Abstract
Background
Emergence of blaKPC and blaNDM co-producing Klebsiella pneumoniae strains have led to the limited therapeutic options for clinical treatment. Understanding the diversity and frequency of resistance and virulence genes of these isolates is of great significance.
Purpose
The aim of this study is to research the diversity and frequency of resistance and virulence genes in the blaKPC and blaNDM co-producing Klebsiella pneumoniae strains.
Methods and Results
In this study, 117 K. pneumonia strains were isolated from China, and among of which, 24 were found to be blaKPC and blaNDM co-producing with significant resistance against almost all the commonly used antibiotics. Additionally, 4 strains were hypermucoviscous and 8 showed high serum resistance. Overall, blaSHV, blaCTX-M, tetA and sul1 resistance genes found in 100% of the isolates, followed by blaTEM (95.8%), oqxA/B (91.7%), qnrB (87.5%), aac(6’)Ib-cr (83.3%), blaDHA (79.2%), rmtB (66.7%), qnrS (54.2%), cat(54.2%), floR (50.0%), sul2 (45.8%) cmlA (20.8%)andblaCMY (8.33%), respectively. What’ more, seven blaCTX-M subtypes [blaCTX-M-14 (n=18), blaCTX-M-3(n=11), blaCTX-M-65 (n=4), blaCTX-M-15 (n=3), blaCTX-M-28 (n=2), blaCTX-M-55 (n=2), blaCTX-M-22 (n=1)] and six blaSHV subtypes [blaSHV-12(n=16), blaSHV-11 (n=4), blaSHV-2a(n=1), blaSHV-1(n=1), blaSHV-38(n=1) and blaSHV-28(n=1)] were detected. The frequency of virulence genes was as follows: 100% for entB, ybtS and irp, 95.8% for mrkD, 91.66% for fimH, 79.2% for iutA, 62.5% for iroBCDE, aerobactin and kfu, 66.7% for allS, 45.8% for wcaG, 37.5% for rmpA, 20.8% for pagO and 16.7% for magA.
Conclusion
From this study, we concluded that the blaKPC and blaNDM co-producing Klebsiella pneumoniae strains have a high diversity and frequency of resistance and virulence genes. This study may offer hospitals important information about the control of infections caused by blaKPC and blaNDM co-producing Klebsiella pneumoniae.
Keywords: Klebsiella pneumoniae, blaNDM, blaKPC, resistance genes, virulence factors
Introduction
Carbapenemase-producing bacteria can hydrolyse carbapenems and most other β-lactam antibiotics which pose significant challenges to clinical diagnosis and treatment. Klebsiella pneumoniae carbapenemase (KPC) and Metallo-B-Lactamases (blaNDM) are the two major groups of carbapenemases that produced by the most of Carbapenemase-Resistant Enterobacteriaceae strains (CRE). The blaKPC and blaNDM genes are commonly found in CRE strains in recent years.1–3 Those type of the carbapenem resistance genes and other resistance genes including the key Extended-Spectrum β-lactamases (ESBLs) genes (blaCTX-M, blaSHV and blaTEM), the fluoroquinolone resistance genes (qnrA, qnrB, qnrS, oqxA/B), aminoglycoside resistance genes (rmtA, rmtB and rmtC), chloramphenicol resistance genes (cat, floR, cmlA, cfr) and tetracycline resistance genes (tetA, tetB, tetC) are carried by the same strain and resulting in high resistance to almost all kinds of antibiotics.4–7 The more worrisome is hypervirulent K. pneumoniae strains (hvKP) emergency sharply in recent years, especially the carbapenemase-producing hvKP related infections in immunocompromised patients which is a serious threat to the patients.8–11
More and more researchers report that HvKP strains are characterized a number of virulence factors including aerobactin (encodes high-affinity iron chelators), rmpA (regulators of mucoid phenotype), wcaG (involved in the biosynthesis of the outer core lipopolysaccharide), allS (associated with allantoin metabolism), kfu (responsible for an iron uptake system), yptS, irp (yersiniabactin biosynthesis) and iroBCDN (salmochelin biosynthesis), entB (catecholate siderophore), fimH and mrkD (fimbrial adhesin, which mediate binding to the extracellular matrix to form the biofilm), pagO (involved in liver abscess formation by liver abscess-Kp.9,12–15
Understanding the diversity and frequency of resistance and virulence genes of these isolates is of great significance to disease prevention and control. For offer hospitals important information about the control of infections caused by blaKPC and blaNDM co-producing K. pneumoniae. In this study, we mainly present the diversity and frequency of resistance and virulence genes in the blaKPC and blaNDM co-producing K. pneumoniae.
Materials and methods
Isolates collection and screening of blaKPC and blaNDM genes
A total of 117 non-repetitive K. pneumonia strains were isolated from sputum, cerebrospinal fluid, wound, and urine samples for routine examination between Aug. 2016 and Sept.2018 at several hospitals in Sichuan, Henan, Fujian province of China. These isolates were identified by VITEK2 Compact System (bioMérieux, France) and 16sRNA sequencing. K. pneumoniae ATCC700603 was used as the control strain for the species identification and antimicrobial susceptibility test. The blaKPC and blaNDM detection were performed according to our previous work by PCR.9,16
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing of the blaKPC and blaNDM co-producing K. pneumoniae strains were performed according to the recommendations of the Clinical Laboratory Standards Institute (CLSI) guidelines (CLSI, 2017). Antimicrobial agents (Oxoid, England) used used in this study included CXM (cefuroxime axetil), TZP (piperacillin-tazobactam), CAZ (ceftazidime), CRO (ceftriaxone), IPM (imipenem), MEM (meropenem), ATM (aztreonam), AMK (amikacin), CIP (ciprofloxacin), CHL (chloramphenicol), TMP-SMZ (trimethoprim/sulfamethoxazole). E. coli strain ATCC 25922 was used as quality control.17
Hypermucoviscosity, biofilm formation and serum killing assay
The hypermucoviscosity phenotype of 24 K. pneumonia was detected by string test.18 The colonies were cultured on blood agar plate overnight at 37°, stretched by a bacteriology inoculation loop. The strain formed a viscous string of >5 mm was designated as hypermucoviscous. Biofilm formation assay was performed by crystal violet staining assay.9 Biofilm formation in each well was measured by microplate reader (Bio-Rad, US) at optical density (OD) 595 nm. The susceptibility of the K. pneumoniae isolates to human serum was explored by an established method.19 Briefly, K. pneumoniae strains were inoculated into LB Broth Medium and incubated at 37 °C with shaking until the logarithmic phase was reached (T=4 h, OD600=0.6). 25 μL of diluted culture (containing 106 CFU of bacteria) and 75 μL human serum were then added into a 10×75 mm Falcon polypropylene tube and incubated at 37 °C with shaking. Viable counts were checked at 0, 1, 2, and 3 h. The response to serum killing in terms of viable counts was scored on six grades as described previously method.20
ERIC-PCR
Enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) method was used to evaluate the genetic diversity of the 24 isolates, as previously described using the primers.21 The PCR products were loaded on a 1% agarose gel with the gelred at 90 V for 40 mins, and the banding patterns were analyzed by gel imaging and analysis system. To determine the similarity rate among the acquired outcomes, Genetic diversity were analyzed using the unweighted pair-group method with arithmetic mean (UPGMA) and isolates with ≥80% similarity were treated as a single cluster.22
Detection of resistance and virulence genes
By using PCR, the carriage of carbapenemase-encoding genes (blaVIM, blaGES, blaDIM, blaGIM, blaSPM and blaAIM),23 ESBL-encoding genes (blaTEM, blaSHV, blaCTX-M, blaCTX-M-1, blaCTX-M-2 and blaCTX-M-9),7 AmpC β-lactamase genes (blaDHA, blaCMY),24 16 s rRNA methylase genes (rmtA, rmtB and rmtC),25 sulfonamides resistance genes (sul1, sul2 and sul3), chloramphenicol resistance genes (cmlA, floR and catB), multiresistance gene (cfr), tigecycline resistance gene (tetA, tetB and tetC)26,27 and quinolone resistance genes (qnrA, qnrB, qnrS, aac(6ʹ)-Ib-cr, qepA and oqxAB)28–30 were detected as described previously. PCR assays were also used to assess the capsular serotypes (K1, K2, K5, K20, K54 and K57)31 and fourteen virulence genes (magA, rmpA, allS, wcaG, ybtS, kfu, iroBCDE, entB, irp, iutA, aerobactin, mrkD, fimH and pagO).12–14,31,32 PCR amplicons were sequenced by Shanghai Sangon Bioengineering Company. Sequences were analyzed by the BLAST programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The primers used were shown in Table S1.
Results
Antimicrobial susceptibility, hypermucoviscosity, serotyping, biofilm, serum resistance assay and ERIC-PCR typing
A total of 24 blaKPC and blaNDM co-producing strains were screen from117 non-repetitive K. pneumonia strains. All the isolates were resistant to piperacillin-tazobactam, cefuroxime axetil, ceftazidime, ceftriaxone, imipenem, meropenem and aztreonam (Table 1). Among the 24 blaKPC and blaNDM co-producing strains, 16.7% (n=4) were the K1 type, while the K2, K5, K20, K57 and K54 serotype were not found (Figure 1). String test showed that 4 (KP103L, KP48L, KP97L, KP36L) blaKPC and blaNDM co-producing K. pneumoniae isolates were hypermucoviscous. Biofilm formation was observed in all the 24 strains, with values of OD595 nm ranged from 0.33 to 2.70, whereas the mean value of the negative control wells is 0.168. Serum killing assay showed that 33.3% (n=8) of the strains were high serum resistance (Grade 5 or Grade 6). Analysis of genetic linkage among isolates by ERIC-PCR showed 34–100% similarity among 24 isolates (Table 2). Genetic diversity was established among 24 blaKPC and blaNDM co-producing K. pneumoniae isolates by detecting 15 different ERIC fingerprints with the similarity cutoff of 80% (Table 2).
Table 1.
The antibiotic resistance phenotype profile and positive rate of the resistance gene of the isolates
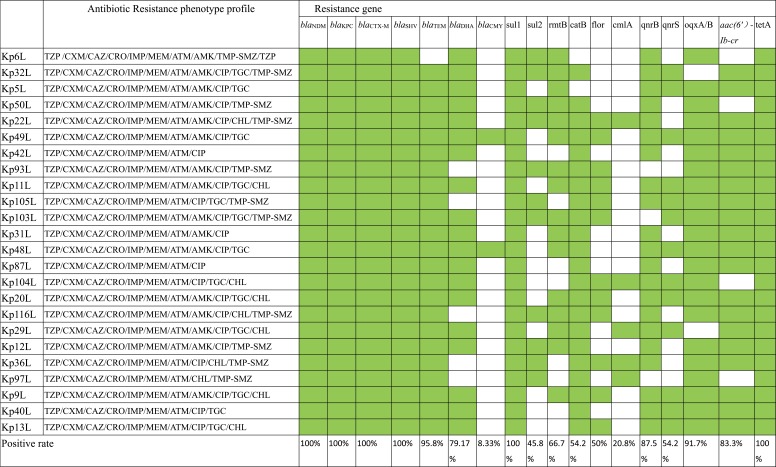 |
Note: The green check represent the positive while the blank is the negative.
Abbreviations: TZP, piperacillin-tazobactam; CXM, cefuroxime axetil; CAZ, ceftazidime; CRO, ceftriaxone; IPM, imipenem; MEM, meropenem; ATM, aztreonam; AMK, amikacin; CIP, ciprofloxacin; CHL, chloramphenicol; TMP-SMZ, trimethoprim/sulfamethoxazole.
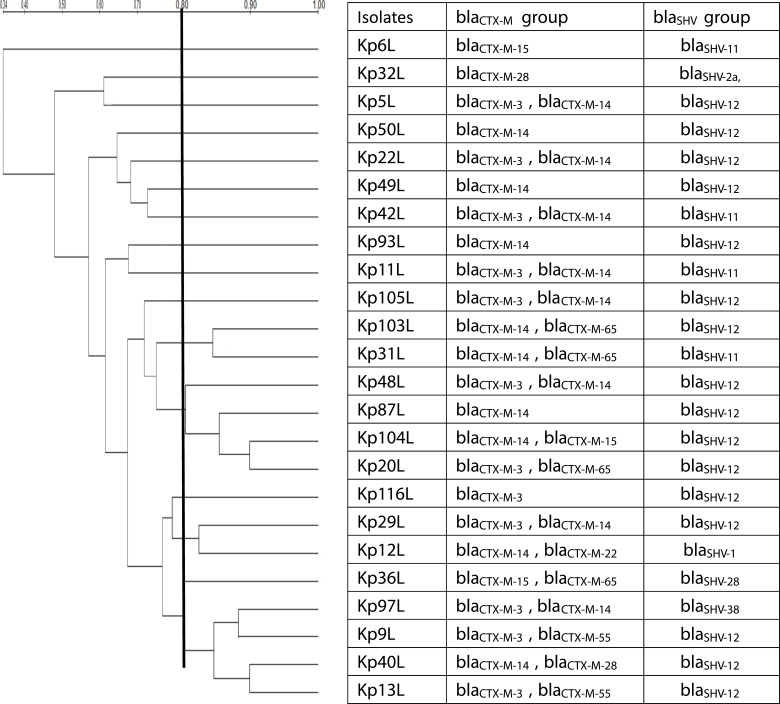
Figure 1.
The dendrogram of ERIC-PCR fingerprints and diversity of the ESBLs genotypes.
Notes: The dendrogram of ERIC-PCR fingerprints was constructed using the Dice coefficient and the unweighted pair-group method with arithmetic mean (UPGMA) and the diversity of the ESBLs (blaCTX-M group and blaSHV group) genotypes.
Table 2.
The string test, serotyping, Serum killing and biofilm formation assay and diversity and frequence of the virulence factors of the blaKPC and blaNDM co-producing Klebsiella pneumoniae
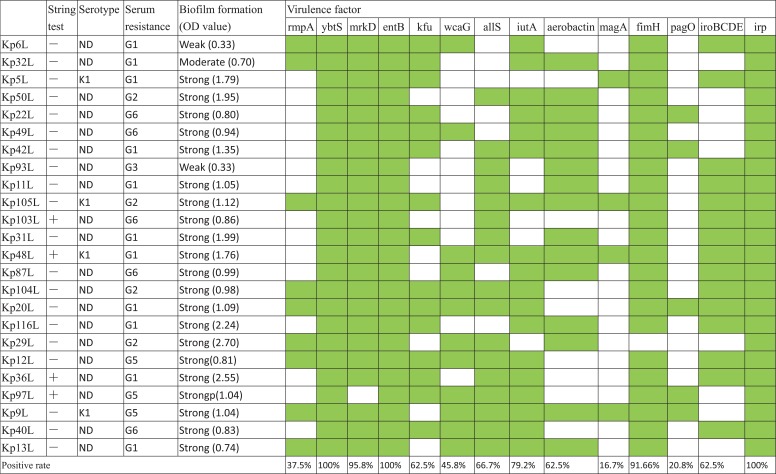 |
Notes: “+”: positive, “-”: negative; The green check represent the positive while the blank is the negative. Biofilm formation expressed as crystal violet optical density value (OD at 595 nm).
Abbreviations: ND, Not Determination; OD, optical density; G, grade.
Diversity and frequency of resistance and virulence gene
As shown in Table 1, all isolates (100%, n=24) carried the resistance gene blaSHV, blaCTX-M, tetA and sul1, followed by blaTEM (95.8%), oqxA/B (91.7%), qnrB (87.5%), aac(6ʹ)Ib-cr (83.3%), blaDHA(79.2%), rmtB (66.7%), qnrS (54.2%), cat (54.2%), floR (50.0%), sul2 (45.8%) cmlA (20.8%) and blaCMY (8.33%), respectively. While the carbapenemase encoding genes blaGES, blaVIM, blaAIM, blaGIM, blaDIM were not detected in any of those strains. Regarding the blaCTX-M group (Table 2; Supplement Sequences), the most widespread subtype was blaCTX-M-14, which was found in 75% (n=18) of the tested isolates, followed by blaCTX-M-3 in 45.8% (n=11), blaCTX-M-65 in 16.7% (n=4), blaCTX-M-15 in 12.5% (n=3), blaCTX-M-28 in 8.3% (n=2), blaCTX-M-55 in 8.3% (n=2), blaCTX-M-22 in 4.2% (n=1). In addition, there are 17 isolates carried two subtypes of blaCTX-M. And the majority of the 8 isolates carried blaCTX-M-14 co-existing with blaCTX-M-3, while 2 isolates co-carried blaCTX-M-14 and blaCTX-M-65 (Table 2). Regarding the blaSHV group, blaSHV-12 (66.7%; n=16) was the most prevalent blaSHV in those 24 blaKPC and blaNDM co-producing strains, followed by blaSHV-11 in 16.7% (n=4), blaSHV-2a, blaSHV-1, blaSHV-38 and blaSHV-28 in 4.2% (n=1) (Table 2).
Diversity and frequency of virulence genes
The prevalence and distribution of virulence factors are given in Table 2. All strains carried the ybtS, entB and irp gene. 95.8% (n=23) strains harbored mrkD gene, 91.6% (n=22) strains harbored fimH gene,79.2% (n=19) strains contained iutA gene, 66.7% (n=16) strains carried allS gene, 62.5% (n=15) strains carried iroBCDE, aerobactin and kfu gene, 45.8% (n=11) strains contained wcaG gene, 37.5% (n=9) strains involved rmpA gene, 20.8% (n=5) strains involved pagO gene and 16.7% (n=4) carried magA gene.
Discussion
The prevalence of co-carried blaNDM and bla KPC in a single bacterial isolate in hospitals has led to heightened concerns because often makes the isolate an extremely drug-resistant variant.2,3 In this study, 117 non-repetitive K. pneumonia strains were isolated from China, and among of which, 24 were found to be blaKPC and blaNDM co-producing with significant resistance against almost all the commonly used antibiotics. This results showed that the positive incidence of the blaNDM and blaKPC co-producing K. pneumonia is increasing. The results were expected that all 24 isolates resist almost the all test antibiotic and biofilm formation was observed in all the 24 strains. This is a dangerous situation for antibiotic treatment because the high biofilm formation pathogenic bacteria often involved in hospital infections and always lead to the failure of antibiotic treatments.9 Additionally, 4 strains were hypermucoviscous and 8 strains showed high serum resistance. To our knowledge, the phenotype of hypermucoviscous, biofilm formation ability and serum resistance were as the virulence evaluation criterion.18,20 Those results indicated that there are harboring hypervirulent variant of Klebsiella pneumonia (hvKp) among the 24 blaNDM and blaKPC co-producing strains. This results suggest that urgent need to enhance clinical awareness and epidemiologic surveillance. Although the genetic diversity was established among 24 blaKPC and blaNDM co-producing K. pneumoniae isolates by detecting 15 different ERIC fingerprints with the similarity cutoff of 80%, we should pay more attention about this like strains clonal spread in the hospital.
In recent years, more and more researchers report that the co-carried blaNDM and blaKPC K. pneumoniae strains carried a large number of resistance genes, making this isolate highly resistance against almost all the commonly used antibiotics. For example, the blaKPC-2 and blaNDM-1 co-carriage strain C. freundii 112298 existance many resistance genes including the blaSHV-12, blaCTX-M-14, aac (6′)-Ib-cr, blaOXA-1, catB3, arr-3, fosA3 and sul1.1 The blaKPC-2 and blaNDM-5 co-carriage strain ZSH6 carried twenty resistance genes blaKPC-2, blaNDM-5, blaCTX-M-3, blaCTX-M-65, blaTEM-1, floR, tet(A), tet(B), dfrA17, aadA5, sul1, mdf(A), mph(A), erm(B), aph(3′)-Ia, aph(3′)-Ib, aph(4)-Ia, aph(6)-Id, aac(3)-Iva, aac(3)-IId.3 In this study, we also found that the high frequency and diversity of the resistance gene were emergency in the blaKPC-2 and blaNDM-1 co-carriage strains. All 24 isolates carried the blaSHV, blaCTX-M, tetA and sul1, followed by blaTEM (95.8%), oqxA/B(91.7%), qnrB(87.5%), aac(6ʹ)Ib-cr (83.3%), blaDHA (79.2%), rmtB (66.7%), qnrS (54.2%), cat (54.2%), floR(50.0%), sul2 (45.8%) and cmlA(20.8%). Particularly the high frequency and diversity of the ESBLs. (blaCTX-M group and blaSHV group) gene. For the blaCTX-M group, there are seven blaCTX-M subtypes including (blaCTX-M-14, blaCTX-M-3, blaCTX-M-65, blaCTX-M-15, blaCTX-M-28, blaCTX-M-55 and blaCTX-M-22) in all 24 strains. Our study showed that blaCTX-M −14 was the most frequent. In addition, there are 17 isolates carried two subtypes of blaCTX-M. And the majority of the 8 isolates carried blaCTX-M-14 co-existing with blaCTX-M-3, while 2 isolates co-carried blaCTX-M-14 and blaCTX-M-65 (Table 1). Regarding the blaSHV group, blaSHV-12 (66.7%, n=16) was the most prevalent blaSHV subtype in 24 blaKPC and blaNDM co-producing strains. The threat of the high frequency and diversity of the resistance gene emergency in the blaKPC-2 and blaNDM-1 co-carriage strains should be strict surveillance and management, although its resist almost all the commonly used antibiotics.2
Besides of the high frequency and diversity of the resistance gene, the virulence genes were also high emergency in 24 K. pneumoniae strains. In this study, we found that the frequency of virulence genes (ybtS, entB, irp, mrkD, fimH) was similar to most of others researcher’s reports. However, the frequency of wcaG (45.8%), allS (66.7%) and pagO (20.8%) gene was slightly higher than our previous work. This results indicated the frequency of some virulence is rising. The high frequency of virulence factors found in these blaNDM and blaKPC bacteria is a problem for treatment. Some researchers suggested that molecular typing and virulence gene analysis are powerful tools that can shed light on Klebsiella pneumonia infections.12,15,33,34 However, in this study, we found that some isolates were high serum resistance (Grade 5 or Grade 6) but the number of the virulence factors was less to some serum resistance strains. This results showed that how to identify the hvKP is still unknown. We suspect that the comprehensive analysis the frequency of the virulence factors, phenotype (biofilm, sting test and serum killing assay) and clinical characteristics maybe a preferable method to identitfy the hvKP strains.
In conclusion, this study demonstrated that the high frequency and diversity of the resistance and virulence factors was in the blaNDM and blaKPC co-producing K. pneumoniae making this strain resistant to almost all antibiotics. This study may offer hospitals important information about the control of infections caused by blaKPC and blaNDM co-producing Klebsiella pneumoniae.
Acknowledgment
This research was funded by the National Natural Science Foundation of China (31500114) and by a grant from the Sichuan Province Science and Technology project (2016JY0223) and Luzhou and Southwest Medical University Natural Science Foundation [2018LZXNYD-ZK51] and Southwest Medical University Science Park funding [2019005].
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
- 1.Feng J, Qiu Y, Yin Z, et al. Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J Antimicrob Chemother. 2015;70:2987. doi: 10.1093/jac/dku445 [DOI] [PubMed] [Google Scholar]
- 2.Zheng B, Xu H, Yu X, et al. Identification and genomic characterization of a KPC-2-, NDM-1- and NDM-5-producing Klebsiella michiganensis isolate. J Antimicrob Chemother. 2017;73:536–538. [DOI] [PubMed] [Google Scholar]
- 3.Fu L, Wang S, Zhang Z, et al. Co-carrying of KPC-2, NDM-5, CTX-M-3 and CTX-M-65 in three plasmids with serotype O89: H10 Escherichia coli strain belonging to the ST2 clone in China. Microb Pathog. 2019;128:1–6. doi: 10.1016/j.micpath.2018.12.033 [DOI] [PubMed] [Google Scholar]
- 4.Freire Martin I, AbuOun M, Reichel R, La Ragione RM, Woodward MJ. Sequence analysis of a CTX-M-1 IncI1 plasmid found in Salmonella 4,5,12: i:-,Escherichia coli and Klebsiella pneumoniae on a UK pig farm. J Antimicrob Chemother. 2014;69:2098–2101. doi: 10.1093/jac/dku098 [DOI] [PubMed] [Google Scholar]
- 5.Peirano G, Schreckenberger PC, Pitout JD. Characteristics of NDM-1-producing Escherichia coli isolates that belong to the successful and virulent clone ST131. Antimicrob Agents Chemother. 2011;55:2986. doi: 10.1128/AAC.01763-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan YS, Zong ZY, Yuan L, et al. Complete sequence of pEC012, a multidrug-resistant IncI1 ST71 plasmid carrying blaCTX-M-65, rmtB, fosA3, floR, and oqxAB in an Avian Escherichia coli ST117 strain. Front Microbiol. 2016;7:1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian GB, Wang HN, Zou LK, et al. Detection of CTX-M-15, CTX-M-22, and SHV-2 extended-spectrum beta-lactamases (ESBLs) in Escherichia coli fecal-sample isolates from pig farms in China. Foodborne Pathog Dis. 2009;6:297. doi: 10.1089/fpd.2008.0164 [DOI] [PubMed] [Google Scholar]
- 8.Chao L, Shi J, Guo J. High prevalence of hypervirulentKlebsiella pneumoniaeinfection in the genetic background of elderly patients in two teaching hospitals in China. Infecti Drug Resist. 2018;11:1031–1041. doi: 10.2147/IDR.S161075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu L, Huang M, Zhang X, et al. Frequency of virulence factors in high biofilm formation blaKPC-2 producing Klebsiella pneumoniae strains from hospitals. Microb Pathog. 2018;116:168–172. doi: 10.1016/j.micpath.2018.01.030 [DOI] [PubMed] [Google Scholar]
- 10.Xu M, Fu Y, Fang Y, et al. High prevalence of KPC-2-producing hypervirulent Klebsiella pneumoniae causing meningitis in Eastern China. Infect Drug Resist. 2019;12:641–653. doi: 10.2147/IDR.S191892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Struve C, Roe CC, Stegger M, et al. Mapping the evolution of Hypervirulent Klebsiella pneumoniae. mBio. 2015;6:e00630. doi: 10.1128/mBio.00630-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. 2008;62:1. doi: 10.1016/j.diagmicrobio.2008.04.007 [DOI] [PubMed] [Google Scholar]
- 13.Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016;6:38929. doi: 10.1038/srep38929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye M, Tu J, Jiang J, et al. Clinical and genomic analysis of liver abscess-causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front Cell Infect Microbiol. 2016;6:165. doi: 10.3389/fcimb.2016.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 2019; 32:e00001–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Zhang H, Zhang X, et al. Characterization of an NDM-19-producing Klebsiella pneumoniae strain harboring 2 resistance plasmids from China. Diagn Microbiol Infect Dis. 2019;93:355–361. doi: 10.1016/j.diagmicrobio.2018.11.007 [DOI] [PubMed] [Google Scholar]
- 17.Fu L, Tang L, Wang S, et al. Co-location of the blaKPC-2, blaCTX-M-65, rmtB and virulence relevant factors in an IncFII plasmid from a hypermucoviscous Klebsiella pneumoniae isolate. Microb Pathog. 2018;124:301–304. doi: 10.1016/j.micpath.2018.08.055 [DOI] [PubMed] [Google Scholar]
- 18.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–118. doi: 10.4161/viru.22718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podschun R, Sievers D, Fischer A, Ullmann U. Serotypes, hemagglutinins, siderophore synthesis, and serum resistance of klebsiella isolates causing human urinary tract infections. J Infect Dis. 1993;168:1415–1421. doi: 10.1093/infdis/168.6.1415 [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhao C, Wang Q, et al. High prevalence of hypervirulent klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60:6115–6120. doi: 10.1128/AAC.01127-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan H, Chai T, Liu J, et al. Source identification of airborne Escherichia coli of swine house surroundings using ERIC-PCR and REP-PCR. Environ Res. 2009;109:511–517. doi: 10.1016/j.envres.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119. doi: 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 24.Pérezpérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153. doi: 10.1128/JCM.40.6.2153-2162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Ling B, Zhou L. Prevalence of 16S rRNA methylase, modifying enzyme, and extended-spectrum beta-lactamase genes among Acinetobacter baumannii isolates. J Chemother. 2015;27:207–212. doi: 10.1179/1973947814Y.0000000190 [DOI] [PubMed] [Google Scholar]
- 26.Aminov RI, Chee-Sanford JC, Garrigues N, Mehboob A, Mackie RI. Detection of tetracycline resistance genes by PCR methods. Methods Mol Biol. 2004;268:3–13. [DOI] [PubMed] [Google Scholar]
- 27.Zhang AY, Wang HN, Tian GB, et al. Phenotypic and genotypic characterisation of antimicrobial resistance in faecal bacteria from 30 Giant pandas. Int J Antimicrob Agents. 2009;33:456. doi: 10.1016/j.ijantimicag.2008.10.030 [DOI] [PubMed] [Google Scholar]
- 28.Wu JJ, Ko WC, Tsai SH, Yan JJ. Prevalence of plasmid-mediated quinolone resistance determinants QnrA, QnrB, and QnrS among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob Agents Chemother. 2007;51:1223–1227. doi: 10.1128/AAC.01195-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andres P, Lucero C, Soler-Bistue A, et al. Differential distribution of plasmid-mediated quinolone resistance genes in clinical enterobacteria with unusual phenotypes of quinolone susceptibility from Argentina. Antimicrob Agents Chemother. 2013;57:2467–2475. doi: 10.1128/AAC.01615-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:3582–3584. doi: 10.1128/AAC.01574-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turton JF, Perry C, Elgohari S, Hampton CV. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;59:541–547. doi: 10.1099/jmm.0.015198-0 [DOI] [PubMed] [Google Scholar]
- 32.Compain F, Babosan A, Brisse S, et al. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol. 2014;52:4377–4380. doi: 10.1128/JCM.02316-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min X, Fu Y, Kong H, et al. Bloodstream infections caused by Klebsiella pneumoniae: prevalence of bla KPC, virulence factors and their impacts on clinical outcome. BMC Infect Dis. 2018;18:358. doi: 10.1186/s12879-018-3109-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Xie Y, Li G, et al. Whole-Genome-Sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence. 2018;9:510–521. doi: 10.1080/21505594.2017.1421894 [DOI] [PMC free article] [PubMed] [Google Scholar]