ABSTRACT
Rap1 belongs to the Ras family of small GTPases, which are involved in a multitude of cellular signal transduction pathways and have extensively been linked to cancer biogenesis and metastasis. The small GTPase is activated in response to various extracellular and intracellular cues. Rap1 has conserved functions in Dictyostelium discoideum amoeba and mammalian cells, which are important for cell polarity, substrate and cell-cell adhesion and other processes that involve the regulation of cytoskeletal dynamics. Moreover, our recent study has shown that Rap1 is required for the formation of the replication-permissive vacuole of an intracellular bacterial pathogen. Here we review the function and regulation of Rap1 in these distinct processes, and we discuss the underlying signal transduction pathways.
KEYWORDS: adhesion, cell migration, chemotaxis, Dictyostelium, host-pathogen interactions, Legionella, macrophage, motility, pathogen vacuole, small GTPase, type IV secretion
A conserved role for Rap1 in essential cell processes
Small GTPases represent molecular switches that control essential cellular processes such as signal transduction, cell adhesion, chemotaxis and motility, cell growth and division, membrane dynamics and vesicle trafficking, as well as interactions with pathogens. To this end, the small GTPases cycle between an inactive GDP-bound and an active GTP-bound state. The switch between these states is tightly controlled by cognate guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs). GEFs facilitate the release of the bound nucleotide and allow the more abundant GTP to rebind, whereas GAPs stimulate the low intrinsic GTPase activity to stimulate the hydrolysis of the bound GTP to complete the cycle.1
Rap1 belongs to the Ras superfamily of small GTPases.2 The switch between its GDP-bound and GTP-bound form is controlled by several specific GEFs and GAPs.3 Rap1 is conserved in mammalian cells as well as in the haploid social soil amoeba Dictyostelium discoideum.4,5 In mammalian cells Rap1 plays a pivotal role for cell growth, proliferation and survival.5,6 In Dictyostelium antisense rapA RNA induction leads to a gradually decreased growth rate and cell viability, and in particular, Rap1-depleted amoeba show a reduced viability in response to osmotic stress.7,8 Rap1 is likely essential for the amoeba, as attempts at generating the corresponding null mutant strain were unsuccessful. Despite extensive studies in various biologic systems, the Rap1 signaling pathways regulating these important processes are still not completely identified and characterized.9 Dictyostelium is an excellent model for studying Rap1-dependent processes, because of its genetic tractability, as well as the evolutionary conservation of the Rap protein and the downstream signaling pathways that govern cytoskeletal rearrangements.4,5 Here we highlight recent findings in Dictyostelium and mammalian cells that implicate Rap1 in the regulation of cytokinesis, cell adhesion, chemotaxis, and pathogen vacuole formation.
Rap1 governs cell adhesion and phagocytosis
Rap1 has been intricately linked to pathways regulating cell adhesion. In mammalian cells activation of Rap1 is triggered by adhesion molecules, cytokines, growth factors like tumor necrosis factor α (TNFα) and interferon γ (IFNγ), or second messengers that are coupled to GEFs.5,10 During phagocytosis, Rap1 (and also the small GTPase Ras) is activated by diacylglycerol, which recruits the Rap1/Ras GEF RasGRP3.11 Rap1 controls cell adhesion dynamics and phagocytosis, especially by mediating the functions of integrins and cadherins.12,13 In this pathway Rap1 acts upstream of the integrin-associated factor talin, and controls the recruitment of the cytoskeletal protein to sites of particle binding and phagocytosis.14 The activation of Rap1 indirectly activates talin, as active Rap at the cell membrane recruits the scaffold protein RIAM (Rap1-GTP-interacting adaptor molecule), which subsequently binds talin and stimulates integrin activation and formation of adhesion complexes.15,16
Rap1 is also essential for cell adhesion in Dictyostelium (Fig. 1).8,17 The GEF GbpD is primarily responsible for activation of Rap1 during substrate attachment.18,19 Active Rap1 mediates cell adhesion via the Ser/Thr kinase Phg2 and talin. Interestingly, in addition to indirectly regulating talin function, Dictyostelium Rap1 also directly binds and activates the cytoskeletal protein. Recent data from our laboratory revealed that the direct interaction of active Dictyostelium Rap1 with the RA domain of talin provides additional strength, which is essential for processes demanding high adhesive forces, such as morphogenesis.17 A direct interaction between Rap1 and talin has also been reported in mice;20 however, the biologic significance of this interaction is not clear.
Figure 1.
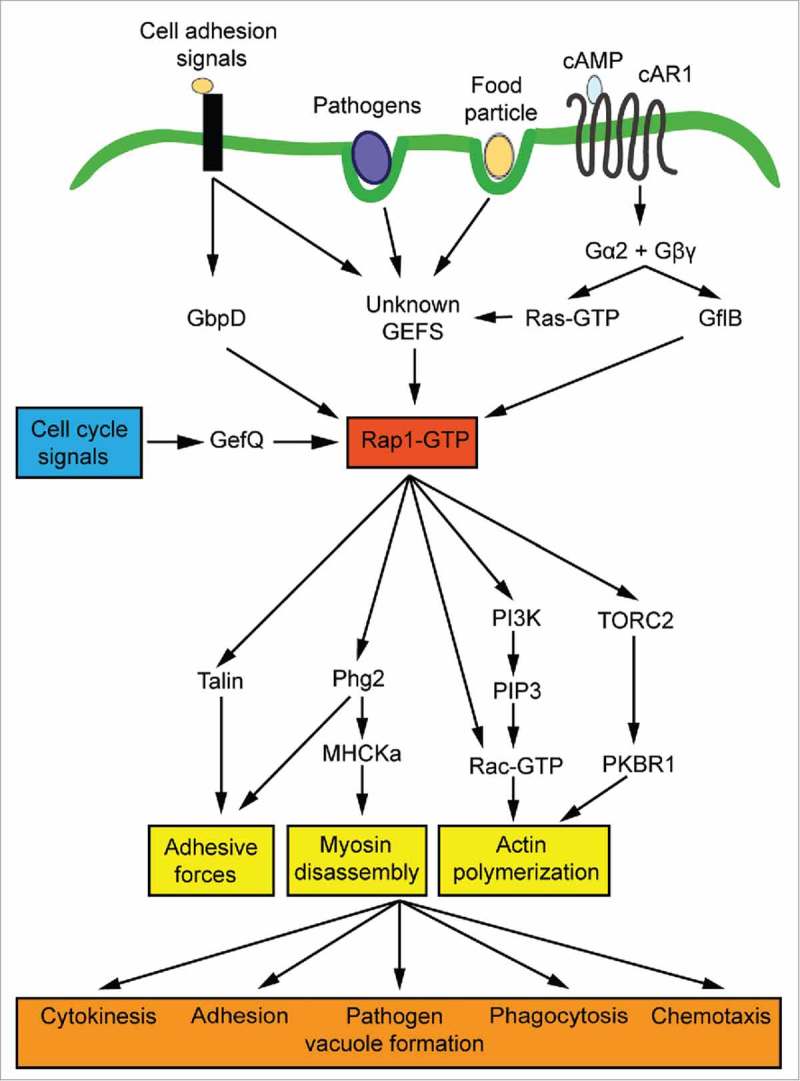
Role of the small GTPase Rap1 in signal transduction, cell dynamics and bacterial infection. Rap1 is activated in response to various extracellular and intracellular stimuli. During substrate attachment and cytokinesis, Rap1 is primarily activated by the GEFs GbpD and GefQ, respectively. In the course of chemotaxis, the Gα2-stimulated RapGEF GflB determines the balance between Rap1 and Ras activation at the leading edge of Dictyostelium cells. Other GEFs transducing the signals to Rap1 remain to be identified. Activated Rap1 is a major regulator of cytoskeletal dynamics and stimulates cellular adhesion, actin filament formation and myosin disassembly via the indicated pathways. Together, these cytoskeleton rearrangements are key for cytokinesis, adhesion, pathogen vacuole formation, phagocytosis and chemotaxis.
Rap1 coordinates cytoskeletal rearrangements during cytokinesis and chemotaxis
In Dictyostelium Rap1 functions as a general regulator of cytoskeletal dynamics (Fig. 1).4,7 To regulate the cytoskeletal dynamics during cell division, Rap1 is uniformly activated in the cell cortex during the early stages of cytokinesis.8 In contrast, at the final stages of the process, the small GTPase is restricted to the cell poles. GefQ appears to be important for regulating Rap1 activation during cytokinesis.8 Furthermore, a recent study suggests that also RapGAP9 is crucial for Rap-mediated cytokinesis progression.21 Decreased or increased Rap1 activation impairs the growth rate and cytoskeletal dynamics. Thus, Dictyostelium Rap1 drives cytokinesis progression, likely by coordinating the major cytoskeletal components, microtubules, actin and myosin II.8 Similar to Dictyostelium, levels of Rap1 activation are tightly controlled during cell division in Hela cells.22 Furthermore, hyper-activation of Rap1 in various human cell lines and Drosophila results in severe cytokinesis defects.23,24 Together, this strongly suggests a conserved and essential role for Rap in the regulation of cytokinesis.
Rap1 and Ras also regulate the balance between F-actin and myosin dynamics during chemotaxis of Dictyostelium amoeba and mammalian leukocytes (Fig. 1).25,26 In response to the chemoattractant cAMP, both Rap1 and Ras are rapidly activated at the leading edge of migrating Dictyostelium.4,27,28 The Gα2-stimulated RapGEF, GflB, is an important regulator of the balance between Rap1 and Ras activation during chemotaxis.29 In addition, Rap1 activity at the leading edge is regulated by an unknown GEF that acts downstream of active Ras.30 Rap1 and Ras can activate the Rac, PI3K and TORC2 pathways, which subsequently results in actin polymerization and pseudopod extension from the front of the cell28,31-40 (Fig. 1). Simultaneously, Rap1 inhibits myosin assembly at the leading edge through activation of its effector Phg2, while low levels of active Rap1 at the side and back of the cell allow myosin filament formation.25,26 Taken together, the spatial and temporal Rap1- and Ras-mediated control of actin and myosin rearrangement is essential for proper chemotaxis.41
Rap1 localizes to the Legionella pathogen vacuole and controls infection
Pathogenic bacteria intimately interact with eukaryotic host cells to subvert immune functions and create a replication-permissive niche. Legionella pneumophila is an environmental Gram-negative bacterium, which can cause a severe pneumonia termed “Legionnaires' disease”.42 The facultative intracellular pathogen uses a seemingly conserved mechanism to replicate in environmental protozoa or immune system phagocytes within a unique membrane-bound compartment, the Legionella-containing-vacuole (LCV).43 Host cells of L. pneumophila include free-living amoeba such as Acanthamoeba or Dictyostelium spp. as well as mammalian macrophages.
LCV formation requires the bacterial Icm/Dot (intracellular multiplication/defective organelle trafficking) type IV secretion system (T4SS), which translocates approximately 300 (!) different “effector proteins” into host cells, where they modulate specific components of the machineries catalyzing transcription, translation, signal transduction or vesicle trafficking.44-46 Some of these effectors target phosphoinositide lipids,47 the retromer complex,48 or small GTPases of the Arf,49 Rab,50,51 or Ran family.52,53 LCVs avoid fusion with lysosomes, but extensively communicate with the host endosomal, secretory and retrograde trafficking pathways, as well as with the endoplasmic reticulum (ER).44,54,55
Intact LCVs can be isolated and purified by a 2-step procedure, including an immuno-affinity purification step and density gradient separation.56,57 To this end, the distinct and specific LCV localization of the Icm/Dot-secreted effector protein SidC is exploited.58-60 Upon treatment with an anti-SidC antibody and a secondary antibody coupled to magnetic microbeads, LCVs are retained in a magnetic field, washed, eluted and further enriched by Histodenz density gradient centrifugation. Using this protocol, pathogen vacuoles harboring L. pneumophila have been isolated from Dictyostelium,61 murine RAW 264.7 macrophages,62 and bone marrow-derived macrophages (BMM) of infection-permissive A/J mice.63 Proteomics analysis of these LCVs revealed more than 1150 host cell factors,64 including 13 Rab GTPases, Ran and Rap1.61,62 The localization to the LCV membrane and impact on intracellular growth of L. pneumophila of some of the Rab GTPases, as well as of Ran and Rap1, was validated by fluorescence microscopy and RNA interference, respectively.52,62,65
The presence of active Rap1 on LCVs was recently found to correlate with intracellular replication of L. pneumophila65 (Fig. 1). In a comparative proteomics approach the proteome of isolated pathogen vacuoles from Dictyostelium amoeba or RAW 264.7 macrophages infected with either the parental L. pneumophila strain Lp02 or the “pentuple” mutant (“Δpentuple”) was determined. The Δpentuple strain, which lacks 5 gene clusters comprising ca. 13% of the genome and at least 31% of the effector proteins, is defective for intracellular replication in Acanthamoeba and Dictyostelium, but grows in BMM derived from the A/J mouse strain.66 In the comparative proteomics study, Rap1 was identified on Dictyostelium LCVs containing the parental strain Lp02 but not the Δpentuple mutant and on macrophage LCVs containing either strain.65 The localization pattern of active Rap1 was validated by fluorescence microscopy and quantitative imaging flow cytometry using Dictyostelium strains producing GFP-Rap1,8,67 or the Rap1-GTP probe RalGDSRBD-GFP.26,67 In these experiments, GTP-bound Rap1 preferentially localized to LCVs harbouring strain Lp02 rather than to vacuoles containing Δpentuple mutant bacteria. Therefore, the accumulation of active Rap1 correlates with the formation of a replication-permissive pathogen vacuole. In agreement with this notion, the depletion of Rap1 by RNA interference reduced intracellular growth of L. pneumophila. In summary, Rap1 was found to represent a novel LCV host component that localizes preferentially to replication-permissive pathogen compartments and is implicated in intracellular bacterial replication (Fig. 1).65
Strikingly, the LCV localization pattern of a downstream target of Rap1, integrin-associated talin, mirrored that of the small GTPase. In Dictyostelium talin was exclusively identified in the Lp02 LCV proteome and not in the Δpentuple LCV proteome, whereas in macrophages talin was present in both proteomes. Taken together, the accumulation of Rap1 and talin on LCVs correlates with intracellular replication of L. pneumophila, and thus, the 2 host factors likely interact with each other not only during phagocytosis, but also in the context of bacterial infection and pathogen vacuole formation.
Concluding remarks
Rap1 is conserved in Dictyostelium amoeba as well as in mammalian cells, and its activation is regulated by various extracellular and intracellular stimuli. Rap1 functions mainly in controlling cytoskeleton rearrangements; it stimulates cellular adhesion, actin filament formation, and myosin disassembly. Rap1-mediated pathways are crucial for pleiotropic cellular processes, including cell adhesion, chemotaxis and motility, cell growth and division, membrane dynamics and vesicle trafficking. Moreover, our recent study has shown that the small GTPase is also required for pathogen vacuole formation and intracellular replication of a bacterial pathogen in Dictyostelium and macrophages.
Abbreviations
- ER
endoplasmic reticulum
- Icm/Dot
intracellular replication/defective organelle trafficking
- LCV
Legionella-containing vacuole
- T4SS
type IV secretion system
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Work in the laboratory of H.H. was supported by the Institute of Medical Microbiology, University of Zürich, the Swiss National Science Foundation (SNF; 31003A_153200), the German Research Foundation (DFG; HI 1511/3–1, SPP 1580), and the Bundesministerium für Bildung und Forschung (BMBF; 031A410A; Infect-ERA project EUGENPATH). The work of A.K. is funded by a NWO-VIDI grant.
References
- [1].Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science 2001; 294:1299-304; PMID:11701921; https://doi.org/ 10.1126/science.1062023 [DOI] [PubMed] [Google Scholar]
- [2].Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE 2004; 2004:RE13; PMID:15367757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gloerich M, Bos JL. Regulating Rap small G-proteins in time and space. Trends Cell Biol 2011; 21:615-23; PMID:21820312; https://doi.org/ 10.1016/j.tcb.2011.07.001 [DOI] [PubMed] [Google Scholar]
- [4].Kortholt A, van Haastert PJ. Highlighting the role of Ras and Rap during Dictyostelium chemotaxis. Cell Signal 2008; 20:1415-22; PMID:18385017; https://doi.org/ 10.1016/j.cellsig.2008.02.006 [DOI] [PubMed] [Google Scholar]
- [5].Caron E. Cellular functions of the Rap1 GTP-binding protein: a pattern emerges. J Cell Sci 2003; 116:435-40; PMID:12508104; https://doi.org/ 10.1242/jcs.00238 [DOI] [PubMed] [Google Scholar]
- [6].Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res 2002; 12:9-18; PMID:11942415; https://doi.org/ 10.1038/sj.cr.7290105 [DOI] [PubMed] [Google Scholar]
- [7].Kang R, Kae H, Ip H, Spiegelman GB, Weeks G. Evidence for a role for the Dictyostelium Rap1 in cell viability and the response to osmotic stress. J Cell Sci 2002; 115:3675-82; PMID:12186953; https://doi.org/ 10.1242/jcs.00039 [DOI] [PubMed] [Google Scholar]
- [8].Plak K, Keizer-Gunnink I, van Haastert PJ, Kortholt A. Rap1-dependent pathways coordinate cytokinesis in Dictyostelium. Mol Biol Cell 2014; 25:4195-204; PMID:25298405; https://doi.org/ 10.1091/mbc.E14-08-1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Frische EW, Zwartkruis FJ. Rap1, a mercenary among the Ras-like GTPases. Dev Biol 2010; 340:1-9; PMID:20060392; https://doi.org/ 10.1016/j.ydbio.2009.12.043 [DOI] [PubMed] [Google Scholar]
- [10].Alsayed Y, Uddin S, Ahmad S, Majchrzak B, Druker BJ, Fish EN, Platanias LC. IFN-gamma activates the C3G/Rap1 signaling pathway. J Immunol 2000; 164:1800-6; PMID:10657627; https://doi.org/ 10.4049/jimmunol.164.4.1800 [DOI] [PubMed] [Google Scholar]
- [11].Botelho RJ, Harrison RE, Stone JC, Hancock JF, Philips MR, Jongstra-Bilen J, Mason D, Plumb J, Gold MR, Grinstein S. Localized diacylglycerol-dependent stimulation of Ras and Rap1 during phagocytosis. J Biol Chem 2009; 284:28522-32; PMID:19700408; https://doi.org/ 10.1074/jbc.M109.009514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Retta SF, Balzac F, Avolio M. Rap1: a turnabout for the crosstalk between cadherins and integrins. Eur J Cell Biol 2006; 85:283-93; PMID:16546572; https://doi.org/ 10.1016/j.ejcb.2005.09.007 [DOI] [PubMed] [Google Scholar]
- [13].Chung J, Serezani CH, Huang SK, Stern JN, Keskin DB, Jagirdar R, Brock TG, Aronoff DM, Peters-Golden M. Rap1 activation is required for Fc gamma receptor-dependent phagocytosis. J Immunol 2008; 181:5501-9; PMID:18832707; https://doi.org/ 10.4049/jimmunol.181.8.5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lim J, Dupuy AG, Critchley DR, Caron E. Rap1 controls activation of the alpha(M)beta(2) integrin in a talin-dependent manner. J Cell Biochem 2010; 111:999-1009; PMID:20665668; https://doi.org/ 10.1002/jcb.22788 [DOI] [PubMed] [Google Scholar]
- [15].Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to Ras GTPase membrane-targeting sequences. J Biol Chem 2009; 284:5119-27; PMID:19098287; https://doi.org/ 10.1074/jbc.M807117200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wynne JP, Wu J, Su W, Mor A, Patsoukis N, Boussiotis VA, Hubbard SR, Philips MR. Rap1-interacting adapter molecule (RIAM) associates with the plasma membrane via a proximity detector. J Cell Biol 2012; 199:317-30; PMID:23045549; https://doi.org/ 10.1083/jcb.201201157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Plak K, Pots H, Van Haastert PJ, Kortholt A. Direct Interaction between TalinB and Rap1 is necessary for adhesion of Dictyostelium cells. BMC Cell Biol 2016; 17:1; PMID:26744136; https://doi.org/ 10.1186/s12860-015-0078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bosgraaf L, Waijer A, Engel R, Visser AJ, Wessels D, Soll D, van Haastert PJ. RasGEF-containing proteins GbpC and GbpD have differential effects on cell polarity and chemotaxis in Dictyostelium. J Cell Sci 2005; 118:1899-910; PMID:15827084; https://doi.org/ 10.1242/jcs.02317 [DOI] [PubMed] [Google Scholar]
- [19].Kortholt A, Rehmann H, Kae H, Bosgraaf L, Keizer-Gunnink I, Weeks G, Wittinghofer A, Van Haastert PJ. Characterization of the GbpD-activated Rap1 pathway regulating adhesion and cell polarity in Dictyostelium discoideum. J Biol Chem 2006; 281:23367-76; PMID:16769729; https://doi.org/ 10.1074/jbc.M600804200 [DOI] [PubMed] [Google Scholar]
- [20].Goult BT, Bouaouina M, Elliott PR, Bate N, Patel B, Gingras AR, Grossmann JG, Roberts GC, Calderwood DA, Critchley DR, et al.. Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. EMBO J 2010; 29:1069-80; PMID:20150896; https://doi.org/ 10.1038/emboj.2010.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mun H, Lee MR, Jeon TJ. RapGAP9 regulation of the morphogenesis and development in Dictyostelium. Biochem Biophys Res Commun 2014; 446:428-33; PMID:24513283; https://doi.org/ 10.1016/j.bbrc.2014.01.196 [DOI] [PubMed] [Google Scholar]
- [22].Dao VT, Dupuy AG, Gavet O, Caron E, de Gunzburg J. Dynamic changes in Rap1 activity are required for cell retraction and spreading during mitosis. J Cell Sci 2009; 122:2996-3004; PMID:19638416; https://doi.org/ 10.1242/jcs.041301 [DOI] [PubMed] [Google Scholar]
- [23].Carmena A, Makarova A, Speicher S. The Rap1-Rgl-Ral signaling network regulates neuroblast cortical polarity and spindle orientation. J Cell Biol 2011; 195:553-62; PMID:22084305; https://doi.org/ 10.1083/jcb.201108112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lancaster OM, Le Berre M, Dimitracopoulos A, Bonazzi D, Zlotek-Zlotkiewicz E, Picone R, Duke T, Piel M, Baum B. Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev Cell 2013; 25:270-83; PMID:23623611; https://doi.org/ 10.1016/j.devcel.2013.03.014 [DOI] [PubMed] [Google Scholar]
- [25].Jeon TJ, Lee DJ, Lee S, Weeks G, Firtel RA. Regulation of Rap1 activity by RapGAP1 controls cell adhesion at the front of chemotaxing cells. J Cell Biol 2007; 179:833-43; PMID:18039932; https://doi.org/ 10.1083/jcb.200705068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jeon TJ, Lee DJ, Merlot S, Weeks G, Firtel RA. Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J Cell Biol 2007; 176:1021-33; PMID:17371831; https://doi.org/ 10.1083/jcb.200607072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kae H, Lim CJ, Spiegelman GB, Weeks G. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep 2004; 5:602-6; PMID:15143344; https://doi.org/ 10.1038/sj.embor.7400151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sasaki AT, Firtel RA. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur J Cell Biol 2006; 85:873-95; PMID:16740339; https://doi.org/ 10.1016/j.ejcb.2006.04.007 [DOI] [PubMed] [Google Scholar]
- [29].Liu Y, Lacal J, Veltman DM, Fusetti F, van Haastert PJ, Firtel RA, Kortholt A. A Gα-stimulated RapGEF is a receptor-proximal regulator of Dictyostelium chemotaxis. Dev Cell 2016; 37:458-72; PMID:27237792; https://doi.org/ 10.1016/j.devcel.2016.05.001 [DOI] [PubMed] [Google Scholar]
- [30].Bolourani P, Spiegelman GB, Weeks G. Rap1 activation in response to cAMP occurs downstream of Ras activation during Dictyostelium aggregation. J Biol Chem 2008; 283:10232-40; PMID:18180289; https://doi.org/ 10.1074/jbc.M707459200 [DOI] [PubMed] [Google Scholar]
- [31].Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 2002; 109:611-23; PMID:12062104; https://doi.org/ 10.1016/S0092-8674(02)00755-9 [DOI] [PubMed] [Google Scholar]
- [32].Arthur WT, Quilliam LA, Cooper JA. Rap1 promotes cell spreading by localizing Rac guanine nucleotide exchange factors. J Cell Biol 2004; 167:111-22; PMID:15479739; https://doi.org/ 10.1083/jcb.200404068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol 2004; 167:505-18; PMID:15534002; https://doi.org/ 10.1083/jcb.200406177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bolourani P, Spiegelman GB, Weeks G. Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol Biol Cell 2006; 17:4543-50; PMID:16885420; https://doi.org/ 10.1091/mbc.E05-11-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Takeda K, Sasaki AT, Ha H, Seung HA, Firtel RA. Role of phosphatidylinositol 3-kinases in chemotaxis in Dictyostelium. J Biol Chem 2007; 282:11874-84; PMID:17331950; https://doi.org/ 10.1074/jbc.M610984200 [DOI] [PubMed] [Google Scholar]
- [36].Charest PG, Shen Z, Lakoduk A, Sasaki AT, Briggs SP, Firtel RA. A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev Cell 2010; 18:737-49; PMID:20493808; https://doi.org/ 10.1016/j.devcel.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kortholt A, Kataria R, Keizer-Gunnink I, Van Egmond WN, Khanna A, Van Haastert PJ. Dictyostelium chemotaxis: essential Ras activation and accessory signalling pathways for amplification. EMBO Rep 2011; 12:1273-9; PMID:22081140; https://doi.org/ 10.1038/embor.2011.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mun H, Jeon TJ. Regulation of actin cytoskeleton by Rap1 binding to RacGEF1. Mol Cells 2012; 34:71-6; PMID:22644079; https://doi.org/ 10.1007/s10059-012-0097-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Plak K, Veltman D, Fusetti F, Beeksma J, Rivero F, Van Haastert PJ, Kortholt A. GxcC connects Rap and Rac signaling during Dictyostelium development. BMC Cell Biol 2013; 14:6; PMID:23363311; https://doi.org/ 10.1186/1471-2121-14-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Khanna A, Lotfi P, Chavan AJ, Montaño NM, Bolourani P, Weeks G, Shen Z, Briggs SP, Pots H, Van Haastert PJ, et al.. The small GTPases Ras and Rap1 bind to and control TORC2 activity. Sci Rep 2016; 6:25823; PMID:27172998; https://doi.org/ 10.1038/srep25823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Artemenko Y, Lampert TJ, Devreotes PN. Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell Mol Life Sci 2014; 71:3711-47; PMID:24846395; https://doi.org/ 10.1007/s00018-014-1638-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Newton HJ, Ang DK, van Driel IR, Hartland EL. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 2010; 23:274-98; PMID:20375353; https://doi.org/ 10.1128/CMR.00052-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hoffmann C, Harrison CF, Hilbi H. The natural alternative: protozoa as cellular models for Legionella infection. Cell Microbiol 2014; 16:15-26; PMID:24168696; https://doi.org/ 10.1111/cmi.12235 [DOI] [PubMed] [Google Scholar]
- [44].Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 2009; 7:13-24; PMID:19011659; https://doi.org/ 10.1038/nrmicro1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Ann Rev Cell Dev Biol 2010; 26:261-83; https://doi.org/ 10.1146/annurev-cellbio-100109-104034 [DOI] [PubMed] [Google Scholar]
- [46].Finsel I, Hilbi H. Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol 2015; 17:935-50; PMID:25903720; https://doi.org/ 10.1111/cmi.12450 [DOI] [PubMed] [Google Scholar]
- [47].Haneburger I, Hilbi H. Phosphoinositide lipids and the Legionella pathogen vacuole. Curr Top Microbiol Immunol 2013; 376:155-73; PMID:23918172 [DOI] [PubMed] [Google Scholar]
- [48].Finsel I, Ragaz C, Hoffmann C, Harrison CF, Weber S, van Rahden VA, Johannes L, Hilbi H. The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe 2013; 14:38-50; PMID:23870312; https://doi.org/ 10.1016/j.chom.2013.06.001 [DOI] [PubMed] [Google Scholar]
- [49].Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 2002; 295:679-82; PMID:11809974; https://doi.org/ 10.1126/science.1067025 [DOI] [PubMed] [Google Scholar]
- [50].Itzen A, Goody RS. Covalent coercion by Legionella pneumophila. Cell Host Microbe 2011; 10:89-91; PMID:21843863; https://doi.org/ 10.1016/j.chom.2011.08.002 [DOI] [PubMed] [Google Scholar]
- [51].Sherwood RK, Roy CR. A Rab-centric perspective of bacterial pathogen-occupied vacuoles. Cell Host Microbe 2013; 14:256-68; PMID:24034612; https://doi.org/ 10.1016/j.chom.2013.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rothmeier E, Pfaffinger G, Hoffmann C, Harrison CF, Grabmayr H, Repnik U, Hannemann M, Wölke S, Bausch A, Griffiths G, et al.. Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. PLoS Pathog 2013; 9:e1003598; PMID:24068924; https://doi.org/ 10.1371/journal.ppat.1003598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Simon S, Wagner MA, Rothmeier E, Müller-Taubenberger A, Hilbi H. Icm/Dot-dependent inhibition of phagocyte migration by Legionella is antagonized by a translocated Ran GTPase activator. Cell Microbiol 2014; 16:977-92; PMID:24397557 [DOI] [PubMed] [Google Scholar]
- [54].Hilbi H, Hoffmann C, Harrison CF. Legionella spp. outdoors: colonization, communication and persistence. Environ Microbiol Rep 2011; 3:286-96; PMID:23761274; https://doi.org/ 10.1111/j.1758-2229.2011.00247.x [DOI] [PubMed] [Google Scholar]
- [55].Personnic N, Bärlocher K, Finsel I, Hilbi H. Subversion of retrograde trafficking by translocated pathogen effectors. Trends Microbiol 2016; 24:450-62; PMID:26924068; https://doi.org/ 10.1016/j.tim.2016.02.003 [DOI] [PubMed] [Google Scholar]
- [56].Urwyler S, Finsel I, Ragaz C, Hilbi H. Isolation of Legionella-containing vacuoles by immuno-magnetic separation. Curr Protoc Cell Biol 2010; Chapter 3: Unit 3.34; PMID:20235103 [DOI] [PubMed] [Google Scholar]
- [57].Hoffmann C, Finsel I, Hilbi H. Pathogen vacuole purification from Legionella-infected amoeba and macrophages. Methods Mol Biol 2013; 954:309-21; PMID:23150404; https://doi.org/ 10.1007/978-1-62703-161-5_18 [DOI] [PubMed] [Google Scholar]
- [58].Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A 2004; 101:841-6; PMID:14715899; https://doi.org/ 10.1073/pnas.0304916101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog 2006; 2:e46; PMID:16710455; https://doi.org/ 10.1371/journal.ppat.0020046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ragaz C, Pietsch H, Urwyler S, Tiaden A, Weber SS, Hilbi H. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol 2008; 10:2416-33; PMID:18673369; https://doi.org/ 10.1111/j.1462-5822.2008.01219.x [DOI] [PubMed] [Google Scholar]
- [61].Urwyler S, Nyfeler Y, Ragaz C, Lee H, Mueller LN, Aebersold R, Hilbi H. Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic 2009; 10:76-87; PMID:18980612; https://doi.org/ 10.1111/j.1600-0854.2008.00851.x [DOI] [PubMed] [Google Scholar]
- [62].Hoffmann C, Finsel I, Otto A, Pfaffinger G, Rothmeier E, Hecker M, Becher D, Hilbi H. Functional analysis of novel Rab GTPases identified in the proteome of purified Legionella-containing vacuoles from macrophages. Cell Microbiol 2014; 16:1034-52; PMID:24373249 [DOI] [PubMed] [Google Scholar]
- [63].Naujoks J, Tabeling C, Dill BD, Hoffmann C, Brown AS, Kunze M, Kempa S, Peter A, Mollenkopf HJ, Dorhoi A, et al.. IFNs modify the proteome of Legionella-containing vacuoles and restrict infection via IRG1-derived itaconic acid. PLoS Pathog 2016; 12:e1005408; PMID:26829557; https://doi.org/ 10.1371/journal.ppat.1005408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Herweg JA, Hansmeier N, Otto A, Geffken AC, Subbarayal P, Prusty BK, Becher D, Hensel M, Schaible UE, Rudel T. Purification and proteomics of pathogen-modified vacuoles and membranes. Front Cell Infect Microbiol 2015; 5:48; PMID:26082896; https://doi.org/ 10.3389/fcimb.2015.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Schmölders J, Manske C, Otto A, Hoffmann C, Steiner B, Welin A, Becher D, Hilbi H. Comparative proteomics of purified pathogen vacuoles correlates intracellular replication of Legionella pneumophila with the small GTPase Ras-related protein 1 (Rap1). Mol Cell Proteomics 2017; 16:622-41; PMID:28183814; https://doi.org/ 10.1074/mcp.M116.063453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].O'Connor TJ, Adepoju Y, Boyd D, Isberg RR. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A 2011; 108:14733-40; PMID:21873199; https://doi.org/ 10.1073/pnas.1111678108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kortholt A, Bolourani P, Rehmann H, Keizer-Gunnink I, Weeks G, Wittinghofer A, Van Haastert PJ. A Rap/phosphatidylinositol 3-kinase pathway controls pseudopod formation. Mol Biol Cell 2010; 21:936-45; PMID:20089846; https://doi.org/ 10.1091/mbc.E09-03-0177 [DOI] [PMC free article] [PubMed] [Google Scholar]


