Abstract
The Bruton tyrosine kinase (BTK) inhibitor ibrutinib has substantially improved therapeutic options for chronic lymphocytic leukemia (CLL). While ibrutinib is not curative, it has a profound effect on CLL cells and may create new pharmacologically exploitable vulnerabilities. To identify such vulnerabilities, we developed a systematic approach that combines epigenome profiling (charting the gene-regulatory basis of cell state) with single-cell chemosensitivity profiling (quantifying cell-type-specific drug response) and bioinformatic data integration. Applying our method to a cohort of matched patient samples collected before and during ibrutinib therapy, we identified characteristic ibrutinib-induced changes that provide a starting point for the rational design of ibrutinib combination therapies. Specifically, we observed and validated preferential sensitivity to proteasome, PLK1, and mTOR inhibitors during ibrutinib treatment. More generally, our study establishes a broadly applicable method for investigating treatment-specific vulnerabilities by integrating the complementary perspectives of epigenetic cell states and phenotypic drug responses in primary patient samples.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western world, predominantly affecting the elderly. It is driven by constitutively activated B cell receptor (BCR) signaling, which promotes clonal proliferation and accumulation of malignant B lymphocytes (CLL cells) in blood, bone marrow, and secondary lymphoid organs1–3. Pharmacological interference with BCR signaling has therapeutic benefit in the treatment of CLL and other B cell malignancies. Specifically, targeting BCR signaling with ibrutinib, a first-in-class Bruton Tyrosine Kinase (BTK) inhibitor, has demonstrated significant clinical efficacy in CLL4,5.
Due to ibrutinib’s high efficacy and acceptable toxicity, the drug has been approved not only for relapsed and refractory CLL, but also as a single-agent frontline therapy6. In addition to interfering with BCR signaling pathways as its primary mechanism of action, ibrutinib appears to block survival signals delivered by the microenvironment, which may include cell-cell contact and cytokines that modulate cell migration, trafficking, and proliferation7–9. Interestingly, ibrutinib treatment induces a redistribution of CLL cells from protected niches to the peripheral blood10,11, causing transient lymphocytosis that eventually resolves as the result of ibrutinib-mediated apoptosis and decreased proliferation of CLL cells. Little is known about the epigenomic changes and gene-regulatory dynamics that ibrutinib induces in CLL cells, although recent studies have started to characterize clonal evolution12, signaling pathways13, miRNA expression14, and transcriptomes15 in response to ibrutinib treatment.
Despite the clinical success of ibrutinib therapy, cellular response to ibrutinib is slow and often incomplete. There is currently no evidence that a cure can be achieved by ibrutinib alone, and drug discontinuation (e.g., due to toxicity16) is associated with rapid disease progression17. Moreover, among those patients that tolerate long-term treatment with ibrutinib, a considerable number eventually develop drug resistance (e.g., due to mutations in the BTK gene18), BTK-independent disease progression, or Richter’s transformation17. Combination therapies could potentially overcome these issues and provide better disease control at reduced toxicity. Based on clinical and pharmacological considerations, recent studies have explored the combined use of ibrutinib with the proteasome inhibitor carfilzomib19, the BCL-2 inhibitor venetoclax20, and the HDAC inhibitor abexinostat14 in preclinical models, and initial clinical trials for ibrutinib-based combination therapies are underway.
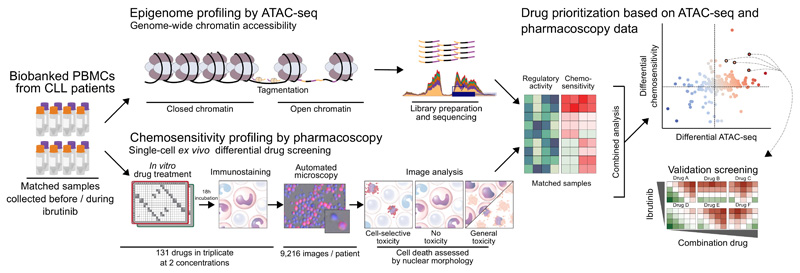
To establish a basis for the rational design of ibrutinib-based combination therapies, we piloted a high-throughput approach that detects and prioritizes vulnerabilities specific to ibrutinib-treated CLL cells, combining epigenetic/regulatory mapping with cellular/phenotypic profiling in primary samples from CLL patients who undergo ibrutinib therapy (Figure 1). We performed chromatin accessibility mapping by ATAC-seq21 on matched CLL samples collected before and during ibrutinib treatment, thus creating a genome-wide map of ibrutinib’s effect on gene regulation and pathway activity. We complemented this epigenetic/regulatory perspective by CLL-cell-specific chemosensitivity profiling for 131 promising drugs and small molecules using pharmacoscopy22, a single-cell automated imaging assay that allowed us to quantify and compare cell-specific drug responses in samples collected before and during ibrutinib treatment. These two assays provided complementary information on ibrutinib-induced changes in CLL cells, enabling us to systematically identify ibrutinib-induced, pharmacologically exploitable vulnerabilities, and to prioritize the translational potential of individual drugs, drug classes, and targetable molecular pathways for ibrutinib combination therapy.
Figure 1. Integrative analysis of epigenetic cell state and cell-selective chemosensitivity in ibrutinib-treated CLL patients.
Biobanked peripheral blood mononuclear cells (PBMCs) from chronic lymphocytic leukemia (CLL) patients isolated before and during ibrutinib treatment were subjected to chromatin accessibility mapping by ATAC-seq and to chemosensitivity profiling using pharmacoscopy, a single-cell automated imaging method for quantifying cell-selective drug response. To connect ibrutinib-induced changes in cell state to induced drug vulnerabilities, we mapped the ATAC-seq and pharmacoscopy data into the shared space of molecular pathways, which provide a joint basis for integrative analysis and prioritization of ibrutinib-based drug combinations for the treatment of CLL and potentially other hematopoietic malignancies.
Integrative analysis of the resulting datasets identified characteristic gene-regulatory changes induced by ibrutinib treatment, including modulation of proteasome, autophagy, and FoxO signaling pathways, which coincided with altered patterns of CLL-cell-selective chemosensitivity. Most notably, we observed preferential killing of CLL cells from patients under ibrutinib therapy for the proteasome inhibitors bortezomib and carfilzomib, the PLK1 inhibitors volasertib and BI2536, the mTOR inhibitor everolimus, and the Hsp90 inhibitor PU-H71. In summary, our study showcases the power of combined epigenome mapping and chemosensitivity profiling in primary patient samples collected before and during in vivo pharmacological therapy, which establishes a method for systematic identification and prioritization of drugs and regulatory pathways for combination therapy.
Results
Ibrutinib induces characteristic changes in chromatin state
To measure the effect of ibrutinib on the epigenetic cell state and gene-regulatory landscape of CLL cells, we performed chromatin accessibility mapping by ATAC-seq21 in matched patient samples collected before and during clinical ibrutinib therapy. Specifically, peripheral blood mononuclear cells (PBMCs) were obtained from 18 patients with relapsed CLL before treatment start and at a median of six weeks after treatment start (see Supplementary Table 1 for patient characteristics and sample details). These samples were subjected to ATAC-seq profiling using a standardized high-throughput workflow (as described in the Online Methods). CLL cell content was verified by flow cytometry and was high for all samples included in the study (median CLL cell content: 91%, sample-specific values are provided in Supplementary Table 1).
Chromatin accessibility mapping of the 18 sample pairs (corresponding to ATAC-seq profiling of 36 primary patient samples) resulted in consistently high data quality (Supplementary Table 2). We identified 119,923 unique chromatin accessible sites (Supplementary Table 3), which showed the expected enrichment for gene promoters, enhancers, and other regulatory elements (Supplementary Figure 1a-b). We observed near-perfect (99.4%) recovery of the chromatin accessible regions we previously described for an unselected CLL patient cohort23, and we identified 18,265 additional chromatin accessible sites that were not found in our previous study (Supplementary Figure 1c), thus establishing a comprehensive regulatory landscape of ibrutinib-treated CLL.
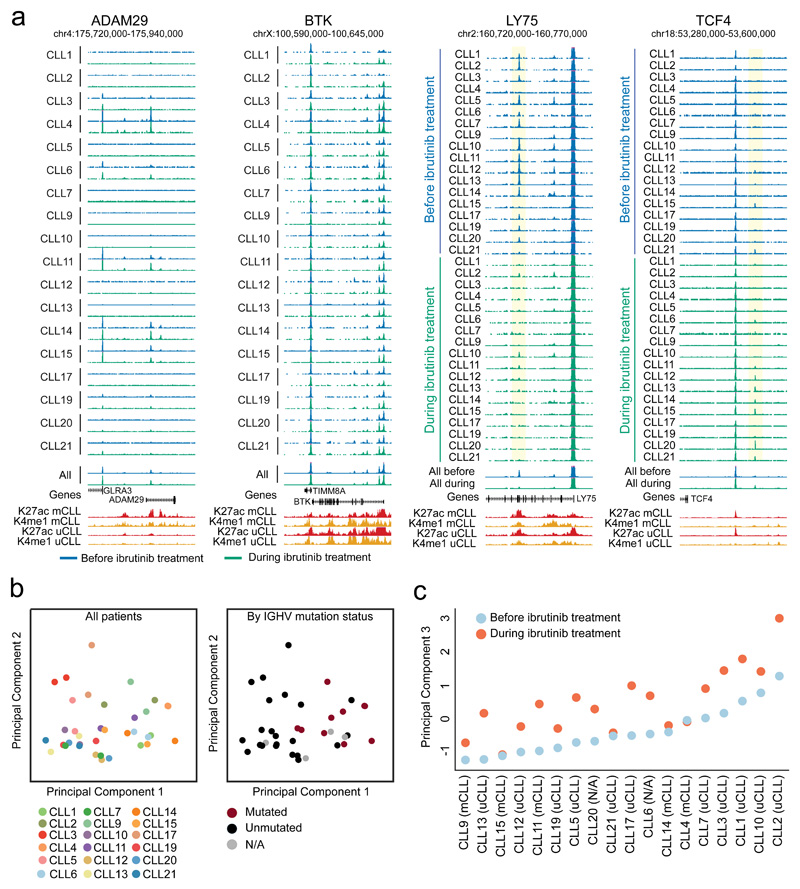
While the genome-wide distribution of chromatin accessibility was consistent across all patients, we identified a subset of regions that showed widespread inter-patient heterogeneity (Figure 2a; Supplementary Website: http://cll-combinations.computational-epigenetics.org). For example, the ADAM Metallopeptidase Domain 29 (ADAM29) gene, whose high expression was previously linked to long treatment-free intervals in Binet Stage A CLL patients24, was characterized by inter-individual heterogeneity independent of ibrutinib treatment. In contrast, we observed little variability at the BTK locus, consistent with the general importance of this gene in CLL and with the relatively homogeneous initial response to BTK inhibition. Comparison among sample pairs identified many regions that lost chromatin accessibility upon ibrutinib treatment, as illustrated by the Lymphocyte Antigen 75 (LY75) locus. Conversely, there were fewer regions with a consistent trend toward higher chromatin accessibility upon ibrutinib treatment, illustrated by a region upstream of the Transcription Factor 4 (TCF4) promoter.
Figure 2. Chromatin accessibility mapping for matched CLL patient samples collected before and during ibrutinib treatment.
(a) UCSC Genome Browser snapshots showing chromatin accessibility data obtained by ATAC-seq for matched samples (n=36) collected before ibrutinib treatment (blue) and during ibrutinib treatment (green). ChIP-seq profiles for two promoter/enhancer associated histone marks (H3K27ac shown in red, H3K4me1 shown in yellow) in IGHV unmutated CLL (uCLL) as well as IGHV mutated CLL (mCLL) are included as an additional reference23. Regions that significantly lose or gain accessibility are highlighted in yellow. (b) Principal component analysis based on ATAC-seq signal intensities of all open chromatin sites in all CLL samples (n=36). Principal component 1 separated the samples by their IGHV mutation status, while there was no obvious correlate of principal component 2. (c) Principal component 3 separated the samples by their ibrutinib treatment status.
For a more systematic, genome-wide comparison, we performed principal component analysis on the patient-specific chromatin accessibility scores of all 119,923 chromatin-accessible sites (Figure 2b). This analysis identified IGHV mutation status as the main factor explaining inter-patient variability in the current cohort, which validates our previous observations in an unrelated CLL cohort23. Furthermore, the third principal component was strongly associated with the ibrutinib treatment status, thus revealing a characteristic genome-wide effect of ibrutinib on the chromatin accessibility landscape in primary CLL samples that affects IGHV mutated and unmutated cases (Figure 2c).
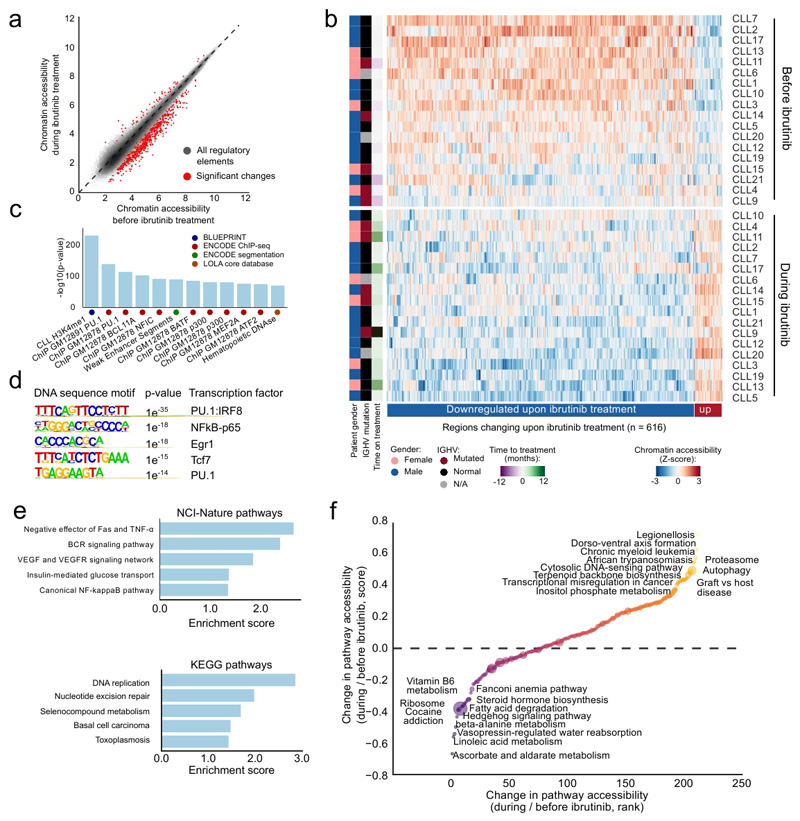
By comparing the chromatin accessibility landscape before and during ibrutinib treatment across all patients, we identified 616 regulatory elements that underwent significant changes in chromatin accessibility (Figure 3a; Supplementary Table 4; http://cll-combinations.computational-epigenetics.org). 92% of these genomic regions lost accessibility upon ibrutinib treatment, while only 8% gained accessibility, unraveling a profound directional effect of ibrutinib on the epigenetic state of CLL cells. While this characteristic loss of chromatin accessibility was shared by all patients, we also observed patient-to-patient differences in the epigenetic response to ibrutinib treatment (Figure 3b). We detected no statistically significant interactions with measured patient characteristics such as age, gender, IGHV mutation status, disease stage, CD38 expression, copy-number aberrations, or TP53 aberrations (Supplementary Figure 1d), which is consistent with the observation that several classical risk markers of CLL progression lose their predictive power with ibrutinib therapy25.
Figure 3. Differential analysis of ibrutinib-induced changes in chromatin accessibility for matched CLL patient samples.
(a) Scatterplot comparing ATAC-seq signal intensities across all open chromatin sites between samples collected before and during ibrutinib treatment. Significant changes correspond to an FDR-adjusted p-value below 0.01 and an absolute log2 fold change above 1 as calculated by the DESeq2 software. The diagonal is shown as a dashed line, as a reference indicating regions with no change in chromatin accessibility upon ibrutinib treatment. (b) Heatmap of normalized chromatin accessibility (Z-scores) for all regions with significantly differential chromatin accessibility according to panel a. (c) Region set enrichment analysis for genomic regions with reduced chromatin accessibility during ibrutinib treatment based on LOLA analysis, showing the twelve most significantly enriched region sets. (d) De novo motif enrichment analysis for regions with reduced chromatin accessibility during ibrutinib treatment. Reported p-values were calculated by the HOMER software using a binomial test. (e) Gene set analysis for enrichment of NCI-Nature and KEGG pathways among genes located in the vicinity of regions with reduced chromatin accessibility during ibrutinib treatment. Enrichment scores were calculated by the Enrichr software and represent the log p-value of a Fisher’s exact test multiplied by a Z-score of deviation from the expected rank. (f) Pathway-centric assessment of changes in chromatin accessibility induced by ibrutinib treatment. Normalized ATAC-seq signals of all genes in each KEGG pathway were aggregated to rank pathways. Yellow/orange dots denote pathways characterized by higher chromatin accessibility during ibrutinib treatment than before ibrutinib treatment, while blue/purple dots indicate pathways with lower chromatin accessibility.
Applying region set enrichment analysis using LOLA26 to those genomic regions that showed reduced chromatin accessibility upon ibrutinib treatment, we identified strong enrichment for CLL-specific regulatory elements marked by histone H3 lysine 4 monomethylation (an enhancer mark), as well as an enrichment for lymphoid-specific binding of transcription factors such as PU.1, NFIC, BCL11A, BATF, p300, MEF2A, and ATF2 (Figure 3c). Moreover, de novo motif discovery using HOMER27 detected NF-κB, EGR, TCF, and PU.1/IRF binding motifs (Figure 3d). When we linked the genomic regions with reduced chromatin accessibility in ibrutinib-treated CLL to their neighboring genes, we observed significant enrichment for genes involved in several signaling pathways (BCR, NF-κB, TNF, VEGF) and in DNA replication (Figure 3e). Together, these results provide a chromatin regulatory basis for previous reports showing reduced BCR and NF-κB signaling13 as well as arrested cell proliferation11 upon ibrutinib treatment. In contrast to the high number of regions with reduced chromatin accessibility identified by ATAC-seq (n=565), there were far fewer regions with increased chromatin accessibility upon ibrutinib treatment (n=51) (Figure 3a, b). These regions were associated with genes such as NFKBIA, FOXO3, and FASLG, which are all involved in cell signaling.
Finally, to quantify global effects of ibrutinib on gene regulation and epigenetic cell state, we calculated the aggregated regulatory activity of all KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways28 based on the ATAC-seq data for each sample (Supplementary Figure 2a, see Online Methods for details). We observed increased chromatin accessibility upon ibrutinib treatment for fundamental cellular processes such as proteasome regulation and autophagy, for cancer-associated transcriptional deregulation, for pathways with a role in inflammation, and for metabolic pathways such as terpenoid backbone synthesis (Figure 3f; Supplementary Figure 2b). Pathways that lost chromatin accessibility upon ibrutinib treatment were associated with metabolic processes (e.g., steroid synthesis, fatty acid degradation) and DNA repair. Collectively, these results uncover characteristic regulatory changes in CLL cells upon ibrutinib treatment, and they pinpoint relevant pathways and transcription regulators that appear to drive the observed changes in epigenetic cell state.
Chemosensitivity profiling detects ibrutinib-induced changes
Toward rational prioritization of ibrutinib-based combination therapies, we complemented the epigenetic/regulatory perspective of ATAC-seq with single-cell chemosensitivity (pharmacoscopy) data on a subset of the CLL sample pairs (Supplementary Table 1). In pharmacoscopy, patient-specific PBMCs are subjected to short-term ex vivo culture in 384-well screening plates that contain a collection of drugs as well as negative controls (DMSO only). After overnight incubation, cell monolayers are fixed, stained for cell type specific diagnostic markers (here we stained for the B-cell / CLL-cell surface receptor CD19), and imaged using automated confocal microscopy. Computational image analysis is used to quantify the number of marker-positive viable cells, as well as the total number of viable cells in each well22. By comparing the number of viable CD19+ cells to the number of viable marker-negative cells in the same well, we can accurately quantify the cell-selective effect on CD19+ cells for each drug. In addition, we measured CD19+ cell cytotoxicity, which was obtained by comparing the number of viable CD19+ cells in each drug-treated well to that of the negative control (DMSO only) wells. This setup also allowed us to effectively control and normalize for differences in CD19+ cell viability during short-term ex vivo culture, although this was not a major issue in our experiments.
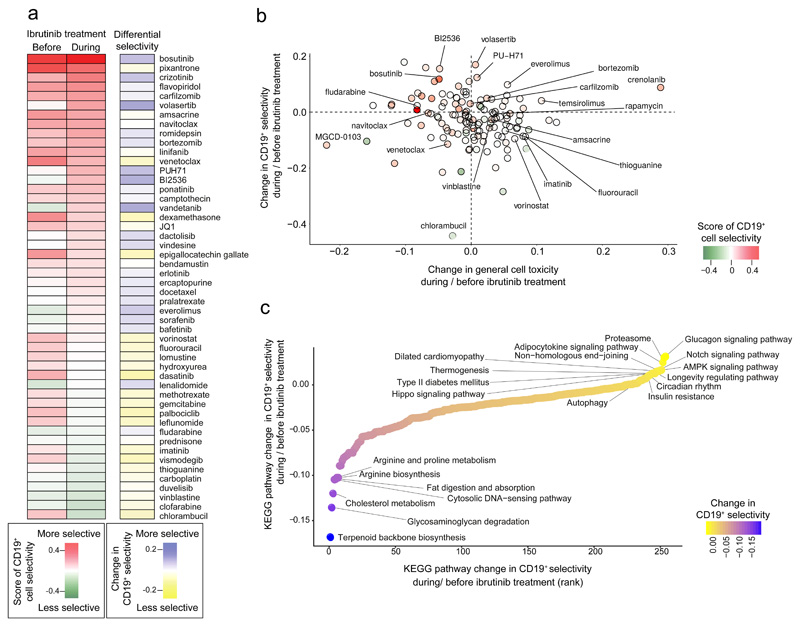
Using pharmacoscopy, we measured the CD19+ cell-selective chemosensitivity of 131 drugs and small molecules in 20 matched PBMC samples from CLL patients undergoing ibrutinib therapy. These 131 compounds (Supplementary Table 5) were selected for inclusion based on their potential relevance in leukemia therapy. Comparing the CD19+ chemosensitivity profiles between samples collected before and during ibrutinib treatment (Figure 4a; Supplementary Figure 3; Supplementary Table 6), we observed the following patterns: (i) drugs that were more selective for CD19+ cells during ibrutinib treatment, which included the PLK1 inhibitors volasertib and BI2536, the mTOR inhibitor everolimus, the dual PI3K/mTOR inhibitor dactolisib, the kinase inhibitors bosutinib and vandetanib, and the Hsp90 inhibitor PU-H71; (ii) drugs that selectively targeted CD19+ cells independent of ibrutinib treatment status, such as the proteasome inhibitors bortezomib and carfilzomib, the anti-neoplastic agent amsacrine, and the BCL-2 inhibitor navitoclax; (iii) drugs that lost CD19+ cell selectivity during ibrutinib treatment, including the base analogue fluorouracil, the HDAC inhibitor vorinostat, and the alkylating agent chlorambucil; and (iv) drugs with low CD19+ cell selectivity independent of ibrutinib treatment, which included classical chemotherapeutic drugs (vinblastine, fludarabine, thioguanine) that showed strong but unspecific cytotoxicity.
Figure 4. Single-cell chemosensitivity profiling for matched CLL patient sample pairs collected before and during ibrutinib treatment.
(a) Heatmap of CD19+ cell-selective cytotoxicity for a subset of the 131 tested drugs (full results are shown in Supplementary Figure 3) in CLL samples collected before and during clinical ibrutinib treatment, averaged across patients. In the first two rows, red indicates drugs that were selective for the CD19+ cell fraction, while green indicates drugs that were anti-selective. The third row depicts the difference in CD19+ cell-selective cytotoxicity for samples collected before and during ibrutinib treatment, where blue is more selective, yellow is less selective, and white indicates no change. (b) All 131 drugs plotted according to their difference in CD19+ cell-selective cytotoxicity (y-axis) versus the difference in general cytotoxicity toward all PBMC populations (x-axis), before and after ibrutinib. The dot color indicates CD19+ cell-selective cytotoxicity during ibrutinib treatment. (c) Ranking of the observed change in CD19+ cell-selective cytotoxicity at the level of KEGG pathways, aggregating the data across all drugs annotated with the respective KEGG pathway. The pharmacoscopy results were based on data for 10 CLL patients with matched samples collected before and during ibrutinib (for one patient, the sample collected during ibrutinib treatment was excluded due to low data quality as the result of poor cell viability after thawing, and the patient was removed from the pharmacoscopy data analysis). Drugs were screened over two 384-well plates per sample in two concentrations (10 μm and 1 μm), where each concentration point was performed in triplicate (10 μM) or in duplicate (1 μM). Drug sensitivities were normalized to DMSO, and there were approximately 40 DMSO control wells on each plate.
Importantly, the single-cell readout provided by pharmacoscopy allowed us to quantify the CD19+ cell selectivity of each drug and thereby distinguish between general cytotoxicity (i.e., affecting most or all cell types in the PBMC sample) and cell-selective effects on CD19+ cells. Such effects would be more challenging to detect and quantify using conventional chemosensitivity assays that measure only the total number of viable cells, without distinguishing between CD19+ cells and other cell populations. Indeed, we observed almost no correlation (Pearson’s r = -0.08) between the ibrutinib-dependent differential CLL cell selectivity and the ibrutinib-dependent differential general cytotoxicity (Figure 4b). This observation underlines the relevance of our single-cell imaging-based method for cell-selective chemosensitivity profiling, allowing us to distinguish with high precision between on-target and off-target cytotoxicity.
To connect the ibrutinib-induced phenotypic changes in chemosensitivity to differences in gene regulation and pathway activity, we aggregated the selectivity scores for each drug according to KEGG annotated target pathways (Figure 4c). This analysis identified increased efficacy of drugs targeting molecular pathways and gene sets including proteasome, autophagy, Notch signaling, Hippo signaling, and insulin signaling (glucagon, type II diabetes mellitus, insulin resistance, adipocytokine signaling) for samples collected during ibrutinib treatment. Moreover, having mapped ATAC-seq epigenetic/regulatory profiles (Figure 3f) as well as pharmacoscopy cellular/phenotypic profiles (Figure 4c) into the shared space of molecular pathways, we now have a common basis for integrating both datasets and for systematically prioritizing ibrutinib combination therapies.
Integrated analysis prioritizes ibrutinib drug combinations
Combining the aggregated results of ATAC-seq (Figure 3f) and pharmacoscopy (Figure 4c) at the pathway level, we ranked and prioritized all identified molecular pathways by their potential for ibrutinib-based combination therapy (Figure 5a; Supplementary Figure 4). Most notably, pathways identified by both approaches included proteasome activity, FoxO signaling, and autophagy. Several drugs and small molecules are available that modulate these pathways, allowing us to validate these potential interactions with ibrutinib therapy. Based on the pharmacoscopy data, we selected for further experimental validation the clinically approved proteasome inhibitors bortezomib and carfilzomib, the PLK1 inhibitors volasertib and BI2536 (which might target FoxO signaling via PLK129), and the mTOR inhibitor everolimus; and we included the BCL-2 inhibitor navitoclax as an additional control.
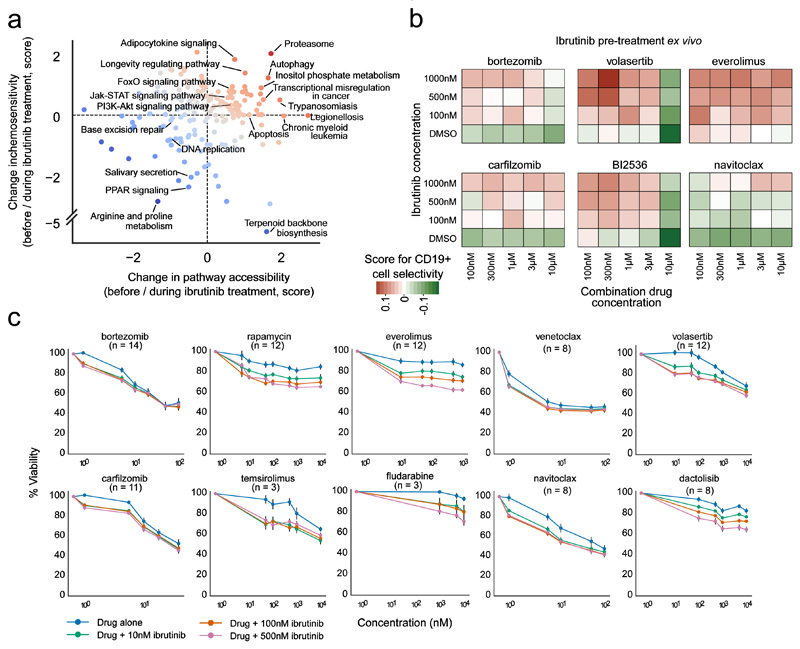
Figure 5. Prioritization and validation of ibrutinib-based drug combinations based on combined chemosensitivity and chromatin profiling.
(a) Integrative analysis of differential chromatin accessibility (x-axis) and differential cell-selective chemosensitivity (y-axis) at the pathway level. Red dots denote pathways characterized by higher chromatin accessibility and/or higher chemosensitivity during ibrutinib treatment than before ibrutinib treatment, while blue dots indicate lower chromatin accessibility and/or lower chemosensitivity. (b) Heatmap of CD19+ cell-selective cytotoxicity for combination matrixes of ibrutinib (y-axis) and six partner drugs (x-axis). Red indicates drug combinations that were selective for the CD19+ cell fraction, while green indicates combinations that were anti-selective. Results shown are averages across five patient samples, where each concentration point was measured in triplicate for each patient sample. (c) Drug responses (% viability) of primary CLL cells pretreated with different concentrations of ibrutinib in a co-culture model using primary bone marrow stromal cells. Viability was normalized to the effect of ibrutinib as a single agent. Error bars indicate the standard error of the mean (SEM) calculated across samples (numbers in brackets).
First, we evaluated the combinatorial effect of ibrutinib with each of the six selected drugs in primary, ibrutinib-naïve, CLL samples collected at diagnosis (n=5) using pharmacoscopy (i.e., short-term ex vivo culture in screening plates, immunostaining for the CLL markers CD5 and CD19, and automated imaging to measure cell abundance). We quantified CD19+/CD5+ cell-selective chemosensitivity using dose-response matrices with three concentrations of ibrutinib and five concentrations of each of the six drugs (Figure 5b; Supplementary Figure 5), done in triplicate. In the absence of ibrutinib, most drugs showed general cytotoxic effects (white/green bottom row in each matrix), while we observed an ibrutinib dose-dependent gain of selectivity for CD19+/CD5+ cells (red box coloring) for all tested drugs except for the BCL-2 inhibitor navitoclax (which was included as a control), as predicted by the chemosensitivity profiles for the CLL samples collected before and during ibrutinib treatment.
Second, we further validated these results in a standardized co-culture model that seeks to mimic the in vivo conditions in the bone marrow niche, growing primary CLL cells on primary bone marrow stromal cells30. The prioritized drugs, along with a selection of related compounds, were effective as single agents, and their efficacy was generally enhanced (already at low concentrations) for cells that had been pre-treated with ibrutinib (Figure 5c). The results also recapitulated differences that we observed in the pharmacoscopy profiling – for example, the combinatorial effect with ibrutinib was stronger for the mTOR and PLK1 inhibitors than for proteasome and BCL-2 inhibitors. However, the proteasome and BCL-2 inhibitors (including bortezomib, carfilzomib, navitoclax, and venetoclax) were extremely effective already as single agents (Figure 5c), which may mask stronger combinatorial effects in these experiments. Finally, the classical chemotherapeutic drug fludarabine, which showed strong but unspecific cytotoxicity in the pharmacoscopy data and which was included as a control in these validations, exhibited a limited additive cytotoxic effect at high concentrations.
Third, to better connect the data from the suspension-based pharmacoscopy experiments with those of the co-culture experiments, we exposed PBMCs from ibrutinib-naive CLL patients (n=2) to ibrutinib in co-culture and subsequently performed pharmacoscopy with immunostaining for the CLL markers CD5 and CD19 (Supplementary Figure 6). Again, we observed an ibrutinib dose-dependent loss of the specific CD19+/CD5+ population after treatment with proteasome inhibitors, PLK1 inhibitors, and mTOR inhibitors, underlining the consistency of these observations across the various models.
In summary, we observed an ibrutinib-induced gain of CLL cell selectivity for proteasome inhibitors, PLK1 inhibitors, and mTOR inhibitor, which was validated in several models and further supported by the high ranking of the (interconnected) proteasome and autophagy pathways in our chromatin data. These results suggest that ibrutinib renders CLL cells more sensitive to the pharmacological disruption of protein turnover and cellular homeostasis, and they provide a basis for further mechanistic dissection and/or clinical evaluation in CLL patients undergoing ibrutinib therapy. Moreover, the results validate our approach for prioritizing drug combinations by integrated analysis of chemosensitivity and chromatin profiles, with broad applicability in CLL and in other areas of medical oncology.
Discussion
Molecularly targeted cancer drugs, of which ibrutinib is a prominent example, have substantially improved the treatment for leukemia and other cancers. Yet these drugs typically fail to eradicate the disease, and patients suffer from the toxicities, evolving resistance, and economic burden of long-term treatment. Rational development of drug combinations could potentially overcome these limitations, in much the same way as it has revolutionized HIV therapy31, but new methods are needed to screen for promising combination therapies systematically and in high-throughput. Toward this goal, here we prototyped an approach that combines epigenetic/regulatory profiling by ATAC-seq with cellular/phenotypic profiling by pharmacoscopy, in order to identify and prioritize drug-induced vulnerabilities. Importantly, ATAC-seq and pharmacoscopy contribute highly complementary perspectives. While ATAC-seq focuses on epigenetic cell state and gene-regulatory mechanisms, which identifies drug-induced molecular rewiring, pharmacoscopy focuses on chemosensitivity and cellular phenotypes, thereby providing a direct functional assessment of drug-induced vulnerabilities.
Applying our method to CLL patients scheduled for ibrutinib therapy, we performed ATAC-seq and pharmacoscopy on PBMCs collected before and during ibrutinib treatment in vivo. We indeed observed characteristic differences, which converged on an interesting set of pathways and drugs. For samples collected during ibrutinib treatment, the ATAC-seq data revealed widespread downregulation of NF-κB and BCR signaling, consistent with recent research13,15, and we observed specific enrichment for proteasome regulation and autophagy at the pathway level. The pharmacoscopy assay, with its single-cell viability readout obtained by automated microscopy, identified drug candidates for these pathways that showed preferential killing of CD19+ and/or CD5+ cells collected from CLL patients during ibrutinib therapy. Most notably, we observed selective targeting of CD19+ and/or CD5+ cells for the proteasome inhibitors bortezomib and carfilzomib, the PLK1 inhibitors volasertib and BI2536, the mTOR inhibitor everolimus, and the Hsp90 inhibitor PU-H71. One of these drugs, carfilzomib, was recently suggested for a potential combination therapy with ibrutinib in CLL19, and the same drug combination has already shown encouraging results in a phase I clinical trial for relapsed and refractory multiple myeloma32. The other prioritized drug combinations for the treatment of CLL are (to our knowledge) new and have not yet been tested in pre-clinical or clinical studies.
While we observed a characteristic ibrutinib-dependent increase in CD19+ and/or CD5+ cell-selective chemosensitivity in CLL samples (which validates our hypothesis of ibrutinib-induced vulnerabilities that can be pharmacologically exploited), these effects were restricted to a handful of drugs. Among the drugs that have previously been proposed and/or evaluated in the context of CLL therapy, we observed more diverse patterns, including reduced target cell-killing selectivity during ibrutinib therapy (e.g., imatinib), lack of selectivity (e.g., fludarabine), or high selectivity independent of ibrutinib treatment status (e.g. bendamustine, navitoclax). The ability of pharmacoscopy to distinguish between general cytotoxicity and cell-selective chemosensitivity thus provides not only a way to prioritize new drug candidates for combination therapy, but can also be used to identify similarities and differences in the cell-specific response to existing candidates for combination therapy in CLL.
A potential limitation of our method lies in its reliance on suspension culture for efficient profiling of a large number of drugs and small molecules, which does not incorporate the protective effects of the microenvironment. Nevertheless, a recent study provided convincing proof-of-concept for chemosensitivity profiling using suspension culture in various hematopoietic malignancies33. Moreover, we implemented three complementary measures to mitigate the drawbacks of suspension culture. First, the imaging-based readout in pharmacoscopy enabled us to detect cell-specific cytotoxicity early and sensitively, such that it was possible to restrict the incubation time to 18 hours. Second, with pharmacoscopy we can use non-CLL PBMCs in each well as internal controls against which we quantified the cell-selective chemosensitivity, allowing us to normalize for different levels of cell death in different wells. Third, we performed validation experiments measuring cytotoxicity in a stromal co-culture model that we have previously published30 and confirmed key results. Further limitations of our study lie in the small sample size and in the lack of validation in preclinical models and clinical trials, which will be needed to assess the relevance of the identified drug combinations for CLL therapy. We did, however, observe promising clinical results in a recent pilot study using pharmacoscopy in a range of aggressive hematopoietic malignancies22, and we are confident that adding the regulatory information provided by ATAC-seq can help refine such predictions.
While we see the value of the study primarily in the development and validation of a systematic, widely applicable technology for prioritizing drug combinations, the convergence of our data on proteasome, autophagy, and mTOR pathways is intriguing. It was recently described that CLL cell survival under reduced BCR activity depends on alternative pathways including mTOR33, and we speculate that the inhibition of BCR signaling by ibrutinib pushes CLL cells into a state that renders them more dependent on mTOR and on the interconnected proteasome and autophagy pathways – which would explain the observed combinatorial effects of the PLK1, Hsp90, proteasome and mTOR inhibitors. In support of this concept, inhibition of PLK1 has been shown to induce autophagy by reducing mTOR phosphorylation in acute myeloid leukemia34; and Hsp90 inhibition by PU-H71 in Burkitt lymphoma suppressed several components of PI3K/AKT/mTOR signaling35. Consistent with our observation that bortezomib and carfilzomib effectively combined with ibrutinib, recent research showed synergy between proteasome inhibitors and ibrutinib both in CLL19 and multiple myeloma36. Finally, inhibition of the autophagy gatekeeper mTOR by everolimus showed striking cytotoxicity in combination with ibrutinib, consistent with ex vivo data in models of diffuse large B-cell lymphoma37,38. A detailed dissection of the interplay between proteasome, autophagy, and mTOR signaling in ibrutinib-treated CLL was beyond the scope of this study but would merit further investigation, in order to unravel the precise mechanism of action for these promising drug combinations.
Taken together, we have demonstrated the combination of epigenetic/regulatory profiling by ATAC-seq with cellular/phenotypic profiling by pharmacoscopy as an effective method for identifying and prioritizing drugs and targetable pathways for ibrutinib-based combination therapy. This approach is readily transferable to other cancers, depending only on the feasibility of obtaining matched patient samples before and after induction of targeted therapy, and it provides a widely applicable tool for the systematic discovery and rational development of drug combinations in precision oncology.
Online Methods
Sample collection
Heparinized peripheral blood was obtained from CLL patients with informed consent. Recruitment and collection protocols were approved by the Ethics Commission of the Medical University of Vienna: EK:36/2007, EK:365/2009, EK:1830/2015, and all relevant laws applicable to this study were followed. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-Hypaque (GE Healthcare) centrifugation. Patients were screened for chromosomal aberrations including deletions on 13q14, 11q22, and 17p13 as well as trisomy 12 by FISH analysis. The IGHV and TP53 mutational status was determined by Sanger sequencing (LGC Genomics). PBMCs from CLL patients were cryopreserved in RPMI 1640 supplemented with 20% FCS and 10% DMSO. Cell viability and percentage of CLL cells (CD19+/CD5+), T cells (CD3+), and monocytes (CD14+) were assessed by flow cytometry using antibodies against CD3 (CD3-APC, Biolegend #300439, clone UCHT1, dilution 1:20), CD5 (CD5-FITC, Biolegend #300606, clone UCHT2, dilution 1:50), CD14 (CD14-FITC, eBioscience #11-0149-42, clone 61D3, dilution 1:50), and CD19 (CD19-APC, eBioscience #17-0198-42, clone SJ25C1, dilution 1:50), with the gating strategy illustrated in Supplementary Figure 7. Patient and sample annotations are provided in Supplementary Table 1.
Assay for transposase accessible chromatin (ATAC-seq)
Chromatin accessibility mapping by ATAC-seq was performed as previously described21,23, with minor adaptations. In each experiment, ~50,000 cells were pelleted by centrifuging for 5 min at 4 °C at 300 × g. After centrifugation, the pellet was carefully resuspended in the transposase reaction mix (12.5 ml 2xTD buffer, 2 ml TDE1 (Illumina), 10.25 ml nuclease-free water, 0.25 μl 5% Digitonin (Sigma)) for 30 min at 37 °C. Following DNA purification with the MinElute kit eluting in 11 μl, 1 μl of the eluted DNA was used in a quantitative PCR (qPCR) reaction to estimate the optimum number of amplification cycles. The remaining 10 μL of each library were amplified for a number of cycles corresponding to the Cq-value (i.e., the cycle number at which fluorescence has increased above background levels) from the qPCR (rounded up). Library amplification was followed by SPRI size selection to exclude fragments larger than 1,200 bp. DNA concentration was measured with a Qubit fluorometer (Life Technologies). Library amplification was performed using custom Nextera primers21. The libraries were sequenced by the Biomedical Sequencing Facility at CeMM using the Illumina HiSeq 3000/4000 platform and the 25 bp paired-end configuration. Sequencing statistics are provided in Supplementary Table 2.
Imaging-based single-cell functional drug screening (pharmacoscopy)
Single-cell differential drug screening by pharmacoscopy was performed as described previously22. Briefly, 50 nl of each drug in DMSO as well as DMSO controls were transferred into 384-well tissue culture treated clear-bottom plates (Corning or PerkinElmer Cell Carrier Ultra) using a Labcyte Echo liquid handler attached to a PerkinElmer cell::explorer workstation. A library comprising 131 drugs and small molecules was designed in-house (Supplementary Table 5) and sourced by Sigma Aldrich Select. KEGG drug activity annotations of the library were gathered using KEGGREST39 (August 2017) or manually annotated where necessary. All drugs used in the validation experiments were purchased from Selleckchem. Previously purified PBMCs frozen in DMSO were rapidly thawed and incubated with 30 units DNase (Sigma) for 30 minutes at room temperature (22–25 °C) in RPMI supplemented 10% FBS and penicillin-streptomycin. 50 μl of culture medium containing approximately 40,000 cells was pipetted into each well of a 384-well plate containing drugs and incubated at 37 °C with 5% CO2 for 18 hours. For the initial screens (Figure 4a) and for secondary co-culture screening (Figure 5b and Supplementary Figure 6), each drug was assayed in triplicate (10 μM) and duplicate (1 μM), randomized across two 384-well screening plates. The combination screening (Figure 5b and Supplementary Figure 5) was performed in triplicate at each concentration point. All screens were stopped by fixing and permeabilizing the cells with a solution of 4% formaldehyde and 0.01% Triton-X114 in PBS. Fixative-containing medium was removed, and a cocktail of CD19 (clone HIB19, Phycoerythrin (12-0199-42) or Alexa488 (53-0199-42) conjugated, eBiosciences) and/or CD5 (clone UCHT2, Phycoerythrin (12-0059-42) conjugated, eBiosciences) was added. Antibodies were diluted at 1:300 in PBS for use. DAPI (at 10 μM; Sigma) was used for the detection of nuclei. Each well of a 384-well plate was imaged in its entirety at 10× magnification with 2×2 non-overlapping images, such that approximately 95% of the well surface area was covered within the 2×2 square. The images were taken sequentially with lasers and bandpass filter sets set to avoid channel overlap. A PerkinElmer Opera Phenix or PerkinElmer Operetta CLS automated spinning-disk confocal microscope was used. The raw TIFF images were exported from the microscope and stored for analysis.
Co-culture experiments
PBMCs from four previously untreated CLL patients were suspended in RPMI 1640 supplemented with 10% heat inactivated fetal calf serum (FCS), 2 mM Glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin (all reagents were obtained from Gibco). PBMCs were incubated with the indicated concentrations of drugs or with equal amounts of solvent. Co-culture experiments of CLL cells with bone marrow stromal cells (BMSCs) were performed as previously described30. Briefly, bone marrow mononuclear cells (BMMCs) were isolated from heparinized bone marrow aspirates. BMMCs were resuspended and cultured in alpha-MEM culture medium supplemented with 20% FCS, L-glutamine, and antibiotics in 75 cm2 culture flasks. Non-adherent cells were removed after 24 h, and cell culture was continued to expand the BMSCs. Cells from fifth passage or lower were used in the co-culture experiments. To initiate co-culture, culture medium from 90% confluent BMSC monolayer was removed, and cells were washed twice with alpha-MEM. CLL cells were suspended in RPMI containing 10% FCS at cell density of 3 × 106 / mL and added to the 6-well culture plates coated with BMSCs. Co-cultures were continued at 37 °C in 5% CO2 either untreated or treated with the drugs. For drug combination experiments, CLL cells were pre-treated in co-culture with ibrutinib for 24 hours followed by incubation with the tested drug for another 24 hours. Cells were then harvested and processed for cell viability assessment.
Cell viability assessment for CLL cells treated under co-culture conditions
To determine the overall effect of the drugs on cell viability, MTT assays (EZ4U-Biomedica) were performed. 3 × 105 CLL cells/well were seeded in 96-well cell culture plates in RPMI containing 10% FCS. After treatment with the drugs, CLL cells were incubated with substrate according to the manufacturer’s instructions, and the conversion of the tetrazolium substrate to formazan was measured with Tecan Infinite M200 Pro ELISA reader using Magellan software. For evaluating the effect of drugs on the induction of apoptosis, CLL cells were collected and resuspended in Annexin-binding buffer containing AnnexinV-FITC (eBioscience) and incubated for 10 min at room temperature (22–25 °C) in the dark. Cells were then washed with PBS and resuspended in Annexin-binding buffer containing 1 μg/mL propidium iodide (PI), followed by analysis on a FACSCalibur (Becton Dickinson) using CellQuestPro (Becton Dickinson) software. Apoptosis was classified as early apoptotic (AnnexinV single positive; Ax+/PI-) and late apoptotic/necrotic (AnnexinV/propidium iodide double positive; Ax+/PI+). The effect of the drugs on cell viability and apoptosis was calculated as the percentage of cells compared to the untreated control samples.
Preprocessing of the ATAC-seq data
ATAC-seq read data were processed as described previously23. Reads were trimmed using Skewer40 and aligned to the GRCh37/hg19 assembly of the human genome using Bowtie241 with the ‘-very-sensitive’ parameter. Duplicate reads were removed using sambamba markdup42, and only properly paired reads with mapping quality >30 and alignment to the nuclear genome were kept. All downstream analyses were performed on the filtered reads. Peak calling was done with MACS243 using the ‘-nomodel’ and ‘-extsize 147’ parameters, and peaks overlapping blacklisted features defined by the ENCODE project44 were discarded.
Bioinformatic analysis of chromatin accessibility data
We created a consensus set of chromatin accessible regions in CLL cells by merging the called peaks from all samples, and we quantified the accessibility of each region in each sample by counting the number of reads from the filtered BAM file that overlapped each region. To normalize read counts across samples, we performed quantile normalization using the normalize.quantiles function from the preprocessCore package in R. Regulatory elements were annotated with the nearest transcription start site from Ensembl and with chromatin states from the 15-state segmentation for CD19+ B cells from the Roadmap Epigenomics Project45 (identifier: E032). Principal component analysis was performed with the scikit-learn library (sklearn.decomposition.PCA) applied to the chromatin accessibility values for the merged regions across the CLL cohort. We used DESeq246 on the raw count values for each sample and regulatory element to identify differential chromatin accessibility between samples collected before and during ibrutinib treatment. Significant regions were defined as having an FDR-corrected p-value below 0.01 and an absolute log2 fold change above 1. Region set enrichment analysis was performed on the downregulated regions using LOLA26 with its core databases. Motif enrichment analysis was done with HOMER27 findMotifsGenome using “-mask -size 150 -length 8,10,12,14,16 -S 12” parameters.
Bioinformatic analysis of pharmacoscopy data
Illumination correction, image quality evaluation, image analysis, and downstream analysis were performed as described previously47. Briefly, images from each screen were analyzed using Cell Profiler (Broad Institute). Differential phenotypes were calculated as the fraction of marker-positive viable cells after drug treatment22. Viability of each cell was determined by quantifying nuclear intensity, size, and existence of a nucleus using image analysis (cells with visible nuclei were considered viable). Image quality control included analysis of nuclear segmentation and detection, nuclear size and intensity, fluorescent staining foreground-to-background determinations, and vehicle (DMSO) marker positive populations per sample. Wells or images containing no viable cells (based on nuclear shape, size, and intensity) automatically failed quality control and were removed from the analysis, along with wells containing fluorescent drugs, wells with less than 85% cell death compared to DMSO, and wells with debris resulting in high background signal. Images containing focus failures or other microscope failures, as determined by the PerkinElmer Harmony microscope driver, were automatically removed from the analysis. The relative cell fraction was calculated as the number of viable marker-positive cells over all viable cells detected for a particular well, drug, and concentration, normalized to the fraction of marker-positive cell contained in the DMSO wells. The selectivity score was calculated as the area over the curve of the averaged relative cell fractions of a given drug at each concentration. The difference in CD19+ and/or CD5+ cell selective killing was calculated as the difference between the selectivity score for samples collected during versus those collected before ibrutinib treatment (Figure 4a). Each screen contained at least three technical replicates, and each patient sample was considered a biological replicate. All replicates were aggregated per patient sample by taking the mean.
Integrative data analysis
To integrate chromatin accessibility and chemosensitivity data for CLL sample pairs, we exploited the fact that both data types reflect aspects of cell state, and we mapped the data into a shared space of molecular pathways to facilitate integrative analysis. For ATAC-seq, we used the same assignments between regulatory elements and genes as described above, while obtaining pathway-gene mappings from the KEGG database. Quantile-normalized values of chromatin accessibility for all regulatory elements associated with each gene in KEGG pathways were aggregated and reduced to their mean chromatin accessibility in each sample. These pathway-level values were transformed into sample-wise Z-scores, and the mean change upon ibrutinib treatment was calculated. A similar procedure was applied to the sample-wise pharmacoscopy data, with the exception that we used the Drug Gene Interaction Database48 version 3.0 to retrieve known drug-gene interactions based on the common/commercial drug name and kept only pathways with more than one annotated drug. Integration of chemosensitivity profiles with the ATAC-seq data as shown in Figure 5a was done by calculating the mean of each pathway across all samples collected before ibrutinib treatment and, separately, during ibrutinib treatment, and calculating log2 fold-changes for the two values, standardized for each data type with a Z-score. Combined changes between data types were calculated as the mean of the Z-scores, as displayed in Supplementary Figure 4.
Supplementary Material
Summary.
Epigenome profiling in combination with imaging-based chemosensitivity and integrative bioinformatic analysis establishes a powerful method for detecting therapy-induced vulnerabilities, as shown here for ibrutinib-treated chronic lymphocytic leukemia.
Acknowledgements
We thank all patients who have donated their samples for this study. We also thank the Biomedical Sequencing Facility at CeMM for assistance with next generation sequencing and J. Bigenzahn, M. Rebsamen as well as the GSF and CB labs for help and advice. This work was performed in the context of the following grants and fellowships: WWTF LS16-034 to GSF and UJ; FWF SFB F 4711-B20 to GSF; EMBO Long-Term Fellowship 1543-2012 to GIV and 733-2016 to TP; Swiss National Science Foundation Fellowship P300P3_147897 and PP00P3_163961 to BS; Marie-Sklodowska Curie Action Fellowship 703668 to NK; Feodor Lynen Fellowship of the Alexander von Humboldt Foundation to C. Schmidl; Marie Curie Action International Outgoing Fellowship (PIOF-2013-624924) to MG; Initiative Krebsforschung (UE71104017, UE71104005, UE71504001, and UE711043037), Austrian Society of Hematology and Oncology (ÖGHO AP00359OFF), and Anniversary Fund of the Austrian National Bank (OeNB AP130120ONB) to MS; Austrian Academy of Sciences New Frontiers Group Award and ERC Starting Grant (European Union’s Horizon 2020 research and innovation programme) 679146 to CB.
Footnotes
Author contributions
CSc, TK performed ATAC-seq experiments; GIV, CT, AR, KR performed image-based chemosensitivity experiments; CSc, GIV, AFR, NK, BS, OLF, SK analyzed the ATAC-seq and image-based chemosensitivity data; SS, CT, TP, MA, RH, DD, MH, MS handled patient samples and performed validation experiments; MS and UJ were responsible for study ethics; CSk, EP, MG, GH, PBS, MS, UJ provided and analyzed clinical data or oversaw patient care and ethics; CSc, GIV, AFR, TP, MS, GSF, UJ, CB wrote the manuscript; MS, GSF, UJ, CB oversaw the project.
Competing Financial Interests Statement
GIV, NK, BS, GSF are co-founders of Allcyte GmbH, which has licensed the pharmacoscopy technology, and they are listed as inventors on patent applications for the the pharmacoscopy / single-cell imaging methodology. GIV and NK have become employees of Allcyte GmbH during the course of this study. UJ received research grants and honoraria from Janssen Cilag, Abbvie, Novartis, and Roche Austria.
Data availability
The ATAC-seq and pharmacoscopy data are available from the Supplementary Website (http://cll-combinations.computational-epigenetics.org). The ATAC-seq data are also available from NCBI GEO under accession number GSE100672. The source code for ATAC-seq data processing is available from a Github repository linked on the above website.
References
- 1.Woyach JA, Johnson AJ, Byrd JC. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120:1175–84. doi: 10.1182/blood-2012-02-362624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevenson FK, Krysov S, Davies AJ, Steele AJ, Packham G. B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2011;118:4313–20. doi: 10.1182/blood-2011-06-338855. [DOI] [PubMed] [Google Scholar]
- 3.Burger JA, Chiorazzi N. B cell receptor signaling in chronic lymphocytic leukemia. Trends Immunol. 2013;34:592–601. doi: 10.1016/j.it.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrd JC, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Brien S, et al. Ibrutinib for patients with relapsed or refractory chronic lymphocytic leukaemia with 17p deletion (RESONATE-17): a phase 2, open-label, multicentre study. Lancet Oncol. 2016;17:1409–1418. doi: 10.1016/S1470-2045(16)30212-1. [DOI] [PubMed] [Google Scholar]
- 6.Burger JA, et al. Ibrutinib as Initial Therapy for Patients with Chronic Lymphocytic Leukemia. N Engl J Med. 2015;373:2425–37. doi: 10.1056/NEJMoa1509388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herman SE, et al. Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765. Blood. 2011;117:6287–96. doi: 10.1182/blood-2011-01-328484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponader S, et al. The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood. 2012;119:1182–9. doi: 10.1182/blood-2011-10-386417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Rooij MF, et al. The clinically active BTK inhibitor PCI-32765 targets B-cell receptor- and chemokine-controlled adhesion and migration in chronic lymphocytic leukemia. Blood. 2012;119:2590–4. doi: 10.1182/blood-2011-11-390989. [DOI] [PubMed] [Google Scholar]
- 10.Chen SS, et al. BTK inhibition results in impaired CXCR4 chemokine receptor surface expression, signaling and function in chronic lymphocytic leukemia. Leukemia. 2016;30:833–43. doi: 10.1038/leu.2015.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burger JA, et al. Leukemia cell proliferation and death in chronic lymphocytic leukemia patients on therapy with the BTK inhibitor ibrutinib. JCI Insight. 2017;2:e89904. doi: 10.1172/jci.insight.89904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burger JA, et al. Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat Commun. 2016;7 doi: 10.1038/ncomms11589. 11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman SE, et al. Ibrutinib inhibits BCR and NF-kappaB signaling and reduces tumor proliferation in tissue-resident cells of patients with CLL. Blood. 2014;123:3286–95. doi: 10.1182/blood-2014-02-548610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bottoni A, et al. Targeting BTK through microRNA in chronic lymphocytic leukemia. Blood. 2016;128:3101–3112. doi: 10.1182/blood-2016-07-727750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landau DA, et al. The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02329-y. 2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maddocks K, Jones JA. Bruton tyrosine kinase inhibition in chronic lymphocytic leukemia. Semin Oncol. 2016;43:251–9. doi: 10.1053/j.seminoncol.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Maddocks KJ, et al. Etiology of Ibrutinib Therapy Discontinuation and Outcomes in Patients With Chronic Lymphocytic Leukemia. JAMA Oncol. 2015;1:80–7. doi: 10.1001/jamaoncol.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woyach JA, et al. Resistance mechanisms for the Bruton's tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–94. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamothe B, et al. Proteasome inhibitor carfilzomib complements ibrutinib's action in chronic lymphocytic leukemia. Blood. 2015;125:407–10. doi: 10.1182/blood-2014-07-585364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng J, et al. Bruton's tyrosine kinase inhibition increases BCL-2 dependence and enhances sensitivity to venetoclax in chronic lymphocytic leukemia. Leukemia. 2017 doi: 10.1038/leu.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snijder B, et al. Image-based ex-vivo drug screening for patients with aggressive haematological malignancies: interim results from a single-arm, open-label, pilot study. Lancet Haematol. 2017;4:e595–e606. doi: 10.1016/S2352-3026(17)30208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rendeiro AF, et al. Chromatin accessibility maps of chronic lymphocytic leukaemia identify subtype-specific epigenome signatures and transcription regulatory networks. Nat Commun. 2016;7 doi: 10.1038/ncomms11938. 11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nuckel H, et al. Lipoprotein lipase expression is a novel prognostic factor in B-cell chronic lymphocytic leukemia. Leuk Lymphoma. 2006;47:1053–61. doi: 10.1080/10428190500464161. [DOI] [PubMed] [Google Scholar]
- 25.O'Brien S, et al. Single-agent ibrutinib in treatment-naive and relapsed/refractory chronic lymphocytic leukemia: a 5-year experience. Blood. 2018;131:1910–1919. doi: 10.1182/blood-2017-10-810044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheffield NC, Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016;32:587–9. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bucur O, et al. PLK1 is a binding partner and a negative regulator of FOXO3 tumor suppressor. Discoveries (Craiova) 2014;2 doi: 10.15190/d.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shehata M, et al. Reconstitution of PTEN activity by CK2 inhibitors and interference with the PI3-K/Akt cascade counteract the antiapoptotic effect of human stromal cells in chronic lymphocytic leukemia. Blood. 2010;116:2513–21. doi: 10.1182/blood-2009-10-248054. [DOI] [PubMed] [Google Scholar]
- 31.Bock C, Lengauer T. Managing drug resistance in cancer: lessons from HIV therapy. Nat Rev Cancer. 2012;12:494–501. doi: 10.1038/nrc3297. [DOI] [PubMed] [Google Scholar]
- 32.Chari A, et al. Phase 1 trial of ibrutinib and carfilzomib combination therapy for relapsed or relapsed and refractory multiple myeloma. Leuk Lymphoma. 2018:1–7. doi: 10.1080/10428194.2018.1443337. [DOI] [PubMed] [Google Scholar]
- 33.Dietrich S, et al. Drug-perturbation-based stratification of blood cancer. J Clin Invest. 2018;128:427–445. doi: 10.1172/JCI93801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao YF, et al. Inhibiting PLK1 induces autophagy of acute myeloid leukemia cells via mammalian target of rapamycin pathway dephosphorylation. Oncol Rep. 2017;37:1419–1429. doi: 10.3892/or.2017.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giulino-Roth L, et al. Inhibition of Hsp90 Suppresses PI3K/AKT/mTOR Signaling and Has Antitumor Activity in Burkitt Lymphoma. Mol Cancer Ther. 2017;16:1779–1790. doi: 10.1158/1535-7163.MCT-16-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xian L, Huff CA, Resar LM. IBRUTinib: BRUTe force against bortezomib-resistant myeloma cells. Cell Cycle. 2015;14:1349–50. doi: 10.1080/15384101.2015.1022058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathews Griner LA, et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci U S A. 2014;111:2349–54. doi: 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ezell SA, et al. Synergistic induction of apoptosis by combination of BTK and dual mTORC1/2 inhibitors in diffuse large B cell lymphoma. Oncotarget. 2014;5:4990–5001. doi: 10.18632/oncotarget.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenenbaum D. KEGGREST: Client-side REST access to KEGG. R package version 1.14.1. 2017 [Google Scholar]
- 40.Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–4. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffman MM, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–41. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ernst J, Kellis M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat Biotechnol. 2015;33:364–76. doi: 10.1038/nbt.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vladimer GI, et al. Global survey of the immunomodulatory potential of common drugs. Nat Chem Biol. 2017 doi: 10.1038/nchembio.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Griffith M, et al. DGIdb: mining the druggable genome. Nat Methods. 2013;10:1209–10. doi: 10.1038/nmeth.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.