BAP1 regulates developmental switch in lineages commonly affected by BAP1-mutant cancers.
Abstract
The BAP1 tumor suppressor is mutated in many human cancers such as uveal melanoma, leading to poor patient outcome. It remains unclear how BAP1 functions in normal biology or how its loss promotes cancer progression. Here, we show that Bap1 is critical for commitment to ectoderm, mesoderm, and neural crest lineages during Xenopus laevis development. Bap1 loss causes transcriptional silencing and failure of H3K27ac to accumulate at promoters of key genes regulating pluripotency-to-commitment transition, similar to findings in uveal melanoma. The Bap1-deficient phenotype can be rescued with human BAP1, by pharmacologic inhibition of histone deacetylase (HDAC) activity or by specific knockdown of Hdac4. Similarly, BAP1-deficient uveal melanoma cells are preferentially vulnerable to HDAC4 depletion. These findings show that Bap1 regulates lineage commitment through H3K27ac-mediated transcriptional activation, at least in part, by modulation of Hdac4, and they provide insights into how BAP1 loss promotes cancer progression.
INTRODUCTION
BAP1 [breast cancer type 1 (BRCA1)–associated protein 1] is emerging as an important tumor suppressor in human cancer (1), as it frequently sustains inactivating mutations in uveal melanoma, renal cell carcinoma, mesothelioma, and other malignancies (2–4). However, the role of BAP1 in normal and tumor biology remains unclear. BAP1 encodes a conserved nuclear-localizing ubiquitin C-terminal hydrolase that deubiquitinates multiple substrates, including histone H2A, host cell factor 1 (HCF1; also known as HCFC1), O-linked N-acetylglucosamine transferase, BRCA1, and the transcription factor Kruppel-like factor 5 (1, 5). Bap1 protein is found in nuclear complexes containing HCF1, the Polycomb group (PcG) proteins ASXL1-3 (“ASXLs”), the forkhead transcription factors FoxK1 and FoxK2 (FOXK1-2), the lysine-specific demethylase 1B (KDM1B), and the transcription factor Yin Yang 1 (YY1) (1), suggesting a role for BAP1 in epigenetic regulation of gene expression. Early cell culture experiments suggested a role for BAP1 in cell cycle regulation (5–7), but in vivo studies have revealed a more complex biological role for BAP1 involving development and differentiation. In Drosophila, the BAP1 ortholog calypso is required for embryonic development and cooperates with Asx to protect transcriptionally active developmental genes against silencing by the Polycomb repressive complex 1 (PRC1) (8, 9). In mice, BAP1 loss in the germ line is embryonic lethal (10), and BAP1 loss in the developing kidney causes differentiation failure and tumorigenesis (11). In human uveal melanoma and renal cell carcinoma, BAP1 loss leads to a dedifferentiated stem-like phenotype associated with aggressive cancer behavior and poor patient outcome (2, 3). To gain new insights into the role of BAP1 in development and tumorigenesis, we investigated its role in vertebrate embryogenesis using Xenopus laevis as a model system.
BAP1 expression is restricted to neural crest progenitor lineages early in embryogenesis
The Xenopus Bap1 protein shares 92% similarity and 71% identity with human BAP1, compared to 85 and 66%, respectively, for zebrafish (fig. S1A). Amino acid identity exceeds 90% in conserved regions, such as the catalytic domain and the binding motifs for HCF1 and ASXLs (fig. S1B). Maternally derived bap1 mRNA is abundant in Xenopus oocytes, and embryonic bap1 transcription commences at the midblastula transition (fig. S1C). In contrast, Bap1 protein was not detected until the 32-cell stage (stage 6), with progressively increasing protein levels thereafter (fig. S1D), suggesting a silencing period in which bap1 transcripts are translationally repressed. By the gastrulation stage, bap1 mRNA is expressed predominantly in ectoderm/mesoderm and becomes restricted to neural plate later, during early neurulation (fig. S2, A to C). By midneurula, bap1 mRNA was detected in the midbrain region and the lateral and anterior neural folds, areas that give rise to neural crest cells and sensorial placodes, including the early eye field (fig. S2, D and E). Later in development, bap1 mRNA expression is restricted to migrating cranial neural crest cells, branchial arches, otic vesicle, and eye fields (fig. S2, F and G). A similar pattern was observed for Bap1 protein expression (fig. S2, H to L).
Loss of Bap1 during Xenopus development produces a distinctive phenotype
To investigate the effects of Bap1 loss during development, we designed an antisense morpholino oligonucleotide that binds the 5′ untranslated region (5′UTR) of bap1 (Bap1MO) and efficiently blocks translation of Bap1 protein (fig. S3, A and B, and data file S1). Injection of Bap1MO into one blastomere in two-cell–stage embryos interfered with blastopore closure, leading to a delay or arrest in gastrulation on the injected side, compared to the uninjected control side (Fig. 1, A and B). This early phenotype may explain why homozygous germline deletion of Bap1 in mice is associated with embryonic lethality (10). The same phenotype was observed, albeit at a lower frequency, using a different morpholino that only targets the bap1 open reading frame, thereby causing less efficient knockdown of Bap1 compared to the 5′UTR morpholino (fig. S3A, data file S1). In contrast, a base pair mismatch control morpholino (Bap1MO-Ctrl) that is unable to bind wild-type bap1 mRNA (fig. S3A and data file S1) caused no phenotypic abnormalities (Fig. 1, A and B), confirming the specificity of the morpholino-driven Bap1 depletion phenotype. About 75% of Bap1MO-injected embryos exhibited gastrulation abnormalities, yet ~60% eventually completed gastrulation and proceeded through development, where they demonstrated additional malformations, including axial foreshortening (Fig. 1C), microphthalmia or anophthalmia (Fig. 1, D to J), and proliferation of immature melanoblasts with an altered migration pattern during late tail bud and early tadpole stages (Fig. 1D). To confirm that the eye phenotype is a direct effect of Bap1 loss, we knocked down Bap1 specifically in cells destined to form the future eye field by injecting Bap1MO into blastomere D1.2—which gives rise to retina, lens, and other eye structures (12)—in 16-cell stage embryos. In the absence of Bap1, the dorsal eye structures appeared morphologically normal, but the medial and ventral eye structures failed to form properly (Fig. 1, E to I). Consistent with morphologic findings, the dorsal eye marker vent2 was unaffected by Bap1 loss, whereas medial and ventral eye markers were depleted or absent (Fig. 1J). The Bap1MO phenotype was rescued by coinjection of Bap1MO with either human BAP1 mRNA (fig. S4, A to C) or a morpholino-resistant Xenopus bap1 mRNA (bap1-MM) containing conservative nucleotide substitutions that abolish morpholino binding (fig. S3A).
Fig. 1. Loss of Bap1 during Xenopus development produces a distinctive phenotype.
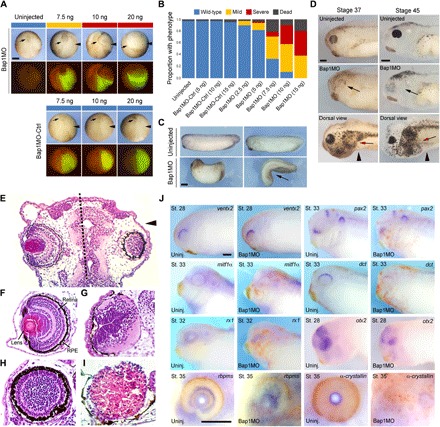
(A) Representative embryos analyzed at late gastrula (stage 12) following injection into one blastomere at the two-cell stage of escalating doses (7.5, 10, and 20 ng) of a morpholino targeting the 5′UTR of bap1 mRNA (Bap1MO) or a bap1 base pair mismatch control morpholino (Bap1MO-Ctrl). Below each bright-field image is a corresponding fluorescence image demonstrating fluorescein isothiocyanate as a lineage tracer for the injected side (green color). Depletion of Bap1 produces gastrulation failure, as evidenced by incomplete blastopore closure (arrows). Arrowheads indicate injected side. Panels show dorsal view, anterior down. (B) Summary of results of experiments described in (A), showing that depletion of Bap1 produces gastrulation failure ranging from mild (yellow) to severe (red) in a dose-dependent manner. (C) Representative embryos treated as above with 7.5 ng of Bap1MO, which eventually completed gastrulation and developed axial foreshortening and bending (arrow) starting at early tail bud stages, compared to uninjected sibling embryos. Panels show lateral view, anterior left, except the lower right panel, which shows dorsal view, anterior down. (D) Representative embryos treated as above with 7.5 ng of Bap1MO, which eventually completed gastrulation and were evaluated at stage 37 or 45, showing microphthalmia or anophthalmia (black arrows) starting at late tail bud stages, and proliferation of morphologically immature melanoblasts with altered migration pattern (red arrows) starting at late tail bud and early tadpole stages. Panels show lateral view (except panels labeled dorsal view), anterior left. Arrowheads indicate the injected side. (E) Transverse sections through the head of a representative early–tadpole stage embryo stained with hematoxylin and eosin, following injection into one blastomere (D1.2) at the 16-cell stage with 7.5 ng of Bap1MO, showing disruption of eye development on the side injected with Bap1MO (right side, arrowhead), compared to normal eye development on the uninjected control side (left side). Dotted line indicates midline. (F) Normal histologic appearance of the eye at early–tadpole stage embryo [same orientation as (E)], with ocular structures indicated. RPE, retinal pigment epithelium. (G to I) Representative eyes at late tail bud/early–tadpole stage embryos showing mild (G), moderate (H), and severe (I) ocular malformation associated with injection of 7.5 ng of Bap1MO into blastomere D1.2 (which gives rise to retina, lens, and other eye structures) at the 16-cell stage. In severe cases, such as the example in (I), retinal tissue does not form, and the eye remains filled with yolk platelets (pink). (J) Whole-mount in situ hybridization (WISH) of indicated eye markers in embryos injected with 7.5 ng of Bap1MO and a lineage tracer into D1.2 at the 16-cell stage and analyzed at the indicated stages, showing aberrant development of ocular tissues in the absence of Bap1. Markers include ventx2 (dorsal retina, unaffected), pax2 (ventral optic stalk), mitfα and dct [retinal pigment epithelium and uveal melanocytes (UMCs)], rx1 (ciliary marginal zone and photoreceptor), otx2 and rbpms (retinal ganglion cell layer), and α-crystallin (lens). Panels show lateral view, anterior side left. Uninj., uninjected. St., stage. Scale bars, 250 μm.
Bap1 loss deregulates expression of pluripotency and lineage commitment genes
To identify embryonic lineages affected by Bap1 loss, we assessed spatial mRNA expression of key developmental genes using whole-mount in situ hybridization (WISH). During gastrulation and early neurulation, Bap1-depleted embryos not only failed to silence the pluripotency factors vent1 and vent2 (orthologs of mammalian Nanog) and oct25 and oct91 (orthologs of mammalian Oct4) but also failed to switch on expression of lineage-specific factors, such as fzd7 (dorsal mesoderm and ectoderm), vegT, brachyury, and myoD (mesoderm), keratin1 (non-neural ectoderm), sox2 (neural ectoderm), rx1 (presumptive eye field), zic1 and msx1 (neural fold/prospective neural crest), and foxD3 and sox10 (neural crest) (Fig. 2). The expression of these molecular markers was restored to wild type by coinjection of human BAP1 mRNA or morpholino-resistant bap1 (fig. S4D). The pattern of transcriptomic disruption observed in Bap1-deficient embryos is evocative of our findings in human uveal melanoma, where BAP1 loss was associated with up-regulation of pluripotency markers and silencing of neural crest/melanocyte lineage markers (2). These findings validated the specificity of Bap1MO, demonstrated the conserved function of Bap1 across vertebrate species, confirmed that Xenopus Bap1 is a bona fide ortholog of human BAP1, and indicated that Bap1 is required for normal development by promoting the transcriptional switch from pluripotency to commitment in multiple lineages.
Fig. 2. Bap1 loss deregulates expression of pluripotency and lineage commitment genes.
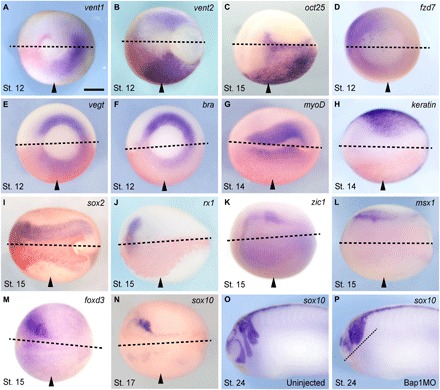
(A to P) Representative embryos injected with 7.5 ng of the Bap1MO morpholino into one blastomere at the two-cell stage (arrowheads indicate injected side) and then fixed and analyzed for mRNA expression of the indicated developmental genes by WISH at the specified stages (gastrula, stage 12; midneurula, stages 14 to 17; early tail bud, stage 24). Bap1-depleted embryos fail to silence pluripotency factors such as vent1/2 (orthologs of mammalian Nanog) and oct25 (ortholog of mammalian Oct4) (A to C) and fail to activate lineage commitment factors such as fzd7 (dorsal mesoderm and ectoderm), vegT and bra (axial mesoderm), myoD (muscle), keratin1 (non-neural ectoderm), sox2 (neural ectoderm), rx1 (early eye field), zic1 and msx1 (neural fold/prospective neural crest), and foxD3 and sox10 (neural crest) (D to N). Bap1 loss results in a failure of neural crest cell migration in sox10-expressing cells compared to the uninjected control (O and P). Panels show the dorsal-caudal view (A, B, and D to F), the dorsal view (C and G to N), or the lateral view (O and P), anterior side left. Dotted lines indicate midline. Arrowheads indicate injected side. Scale bar, 250 μm.
Bap1 function during development is dependent on its catalytic activity and interaction with Asxls
To explore how Bap1 regulates these transcriptional programs, we first asked whether it requires interaction with Asxls, which are among the most abundant Bap1-interacting proteins in Xenopus embryos (data file S2A) and human uveal melanoma cells, as well as other vertebrate cells (6, 10). The predominant family member expressed during Xenopus development is asxl1, and its pattern of mRNA expression closely parallels that of bap1 (fig. S5, A and B). To analyze Asxl1 function in vivo, we designed two specific morpholinos that target asxl1.S and asxl1.L, respectively, which were used together and referred to hereafter as Asxl1MO (data file S1). Asxl1 depletion resulted in morphogenetic defects identical to those observed in Bap1-deficient embryos (fig. S5C). Asxls interact with Bap1 through a highly conserved Asxl-binding motif (ABM) comprising amino acid residues 618 to 641 located in the Bap1 C-terminal region (fig. S1B) (13). This interaction promotes histone H2A deubiquitination by increasing the affinity of Bap1 for ubiquitinated Lys119 (8, 13, 14). We generated a complementary DNA (cDNA) expression construct encoding a Bap1 protein containing a four–amino acid deletion within the ABM (Bap1ΔABM) to abrogate Bap1-Asxl1 binding (fig. S3, C and D). In contrast to wild-type Bap1 and human BAP1, Bap1ΔABM failed to rescue the Bap1-deficient morphogenetic phenotype (fig. S5D). Similarly, a Bap1 mutant containing a single amino acid substitution at the conserved catalytic Cys91 residue (Bap1-C91W), corresponding to a human cancer–derived missense mutation that abrogates ubiquitin hydrolase activity (15), also failed to rescue the Bap1MO phenotype (fig. S3C and data not shown). Since most BAP1 missense mutations in human tumors cluster around the ABM and catalytic domain (1, 2, 13), the functions of Bap1 in development seem to parallel those in tumor suppression.
Loss of Bap1 abrogates the assembly of H3K27ac at promoters of key genes regulating lineage commitment and differentiation
Bap1 and Asxls form the catalytic and regulatory subunits, respectively, of the polycomb repressive deubiquitinase (PR-DUB) that deubiquitinates histone H2A at Lys119, thereby opposing the H2AK119 ubiquitin ligase activity of the PRC1 complex (8, 14) that, in turn, promotes the repressive PRC2-mediated trimethylation of H3K27 (16, 17). To explore in further detail the transcriptional and epigenetic programs disrupted by Bap1 loss, we performed RNA sequencing (RNA-seq) and chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) in late gastrula stage embryos (stage 12), injected with either Bap1MO or control morpholino at one-cell stage (data file S2, B and C). Consistent with WISH experiments, RNA-seq shows that depletion of Bap1 abrogated the induction of genes regulating ectoderm, mesoderm, and neural crest commitment, including components of the Wnt and Hippo pathways (Fig. 3, A and B). As expected, depletion of Bap1 resulted in a global increase in H2AK119ub and H3K27me3 around transcription start sites across the genome (Fig. 3C). Unexpectedly, however, the histone mark that is most strongly associated with failed expression of lineage commitment genes in Bap1-depleted embryos is H3K27ac (Fig. 3D), with promoters around many of these genes failing to assemble H3K27ac in the absence of Bap1 (Fig. 3E and data file S2D).
Fig. 3. Loss of Bap1 abrogates the assembly of H3K27ac at promoters of key genes regulating lineage commitment and differentiation.
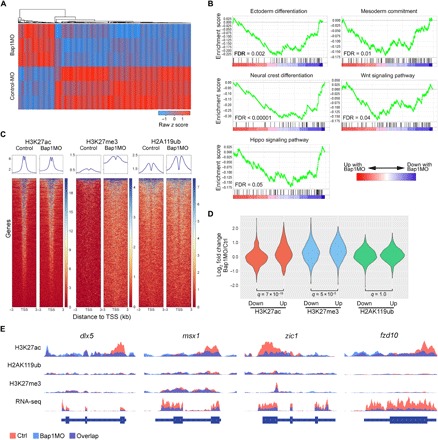
(A) Heat map demonstrating the top 1000 most differentially expressed genes by RNA-seq between embryos at one-cell stage with 15 ng of either Bap1MO or Bap1MO-Ctrl and collected at late gastrulation (stage 12), when morphologic effects of Bap1 loss are first evident. (B) Gene set enrichment analysis (GSEA) plots demonstrating the most highly significant pathways represented by the differentially expressed genes associated with Bap1 loss. FDR, false discovery rate. (C) Heat maps of ChIP-seq data demonstrating global genomic occupancy of the indicated histone marks across all annotated genes in embryos treated as above with either Bap1MO or Bap1MO-Ctrl. TSS, transcription start site. (D) Violin plots summarizing ChIP-seq data restricted to differentially expressed genes. (E) ChIP-seq and RNA-seq tracks of representative lineage commitment genes that fail to assemble H3K27ac at promoters and to activate mRNA expression in Bap1-deficient embryos.
Pharmacologic inhibition of histone deacetylase activity rescues Bap1-deficient gastrulation phenotype
Since Bap1 orchestrates the assembly of H3K27ac at particular lineage commitment genes, we reasoned that the abnormalities resulting from Bap1 loss may be rescued, at least in part, by opposing histone deacetylase (HDAC) activity. Treatment of Bap1-depleted embryos starting at the 32-cell stage (stage 6) with the pan-HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) rescues gastrulation arrest, allowing embryos to complete blastopore closure (fig. S6A). In contrast, no rescue was observed with pharmacologic inhibition of Ring1, the catalytic subunit of the PRC1 complex that ubiquitinates H2AK119, nor with depletion of the PRC1 core component Bmi1 (fig. S6C). Addition of a Bmi1-directed morpholino exacerbates the gastrulation defect caused by the Bap1-directed morpholino (data file S1 and data not shown), suggesting that PRC1 and the Bap1-containing PR-DUB complex do not function in a purely antagonistic relationship, similar to findings in Drosophila (8). Similarly, no rescue was observed with a pharmacological inhibition of the PRC2 catalytic subunit enhancer of zeste homolog 2 (Ezh2), which catalyzes trimethylation of H3K27 (fig. S6C).
Hdac4 is a key mediator of the Bap1-deficient phenotype
Since SAHA inhibits numerous HDACs, we questioned whether there may be a specific HDAC that is up-regulated by Bap1 loss and mediates the Bap1-deficient phenotype. By comparing RNA-seq data from our embryo experiments to the publicly available human uveal melanoma RNA-seq data from The Cancer Genome Atlas (TCGA) (18), we observed that HDAC4 is the only HDAC that is significantly up-regulated [false discovery rate (FDR) < 0.001] by BAP1 loss in both datasets (fig. S7A). Therefore, we designed two specific morpholinos that target hdac4.S and hdac4.L, respectively, which were used together and referred to hereafter as Hdac4MO (fig. S3A and data file S1). Concomitant depletion of Bap1 and Hdac4 in whole embryos using Bap1MO and Hdac4MO rescues the Bap1-deficient phenotype, as evidenced by restoration of normal morphologic development (Fig. 4, A and B); transcriptional activation of lineage commitment genes such as sox2, bra, msx1, and fzd7 (Fig. 4C); and accumulation of H3K27ac at the promoters of lineage commitment genes (Fig. 4D). Together, these findings implicate Hdac4 as an important mediator of Bap1 function by controlling the timely assembly of H3K27ac at genes regulating induction of multiple embryonic lineages.
Fig. 4. Hdac4 is a key mediator of the Bap1-deficient phenotype.
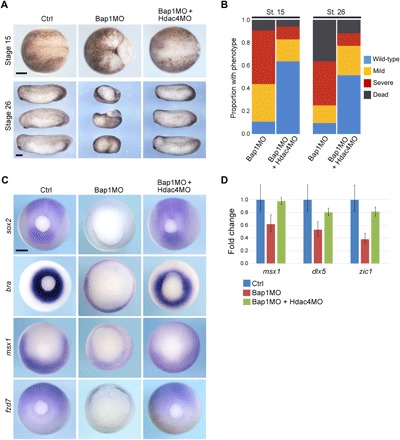
(A) Representative embryos injected at the one-cell stage with 7.5 ng of Bap1MO with or without 16 ng of a morpholino directed against Hdac4 (Hdac4MO) and analyzed at midneurula stage (stage 15, dorsal view, anterior to left) and early tail bud stage (stage 26, lateral view, anterior to left). (B) Summary of results at stages 15 and 26, showing substantial rescue of the Bap1-deficient phenotype with Hdac4MO. (C) Representative embryos treated as above and analyzed by WISH, demonstrating that failed induction of the indicated developmental genes in Bap1-deficient embryos is rescued by Hdac4MO. Caudal view, dorsal up. (D) ChIP–quantitative polymerase chain reaction (qPCR) for indicated gene promoters following ChIP for H3K27ac, confirming that failure to assemble H3K27ac at promoters in Bap1-deficient embryos can be rescued by depletion of Hdac4. Scale bars, 250 μm.
Hdac4 acts independently of the Smrt/Ncor1-Hdac3 complex
Since HDAC4 has been shown to interact with the SMRT/NCoR-HDAC3 repressor complex (19), we wondered whether the Smrt/Ncor1 complex plays a role in the Bap1-deficient phenotype. To address this possibility, we first compared the changes in gene expression associated with depletion of Bap1 to those associated with depletion of both Bap1 and Hdac4. However, differentially expressed genes were not enriched for a curated list of SMRT/NCoR gene targets (JEPSEN_SMRT_TARGETS) using gene set enrichment analysis (GSEA) (data not shown). Next, we investigated whether the Bap1-deficient phenotype could be rescued using an Hdac3-directed morpholino (Hdac3MO). In contrast to Hdac4, depletion of Hdac3 had no effect on Bap1-deficient embryos (fig. S7, B and C). These findings suggest that regulation of Hdac4 by Bap1 is not mediated through Smrt/Ncor1-Hdac3, at least during the stages of development under investigation here.
Antagonistic effect of HDAC4 and BAP1 in human uveal melanocytic cells
Hdac4 was not found among Bap1-interacting proteins in stage 12 frog embryos analyzed by mass spectrometry (data file S2A), suggesting that Bap1 and Hdac4 may interact functionally rather than through direct physical interaction. Consequently, we hypothesized that Bap1 may regulate Hdac4 by modulating its nuclear-cytoplasmic shuttling, an important mechanism for regulating its activity (20). Hdac4 is largely restricted to the cytoplasm in BAP1 wild-type uveal melanoma cells and in normal human uveal melanocytes (UMCs), whereas it localized to the nucleus in BAP1-mutant uveal melanoma cells and in UMCs in which a BAP1 mutation was introduced using CRISPR-Cas9 (fig. S7D). Thus, Bap1 appears to inhibit the epigenetic function of Hdac4, at least in part, by restricting its localization to the nucleus. Moreover, short hairpin RNA (shRNA)–mediated depletion of HDAC4 in BAP1-mutant uveal melanoma cells significantly impaired cell proliferation (fig. S8). Future work will be needed to elucidate the mechanism by which Bap1 carries out this function.
DISCUSSION
Disruption of Bap1 during development results in abnormalities largely affecting ectoderm, mesoderm, and neural crest, which may help to explain the spectrum of cancers associated with BAP1 mutations, which are mostly derived from those lineages (1). At the organismal level, BAP1 primarily appears to regulate cell identity and differentiation, rather than cell cycle and proliferation, which may explain why BAP1 mutations typically are not initiating events but, rather, later events associated with cancer progression (3, 21). Germline BAP1 mutations usually do not result in tumor formation unless accompanied by an initiating mutation, such as a Gq mutation in uveal melanoma or a BRAF mutation in cutaneous melanoma (21, 22). This phenomenon may explain why families with germline BAP1 mutations display reduced penetrance for any specific cancer type (23). While BAP1 can exert diverse effects through multiple interacting partners (1), our findings indicate that a critical function of Bap1 in development is to prevent deacetylation of H3K27 at genes regulating lineage commitment in a manner dependent on its deubiquitinase activity. Consistently, H3K27 deacetylation and H2AK119 ubiquitination represent two early chromatin alterations that enforce gene silencing during embryonic development (24). When Bap1 is lost in susceptible cell types, H3K27ac fails to accumulate at key promoters, at least in part, because of unrestrained Hdac4 activity, which may explain why BAP1 loss sensitizes uveal melanoma and mesothelioma cells to treatment with HDAC inhibitors (25, 26). These findings provide novel insights into the role of BAP1 in development and cancer, and they suggest HDAC inhibitors as potential therapeutic agents for treating BAP1-mutant cancers.
MATERIALS AND METHODS
Embryo manipulation
To obtain X. laevis embryos, female frogs were induced to ovulate by injecting 500 to 700 U of human chorionic gonadotropin subcutaneously, directly above the dorsal lymph sacs. After injection, females were maintained in the dark at 18°C overnight. After females began ovulating the following morning, one male was euthanized by placing it in 0.1% tricaine solution buffered with sodium bicarbonate for 30 min. Both testes were dissected and placed into a 35-mm dish containing 1.5 ml of 1× Mark’s modified ringer (MMR) solution [0.1 M NaCl, 2 mM KCl, 1 mM MgSO4, 2 mM CaCl2, and 5 mM Hepes (pH 7.8)]. Eggs were gently squeezed from females into 10-cm plates. Sperm were released to fertilize the eggs by mincing a small portion of the testes in a few drops of 1× MMR placed beside the eggs followed by gentle mixing of eggs and sperm together. After 10 min at room temperature (RT), the fertilized eggs were flooded with 0.1× MMR solution (pH 7.4). Embryos were dejellied using 2% l-cysteine solution (pH 7.8) for 2 min. Embryos were washed thrice with 0.1× MMR solution and placed in 0.1× MMR. At the desired developmental stage, embryos were injected with a Nanoject II Auto-Nanoliter Injector (Drummond Scientific Inc.). Injected embryos were maintained in 4% Ficoll/0.1% MMR solution at 18°C until they reached stage 9 and then transferred to 0.1× MMR buffer.
Histological analysis, WISH, and immunohistochemistry
X. laevis embryos were injected with morpholinos and/or mRNAs into one blastomere at the 1-, 2-, or 16-cell stage and collected at different stages; staging was according to Nieuwkoop and Faber (27). For histologic analysis, embryos were embedded into Paraplast, and 12-μm sections were cut on a rotary microtome and stained with hematoxylin and eosin. Immunohistochemistry for Bap1 was performed as previously described (28) using an antibody raised against human BAP1 (H300; Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:100 dilution. Embryos were incubated with the primary antibody for 24 hours and then incubated with alkaline phosphatase (AP)–conjugated secondary antibody (ab6729, Abcam, Cambridge, MA). Color was developed using a combination of nitro blue tetrazolium (NBT) chloride and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Promega, Madison, WI). WISH was performed as described (29). Embryos were fixed in 4% MEMFA [100 mM MOPS (pH 7.4), 2 mM EGTA, and 1 mM MgSO4, and 3.7% (v/v) formaldehyde] 1 hour at RT, dehydrated with 100% methanol in three 10-min washes, and either kept at −20°C in methanol or rehydrated through a methanol wash series {75, 50, and 25% in PTw [phosphate-buffered saline (PBS) buffer and Tween 20]} for 5 min each step, followed by a 5-min PTw wash. Embryos were rinsed for 5 min, prehybridized with hybridization buffer at 65°C for 5 to 6 hours, and then hybridized overnight in hybridization buffer containing the appropriate amount of antisense probe at 65°C. Next, embryos were sequentially washed in 50% formamide/2× SSC and 0.1% Tween 20 (10 min/60°C), 25% formamide/2× SSC and 0.1% Tween 20 (10 min/60°C), 12.5% formamide/2× SSC and 0.1% Tween 20 (10 min/60°C), 2× SSC and 0.1% Tween 20 (10 min/60°C), and 0.2× SSC and 0.1% Tween 20 (30 min/60°C). After three 5-min washes with PTw and two 5-min washes with maleic acid buffer (MAB), embryos were placed in MAB/2% Boehringer Blocking Reagent (BMB) blocking reagent (Roche) for 2 hours and then incubated in MAB/2% BMB containing 1:3000 diluted anti-digoxigenin/AP (Roche, Basel, Switzerland) overnight. After incubation, embryos were washed six times with MAB for 30 min each and twice with fresh AP buffer for 5 min, transferred to a precooled color reaction solution (AP buffer containing NBT chloride and BCIP), and incubated at RT in the dark until sufficient staining was reached. The staining reaction was stopped by replacing the staining solution with methanol, twice for 5 min. Embryos were left in a third methanol wash for 40 min at 62°C and then washed three to five times with water for rehydration. To bleach pigments, embryos were treated with 2% H2O2 in 1× PBS (stocks: 30% H2O2 and PBS 20×) under fluorescent light for 4 to 6 hours, washed several times with water, and stored in 4% formaldehyde in 1× PBS at 4°C. In all experiments in which embryos were injected past the one-cell stage, fluorescein isothiocyanate was coinjected to mark the injected side.
Morpholinos and DNA constructs
Morpholino antisense oligonucleotides were designed and synthesized against X. laevis bap1, asxl, and hdac4, targeting both homologs, from the long and short chromosomes (Gene Tools LLC). Morpholino against hdac3 was designed as previously described (data file S1). Doses of 5 to 15 ng per embryo were microinjected as mentioned above. Plasmids containing cDNA clones that were used to generate WISH RNA probes were obtained from previously described sources (data file S1). The following constructs were purchased from GE Dharmacon (Lafayette, CO): asxl1 (ID446414), bap1 (ID493568), and fzd7 (ID378787). Mutations were introduced to Xenopus bap1 and asxl1 cDNA using a Q5 site-directed mutagenesis kit (New England Biolabs, Ipswich, MA).
Mass spectrometry
FLAG-tagged Xenopus bap1 mRNA was synthesized from Not I–linearized pCS2+ plasmids using mMESSAGE mMACHINE SP6 transcription kit (Thermo Fisher Scientific, Waltham, MA). Embryos were injected at the one-cell stage with 0.5 ng of the synthetic mRNA and maintained at 22°C until they reached stage 12. Then, ~250 injected embryos were collected and lysed in 2.5 ml of mammalian cell PE LB buffer (G-Biosciences, St. Louis, MO) containing a complete protease inhibitor cocktail (Roche, Indianapolis, IN) by passing embryos through a 21-gauge needle ~20 times, placed on ice for 30 min, and then centrifuged at 12,000g for 10 min to obtain clear lysates. An equal number of uninjected sibling embryos were processed in an identical fashion, and then 1× FLAG peptide (Sigma-Adrich, St. Louis, MO) was added to the clear lysate to the final concentration of 10 ng/ml to be used as a control. Both FLAG-Bap1 and FLAG-containing control lysates were precleared by coincubation with magnetic protein G beads (Thermo Fisher Scientific, Waltham, MA) and mouse immunoglobulin G (2 μg/ml) at 4°C for 2 hours. Final coimmunoprecipitation was performed overnight at 4°C using FLAG-M2 mouse antibody, followed by a 10-min wash with coimmunoprecipitation buffer. The magnetic beads containing coimmunoprecipitated complexes were sent for further processing and analysis to the Proteomics and Metabolomics Shared Resource at The Wistar Institute (Philadelphia, PA). Mass spectrometry findings were filtered in the following manner. Any protein with a label-free quantification (LFQ) intensity greater in the control than the FLAG-Bap1 sample was discarded. In addition, all proteins were removed where the control LFQ intensity was >1% of the corresponding FLAG-Bap1 LFQ intensity. Furthermore, proteins were required to have at least an LFQ intensity of 5,000,000 and two unique peptide reads. Last, results were filtered for known mass spectrometry contaminants of FLAG-tagged experiments using the CRAPome (www.nature.com/articles/nmeth.2557).
In vitro translation
In vitro translation was carried out using wheat germ extracts according to the manufacturer’s instructions (Promega, Madison, WI). A total of 1.0 μg of capped-mRNA and different amounts of morpholinos were used per reaction. Protein expression was then analyzed by Western blot.
SAHA rescue experiments
Two-cell stage embryos were injected into one blastomere with 10 ng of Bap1MO and maintained in 4% Ficoll/1× MMR solution at 22°C until maturing to stage 6. SAHA was dissolved in 100% dimethyl sulfoxide (DMSO) to make a 1 mM stock solution and kept at −20°C. SAHA was added to medium containing the embryos to a final concentration of 20 μM. Control sibling embryos were treated with an equal volume of DMSO. Upon reaching stage 10, embryos were transferred to 0.1 MMR solution supplemented with fresh SAHA every 4 hours. Embryos were collected and fixed at different stages for morphological and molecular analysis.
ChIP-seq and ChIP–quantitative polymerase chain reaction
ChIP-seq and ChIP–quantitative polymerase chain reaction (qPCR) were performed on 500 and 100 stage 12 embryos, respectively, according to a protocol from the Cold Spring Harbor Laboratory (30), with 35 min of cross-linking. Sodium N-lauroylsacrosine in buffer E3 was replaced by 0.1% SDS, as suggested by the authors to increase specificity for certain antibody types. Chromatin was sonicated to an average fragment size of 200 base pairs (bp) with a Diagenode Bioruptor Pico before ChIP. A total of 5 μg of antibody was used for each ChIP, with the exception of H2AubK119, where 2 μg was used. The following antibodies were used to perform ChIP: H2AK199ub (Cell Signaling Technology, D27C4, no. 8240), H3K27me3 (Cell Signaling Technology, C36B11, 9733BF), H3K27AC (Active Motif, no. 39133), and H3K4me3 (Active Motif, no. 39915). Libraries were prepared using the NEBNext Ultra kit and sequenced on the Illumina NextSeq 500 (1 × 75 bp). ChIP-seq data were marked for duplicates using Picard (v1.128) and filtered for mapping quality using SAMtools (v1.2). Normalization of tracks was conducted using THOR, with binning set to 10/5 (31). Plotting of the average ChIP-seq signals was performed using a custom script. Heat maps were generated with deeptools (32), and kmeans clustering selected to compute the matrices. Quantification of ChIP-seq data for specific regions was done using bedtools (33). Genes that did not have National Center for Biotechnology Information (NCBI) annotations were identified via a custom-built script, which performed automated alignment using BLASTN online alignment tool against the NCBI genome database. The unidentified genes were assigned a name based on the alignment score ≥200. ChIP-qPCR primer sequences were deposited in ChIPprimersDB (34) and are listed in data file S1.
RNA sequencing
Embryos were injected at the one-cell stage with 15 ng of Bap1MO-Ctrl or Bap1MO and maintained at 22°C until they reached stage 12. RNA was isolated with TRIzol/chloroform, and yolk and pigment were further removed from samples using RNeasy MinElute Cleanup Kit (QIAGEN), according to the manufacturer’s protocol. Libraries were prepared using the NEBNext Ultra kit and sequenced on the Illumina NextSeq 500 (1 × 75 bp). Next generation sequencing quality was assessed using FastQC (v0.11.3). Reads were trimmed (if required) and aligned to the X. laevis 9.1 genome (www.xenbase.org/) using STAR (v2.5.2a) (35). Read counts of RNA-seq data were obtained using HTSeq (36) and analyzed for differential expression using EdgeR (37) with batch correction for paired experimental data. X. laevis gene names were changed to the names of human homologs to allow GSEA (38), which was conducted with recommended parameters and annotated gene sets obtained from the Molecular Signatures Database (39). To compare the differential expression of HDACs with respect to BAP1 status in Xenopus embryos and human uveal melanoma samples, we analyzed RNA-seq data from 80 primary human UMs generated by the TCGA (http://cancergenome.nih.gov/) (21).
Cell culture
This study was approved by the Institutional Review Board, and written informed consent was obtained from the patients. A BAP1 wild-type uveal melanoma cell line (UMM055) and a BAP1-mutant uveal melanoma cell line (UMM061) were generated from patient-derived xenografts (PDXs) generated from patients undergoing enucleation. PDXs were expanded in the intrascapular fat pad of NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ JAX immunodeficient (NSG) mice for up to 3 months. The tissue was digested with collagenase IV for 3 hours at 37°C, dissected into a single-cell suspension by pipetting, and cultured in 5% partial pressure of oxygen (pO2) in the UMM media containing Dulbecco’s modified essential medium (DMEM)/F12 with 5% heat-inactivated fetal bovine serum (HI-FBS), B-27 minus vitamin A (Life Technologies), 1% penicillin-streptomycin, 2 mM Glutamax, basic fibroblast growth factor (bFGF) (10 ng/ml) (PeproTech), recombinant stem cell factor (rSCF) (10 ng/ml) (PeproTech), and epidermal growth factor (20 ng/ml) (PeproTech). Cell culture surface was coated with 0.1% porcine gelatin (Millipore Sigma) before plating. PDX-derived BAP1-mutant UM cell line MP38 and BAP1 wild-type UM cell line MP41 were provided by S. Roman-Roman (40) and maintained in 5% pO2 in DMEM/F12 with 10% HI-FBS, 1% penicillin-streptomycin, 2 mM Glutamax, and 0.5% insulin-transferrin-selenium (Thermo Fisher Scientific). A cell line was generated from a normal UMC sample derived from unaffected normal intraocular uveal tissue in a patient undergoing enucleation. The uveal tissue was digested with collagenase IV for 3 hours at 37°C, dissected into a single-cell suspension by pipetting, and cultured in 5% pO2 in the UMC media containing Ham’s F12 with 10% HI-FBS, 1% penicillin-streptomycin, 2 mM Glutamax, 100 μM 3-isobutyl-1-methylxanthine, and 10 ng/ml of each: bFGF (PeproTech), rSCF (PeproTech), and cholera toxin (Sigma-Aldrich). UMCs were enriched relative to other cell types by addition of G418 (100 μg/ml) (Thermo Fisher Scientific) to the media for the first 5 days in culture. UMCs were then immortalized by retroviral expression of human TERT (Addgene plasmid no. 1773), followed by hygromycin selection. UMCs with knockout of BAP1 were created by clonally isolating UMCs stably transduced with lentiviral particles encoding spCAS9 (Addgene plasmid no. 50661) and guide RNA against BAP1 (Addgene plasmid no. 64114) directing the CRISPR-mediated deletion of the first exon of the BAP1 gene. Immunofluorescence microscopy was performed on UM and UMC cells using Santa Cruz Biotechnology antibodies against HDAC4 (sc-46672) and BAP1 (sc-28236). shRNA CGACAGGCCTCGTGTATGATT and AAATTACGGTCCAGGCTAATT sequences targeting human HDAC4 cDNA (shHDAC4) were individually cloned into a pLH-spsgRNA2 vector (Addgene plasmid no. 64114) and stably integrated into UM cells via lentiviral transduction and selection with hygromycin B (50 μg/ml). Nonspecific shRNA sequence AACAGCCACAACGTCTATATC (shSCR) was used as control. Equal amount of shSCR- or shHDAC4-expressing cells was plated into 12-well plate, and after 2 weeks, the cells were stained using 0.5% crystal violet and 6% glutaraldehyde solution and counted using ImageJ.
Real-time reverse transcription PCR
Total RNA from three embryos was extracted using TRIzol (Invitrogen), and cDNA synthesis was carried out using random hexamer priming and the StrataScript Reverse Transcriptase. Quantitative reverse transcription PCR was performed on the 7900HT Fast Real-Time PCR TaqMan System (Applied Biosystems) using the Brilliant SYBR Green qPCR Master Mix (Stratagene). Measurements were performed in triplicates and normalized to the expression levels of eukaryotic translation elongation factor 1 α (ef1a). Bars indicate SE. The primer sequences are described in data file S1.
Ethical use of animals
The animal protocols used in this work were evaluated and approved by the Institutional Animal Care and Use Committee of the University of Miami (no. 12-276). All activities were performed in compliance with federal state and institutional regulations. The University was granted full accreditation by the Association for Assessment and Accreditation of Laboratory Animal Care, International in February 2005 and received its current reaccreditation in 14 March 2019. In addition, University of Miami is licensed by the U.S. Department of Agriculture and has filed a Letter of Assurance with the Office of Laboratory Animal Welfare, U.S. Department of Health and Human Services.
Statistical analysis
For GSEA, significance was assessed using FDR (38). Two-tailed t test was applied unless otherwise specified.
Supplementary Material
Acknowledgments
We thank L. Morey for helpful comments on the manuscript, the Sylvester Comprehensive Cancer Center Biostatistics and Bioinformatics and Onco-Genomics Shared Resources, and the University of Miami Center for Computational Science. Funding: This work was supported by the Department of Defense grant W81XWH-15-1-0578 (J.W.H.); National Institutes of Health (NIH) grants R01 CA125970 (J.W.H.), GM102397 (M.L.K.), and F30 CA206430 (M.G.F.); Research to Prevent Blindness Inc. Senior Scientific Investigator Award (J.W.H.), Alcon Research Institute (J.W.H.), University of Miami Sheila and David Fuente Graduate Program in Cancer Biology (M.G.F. and M.A.D.), AACR-Ocular Melanoma Foundation Fellowship (S.K.), and a gift from M. J. Daily (J.W.H.). Bascom Palmer Eye Institute also received funding from NIH Core Grant P30EY014801, Department of Defense Grant W81XWH-13-1-0048, and a Research to Prevent Blindness Inc. Unrestricted Grant. Author contributions: J.N.K., T.H.A., D.A.O., S.K., M.G.F., M.A.D., M.L.K., and J.W.H. designed experiments and analyzed the data; J.N.K., T.H.A., D.A.O., S.K., and D.A.R. performed experiments. S.K., M.G.F., and M.A.D. performed bioinformatic analysis. J.W.H. wrote the manuscript. M.L.K. and J.W.H. oversaw the work. Competing interests: J.W.H. is the inventor of intellectual property related to uveal melanoma and receives royalties from its commercialization (patent no. US 9,809,856, issued: 7 November 2017; patent no. US 9,441,277, issued: 13 September 2016). He is a paid consultant for Castle Biosciences, licensee of this intellectual property. All other authors declare no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All raw and processed sequencing data in this study have been submitted to the NCBI Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE126599. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaax1738/DC1
Fig. S1. X. laevis and human Bap1 proteins show extensive sequence homology.
Fig. S2. Anatomic distribution of Xenopus bap1 mRNA and Bap1 protein expression during early development.
Fig. S3. Bap1 morpholinos and expression constructs.
Fig. S4. Developmental abnormalities in Bap1-deficient embryos are rescued by exogenous human BAP1.
Fig. S5. Loss of Asxl1 phenocopy loss of Bap1 during Xenopus development.
Fig. S6. Pharmacologic rescue of Bap1-deficient phenotype.
Fig. S7. Hdac4 is a key mediator of Bap1-deficient phonotype and acts independently of Hdac3 during development.
Fig. S8. Inhibition of HDAC4 significantly impairs proliferation of uveal melanoma cells in a BAP1-dependent manner.
Data file S1. Oligonucleotides and plasmids used in this study.
Data file S2. Proteomic, transcriptomic, and epigenomic analyses.
REFERENCES AND NOTES
- 1.Carbone M., Yang H., Pass H. I., Krausz T., Testa J. R., Gaudino G., BAP1 and cancer. Nat. Rev. Cancer 13, 153–159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harbour J. W., Onken M. D., Roberson E. D., Duan S., Cao L., Worley L. A., Council M. L., Matatall K. A., Helms C., Bowcock A. M., Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330, 1410–1413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peña-Llopis S., Vega-Rubín-de-Celis S., Liao A., Leng N., Pavía-Jiménez A., Wang S., Yamasaki T., Zhrebker L., Sivanand S., Spence P., Kinch L., Hambuch T., Jain S., Lotan Y., Margulis V., Sagalowsky A. I., Summerour P. B., Kabbani W., Wong S. W., Grishin N., Laurent M., Xie X.-J., Haudenschild C. D., Ross M. T., Bentley D. R., Kapur P., Brugarolas J., BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 44, 751–759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bott M., Brevet M., Taylor B. S., Shimizu S., Ito T., Wang L., Creaney J., Lake R. A., Zakowski M. F., Reva B., Sander C., Delsite R., Powell S., Zhou Q., Shen R., Olshen A., Rusch V., Ladanyi M., The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 43, 668–672 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin J., Zhou Z., Chen W., Wang C., Zhang H., Ge G., Shao M., You D., Fan Z., Xia H., Liu R., Chen C., BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat. Commun. 6, 8471 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machida Y. J., Machida Y., Vashisht A. A., Wohlschlegel J. A., Dutta A., The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J. Biol. Chem. 284, 34179–34188 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misaghi S., Ottosen S., Izrael-Tomasevic A., Arnott D., Lamkanfi M., Lee J., Liu J., O’Rourke K., Dixit V. M., Wilson A. C., Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol. Cell. Biol. 29, 2181–2192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheuermann J. C., de Ayala Alonso A. G., Oktaba K., Ly-Hartig N., McGinty R. K., Fraterman S., Wilm M., Muir T. W., Müller J., Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campagne A., Lee M.-K., Zielinski D., Michaud A., Le Corre S., Dingli F., Chen H., Shahidian L. Z., Vassilev I., Servant N., Loew D., Pasmant E., Postel-Vinay S., Wassef M., Margueron R., BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat. Commun. 10, 348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dey A., Seshasayee D., Noubade R., French D. M., Liu J., Chaurushiya M. S., Kirkpatrick D. S., Pham V. C., Lill J. R., Bakalarski C. E., Wu J., Phu L., Katavolos P., LaFave L. M., Abdel-Wahab O., Modrusan Z., Seshagiri S., Dong K., Lin Z., Balazs M., Suriben R., Newton K., Hymowitz S., Garcia-Manero G., Martin F., Levine R. L., Dixit V. M., Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S.-S., Gu Y.-F., Wolff N., Stefanius K., Christie A., Dey A., Hammer R. E., Xie X.-J., Rakheja D., Pedrosa I., Carroll T., McKay R. M., Kapur P., Brugarolas J., Bap1 is essential for kidney function and cooperates with Vhl in renal tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 111, 16538–16543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moody S. A., Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev. Biol. 119, 560–578 (1987). [DOI] [PubMed] [Google Scholar]
- 13.Peng H., Prokop J., Karar J., Park K., Cao L., Harbour J. W., Bowcock A. M., Malkowicz S. B., Cheung M., Testa J. R., Rauscher F. J. III, Familial and somatic BAP1 mutations inactivate ASXL1/2-mediated allosteric regulation of BAP1 deubiquitinase by targeting multiple independent domains. Cancer Res. 78, 1200–1213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahtoe D. D., van Dijk W. J., Ekkebus R., Ovaa H., Sixma T. K., BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat. Commun. 7, 10292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishikawa H., Wu W., Koike A., Kojima R., Gomi H., Fukuda M., Ohta T., BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res. 69, 111–119 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Cooper S., Grijzenhout A., Underwood E., Ancelin K., Zhang T., Nesterova T. B., Anil-Kirmizitas B., Bassett A., Kooistra S. M., Agger K., Helin K., Heard E., Brockdorff N., Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 7, 13661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaFave L. M., Béguelin W., Koche R., Teater M., Spitzer B., Chramiec A., Papalexi E., Keller M. D., Hricik T., Konstantinoff K., Micol J.-B., Durham B., Knutson S. K., Campbell J. E., Blum G., Shi X., Doud E. H., Krivtsov A. V., Chung Y. R., Khodos I., de Stanchina E., Ouerfelli O., Adusumilli P. S., Thomas P. M., Kelleher N. L., Luo M., Keilhack H., Abdel-Wahab O., Melnick A., Armstrong S. A., Levine R. L., Loss of BAP1 function leads to EZH2-dependent transformation. Nat. Med. 21, 1344–1349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson A. G., Shih J., Yau C., Gibb E. A., Oba J., Mungall K. L., Hess J. M., Uzunangelov V., Walter V., Danilova L., Lichtenberg T. M., Kucherlapati M., Kimes P. K., Tang M., Penson A., Babur O., Akbani R., Bristow C. A., Hoadley K. A., Iype L., Chang M. T.; TCGA Research Network, Cherniack A. D., Benz C., Mills G. B., Verhaak R. G. W., Griewank K. G., Felau I., Zenklusen J. C., Gershenwald J. E., Schoenfield L., Lazar A. J., Abdel-Rahman M.-H., Roman-Roman S., Stern M. H., Cebulla C. M., Williams M. D., Jager M. J., Coupland S. E., Esmaeli B., Kandoth C., Woodman S. E., Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 32, 204–220.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischle W., Dequiedt F., Hendzel M. J., Guenther M. G., Lazar M. A., Voelter W., Verdin E., Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9, 45–57 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Wang A. H., Yang X. J., Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol. 21, 5992–6005 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Field M. G., Durante M. A., Anbunathan H., Cai L. Z., Decatur C. L., Bowcock A. M., Kurtenbach S., Harbour J. W., Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat. Commun. 9, 116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiesner T., Obenauf A. C., Murali R., Fried I., Griewank K. G., Ulz P., Windpassinger C., Wackernagel W., Loy S., Wolf I., Viale A., Lash A. E., Pirun M., Socci N. D., Rutten A., Palmedo G., Abramson D., Offit K., Ott A., Becker J. C., Cerroni L., Kutzner H., Bastian B. C., Speicher M. R., Germline mutations in BAP1 predispose to melanocytic tumors. Nat. Genet. 43, 1018–1021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walpole S., Pritchard A. L., Cebulla C. M., Pilarski R., Stautberg M., Davidorf F. H., de la Fouchardière A., Cabaret O., Golmard L., Stoppa-Lyonnet D., Garfield E., Njauw C. N., Cheung M., Turunen J. A., Repo P., Jarvinen R. S., van Doorn R., Jager M. J., Luyten G. P. M., Marinkovic M., Chau C., Potrony M., Hoiom V., Helgadottir H., Pastorino L., Bruno W., Andreotti V., Dalmasso B., Ciccarese G., Queirolo P., Mastracci L., Wadt K., Kiilgaard J. F., Speicher M. R., van Poppelen N., Kilic E., Al-Jamal R. T., Dianzani I., Betti M., Bergmann C., Santagata S., Dahiya S., Taibjee S., Burke J., Poplawski N., O'Shea S. J., Newton-Bishop J., Adlard J., Adams D. J., Lane A. M., Kim I., Klebe S., Racher H., Harbour J. W., Nickerson M. L., Murali R., Palmer J. M., Howlie M., Symmons J., Hamilton H., Warrier S., Glasson W., Johansson P., Robles-Espinoza C. D., Ossio R., de Klein A., Puig S., Ghiorzo P., Nielsen M., Kivela T. T., Tsao H., Testa J. R., Gerami P., Stern M. H., Paillerets B. B., Abdel-Rahman M. H., Hayward N. K., Comprehensive study of the clinical phenotype of germline BAP1 variant-carrying families worldwide. J. Natl. Cancer Inst. 110, 1328–1341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Żylicz J. J., Bousard A., Žumer K., Dossin F., Mohammad E., da Rocha S. T., Schwalb B., Syx L., Dingli F., Loew D., Cramer P., Heard E., The implication of early chromatin changes in X chromosome inactivation. Cell 176, 182–197.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landreville S., Agapova O. A., Matatall K. A., Kneass Z. T., Onken M. D., Lee R. S., Bowcock A. M., Harbour J. W., Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin. Cancer Res. 18, 408–416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sacco J. J., Kenyani J., Butt Z., Carter R., Chew H. Y., Cheeseman L. P., Darling S., Denny M., Urbé S., Clague M. J., Coulson J. M., Loss of the deubiquitylase BAP1 alters class I histone deacetylase expression and sensitivity of mesothelioma cells to HDAC inhibitors. Oncotarget 6, 13757–13771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.P. D. Nieuwkoop, J. Faber, Normal Table of Xenopus Laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis (North Holland Publishing Co., ed. 2, 1967). [Google Scholar]
- 28.Khokha M. K., Chung C., Bustamante E. L., Gaw L. W., Trott K. A., Yeh J., Lim N., Lin J. C., Taverner N., Amaya E., Papalopulu N., Smith J. C., Zorn A. M., Harland R. M., Grammer T. C., Techniques and probes for the study of Xenopus tropicalis development. Dev. Dyn. 225, 499–510 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Agüero T. H., Fernández J. P., López G. A., Tríbulo C., Aybar M. J., Indian hedgehog signaling is required for proper formation, maintenance and migration of Xenopus neural crest. Dev. Biol. 364, 99–113 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Gentsch G. E., Smith J. C., Investigating physical chromatin associations across the Xenopus genome by chromatin immunoprecipitation. Cold Spring Harb. Protoc. 2014, pdb.prot080614 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Allhoff M., Sere K., Pries J. F., Zenke M., Costa I. G., Differential peak calling of ChIP-seq signals with replicates with THOR. Nucleic Acids Res. 44, e153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinlan A. R., BEDTools: The Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.11–11.12.34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurtenbach S., Reddy R., Harbour J. W., ChIPprimersDB: A public repository of verified qPCR primers for chromatin immunoprecipitation (ChIP). Nucleic Acids Res. 47, D46–D49 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J. P., Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurent C., Gentien D., Piperno-Neumann S., Némati F., Nicolas A., Tesson B., Desjardins L., Mariani P., Rapinat A., Sastre-Garau X., Couturier J., Hupe P., de Koning L., Dubois T., Roman-Roman S., Stern M. H., Barillot E., Harbour J. W., Saule S., Decaudin D., Patient-derived xenografts recapitulate molecular features of human uveal melanomas. Mol. Oncol. 7, 625–636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaax1738/DC1
Fig. S1. X. laevis and human Bap1 proteins show extensive sequence homology.
Fig. S2. Anatomic distribution of Xenopus bap1 mRNA and Bap1 protein expression during early development.
Fig. S3. Bap1 morpholinos and expression constructs.
Fig. S4. Developmental abnormalities in Bap1-deficient embryos are rescued by exogenous human BAP1.
Fig. S5. Loss of Asxl1 phenocopy loss of Bap1 during Xenopus development.
Fig. S6. Pharmacologic rescue of Bap1-deficient phenotype.
Fig. S7. Hdac4 is a key mediator of Bap1-deficient phonotype and acts independently of Hdac3 during development.
Fig. S8. Inhibition of HDAC4 significantly impairs proliferation of uveal melanoma cells in a BAP1-dependent manner.
Data file S1. Oligonucleotides and plasmids used in this study.
Data file S2. Proteomic, transcriptomic, and epigenomic analyses.


