubiquitin-like protein ISG15 (interferon-stimulated gene of 15 kDa) in host defense against heart failure in a mouse model of virus-induced cardiomyopathy. Circulation. 2014;130:1589–1600. doi: 10.1161/CIRCULATIONAHA.114.009847. [DOI] [PubMed] [Google Scholar]
- 38.Zhao C., Sridharan H., Chen R., Baker D.P., Wang S., Krug R.M., Loeb K.R., Haas A.L., Sridharan H., Zhao C., Krug R.M., Skaug B., Chen Z.J., Malakhov M.P., Malakhova O.A., Kim K.I., Ritchie K.J., Zhang D.E., Zhao C., Collins M.N., Hsiang T.Y., Krug R.M., Yuan W., Krug R.M., Guerra S., Caceres A., Knobeloch K.P., Horak I., Esteban M., Frias-Staheli N., Lenschow D.J., Lai C., Morales D.J., Hsiang T.Y., Zhao C., Krug R.M., Zhao C., Hsiang T.Y., Kuo R.L., Krug R.M., Tang Y., Durfee L.A., Lyon N., Seo K., Huibregtse J.M., Werneke S.W., Bogunovic D., Zhang X., Malakhova O.A., Speer S.D., Versteeg G.A., Guan R., Ng A.K., Sherry L., Smith M., Davidson S., Jackson D., Ketscher L., Eisfeld A.J., Neumann G., Kawaoka Y., Reich S., Zheng W., Tao Y.J., Ye Q., Krug R.M., Tao Y.J., Mondal A., Potts G.K., Dawson A.R., Coon J.J., Mehle A., Turrell L., Lyall J.W., Tiley L.S., Fodor E., Vreede F.T., Shen Y.F., Schneider J., Dauber B., Melen K., Julkunen I., Wolff T., Ye Q., Hoffmann E., D'Cunha J., Knight E., Haas A.L., Truitt R.L., Borden E.C., Huber V.C., Kleimeyer L.H., McCullers J.A., Fodor E. Influenza B virus non-structural protein 1 counteracts ISG15 antiviral activity by sequestering ISGylated viral proteins. Nat. Commun. 2016;7:12754. doi: 10.1038/ncomms12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frias-Staheli N., Giannakopoulos N.V., Kikkert M., Taylor S.L., Bridgen A., Paragas J., Richt J.A., Rowland R.R., Schmaljohn C.S., Lenschow D.J., Snijder E.J., García-Sastre A., Virgin H.W. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swatek K.N., Aumayr M., Pruneda J.N., Visser L.J., Berryman S., Kueck A.F., Geurink P.P., Ovaa H., van Kuppeveld F.J.M., Tuthill T.J., Skern T., Komander D. Irreversible inactivation of ISG15 by a viral leader protease enables alternative infection detection strategies. Proc. Natl. Acad. Sci. U. S. A. 2018;115:2371–2376. doi: 10.1073/pnas.1710617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Kasteren P.B., Bailey-Elkin B.A., James T.W., Ninaber D.K., Beugeling C., Khajehpour M., Snijder E.J., Mark B.L., Kikkert M. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E838–E847. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scholte F.E.M., Zivcec M., Dzimianski J.V., Deaton M.K., Spengler J.R., Welch S.R., Nichol S.T., Pegan S.D., Spiropoulou C.F., Bergeron É. Crimean-Congo hemorrhagic fever virus suppresses innate immune responses via a ubiquitin and ISG15 specific protease. Cell Rep. 2017;20:2396–2407. doi: 10.1016/j.celrep.2017.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niemeyer D., Mösbauer K., Klein E.M., Sieberg A., Mettelman R.C., Mielech A.M., Dijkman R., Baker S.C., Drosten C., Müller M.A. The papain-like protease determines a virulence trait that varies among members of the SARS-coronavirus species. PLoS Pathog. 2018;14:e1007296. doi: 10.1371/journal.ppat.1007296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindner H.A., Lytvyn V., Qi H., Lachance P., Ziomek E., Ménard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem. Biophys. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capodagli G.C., McKercher M.A., Baker E.A., Masters E.M., Brunzelle J.S., Pegan S.D. Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J. Virol. 2011;85:3621–3630. doi: 10.1128/JVI.02496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Kasteren P.B., Beugeling C., Ninaber D.K., Frias-Staheli N., van Boheemen S., Garcia-Sastre A., Snijder E.J., Kikkert M. Arterivirus and nairovirus ovarian tumor domain-containing deubiquitinases target activated RIG-I to control innate immune signaling. J. Virol. 2011;86:773–785. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel D., Nan Y., Shen M., Ritthipichai K., Zhu X., Zhang Y.-J. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J. Virol. 2010;84:11045–11055. doi: 10.1128/JVI.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Z., Li Y., Ransburgh R., Snijder E.J., Fang Y. Nonstructural protein 2 of porcine reproductive and respiratory syndrome virus inhibits the antiviral function of interferon-stimulated gene 15. J. Virol. 2012;86:3839–3850. doi: 10.1128/JVI.06466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deaton M.K., Spear A., Faaberg K.S., Pegan S.D. The vOTU domain of highly-pathogenic porcine reproductive and respiratory syndrome virus displays a differential substrate preference. Virology. 2014;454–455:247–253. doi: 10.1016/j.virol.2014.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bester S.M., Daczkowski C.M., Faaberg K.S., Pegan S.D. Insights into the porcine reproductive and respiratory syndrome virus viral ovarian tumor domain protease specificity for ubiquitin and interferon stimulated gene product 15. ACS Infect Dis. 2018;4:1316–1326. doi: 10.1021/acsinfecdis.8b00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narasimhan J., Wang M., Fu Z., Klein J.M., Haas A.L., Kim J.J. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J. Biol. Chem. 2005;280:27356–27365. doi: 10.1074/jbc.M502814200. [DOI] [PubMed] [Google Scholar]
- 54.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Catic A., Fiebiger E., Korbel G.A., Blom D., Galardy P.J., Ploegh H.L. Screen for ISG15-crossreactive deubiquitinases. PLoS One. 2007;2:e679. doi: 10.1371/journal.pone.0000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durfee L.A., Kelley M.L., Huibregtse J.M. The basis for selective E1–E2 interactions in the ISG15 conjugation system. J. Biol. Chem. 2008;283:23895–23902. doi: 10.1074/jbc.M804069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pattyn E., Verhee A., Uyttendaele I., Piessevaux J., Timmerman E., Gevaert K., Vandekerckhove J., Peelman F., Tavernier J. HyperISGylation of Old World monkey ISG15 in human cells. PLoS One. 2008;3:e2427. doi: 10.1371/journal.pone.0002427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Versteeg G.A., Hale B.G., van Boheemen S., Wolff T., Lenschow D.J., Garcia-Sastre A. Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. J. Virol. 2010;84:5423–5430. doi: 10.1128/JVI.02395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Basters A., Geurink P.P., Rocker A., Witting K.F., Tadayon R., Hess S., Semrau M.S., Storici P., Ovaa H., Knobeloch K.P., Fritz G. Structural basis of the specificity of USP18 toward ISG15. Nat. Struct. Mol. Biol. 2017;24:270–278. doi: 10.1038/nsmb.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Basters A., Knobeloch K.-P., Fritz G. How USP18 deals with ISG15-modified proteins: structural basis for the specificity of the protease. FEBS J. 2018;285:1024–1029. doi: 10.1111/febs.14260. [DOI] [PubMed] [Google Scholar]
- 61.James T.W., Frias-Staheli N., Bacik J.-P., Levingston Macleod J.M., Khajehpour M., García-Sastre A., Mark B.L. Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2222–2227. doi: 10.1073/pnas.1013388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guan R., Ma L.C., Leonard P.G., Amer B.R., Sridharan H., Zhao C., Krug R.M., Montelione G.T. Structural basis for the sequence-specific recognition of human ISG15 by the NS1 protein of influenza B virus. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13468–13473. doi: 10.1073/pnas.1107032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li L., Wang D., Jiang Y., Sun J., Zhang S., Chen Y., Wang X. Crystal structure of human ISG15 protein in complex with influenza B virus NS1B. J. Biol. Chem. 2011;286:30258–30262. doi: 10.1074/jbc.C111.257899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daczkowski C.M., Dzimianski J.V., Clasman J.R., Goodwin O., Mesecar A.D., Pegan S.D. Structural insights into the interaction of coronavirus papain-like proteases and interferon-stimulated gene product 15 from different species. J. Mol. Biol. 2017;429:1661–1683. doi: 10.1016/j.jmb.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang Y., Wang X. Structural insights into the species preference of the influenza B virus NS1 protein in ISG15 binding. Protein Cell. 2018 doi: 10.1007/s13238-018-0598-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langley C., Goodwin O., Dzimianski J.V., Daczkowski C.M., Pegan S.D. Structure of interferon-stimulated gene product 15 (ISG15) from the bat species Myotis davidii and the impact of interdomain ISG15 interactions on viral protein engagement. Acta Crystallogr. Sect. D Struct. Biol. 2019;75 doi: 10.1107/S2059798318015322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glaser F., Pupko T., Paz I., Bell R.E., Bechor-Shental D., Martz E., Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. http://www.ncbi.nlm.nih.gov/pubmed/12499312 [DOI] [PubMed] [Google Scholar]
- 68.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Celniker G., Nimrod G., Ashkenazy H., Glaser F., Martz E., Mayrose I., Pupko T., Ben-Tal N. ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 2013;53:199–206. doi: 10.1002/ijch.201200096. [DOI] [Google Scholar]
- 71.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sorensen C.M., Rempel L.A., Nelson S.R., Francis B.R., Perry D.J., Lewis R.V., Haas A.L., Hansen T.R. The hinge region between two ubiquitin-like domains destabilizes recombinant ISG15 in solution. Biochemistry. 2007;46:772–780. doi: 10.1021/bi061408x. [DOI] [PubMed] [Google Scholar]
- 73.Yin C., Aramini J.M., Ma L.C., Cort J.R., Swapna G.V., Krug R.M., Montelione G.T. Backbone and Ile-delta1, Leu, Val methyl 1H, 13C and 15N NMR chemical shift assignments for human interferon-stimulated gene 15 protein. Biomol NMR Assign. 2011;5:215–219. doi: 10.1007/s12104-011-9303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang Y.G., Yan X.Z., Xie Y.Y., Gao X.C., Song A.X., Zhang D.E., Hu H.Y. Different roles for two ubiquitin-like domains of ISG15 in protein modification. J. Biol. Chem. 2008;283:13370–13377. doi: 10.1074/jbc.M800162200. [DOI] [PubMed] [Google Scholar]
- 75.Lai C., Struckhoff J.J., Schneider J., Martinez-Sobrido L., Wolff T., Garcia-Sastre A., Zhang D.E., Lenschow D.J. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J. Virol. 2009;83:1147–1151. doi: 10.1128/JVI.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sridharan H., Zhao C., Krug R.M. Species specificity of the NS1 protein of influenza B virus: NS1 binds only human and non-human primate ubiquitin-like ISG15 proteins. J. Biol. Chem. 2010;285:7852–7856. doi: 10.1074/jbc.C109.095703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Capodagli G.C., Deaton M.K., Baker E.A., Lumpkin R.J., Pegan S.D. Diversity of ubiquitin and ISG15 specificity among nairoviruses' viral ovarian tumor domain proteases. J. Virol. 2013;87:3815–3827. doi: 10.1128/JVI.03252-12. %! Diversity of Ubiquitin and ISG15 Spec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deaton M.K., Dzimianski J.V., Daczkowski C.M., Whitney G.K., Mank N.J., Parham M.M., Bergeron E., Pegan S.D. Biochemical and structural insights into nairoviral deISGylases preference for interferon-stimulated-gene-product 15 originating from certain species. J. Virol. 2016 doi: 10.1128/JVI.00975-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ratia K., Kilianski A., Baez-Santos Y.M., Baker S.C., Mesecar A. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014;10:e1004113. doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Daczkowski C.M., Goodwin O.Y., Dzimianski J.V., Farhat J.J., Pegan S.D. Structurally guided removal of DeISGylase biochemical activity from papain-like protease originating from Middle East respiratory syndrome coronavirus. J. Virol. 2017;91 doi: 10.1128/JVI.01067-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Kasteren P.B., Knaap R.C.M., van den Elzen P., Snijder E.J., Balasuriya U.B.R., van den Born E., Kikkert M. In vivo assessment of equine arteritis virus vaccine improvement by disabling the deubiquitinase activity of papain-like protease 2. Vet. Microbiol. 2015;178:132–137. doi: 10.1016/j.vetmic.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bergeron É., Zivcec M., Chakrabarti A.K., Nichol S.T., Albariño C.G., Spiropoulou C.F. Recovery of recombinant Crimean Congo hemorrhagic fever virus reveals a function for non-structural glycoproteins cleavage by furin. PLoS Pathog. 2015;11:e1004879. doi: 10.1371/journal.ppat.1004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akutsu M., Ye Y., Virdee S., Chin J.W., Komander D. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2228–2233. doi: 10.1073/pnas.1015287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coleman C.M., Sisk J.M., Halasz G., Zhong J., Beck S.E., Matthews K.L., Venkataraman T., Rajagopalan S., Kyratsous C.A., Frieman M.B. CD8 + T cells and macrophages regulate pathogenesis in a mouse model of Middle East respiratory syndrome. J. Virol. 2017;91 doi: 10.1128/JVI.01825-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cockrell A.S., Yount B.L., Scobey T., Jensen K., Douglas M., Beall A., Tang X.-C., Marasco W.A., Heise M.T., Baric R.S. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat. Microbiol. 2016;2:16226. doi: 10.1038/nmicrobiol.2016.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li K., Wohlford-Lenane C.L., Channappanavar R., Park J.-E., Earnest J.T., Bair T.B., Bates A.M., Brogden K.A., Flaherty H.A., Gallagher T., Meyerholz D.K., Perlman S., McCray P.B. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Douglas M.G., Kocher J.F., Scobey T., Baric R.S., Cockrell A.S. Adaptive evolution influences the infectious dose of MERS-CoV necessary to achieve severe respiratory disease. Virology. 2018;517:98–107. doi: 10.1016/j.virol.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]