ABSTRACT
Post-translational modifications, such as the phosphorylation of tyrosines, are often the initiation step for intracellular signaling cascades. Pan-reactive antibodies against modified amino acids (e.g., anti-phosphotyrosine), which are often used to assay these changes, require isolation of the specific protein prior to analysis and do not identify the specific residue that has been modified (in the case that multiple amino acids have been modified). Phosphorylation state-specific antibodies (PSSAs) developed to recognize post-translational modifications within a specific amino acid sequence can be used to study the timeline of modifications during a signal cascade. We used the FcϵRI receptor as a model system to develop and characterize high-affinity PSSAs using phage and yeast display technologies. We selected three β-subunit antibodies that recognized: 1) phosphorylation of tyrosines Y218 or Y224; 2) phosphorylation of the Y228 tyrosine; and 3) phosphorylation of all three tyrosines. We used these antibodies to study the receptor activation timeline of FcϵR1 in rat basophilic leukemia cells (RBL-2H3) upon stimulation with DNP24-BSA. We also selected an antibody recognizing the N-terminal phosphorylation site of the γ-subunit (Y65) of the receptor and applied this antibody to evaluate receptor activation. Recognition patterns of these antibodies show different timelines for phosphorylation of tyrosines in both β and γ subunits. Our methodology provides a strategy to select antibodies specific to post-translational modifications and provides new reagents to study mast cell activation by the high-affinity IgE receptor, FcϵRI.
Keywords: Phosphorylation state-specific antibodies, phage display, yeast display, FcϵR1 receptor subunits, post-translational modifications
Introduction
Post-translational modifications of transmembrane receptors regulate cell responses to external stimuli. Phosphorylation of serine, threonine, and tyrosine residues in the intracellular domains of various cell surface receptors by cellular kinases is often the initiation step for many signal cascade pathways.1 These receptors are subsequently dephosphorylated by phosphatases based on the concentration and duration of the external stimuli.2 The transient phosphorylation of transmembrane proteins and receptors may induce conformational changes that alter protein function, as occurs with the cystic fibrosis transmembrane conductance regulator (CFTR) protein.3 Transmembrane receptor phosphorylation may also generate binding sites for other intracellular proteins, leading to the assembly of new functional complexes.4,5 Receptor tyrosine kinases such as Kit, FGFR, and the insulin receptor function similarly by promoting the formation of intracellular signaling complexes.6
Aberrant changes in the balance of phosphorylation and dephosphorylation of cellular signaling proteins contribute to pathogenesis in a wide range of cancers,7,8 immunological disorders9, and neurodegenerative diseases.10,11 The phosphorylation sites of mammalian transmembrane proteins may be subject to, or affected by, disease-causing mutations. Molecular tools capable of interrogating protein phosphorylation in a site- and state-specific manner have proven invaluable in elucidating the dynamics of numerous disease processes.
Signaling studies often rely on “pan-reactive” monoclonal and polyclonal antibodies to phosphorylated amino acids. Commercially available anti-phosphotyrosine (anti-pY) reagents include pY20, pY99, and P-Tyr-1000; these reagents recognize pY residues in the context of a relatively broad peptide sequence backbone.12–15 A smaller group of anti-phospho-threonine and phospho-serine antibodies is also commercially available.16,17 The utility of these generic antibodies is somewhat limited, since they must typically be combined with immunoprecipitation or an alternative protocol to ensure specificity of labeling. Pan-reagents fail to identify specific phosphorylation sites, which is particularly problematic if target proteins are substrates for multiple kinases.
Since the 1990s, the field has been improved by the development of sequence-specific reagents, referred to by Mandell4 as phosphorylation state-specific antibodies (PSSAs). The early generations of PSSAs were typically obtained by immunization of host animals (rabbits, goats) with phosphorylated peptides to obtain polyclonal antisera or generate hybridomas expressing PSSAs.4,18 Despite serious concerns regarding the specificity of research-grade antibodies in general,19–24 including PSSAs25-29 and antibodies targeting other post-translational modifications,28,29 there are examples of phosphospecific antibodies that perform well.27–29
Recently, highly specific PSSAs recognizing specific-phosphorylated residues in the tau protein were generated by chicken immunization with phosphorylated peptides, followed by selection from single-chain variable fragment (scFv) libraries created from the immunized spleen and bone marrow.30 The structure of one of these antibodies was determined and showed novel complementarity-determining region (CDR) structures with a “bowl-like” conformation in CDR-H2 that tightly and specifically interacts with the phospho-Thr-231 phosphate group, as well as a long, disulfide-constrained CDR-H3 that mediates peptide recognition.14 The success of this strategy has led to production of additional PSSAs to other phosphorylation sites based upon screening of specifically designed scFv libraries.31 The antibodies obtained using these methods31 showed phospho-peptide specificity, but required prior target protein immunoprecipitation to demonstrate recognition of phosphorylation sites in proteins on western blots. While the low affinity of these new reagents has been a limitation to date, recombinant antibodies can be modified by in vitro evolution to improve affinity or specificity.32 Furthermore, reproducibility is a hallmark of recombinant probes. In general, two reduced-size antibody formats, scFvs33 and antigen-binding fragments (Fabs), both of which comprise the essential antibody binding portions, are used in in vitro display systems.
Here, we describe an in vitro method to develop recombinant PSSAs. We initially used a library of scFv antibodies displayed on M13 bacteriophage particles. The outputs of these phage selections were used as inputs to flow cytometry-based yeast sorting, as previously described.34–36 Strategies for phage display selection, yeast display sorting, affinity maturation, and characterization of phospho-specific binding are presented.
As a model system, we chose critical phospho-peptides derived from signaling subunits of the high-affinity IgE receptor (FcϵRI) expressed on mast cells, basophils, and antigen-presenting cells.37 FcϵRI is an αβγ2 tetrameric complex, where the extracellular domain of α binds IgE and the βγ2 chains mediate intracellular signaling through immunoreceptor tyrosine-based activation motifs (ITAMs) in their cytoplasmic tails. ITAMs are short peptide sequences composed of two tyrosines separated by 10 amino acid residues.38 The FcϵRI γ ITAM sequence, with tyrosine residues at position 65 and 76 in the tail, is typical of the ITAM family. The β subunit ITAM is non-canonical, in that there are three tyrosines (Y218, Y224, and Y228) in its sequence. In the classical model of FcϵRI signaling, antigen-driven aggregation of IgE-FcϵRI complexes leads to phosphorylation of the two canonical tyrosines in the γ subunit, which in turn serve as docking sites for the dual SH2 domains of Syk tyrosine kinase to initiate downstream signaling.39–41 Studies of the B-cell receptor (BCR), an ITAM-bearing immunoreceptor critical for B lymphocyte signaling, have suggested distinct signaling outcomes for mono-phosphorylated or dually phosphorylated ITAMs.42 Similarly, the aggregation state of FcϵRI is a critical factor in determining the balance between positive and negative signaling regulation,43 which may be explained in part by differential recruitment of signaling molecules based on specific ITAM phosphorylation patterns.44–46 Differential phosphorylation of the three β ITAM tyrosines allows for eight different phosphorylation patterns, while the two γ-ITAM tyrosines lead to four different phosphorylation patterns. We consider the complexity of this analysis in the screening and development of PSSAs for FcϵRI β and γ ITAMs. These new reagents could enable sensitive readouts of allergen-induced FcϵRI activation, which in turn may facilitate the potential development of therapeutic strategies to manage allergic responses.47 Our methods provide a pathway for developing phospho-specific antibodies to other target proteins with complex phosphorylation patterns, as well as troubleshooting strategies for peptide-based selection. These methods should also be broadly applicable to evaluating other post-translational modifications, including ubiquitination,48 sulfation,49 and methylation.50
Results
Our model system, the tetrameric FcϵRI, with its two ITAM-bearing β and γ subunits,51,52 is shown in Figure 1(a). The complexity of phosphorylation patterns in these two chains is illustrated in Figure 1(b-c), which presents the eight different phospho-peptide combinations for the three tyrosines in the β ITAM composed of residues 211–236 (KVPDDRLYEELHVYSPIYSALEDTR) versus the four phospho-peptide combinations for the two tyrosines in the γ ITAM that are contained in residues 56–83 (ASREKSDAVYTGLNTRNQETYETLKHE). In this illustration, phosphorylated tyrosines are shown in red.
Figure 1.
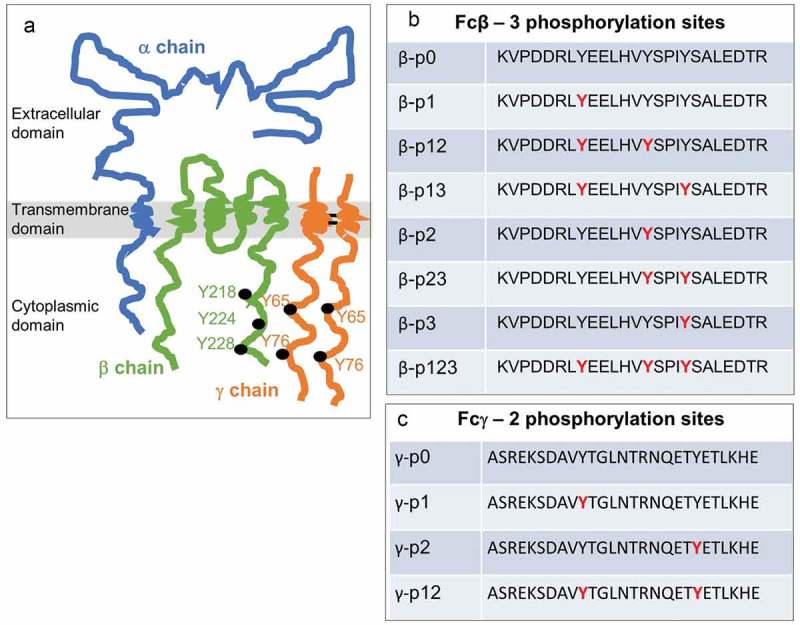
FcϵR1 receptor subunits and corresponding ITAMs with associated phosphorylation patterns.
a) Cartoon of the oligomeric (α,β,γ2) FcϵR1 complex. b-c) ITAMs in the β and γ subunits have three and two tyrosine phosphorylation sites (red), respectively. Panels b-c illustrate the set of combinations that can arise from partial or complete phosphorylation at these sites.
Antibodies with varying specificities were obtained after three rounds of phage display selection from a well-characterized antibody library.53 After three rounds of phage panning, the scFv genes were subcloned into a yeast display system for additional screening. This approach enabled us to use flow-based assays that allowed sorting of the top 1% clones, as well as the ability to perform peptide and phosphate specificity assays in a multiplex format using biotinylated and unlabeled peptides. Three rounds of yeast fluorescence-activated cell sorting under different conditions were used to identify specific populations of yeast-displayed antibodies. Examples of scFvs obtained using the combination of phage and yeast display technologies are shown in Figure 2. More than 20 different scFvs with varying phosphate specificities were obtained. Some scFvs were specific to a single phosphorylation pattern (e.g., p123, p12); other scFvs recognized phosphorylation of a specific tyrosine (e.g., Y228 or p3). Many scFvs obtained recognized multiple phospho-peptides and did not have a specific recognition pattern. ScFvs with “interesting” phosphate specificities were further assayed for their ability to recognize non-biotinylated peptides using a competition assay (Figure 2(b)). Our results showed that scFvs that recognized only the biotinylated peptides failed to recognize the FcϵR1 subunit in subsequent assays. ScFvs chosen based on these assays were affinity matured. Here, we describe development of each of the affinity-matured scFvs and their characterization.
Figure 2.
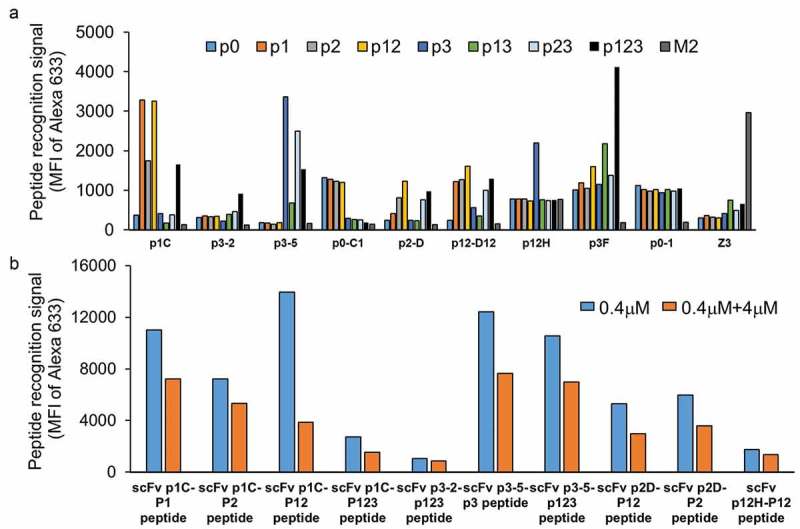
ScFv peptide and phosphate specificity assays upon completion of the initial selection process for the β ITAM peptides.
a) Nine example scFvs obtained following three rounds of phage selection and three rounds of yeast sorting. Yeast displaying scFvZ3 and its antigen M2 peptide of Influenza A were used as negative controls in this flow-based assay. Binding assays were conducted at 100 nM peptide concentration. Data show varying levels of specificity of individual scFvs for different phospho-peptide(s). b) Five scFvs (p1C, p3-2, p3-5, p2D, and p12H) from panel A were evaluated for recognition of non-biotinylated (native) peptide using a competition assay. Paired plots show relative binding of biotinylated peptides to scFv displayed on yeast in the absence (blue) or presence (orange) of 10X non-biotinylated peptide.
ScFv p1-1 (anti-β p1 and/or p2)
The bio-p1 peptide in the presence of 10X competitive non-biotinylated peptides (Set 4 – see materials and methods) was used for phage antibody selection. Competitive yeast sorting was also performed in the presence of the four non-biotinylated peptides (p0, p2, p3, p23). The final-selected antibody (scFv p1C) recognized p1, p2, and p12 peptides and had KD values ranging from 478 nM to 1214 nM. The scFv p1C was also able to recognize the non-biotinylated p1, p2 and p12 peptides in the competition assay (Figure 2(b)), indicating its ability to recognize the phosphorylated FcϵR1 β-subunit. The ability of this low-affinity antibody to recognize the β-subunit was evaluated following immunoprecipitation of the β-subunit from cells stimulated with DNP24-BSA and its corresponding IgE. Upon successful recognition of the phosphorylated β-subunit, the antibody was affinity matured using error-prone polymerase chain reaction (PCR) followed by yeast sorting at sequentially lowered peptide antigen concentrations. The final derivative, scFv p1-1, recognized phosphorylation of tyrosines 218 and/or 224 (p1/p2). Phosphorylation of Y228 (p3) reduced the binding signal for this scFv by ~100-fold (Figure 3(a) and Table 1). The KD values for βp13, βp23, and βp123 peptides range from 148 nM to 194 nM, and the KD value for p0 peptide is 317 nM (Table 1), compared to 1–2 nM for the p1, p12, and p2 peptides.
Figure 3.
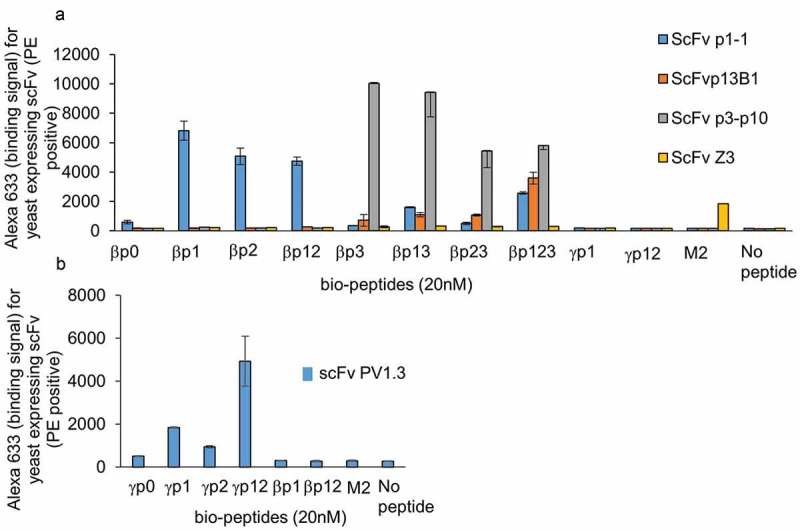
Peptide and phosphate specificity assays for affinity-matured scFvs.
a) Specificity assay for scFvs p1-1, p13B1, p3p10 against the eight β-phospho-peptides using yeast displayed scFvs in a flow-based assay. FcϵR1 γp1 and γp12 peptides and influenza M2 peptides were used as controls. The data also show binding profile for scFvZ3, which was used as negative control antibody. b) Specificity assay for anti-γ scFv PV1.3 against the four γ phospho-peptides with βp1, βp12, and M2 peptides used as controls.
Table 1.
Summary of antibody selection strategy for the best scFv clones isolated. Phage selection method, yeast sorting strategy and the affinities of starting clone for four different antibodies are given. Phosphate specificity and KD values for the affinity-matured clone is given in the last column.
| Antibody name | Phage selection method | Yeast sorting strategy | Starting affinity (nM) | Phosphate specificity and KD (nM) of the affinity matured clones for different peptide targets | |
|---|---|---|---|---|---|
| scFv p1-1 | Competitive selection with p0, p2, p3, p23 | Competitive sorting with p0, p2, p3, p23 | 478-1214nM | βp0 | 317.1 |
| βp1 | 2.15 | ||||
| βp2 | 2.21 | ||||
| βp12 | 1.45 | ||||
| βp13 | 176.00 | ||||
| βp23 | 193.80 | ||||
| βp123 | 148.00 | ||||
| scFv p13B1 | Competitive selection with p0 alone | Competitive sorting with p0 | 500-1600nM | βp0 | 6183.23 |
| βp123 | 0.50 | ||||
| βp13 | 10.57 | ||||
| βp23 | 21.50 | ||||
| scFv p3-10 | Competitive selection with p0 alone | Competitive sorting with p0 | >1000nM | βp0 | ~6469.31 |
| βp3 | 3.77 | ||||
| βp13 | 6.89 | ||||
| βp23 | 3.95 | ||||
| βp123 | 2.50 | ||||
| scFv PV1.3 | No competition | No competition | 800-2000nM | γp0 | 668.34 |
| γp12 | 9.43 | ||||
| γp1 | 19.68 | ||||
| γp2 | 68.20 | ||||
ScFv p13B1 (anti-β p123)
This antibody was isolated during p3-specific selection. A newly synthesized bio-p3 peptide was used as a selection target in phage display experiments. Only the non-biotinylated p0 peptide was used as a competitor in both phage display and yeast sorting experiments. Specific elution with 10X non-biotinylated peptides was used during phage display selection. ScFv p3-2 showed the highest binding signal for the p123 peptide (Figure 2(a)) and recognized the non-biotinylated βp123 peptide (Figure 2(b)). Upon successful detection of immunoprecipitated β-subunit, the parental antibody was affinity matured to recognize the triple phosphate peptide with a binding affinity of 0.5 nM (Figure 3(a) and Table 1). The affinities of p13B1 for βp23 and βp13 peptides are 10.57 nM and 21.5 nM, respectively. This antibody does not recognize p0, p1, p2, and p12 peptides; the KD value for p0 peptide is around 6000 nM.
ScFv p3-p10 (anti-β p3)
This antibody was isolated using a p3-specific antibody selection. ScFv p3-p10, which recognizes all four peptides with p3 phosphorylation and does not recognize any of the peptides without phosphorylation at the third tyrosine, was evolved from scFv p3-5 (Figure 2(a)). The p3-5 scFv also recognized non-biotinylated p3 and non-biotinylated p123 peptides in competition assays (Figure 2(b)). Two sets of error-prone libraries were constructed, and six rounds of yeast sorting were performed to isolate the p3-p10 antibody, which had KD values ranging from 2 to 6 nM for the peptides with p3 phosphorylation (Figure 3(a) and Table 1). The approximate KD value for the p0 peptide was measured to be ~6500 nM, 1000x lower than specific recognition of peptides phosphorylated at the p3 position.
ScFv PV1.3 (anti γ-p1/p2)
ScFv PV1 was isolated during selection on γ-p0 peptide. Competitive peptides were not used during the phage selection or the yeast sorting. Despite extensive efforts, this was the only successful antibody selected against the γ-peptide. All the other scFvs selected against the γ-peptide failed to recognize the non-biotinylated peptide in the competition assay (as shown in Figure 2(b)) and/or the immunoprecipitated γ-subunit from stimulated cell lysates. An error-prone PCR library was prepared from scFv PV1, and the high-affinity PV1.3 clone was isolated using yeast sorting by lowering the amounts of peptide antigen after each sorting round. ScFv PV1.3 recognizes γp12 with ~10 nM affinity and the γp1 peptide with ~20 nM affinity. The KD values for the γp2 peptide are considerably lower at 68.2 nM, and this antibody recognizes γP0 peptide with KD value of 668 nM (Figure 3(b) and Table 1).
ScFv C1 (anti-β p0), scFv p2D (anti- β p2) and scFv p12H
We isolated several antibodies that were not further characterized but had unique binding specificities. ScFv C1 was isolated from a selection on β-p0 peptide. This antibody recognized peptides without p3 phosphorylation (Figure 2(a)). However, affinity maturation and sorting did not provide an antibody that recognized only the p0 peptide. None of the derivatives from this antibody were further characterized. ScFv p2D (Figure 2(a and b)) recognized peptides with phosphorylation of the second tyrosine (p2, p12, p23, p123). However, the best affinity-matured clones originating from p2D had similar binding specificities to scFv p1-1, which had higher recognition signals. Therefore, none of the p2D derivative antibodies were further studied. ScFv p12H recognized the p12 peptide specifically (Figure 2(a and b)); however, upon affinity maturation, it also had similar specificities to scFv p1-1, but with a lower binding signal.
The sequences of the final affinity-matured antibodies are provided in Table 2. Affinity-matured clones differ from the original clones in 1–8 amino acids (red letters) depending upon the number of rounds of error-prone PCR. The vH CDR2 (underlined) was modified for all three anti-β scFvs. The vH CDR3 remained unchanged for all scFvs. Sequence comparison of the original and affinity-matured scFvs are provided in supplementary results.
Table 2.
Sequences of affinity-matured scFvs. Amino acid sequences for the affinity matured anti-β and anti-γ ITAM scFvs are given here. Complementary determining regions (CDRs) of the vL and vH of the scFv are highlighted in yellow. The linker sequence between the vL and vH is highlighted in grey. Red letters indicate amino acid changes during affinity maturation. ScFv p1-1 and p13B1 each had three changes resulting from error prone PCR. ScFv p3-p10 due to two rounds of error prone PCR and sorting contained eight amino acid modifications. All three scFv acquired changes in vH CDR2 and vH CDR3 remained unchanged. The anti-γ scFv PV1.3 had only one amino acid change from serine to asparagine.
| scFv p1-1 (anti-β p1/p2) – affinity matured clone of p1C |
| VL |
| GAHADIQMTQSPSFLSASVGDRVTITCRASQGISSWLAWYQQKPGKAPKLLIYAASSLQSGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQANSFPLTFGGGTRLEIKS |
| Linker |
| GGSTITSYNVYNTKLSSSGT |
| VH |
| QVQLVESGGGLVQPGGSLRLSCAASGLTFSNYAMNWVRQAPGKGLEWVSSISSRGDTTYYADSVKGRFTISRDNSKSTLYLQMNSLRAEDAAVYYCARLRNSGGMDVWGPGTTVTVSSAS |
| scFv p13B1 (anti-β p3+1p) – affinity matured from p3-2 |
| VL |
| GAHAQSVLTQPLSASGTPGQGVTISCSGGSSNIGSHNVYWYQHLPGTAPNLLLYKSNQRPSGVPDRFSGSKSGTSASLAISGLRSEDEADYYCAAWDDSLSGWVFGGGTKLTVLS |
| Linker |
| GGSTITSSNVYYTKLSSSGT |
| VH |
| QVQLQQSGPGLVKPSQTLSLTCAISGDSVSSNSAAWNWIRQSPSGGLEWLGRTYYRSKWYSDYAVSVKSRITINPDTSKNQISLQLNSVTPEDTAVYYCARSLAGIRAFDIWGQGTMVTVSSAS |
| scFv p3-p10 (anti-β p3) – affinity matured from p3-5 (two EP libraries total of 2x4 sorts from original clone) |
| VL |
| GAHAQSVLTQPPSVSGAPGQRVTISCTGSSSNIGAGYDVHWYQRLPGTAPKLLIYGNSNRPSGVPDRFSGSKSGTSASLAITGLQAEDEAEYYCAAWDDSLKGYVFGTGTKLTVLS |
| Linker |
| GGSTRSSYNVYYTKLSSSGT |
| VH |
| QVQLVESGGGVAQPGRSLSLSCAASGFTFRSYAMHWVRQAPGRGLEWVAVISYDGTYKYYADSVKGRFTISRDNSKNTLYLQMNSLRAEDTAVYYCARVQTGMVRGGNGMDVWGQGTTVTVSSAS |
| scFv – PV1.3 (anti-γ p1/p12) – affinity matured from PV1 |
| VL |
| GAHASSELTQEPAVSVALGQTVRITCQGDSLRSYYANWYQQKPGQAPVLVIYGKNNRPSGIPDRFSGSSSGDTASLTITGAQAEDEADYYCNSRDSSGNHVVFGGGTKVTVLS |
| Linker |
| GGSTITSYNVYYTKLSSSGT |
| VH |
| QVQLVQSGAEVKKPGASVKVSCKASGYTFTGYYMHWVRQAPGQGLEWMGWINPNSGGTNYAQKFQGRVTMTRDTSISTAYMELSRLRSDDMAVYYCAVGKNAFDIWGQGTMVTVSSAS |
Recognition of the β and γ-subunits from cell lysate
Figure 4 shows the recognition of the β and γ subunits from the rat basophilic leukemia (RBL) cell lysate for the four high-affinity antibodies (p1-1, p3p10, p13B1, and PV1.3). For these experiments, the RBL-2H3 cells were primed with DNP-specific IgE and stimulated with 2 nM DNP24-BSA for 30 s, 2 min, and 5 min. Rat kidney cells and antibody Z3, which recognizes the M2 protein of influenza, were used as negative controls.
Figure 4.
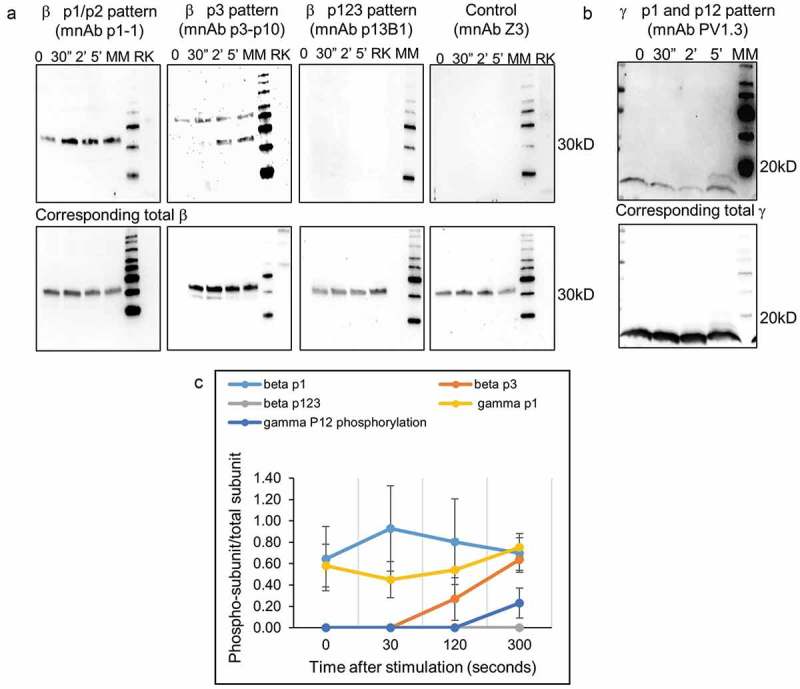
Phosphorylation pattern analysis of FcϵR1 subunits.
a) Assessment of phosphorylation patterns of β subunits after stimulation with DNP-BSA is shown using three antibodies p1-1, p13B1, and p3p10 utilizing western blotting with antibodies in minibody format. The FcϵR1 receptor was stimulated with 2 nM DNP-BSA and the data show phosphorylation patterns at various time points post stimulation. Phosphorylation at resting (basal) is also given. Antibody Z3 and rat kidney cell lysate (RK) were used as negative controls. The lower panel shows the total β detection used as a loading control. b) Similar analysis performed for anti-γ PV1.3. c) Patterns of phosphorylation graphed with data from multiple experiments (3–8 cell lysates prepared at different times).
The results obtained using antibody p1-1 indicated that Y218/Y224 (p1/p2) are already phosphorylated prior to stimulation and undergo a modest increase after 30 s of stimulation, followed by a decrease to return to roughly basal levels within 5 min. The Y228 (p3) phosphorylation detected by the p3p10 antibody became elevated only after 2 min of stimulation and was sustained at 5 min as well. The analysis performed with antibody p13B1 showed no recognition signal, suggesting that all three tyrosines are never simultaneously phosphorylated during stimulation. Total β-subunit detection was performed and used to normalize the signal intensities for quantification shown in Figure 4(c) to account for potential unequal sample loading on the gel.
Our experiments using scFv PV1.3 revealed that, similar to the β-subunit, the γ-subunit is also already phosphorylated prior to addition of the DNP24-BSA. When using 2 nM DNP24-BSA for stimulation, the phosphorylation initially decreases at 30 s to 2 min, prior to a dramatic increase at 5 min. Appearance of a second phospho-γ band at 5 min suggestes that the γ-subunit could be phosphorylated at both tyrosines simultaneously. These data show that our antibodies detect distinct phosphorylation patterns for the tyrosine residues of the FcϵR1 receptor’s β and γ subunits upon stimulation with 2 nM DNP24-BSA.
The phosphate specificity of three of these antibodies (p1-1, p13B1, PV1.3) were further assessed using a phosphatase assay to evaluate whether dephosphorylation resulted in a reduction in recognition signal. The results showed that the phosphatase treatment significantly reduced the total phospho-tyrosine in the cell lysate (Figure 5(a)), and that total phospho-tyrosine analyses cannot distinguish individual phosphorylated proteins in a cell lysate. Data obtained with anti-β antibodies p1-1 and p3p10 are shown in Figure 5(b). The data show a reduction of recognition signal upon phosphatase treatment for the p1-1 antibody and complete elimination of recognition of the anti-p3 antibody. Total β in the cell lysates was assayed as a loading control. Similar results were obtained for the anti-γ p1/p12 antibody (Figure 5(c)). The phosphatase treatment drastically decreases the recognition by antibody PV1.3, indicating that all three antibodies require phosphorylation of specific FcϵR1 receptor tyrosines for recognition.
Figure 5.
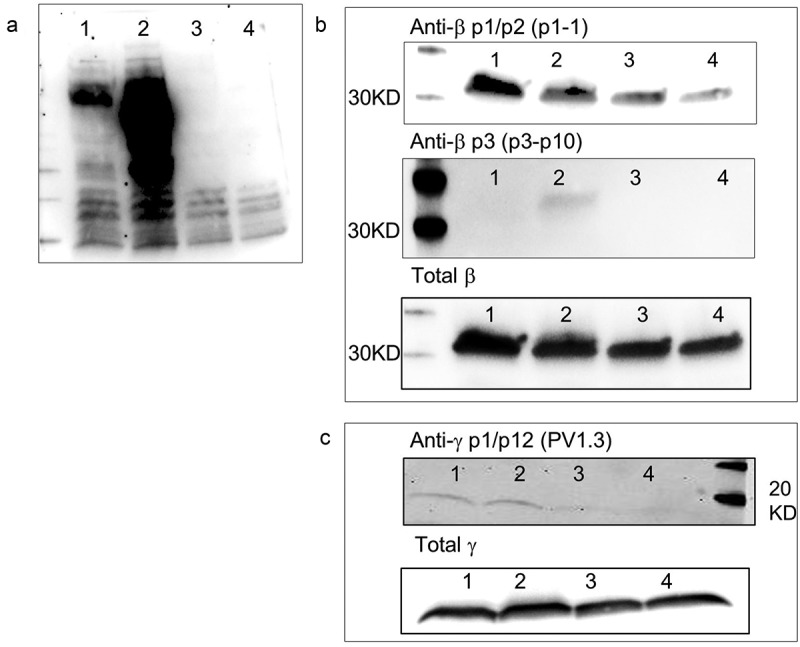
Antibody recognition FcϵR1 subunits of untreated and phosphatase treated cell lysates.
a) Presence of phospho-tyrosines in whole cell lysates at resting (#1), stimulated with DNP-BSA for 5 min (#2) and after these samples have been treated with tyrosine phosphatase for 30 min (#3 & 4), assayed using pan-reactive pY antibodies utilizing western blotting. b) Specificity of the p1-1 and p3p10 antibodies for phosphorylated β subunits were assessed by comparing recognition of pattern of cell lysates before and after phosphatase treatment. Total β subunit western analysis was performed as a loading control. c) Similar analysis performed for the anti-γ antibody PV1.3.
Discussion
The role of post-translational modifications in cell signaling cascades is well established.1,2 However, the availability of reagents to study these changes is limited. While antibodies to a modified amino acid residue (e.g., phosphotyrosine) are commercially available, phosphorylation and sequence-specific antibodies (PSSA) are difficult to obtain.4,5 This has generally resulted in a two-stage procedure in which proteins are first immunoprecipitated with antibodies recognizing the target, and are then tested for the presence of phosphorylation in the immunoprecipitated protein. In addition to the additional work required, this approach only indicates that one or more sites are phosphorylated, without identifying the specific sites modified. The value of PSSAs is the ability to assess the phosphorylation of specific sites without carrying out a time-intensive, two-stage procedure.
To address these shortcomings, we developed strategies to select, improve, and validate PSSAs using the FcϵR1 β and γ-subunits as a model. Multiple selection and sorting strategies were evaluated using a β-p1 peptide as an antigen. We found that using mass spectrometry analysis to evaluate the purity and phosphorylation state of the peptides prior to initiating selection is essential for obtaining specific antibodies. Our strategy of performing antibody selection using biotinylated peptide, followed by elution using non-biotinylated peptides is an effective method to obtain specific antibodies. Furthermore, our results show that using non-biotinylated peptides as competition during the selection process improves the specificity of the antibodies obtained (Supplementary Figure 1, 2, 3 and Supplementary Table 1). While we used multiple competition peptides in earlier experiments, later experiments with β-p3 and γ-peptides showed that the use of a single non-phosphorylated competitor peptide may be sufficient to obtain specific antibodies.
Yeast display-based flow cytometry in combination with phage display was used to assess specificity and affinity of the antibodies selected. This assay platform in combination with the array of differentially phosphorylated biotinylated peptides was exquisitely suited to assay specificity of antibodies to the various phosphorylation patterns using different peptides. The use of the competition assay is an important step in identifying antibodies that will recognize the natural receptor subunit, since during our selection and sorting protocols, we obtained many antibody clones that recognized only the biotinylated peptide in combination with streptavidin. This problem was particularly pronounced in the γ-p12 peptide selection. Despite numerous attempts using many variations on the overall strategy, all the γ-p12 antibodies we obtained only recognized the biotinylated peptides. The competition assay we optimized ensures the rapid identification of antibodies that will be unable to recognize cell surface receptors. In our hands, all antibodies that recognized the non-biotinylated peptide during the competition assay successfully recognized the native receptor subunit and none of the antibodies that failed the competition assay were able to recognize the FcϵR1 subunits. Consequently, this assay saves the extensive time and expense associated with cell culture-based validations.
We incorporated a set of experiments into our workflow to evaluate the ability of selected antibodies to recognize the phosphorylated receptor subunit. In our strategy, low-affinity antibodies (0.5–1 mM) obtained after initial phage selection were evaluated using immunoprecipitated receptor. This avoided the further development of antibodies that did not recognize the natural substrate. Suitable antibodies were affinity matured using error-prone PCR and yeast display-based flow cytometry.
Using this strategy, we were able to obtain antibodies with affinities of 0.5–200 nM, with antibody affinity varying according to the phosphorylation status of the substrate. For example, the p1-1 antibody recognizes the p1, p2 and p12 peptides with affinities 1–2 nM, with 100-fold worse recognition upon addition of the p3 phosphorylation in p13, p23, and p123 peptides. Antibodies against β-p3 (Y228) were not obtained when multiple peptides were used as competition, and therefore we developed an alternate strategy using only the β-p0 peptide as competition. The selection and sorting strategy described for the p3 peptide yielded an antibody that recognized the β-subunit when Y228 was phosphorylated and was unaffected by phosphorylation on the other tyrosines. Antibody p13B1, which recognizes the triple phosphorylated β-peptide, had a sub-nanomolar affinity to its target peptide.
While the affinity maturation improved scFv affinity as expected, the specificity of the antibodies to various peptides were not vastly altered by the error-prone PCR. However, antibodies that recognized the p12 peptide also recognized p1 and p2 peptides upon affinity maturation. Interestingly, three of the four affinity matured antibodies accumulated mutations in HCDR2, which was previously identified as being important in recognition of phosphopeptides.31 We used three separate assays to evaluate the recognition of FcϵR1 subunits by the antibodies selected. The initial evaluation used the immunoprecipitated subunit, as described in the Materials and Methods section. Once we obtained the high-affinity antibodies, cell lysates could be used directly in western blot experiments to study the phosphorylation pattern of the subunits at various times following cell stimulation. We also developed a phosphatase-based assay to further evaluate the phosphate specificity of the antibodies selected.
The results obtained show differential phosphorylation patterns for the different tyrosines, indicating exquisite regulation of the signal cascade. The data obtained indicate that one of the tyrosines is phosphorylated at basal (resting) stage (Figure 4). Phosphorylation of an additional tyrosine occurs upon stimulation, and this facilitates the initiation of the signal cascade. The results obtained here also show that the phosphorylation of the β-subunit precedes γ initiation. At the resting stage, both β and γ subunits are singly phosphorylated at the p1 or p2 position. The third tyrosine of the β-subunit becomes phosphorylated in the next step after 2 min. The γ-subunit initiation occurs as a third step at 5 min as both tyrosines become phosphorylated, changing the status to double phosphorylation (p12).
This level of granularity of information on FcϵR1 signal cascade initiation has not been previously reported. Previous studies have been limited by examining only total phosphorylation of β or γ subunits upon stimulation with a ligand using generic anti-phospho-tyrosine antibodies in immunoprecipitated protein,43 providing no site-specific information. To the best of our knowledge, this is the first study demonstrating successful characterization of antibodies that identify different sites of ITAM phosphorylation and bind with high affinity to exclude the need to immunoprecipitate the subunits prior to further analysis. This can lead to better understanding of the regulation of positive and negative signaling through this receptor, where the recruitment of signaling molecules to FcϵRI may depend on differential site-specific ITAM phosphorylation.
We experienced greater difficulty obtaining antibodies for γ-subunit peptides as compared to β-subunit peptides. After numerous attempts, we were able to obtain one antibody that recognized the phosphorylated γ-subunit. Results from evaluation of a bio-γp0 (scFv B9) and bio-γp12 (scFv C7) are provided in Supplementary Figure 4. It is not entirely clear why obtaining PSSA to the γ-subunit was more difficult than the β-subunit. Another challenge we were unable to overcome was selection of antibodies that differentiated the non-phosphorylated subunit from phosphorylated subunit. Despite employing multiple variations to the selection and sorting strategies, antibodies recognizing only β-p0 or γ-p0 peptides could not be obtained. Nonetheless, our study allowed us to identify strategies for selecting, affinity-maturing, and characterizing PSSAs against a broad range of targets. In Table 3, we provide guidelines for researchers wishing to generate additional PSSAs against other targets, culled from the experience obtained during the course of the studies described here.
Table 3.
Lessons learned and successful strategies for selection of PSSA antibodies. This table provides strategies to other researchers interested in developing phosphorylation state- and sequence specific antibodies.
| Guidelines for the selection of phospho site specific antibodies from naïve antibody libraries |
|---|
| Do not use phosphate buffered saline |
| Use a combination of phage and yeast display |
| Biotinylate the target peptide and select in the presence of non-biotinylated competitor peptides (either other phosphorylated sites or non-phosphorylated peptides) |
| Elute with non-biotinylated target peptide |
| Use of excess bio-peptide to coat streptavidin magnetic beads reduces streptavidin binders |
| Three rounds of phage selection was ideal. Two rounds gave lower affinity antibodies. 4 or 5 rounds of selection resulted in overabundance of streptavidin binders. |
| Test antibody specificity for target peptide with competition assays with non-biotinylated competitors (competition assay). In this assay it is important to pre-mix the bio-peptide with the 10X excess nonbio-peptide prior to antibody addition (e.g. before adding yeast displaying scFv). |
| Test selected antibodies against real substrates as early as possible to avoid the selection of peptide-specific antibodies |
| Selection against one peptide does not guarantee non-reactivity against a different related peptide |
| Presence of protease and phosphatase inhibitors are crucial during cell lysate preparation |
| Lowering/absence of recognition with phosphatase treated samples is an excellent test for antibody specificity. |
Methods described here should also apply to the development of antibodies against a diverse set of protein targets with post-translational modifications. Not only are the antibodies described here exquisitely specific for the sites described, but, being recombinant (with sequences provided), researchers can easily produce the antibodies themselves. The ability to refer to a specific (publicly available) sequence is expected to increase reproducibility.20,21 These scFv genes can be cloned into different vectors to produce the binding moiety with different properties (e.g., fluorescence, nucleic acid binding, artificial scaffolds). These reagents can subsequently be used to study in situ phosphorylation dynamics. Post-translational modifications are known to play key roles in carcinogenesis; site- and modification-specific antibodies linked to chemotherapeutic agents could prove to be powerful treatment options.
Material and methods
Preparation of the phage library
The phage display library was prepared as described previously,53 with the exception that phages were PEG precipitated and reconstituted in a 30 mM Tris buffer pH 8.0 to allow selection of phosphorylation-specific antibodies. Phage titers were used to assess the quality and quantity of the library. The phage library was blocked with 5% bovine serum albumin (BSA) for 30 min at room temperature, and 200 μL was used per selection.
Preparation of peptide antigens
Peptide antigens were synthesized by Genscript Inc. or by Macromolecular Resources Inc. The peptide quality was evaluated by mass spectrometric analyses either by the manufacturer or in-house to ensure that the phosphorylation pattern matched the atomic mass. Peptide powder was dissolved in 30 mM Tris buffer pH 8.0 for 10 min, rotating. The solution was centrifuged at 14,000 rpm for 5 min and the supernatant transferred to a fresh tube. Absorbance at 280 nm was used to calculate the concentration of stock peptides. 20 μM peptide stocks were prepared and used in daily experiments. The binding capacity of 10 μL of Dynabeads M-80 streptavidin (Invitrogen Inc. cat#11205D) is 200 pmoles for a 3.5 KDa biotinylated peptide (bio-peptide), which we calculated to be ~0.7 μg. 100 mM stock peptides were equal to a concentration of 0.35 mg/mL. Therefore, 2 μL of peptide was added to 200 μL of blocked phage library. Per this calculation, the first round of selection was done with 1 μM bio-peptide, second round with 500 nM, and third round with 200 nM.
Phage selection strategy – four sets
We explored four selection strategies with four β peptides (p0, p1, p2, and p3). Set 1 was a standard selection with bio-peptides and acid elution. In set 2, 10x excess of non-biotinylated (nb) peptides was added at the phage-library incubation step to increase the stringency of the selection. Set 3 had no competition during selection; however, 10x excess of the specific non-biotinylated peptide was used to elute the bound phages after the washes. In set 4, both competition and peptide elution were used to further increase the stringency. An example of the four selection strategies for bio-p1 selection at 1 μM bio-peptide concentration is given in Table 4. Concentration of the competition and elution was subsequently reduced during second and third selection rounds to match the bio-peptide concentration.
Table 4.
Four sets of selection strategy is shown using p1 peptide selection as an example. The four selection strategies are 1) no competition with acid elution 2) competition with 10X excess of non-specific peptides with acid elution 3) no competition, but elution by 10X excess of specific peptide 4) both competition and specific elution strategies were combined.
| antigen | competition | elution | |
|---|---|---|---|
| Set 1 | Bio-p1 1μM | 0.1M HCl | |
| Set 2 | Bio-p1 1μM | 10μM nb-p0 + 10μM nb-p2 + 10μM nb-p3 + 10μM nb-p23 | 0.1M HCl |
| Set 3 | Bio-p1 1μM | 10μM nb-p1 | |
| Set 4 | Bio-p1 1μM | 10μM nb-p0 + 10μM nb-p2 + 10μM nb-p3 + 10μM nb-p23 | 10μM nb-p1 |
The blocked phage library was incubated with bio-peptides or with bio-peptide +10x competition as described above. Binding to streptavidin magnetic beads and washes were performed using the 24-well KingFisher magnetic particle separators (ThermoFisher Inc.). Streptavidin bead-bio-peptide interaction was allowed for 10 min, followed by a total of six washes (three washes with tris – tween buffer [30 mM Tris buffer pH 8.0 + 0. 1% tween 20] and three washes in tris buffer [30 mM Tris buffer pH 8.0]). Each well had 200 μL of wash buffer. Each wash step was for 30 s for the first round and 5 min for the second and third rounds. Acid elution was carried out for 4 min using 150 μL of 0.1 M HCl. The reaction was quenched with 50 μL of 1.5 M Tris pH 8.8. Peptide elutions were carried out for 30 min using a 10x concentration of the nb-peptide. This was followed by phage propagation and titrations as described in Sblattero et. al.53
Modifications implemented in the second set of β p3 peptide selections included: 1) an excess of target bio-peptide (5 μM) used to coat streptavidin magnetic beads, and 2) only nb-p0 peptide used as competition (instead of all competitive peptides), both during phage selections and yeast sorting. Antibody selections against γ-peptides were carried out using a similar protocol. Excess amounts of bio-peptide (5 μM) were used to maximize antigen concentration on the streptavidin magnetic beads. Selections were carried out according to conditions detailed in the set 3 and set 4 selection protocols.
PCR amplification of scFv insert and sub-cloning to yeast vector
Plasmid DNA was prepared from second and third round phage selection outputs, and the scFv genes were amplified using specific primers containing regions of DNA overlapping with the yeast display vector pDNL6.29 The yeast vector was digested with restriction enzymes BssH II, Nhe I and Nco I and purified using Qiagen PCR purification columns (Qiagen Inc. cat# 28104). The vector and scFvs were co-transformed into yeast cells using yeast 1Kit (Sigma Aldrich Inc. cat# YEAST1) to allow cloning by gap repair.54,55
Optimization of yeast library sorting conditions
We explored two strategies to sort positive antibodies after phage antibody selection: competitive sorting, and subtractive sorting followed by competitive sorting. The strategy is illustrated in Supplementary Figure 1 using the β p1 antibody selection as an example. Yeast cells were labeled with anti-SV5-PE to assess scFv display levels, and streptavidin labeled with Alexa 633 (ThermoFisher cat#S21375) was used to detect binding of biotinylated peptides. Double-positive (expression and binding) yeast clones were sorted (Quadrant 2). Sorted yeast was grown at 30°C for two nights and induced at 20°C for 16 h prior to the next round of analyses/sorting. All sorting experiments were performed using a FACSAria (Becton Dickinson) flow cytometer.
With the competitive sorting strategy, yeast libraries were screened for specific binders to each bio-peptide at 1 μM concentration in the presence of 10x excess of non-biotinylated competition peptides. The bio-peptide concentration was reduced to 500 nM and 200 nM for second and third rounds of sorting, and the concentrations of non-biotinylated peptides were also lowered correspondingly. Specific binders were sorted from the Q2 population.
For subtractive sorting, the yeast mini-library was stained with biotin-labeled versions of all the non-specific competition peptides at 1 μM, and the yeast population that did not recognize any of the competition was sorted (Q4) (Supplementary Figure 1). The concentration of the antigen was lowered for subsequent rounds of sorting as described above. This yeast was re-grown, and specific binders were once again sorted with competition. All 16 selections (four strategies x four peptides) were subjected to three rounds of sorting based on both strategies.
Yeast monoclonal analyses
Single colonies of yeast were analyzed for recognition of specific peptides. Twelve yeast clones that gave the highest signals were chosen for specificity analysis across all 8 β peptides and with negative-control non-ITAM peptide targets. Single clone experiments were performed using the LSR II (Becton Dickinson) cytometer. The selected specific clones were assayed for recognition of unlabeled peptide using a competition assay, in which excess non-biotinylated target was incubated with biotinylated target peptide prior to binding to yeast. The same strategy was also employed during selection of antibodies against the γ subunit.
The competition assay experiment was initially performed with 1 μM of bio-peptide and 10 μM of unlabeled peptide. During optimization of the competition assay, we tested the signal reduction with bio-peptide concentrations at 4 μM, 400 nM and 40 nM with corresponding 10x excess of non-labeled peptide. Competition assays were performed for β-p3 and γ-peptide selections at 400 nM bio-peptide and 4 μM non-bio peptide. The labeled and the excess unlabeled peptide were pre-mixed prior to addition to yeast cells. A sample containing only bio-peptide staining was also prepared for signal competition comparison. Peptide-bound yeast clones were stained with anti-SV5-PE and streptavidin Alexa 633 as previously described. Selected clones were assessed for restriction fragment length polymorphism with BstN1 enzyme and sequenced to identify unique scFvs.
Yeast minibody (scFv-Fc) cloning and expression
Unique scFv clones were amplified by PCR and subcloned into a yeast scFv-Fc expression vector, pDNL9-Rabbit Fc. ScFvs expressed from this vector are fused to rabbit Fc domains. Sub-cloning was performed either by homologous recombination or by restriction enzyme-based cloning. YVH10 yeast cells were used for the expression using SGT media following yeast secretion protocols described by Wentz and Shusta.55,56 The protein production was allowed to proceed for three days at 20°C. Culture supernatants were used directly as reagents in enzyme-linked immunosorbent assays and western blot assays.
Affinity maturation using error-prone PCR
Error-prone PCR was performed with a differential ratio of deoxynucleoside triphosphates (1 mM T/C and 200 µM A/G) in the presence of dimethysulfoxide (10%), MnCl2 (0.2 mM) and MgSO4 (2 mM), using Taq polymerase (New England Biolabs cat# M0273L). The PCR product was column purified using the Qiagen PCR purification kit (Qiagen Inc. cat# 28104) and cloned into the yeast display vector pDNL6 using yeast homologous recombination as described above. The top 1% of binders from the error-prone library were sorted as described above. Three or four rounds of sorting were carried out until sorting at 1 nM antigen concentration was possible. For the β-p3-specific binder, two rounds of error-prone PCR and sorting were performed. Affinity maturation of the anti-γ p1 was carried out similarly. Monoclonal analyses were performed to identify the best binder and to assess specificity. The best binders were cloned into the scFv-Fc yeast expression vector as described above. The minibodies were used for cell lysate western analyses. Detailed protocol for error-prone PCR, yeast library creation, and affinity measurement are given in Supplementary Materials and Methods.
Detection of FcϵR1 β and γ subunits
RBL cell monolayers were grown in tissue culture plates for 24 h and sensitized overnight with 5 nM anti-DNP IgE in suspension. The cells were washed with complete HANKs buffer43 and stimulated with 0.1 µg/mL (2 nM) DNP24-BSA for the specified amount of time. Cells were placed on ice and lysed using 1% NP-40 in presence of protease and phosphatase inhibitors.57 Total protein concentrations were measured using the BCA protein assay kit (ThermoFisher Inc. cat#23227) and equal amounts of total protein (maximum volume 30 µL/well) were used for cell lysate western experiments.
All protein electrophoresis was performed using 4–20% mini-protean TGX gels (Biorad, cat# 456–8094) and transfer of protein to nitrocellulose was performed using the Biorad transfer blot SD semi-dry transfer cell. The blots were blocked with 1% milk plus 1% BSA in 30mM Tris buffer pH 8.0. 2 mL of yeast culture supernatant containing scFv-RFc was incubated with the blot for 2 h or overnight. Tris-tween and tris buffers were used for washing and bound antibodies were detected using anti-rabbit HRP (Santa Cruz Biotechnology Inc.cat# sc-2030) Chemiluminescence signal was detected using Supersignal westdura substrate system (ThermoFisher cat# 34075) and Chemidoc instrument system (Biorad Inc.). Band quantification was performed using GeneTools analysis software (Syngene Inc.). Experiments were repeated 3 to 8 times for each blot and signals were normalized to the signal for the corresponding total β.
Immunoprecipitation of FcϵR1 β and γ subunits
Immunoprecipitation of FcϵR1 β-subunit was performed using goat polyclonal IgG (Santa Cruz Biotechnology, cat# sc33491) against this subunit bound to protein G agarose beads (Sigma Inc., Cat #11719416001). 2 μg of IgG was used per 1 mL of cell lysate for overnight immunoprecipitation. 1% NP40, 0.1% NP40 and water were used for washes, and the beads were re-suspended in 60 mL of Laemmli sample buffer. 20–30 μL of sample per lane was used in immunoprecipitation western blotting experiments. The γ-subunit immunoprecipitation was performed using an anti-γ antibody purchased from Upstate Inc. (cat# 06–727) using the same protocol.
Phosphatase treatment of cell lysates
Phosphatase treatment of cell lysates was performed using rat recombinant phosphatase (Sigma Inc. cat# SRP0214). Cell lysates were treated with 10 nM phosphatase for 30 min at 30°C and phosphatase reaction was terminated by addition of Laemmli sample buffer and boiling of samples prior to protein electrophoresis. The effect of phosphatase treatment was evaluated using two commercial antibodies selective for phosphotyrosine (Santa Cruz Biotechnolgy Inc. cat# sc-7020 and sc-508). Western analysis to determine the ability of anti-phospho β and γ antibodies to distinguish between phosphatase treated and untreated samples were performed as described above in the section titled detection of FcϵR1 β and γ subunits.
Funding Statement
This work was supported by National Institutes of Health Grant P50GM085273 (BW)Foundation for the National Institutes of Health [P50GM085273]; Los Alamos National Laboratory's Laboratory Directed Research & Development grant (20180005DR).
Author contributions
NV, AM, LC, PV, NT, NA, CFH, TG, and SC conducted the experimental work. NV, AM, and ARMB wrote initial drafts of the paper. CFH and BW provided critical input during manuscript revisions. All authors provided important revisions. ARMB led the antibody selection project, and the overall project PI was BW.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Sibley DR, Benovic JL, Caron MG, Lefkowitz RJ.. Phosphorylation of cell surface receptors: A mechanism for regulating signal transduction pathway. Endocr Rev. 1988;9(1):38–56. doi: 10.1210/edrv-9-1-38. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P. The origins of protein phosphorylation. Nat Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 3.Rabeh WM, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. Correction of both NBD1 energetics and domain interface is required to restore ΔF508 CFTR folding and function. Cell. 2012;148:150–63. doi: 10.1016/j.cell.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandell JW. Phosphorylation state-specific antibodies: applications in investigative and diagnostic pathology. Am J Pathol. 2003;163:1687–98. doi: 10.1016/S0002-9440(10)63525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandell JW. Immunohistochemical assessment of protein phosphorylation state: the dream and the reality. Histochem Cell Biol. 2008;130:465–71. doi: 10.1007/s00418-008-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemmon MA, Schlessinger J. cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Appella E, Anderson CW. Post‐translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–72. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 8.Danilkovitch-Miagkova and Zbar Dysregulation of Met receptor tyrosine kinase activity in invasive tumors. J Clin Invest. 2002;109(7):863–67. doi: 10.1172/JCI15418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiorillo E1, Orrú V, Stanford SM, Liu Y, Salek M, Rapini N, Schenone AD, Saccucci P, Delogu LG, Angelini F, et al. Autoimmune-associated PTPN22 R620W variation reduces phosphorylation of lymphoid phosphatase on an inhibitory tyrosine residue. J Biol Chem. 2010;285(34):26506–18. doi: 10.1074/jbc.M110.111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong CX1, Liu F, Grundke-Iqbal I, Iqbal K. Dysregulation of protein phosphorylation/dephosphorylation in Alzheimer’s disease: a therapeutic target. J Biomed Biotechnol. 2006. ;2006(3):31825. doi: 10.1155/JBB/2006/31825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F1, Zeltwanger S, Hu S, Hwang TC. Deletion of phenylalanine 508 causes attenuated phosphorylation-dependent activation of CFTR chloride channels. J Physiol. 2000;3:637–48. doi: 10.1111/j.1469-7793.2000.00637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okiyoneda T, Veit G, Dekkers JF, Bagdany M, Soya N, Xu H, Roldan A, Verkman AS, Kurth M, Simon A, et al. Mechanism-based corrector combination restores ΔF508-CFTR folding and function. Nat Chem Biol. 2013;9(7):444–54. doi: 10.1038/nchembio.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schorb W, Peeler TC, Madigan NN, Conrad KM, Baker KM. Angiotensin II-induced protein tyrosine phosphorylation in neonatal rat cardiac fibroblasts. J Biol Chem. 1994;269:19626–32. [PubMed] [Google Scholar]
- 14.Roy B, Cathcart MK. Induction of 15-lipoxygenase expression by IL-13 requires tyrosine phosphorylation of Jak2 and Tyk2 in human monocytes. J Biol Chem. 1998;273(48):32023–29. doi: 10.1074/jbc.273.48.32023. [DOI] [PubMed] [Google Scholar]
- 15.Worboys JD, Sinclair J, Yuan Y, Jørgensen C. Systematic evaluation of quantotypic peptides for targeted analysis of the human kinome. Nat Methods. 2014;11(10):1041–44. doi: 10.1038/nmeth.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davison G, Marchbank T, March DS, Thatcher R, Playford RJ. Zinc carnosine works with bovine colostrum in truncating heavy exercise-induced increase in gut permeability in healthy volunteers. Am J Clin Nutr. 2016;104(2):526–36. doi: 10.3945/ajcn.116.134403. [DOI] [PubMed] [Google Scholar]
- 17.Papaspyropoulos A, Bradley L, Thapa A, Leung CY, Toskas K, Koennig D, Pefani DE, Raso C, Grou C, Hamilton G, et al. RASSF1A uncouples Wnt from Hippo signalling and promotes YAP mediated differentiation via p73. Nat Commun. 2018;9(1):424. doi: 10.1038/s41467-017-02786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glenney JR, Zokas L, Kamps MP. Monoclonal antibodies to phosphotyrosine. J Immunol Methods. 1988;109(2):277–85. doi: 10.1016/0022-1759(88)90253-0. [DOI] [PubMed] [Google Scholar]
- 19.Bradbury AM, Plückthun A. Antibodies: validate recombinants once. Nature. 2015;520:295. doi: 10.1038/520295b. [DOI] [PubMed] [Google Scholar]
- 20.Bradbury A, Plückthun A. Reproducibility: standardize antibodies used in research. Nature. 2015;518:27–29. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- 21.Bradbury AR, Plückthun A. Getting to reproducible antibodies: the rationale for sequenced recombinant characterized reagents. Protein Eng Des Sel. 2015;28:303–05. doi: 10.1093/protein/gzv051. [DOI] [PubMed] [Google Scholar]
- 22.Schonbrunn A. Editorial: antibody can get it right: confronting problems of antibody specificity and irreproducibility. Mol Endocrinol. 2014;28:1403–07. doi: 10.1210/me.2014-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker M. Reproducibility crisis: blame it on the antibodies. Nature. 2015;521:274–76. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- 24.Bordeaux J, Welsh A, Agarwal S, Killiam E, Baquero M, Hanna J, Anagnostou V, Rimm D. Antibody validation. Biotechniques. 2010;48(3):197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campa VM, Kypta RM. Issues associated with the use of phosphospecific antibodies to localise active and inactive pools of GSK-3 in cells. Biol Direct. 2011;6:4. doi: 10.1186/1745-6150-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panse S, Dong L, Burian A, Carus R, Schutkowski M, Reimer U, Schneider-Mergener J. Profiling of generic anti-phosphopeptide antibodies and kinases with peptide microarrays using radioactive and fluorescence-based assays. Mol Divers. 2004;8:291–99. doi: 10.1023/B:MODI.0000036240.39384.eb. [DOI] [PubMed] [Google Scholar]
- 27.Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J Biol Chem. 2004;279:24957–64. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bock I, Dhayalan A, Kudithipudi S, Brandt O, Rathert P, Jeltsch A. Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics. 2011;6:256–63. doi: 10.4161/epi.6.2.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shih HH, Tu C, Cao W, Klein A, Ramsey R, Fennell BJ, Lambert M, Ni Shuilleabhain D, Autin B, Kouranova E, et al. An ultra-specific avian antibody to phosphorylated tau protein reveals a unique mechanism for phosphoepitope recognition. J Biol Chem. 2012;287:44425–34. doi: 10.1074/jbc.M112.415935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koerber JT, Thomsen ND, Hannigan BT, Degrado WF, Wells JA. Nature-inspired design of motif-specific antibody scaffolds. Nat Biotechnol. 2013;31:916–21. doi: 10.1038/nbt.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradbury ARM, Sidhu S, Dübel S, McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol. 2011;29:245–54. doi: 10.1038/nbt.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U.S.A.. 1998;85:5879–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrara F, D’Angelo S, Gaiotto T, Naranjo L, Tian H, Graslund S, Dobrovetsky E, Hraber P, Lund-Johansen F, Saragozza S, et al. Recombinant renewable polyclonal antibodies. MAbs. 2015;7:32–41. doi: 10.4161/19420862.2015.989047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glanville J, D’Angelo S, Khan TA, Reddy ST, Naranjo L, Ferrara F, Bradbury A. Deep sequencing in library selection projects: what insight does it bring?. Curr Opin Struct Biol. 2015;(33):146–60. doi: 10.1016/j.sbi.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrara F, Naranjo LA, Kumar S, Gaiotto T, Mukundan H, Swanson B, Bradbury AR. Using phage and yeast display to select hundreds of monoclonal antibodies: application to antigen 85, a tuberculosis biomarker. PLoS One. 2012;7:e49535. doi: 10.1371/journal.pone.0049535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Youssef LA, Schuyler M, Wilson BS, Oliver JM. Roles for the high affinity IgE receptor, FcϵRI, of human basophils in the pathogenesis and therapy of allergic asthma: disease promotion, protection or both? Open Allergy J. 2010;3:91–101. doi: 10.2174/1874838401003010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Getahun A, Cambie JC, ITIMs O. ITAMs, and ITAMis: revisiting immunoglobulin Fc receptor signaling. Immunol Rev. 2015;268:66–73. doi: 10.1111/imr.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benhamou M, Ryba NJ, Kihara H, Nishikata H, Siraganian RP. Protein-tyrosine kinase p72syk in high affinity IgE receptor signaling. Identification as a component of pp72 and association with the receptor gamma chain after receptor aggregation. J Biol Chem. 1993;268:23318–24. [PubMed] [Google Scholar]
- 40.Kihara H, Siraganian RP. Src homology 2 domains of Syk and Lyn bind to tyrosine-phosphorylated subunits of the high affinity IgE receptor. J Biol Chem. 1994;269:22427–32. [PubMed] [Google Scholar]
- 41.Rivera J, Gilfillan AM. Molecular regulation of mast cell activation. J Allergy Clin Immunol. 2006;117:1214–25. doi: 10.1016/j.jaci.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 42.O’Neill SK, Getahun A, Gauld SB, Merrell KT, Tamir I, Smith MJ, Dal Porto JM, Li QZ, Cambier JC. Monophosphorylation of CD79a and CD79b ITAM motifs initiates a SHIP-1 phosphatase-mediated inhibitory signaling cascade required for B cell anergy. Immunity. 2011;35(5):746–56. doi: 10.1016/j.immuni.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahajan A, Barua D, Cutler P, Lidke DS, Espinoza FA, Pehlke C, Grattan R, Kawakami Y, Tung CS, Bradbury AR, et al. Optimal aggregation of FcepsilonRI with a structurally defined trivalent ligand overrides negative regulation driven by phosphatases. ACS Chem Biol. 2014;9:1508–19. doi: 10.1021/cb500134t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furumoto Y, Nunomura S, Terada T, Rivera J, Ra C. The FcepsilonRIbeta immunoreceptor tyrosine-based activation motif exerts inhibitory control on MAPK and IkappaB kinase phosphorylation and mast cell cytokine production. J Biol Chem. 2004;279:49177–87. doi: 10.1074/jbc.M404730200. [DOI] [PubMed] [Google Scholar]
- 45.Kimura T, Kihara H, Bhattacharyya S, Sakamoto H, Appella E, Siraganian RP. Downstream signaling molecules bind to different phosphorylated immunoreceptor tyrosine-based activation motif (ITAM) peptides of the high affinity IgE receptor. J Biol Chem. 1996;271:27962–68. doi: 10.1074/jbc.271.44.27962. [DOI] [PubMed] [Google Scholar]
- 46.On M, Billingsley JM, Jouvin MH, Kinet JP. Molecular dissection of the FcRbeta signaling amplifier. J Biol Chem. 2004;279:45782–90. doi: 10.1074/jbc.M404890200. [DOI] [PubMed] [Google Scholar]
- 47.Manikandan J, Kothandaraman N, Hande MP, Pushparaj PN. Deciphering the structure and function of FcepsilonRI/mast cell axis in the regulation of allergy and anaphylaxis: a functional genomics paradigm. Cell Mol Life Sci. 2012;69:1917–29. doi: 10.1007/s00018-011-0886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkinson KD. The discovery of ubiquitin-dependent proteolysis. Proc Natl Acad Sci U S A. 2005;102(43):15280–82. doi: 10.1073/pnas.0504842102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang YS, Wang CC, Chen BH, Hou YH, Hung KS, Mao YC. Tyrosine sulfation as a protein post-translational modification. Molecules. 2015;20(2):2138–64. doi: 10.3390/molecules20022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murn J, Shi Y. The winding path of protein methylation research: milestones and new frontiers. Nat Rev Mol Cell Biol. 2017;18:517–27. doi: 10.1038/nrm.2017.35. [DOI] [PubMed] [Google Scholar]
- 51.Metzger H. The receptor with high affinity for IgE. Immunol Rev. 1992;125:37–48. doi: 10.1111/j.1600-065X.1992.tb00624.x. [DOI] [PubMed] [Google Scholar]
- 52.Metzger H, Alcaraz G, Hohman R, Kinet JP, Pribluda V, Quarto R. The receptor with high affinity for immunoglobulin E. Annu Rev Immunol. 1986;4(1):419–70. doi: 10.1146/annurev.iy.04.040186.002223. [DOI] [PubMed] [Google Scholar]
- 53.Sblattero D, Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat Biotechnol. 2000;18:75–80. doi: 10.1038/71958. [DOI] [PubMed] [Google Scholar]
- 54.Boder ET, Wittrup KD. Optimal screening of surface-displayed polypeptide libraries. Biotechnol Prog. 1998;14:55–62. doi: 10.1021/bp970144q. [DOI] [PubMed] [Google Scholar]
- 55.Wentz AE, Shusta EV. Enhanced secretion of heterologous proteins from yeast by overexpression of ribosomal subunit RPP0. Biotechnol Prog. 2008;24:748–56. doi: 10.1021/bp070345m. [DOI] [PubMed] [Google Scholar]
- 56.Wentz AE, Shusta EV. A novel high-throughput screen reveals yeast genes that increase secretion of heterologous proteins. Appl Environ Microbiol. 2007;73:1189–98. doi: 10.1128/AEM.02427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson BS, Kapp N, Lee RJ, Pfeiffer JR, Martinez AM, Platt Y, Letourneur F, Oliver JM. Distinct functions of the Fc epsilon R1 gamma and beta subunits in the control of Fc epsilon R1-mediated tyrosine kinase activation and signaling responses in RBL-2H3 mast cells. J Biol Chem. 1995;270:4013–22. doi: 10.1074/jbc.270.8.4013. [DOI] [PubMed] [Google Scholar]
- 58.Cirino CK, Mayer KM, Umeno D. Generating mutant libraries using error-prone PCR. Directed Evol Lib Creation. 2003. doi: 10.1385/1-59259-395-X:3. [DOI] [PubMed] [Google Scholar]
- 59.Zhou Y, Drummond DC, Zou H, Hayes ME, Adams GP, Kirpotin DB, Marks JD. Impact of single-chain Fv antibody fragment affinity on nanoparticle targeting of epidermal growth factor receptor-expressing tumor cells. J Mol Biol. 2007;371(4):934–47. doi: 10.1016/j.jmb.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weaver-Feldhaus JM, Miller KD, Feldhaus MJ, Siegel RW. Directed evolution for the development of conformation-specific affinity reagents using yeast display. Protein Eng Des Sel. 2005;18(11):527–36. doi: 10.1093/protein/gzi060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


