Abstract
One fundamental, but understudied mechanism of gene regulation in disease is allele specific expression (ASE), the preferential expression of one allele. We leveraged RNA-seq from human brain to assess ASE in autism spectrum disorder (ASD). When ASE is observed in ASD, the allele with lower population frequency (minor allele) is preferentially more highly expressed than the major allele, opposite than the canonical pattern. Importantly, genes showing monoallelic expression (MAE) in ASD are enriched in those down-regulated in ASD postmortem brain and in genes harboring de novo mutations in ASD. Two regions, 14q32 and 15q11, containing all known orphan C/D box snoRNAs, are particularly enriched in shifts to higher minor allele expression. We demonstrate that this allele shifting enhances snoRNA targeted splicing changes in ASD-related target genes in idiopathic ASD and 15q11-q13 duplication syndrome. Together, these results implicate allelic imbalance and dysregulation of orphan C/D box snoRNAs in ASD pathogenesis.
Autism spectrum disorder (ASD) is defined by deficits in social interaction and repetitive behaviors1,2 and has a global prevalence of approximately 1 case per 68 people1. Although genetic factors contribute to a substantial proportion of disease liability the genetics of ASD are complex with contributions from both rare and common alleles, including de novo mutations and polygenic inheritance2. Despite this heterogeneity and polygenicity, we have previously identified a reproducible shared pattern of gene expression alterations in human postmortem cortex from subjects with idiopathic ASD3 and replicated this signal in independent subjects with idiopathic ASD and dup15q4. This gene expression signal may be mediated by epigenetic factors5,6, such as DNA methylation7 or histone modification6, in addition to underlying genetic variation.
Allele specific expression (ASE) is a form of genetic regulation in which expression of mRNA at a specific locus is biased towards a specific allele8–10. An extreme case of ASE is genomic imprinting, whereby a particular allele is completely silenced8,11. But, most ASE represents more subtle shifts in the allelic ratio rather than complete silencing of one allele9,12 as evidenced by the observations of preferential reduced expression of mutant disease alleles, and a substantial proportion of cis-acting expression quantitative trait loci (cis-eQTL) are mediated by ASE12–14. Indeed, cis-eQTL and ASE are complementary mechanisms involved in the regulation of gene expression15. One advantage of analyzing ASE in patient tissues is that it is less influenced by genetic, environmental, and technical confounders because ASE relies primarily on within-subject expression comparisons, rather than comparisons between subjects15,16.
A number of diseases can be attributed to abnormalities in ASE17,18. For example, Prader-Willi syndrome (PWS) and Angelman syndrome (AS) are rare neurodevelopmental disorders resulting from deletions within the highly-imprinted region, 15q11-q13. Conversely, recurrent maternally-derived duplications of this region lead to dup15q, a rare, but penetrant syndromic form of ASD comprising approximately 1% of cases19. Although duplications generally increase gene expression19, varied patterns are observed across genes in the region impacted by dup15q due to complex local regulatory mechanisms4,19–21.
In addition to known imprinted regions, ASE extends to over 5% of autosomal genes and is especially enriched in transmembrane receptors and cell surface molecules9. When not associated with parent-of-origin imprinting, such monoallelic expression (MAE) is random MAE (RMAE), which may be important for cell-type specific gene dosage effects10,22. Moreover, emerging evidence suggests that RMAE is involved in neurodevelopmental disorders, as recently observed for developmental dyspraxia caused by RMAE in FOXP223. In this case, ASE may contribute to disease susceptibility by exacerbating the deleterious effects of a mutation via haploinsufficiency, or mosaic somatic expression.
Except for one notable study, which showed that ASE in several specific genes may play a role in a subset of ASD cases8, the role of genome-wide ASE has not been explored in a sufficiently powered cohort. The overall paucity of ASE studies in brain disorders, such as ASD, is likely due to several challenges including limited access to human brain tissue, small sample sizes, incomplete transcriptome-wide genotyping data, and reference bias in mapping transcripts8,12,13,24,25. Reference bias is attributable to reduced mapping of alternative-allele-containing RNA-seq reads (due to a greater number of mismatches) and can generate up to 40% false positive signals if not properly addressed9,13,25. However, when these issues are properly addressed, ASE can be a highly robust approach to investigate the relationship between epigenetic variation and gene expression underlying diseases15. Here, we applied an optimized ASE pipeline in a relatively large sample of postmortem brains from idiopathic ASD and control subjects. We found that most loci showing ASE are shared between ASD and control, which is contrast to ASE between the two brain regions, cerebral cortex and cerebellum, which shows large differences24. A small proportion of loci do manifest significant reproducible ASE changes between ASD and controls, which may represent a novel form of genetic regulation related to disease pathogenesis.
Results
ASE identification in brain
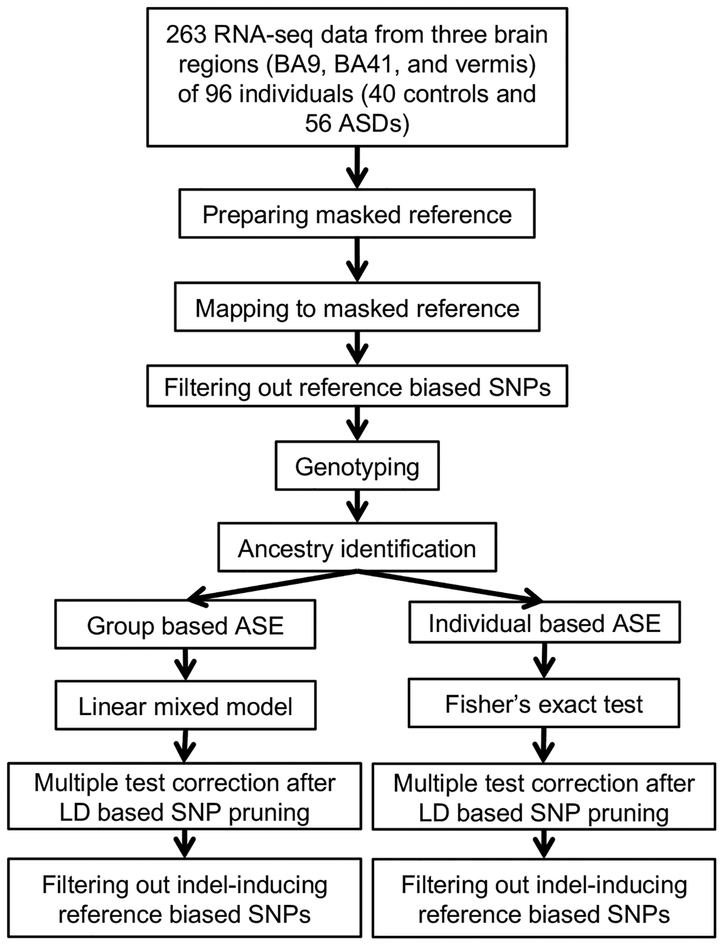
We first developed an optimized ASE identification pipeline (Fig. 1; Methods) to overcome potential challenges, such as reference bias in mapping transcripts, lack of transcriptome-wide genotyping data, and allelic expression bias introduced by ethnicity. We collected 263 postmortem brain tissue samples from frontal cortex (Brodmann area (BA)9), temporal cortex (BA41-42-22 (BA41)), and cerebellar vermis from 96 individuals (40 controls and 56 ASD cases including 8 cases with dup15q). Gene expression profiling was performed using RNA-seq4, and genotyping was conducted on arrays (Methods).
Figure 1.
A schematic of the pipeline for analysis of ASE. RNA-seq data were generated from 263 brain samples across three brain regions from subjects with 56 ASDs and 40 controls. After preparing a masked reference file, RNA-seq data were mapped, and reference biased SNPs were removed (Methods). Genotyping data from SNP array, imputation, and RNA-seq based genotyping were used for ancestry identification and heterozygous SNP identification for ASE investigation. Group based ASE was investigated across case and control groups using a linear mixed model within each brain region separately. Individual based ASE analysis was performed within each individual sample using Fisher’s exact test. The results underwent LD pruning followed by multiple test correction (Bonferroni). As post hoc analysis, we filtered out reference biased SNPs generated by proximal indels.
To overcome reference bias, we generated a novel reference genome masked for single nucleotide polymorphisms (SNPs) that could contribute to mapping bias. We compiled ~40 million possible variants based on the union of SNPs called from RNA-seq data (Supplementary Fig. 1a) and SNPs already identified from microarray probe sets and the 1000 Genomes26,27 (Methods). We prepared a custom human reference genome after masking these SNPs (Methods). Using this masked reference, we identified and removed 11.13% of SNPs that showed biased mapping from a simulated dataset18 (Methods). To collect transcriptome wide genotyping data from our brain samples, we performed imputation from genotyping array and RNA-seq based genotyping (Methods; Supplementary Fig. 1b and 1c), and ancestry was identified for all samples (Methods; Supplementary Fig. 1d).
For investigation of group-level ASE patterns across cases and brain regions, 1,163,249 heterozygous SNPs in 21,929 genes were used following quality control (Methods), and their significance was quantified using a linear mixed model to account for sample variation and covariates (Supplementary Table 1; Methods). The values of excluded covariates showed balanced distributions between control and ASD (Supplementary Fig. 1e; Methods).
We filtered out all SNPs showing ancestry-associated variation to avoid any potential effects of population stratification (Methods). ASE was also assessed within each individual sample for quality-controlled SNPs using Fisher’s exact test (Methods). Both individual and group-level ASE studies underwent multiple testing correction for independent SNPs after linkage disequilibrium (LD)-based SNP pruning (Methods). Among the alleles showing ASE, we also filtered out reference biased SNPs related to proximal indels (Methods). As expected, SNPs within the same haplotype blocks show similar allelic expression patterns (Supplementary Fig. 1f and 1g).
Common ASE patterns in brain tissues
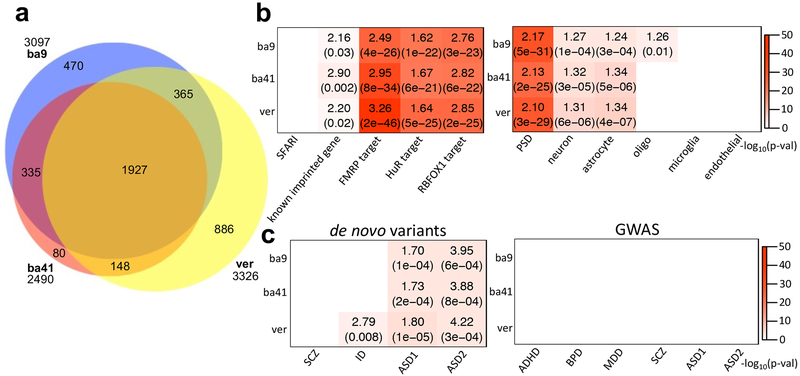
For external validation, we performed two analyses. First, we compared genes with evidence of ASE with those in dbMAE, an integrated database compiled from ASE analyses across human and mouse tissues22. Although this database contains data from mostly non-neural tissues, genes manifesting ASE in our study showed significant overlap with those in dbMAE (Supplementary Fig. 2a). Furthermore, although assessed separately in each brain region, there was strong concordance in common ASE genes between regions in our data (Fig. 2a and Supplementary Table 2). For example, the overlap between genes showing ASE in the two cortical regions, BA9 and BA41 is extremely high (odds ratio (OR)=221.00 and p-value<2.2e-16; Supplementary Fig. 2b), whereas the degree of overlap between two more developmentally and evolutionarily distinct brain regions, cortex and cerebellum, is substantial, but lower than in the two more similar cortical regions (between BA9 and vermis: OR=49.00 and p-value<2.2e-16; between BA41 and vermis: OR=72.54 and p-value<2.2e-16). This is consistent with recent GTEx data analysis showing that the cerebellum has a distinct ASE pattern relative to other brain tissues24.
Figure 2.
ASE patterns shared among cases and controls. (a) Venn diagram comparing the overlap of ASE genes from three brain tissues, BA9, BA41, and vermis. (b) Enrichment analyses of the ASE genes with ASD-relevant (SFARI gene list36; Methods) and cell-type specific gene lists (Methods)31. Across all three brain regions, ASE genes showed strong overlap with known imprinted genes (Methods) as well as targets of FMRP28, HuR29, and RBFOX130. Plot showed ORs and FDR corrected p-values for enrichment, if significant. (c) Gene set enrichment study of ASE genes with risk variants in psychiatric disease dataset. From de novo variant datasets45,33, we considered SCZ, intellectual disorder (ID), and ASD. The dataset for SCZ, ID, and ASD1 were gene lists containing de novo likely gene disrupting mutations from the previous study of Iossifov et al.45, and the data for ASD2 represents risk genes integrating de novo copy number variations (FDR≦0.01) from the study of Sanders et al.33 (Methods). The risk variants from GWAS34,35 were considered for ADHD, BPD, MDD, SCZ, and ASD (Methods). Here, ASD1 and ASD2 represent the GWAS datasets from the Cross-Disorder Group of the Psychiatric Genomics34 and Grove et al.35, respectively. If significant, the plot shows ORs and FDR corrected p-values for de novo variant datasets and FDR corrected p-values for GWAS. The sample numbers of BA9, BA41, and vermis are 67, 64, and 64, respectively.
For further external validation, we applied the specific methods and pipeline used in the primary analysis of GTEx24 (Supplementary Fig. 2c). Both the GTEx pipeline and our modified approach showed similar patterns of distinct ASE between the cortex and cerebellum. In the GTEx data, the mean of ASE rates from the two cerebral cortical regions and cerebellum show significant correlation with our data (R2=0.9995 and p-value=0.0098; Methods).
The distribution of expression of imbalanced alleles was significantly shifted lower than balanced alleles (Kolmogorov-Smirnov (KS) test, p-value<2.0e-16, the means of log2(RPSM): 0.37 vs 0.71; Supplementary Fig. 2d). ASE was also enriched at known imprinted genes including the targets of RNA binding proteins, FMRP28, ELAVL1 (HuR)29, and RBFOX130 (Fig. 2b; Methods). Genes showing evidence for ASE were also preferentially enriched for neuronal31 and astrocyte markers31 (Fig. 2b and Supplementary Fig. 2e). Consistent with a strong neuronal signal, they were also enriched for genes encoding proteins of the postsynaptic density (PSD)32, similar to previous observations9. Remarkably, across all three brain regions, ASE genes are enriched for genes previously shown to be down-regulated in postmortem brain from patients with ASD4 (Supplementary Fig. 2f). In cerebral cortex, genes down-regulated in ASD postmortem brain were enriched among those showing ASE. Those up-regulated in ASD were depleted in those showing ASE; 37% of down-regulated genes were subject to ASE. In cortex, genes up-and down-regulated in ASD were 448 and 503, respectively. However, among ASE genes, 84 and 181 genes were respectively up-and down-regulated in ASD (OR=1.92 and p-value=7.03e-06). Both ASE genes and those down-regulated in ASD are involved in synaptic transmission and vesicular transport4, consistent with their enrichment in PSD and targets of FMRP28, HuR29, and RBFOX1 genes previously implicated in ASD4. Importantly, genes showing ASE also showed significant enrichment in genes harboring de novo ASD risk variants identified in two different studies32,33 (Fig. 2c; Methods). However, genes manifesting ASE did not show any significant enrichment with GWAS data from several major psychiatric diseases including attention-deficit/hyperactivity disorder (ADHD)34, bipolar disorder (BPD)34, major depression disorder (MDD)34, schizophrenia (SCZ)34, and ASD34,35.
ASE patterns shared across idiopathic ASD cases and controls
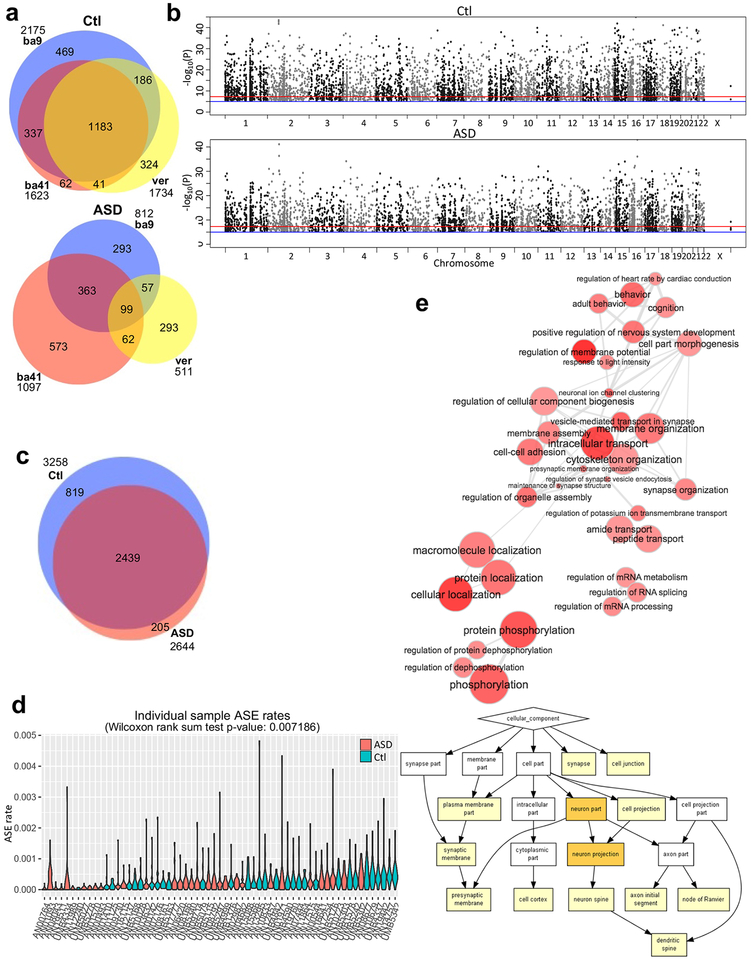
Similar to controls, idiopathic ASD cases showed tissue specific ASE across the three brain regions (Fig. 3a, Supplementary Fig. 3a, and Supplementary Fig. 3b). Since the two cortical regions showed strong overlap in ASE and cortical manifestations of transcriptional dysregulation in ASD are more prominent than those in cerebellum4, we focused on the cerebral cortex for most of the subsequent case-control comparisons. We found that SNPs showing ASE are broadly and evenly distributed throughout the genome in both control and idiopathic ASD (Fig. 3b). The majority of genes exhibiting ASE (70.41%) are shared across the case and control groups providing further confidence in the reproducibility of these results (Fig. 3c and Supplementary Table 3). Nevertheless, fewer genes showing ASE are observed in idiopathic ASD compared to controls (Fig. 3c), and this was recapitulated when studying the rate of ASE at the level of each individual sample (Fig. 3d).
Figure 3.
ASE patterns distinguish control and idiopathic ASD brain. (a) The comparison of ASE genes from BA9, BA41, and vermis within control and idiopathic ASD groups. (b) Manhattan plots for control and idiopathic ASD cortex show broad distribution of genomic-loci exhibiting ASE. Sample numbers of control and idiopathic ASD brains are 69 and 62, respectively. (c) Venn diagram showing comparison of ASE genes across groups in cortex. (d) ASE rates for individual samples from ASD and control groups (Methods). Idiopathic ASD samples (n=32) show lower overall rates of ASE compared with controls (n=31). For the violin plots, ASE rates were calculated per chromosome form each cortical sample (Methods). The Wilcoxon rank sum test was two-sided. (e) GO analyses are shown for common ASE genes between control (n=69) and idiopathic ASD groups (n=62). At the above interactive graph, the bubble color indicates the p-value of the GO term, and bubble size indicates its frequency49. The p-values and other results of the GO analysis are at Supplementary Table 4. Highly similar GO terms are linked by edges, and the line width indicates the degree of similarity. At the below figure, the white, yellow, and orange boxes represent p-value>10−3, 10−5<p-value≦10−3, and 10−7<p-value≦10−5, respectively.
Comparing biological pathway enrichment in cases and controls revealed overlap and differences in ASE gene function. Genes exhibiting ASE in both idiopathic ASD and control are enriched in phosphoprotein, RNA splicing, and neuron projection pathways (Fig. 3e and Supplementary Table 4). Genes exhibiting control-specific ASE are significantly enriched for pathways related to growth cone and synapse (Supplementary Fig. 3c and Supplementary Table 4). Though there are fewer idiopathic ASD-specific ASE genes than control-specific ASE genes, those showing idiopathic ASD-specific ASD are significantly enriched for pathways involved in transport (Supplementary Fig. 3d and Supplementary Table 4). Genes exhibiting ASE in both idiopathic ASD and controls show significant enrichment in genes harboring de novo ASD risk variants (Supplementary Fig. 3e; OR=1.68, p-value=5e-04 for ASD1; OR=3.83, p-value=0.001 for ASD2). However, control-specific and idiopathic ASD-specific ASE genes do not show any enrichment with either the rare de novo ASD risk variants or common variants from GWAS data (Supplementary Fig. 3e).
Quantitative allelic imbalance in idiopathic ASD
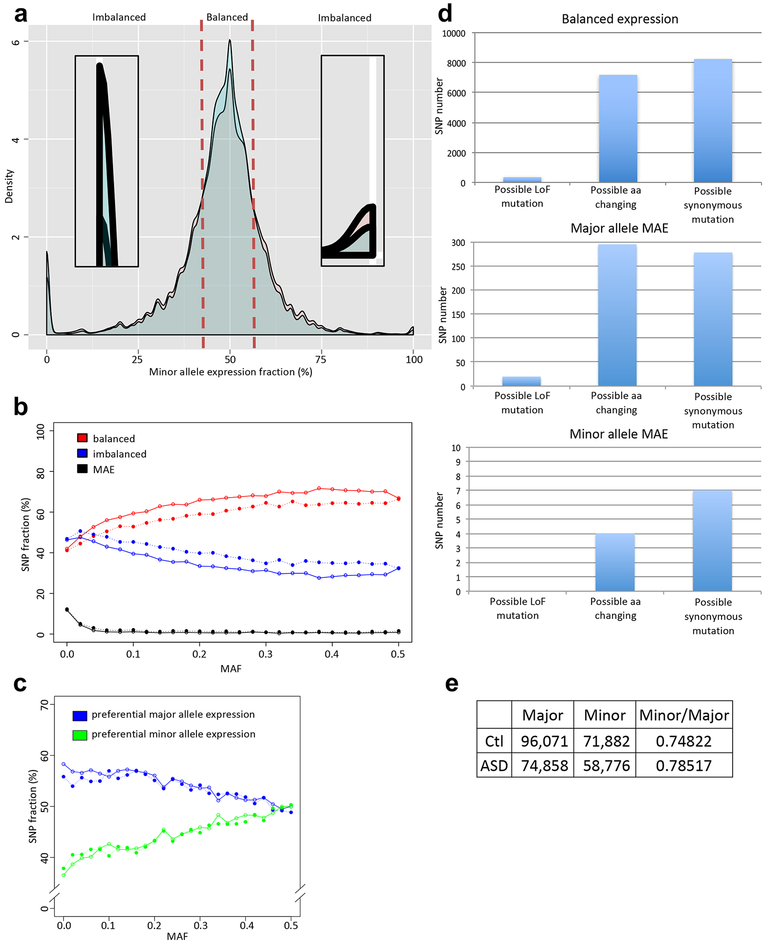
ASE is not always completely monoallelic and may involve subtle shifts in allelic balance, whereby one allele is quantitatively favored over the other8,15. To investigate this full spectrum of ASE in control and idiopathic ASD, we compared the distributions of the minor allele expression fraction for SNPs in which these calls are high quality (Methods; Fig. 4a). As expected, the majority of heterozygous alleles show no evidence of ASE (herein called “balanced”) with the fraction near 50%. A much smaller number of alleles show complete MAE and accordingly their fractions are either 0% or 100%. Finally, we define “imbalanced” alleles as those that lie between the balanced and MAE distribution ranges. Controls had a higher density of balanced alleles than ASD cases while idiopathic ASD cases showed more imbalanced alleles (Supplementary Fig. 4a and Fig. 4a). However, controls showed significantly more major allele MAE for low frequency alleles (minor allele frequency (MAF)<0.05) compared to idiopathic ASD (Supplementary Fig. 4b). These patterns were recapitulated in the pseudoautosomal region (PAR) of the chromosome X (Supplementary Fig. 4c).
Figure 4.
Quantitative allelic imbalance in idiopathic ASD and control cortex. (a) The distribution of minor allele expression fraction for idiopathic ASD (red) and control (blue) groups for autosomal SNPs. The majority of loci show balanced expression. The density plot near 0% and 100% are zoomed in to show patterns of monoallelic expression. (b) SNP fractions of balanced, imbalanced, or MAE expression per MAF. (c) SNP fractions showing preferential major or minor allele expression per MAF. At (b) and (c), open circles are for control, and close circles are for idiopathic ASD. (d) The comparison of SNP numbers, which possibly can cause LoF mutation, amino acid change, or synonymous mutation at control cortex. Major allele MAE has a role to prevent deleterious LoF and missense mutations. Y-axes show SNP numbers. (e) SNP counts showing preferential expression of the major and minor alleles in control and idiopathic ASD. Their ratio is expressed as the Minor/Major.
We next investigated whether the differential ASE pattern was related to the MAF genome-wide (Fig. 4b and Supplementary Fig. 4b). The disease specific patterns (balanced alleles: control > idiopathic ASD; imbalanced alleles: control < idiopathic ASD) were largely stable across the range of MAFs. However, we observed that complete MAE was strictly limited to rare alleles. Furthermore, preferential major allele expression becomes stronger as the minor allele frequency decreases (Fig. 4c). Major alleles exhibiting MAE are enriched in genes that are more tolerant of loss of function12 (LoF; Methods) and missense mutations compared to those showing balanced expression (chi-square test p-value=0.0134; Fig. 4d). This suggests that a bias towards the major allele expression may provide a buffer against the consequences of potentially deleterious mutations. Remarkably, this preferential pattern of major allele expression shows a significant interaction with idiopathic ASD (Fig. 4e). On average, minor alleles are preferentially more highly expressed in idiopathic ASD than in controls (Minor/major allele ratio: 0.79 in ASD and 0.75 in control; chi-square test p-value=7.33e-11). Together, these results indicate that rare and potentially pathogenic alleles are more likely to be unmasked in idiopathic ASD brain by the patterns of ASE.
Monoallelic expression occurs primarily from the major allele
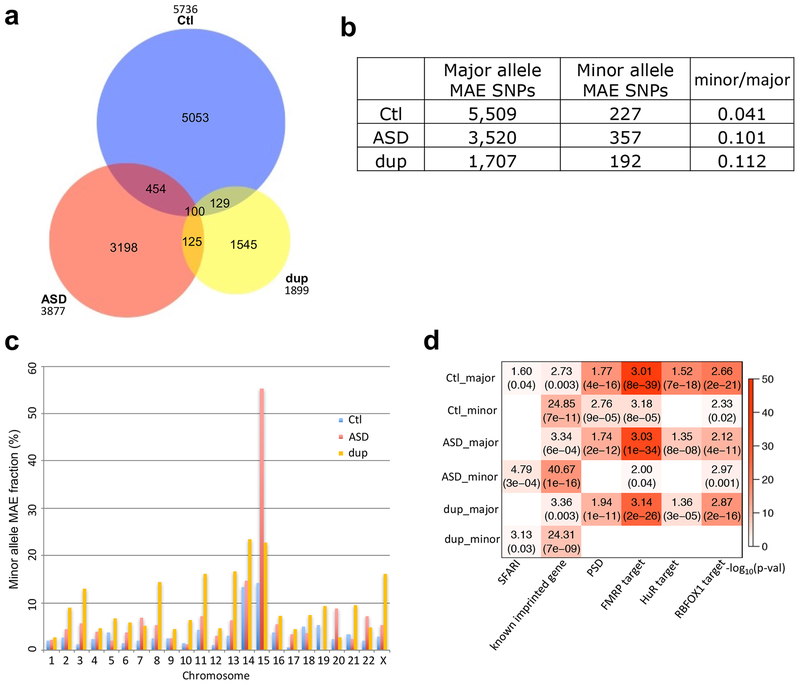
Overall, we observed a strong bias towards expression of the major allele for sites in genes harboring rare minor alleles and MAE alleles. However, unlike global ASE patterns (Fig. 3c) MAE patterns differed between cases and controls (Fig. 5a). Although MAE SNPs were observed across the genome (Supplementary Fig. 5), there was an approximately 2-fold enrichment on chromosome 15 harboring a number of known imprinted loci (Supplementary Table 5). Reasoning that this could provide a convergent mechanism underlying the co-occurrence of dup15q syndrome and idiopathic ASD, we next investigated MAE in 8 cases of dup15q. We observed a substantial enrichment of minor allele MAE in ASD and dup15q compared to controls (Fig. 5b; 2.5 and 2.7fold, respectively). This genome-wide enrichment pattern persisted when SNPs were analyzed at individual genes (Supplementary Fig. 6a and 6b).
Figure 5.
MAE SNPs across control, ASD, and dup15q groups. (a) Venn diagram showing overlap of MAE SNPs across groups. Less than 15% of MAE SNPs were overlapped between cases and controls. (b) The number of major allele MAE SNPs and minor allele MAE SNPs across groups. Both chi-square test p-values in ASD and dup15q compared to control are <2.2e-16. (c) Minor allele MAE fraction (minor / (major + minor)) across chromosomes for control, ASD, and dup15q. (d) Gene set enrichment for major and minor allele MAE genes in control (n=69), ASD (n=62), and dup15q (n=15) groups (Methods). Enrichment was assessed for known ASD risk genes (SFARI36; Methods), known imprinted (Methods), PSD32, FMRP target28, HuR target29, and RBFOX1 target genes30. Enrichment was assessed separately for genes showing major allele and minor allele MAE. Dup is for dup15q.
Overall, minor allele predominant MAE was significantly enriched on chromosomes 14 and 15 (Fig. 5c) with ASD and dup15q showing stronger patterns compared to controls. Compared to chromosome 14, chromosome 15 exhibited a substantially higher minor allele MAE fraction in both ASD and dup15q than controls (3.9- and 1.6-fold, respectively; chi-square test p-values<2.2e-16 and 0.0184, respectively; Fig. 5c and Supplementary Table 5). Chromosomes 3, 6, and 7 also showed higher fractions of minor allele MAE in ASD and dup15q subjects than controls, of which only chromosome 3 was significant in both (4.2- and 9.8-fold, respectively; chi-square p-values=0.0022 and 8.89e-08, respectively). However, we focused on the chromosome 14 and 15 for further analysis since chromosome 3 had an overall lower degree of minor allele MAE.
Pathway analysis of minor allele MAE genes showed strong enrichment for known imprinted genes, as expected (Fig. 5d). Interestingly, these MAE genes were enriched for distinct pathways from typical ASE genes that favor major allele expression (Fig. 2b) suggesting that orthogonal biological processes are involved. For example, genes showing minor allele MAE showed no overlap with astrocyte markers31, PSD32, or ASD-specific down-regulated genes4 (Supplementary Fig. 6c). However, they did show enrichment for genes harboring known rare mutations increasing risk for ASD (SFARI genes36; Methods) whereas other classes of genes showing ASE did not. Like genes that manifest ASE (Fig. 2c), major allele MAE genes show significant enrichment with genes associated with the rare de novo ASD risk variants (Supplementary Fig. 6d). However, MAE genes in control and ASD showed some enrichment with common variants from GWAS data of BPD and SCZ (Supplementary Fig. 6d).
Allele shift regions enriched in minor allele MAE in ASD and dup15q and snoRNAs
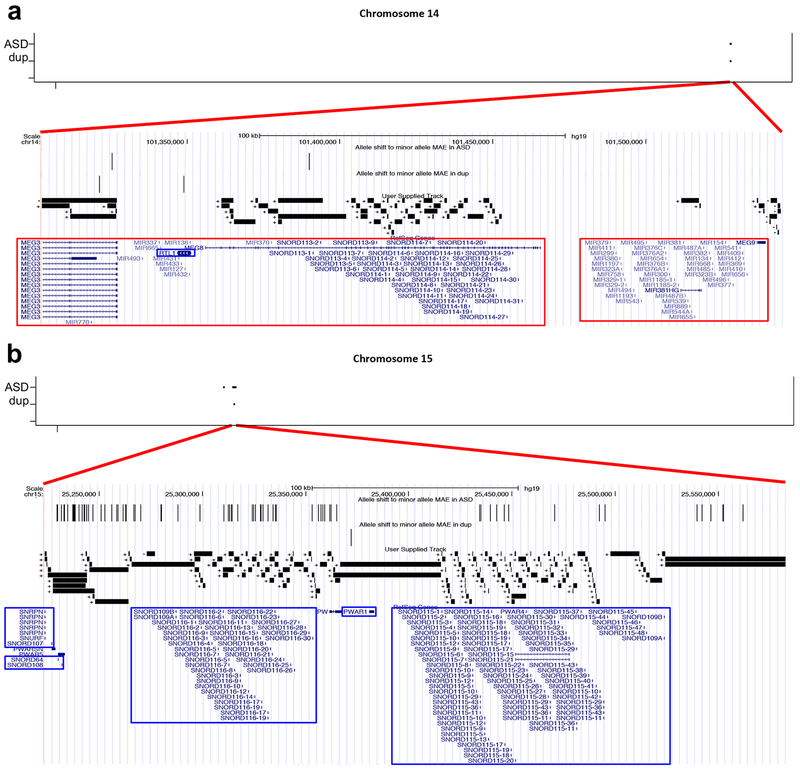
We have so far demonstrated that ASD and dup15q are associated with an increased rate of genes showing minor allele MAE that are enriched on chromosomes 14 and 15 and overlap known ASD risk genes, especially those harboring deleterious mutations that act via haploinsufficiency. Interestingly, the genomic regions showing MAE allele shifts from major to minor allele in both ASD and dup15q relative to controls are located on just a few chromosomes (Table 1a and 1b) and overlap regions known to harbor ASD-associated CNVs37. Among commonly existing MAE alleles, the frequencies of the allele shift in ASD and dup15q are 15.52% and 2.62%, respectively. To understand the effect of this apparent MAE allele shift in ASD, we compiled a set of high confidence genomic regions which we called “allele shift rich regions”. We defined these regions as having such allele shifts in both ASD and dup15q relative to controls. This analysis identities only two genomic regions, 14q32 (chr14:101302,638–101,544,745) and 15q11 (chr15:25,223,730–25,582,395), which are known imprinted loci that have many small splice junctions (Fig. 6) and show continuous high expression (Supplementary Fig. 7) as single transcript units38–40. Both regions also contain tandem repeats of multiple orphan C/D box snoRNA genes39,40, which are located within repeated introns of MEG840, SNURP-SNFPN, and SNHG14 transcripts. Indeed, all known orphan C/D box snoRNA genes are located within these two regions, including SNORD113 and SNORD114 at 14q32 (Fig. 6a), and SNORD64, SNORD107, SNORD108, SNORD109, SNORD115, and SNORD116 at 15q11 (Fig. 6b). These snoRNA genes are highly expressed in the brain39,40 and control alternative splicing of specific target genes without complementarity to rRNA within their sequences39–41.
Table 1.
Allele shift regions to minor allele MAE in ASD (a) and dup15q (b) and previously reported relevant ASD related mutations. If there are more than two allele shifts, we defined its boundaries using their maximum and minimum coordinates.
| Coordinate | Region | Size (bp) | SNP # | Previous ASD report |
|---|---|---|---|---|
| chr3:66,429,475 | 3p14.1 | 1 | 1 | 3p14.1 de novo microdeletion |
| chr14:101,325,640–101,390,093 | 14q32.2 | 64,454 | 2 | 14q32.2 duplication |
| chr15:23,889,739–25,561,958 | 15q11.2 | 1,672,220 | 83 | 15q11.2-q13.1 duplication |
| Coordinate | Region | Size (bp) | SNP # | Previous ASD report |
|---|---|---|---|---|
| chr2:207,173,390–207,175,070 | 2q33.3 | 1,681 | 2 | 2q33.3-q34 interstitial deletion, 2q32.3-q37.3 duplication |
| chr11:2,690,293 | 11p15.5 | 1 | 1 | 11p15.5-p15.4 duplication |
| chr14:101,321,515–101,349,017 | 14q32.2 | 27,503 | 2 | 14q32.2 duplication |
| chr15:25,372,247 | 15q11.2 | 1 | 1 | 15q11.2-q13.1 duplication |
Figure 6.
Allele shift rich regions at ASD and dup15q. (a) and (b) The allele shift rich regions on chromosome 14 (chr14:101,302,638–101,544,745; 242,108bp) (a) and on chromosome 15 (chr15:25,223,730–25,582,395; 358,666bp) (b). Chromosomal locations of allele shifting to minor allele MAE are shown on top. Allele shift to minor allele MAE in ASD and dup15q tracks shows the allele shifts. We visualized all splice junctions identified from RNA-seq mapping data and their chromosomal directions, which are shown with + and −. The RefSeq Genes model is shown on the bottom, indicating known imprinted genes, maternally (outlined in red rectangles) and paternally (outlined in blue rectangles) expressed genes. Dup represent dup15q.
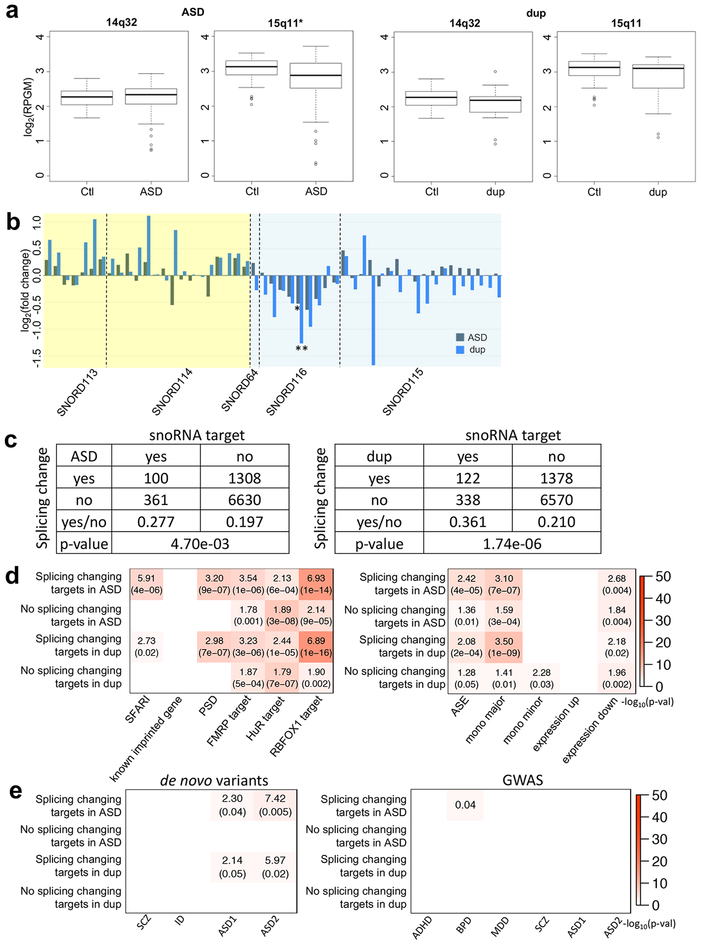
To begin to understand the impact of the allele shift at the regions, we next assessed whether they were differentially expressed in ASD or dup15q cases versus controls. Analysis of RNA-seq data showed regional downregulation of 15q11 in ASD (p-value=0.0016; Fig. 7a). SNORD116–24 downregulation was observed in ASD and dup15q from small non-coding RNA-seq data (sncRNA-seq) data42 (p-values=0.0347 and 0.0019, respectively; Fig. 7b). As snoRNA genes regulate the splicing of downstream targets, allele shifts would change potential binding and may alter the splicing patterns of snoRNA targets. To test this hypothesis, we used a known splicing target list41 of the snoRNAs to investigate whether these genes show splicing changes based on RNA-seq data from postmortem ASD brain4. The accuracy of these splicing target predictions has been confirmed in previously published studies43,44 providing experimentally validated snoRNA targets including some studies in neuronal cells43. From receiver operating characteristic analysis44, the splicing target prediction showed a 90–100% true positive rate within a 0.02–0.3% false positive rate range. Indeed, we find that the snoRNA target genes show more splicing changes in ASD and dup15q compared with controls (Fisher’s exact tests: OR=1.40 and 1.72, respectively; Fig. 7c). The snoRNA targeting sites were located significantly proximal to alternatively spliced junctions of their target genes in ASD (Supplementary Fig. 8a), and their splicing changes show significant correlations with the expression changes of their specific snoRNAs (Supplementary Fig. 8b and 8c). When one snoRNA has two different targets, their splicing changes also show strong correlation (Supplementary Fig. 8d).
Figure 7.
The characterization of the allele shift rich regions in ASD and dup15q. (a) Regional expression changes of the common allele shift rich regions in ASD and dup15q. The X axes are log2(reads per kilobase of gene model per million mapped reads) (log2(RPGM)), and two tailed unpaired t-test p-values are 0.6397, 0.0016, 0.1538, and 0.1525 for 14q32 in ASD, 15q11 in ASD, 14q32 in dup15q, and 15q11 in dup15q, respectively. Significantly down-regulated regions are marked with an asterisk. 14q32 in ASD also has less mean than control like the others. The minimum, 1st quantile, median, 3rd quantile, and maximum values of the boxplots show at 14q32 (control: 1.671, 2.048, 2.270, 2.442, and 2.806; ASD: 0.7351, 2.0643, 2.3355, 2.5048, and 2.9375; dup15q: 0.9286, 1.8464, 2.1859, 2.2984, and 3.0108, respectively) and 15q11 (control: 2.039, 2.890, 3.130, 3.302, and 3.524; ASD: 0.3255, 2.5385, 2.8784, 3.2264, and 3.7170; dup15q: 1.114, 2.537, 3.105, 3.208, and 3.430, respectively). (b) snoRNA gene expression changes in ASD and dup15q. Yellow and blue backgrounds indicated 14q32 and 15q11, respectively. Among snoRNA genes, 51 genes (RPKM≧1) are selected. From linear mixed model based differential expression gene study, significantly down-regulated SNORD116–24 genes (ASD: p-value=0.0347; dup15q: p-value=0.0019) are marked with asterisks (*: p-value≦0.05; **: p-value≦0.001). (c) The number of snoRNA target genes and genes with splicing changes in ASD and dup15q. Two-sided Fisher’s exact test p-values were shown at the bottom of tables. (d) Gene set enrichment analysis for snoRNA target genes. Among snoRNA target genes, we compared splicing change and the other genes in ASD and dup15q brain. The labels “mono major” and “mono minor” are major and minor allele MAE genes, respectively. The labels “expression up” and “expression down” represent significantly up- and down-regulated genes in idiopathic ASD4. (e) Gene set enrichment study of snoRNA target genes with risk variants in psychiatric diseases. For de novo variant datasets45,33, SCZ, ID, and ASD1 gene lists were de novo likely gene disrupting mutations45, and ASD2 represents ASD risk genes integrating de novo copy number variations (FDR≦0.01)33 (Methods). Plot showed ORs and the p-values if significant. The GWAS datasets were considered for ADHD34, BPD34, MDD34, SCZ34, and ASD34,35. Here, among ASD GWAS datasets, ASD1 and ASD2 represent the Cross-Disorder Group of the Psychiatric Genomics34 and Grove et al.35, respectively. If significant, the plots show FDR corrected p-values for GWAS (Methods). Dup is dup15q patients. For (a), (b), (d), and (e), RNA-seq sample numbers of control, ASD, and dup15q are 69, 62, and 15, respectively.
Furthermore, among snoRNA targets, genes showing altered splicing changes in ASD and dup15q are significantly enriched for known ASD risk genes (SFARI) and genes encoding PSD proteins compared with the other target genes without splicing changes (Fig. 7d; Methods). These results are consistent with the bioinformatic predictions above and potentially implicate disruption of snoRNA-mediated splicing of synaptic ASD risk genes in the pathophysiology of ASD. Moreover, the shared patterns of MAE allele shifting in ASD and dup15q provide a potential convergent biological mechanism linking idiopathic ASD and dup15q syndrome. Like ASE genes (Fig. 2c), the splicing changing snoRNA targets in ASD and dup15q showed significant enrichment with two different datasets of de novo ASD risk variants (in ASD, OR=2.30 (p-value=0.04) for the first and OR=7.42 (p-value=0.005) for the second; in dup15q, OR=2.14 (p-value=0.05) for the first and OR=5.97 (p-value=0.02) for the second; Fig. 7e).
Discussion
This study provides the first large-scale, genome-wide investigation of ASE patterns in human brain samples from subjects with ASD and dup15q. We identify different patterns of ASE in ASD than in controls including overall fewer sites showing ASE in ASD. However, when ASE does occur, ASD subjects show a preferential minor allele predominance, rather than the usual pattern of major allele predominance particularly in instances of pure monoallelic expression. Genes exhibiting ASE were also enriched in genes that harbor de novo ASD risk variation, consistent with the expectation that such genes could be highly dosage sensitive. It follows from this that ASE could increase ASD risk from the de novo mutations in highly dosage sensitive genes. Moreover, since snoRNA target genes showing splicing changes are enriched in genes showing ASE and de novo ASD risk variation, snoRNA mediated splicing changes could enhance ASD risk in dosage sensitive genes that harbor de novo risk variants. In ASD, loci showing minor allele MAE were enriched for known ASD-risk genes and the post-synaptic density genes. Furthermore, we identified two orphan C/D box snoRNA rich regions at 14q32 and 15q11 strongly enriched for ASD-related MAE change. These loci were also enriched for minor allele MAE in dup15q demonstrating convergence between known genetic risk factors and DNA methylation changes in brain. The allele shift towards minor allele MAE in ASD highlights the importance of understudied mode of gene regulation in ASD pathogenesis, and the snoRNA-mediated splicing changes point to novel potential biological disease mechanisms in brain.
Our results were enabled by an optimized pipeline that we developed to perform a large-scale genome-wide ASE investigation in ASD overcoming multiple potential challenges. To maximize transcriptome-wide SNP collection, we combined results from multiple methods including SNP array, imputation, and RNA-seq based genotyping. This is especially important for identifying SNPs with MAE, which often are missed by RNA-seq based genotyping. We generated a masked reference genome to filter out all potential reference biased SNPs during mapping to increase the accuracy of ASE measurement and to reduce false positive results. We further filtered SNPs showing association with ethnicity to identify generalizable ASE patterns across populations. We used the largest number of human postmortem brain samples from subjects with ASD4 including multiple brain regions per individual to bolster reproducibility of results. Finally, we employed a novel linear mixed-model approach that accounts for sample-level variation in order to quantify ASE patterns across ASD and control groups. This optimized work-flow can guide future large-scale genome-wide ASE studies in disease.
Preferential major allele expression is the most common pattern observed in most cases of MAE presumably because it can provide a buffer against unexpected, potentially deleterious rare SNP risks that might increase risk for disease12. Consistent with this interpretation, rare SNPs show more preferential major allele expression than common SNPs. Common SNPs, in general, show more balanced expression patterns as expected for non-deleterious alleles that have been through the filter of natural selection. Since rare alleles are more likely to be deleterious than common alleles45, the decrease in preferential major allele expression in ASD and dup15q, which could protect against the effect of deleterious rare alleles, could contribute to the risk of developing ASD. This is supported by the overlap of genes showing this pattern in ASD postmortem brain with genes harboring known ASD risk alleles.
We found a notable convergence between idiopathic ASD and dup15q, both of which show enrichment of allele shift from major allele MAE to minor allele MAE at specific loci on chromosomes 14 and 15. 15q11-q13 duplication causes hypermethylation at 15q11-q1319 as well as a genome-wide increase of minor allele MAE. Both idiopathic ASD and dup15q have increased minor allele MAE compared to control and the same trends of regional and snoRNA expression changes at the two regions showing a high frequency of shifts from the major to minor alleles. Idiopathic ASD shows extensive allele shifting at the 15q11 locus, which suggests that strong allele-specific methylation may be regulating gene expression46 in a similar fashion as the 15q11-q13 duplication itself. Because idiopathic ASD and dup15q share similar expression patterns within both of the allele shift rich regions, the splicing changes targeted by snoRNA may be strongly related to disease pathogenesis.
An allele shift rich region was identified at 14q32 which also contains two dense clusters of over 50 miRNAs within the delta-like 1 homolog-type 3 iodothyronine deiodinase (DLK1-DIO3) domain locus38. These clusters are maternally expressed (Fig. 6) and share an upstream imprinting control region with the other genes at the allele shift rich region38. Several of these miRNAs have been shown to be down-regulated in cancer47 and schizophrenia38, and their targets are enriched in axon guidance pathways. This could represent a point of convergence between ASD and schizophrenia, which are known to have significant phenotypic and genetic overlap38.
These analyses highlight preferential minor allele expression and orphan C/D box snoRNA mediated splicing changes as two novel forms of genetic regulation altered in ASD. Our novel ASE identification approach incorporates both allele frequencies and quantitative measures of allelic imbalance. We identified strong enrichment of minor allele containing transcripts in ASD, which were further enriched for known ASD risk genes, including PSD components32, RBFOX1 targets30, HuR targets29, and FMRP targets28. However, we recognize that the studies of PSD components, and FMRP and RBFOX1 targets rely on postnatal data. Prenatal studies could identify other pathways, including chromatin modifiers. However, we note that even using these postnatal data, we did identify overlap with de novo mutation containing ASD risk genes, which include chromatin modifiers and transcriptional regulators. Furthermore, we identify evidence supporting orphan C/D box snoRNA mediated splicing changes in ASD brain providing the first evidence of an association between snoRNA and ASD.
Among all forms of small non-coding RNAs (sncRNAs), miRNA have received the most focus as a target for ASD and other psychiatric diseases studies38,42. Although the orphan C/D box snoRNA is a small family of snoRNAs existing only at chromosomes 14q32 and 15q11, this family controls the splicing of a unique set of about 400 target genes that are distributed across genome. Although we could not find enrichment for common ASD risk in these targets, it may be due to simple lack of power of current GWAS studies in ASD. The most recent ASD GWAS contained over 45,000 subjects35 and yet identified an order of magnitude lower significant loci than a similar size study of schizophrenia48, consistent with substantially less power in ASD. Once larger more conclusive GWAS results are available, future studies can identify whether this group of genes is enriched in common variation that could contribute to polygenic risk for ASD. The allele shift rich regions in both idiopathic ASD and dup15q are directly overlapping with these orphan C/D box snoRNA repeat loci. Remarkably, we identified splicing changes in the downstream targets of these snoRNA genes providing a potential mechanistic link between genetic, epigenetic, and transcriptomic changes in ASD. These results highlight novel genetic and epigenetic regulations in ASD identifying novel biological mechanisms warranting further investigation.
Methods
Samples, RNA-seq, and genotyping.
As in our previous study4, human postmortem brain samples were acquired from the Autism Tissue Program at the Harvard Brain Bank, the University of Maryland Brain and Tissue Bank (a Brain and Tissue Repository of the NIH NeuroBioBank), the UK Brain Bank for Autism and Related Developmental Research, and the MRC London Neurodegenerative Diseases Brain Bank. A total of 263 brain samples were collected from BA9, BA41, and vermis of 40 controls and 56 ASD cases including 8 cases with dup15q syndrome. Though most sample information was already published4, we provided it again including further information (Supplementary Table 1). After RNA-seq library preparation with ribosomal RNA depletion (Illumina TruSeq v2 with Ribozero Gold (Illumina)), RNA-seq was performed using Illumina HiSeq2000 to generate 50bp paired-end reads as published4. Sample genotyping was performed using Illumina Omni2.5 SNP array.
Preparing masked reference and RNA-seq data mapping.
To avoid reference bias, we first prepared a masked reference genome after collecting all possible SNPs. We compiled the set of SNPs (39,683,000 SNPs) from multiple sources including those present on SNP arrays (Omni2.5 SNP: 2,372,783 SNPs; Affymetrix: 905,721 SNPs), those present in 1000 Genomes (38,270,182 SNPs)26,27, and those identified in the brain samples from RNA-seq based SNP calling (3,163,431 SNPs).
RNA-seq based SNP calling was performed using: GATK50 and Samtools51. To increase their SNP calling accuracy, we optimized bam files following the recommended GATK pipeline50 (Supplementary Fig. 1a). After updating mapping quality scores, we added group headers and read groups. Once reordering chromosomes and marking duplicate reads, we recalibrated quality scores. Possible false positive SNP calling data was filtered out based on the common outputs of GATK and Samtools. We removed indels and retain only single base variants (e.g., SNPs). When multiple SNPs were present at the same genomic coordinate, we randomly kept only one for downstream analysis. Using these collected SNPs, we masked the hg19 reference sequence with randomly selected third alleles to minimize systematic mapping bias. We mapped all RNA-seq data to this masked reference genome using TopHat252 with Ensembl v73 annotations and the following parameters:
Filtering out reference biased SNPs.
For further prevention of potential reference mapping bias by TopHat2, we generated a simulated RNA-seq dataset consisting of all possible 50bp reads overlapping each SNP for both reference and alternative alleles (100 reads total per SNP). We mapped these simulated reads to the masked reference genome as described above. We identified any SNPs that show reference bias (despite mapping to the masked reference genome) using chi-square test (p-value≦0.05 and read depth<10). These reference biased SNPs were removed from downstream analysis.
Genotyping.
SNP array data were imputed to 1000 genomes using Mach/Minimac53,54. Imputed SNPs were filtered out by their output quality (filtered out based on R2 and score values: R2<0.5 and score value>score digit ± 0.2) and Hardy-Weinberg equilibrium (HWE; p-value<1.0X10−6) to yield 10,992,184 high quality imputed SNPs.
To generate additional genotyping coverage, we did RNA-seq based genotyping using the following parameters:
We called the SNPs based on the first-, second-, and third-most abundant allele counts (a1, a2, and a3) in mapping transcript, respectively. We filtered out the genotyped SNPs if they did not pass the HWE filter (p-value<1.0X10−6). Compared with the previously published RNA-seq based genotyping methods by Quinn et al.55, these genotyping parameters show much higher accuracy with sensitivity exceeding 95% (Supplementary Fig. 1b) and specificity in the range of 90–95% (Supplementary Fig. 1c; see Quinn at el.55 for accuracy tests).
We integrated the three genotyping data, SNP array, imputation, and RNA-seq based genotyping. If we observed discordance among these methods, we weighted the data in the following order, favoring SNP array > imputation > RNA-seq based genotyping.
Ancestry Identification.
Using the integrated genotyping data, we identified ancestry by a multidimensional scaling (MDS) plot with HapMap3 populations. We categorized our samples into European, Mexican, Asian, African, and ambiguous groups (Supplementary Fig. 1d).
Group based ASE identification.
We first studied ASE patterns within each tissue group (BA9, BA41, vermis, and cortex (BA9 and BA41)) considering idiopathic ASD and control. In this study, we excluded dup15q samples from the other ASD case since they had clear structural chromosome alteration that causes ASD. We called the other ASD cases as idiopathic ASD for which the causes are unknown. Control brain samples were from someone who were not diagnosed as ASD and whose structural chromosome alternations were unknown.
First, we collected SNPs if “good” SNPs were present in 80% of samples15. SNPs were defined as “good” SNPs if they had the 3rd and 4th allele counts less than 5% of the major allele. We also collected SNPs considered “present” in at least 20% of samples15. “Present” is defined as the percent of individuals that have more than 10 allele counts. “Good” and “present” filters account for expression levels and RNA-seq read mapping error, respectively.
For each heterozygous SNP, we counted the number of reads mapping to each allele. To normalize for differences in library size, results were converted to “reads per kilobase of SNP area length per million mapped reads” (RPSM) values defined as:
The SNP area length is calculated as (read length X 2) - 1, and mapped reads can reach a SNP within the SNP area.
For each group, ASE was quantified at SNP level using a linear mixed model (log2(RPSM) ~ allele + age + sex + sequencing batch + RNA integrity number (RIN) + brain bank + ancestry, rand = ~1|biolrep/subject). Fixed effects included allele, age, sex, sequencing batch, RIN, brain bank, and ancestry. Biological replicate (biolrep) and subject were included as random effects. Biological replicate is RNA-seq data replicates. To ensure if groups were balanced with respect to these covariates, we removed samples based on age (≦7), brain banks (The UK Brain Bank for Autism and Related Developmental Research and the MRC London Neurodegenerative Diseases Brain Banks), and SNP annotation (mysterious) (Supplementary Table 1)4. We removed the other possible covariates (PMI, brain mass, and GC contents) that did not affect this ASE study. GC content was tested using AT and GC dropout values from Picard (https://broadinstitute.github.io/picard/; Supplementary Table 1). The AT and GC dropout values are the percentage of misaligned reads at low GC (GC<50%) or high GC (GC>50%) conditions, respectively. In BA9 samples, the distributions of the covariate values were balanced between control and ASD (Supplementary Fig. 1e). When we identified ASEs from additional linear mixed model including the covariates, the covariates did not show significant p-values without affecting the ASE results.
We investigated both autosome and chromosome X. For no PARs on chromosome X, we considered only female data. PARs were defined as: PAR1(chrX:60,001–2,699,520, chrY:10,001–2,649,520), PAR2 (chrX:154,931,044–155,260,560, chrY:59,034,050–59,363,566), and PAR3 (chrX:88,400,000–92,000,000, chrY:3,440,000–5,750,000)56.
Based on the p-values of the ancestry covariate from the linear mixed model regression, we filtered out SNPs showing ethnicity biased allele expression. After LD based SNP pruning using PLINK (R2 cutoff=0.2)57, we counted the number of LD-independent SNPs and identified ASE SNPs by Bonferroni correction.
To filter out potential indel-inducing reference bias from the identified ASEs, we considered 1,450,137 imputed indels identified during the above genotyping step. Among them, we selected only accurate 1,040,318 indel outputs from Mach/Minimac (deletions (≦3 bp) and inserts (1 bp)). We further filtered them out by the imputation quality (R2<0.5 and score value>score digit ± 0.2), HWE (p-value<1.0X10−6), missing genotype ratio (<0.05), and MAF (<0.01) to yield high quality imputed indels. To test indel-inducing reference bias, we selected 457,817 indels that show heterozygous types at least one sample.
Based on the masked reference genome, we generated a simulated RNA-seq dataset (50mer fastq file), which overlaps each indel area. It contains both indel containing and not containing sequences. We aligned the fastq file to the masked reference genome using TopHat2 and identified possible indel-inducing reference biases (chi-square test p-value≦0.05) from their proximal SNPs (read depth≧10). Once SNPs show the reference biases from at least one sample that has heterozygote genotypes from both indels and SNPs, we filtered them out from the ASE results. To genotype SNPs, we integrated SNP array, imputation, and RNA-seq based genotyping as described above.
To compare ASE across groups, Manhattan plots were generated with the p-values of the allele covariate from the linear mixed model regression using the qqman R package (https://cran.r-project.org/web/packages/qqman/; Fig. 3b). If ASE SNPs were located in gene bodies, we called the genes as ASE genes. The ASE genes were compared using area-proportional Venn diagrams58 (Fig. 3c).
Individual based ASE identification.
We next studied ASE within individual samples comparing differences in allele counts for heterozygous SNPs. We used only “good” SNPs in at least 80% of samples (as defined above) without considering the present filter. P-values for each SNP were calculated using Fisher’s exact test. After LD-based SNP pruning, ASE SNPs were identified by Bonferroni correction (as described above). We filtered out indel-inducing reference biased SNPs as described above.
For each cortical sample, ASE rates were calculated per chromosome as the number of ASE SNPs / total number of tested SNPs. Differences in the ASE rate was tested using Wilcoxon rank sum test (Fig. 3d).
Evaluation of identified ASE.
To test ASE identification accuracy, we identified haplotype blocks using PLINK with the SNPs showing no ethnicity biased allele expression. We calculated deviations of log2(fold change) values (the beta values of allele covariate from the group based ASE identifications) per haplotype block in cortex and compared their distributions with normal distributions (Supplementary Fig. 1f and 1g). For further validation, we compared ASE genes identified in our study with those present in dbMAE (https://mae.hms.harvard.edu/)22 using Fisher’s exact test (Supplementary Fig. 2a). Of note, dbMAE mostly includes non-neural tissues, and we considered its human and mouse data. Nevertheless, we observe strong overlap between them providing validation of our ASE genes. For additional external validation, we generated data as following GTEx data analysis24. Like the method of GTEx data, we considered only heterozygote SNPs and followed the same p-value cutoff of GTEx data. We considered the ASE SNPs once their p-values are below 0.005 at the previous individual based ASE identification. Like GTEx data, we calculated ASE rates per tissue (Supplementary Fig. 2c). To compare the GTEx data24 with our data, we compared the mean of ASE rates from the tissues.
ASE and expression comparison.
We calculated less expressed allele expression fraction per each heterozygote SNP from a sample, UMB5303 (tissue: BA41). The fraction is a2 / (a1 + a2). a1 is more expressed allele read count, and a2 is less expressed allele read count. If the SNP has more than 10 mapped reads, we calculated log2(RPSM). Based on the less expressed allele expression fraction, we grouped the SNPs as imbalance and other groups. The fractions of the imbalance group were equal or less than 30%. The fractions of the other group were greater than 30%. We compared their log2(RPSM) values using KS tests (Supplementary Fig. 2d).
Gene set enrichment.
Gene set enrichment analyses were performed using logistic regression accounting for gene length as a covariate. Heatmaps were prepared as showing OR and FDR corrected p-values for enrichment, if significant. For known ASD risk gene list (SFARI gene), we selected genes from category 1, 2, and 3 corresponding to gene scoring criteria (https://gene.sfari.org/autdb/GS_Home.do); Supplementary Table 6) The gene list was previously used at the previous publication36.
For this gene set enrichment studies, we also used known imprinted genes (www.geneimprint.com), PSD32, FMRP target28, HuR target29, and RBFOX1 target genes30, cell marker genes31 (neuron, astrocyte, oligodendrocyte, microglia, and endothelial), expression up- and down-regulated genes in ASD cortex4 (Supplementary Table 6). HuR target gene list29 was collected from photoactivatable ribonucleoside crosslinking and immunoprecipitation (PAR-CLIP) data instead of RNA binding proteins immunoprecipitation complementary DNA array (RIP-chip) data41 since PAR-CLIP data identified RNA binding proteins binding sites more precisely than RIP-chip (Supplementary Table 6).
For the de novo variant data, we used the risk genes containing rare de novo likely gene disrupting mutations in SCZ, ID, and ASD from the dataset of Iossifov et al.45 (Supplementary Table 6). Since this list is not based on the most stringent statistical thresholding (353 genes for ASD), we used additional risk gene lists from other work to cross reference. This includes Sanders et al.33 who identified risk genes after integrating small de novo deletions using the transmission and de novo association model. For the risk gene list, we analyzed gene set enrichment with different FDR cutoffs (FDR≦0.01, 0.01<FDR≦0.05, and 0.05<FDR≦0.1) and found the highest enrichments using the most stringent FDR cutoff. of FDR≦0.01, which we report in our study (Supplementary Table 6).
For gene set enrichment study with risk variants from psychiatric diseases, we considered GWAS data from Psychiatry Genomics Consortium34,35 and de novo variant data32,33. For the GWAS data, we considered five psychiatric diseases, ADHD, BPD, MDD, SCZ, and ASD. Gene set enrichment analyses were performed by MAGMA version 1.07b59. For the analyses, annotation steps were performed first using hg18 for the dataset of Cross-Disorder Group of the Psychiatric Genomics34 and hg19 for the dataset of Grove et al.35. Genes boundaries were set stringently between transcription start and stop sites. In the next gene analysis steps, LD was calculated using the 1000 Genome European ancestry reference dataset. Gene set analyses were then performed to create aggregate statistics for each gene as considering the LDs.
Gene ontology (GO) analyses were performed using GOrilla60,61 and visualized through REViGO49. For both studies, brain expressed genes were used as background genes that have more than 10 allele counts (Supplementary Table 6).
Quantitative allele imbalance study in idiopathic ASD.
From SNPs genotyped by SNP array or imputation, we collected SNPs based on meeting the criteria of at least 20% present and 80% good15. These criteria included rare SNPs. If SNPs showed the reference biases from at least one sample that has heterozygote genotypes from both proximal indels and SNPs, we filtered them out as described above. We calculated minor allele expression fractions (minor allele expression / total expression) and less expressed allele expression fractions. We visualized their distributions using ggplot2 R package (http://ggplot2.org/) and compared them between control and idiopathic ASD at autosome (Fig. 4a and Supplementary Fig. 4b) and chromosome X (Supplementary Fig. 4c). Although there are three different PARs at chromosome X, we combined their data into one dataset due to their small SNP numbers. At no PARs, we used only female data so that we considered two alleles from chromosome X.
To compare the number of SNPs that can introduce LoF mutation, amino acid change, and synonymous mutation (Fig. 4d), we counted SNPs for three different allele categories. They contain alleles showing balanced expression, major allele MAE, and minor allele MAE. For LoF mutation, we counted alleles located at stop codon generating mutations, 5’ splice site mutations, or 3’ splice site mutations.
MAE study.
For the genome-wide views of MAE SNPs, we used the previously described qqman R package. For major and minor allele MAE SNPs, we visualized them on the different lines (Supplementary Fig. 5). To visualize allele shift to minor allele MAE at UCSC genome viewer, we prepared wig files as assigning constant value (1) for each allele shift (Fig. 6). To visualize splice junctions at UCSC genome browser, we combined all bed files from TopHat2 outputs and prepared a juncs file using bed_to_juncs program39.
sncRNA-seq data process.
Based on RNA sample availability, our previous study prepared 50mer single-end read sncRNA-seq data42. The data were generated by Illumina HiSeq2000 from rRNA depleted libraries. To remove adaptor sequences, the sncRNA-seq read are processed by fastx_clipper of FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) and mapped with bowtie262. From uniquely mapped reads, we counted read number for smaller than 100bp genes using htseq-count63 and calculated their reads per kilobase of transcript per million mapped reads (RPKM)64. We used a linear mixed model (expression ~ condition + age + sex + sequencing batch + RIN + bank, rand = ~1|biological replicate/subject) to identify differentially expressed genes. Expression is log2(RPKM), and the expression could be driven by condition, age, sex, sequencing batch, RIN, and brain bank. Also, as the random factors we used biological replicate and subject. We removed samples based on age (≦7) and brain banks (The UK Brain Bank for Autism and Related Developmental Research and the MRC London Neurodegenerative Diseases Brain Banks).
For snoRNA gene expression change study in the allele shift regions, we calculated the log2(fold change) from the linear mixed model regression outputs. We considered 51 snoRNAs that have PRKMs equal or greater than 1 (Fig. 7b).
Regional expression and splice junction study.
To study regional expression change at the allele shift rich regions, we calculated their log2(RPGMs) values65 from uniquely mapped reads of RNA-seq data and compared them between control and cases (Fig. 7a).
snoRNA target splicing change study.
For orphan C/D Box snoRNAs at the allele shift rich regions, we relied on previously identified splicing targets41. We identified splicing changing genes that show ≧2SD alternative splicing change (beta values from the previously published linear mixed model regression data4) in ASD and dup15q versus controls. Using a Fisher’s exact test, we assessed if the snoRNA target genes show more alternative splicing changes in ASD and dup15q versus other non-snoRNA targets (Fig. 7c). Among the snoRNA target genes, we performed a gene set enrichment study for gene showing significant splicing changes (Fig. 7d).
Summary of statistical methods.
Sample sizes.
No statistical methods were used to pre-determine sample sizes but our sample sizes are larger than to those reported in previous publications that studied ASE8,15.
Normality of data distribution.
For ASE identification and quantitative allele imbalance study, normality was not formally tested, but data distribution was assumed to be normal but this was not formally tested.
Randomization.
To avoid reference bias, we randomized allele selection for a masked reference preparation.
Blinding.
Data analysis was not performed blind to the metadata information of the brain samples.
Code availability.
The R code for the ASE identification using a linear mixed model is provided in Supplementary Software 1.
Data availability.
The detailed description of brain samples is provided in Supplementary Table 1. For each tissue, group-based ASE identification results are available in Supplementary Tables 2 and 3. We also include gene lists that we used for gene set enrichment and GO analysis that includes brain expressed gene that we used for our study (Supplementary Table 6). Raw next generation sequencing data from human postmortem brain samples are available from published RNA-seq4 and sncRNA-seq42 studies. They have been deposited to the PsychENCODE Knowledge Portal (http://dx.doi.org/10.7303/syn4587609).
Supplementary Material
Acknowledgements
We thank Dr. Serghei Mangul for helpful advice about his published ASE data filtering method15. We also thank Drs. Neelroop N. Parikshak, Ye E. Wu, and T. Grant Belgard for providing published RNA-seq4 and sncRNA-seq42 data and helpful advice. We also thank Dr. Katrina Y. Choe for proofreading. This work was supported by Simons Foundation grant 401457 to D.H.G. and National Institutes of Health grants R01MH110927 and R01MH109912 to D.H.G.
Footnotes
Accession codes. Raw RNA-seq and sncRNA-seq data from brain samples were deposited to the PsychENCODE Knowledge Portal (http://dx.doi.org/10.7303/syn4587609).
Competing Interests Statement
The authors declare no competing interests.
References
- 1.Elsabbagh M et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res 5, 160–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geschwind DH & State MW Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol 14, 1109–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voineagu I et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parikshak NN et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nardone S et al. DNA methylation analysis of the autistic brain reveals multiple dysregulated biological pathways. Transl Psychiatry 4, e433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karlic R, Chung HR, Lasserre J, Vlahovicek K & Vingron M Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A 107, 2926–31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong CCY et al. Genome-wide DNA methylation profiling identifies convergent molecular signatures associated with idiopathic and syndromic autism in post-mortem human brain tissue. Hum Mol Genet 28, 2201–2211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-David E, Shohat S & Shifman S Allelic expression analysis in the brain suggests a role for heterogeneous insults affecting epigenetic processes in autism spectrum disorders. Hum Mol Genet 23, 4111–24 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Chess A Monoallelic Gene Expression in Mammals. Annu Rev Genet 50, 317–327 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Gimelbrant A, Hutchinson JN, Thompson BR & Chess A Widespread monoallelic expression on human autosomes. Science 318, 1136–40 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Gregg C, Zhang J, Butler JE, Haig D & Dulac C Sex-specific parent-of-origin allelic expression in the mouse brain. Science 329, 682–5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kukurba KR et al. Allelic expression of deleterious protein-coding variants across human tissues. PLoS Genet 10, e1004304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degner JF et al. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25, 3207–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickrell JK et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464, 768–72 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang EY et al. Discovering Single Nucleotide Polymorphisms Regulating Human Gene Expression Using Allele Specific Expression from RNA-seq Data. Genetics 204, 1057–1064 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castel SE, Levy-Moonshine A, Mohammadi P, Banks E & Lappalainen T Tools and best practices for data processing in allelic expression analysis. Genome Biol 16, 195 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de la Chapelle A Genetic predisposition to human disease: allele-specific expression and low-penetrance regulatory loci. Oncogene 28, 3345–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gicquel C et al. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet 37, 1003–7 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Scoles HA, Urraca N, Chadwick SW, Reiter LT & Lasalle JM Increased copy number for methylated maternal 15q duplications leads to changes in gene and protein expression in human cortical samples. Mol Autism 2, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogart A et al. Chromosome 15q11–13 duplication syndrome brain reveals epigenetic alterations in gene expression not predicted from copy number. J Med Genet 46, 86–93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meguro-Horike M et al. Neuron-specific impairment of inter-chromosomal pairing and transcription in a novel model of human 15q-duplication syndrome. Hum Mol Genet 20, 3798–810 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savova V, Patsenker J, Vigneau S & Gimelbrant AA dbMAE: the database of autosomal monoallelic expression. Nucleic Acids Res 44, D753–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adegbola AA et al. Monoallelic expression of the human FOXP2 speech gene. Proc Natl Acad Sci U S A 112, 6848–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium GT Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeVeale B, van der Kooy D & Babak T Critical evaluation of imprinted gene expression by RNA-Seq: a new perspective. PLoS Genet 8, e1002600 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genomes Project C et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sudmant PH et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darnell JC et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee N et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 43, 327–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weyn-Vanhentenryck SM et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep 6, 1139–1152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34, 11929–47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iossifov I et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders SJ et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–1233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cross-Disorder Group of the Psychiatric Genomics, C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grove J et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 51, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parikshak NN et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook EH Jr. & Scherer SW Copy-number variations associated with neuropsychiatric conditions. Nature 455, 919–23 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Gardiner E et al. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry 17, 827–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cavaille J et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci U S A 97, 14311–6 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavaille J, Seitz H, Paulsen M, Ferguson-Smith AC & Bachellerie JP Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum Mol Genet 11, 1527–38 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Bazeley PS et al. snoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene 408, 172–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu YE, Parikshak NN, Belgard TG & Geschwind DH Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nat Neurosci 19, 1463–1476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kishore S & Stamm S The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311, 230–2 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Kehr S, Bartschat S, Stadler PF & Tafer H PLEXY: efficient target prediction for box C/D snoRNAs. Bioinformatics 27, 279–80 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Kryukov GV, Pennacchio LA & Sunyaev SR Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet 80, 727–39 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tycko B Allele-specific DNA methylation: beyond imprinting. Hum Mol Genet 19, R210–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oberg AL et al. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS One 6, e20465 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Supek F, Bosnjak M, Skunca N & Smuc T REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenna A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim D et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuchsberger C, Abecasis GR & Hinds DA minimac2: faster genotype imputation. Bioinformatics 31, 782–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howie B, Fuchsberger C, Stephens M, Marchini J & Abecasis GR Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44, 955–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinn EM et al. Development of strategies for SNP detection in RNA-seq data: application to lymphoblastoid cell lines and evaluation using 1000 Genomes data. PLoS One 8, e58815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veerappa AM, Padakannaya P & Ramachandra NB Copy number variation-based polymorphism in a new pseudoautosomal region 3 (PAR3) of a human X-chromosome-transposed region (XTR) in the Y chromosome. Funct Integr Genomics 13, 285–93 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Purcell S et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hulsen T, de Vlieg J & Alkema W BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eden E, Navon R, Steinfeld I, Lipson D & Yakhini Z GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eden E, Lipson D, Yogev S & Yakhini Z Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol 3, e39 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mortazavi A, Williams BA, McCue K, Schaeffer L & Wold B Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5, 621–8 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Lee C, Mayfield RD & Harris RA Altered gamma-aminobutyric acid type B receptor subunit 1 splicing in alcoholics. Biol Psychiatry 75, 765–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The detailed description of brain samples is provided in Supplementary Table 1. For each tissue, group-based ASE identification results are available in Supplementary Tables 2 and 3. We also include gene lists that we used for gene set enrichment and GO analysis that includes brain expressed gene that we used for our study (Supplementary Table 6). Raw next generation sequencing data from human postmortem brain samples are available from published RNA-seq4 and sncRNA-seq42 studies. They have been deposited to the PsychENCODE Knowledge Portal (http://dx.doi.org/10.7303/syn4587609).