Abstract Abstract
The genus Beauveria is considered a cosmopolitan anamorphic and teleomorphic genus of soilborne necrotrophic arthropod-pathogenic fungi that includes ecologically and economically important species. Species identification in Beauveria is difficult because of its structural simplicity and the lack of distinctive phenotypic variation. Therefore, the use of multi-locus sequence data is essential to establish robust species boundaries in addition to DNA-based species delimitation methods using genetic distance, coalescent, and genealogical concordance approaches (polyphasic approaches). In this regard, our study used multilocus phylogeny and five DNA-based methods to delimit species in Beauveria using three molecular makers. These polyphasic analyses allowed for the delimitation of 20–28 species in Beauveria, confirming cryptic diversity in five species (i.e. B. amorpha, B. bassiana, B. diapheromeriphila, and B. pseudobassiana) and supporting the description of B. peruviensis as a new taxon from northeastern Peru. The other five species were not evaluated as they did not have enough data (i.e. B. araneola, B. gryllotalpidicola, B. loeiensis, B. medogensis, and B. rudraprayagi). Our results demonstrate that the congruence among different methods in a polyphasic approach (e.g. genetic distance and coalescence methods) is more likely to show reliably supported species boundaries. Among the methods applied in this study, genetic distance, coalescent approaches, and multilocus phylogeny are crucial when establishing species boundaries in Beauveria.
Keywords: Beauveria , fungal diversity, multi-locus phylogeny, Peru, polyphasic approaches, species delimitation
Introduction
Around 1800, a silkworm disease called “calcine”, “real del segno” or “muscardine” was causing great trouble in Italy and France (Redaelli and Visocchi 1940). Experiments developed by Agostino Bassi in Mariago, Italy showed that a parasitic fungus produced this disease (Redaelli and Visocchi 1940). Balsamo (1835) confirmed this discovery and concluded that the incrustation and white efflorescence, which covered the body of a dead silkworm, were a fungus of the genus Botrytis. He first named this species Botrytis paradoxa Balsamo and later Botrytis bassiana Balsamo (Balsamo 1835). Then, this species was transferred to its own genus and Beauveria Vuillemin was established on the basis of B. bassiana Vuillemin as the type species (Vuillemin 1912).
The genus Beauveria is considered a cosmopolitan genus of soilborne necrotrophic arthropod-pathogenic fungi that includes ecologically and economically important species (Rehner et al. 2011, Kepler et al. 2017, Chen et al. 2018). Morphologically, Beauveria genus have been characterized asexually by having conidiogenous cells arising from short, often one-celled, more or less swollen stalk cells, often in dense clusters, or scattered or in whorls from undifferentiated hyphae; they consist of a globose to fusiform basal part, and a geniculate, denticulate rachis. Conidia one-celled, hyaline, smooth, thin-walled, globose to ellipsoidal (de Hoog 1972). The sexual morphs form stromata solitary, paired or gregarious, unbranched, fleshy texture, fertile area apical, cylindrical to clavate, yellowish to orange; perithecia partially immersed, in longitudinal section oval to ovoid; and asci hyaline with cylindrical and filiform ascospores (Kepler et al. 2017).
Based on the end of dual nomenclature for different morphs of the same fungus in 2011 (McNeill et al. 2012), Kepler et al. (2017) phylogenetically established the genetic boundaries in Cordycipitaceae regardless of life-stage or the associated morphological differences. One of the most significant changes was the recognition of Beauveria as a genus separate from Cordyceps. Although direct links between species of Beauveria and cordyceps-like sexual morphs have been demonstrated from molecular data and culture-based experiments (Shimazu et al. 1988, Li et al. 2001, Huang et al. 2002, Shrestha et al. 2014), their respective type species are not congeneric (Kepler et al. 2017). Thereby, the clade composed of Beauveria currently includes the traditional species known from asexual morphs, as well as several taxa previously described for sexual morphs in Cordyceps (Sanjuan et al. 2014, Kepler et al. 2017).
Initially, Beauveria was delimitated based on diagnostic features, and three species were recognized, i.e., B. bassiana, B. brongniartii and B. alba (Limber) Saccas (de Hoog, 1972). New additions were included by de Hoog and Rao (1975), Samson and Evans (1982), Bissett and Widden (1986) and Rehner et al. (2006). Molecular analyses confirmed the monophyly and placement of seven species of Beauveria within Cordycipitaceae (Rehner and Buckley 2005, Sung et al. 2007). More recent molecular studies based on multilocus phylogenetic analysis that included the Bloc nuclear intergenic region, internal transcribed spacer (ITS), translation elongation factor-1α (TEF), and RNA polymerase II largest subunit (RPB1) and second largest subunit (RPB2) demonstrated that Beauveria is composed of 26 species (Rehner et al. 2011, Sanjuan et al. 2014, Kepler et al. 2017, Chen et al. 2018). These phylogenetic studies also revealed that the most commonly reported species, namely, B. bassiana and B. brongniartii, encompass cryptic lineages with worldwide distributions (Rehner et al. 2006, 2011, Ghikas et al. 2010). Although morphologically distinctive as a genus, species identification in Beauveria, especially in the conidiogenic state, is difficult because of its structural simplicity and lack of distinctive phenotypic variation. Thus, numerous registered mycoinsecticide formulations based on B. bassiana and B. brongniartii that are extensively used for the control of insect pests worldwide (Faria and Wraight 2007) are not likely based on these species (Rehner et al. 2006).
In the Amazonian region, a total of five species have been reported (Rehner et al. 2011, Sanjuan et al. 2014). Two of these species B. acridophila (T. Sanjuan & Franco-Mol.) T. Sanjuan, B. Shrestha, Kepler & Spatafora and B. diapheromeriphila (T. Sanjuan & S. Restrepo) T. Sanjuan, B. Shrestha, Kepler & Spatafora, and a lectotype, namely, B. locustiphila (Henn.) B. Shrestha, Kepler & Spatafora were recently described on the basis of molecular data and their sexual stages were characterized (Sanjuan et al. 2014). Additionally, two species of Beauveria were reported from Peru: B. amorpha Samson & Evans and B. bassiana, but only the former has been confirmed by molecular analysis while the latter is extensively used in coffee rust programs to control the expansion of the coffee borer (Rehner et al. 2011).
Given the problems with species delimitation in fungi using morphology, molecular data are becoming the standard for delimiting species and testing their traditional boundaries (Rehner et al. 2011). The recognition of distinct clades in gene trees as species is likely to be misleading in understanding the evolutionary history of taxa (Liu et al. 2016). Therefore, the use of multi-locus sequence data is essential to establish robust species boundaries (Lumbsch and Leavitt 2011). Most researchers, however, did not carefully examine the species boundaries but simply recognized distinct clades in single-gene trees as separate species (Stewart et al. 2014). Estimating the species tree and species delimitation using genetic distance (e.g. automated barcode gap discovery algorithm, ABGD; and statistical parsimony, SPN), coalescent (e.g. generalized mixed Yule coalescent, GMYC; and Bayesian phylogenetics and phylogeography, BPP), and genealogical concordance (genealogical concordance phylogenetic species recognition, GCPSR) methods have proven very useful and have been used for a range of animal and plant taxa (Liu et al. 2016). These methods have otherwise not been used much in fungi, especially in studies of pathogenic fungi (Millanes et al. 2014, Liu et al. 2015). Therefore, the use of several methodologies and data sets to delimit species is recommended, and subsequently, the achievement of congruent results across the methods is likely to prove most useful for framing reliably supported species boundaries (Carstens et al. 2013).
In this study, we analyzed species of the newly circumscribed genus Beauveria, including an unreported species isolated from coffee farms in northeastern Peru, based on morphological observations, phylogenetic inferences, and DNA-species delimitation methods. Three nuclear molecular markers (Bloc, rpb1, and tef1) were used to examine their phylogenetic relationships and to assess species boundaries within the genus Beauveria.
Materials and methods
Collection of specimens and isolation
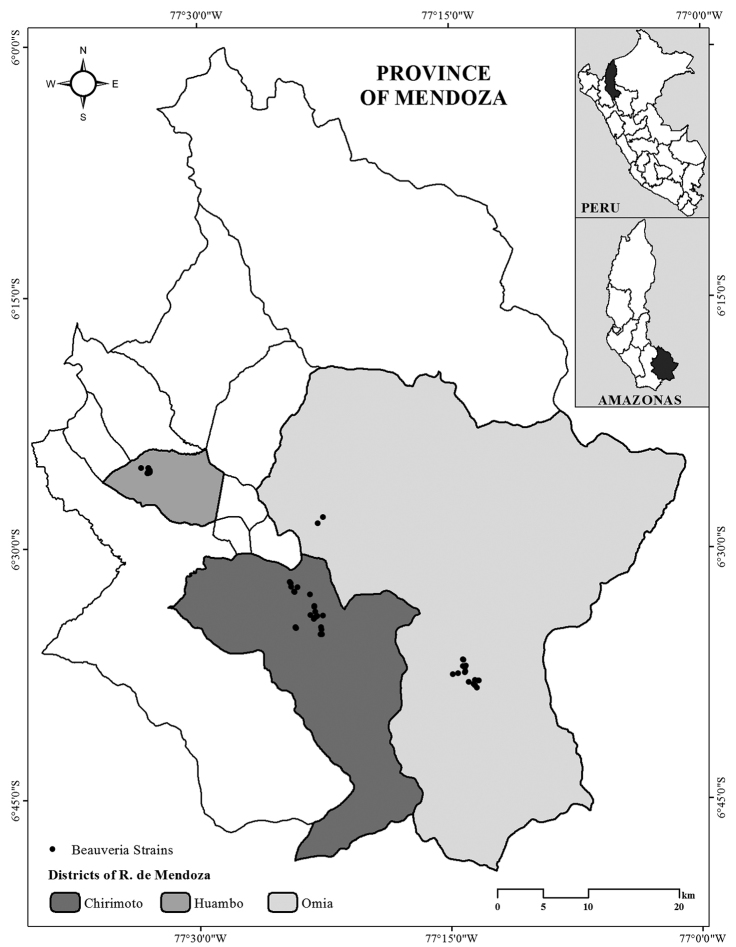
Fungal strains were isolated from infected coffee borers (Hypothenemus hampei) obtained from infected coffee berries according to Gerónimo-Torres et al. (2016). They were collected during July and August 2017 from three districts in the province of Rodriguez de Mendoza, Amazonas, Peru (Fig. 1). Briefly, infected coffee berries were preserved at 5 °C until coffee borers were recovered from them. The coffee borers with signs of fungal infection were cleaned superficially in 0.5% sodium hypochlorite solution and rinsed with sterile distilled water. Then, insects were placed in a humid chamber (90% RH and 25 °C) for 8 days to allow the growth of the entomopathogenic fungus. Once visible mycelia appeared on the borers under observations with a stereo microscope (Nikon SMZ18, Tokyo, Japan), these were transferred to a Petri-dish containing potato dextrose agar (PDA; Merck, Darmstadt, Germany).
Figure 1.
Collections of the 55 strains of B. amazonensis sp. nov. from the Rodriguez de Mendoza Province.
Identification of isolates
Fifty-five fungal strains were incubated as monosporic cultures on PDA at 25 °C for 15 days. Morphological characterization of the fungus was performed as described by Rehner and Buckley (2005). Microscope observations were made from fungal mycelia and other structures stained with methylene blue (0.1–0.5%). Photomicrographs were taken under an inverted microscope (IX83; Olympus, Tokyo, Japan) with an integrated camera (Nikon D810, Tokyo, Japan). Fungal strains were deposited as semisolid and dry material in the herbarium of Toribio Rodriguez de Mendoza National University (UT), Peru.
Molecular phylogenetic analyses
Genomic DNA was extracted from semisolid PDA cultures using the NucleoSpin Plant II Kit (Macherey-Nagel, Düren, Germany), following the manufacturer’s instructions. Three genes were sequenced, i.e., Bloc, rpb1, and tef1. Each gene was amplified using polymerase chain reaction (PCR) with MasterMix (Promega, Wisconsin, USA) in the following reaction mixture: 10 ng of DNA and 0.25–0.5 pmol of forward and reverse primers for a total volume of 10 μl. The PCR protocols and primer combinations for Bloc (B5.1F, B5.4F, B3.1R, B3.3R), rpb1 (RPB1A, RPB1A_VH6R, RPB1B_VH6Fa, RPB1B_G2R), and tef1 (983F, 1567RintB) followed Rehner et al. (2011). The sequences of the forward and reverse strands were determined commercially by Macrogen Inc. (Macrogen, Seoul, Korea). New Bloc, rpb1, and tef1 sequences were deposited in GenBank (Table 1). These sequences and others obtained from GenBank were initially aligned with Muscle algorithms (Thompson et al. 1994) and were adjusted manually with MEGA6 software (Tamura et al. 2013).
Table 1.
List of species used in the molecular analyses.
| Species | Country | Strain | Bloc | RPB1 | tef1 |
|---|---|---|---|---|---|
| B. acridophila | Colombia | HUA 179219 | – | JX003857 | JQ958613 |
| Colombia | HUA 179221 | – | JX003853 | JQ958615 | |
| Colombia | HUA 179220 | – | JX003852 | JQ958614 | |
| Colombia | MCA 1181 | – | MF416628 | – | |
| B. amorpha | Australia | ARSEF4149 | HQ880735 | HQ880876 | HQ881006 |
| USA, Colorado | ARSEF7542 | HQ880736 | HQ880877 | HQ881007 | |
| Chile | B518a | HQ880737 | HQ880878 | HQ881008 | |
| Peru | ARSEF1969 | HQ880738 | HQ880879 | AY531907 | |
| Brazil | ARSEF2641 | HQ880739 | HQ880880 | AY531917 | |
| B. asiatica | China | ARSEF4384 | HQ880716 | HQ880857 | AY531935 |
| China | ARSEF4474 | HQ880717 | HQ880858 | AY531936 | |
| Korea | ARSEF4850 | HQ880718 | HQ880859 | AY531937 | |
| B. australis | Australia | ARSEF4580 | HQ880719 | HQ880860 | HQ880994 |
| Australia | ARSEF4622 | HQ880721 | HQ880862 | HQ880996 | |
| Australia | WCN2015 | KT961698 | HQ880861 | HQ880995 | |
| B. bassiana | Japan | ARSEF1040 | HQ880689 | HQ880830 | AY531881 |
| Australia | ARSEF300 | HQ880690 | HQ880831 | AY531924 | |
| Italy | ARSEF1564 | HQ880692 | HQ880833 | HQ880974 | |
| Japan | ARSEF7518 | HQ880693 | HQ880834 | HQ880975 | |
| Vietnam | ARSEF751 | HQ880694 | HQ880831 | AY531954 | |
| Brazil | ARSEF1478 | HQ880695 | HQ880836 | AY531890 | |
| Morocco | ARSEF1811 | HQ880696 | HQ880837 | AY531901 | |
| B. brongniartii | Japan | ARSEF7516 | HQ880697 | HQ880838 | HQ880976 |
| USA, Oregon | ARSEF10278 | HQ880700 | HQ880841 | HQ880979 | |
| Korea | ARSEF7268 | HQ880703 | HQ880844 | HQ880982 | |
| USA, New York | ARSEF6213 | HQ880706 | HQ880847 | HQ880985 | |
| Japan | ARSEF4363 | HQ880707 | HQ880848 | HQ880986 | |
| Japan | ARSEF4362 | HQ880708 | HQ880849 | HQ880980 | |
| USA, Kentucky | ARSEF2271 | HQ880710 | HQ880851 | HQ880988 | |
| USA, Oregon | ARSEF10277 | HQ880711 | HQ880852 | HQ880989 | |
| France | ARSEF979 | HQ880714 | HQ880855 | HQ880992 | |
| B. caledonica | Switzerland | ARSEF1567 | HQ880747 | HQ880888 | AY531894 |
| Scotland | ARSEF2567 | HQ880748 | HQ880889 | AY531915 | |
| Denmark | ARSEF8024 | HQ880749 | HQ880890 | HQ881012 | |
| Brazil | ARSEF2251 | HQ880750 | HQ880891 | AY531912 | |
| USA, Georgia | ARSEF7117 | HQ880751 | HQ880892 | HQ881013 | |
| Australia | ARSEF4302 | HQ880752 | HQ880893 | HQ881014 | |
| B. diapheromeriphila | Ecuador | QCNE 186272 | – | JX003848 | JQ958610 |
| Ecuador | QCNE 186714 | – | MF416648 | MF416491 | |
| Ecuador | MCA 1557 | – | JX003848 | JQ958610 | |
| B. hoplocheli | Reunion | Bt116 | KM453967 | KM453957 | KC339703 |
| Reunion | Bt121 | KM453968 | KM453956 | KC339704 | |
| Reunion | Bt124 | KM453969 | KM453955 | KC339699 | |
| Reunion | Bt125 | KM453970 | KM453953 | KC339701 | |
| Reunion | Bt128 | KM453972 | KM453952 | KC339705 | |
| Reunion | Bt129 | KM453973 | KM453951 | KC339706 | |
| Madagascar | Bt96 | KM453974 | KM453950 | KC339709 | |
| Reunion | Bt99 | KM453975 | KM453949 | KC339710 | |
| B. kipukae | USA, Hawaii | ARSEF7032 | HQ880734 | HQ880875 | HQ881005 |
| B. lii | China | RCEF5500 | JN689373 | JN689374 | JN689371 |
| B. malawiensis | China | GZU12142 | MG052638 | MG052645 | MG052641 |
| China | GZU12141 | MG052639 | MG052644 | MG052640 | |
| Australia | ARSEF4755 | HQ880754 | HQ880895 | HQ881015 | |
| Australia | BCC17613 | HQ880755 | HQ880896 | HQ881016 | |
| Malawi | ARSEF7760 | HQ880756 | HQ880897 | DQ376246 | |
| B. peruviensis | Peru | UTRF21 | MN094752 | MN100113 | MN094767 |
| Peru | UTRF24 | MN094753 | MN100119 | MN094768 | |
| Peru | UTRF25 | MN094754 | MN100114 | MN094769 | |
| Peru | UTRF26 | MN094758 | MN100120 | MN094770 | |
| Peru | UTRF35 | MN094755 | MN100115 | MN094771 | |
| Peru | UTRF37 | MN094756 | MN100116 | MN094772 | |
| Peru | UTRF38 | MN094759 | MN100121 | MN094773 | |
| Peru | UTRF40 | MN094760 | MN100122 | MN094774 | |
| Peru | UTRF42 | MN094761 | MN100123 | MN094775 | |
| Peru | UTRF58 | MN094762 | MN100124 | MN094776 | |
| Peru | UTRP6 | MN094763 | MN100125 | MN094777 | |
| Peru | UTRP7 | MN094764 | MN100127 | MN094778 | |
| Peru | UTRP13 | MN094765 | MN100126 | MN094779 | |
| Peru | UTRP17 | MN094766 | MN100117 | MN094780 | |
| Peru | UTRP19 | MN094757 | MN100118 | MN094781 | |
| B. pseudobassiana | Portugal | ARSEF3220 | HQ880722 | HQ880863 | AY531928 |
| USA, Kentucky | ARSEF3405 | HQ880723 | HQ880864 | AY531931 | |
| USA, Wisconsin | ARSEF3216 | HQ880725 | HQ880866 | AY531927 | |
| USA, Maryland | ARSEF3529 | HQ880726 | HQ880867 | HQ880998 | |
| France | ARSEF4933 | HQ880726 | HQ880870 | AY531938 | |
| Canada | ARSEF1855 | HQ880727 | HQ880868 | HQ880999 | |
| Canada | ARSEF2997 | HQ880728 | HQ880869 | HQ881000 | |
| China | ARSEF6229 | HQ880730 | HQ880871 | HQ881001 | |
| Korea | ARSEF7242 | HQ880730 | HQ880865 | HQ880997 | |
| B. scarabaeicola | Korea | ARSEF5689 | – | DQ522380 | DQ522335 |
| Japan | ARSEF1685 | HQ880740 | HQ880881 | AY531899 | |
| Korea | ARSEF5689 | HQ880741 | HQ880882 | AY531939 | |
| Korea | ARSEF7043 | HQ880742 | HQ880883 | AY531948 | |
| Korea | ARSEF7044 | HQ880743 | HQ880884 | AY531949 | |
| Korea | ARSEF7279 | HQ880743 | HQ880885 | HQ881009 | |
| Korea | ARSEF7280 | HQ880744 | HQ880886 | HQ881010 | |
| Korea | ARSEF7281 | HQ880746 | HQ880887 | HQ881011 | |
| B. sinensis | China | RCEF3903 | – | JX524283 | HQ270151 |
| B. staphylinidicola | Korea | ARSEF5718 | – | EF468881 | EF468776 |
| B. varroae | France | ARSEF8259 | HQ880732 | HQ880873 | HQ881003 |
| Switzerland | ARSEF2694 | HQ880733 | HQ880874 | HQ881004 | |
| France | ARSEF8257 | HQ880733 | HQ880872 | HQ881002 | |
| B. vermiconia | Chile | ARSEF2922 | HQ880753 | HQ880894 | AY531920 |
| Cordyceps cicadae | Korea | ARSEF7260 | HQ880757 | HQ880898 | HQ881017 |
| Blackwiella cardinalis | USA | OSC93610 | – | EF469088 | EF469059 |
| Ascopolyporus polychrous | – | PC546 | – | DQ127236 | DQ118745 |
The phylogeny was based on concatenated data combining Bloc, rpb1, and tef1 (101 sequences, Table 1). Selection of the best-fitting nucleotide substitution model was conducted using the program PartitionFinder (Lanfear et al. 2012) with three partitions (Bloc, rpb1, and tef1). The best partition strategy and model of sequence evolution were selected based on the Bayesian Information Criterion (BIC). The general time reversible nucleotide substitution model with a gamma distribution and a proportion of invariable sites (GTR + Γ + I) was selected for all partitions. Maximum likelihood (ML) analyses were conducted with the RAxML HPC-AVX program (Stamatakis 2014) implemented in the raxmlGUI 1.3.1 interface (Silvestro and Michalak 2012) using a GTRGAMMAI model with 1000 bootstrap replications. Bayesian inference (BI) was performed with MrBayes v. 3.2.6 software (Ronquist et al. 2012) using Metropolis-coupled MCMC and the GTR + Γ + I model. We conducted two runs each with four chains (three hot and one cold) for 10,000,000 generations, sampling trees every 1,000 generations. We plotted likelihood vs. generation using the Tracer Version v. 1.6 program (Rambaut et al. 2014) to reach a likelihood plateau and set the burn-in value.
DNA-based species delimitation
Although 26 species have been molecularly confirmed in Beauveria (Rehner et al. 2011, Kepler et al. 2017, Chen et al. 2018), only 21 of these species and Beauveria sp. from Peru were used in the DNA-based delimitation methods. Beauveria araneola W.H. Chen, Y.F. Han, Z.Q. Liang & D.C. Jin, B. gryllotalpidicola Luangsa-ard, Ridkaew & Tasanathai, B. loeiensis Luangsa-ard, Ridkaew & Tasanathai, B. medogensis Imoulan & Y.J. Yao, and B. rudraprayagi Y. Agrawal, P. Mual & B.D. Shenoy were not used due to abundant missing data and short sequences for the three markers (e.g. ~731 bp for rpb1 and ~720 bp for tef1).
We explored five different DNA-based delimitation methods using Bloc, rpb1, and tef1 data sets to assess species boundaries in Beauveria. Although B. acridophila, B. blattidicola M. Chen, Aime, T.W. Henkel & Spatafora, B. diapheromeriphila, B. locustiphila, and B. staphylinidicola (Kobayasi & Shimizu) B. Shrestha, Kepler & Spatafora lack Bloc sequences, these species were used in the analysis to evaluate its status in the new circumscribed Beauveria. Two of these DNA-based delimitation methods are based on genetic distance [statistical parsimony network analysis (SPN) (Hart and Sunday 2007) and automatic barcoding gap detection (ABGD) (Puillandre et al. 2012)], two in coalescence [generalized mixed Yule coalescent method (GMYC) (Pons et al. 2006) and Bayesian phylogenetics and phylogeography (BPP) (Rannala and Yang 2003)], and one in genealogical concordance [genealogical concordance phylogenetic species recognition (GCPSR) (Quaedvlieg et al. 2014)]. For the SPN analyses of Bloc, rpb1, and tef1, data sets were generated in TCS 1.21 (Clement et al. 2000) with a maximum connection probability set at 95% statistical confidence. The ABGD method was tested via a web interface (ABGD web, http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html). Before analysis, the model criteria were set as follows: variability (P) between 0.001 (Pmin) and 0.1 (Pmax), minimum gap width (X) of 0.1, Kimura-2-parameters and 50 screening steps.
To perform the GMYC delimitation method, an ultrametric tree was constructed in BEAST v.2.0.2 (Drummond et al. 2012), relying on the uncorrelated lognormal relaxed clock, the GTR + Γ + I model, and a coalescent tree prior. Bayesian Markov chain Monte Carlo was run for 50 million generations, and trees and parameters were sampled every 1000 generations. Log files were visualized in Tracer v.1.6 (Rambaut et al. 2014) for assessing the stationary state of parameters on the basis of the value of estimate-effective sample size (ESS). After removing 25% of trees as burn-in, the remaining trees were used to generate a single summarized tree in TreeAnnotator v.2.0.2 (part of the BEAST v.2.0.2 package) as an input file for GMYC analyses. The GMYC analyses with a single threshold model were performed in R (R Development Core Team, http://www.R-project.org) under the ‘splits’ package using the ‘gmyc’ function (R-Forge, http://r-forge.r-project.org/projects/splits/).
To validate the outcomes of single locus species delimitation, a multilocus BPP was applied using the program BP&P v.2.0 (Rannala and Yang 2003, Yang and Rannala 2010, Liu et al. 2015). The three-gene data (Bloc, rpb1, and tef1) were used as input for BPP under the A11 model (A11: species delimitation = 1, species tree = 1). Specimens were a priori assigned to species based only on the minimum number of species from the results of the phylogenetic analysis. The guide tree derived from the three-gene ML analysis was used. Five variables (ε1~ε5) were automatically fine-tuned following the instructions of BP&P (Rannala and Yang 2003, Yang and Rannala 2010). The prior distribution of θ and τ could have influenced the posterior probabilities for different models (Yang and Rannala 2010). Analyses were run with three different prior combinations (Leaché and Fujita 2010). Each analysis was run three times to confirm consistency between runs. Two independent MCMC analyses were run for 100,000 generations with the ‘burn-in’ = 20,000.
GCPSR was implemented by identifying independent evolutionary lineages (IELs) and by exhaustive subdivision of strains into phylogenetic species. The criteria used to identify IELs and exhaustive subdivision were the same as those used by Brankovics et al. (2018). These were implemented using Perl scripts developed by Brankovics et al. (2018) and available at GitHub (https://github.com/b-brankovics/GCPSR).
Results
Molecular phylogeny
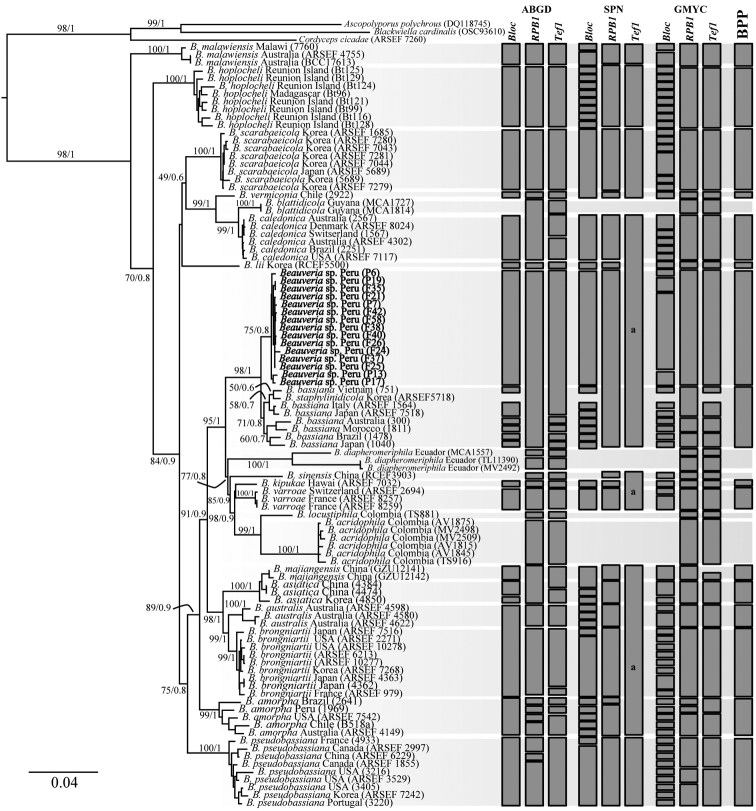
In the phylogeny of Beauveria species, the analyzed data matrix included 1592 base pairs (bp) for Bloc, 2890 bp rpb1, and 1181 bp for tef1 of 101 individuals. Phylogenetic trees obtained from ML and BI analyses confirmed the robustly supported monophyly of the genus Beauveria (Fig. 2). The tree topologies for the individual genes (tef1, Bloc, and rpb1) did not show congruence (Suppl. material 1: Figs S1–S3). These trees showed topological differences, especially in the clades composed of B. asiatica / B. majiangensis and by B. bassiana / B. staphylinidicola / Beauveria sp. from Peru. Although the individual gene trees did not show congruence with the combined data, the latter resolved these clades, suggesting conspecificity in the first clade and sister relationship in the second. Moreover, the multilocus phylogeny showed well-supported clades in both the ML and BI analyses except in B. lii, B. majiangensis, and B. staphylinidicola. The genetic divergence comparisons showed that the minimum threshold (p-distance) to distinguish genetic species in Beauveria was 1.3%, 0.4%, and 0.2% for Bloc, rpb1, and tef1, respectively, as occurred between B. australis and B. asiatica.(Table 2).
Figure 2.
Phylogenetic tree based on maximum likelihood inference of combined Bloc, RPB1, Tef1 data. Value above branches = Maximum likelihood bootstrap values (BS) / Bayesian posterior probabilities. Grey bars represent species delimitation results from ABGD-, SPN-, GMYC- and BPP based algorithmic methods based on Bloc, RPB1, and Tef1 sequences. Scale bar indicates the number of nucleotide substitution per site. a: delimited as the same species. B. araneola, B. gryllotalpidicola, B. loeiensis, B. medogensis, and B. rudraprayagi were not delimited by any DNA-based algorithm due to abundant missing data in their sequences.
Table 2.
Genetic distance (p-distances) in percentage for species of Beauveria for three markers.
| Taxa | Markers | ||
| Bloc | RPB1 | tef1 | |
| B. australis – B. asiatica | 1.3 | 0.4 | 0.2 |
| B. bassiana – B. staphylinidicola | 3.1 | 0.5 | 0.2 |
| B. bassiana – B. peruviensis | 3.5–4.1 | 0.3–0.5 | 0.2–0.4 |
| B. peruviensis – B. staphylinidicola | 4.1–4.7 | 0.7–1.1 | 0.2 |
Species delimitation
The species-delimitation methods based on genetic distance (ABGD, SPN), coalescence (GMYC, BPP), and genealogical concordance (GCPSR) showed incongruent results for the three genes (Fig. 2, Table 3). Among these methods, the highest number of species was delimited in the GMYC analysis for the Bloc gene, whereas conservative results were observed in BPP. The species delimitations by SPN and GCPSR have inadequate and contradictory results. The genetic distance method based on the barcode gap (ABGD) found similar species numbers for Bloc, rpb1, and tef1, differing only in the species recognized in the clades B. asiatica / B. majiangensis and B. bassiana / B. staphylinidicola / Beauveria sp. from Peru. In the former clade, there were 3, 1 and 2 species for Bloc, rpb1, and tef1; whereas in the latter clade, there were 7, 1, and 5 species for Bloc, rpb1, and tef1. The GMYC identified relatively conserved results in RPB1 (28) and tef1 (26) and plenty of species in the Bloc data set (63). This high number of species for the Bloc data set is a consequence of the splitting of the main clades into different species but lacking significance (Suppl. material 1: Table S1, Fig. S4). Regarding the multi-locus coalescent species validation (BPP), the highest posterior probabilities for Bloc, rpb1, and tef1 were found by recognizing 16 species based on the results from the phylogenetic analysis and single species delimitation methods (Suppl. material 1: Table S2). Conversely, the BPP analyses with the maximum number of species (39 and 62) were not used based on the inadequate results from SPN and GCPSR. Although there were incongruent results among different methods, the conservative results from species delimitation methods (ABGD and GMYC) and phylogenetic analysis suggest that Beauveria is composed of 20–28 and 26 species, respectively. These results also suggest that the clade composed of B. asiatica / B. majiangensis, B. diapheromeriphila, and B. bassiana / B. staphylinidicola / Beauveria sp. from Peru were genetically composed of more than one species. Our analysis also revealed that Beauveria sp. from Peru was supported as a distinct species by ABGD (Bloc gene), GMYC, BPP, and phylogeny. Thereby, the description of Beauveria sp. as a new species is proposed.
Table 3.
Species number in Beauveria identified under DNA-based species-delimitations methods and phylogeny.
| Taxa | Genetic distance | Coalescence | Genealogical concordance | Phylogeny | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABGD | SPN | GMYC | BPP | GCPSR | ||||||||
| Bloc | RPB1 | Tef1 | Bloc | RPB1 | Tef1 | Bloc | RPB1 | Tef1 | ||||
| B. acridophila | – | 1 | 1 | – | x | x | – | 1 | 1 | – | 1 | 1 |
| B. amorpha | 1 | 4 | 3 | 5 | 2 | x | 5 | 3 | 2 | 1 | 1 | |
| B. asiatica | 2 | x | 1 | 2 | 1 | x | 2 | 1 | 1 | 1 | 1 | |
| B. australis | x | 1 | 1 | 3 | 1 | x | 2 | 1 | 1 | 1 | 1 | |
| B. bassiana | 6 | x | 5 | 6 | x | x | 6 | 3 | 3 | 1 | 1 | |
| B. blattidicola | – | x | 1 | – | x | x | – | 1 | 1 | – | 1 | |
| B. brongniartii | x | 1 | 2 | 2 | 1 | x | 8 | 1 | 1 | 1 | 1 | |
| B. caledonica | 1 | 1 | 2 | 1 | 1 | x | 6 | 1 | 1 | 1 | 1 | |
| B. diapheromeriphila | – | 2 | 2 | – | x | x | – | 2 | 2 | – | 1 | |
| B. hoplocheli | 1 | 1 | 1 | 8 | 1 | 1 | 8 | 1 | 1 | 1 | 1 | |
| B. kipukae | 1 | 1 | 1 | 1 | 1 | x | 1 | 1 | 1 | 1 | 1 | |
| B. lii | 1 | 1 | 1 | 1 | 1 | x | 1 | 1 | 1 | 1 | 1 | |
| B. locustiphila | – | 1 | 1 | – | x | x | – | 1 | 1 | – | 1 | |
| B. majiangensis | 1 | x | x | x | 1 | x | 1 | 1 | 1 | 1 | 1 | |
| B. malawiensis | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | |
| B. pseudobassiana | 1 | 3 | 1 | 2 | 1 | 1 | 9 | 3 | 2 | 1 | 1 | |
| B. scarabaeicola | 1 | 1 | 1 | x | 1 | x | 6 | 1 | 1 | 1 | 1 | |
| B. sinensis | – | 1 | 1 | – | 1 | x | 1 | 1 | 1 | – | 1 | |
| B. staphylinidicola | – | x | x | – | x | x | – | x | x | – | 1 | |
| B. varroae | 1 | 1 | 1 | 1 | 1 | x | 2 | 1 | 1 | 1 | 1 | |
| B. vermiconia | 1 | 1 | 1 | x | 1 | x | 1 | 1 | 1 | 1 | 1 | |
| B. peruviensis | 1 | x | x | 1 | x | x | 2 | 1 | 1 | 1 | 1 | |
| Total | 20 | 22 | 28 | 35 | 16 | 3 | 63 | 28*** | 26*** | 16* | 1 | 22 |
x = non recognized as species, - = not evaluated, * = posterior probabilities higher or equal than 0.53, *** =highly significant
Morphological observations
Beauveria peruviensis
D.E.Bustamante, M.S.Calderon, M.Oliva, S.Leiva sp. nov.
0499423D9E175E25A78F332AB62C432D
MycoBank No: 829032
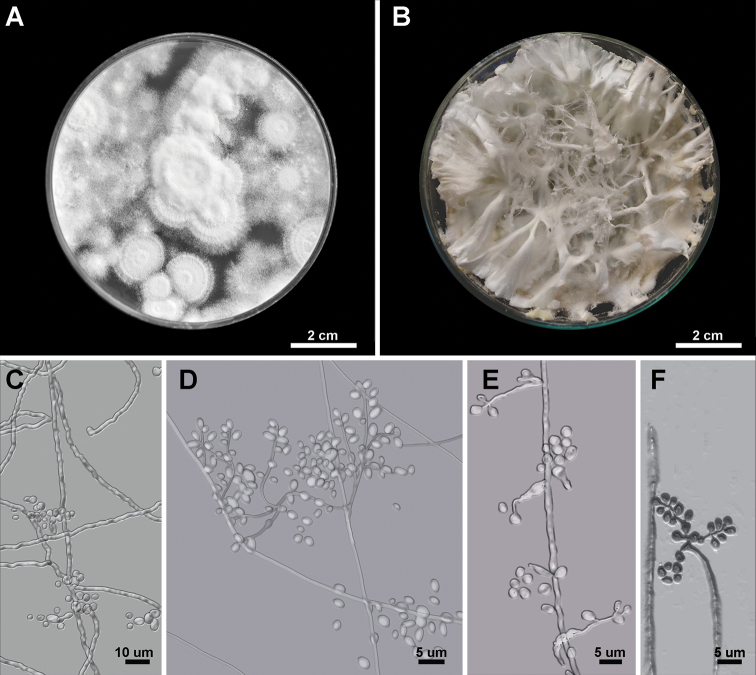
Figure 3.
Morphology of Beauveria amazonensis. A, B Colony growth on PDA showing the habit C–F conidiogenous cells and conidia.
Diagnosis.
Species very similar morphologically to Beauveria bassiana, but differing in the sister phylogenetic relationship with this species (Fig. 2). The sequence divergence between B. peruviensis and B. bassiana is 3.5–4.1% for Bloc, 0.3–0.5% for rpb1, and 0.2–0.4% for tef1. B. peruviensis is occurring in coffee plantations located in the middle altitudes of the Amazon region of Peru.
Type.
PERU. Amazonas: Prov. Rodríguez de Mendoza, Dist. Huambo, latitude -6.469, longitude -77.376, elev. 1642 m, entomopathogenic, 08 Nov. 2017, G. Ángulo, UTRP19 (holotype: UFV5609; isotype: ARSEF14196).
Description.
Colony growth on PDA, 15–38 mm diam. after 15 d at 25 C, 1.4–1.9 daily rate of radial growth, velutinous and closely appressed to agar surface, up to 3.5 mm thick, white, changing to yellowish white in older sections of the colony. Conidia aggregated as ca. 0.1 mm spherical clusters and white in mass. Colony reverse colorless or yellowish white to grayish white. Odor indistinct. Vegetative hyphae septate, branched, hyaline, smooth-walled, 1–1.5 μm wide. Conidiogenous cells, phialidic, solitary or occurring in dense lateral clusters, base subsphaerical, 3–6 μm wide, sympodially branched neck tapering into a long slender denticulate rachis, geniculate or irregularly bent, 2.0–3.5 × 1.5–2.5 μm. Conidia, 2–3 × 1–3 μm, Q = 1.0–1.8 (Lm = 2.5 μm, Wm = 2.2 μm, Qm = 1.6), mainly globose, slightly ellipsoid, oblong or cylindrical, hyaline, aseptate, walls smooth and thin. Mycelium on the host is granular-pulverulent, sometimes funiculose or rarely producing synnemata, white, rarely yellowish. Hyphae of the aerial mycelium bearing a conidial apparatus as described above. Basal parts of the conidiogenous cells globose, subglobose or somewhat flask-shaped.
Distribution.
This species is widely spread on coffee plantations in the middle altitudes of the Amazon region in northeastern Peru.
Ecology.
B. peruviensis was isolated from coffee borers (Hypothenemus hampei) obtained from coffee grains. Only the asexual stage was found.
Etymology.
The specific epithet ‘peruviensis’ is derived from the country where the samples were collected.
Additional specimens examined.
PERU. Amazonas: Prov. Rodríguez de Mendoza, Dist. Chirimoto, Achamal, -6.535, -77.408, 1351 m alt., 26 Jul. 2017, G. Angulo UTRF21 (UTR) ; -6.534, -77.409, 1345 m alt., 26 Jul. 2017, G. Angulo UTRF22 (UTR) ; -6.544, -77.404, 1435 m alt., 26 Jul. 2017, G. Angulo UTRF23 (UTR); -6.539, -77.401, 1374 m alt., 26 Jul. 2017, G. Angulo UTRF24 (UTR); -6.539, -77.407, 1386 m alt., 26 Jul. 2017, G. Angulo UTRF25 (UTR); -6.543, -77.405, 1428 m alt., 26 Jul. 2017, G. Angulo UTRF26 (UTR); Paraiso, -6.569, -77.383, 1218 m alt., 26 Jul. 2017, G. Angulo UTRF37 (UTR); -6.568, -77.382, 1197 m alt., 26 Jul. 2017, G. Angulo UTRF38 (UTR); -6.567, -77.389, 1387 m alt., 26 Jul. 2017, G. Angulo UTRF39 (UTR); -6.571, -77.385, 1250 m alt., 26 Jul. 2017, G. Angulo UTRF40 (UTR); -6.579, -77.403, 1427 m alt., 10 Aug. 2017, G. Angulo UTRP12 (UTR); -6.58, -77.403, 1444 m alt., 10 Aug. 2017, G. Angulo UTRP13 (UTR); -6.579, -77.404, 1439 m alt., 10 Aug. 2017, G. Angulo UTRP14 (UTR); Trancapata, -6.546, -77.389, 1255 m alt., 26 Jul. 2017, G. Angulo UTRF31 (UTR); -6.564, -77.384, 1161 m alt., 26 Jul. 2017, G. Angulo UTRF34 (UTR); Virgen del Carmen, -6.586, -77.379, 1313 m alt., 26 Jul. 2017, G. Angulo UTRF42 (UTR); -6.586, -77.378, 1271 m alt., 26 Jul. 2017, G. Angulo UTRF43 (UTR); -6.586, -77.377, 1256 m alt., 26 Jul. 2017, G. Angulo UTRF44 (UTR); -6.581, -77.377, 1138 m alt., 26 Jul. 2017, G. Angulo UTRF46 (UTR); Zarumilla, -6.568, -77.376, 1118 m alt., 26 Jul. 2017, G. Angulo UTRF35 (UTR); -6.58, -77.403, 1461 m alt., 10 Aug. 2017, G. Angulo UTRP15 (UTR); -6.58, -77.403, 1149 m alt., 10 Aug. 2017, G. Angulo UTRP16 (UTR); -6.559, -77.385, 1160 m alt., 10 Aug. 2017, G. Angulo UTRP17 (UTR); -6.558, -77.385, 1160 m alt., 10 Aug. 2017, G. Angulo UTRP18 (UTR); Huambo, Chontapamapa, -6.419, -77.557, 1637 m alt., 27 Jul. 2017, G. Angulo UTRF66 (UTR); Dos Cruces, -6.579, -77.378, 1624 m alt., 27 Jul. 2017, G. Angulo UTRF53 (UTR); -6.424, -77.548, 1668 m alt., 27 Jul. 2017, G. Angulo UTRF58 (UTR); -6.425, -77.55, 1642 m alt., 11 Aug. 2017, G. Angulo UTRP19 (UTR); -6.425, -77.55, 1629 m alt., 11 Aug. 2017, G. Angulo UTRP20 (UTR); -6.424, -77.549, 1661 m alt., 11 Aug. 2017, G. Angulo UTRP21 (UTR); -6.425, -77.548, 1671 m alt., 11 Aug. 2017, G. Angulo UTRP22 (UTR); -6.424, -77.548, 1681 m alt., 11 Aug. 2017, G. Angulo UTRP23 (UTR); -6.423, -77.548, 1682 m alt., 11 Aug. 2017, G. Angulo UTRP24 (UTR); -6.422, -77.548, 1671 m alt., 11 Aug. 2017, G. Angulo UTRP25 (UTR); Escobar, -6.42, -77.549, 1666 m alt., 27 Jul. 2017, G. Angulo UTRF59 (UTR); -6.42, -77.549, 1674 m alt., 27 Jul. 2017, G. Angulo UTRF60 (UTR); Omia, El Tingo, -6.469, -77.376, 1431 m alt., 25 Jul. 2017, G. Angulo UTRF19 (UTR); -6.475, -77.381, 1349 m alt., 25 Jul. 2017, G. Angulo UTRF20 (UTR); La Primavera, -6.634, -77.231, 1283 m alt., 25 Jul. 2017, G. Angulo UTRF5 (UTR); -6.64, -77.224, 1362 m alt., 25 Jul. 2017, G. Angulo UTRF7 (UTR); -6.632, -77.222, 1205 m alt., 3 Aug. 2017, G. Angulo UTRP4 (UTR); -6.632, -77.222, 1209 m alt., 3 Aug. 2017, G. Angulo UTRP5 (UTR); -6.638, -77.225, 1280 m alt., 3 Aug. 2017, G. Angulo UTRP6 (UTR); -6.637, -77.225, 1275 m alt., 3 Aug. 2017, G. Angulo UTRP7 (UTR); -6.636, -77.227, 1255 m alt., 25 Jul. 2017, G. Angulo UTRP8 (UTR); -6.632, -77.225, 1238 m alt., 4 Aug. 2017, G. Angulo UTRP9 (UTR); Libano, -6.623, -77.235, 1174 m alt., 24 Jul. 2017, G. Angulo UTRF2 (UTR); -6.611, -77.237, 1330 m alt., 24 Jul. 2017, G. Angulo UTRF3 (UTR); -6.625, -77.242, 1235 m alt., 24 Jul. 2017, G. Angulo UTRF4 (UTR); -6.612, -77.237, 1307 m alt., 3 Aug. 2017, G. Angulo UTRP1 (UTR); -6.618, -77.234, 1242 m alt., 3 Aug. 2017, G. Angulo UTRP2 (UTR); -6.626, -77.247, 1284 m alt., 3 Aug. 2017, G. Angulo UTRP3 (UTR); -6.62, -77.235, 1226 m alt., 3 Aug. 2017, G. Angulo UTRP10 (UTR); -6.618, -77.237, 1236 m alt., 4 Aug. 2017, G. Angulo UTRP11 (UTR).
Notes.
Beauveria peruviensis is practically indistinguishable in morphology to other Beauveria species. The shape and size of the conidia and the colony color of B. peruviensis among other morphological features have been observed in B. bassiana, B. kipukae, B. pseudobassiana, and B. varroae (Rehner et al. 2011). The lack of diagnostic morphological features to distinguish Beauveria peruviensis was overcome by delimiting this species with DNA-based methodologies.
Discussion
Accurate species identification within the entomopathogenic fungi Beauveria is crucial for disease control and prevention (Liu et al. 2016). This genus has recently been circumscribed, and its taxonomy has been updated with new combinations and the description of new species based mainly on multi-locus phylogenies in the absence of diagnostic features that delimit species (Sanjuan et al. 2014, Shrestha et al. 2014, Kepler et al. 2017, Chen et al. 2017, 2018). In addition to phylogenies, other methodologies and data sets to delimit species are recommended to establish well-supported boundaries among species (Carstens et al. 2013) because most researchers simply recognize distinct clades in either single- or multi-locus trees as species (Stewart et al. 2014). In this regard, our study used phylogeny and five DNA-based methods to delimit species in Beauveria using three molecular makers. Although incongruence among some of these methods was observed in our analyses, a genetic distance (ABGD), a coalescence method (BPP), and the multilocus phylogeny strongly supported 20–28 different species, including the new species B. peruviensis from Peru.
The use of multi-locus sequence data is essential to establish robust species boundaries (Lumbsch and Levitt 2011), and our results for Beauveria showed well-supported clades, although it resulted in incongruence to the single locus phylogenies (Suppl. material 1: Figs S1–S3). This conflict can be a result of incomplete lineage sorting, horizontal gene transfer, gene duplication and loss, hybridization, or recombination (Degnan and Rosenberg 2009). This study cannot determine which of these scenarios are occurring in Beauveria; nevertheless, it serves as a baseline for investigating causes of gene tree discordance that can be identified by further analyses at the genomic level (Patterson et al. 2006, Liu et al. 2016). According to our multilocus phylogeny, 22 of the 26 molecularly confirmed species in Beauveria were recognized. Previous studies have delimited B. araneola, B. gryllotalpidicola, B. loeiensis, B. medogensis, and B. rudraprayagi as valid species on the basis of their phylogenies (Agrawal et al. 2014, Imoulan et al. 2016, Chen et al. 2017); however, our study did not include these sequences because they have abundant missing data, and thus, their status was not evaluated. These species would require further revision to be recognized as supported lineage within the genus Beauveria.
Regarding the genetic distance methods, the ABGD showed similar results when delimiting Beauveria species to those from the multilocus phylogeny. The additional putative species in ABGD is mainly due to the split of B. bassiana. This confirms that B. bassiana encompasses cryptic lineages as proposed initially by Rehner et al. (2011). Therefore, the original B. bassiana should be the clade that includes the specimen from the type locality, namely, Italy (Vuillemin 1912). Additionally, these results delimited B. majiangensis and B. asiatica as different lineages, although the multilocus analysis showed low support. B. majiangensis needs further analysis with additional and longer sequences to confirm its status because one or only a few individuals often fail to represent the species as a whole (Davis and Nixon 1992, Walsh 2000). On the other hand, the SPN method showed conflicting results among the Bloc, RPB1, and tef1 loci, leading to incorrect inferences. The number of species inferred by SPN greatly matched the number of Linnaean species in mitochondrial markers (e.g., COI) (Hart and Sunday 2007). Therefore, our nuclear markers due to indels might generate many reticulations that allow inadequate species delimitation in our data (Paradis 2018).
In the coalescence methods, although 6 species were not included in the BPP analysis due to the lack of their Bloc sequences, this method supports the conservative results obtained from the multilocus phylogeny. BPP supported the status of 16 species (posterior probabilities higher than 0.52), which are not high supportive, but these probabilities are not supportive at all when splitting or merging species in the BPP analysis (Suppl. material 1: Table S2). Zhang et al. (2011) found that the correct species model was inferred with a high posterior probability with only one or two loci when 5 or 10 sequences were sampled from each population or with 50 loci when only one sequence was sampled, and they also demonstrated that the migration rate might affect these results. This suggests that further analysis might need to increase the number of sequences per locus among different populations of species of Beauveria and assess their migration rate to obtain supportive delimitations. Moreover, the highly significant results obtained from the GMYC method for the tef1 and rpb1 loci partially support the ABGD and multilocus analyses. The additional number of putative species in the GMYC analyses, as occurred with the ABGD, is due to the presence of more than one lineage in B. amorpha, B. bassiana, B. diapheromeriphila, and B. pseudobassiana confirming cryptic diversity (Rehner et al. 2011). The performance in empirical studies of the ABGD and GMYC tends to under- and oversplit species, respectively (Luo et al. 2018). However, our results suggest that GMYC and ABGD are appropriate for determining cryptic diversity in Beauveria by splitting well-supported clades from the multi-locus phylogeny.
Regarding B. peruviensis, ABGD (Bloc), SPN (Bloc), GMYC, BPP, and the phylogenetic analyses support this species as a different lineage from B. bassiana and B. staphylinidicola. Additionally, the genetic divergence between B. peruviensis and these species is higher than the minimum threshold observed in species of Beauveria (Table 2). In our study, Beauveria peruviensis showed morphological indistinctiveness to other Beauveria species that produce globose/subglobose/ellipsoid conidia. Additionally, B. peruviensis conidia is also similar in size to other Beauveria, especially B. bassiana. Previously, Rehner et al. (2011) noted that B. bassiana is hardly distinguishable from other species of Beauveria. The lack of diagnostic morphological features to delimit species in Beauveria was overcome by the application of molecular methods in fungal taxonomy. The segregation of B. peruviensis from B. bassiana and B. staphylinidicola confirmed that phylogenetic diversity and DNA-species delimitation methods discover taxa within morphologically defined species (Goldstein et al. 2000, Liu et al. 2016). Ecologically, the segregation of B. peruviensis from B. bassiana and B. staphylinidicola is supported by the specificity of B. peruviensis to the coffee borer from Amazon and the well-supported lineage in the phylogenetic analysis that might indicate the presence of a barrier in gene flow in nature (Van Valen 1976, Liu et al. 2016).
Recently, polyphasic approaches have been used to reflect the natural classification of species within many important fungal genera (Aveskamp et al. 2010, Milic et al. 2012, Liu et al. 2016). These approaches frequently incorporate morphological and phylogenetic analyses and metabolomics, but few of them use genetic distance and coalescent methods (Liu et al. 2016). The use of polyphasic analysis, including DNA-based delimitation methods, allowed the establishment of boundaries among species of morphologically conserved genera such as Beauveria and thus provided support for the description of new taxa (e.g., B. peruviensis) or validated the taxonomic uncertain of others (e.g., B. majiangensis). Although more recent methods avoid arbitrary cut-offs (Knowles and Carstens 2007), our results demonstrate that the congruence among this method and other methods used in a polyphasic approach (e.g., genetic distance, coalescence methods) are more likely to prove reliably supported species boundaries (Carstens et al. 2013). Among the methods applied in this study, ABGD, GMYC, BPP, and multilocus phylogeny are crucial when establishing species boundaries in Beauveria.
Supplementary Material
Acknowledgements
This study was supported by the National Institute of Agrarian Innovation of Peru (Project number: 002-2016-INIA-PNIA-UPMSI/IE).
Citation
Bustamante DE, Oliva M, Leiva S, Mendoza JE, Bobadilla L, Angulo G, Calderon MS (2019) Phylogeny and species delimitations in the entomopathogenic genus Beauveria (Hypocreales, Ascomycota), including the description of B. peruviensis sp. nov. MycoKeys 58: 47–68. https://doi.org/10.3897/mycokeys.58.35764
Funding Statement
National Institute of Agrarian Innovation of Peru (Project number: 002-2016-INIA-PNIA-UPMSI/IE)
Contributor Information
Danilo E. Bustamante, Email: ddanilobm@gmail.com.
Martha S. Calderon, Email: martha.calderon@untrm.edu.pe.
Supplementary materials
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Danilo E. Bustamante, Manuel Oliva, Santos Leiva, Jani E. Mendoza, Leidy Bobadilla, Geysen Angulo, Martha S. Calderon
Tables S1, S2, Figures S1–S4
Data type: molecular data
Explanation note: Table S1. Results of the Generalized Mixed Yule-Coalescent (GMYC) analyses under the single threshold model. Table S2. Highest posterior probabilities of the three-gene Bayesian species delimitation analysis (BPP) by jointing species delimitation and species tree inference. Figure S1. Phylogenetic tree based on maximum likelihood inference of combined Bloc data. Figure S2. Phylogenetic tree based on maximum likelihood inference of combined RPB1 data. Figure S3. Phylogenetic tree based on maximum likelihood inference of combined Tef1 data. Figure S4. Bayesian inference ultrametric gene tree.
References
- Agrawal Y, Mual P, Shenoy BD. (2014) Multi-gene genealogies reveal cryptic species Beauveria rudraprayagi sp. nov. from India. Mycosphere 5: 719–736. 10.5943/mycosphere/5/6/3 [DOI] [Google Scholar]
- Aveskamp MM, De Gruyter J, Woudenberg JHC, Verkley GJM, Crous PW. (2010) Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Studies in Mycology 65: 1–60. 10.3114/sim.2010.65.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsamo G. (1835) Zwei neuen Arten Mucedineen, Botrytis bassiana und Mucor radicans, und uber die Entickelung der ersteren Art im Seidenwurme. Linneae 10: 609–618. [Google Scholar]
- Bissett J, Widden P. (1986) A new species of Beauveria from Scottish moorland soil. Canadian Journal of Botany 66: 361–362. 10.1139/b88-057 [DOI] [Google Scholar]
- Brankovics B, Van Dam P, Rep M, de Hoog S, Van der Lee TAJ, Waalwijk C, Van Diepeningen AD. (2018) Mitochondrial genomes reveal recombination in the presumed asexual Fusarium oxysporum species complex. BMC Genomics 201718: 735. 10.1186/s12864-017-4116-5 [DOI] [PMC free article] [PubMed]
- Carstens BC, Pelletier TA, Reid NM, Satler JD. (2013) How to fail at species delimitation. Molecular Ecology 22: 4369–4383. 10.1111/mec.12413 [DOI] [PubMed] [Google Scholar]
- Chen WH, Han YF, Liang ZQ, Jin DC. (2017) A new araneogenous fungus in the genus Beauveria from Guizhou, China. Phytotaxa 302: 57–64. 10.11646/phytotaxa.302.1.5 [DOI] [Google Scholar]
- Chen WH, Man L, Huang ZX, Yang GM, Han YF, Liang JD, Liang ZQ. (2018) Beauveria majiangensis, a new entomopathogenic fungus from Guizhou, China. Phytotaxa 333: 243–250. 10.11646/phytotaxa.333.2.8 [DOI] [Google Scholar]
- Clement M, Posada D, Crandall KA. (2000) TCS: a computer program to estimate gene genealogies. Molecular Ecology 9: 1657–1659. 10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Davis JI, Nixon KC. (1992) Populations, genetic variation, and the delimitation of phylogenetic species. Systematic Biology 41: 421–435. 10.1093/sysbio/41.4.421 [DOI] [Google Scholar]
- Degnan JH, Rosenberg NA. (2009) Gene tree discordance, phylogenetic inference, and the multispecies coalescent. Trends in Ecology and Evolution 24: 332–340. 10.1016/j.tree.2009.01.009 [DOI] [PubMed] [Google Scholar]
- De Hoog GS. (1972) The genera Beauveria, Isaria, Tritirachium, and Acrodontium gen. nov. Studies in Mycology 1: 1–41. [Google Scholar]
- De Hoog GS, Rao V. (1975) Some new hyphomycetes. Persoonia 8: 207–212. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria MR, Wraight SP. (2007) Mycoinsecticides and mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biological Control 43: 237–256. 10.1016/j.biocontrol.2007.08.001 [DOI] [Google Scholar]
- Gerónimo-Torres JDC, Torres-de-la-Cruz M, Cruz MP, De-la-Cruz-Pérez A, Ortiz-García CF, Cappello-García S. (2016) Caracterización de aislamientos nativos de Beauveria bassiana y su patogenicidad hacia Hypothenemus hampei en Tabasco, México. Revista Colombiana de Entomología. 42: 28–35. 10.25100/socolen.v42i1.6666 [DOI] [Google Scholar]
- Ghikas DV, Kouvelis VN, Typas MA. (2010) Phylogenetic and biogeographic implications inferred by mitochondrial intergenic region analyses and ITS1-5.8S-ITS2 of the entomopathogenic fungi Beauveria bassiana and B. brongniartii BMC Microbiology 10: 174. 10.1186/1471-2180-10-174 [DOI] [PMC free article] [PubMed]
- Goldstein PZ, Desalle R, Amato G, Vogler AP. (2000) Conservation genetics at the species boundary. Conservation Biology 14: 120–131. 10.1046/j.1523-1739.2000.98122.x [DOI] [Google Scholar]
- Hart MW, Sunday J. (2007) Things fall apart: biological species form unconnected parsimony networks. Biology Letters 3: 509–512. 10.1098/rsbl.2007.0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Li CR, Li ZG, Fan MZ, Li ZZ. (2002) Molecular identification of the teleomorph of Beauveria bassiana. Mycotaxon 81: 229–236. [Google Scholar]
- Imoulan A, Wu HJ, Lu WL, Li Y, Li BB, Yang RH, Wang WJ, Wang XL, Kirk PM, Yao YJ. (2016) Beauveria medogensis sp. nov., a new fungus of the entomopathogenic genus from China. Journal of Invertebrate Pathology. 139: 74–81. 10.1016/j.jip.2016.07.006 [DOI] [PubMed] [Google Scholar]
- Kepler RM, Luangsa-ard JJ, Hywel-Jones NL, Quandt CA, Sung GH, Rehner SA, Aime MC, Henkel TW, Sanjuan T, Zare R, Chen M, Li Z, Rossman AY, Spatafora JW, Shrestha B. (2017) A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8, 335–353. 10.5598/imafungus.2017.08.02.08 [DOI] [PMC free article] [PubMed]
- Knowles LL, Carstens BC. (2007) Delimiting species without monophyletic gene trees. Systematic Biology 56: 887–895. 10.1080/10635150701701091 [DOI] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29: 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- Leaché AD, Fujita MK. (2010) Bayesian species delimitation in west African forest geckos (Hemidactylus fasciatus). Proceedings of the Royal Society B 277: 3071–3077. 10.1098/rspb.2010.0662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Lin SM, Chen PC. (2015) Phylogeny, species diversity and biogeographic patterns of the genus Tricleocarpa (Galaxauraceae, Rhodophyta) from the Indo-Pacific region, including T. confertus sp. nov. from Taiwan. European Journal of Phycology 50: 439–456. 10.1080/09670262.2015.1076892 [DOI] [Google Scholar]
- Liu F, Wang M, Damm U, Crous PW, Cai L. (2016) Species boundaries in plant pathogenic fungi: a Colletotrichum case study. BMC Evolutionary Biology 16: 81. 10.1186/s12862-016-0649-5 [DOI] [PMC free article] [PubMed]
- Lumbsch HT, Leavitt SD. (2011) Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Diversity 50: 59–72. 10.1007/s13225-011-0123-z [DOI] [Google Scholar]
- Luo A, Ling C, Ho SYW, Zhu CD. (2018) Comparison of methods for molecular species 465 delimitation across a range of speciation scenarios. Systematic Biology 67: 830–846. 10.1093/sysbio/syy011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill J, Barrie FF, Buck WR, Demoulin V, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Marhold K, Prado J, Prud’homme Van Reine WF, Smith GF, Wiersema JH. (2012) International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). [Regnum vegetabile no. 154.]Königstein: Koeltz Scientific Books.
- Milic N, Kostidis S, Stavrou A, Gonou-Zagou Z, Kouvelis VN, Mikros E, Fokialakis N. (2012) A polyphasic approach (metabolomics, morphological and molecular analyses) in the systematics of Cladobotryum species in Greece. Planta Medica 78:1137. 10.1055/s-0032-1320657 [DOI]
- Millanes AM, Truong C, Westberg M, Diederich P, Wedin M. (2014) Host switching promotes diversity in host-specialized mycoparasitic fungi: uncoupled evolution in the Biatoropsis-Usnea system. Evolution 68: 1576–1593. 10.1111/evo.12374 [DOI] [PubMed] [Google Scholar]
- Paradis E. (2018) Analysis of haplotype networks: The randomized minimum spanning tree method. Methods in Ecology and Evolution 9: 1308–1317. 10.1111/2041-210X.12969 [DOI] [Google Scholar]
- Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. (2006) Genetic evidence for complex speciation of humans and chimpanzees. Nature 441: 1103–1108. 10.1038/nature04789 [DOI] [PubMed] [Google Scholar]
- Pons J, Barraclough T, Gomez-Zurita J, Cardoso A, Duran D, Hazell S, Kamoun S, Sumlin W, Vogler A. (2006) Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Systematic Biology 55: 595–609. 10.1080/10635150600852011 [DOI] [PubMed] [Google Scholar]
- Puillandre N, Lambert A, Brouillet S, Achaz G. (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21: 1864–1877. 10.1111/j.1365-294X.2011.05239.x [DOI] [PubMed] [Google Scholar]
- Quaedvlieg W, Binder M, Groenewald JZ, Summerell BA, Carnegie AJ, Burgess TI. (2014) Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 33: 1–40. 10.3767/003158514X681981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ. (2014) Tracer v1.6. http://beast.bio.ed.ac.uk/Tracer
- Rannala B, Yang Z. (2003) Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164: 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redaelli P, Visocchi V. (1940) Agostino Bassi precursor of comparative mycopathology Mycopathologia 2: 37–42. 10.1007/BF00450241 [DOI]
- Rehner SA, Buckley EP. (2005) A Beauveria phylogeny inferred from ITS and EF1-a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. 10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- Rehner SA, Posada F, Buckley EP, Infante F, Castillo A, Vega FE. (2006) Phylogenetic origins of African and Neotropical Beauveria bassiana sl pathogens of the coffee berry borer, Hypothenemus hampei. Journal of Invertebrate Pathology 93: 11–21. 10.1016/j.jip.2006.04.005 [DOI] [PubMed] [Google Scholar]
- Rehner SA, Minnis AM, Sung GH, Luangsa-ard JJ, Devotto L, Humber RA. (2011) Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia 103: 1055–1073. 10.3852/10-302 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres D, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Evans HC. (1982) Two new Beauveria spp. from South America. Journal of Invertebrate Pathology 39: 93–97. 10.1016/0022-2011(82)90162-8 [DOI] [Google Scholar]
- Sanjuan T, Tabima J, Restrepo S, Læssøe T, Spatafora JW, Franco-Molano AE. (2014) Entomopathogens of Amazonian stick insects and locusts are members of the Beauveria species complex (Cordycepssensu stricto). Mycologia 106: 260–275. 10.3852/13-020 [DOI] [PubMed] [Google Scholar]
- Shrestha B, Hyun MW, Oh J, Han J-G, Lee TH, Cho JY, Kang H, Kim SH, Sung GH. (2014) Molecular evidence of a teleomorph-anamorph connection between Cordyceps scarabaeicola and Beauveria sungii and its implication for the systematics of Cordyceps sensu stricto. Mycoscience 55: 231–239. 10.1016/j.myc.2013.09.004 [DOI] [Google Scholar]
- Shimazu M, Mitsuhashi W, Hashimoto H. (1988) Cordyceps brongniartii sp. nov., the teleomorph of Beauveria brongniartii. Transactions of the Mycological Society of Japan 29: 323–330. [Google Scholar]
- Silvestro D, Michalak I. (2012) RaxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12: 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML Version 8: A tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JE, Timmer LW, Lawrence CB, Pryor BM, Peever TL. (2014) Discord between morphological and phylogenetic species boundaries: incomplete lineage sorting and recombination results in fuzzy species boundaries in an asexual fungal pathogen. BMC Evolutionary Biology14: 38. 10.1186/1471-2148-14-38 [DOI] [PMC free article] [PubMed]
- Sung GH, Hywel-Jones NL, Sung JM, Luangsa-ard JJ, Shrestha B, Spatafora JW. (2007) Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Studies in Mycology 57: 5–59. 10.3114/sim.2007.57.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. (2010) Bayesian species delimitation using multilocus sequence data Proceedings of the National Academy of Sciences 107: 9264–9269. 10.1073/pnas.0913022107 [DOI] [PMC free article] [PubMed]
- Van Valen L. (1976) Ecological species, multispecies, and oaks. Taxon 25: 233–239. 10.2307/1219444 [DOI] [Google Scholar]
- Vuillemin P. (1912) Beauveria, nouveau genre de Verticilliacées. Bulletin de la Société Botanique de France 29: 34–40. 10.1080/00378941.1912.10832379 [DOI] [Google Scholar]
- Walsh PD. (2000) Sample size for the diagnosis of conservation units. Conservation Biology 14: 1533–1537. 10.1046/j.1523-1739.2000.98149.x [DOI] [Google Scholar]
- Zhang DX, Zhu T, Yang Z. (2011) Evaluation of a Bayesian Coalescent Method of Species Delimitation. Systematic Biology 60: 747–761. 10.1093/sysbio/syr071 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Danilo E. Bustamante, Manuel Oliva, Santos Leiva, Jani E. Mendoza, Leidy Bobadilla, Geysen Angulo, Martha S. Calderon
Tables S1, S2, Figures S1–S4
Data type: molecular data
Explanation note: Table S1. Results of the Generalized Mixed Yule-Coalescent (GMYC) analyses under the single threshold model. Table S2. Highest posterior probabilities of the three-gene Bayesian species delimitation analysis (BPP) by jointing species delimitation and species tree inference. Figure S1. Phylogenetic tree based on maximum likelihood inference of combined Bloc data. Figure S2. Phylogenetic tree based on maximum likelihood inference of combined RPB1 data. Figure S3. Phylogenetic tree based on maximum likelihood inference of combined Tef1 data. Figure S4. Bayesian inference ultrametric gene tree.