Abstract
The gut microbiota plays a critical role in human health. Diets could modulate the gut microbiota, which in turn may contribute to altered health outcomes by way of changing the relative risk of chronic diseases. Limonin, widely found in citrus fruits, has been reported to possess multiple beneficial health effects. However, the gastrointestinal fate of limonin and its effect on gut microbiota remain unknown. Herein, mice were fed a diet containing 0.05% limonin (w/w) for 9 weeks. Liquid chromatography-mass spectrum analysis showed that limonin was concentrated along the gastrointestinal tract and reached 523.14 nmol/g in the colon lumen. Compared to control mice, colonic microbiota richness was significantly increased by limonin. Gut microbiota community was also clearly distinct from the control group as shown by Principle Coordinate Analysis. Additionally, the relative abundance of 22 genera (relative abundance > 0.1%) was altered significantly. Among these, generally regarded probiotics (Lactobacillus and Bifidobacterium) were reduced, which was not due to direct inhibitory effect of limonin. According to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, amino acid metabolism, lipid, metabolism and immune system function were predicted to be upregulated, and immune system disease and infectious disease markers were predicted to be suppressed dramatically by limonin based on gut microbiota composition. Within the infectious disease category, bacterial toxin and Staphylococcus aureus infection markers were suppressed significantly with limonin treatment. Collectively, our study provides the first line of evidence that oral intake of limonin could shift gut microbiota composition and its functions, which warrants further investigation to determine its implication in human health.
Keywords: dietary, bioactive, limonin, functional metagenome, gut microbiota
INTRODUCTION
The human gastrointestinal tract (GIT) is colonized by environmental microorganisms rapidly after birth.1 After several years, the GIT microbial community becomes stable, and the bacterial cell number is estimated to be around 1013 to 1014, close to total human body cell count.2 The presence of this gut microbiota community has several host benefits such as energy homeostasis enhancement,3 metabolic function improvement,4 and supplemental immune system regulation.5 Gut microbiota dysbiosis is associated with several host diseases, such as obesity, diabetes, coronary heart disease,6,7 and inflammatory bowel disease,8 and it is also implicated in neurodevelopment and cognitive processes as well.9,10 Aside from genetic factors, emerging evidence has suggested that the gut microbiota community responds to and interacts with several external elements including diet, lifestyle, and intake of xenobiotics (prebiotics or antibiotics).11–13 Among these factors, dietary interventions can be a viable strategy to restore or enhance gut microbiota function depending on the desired outcomes. It was demonstrated that when healthy female rats were fed green tea polyphenols for 3 and 6 months, their colonic microbiota was modified dramatically in a dose-dependent manner.14 The administration of the low molecular weight phytochemical quercetin and trans-resveratrol ameliorated gut microbiota dysbiosis and modulated gut barrier function impairments induced by high-fat sucrose diet in rats 15, suggesting that dietary components have the capacity to modify gut microbiota and benefit host health.
Limonin is widely present in citrus fruit16, 17. It belongs to a group of triterpenoid aglycone derivatives named limonoids.18 Limonin has been reported to possess various functions including anti-carcinogenic, anti-inflammatory, antibacterial, and antiviral activity.19–22 Accordingly, limonoids have been recognized as one of the most beneficial and active components of medicinal foods.23 Limonin has a low bioavailability due to its relatively large molecular size and highly lipophilic nature.24 Thus, limonin may evade rapid absorption during transition through the GI tract. The unabsorbed limonin may reach the colon intact and interact with the gut flora. However, the gastrointestinal fate of limonin and its interaction with gut microbiota is so far unknown. In this study, we examined the gastrointestinal fate of limonin and its effect on the gut microbiota in mice. We hypothesized that limonin would persist in the colon, where it would alter the gut microbiota.
MATERIALS AND METHODS
Animal model and diet construction
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of University of Massachusetts and experiments were approved by the Animal Ethics Committee of University of Massachusetts. Twenty male CD-1 mice (aged 6–8 weeks) from Charles River Laboratories (Wilmington, MA, US) were transported to the animal facility on the University of Massachusetts, Amherst campus. Mice were housed in an air-conditioned room (temperature 23 ± 2 oC, 50 ± 10% humidity, 12-hour light-dark cycle) with free access to water and a standard chow diet. Cage rotation was performed to minimize the individual variation of gut microbiota during the 1-week acclimation by means of distribution. 20 male mice were then assigned to the limonin treatment and control groups randomly (10 mice/group). The control group was fed with AIN-93G diet, while the limonin treatment group was fed with the AIN-93G diet containing 0.05% (w/w) limonin. After 9-weeks of treatment, mice were sacrificed with CO2 asphyxiation and stool from distal colon were collected for fecal flora analysis and limonin quantification. GI components including cecum and colonic mucosa were also harvested from the specimen and stored at −80 oC until later extraction and analysis. This animal study was based on a protocol approved by the University of Massachusetts, Amherst Institutional Animal Care and Use Committee (#2014–0079).
Sample preparation and liquid chromatography-mass spectrometry (LC-MS) conditions
Limonin from colonic digesta and mucosa was extracted based on the methods by Liang et al. 25. The extracts were re-dissolved in 50% acetonitrile for LC-MS analysis (Model 2020, Shimadzu, Kyoto, Japan) with a negative ionization mode on a Zorbax SB-Aq C 18 column (150 mm × 4.6 mm, 5 µm, Agilent Technologies, USA) at a flow rate of 0.80 mL/min. The linear gradient elution condition was: 80% mobile phase A (5% ACN/water, v/v)/20% mobile phase B (100% ACN) (v/v) for 5 min initially, then shifted to 80% B/20% A over 30 min and held at 80% B for an additional 5 min. The elution was monitored on a selected m/z− of 469.
Cecal short chain fatty acids (SCFAs) analysis
Cecum contents were homogenized with 6-fold volume of acidified water, and supernatants were obtained by centrifugation (12,000 rpm, 10 min, 4 °C), and then filtered through a 0.22 μm membrane. A system composed of a 6890N gas chromatograph (Agilent Technologies Inc., Palo Alto, CA, USA) connected to an ion flame detector and a 5973N mass spectrometer detector (Agilent) was used for quantification and identification of cecum short chain fatty acid (SCFA) content as described previously.26
Microbial DNA extraction
Total fecal DNA was extracted using QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instruction with the addition of a Bead Ruptor (Omni, Kennesaw, GA, USA) bead mill homogenization step to increase DNA yield. Extracted DNA quantity was measured with NanoDrop Spectrophotometer (Thermo Scientific, Waltham, MA, US) and quality was verified with agarose gel electrophoresis.
Microbial phylogenetic profiling by sequencing of the 16S rRNA gene amplicon
PCR was performed to amplify the V3 and V4 regions of the16S rRNA gene, which incorporates targeted primers and the Illumina overhang adaptor. The primer set was developed by Illumina (16S Amplicon PCR Forward Primer = 5’TCGTCGGCAGCGTCAGATGTGTATAAGA GACAGCCTACGGGNGGCWGCAG) and (16S Amplicon PCR Reverse Primer = 5’GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC)(Yasir et al., 2015). PCR was performed in a 96 well format on a Veriti thermal cycler (Life technology, Carlsbad, CA, US) with 2x KAPA HiFi Hotstart ReadyMix (KAPA Biosystem, Wilmington, MA, US). After purification on AMPure XP beads (Beckman Coulter, Danvers MA, US), a limited cycle PCR was performed using the Nextera XT Index Kit (Illumina, San Diego, CA, US) to attach dual indices and Illumina sequencing adapters, followed by an additional purification on AMPure XP beads. The quantity and quality of the purified PCR products was measured by Qubit dsDNA BR Assay kit (Life technology, Carlsbad, CA, US) and by ScreenTape Assay on Tape Station 2200 (Agilent Technologies, Santa Clara, CA, US). After quantification and qualification, samples were pooled in equimolar amounts and pair-end 2300bp sequencing was performed on the Illumina MiSeq platform (Illumina, San Diego, CA, US).
Microplate growth assay
Lactobacillus plantarum ATCC BAA-793 (L. plantarum), Bifidobacterium longum subsp. longum ATCC 15707 (B. longum), and Bifidobacterium infantis 272 (B. infantis) were procured from the American Type Culture Collection (ATCC). These three strains were verified in-house by Dr. David Sela’s group.27 The three strains were propagated in de Man-Rogosa-Sharpe (MRS; Oxoid, Hampshire, England) medium supplemented with 0.05% (w/v) L-cysteine (Sigma-Aldrich, St. Louis, MO) 28 at 37 °C in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) overnight. For each studied strain, 2 L of culture was inoculated in 200 L MRS medium with or without limonin of varying concentration (10 M or 100 M) and growth phenotypes were monitored over 48 h in a 96-well microplate held in anaerobic conditions at 37 °C by assessing optical density at 600 nm (OD600) using an automated PowerWave HT microplate spectrophotometer (BioTek Instruments, Inc., Winooski, VT). Each strain was evaluated in biological triplicate with three technical replicates.
Data handling and statistical analysis
The bacterial 16S rRNA gene sequencing data was processed by QIIME software pipeline v1.9.1.29 In general, the high quality (quality score > 25) sequence data was demultiplexed. Sequences were then clustered into operational taxonomic units (OTUs) using open reference OTU picking with 97% similarity threshold and taxonomy was assigned according to the Greengenes bacterial 16S rRNA database (13_8 release).30
α-diversity (diversity metrics within sample community) was determined with ten iterations at a maximal sequence depth where all samples could be included. β-diversity (between sample community dissimilarity) was calculated using weighted and unweighted UniFrac distances.31 To investigate the effect of limonin treatment on relative abundance of taxa, Student’s t-test and linear discriminant analysis effective size (LEfSe) analysis were performed.
Galaxy (Huttenhower Lab) Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to explore the predicted functional metagenome shifts between communities. According to the requirements for the PICRUSt algorithm, operational taxonomic units (OTUs) were aligned to the Greengenes 16S rRNA database using a closed reference picking protocol.32 Statistical analysis was used to compare functional shifts between groups in the STAMP software.33 For all analyses, statistical significance was declared if p < 0.05.
RESULTS
General physiology of limonin-fed mice
There was no difference in initial mouse body weights (results not shown), and after a 9-week intervention period, no observed difference was found between the groups’ final body weights (Control: 39.08 ± 1.83 g, Limonin group: 40.32 ± 3.89 g, p = 0.62) (Table S1). Additionally, no differences were found for the liver or spleen weights, indicating that 0.05% limonin (w/w) in diet had no appreciable toxic effect on mice.
Distribution of limonin in mouse gastrointestinal tract (GIT)
To explore the effect of limonin on gut microbiota, it was critical to ensure that limonin could reach the colon to direct interact with gut microbiota. Herein, GIT contents and tissues were subjected to LC-MS analysis to determine the abundance of limonin. As shown in Figure 1A, the concentration of limonin in the digesta increased following transit through the small intestine (SI). Mouse cecum and colon experienced a higher concentration of limonin in general for both digesta and mucosa. Indeed, the limonin in colon digesta was as high as 523.14 ± 95.67 nmoL/g. However, limonin abundance in the GIT mucosa was markedly lower than that in the digesta (Fig. 1B). Cecum mucosa had the highest concentration (15.02 ± 3.80 nmoL/g tissue), which may be due to its function as a sort of time-gated reservoir for chyme and bacteria during passage from the small to large intestines. Still, compared to the high concentration of limonin in colon digesta, limonin in colon mucosa was detected at a 3.82 ± 1.17 nmoL/g tissue. Consistent with a previous report, the amount of limonin present within other organs was also much lower than that found in the digestive system.25 As shown in Figure 1C, the highest concentration of limonin among the collected organs was 2.760.85 nmoL/g, in the spleen, which is approximately 1.4% of the average concentration found in the GIT digesta (191.57 nmoL/g). Limonin concentration in the liver and plasma were both below 0.5 nmoL/g tissue. Taking the tissue weight into account, the absorbed limonin was no more than 1% of the total administrated limonin (data not show). Therefore, we concluded that most of the limonin was unabsorbed and accumulating in the digesta within the distal colon, where a high density of bacteria exists.
Figure 1.
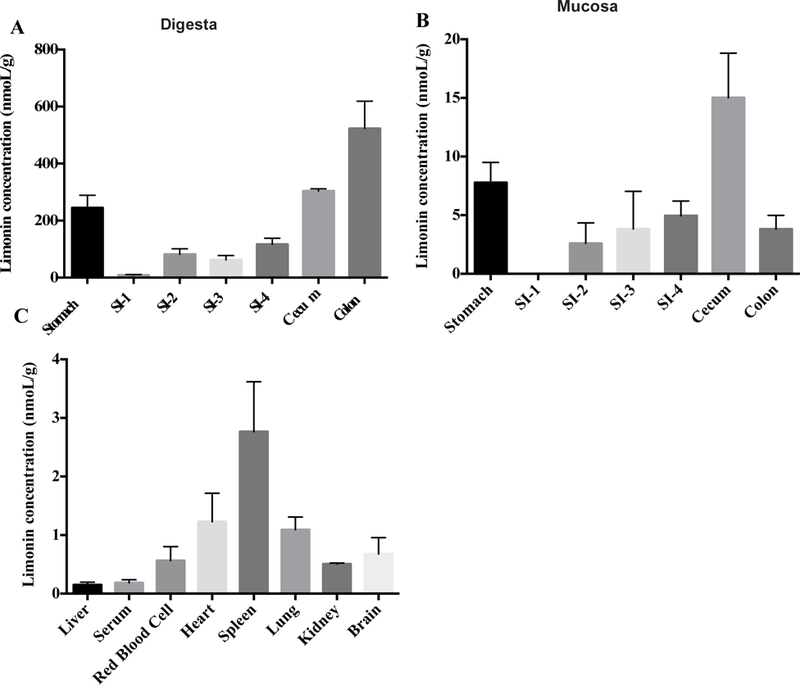
Limonin distribution in mouse digesta, gastrointestinal mucosa, and other tissues. (A) Limonin distribution in the digesta along the gastrointestinal tract (GIT); (B) Limonin distribution in the mucosa along the GIT; (C) Limonin distribution in mice organs.
Mouse fecal microbial activity and community profile
SCFA production in the cecum
SCFAs are the end-products of bacteria fermentation in the cecum and colon. To measure the colonic microbial activity, cecal SCFAs were analyzed to determine the levels of acetate, propionate, isobutyrate, butyrate, isovalerate, and valerate. In agreement with most published research, acetate was the predominant SCFA in the cecum.34, 35 However, no statistical difference was observed in SCFA content between limonin-administered mice and control mice (Fig. 2). Since limonin itself cannot directly serve as a substrate for SCFAs production, the measured yet statistically insignificant changes might be a result of changes to the gut microbiota composition.
Figure 2.
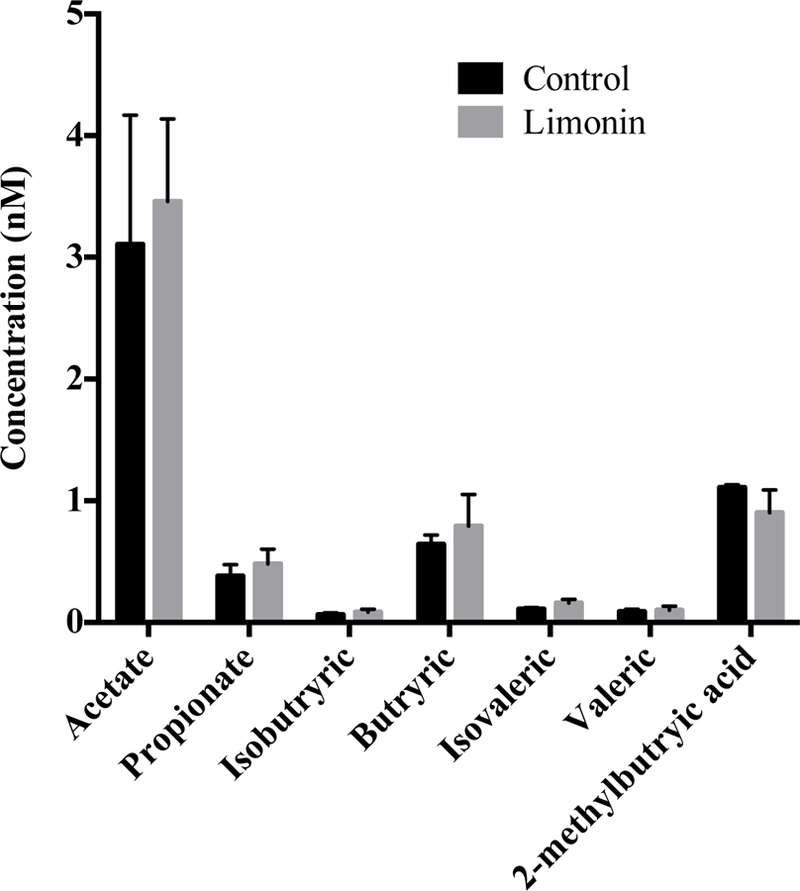
Short chain fatty acid content (SCFA) in control and limonin-treated mice cecum.
Variation of fecal microbial community diversity
To investigate the changes to the mouse gut microbiota generated by dietary limonin intervention, five distal colon fecal samples randomly picked from each group, were subjected to microbial 16S rRNA gene sequencing on the Illumina MiSeq platform. A total of 953,581 counts were obtained, with a mean of 95358.1 counts (range = 56470–151193)/sample. The data set was rarified to a sequence depth of 56470 for diversity analysis.
-diversity including phylogenetic diversity whole tree matrix comparison (PD Whole Tree), Observed OTU richness, Chao1, and Shannon indices were estimated using a linear mixed model. Compared to the control, gut microbiota species richness was increased by limonin treatment remarkably (number of observed species at 97% similar out clusters and Chao1 index) (Table 1). When considering the relative abundance of each species, the Shannon index was obviously increased with limonin diet (Table 1), suggesting that limonin treatment increased mouse gut microbiota diversity.
Table 1:
-diversity of mice fecal microbiota treated with limonin
| Diversity index | Control |
Limonin |
p value | ||
|---|---|---|---|---|---|
| Value | ± SD | Value | ± SD | ||
| PD Whole Tree | 81.31 | 20.92 | 101.06 | 8.76 | 0.09 |
| Observed OTUs | 2305.60 | 622.43 | 3415.80 | 306.51 | 0.01 |
| Chao1 | 5303.89 | 1375.58 | 7005.83 | 578.54 | 0.03 |
| Shannon index | 5.36 | 0.39 | 6.98 | 0.26 | 0.01 |
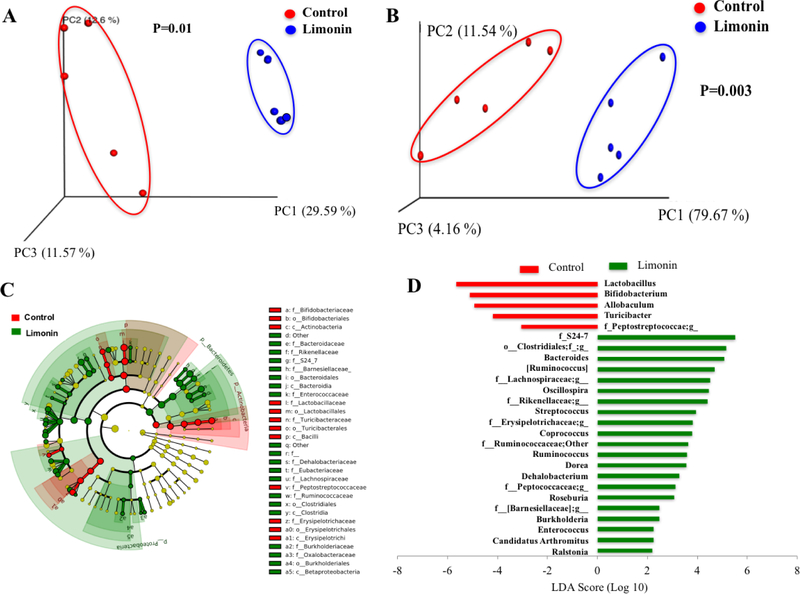
In addition, principal coordinates analysis (PCoA) of weighted and unweighted UniFrac distances performed on the 97% OTU abundance matrix showed a distinct separation (p < 0.05) on the gut microbial community structures (β-diversity) between limonin and control groups (Fig. 3A and 3B, respectively). ANOSIM with 999 permutations was used to test the significant differences between the two groups based on unweighted and weighted UniFrac distances.36 As expected, samples from limonin treatment group clustered far away from the control group (p = 0.01 for unweighted and p = 0.003 for weighted), indicating that limonin treatment altered gut microbiota structure in mice. The main differences in microbiota composition that produced this separation were further investigated by LEfSe as explained below.
Figure 3.
Principal coordinate analysis (PCoA) of unweighted (A) and weighted (B) UniFrac distances of fecal microbial sample communities arranged in an OTU table at 97% similarity threshold. Each dot represents a sample from each mouse fed diets (five out of ten mice in each group was picked randomly for microbiome analysis). Taxonomic difference of colonic microbiota between control and limonin treated groups identified by linear discriminant analysis (LDA) coupled with effect size (LEFSe) analysis. (C) Taxonomic cladogram representing significant features in microbiota profile with respect to limonin treatment. (D) Gut microbiota genera differentially represented between control and limonin treated groups (LDA > 2, p < 0.05). Red indicating taxa suppressed by limonin treatment, green suggesting taxa enriched by limonin diet.
Taxonomic shifts in limonin-treated mice
Version 13.8 of the Greengenes database assigned usable raw reads to 9 phyla, 18 families, and 81 genera among the samples sequenced. As expected, the most abundant phyla in both groups were Firmicutes and Bacteroidetes (Table S2). LEfSe analysis was applied to further explore the differences in taxonomic categories between the limonin-treated and control groups. The phyla Proteobacteria and Bacteroidetes were significantly enriched by limonin treatment, while the phylum Actinobacteria was suppressed (LDA > 2.0, p < 0.05) (Fig. 3C). Meanwhile, relative abundance of Firmicutes decreased by 25% (from 65.39 ± 2.90 to 49.10 ± 6.09%, p = 0.09). Among the 81 identified genera, 18 genera (Unidentified genus of family S24–7, unidentified genus of order Clostridiales, Bacteroides, unidentified genus of family Lachnospiraceae, unidentified genus of family Rikenellaceae, Oscillospira, etc.) were significantly enriched and four genera (Lactobacillus, Bifidobacterium, Allobaculum, and unidentified genus of family Peptostreptococcaceae) were significantly reduced by limonin (LDA > 2.0, p < 0.05) (Fig. 3D). Our data demonstrated that limonin treatment could dramatically impact microbial composition. Genus Oscillospira was increased by ~9-fold (Table S3), which has been associated with leanness in humans37 and decreased incidence of inflammatory bowel disease38. Unexpectedly, the relative abundance of the genera Bifidobacterium and Lactobacillus, which are widely regarded as beneficial bacteria,39, 40 were significantly decreased by limonin (Fig. 3D).
Effect of limonin on bacteria Lactobacillus and Bifidobacterium growth
To potentially explain the decreased relative abundance of Bifidobacterium and Lactobacillus, the effect of limonin on the growth of Lactobacillus and Bifidobacterium was examined. From the growth curve of the three strains, no obvious inhibition was observed (Fig. 4A–C). Conversely, limonin (10 M and 100 M) significantly increased the maximum bacteria optical density of B. longum and B. infantis, while limonin had no effect on L. plantarum growth (Fig. 4D). These findings support the notion that limonin presence did not directly influence the significantly reduced relative abundance of genera Bifidobacterium and Lactobacillus in the mouse gut microbiome that was observed.
Figure 4:
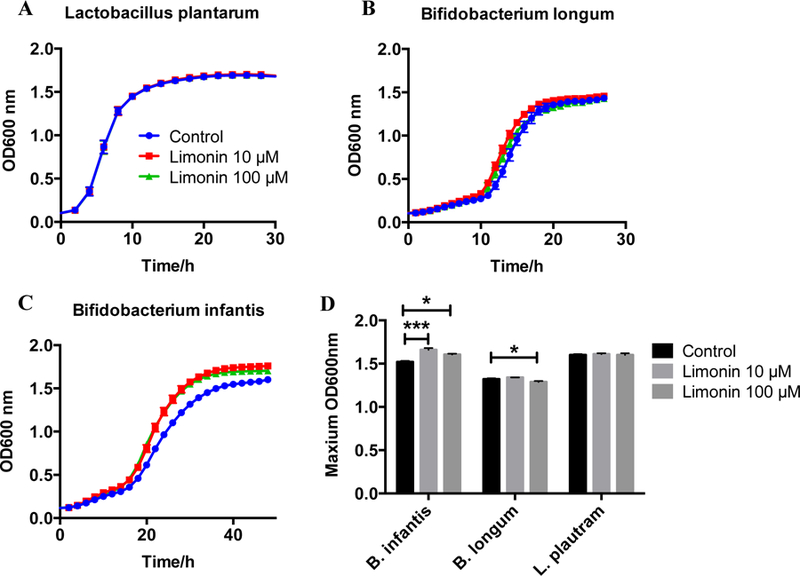
The effect of limonin on probiotic culture growth. The growth curve of (A) L. plantarum, (B) B. longum, and (C) B. infantis with limonin treatment at different concentrations. (D) The maximum OD600nm of the three strains with and without limonin treatment.
Variation of predicted functional metagenomes induced by limonin supplementation
Given the effect of limonin on mouse gut microbiota composition and diversity, Galaxy PICRUSt was applied as an exploratory tool to predict the differences in microbial function between limonin-treated and control groups. Despite the accuracy of such predictions being lower for other mammals than for humans (mean NSTI = 0.03 ± 0.02), it could still provide useful insight on the potential functional properties of mammalian microbiomes.32 The bacterial community corresponding to limonin treatment was suggested to be more abundant in gene families involved in amino acid metabolism, metabolism of cofactors and vitamins, lipid metabolism, biosynthesis of secondary metabolites, and immune system function (Fig. 5A). On the other hand, mouse gut microbiota treated with limonin had lower predicted activities associated with immune system disease and infection disease (LDA > 2, p < 0.05) (Fig. 5A). Specifically, KEGG pathways corresponding to Staphylococcus aureus infection was profoundly reduced by 78% (p = 0.001) by limonin treatment (Fig. 5B). In summary, limonin treatment could potentially influence distal colon microbiota function.
Figure 5:
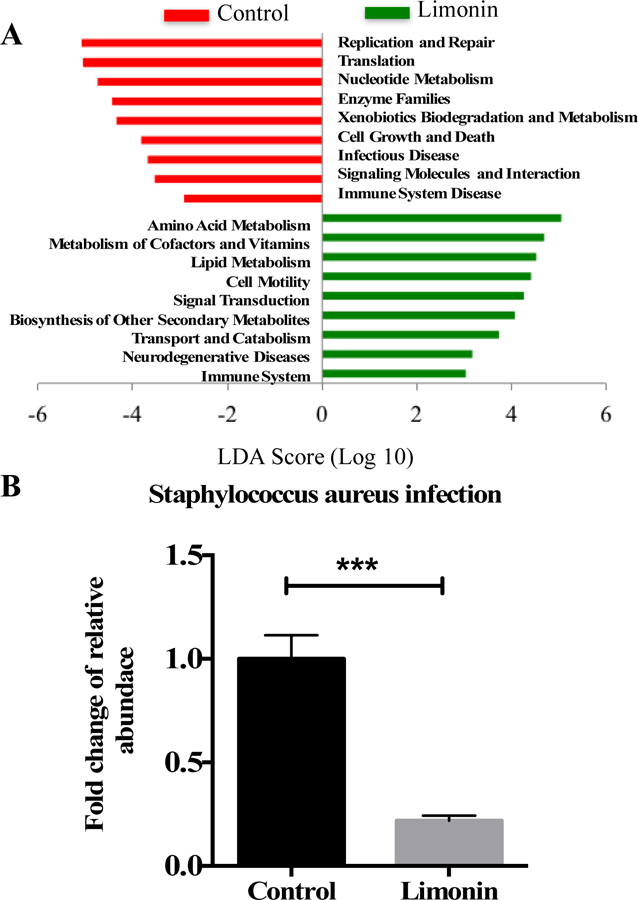
Predicted microbial functional pathways significantly shifted with limonin treatment using predictive metagenomics. (A) Differential gene expression associated with functional pathways determined in PICRUSt. (B) Fold change of pathway relative abundance associated with Staphylococcus aureus infection. The significantly affected functional pathways were identified by LEfSe (LDA>2, p < 0.05). Red box: suppressed by limonin treatment, green box: enriched by limonin treatment.
DISCUSSION
Limonin, a triterpene derived from citrus fruits, has been recognized to have a wide range of bioactivities.19–21 It has been reported to inhibit the proliferation of human colon adenocarcinoma (SW480) cells through mitochondria-mediated intrinsic apoptosis19 and suppress AOM-induced colon cancer in male rats21. Though numerous beneficial functions of limonin have been reported, limited information about the effect of limonin on the gut microbiota in animals is available, an ecosystem that is closely associated with host health. Therefore, we determined tissue distribution of limonin and its impact on gut microbiota in mice after its oral administration. Orally-ingested xenobiotic bioavailability depends on the compound’s physicochemical properties. Based on clinical evidence, the oral bioavailability of xenobiotics with molecular weights (MW) above 400 g/mol was less than 20%.41 As limonin has a MW of 470.52 g/mol and is generally hydrophobic in nature, there are indications that limonin’s in situ bioavailability should be below 20%. As expected, our results showed that a large fraction of the orally administrated limonin was unabsorbed and persisted to the colon, potentially contributing to gut microenvironment and bacterial composition alterations.
Indeed, our results indicated that the mouse gut microbial community was distinctly different after 9-weeks of treatment with 0.05% w/w limonin in the diet. The 16S rRNA gene analysis revealed that the gut microbial diversity (-diversity and -diversity) was significantly shifted by limonin intervention. Microbial species richness (the number of species present in certain microbiota ecosystem) was significantly increased by limonin treatment. This could be interpreted as a beneficial effect, given that communities with higher species richness are more resistant to pathogen invasion, as these communities are generally more efficient at resource utilization and limit viable pathogen competition.42 High species richness could also improve the stability of the host gut microbiota ecosystem overall43 while low diversity was observed in high-fat and high-sugar diet-administered obese mice44, 45.
Additionally, the composition of the colonic microbiota was altered in response to dietary limonin intervention. At the phylum level, the relative abundance of Bacteroidetes and Proteobacteria in mouse gut were significantly higher in the limonin treatment group (Table S2). The alteration in relative abundance of Proerobacteria may result in modifications to host energy accumulation.46–48 The relative abundance of Actinobacteria was decreased dramatically (Table S2) and this alteration could have different effects on host health depending on age and health status. Previously, it was shown that children with autism had lower relative abundance of Actinobacteria in the gut,49 while people with inflammatory bowel disease (IBD) had higher levels of Actinobacteria on average.50 The proportion of Bacteroidetes and Firmicutes were typically reported to be associated with obesity, with a decreasing F/B ratio being highly related with gut microbiota dysbiosis51 and western high-fat diets.52
Three out five genera in the phylum Bacteroidetes were distinctly increased by limonin treatment, including Bacteroides, f_Rikenellaceae;g__, and f_S24–7;g__. Certain commensal Bacteroides species could induce IBD in an ulcerative colitis mouse model (dnKO) with or without antibiotic pretreatment, and innate and adapted immune responses were activated in a host-genotype-specific fashion.53 Increased abundance of f_S24–7;g__ could potentially contribute to increased plant carbohydrate fermentation54 and SCFA production in the cecum. From the phylum Firmicutes, several genera were increased significantly such as: o_Clostridiales;f__;g__, f__Lachnospiraceae;g__, Ruminococcus, Oscillospira, and Ruminococcus. The genus Oscillospira was negative correlated with body mass index (BMI) and inflammatory disease.37, 55 The genus Ruminococcus was increased by ~9-fold, which might enhance the gut microbiota ability in degrading and utilizing carbohydrates from the host’s diet.56
From the taxonomic results, the relative abundance of genera Lactobacillus and Bifidobacterium were significantly reduced by limonin supplementation. Bacterial growth curves with and without limonin treatment showed that limonin had no inhibitory effect on their growth, and even revealed a significant improvement to the growth of the Bifidobacterium strains tested. Therefore, the reduced relative abundance of Lactobacillus and Bifidobacterium may due to the growth and out-competition by other bacterial clades rather than by a direct inhibitory effect. The exact mechanism of reduced relative abundance of genera Bifidobacterium and Lactobacillus with limonin treatment need to be further examined.
The metagenome functional analysis results demonstrated the modulation of KEGG pathways by limonin in mice. Microbiota populations resulting from limonin treatment showed the suppression of gene families associated with infectious disease, which might be further enhanced by general increases in the richness of the gut community.42 Also, gene functions associated with amino acid and lipid metabolism were increased markedly. Certain bacterial taxa were associated with lipid metabolism and their modification might impact host lipid metabolism and presence of signaling molecules.57, 58 Increased amino acid metabolism of bacteria could facilitate protein synthesis or fermentation to promote nutrient metabolism and utilization.59 Considering the limitations of 16S rRNA gene sequencing in metagenomics analysis for non-humans, RNA-seq should be applied in the future to monitor the differential expression of functional genes related with limonin treatment.
CONCLUSION
This study investigated the gastrointestinal fate of orally-administered limonin and its influence on colonic microbiota in mice. Our study revealed that large portion of limonin could evade absorption and metabolism through the GIT and persist to the colon. The gut microbiota profile was distinctly modified, species richness was enhanced by limonin treatment, and the predicted microbial function was altered in response to dietary limonin intervention. This study provided fundamental knowledge for limonin application as a bioactive ingredient in functional foods.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported in part by National Institutes of Health (R01 AT010229) and USDA/NIFA and Hatch Fund.
Abbreviations:
- GIT
gastrointestinal tract
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LEfSe
linear discriminant analysis effective size
- OTUs
operational taxonomic unites
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- RD
red blood cells
- SCFAs
short chain fatty acids
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Tannock GW, What immunologists should know about bacterial communities of the human bowel. Semin Immunol 2007, 19, 94–105. [DOI] [PubMed] [Google Scholar]
- 2.Sender R; Fuchs S; Milo R, Revised Estimates for the Number of Human and Bacteria Cells in the Body. Plos Biol 2016, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backhed F; Ding H; Wang T; Hooper LV; Koh GY; Nagy A; Semenkovich CF; Gordon JI, The gut microbiota as an environmental factor that regulates fat storage. P Natl Acad Sci USA 2004, 101, 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gill SR; Pop M; Deboy RT; Eckburg PB; Turnbaugh PJ; Samuel BS; Gordon JI; Relman DA; Fraser-Liggett CM; Nelson KE, Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow J; Lee SM; Shen Y; Khosravi A; Mazmanian SK, Host-bacterial symbiosis in health and disease. Adv Immunol 2010, 107, 243–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kootte RS; Vrieze A; Holleman F; Dallinga-Thie GM; Zoetendal EG; de Vos WM; Groen AK; Hoekstra JB; Stroes ES; Nieuwdorp M, The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes Metab 2012, 14, 112–20. [DOI] [PubMed] [Google Scholar]
- 7.Tang WH; Hazen SL, The contributory role of gut microbiota in cardiovascular disease. J Clin Invest 2014, 124, 4204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamada N; Seo SU; Chen GY; Nunez G, Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 2013, 13, 321–35. [DOI] [PubMed] [Google Scholar]
- 9.O’Mahony SM; Clarke G; Borre YE; Dinan TG; Cryan JF, Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res 2015, 277, 32–48. [DOI] [PubMed] [Google Scholar]
- 10.Cryan JF; O’Mahony SM, The microbiome-gut-brain axis: from bowel to behavior. Neurogastroenterol Motil 2011, 23, 187–92. [DOI] [PubMed] [Google Scholar]
- 11.Benson AK; Kelly SA; Legge R; Ma FR; Low SJ; Kim J; Zhang M; Oh PL; Nehrenberg D; Hua KJ; Kachman SD; Moriyama EN; Walter J; Peterson DA; Pomp D, Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. P Natl Acad Sci USA 2010, 107, 18933–18938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conlon MA; Bird AR, The Impact of Diet and Lifestyle on Gut Microbiota and Human Health. Nutrients 2015, 7, 17–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Org E; Parks BW; Joo JWJ; Emert B; Schwartzman W; Kang EY; Mehrabian M; Pan C; Knight R; Gunsalus R; Drake TA; Eskin E; Lusis AJ, Genetic and environmental control of host-gut microbiota interactions. Genome Research 2015, 25, 1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J; Tang L; Zhou H; Zhou J; Glenn TC; Shen CL; Wang JS, Long-term treatment with green tea polyphenols modifies the gut microbiome of female sprague-dawley rats. J Nutr Biochem 2018, 56, 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eteberria U; Arias N; Boque N; Macarulla MT; Portillo MP; Martinez JA; Milagro FI, Reshaping faecal gut microbiota composition by the intake of trans-resveratrol and quercetin in high-fat sucrose diet-fed rats. Journal of Nutritional Biochemistry 2015, 26, 651–660. [DOI] [PubMed] [Google Scholar]
- 16.Roy A; Saraf S, Limonoids: overview of significant bioactive triterpenes distributed in plants kingdom. Biol Pharm Bull 2006, 29, 191–201. [DOI] [PubMed] [Google Scholar]
- 17.Manners GD, Citrus limonoids: analysis, bioactivity, and biomedical prospects. J Agric Food Chem 2007, 55, 8285–8294. [DOI] [PubMed] [Google Scholar]
- 18.Langeswaran K; Gowthamkumar S; Vijayaprakash S; Revathy R; Balasubramanian MP, Influence of limonin on Wnt signalling molecule in HepG2 cell lines. J Nat Sci Biol Med 2013, 4, 126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murthy KNC; Jayaprakasha GK; Kumar V; Rathore KS; Patil BS, Citrus Limonin and Its Glucoside Inhibit Colon Adenocarcinoma Cell Proliferation through Apoptosis. J Agr Food Chem 2011, 59, 2314–2323. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu S; Miyamoto S; Fujii G; Nakanishi R; Onuma W; Ozaki Y; Fujimoto K; Yano T; Mutoh M, Suppression of intestinal carcinogenesis in Apc-mutant mice by limonin. J Clin Biochem Nutr 2015, 57, 39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka T; Maeda M; Kohno H; Murakami M; Kagami S; Miyake M; Wada K, Inhibition of azoxymethane-induced colon carcinogenesis in male F344 rats by the citrus limonoids obacunone and limonin. Carcinogenesis 2001, 22, 193–198. [DOI] [PubMed] [Google Scholar]
- 22.Langeswaran K; Kumar SG; Perumal S; Revathy R; Balasubramaniam MP, Limonin – A citrus limonoid, establish anticancer potential by stabilizing lipid peroxidation and antioxidant status against N-nitrosodiethylamine induced experimental hepatocellular carcinoma. Biomedicine & Preventive Nutrition 2013, 3, 165–171. [Google Scholar]
- 23.Ozaki Y; Ayano S; Inaba N; Miyake M; Berhow MA; Hasegawa S , Limonoid Glucosides in Fruit, Juice and Processing by-Products of Satsuma Mandarine (Citrus-Unshiu Marcov). Journal of Food Science 1995, 60, 186–189. [Google Scholar]
- 24.Manners GD; Jacob RA; Breksa AP; Schoch TK; Hasegawa S, Bioavailability of citrus limonoids in humans. J Agr Food Chem 2003, 51, 4156–4161. [DOI] [PubMed] [Google Scholar]
- 25.Liang Y; Xie L; Liu XD; Hu YZ; Lu T; Wang GJ, Gender differences in limonin pharmacokinetics in rats. Eur J Drug Metab Pharmacokinet 2005, 30, 243–8. [DOI] [PubMed] [Google Scholar]
- 26.Salazar N; Lopez P; Valdes L; Margolles A; Suarez A; Patterson AM; Cuervo A; de los Reyes-Gavilan CG; Ruas-Madiedo P; Gonzalez S; Gueimonde M, Microbial Targets for the Development of Functional Foods Accordingly with Nutritional and Immune Parameters Altered in the Elderly. Journal of the American College of Nutrition 2013, 32, 399–406. [DOI] [PubMed] [Google Scholar]
- 27.Ozcan E; Sun J; Rowley DC; Sela DA, A human gut commensal ferments cranberry carbohydrates to produce formate. Appl Environ Microbiol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turroni F; Foroni E; Pizzetti P; Giubellini V; Ribbera A; Merusi P; Cagnasso P; Bizzarri B; de’Angelis GL; Shanahan F; van Sinderen D; Ventura M, Exploring the Diversity of the Bifidobacterial Population in the Human Intestinal Tract. Appl Environ Microb 2009, 75, 1534–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG; Kuczynski J; Stombaugh J; Bittinger K; Bushman FD; Costello EK; Fierer N; Pena AG; Goodrich JK; Gordon JI; Huttley GA; Kelley ST; Knights D; Koenig JE; Ley RE; Lozupone CA; McDonald D; Muegge BD; Pirrung M; Reeder J; Sevinsky JR; Tumbaugh PJ; Walters WA; Widmann J; Yatsunenko T; Zaneveld J; Knight R, QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeSantis TZ; Hugenholtz P; Larsen N; Rojas M; Brodie EL; Keller K; Huber T; Dalevi D; Hu P; Andersen GL, Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb 2006, 72, 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozupone C; Lladser ME; Knights D; Stombaugh J; Knight R, UniFrac: an effective distance metric for microbial community comparison. ISME J 2011, 5, 169–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langille MGI; Zaneveld J; Caporaso JG; McDonald D; Knights D; Reyes JA; Clemente JC; Burkepile DE; Thurber RLV; Knight R; Beiko RG; Huttenhower C , Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013, 31, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parks DH; Tyson GW; Hugenholtz P; Beiko RG, STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cummings JH; Pomare EW; Branch WJ; Naylor CPE; Macfarlane GT, Short Chain Fatty-Acids in Human Large-Intestine, Portal, Hepatic and Venous-Blood. Gut 1987, 28, 1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hijova E; Chmelarova A, Short chain fatty acids and colonic health. Bratisl Med J 2007, 108, 354–358. [PubMed] [Google Scholar]
- 36.Lozupone C; Knight R, UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb 2005, 71, 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tims S; Derom C; Jonkers DM; Vlietinck R; Saris WH; Kleerebezem M; de Vos WM; Zoetendal EG, Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J 2013, 7, 707–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong YD; Marungruang N; Fak F; Nyman M, Effects of two whole-grain barley varieties on caecal SCFA, gut microbiota and plasma inflammatory markers in rats consuming low- and high-fat diets. Brit J Nutr 2015, 113, 1558–1570. [DOI] [PubMed] [Google Scholar]
- 39.Peran L; Camuesco D; Comalada M; Bailon E; Henriksson A; Xaus J; Zarzuelo A; Galvez J, A comparative study of the preventative effects exerted by three probiotics, Bifidobacterium lactis, Lactobacillus casei and Lactobacillus acidophilus, in the TNBS model of rat colitis. J Appl Microbiol 2007, 103, 836–44. [DOI] [PubMed] [Google Scholar]
- 40.Tomasz B; Zoran S; Jaroslaw W; Ryszard M; Marcin G; Robert B; Piotr K; Lukasz K; Jacek P; Piotr G; Przemyslaw P; Michal D, Long-term use of probiotics Lactobacillus and Bifidobacterium has a prophylactic effect on the occurrence and severity of pouchitis: a randomized prospective study. Biomed Res Int 2014, 2014, 208064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou TJ; Wang JM; Zhang W; Xu XJ, ADME evaluation in drug discovery. 6. Can oral bioavailability in humans be effectively predicted by simple molecular property-based rules? J Chem Inf Model 2007, 47, 460–463. [DOI] [PubMed] [Google Scholar]
- 42.Levine JM; D’Antonio CM, Elton revisited: a review of evidence linking diversity and invasibility. Oikos 1999, 87, 15–26. [Google Scholar]
- 43.Tap J; Furet JP; Bensaada M; Philippe C; Roth H; Rabot S; Lakhdari O; Lombard V; Henrissat B; Corthier G; Fontaine E; Dore J; Leclerc M, Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environmental Microbiology 2015, 17, 4954–4964. [DOI] [PubMed] [Google Scholar]
- 44.Turnbaugh PJ; Baeckhed F; Fulton L; Gordon JI, Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host & Microbe 2008, 3, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yatsunenko T; Rey FE; Manary MJ; Trehan I; Dominguez-Bello MG; Contreras M; Magris M; Hidalgo G; Baldassano RN; Anokhin AP; Heath AC; Warner B; Reeder J; Kuczynski J; Caporaso JG; Lozupone CA; Lauber C; Clemente JC; Knights D; Knight R; Gordon JI, Human gut microbiome viewed across age and geography. Nature 2012, 486, 222-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koren O; Goodrich JK; Cullender TC; Spor A; Laitinen K; Backhed HK; Gonzalez A; Werner JJ; Angenent LT; Knight R; Backhed F; Isolauri E; Salminen S; Ley RE, Host Remodeling of the Gut Microbiome and Metabolic Changes during Pregnancy. Cell 2012, 150, 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amato KR; Leigh SR; Kent A; Mackie RI; Yeoman CJ; Stumpf RM; Wilson BA; Nelson KE; White BA; Garber PA, The Role of Gut Microbes in Satisfying the Nutritional Demands of Adult and Juvenile Wild, Black Howler Monkeys (Alouatta pigra). Am J Phys Anthropol 2014, 155, 652–664. [DOI] [PubMed] [Google Scholar]
- 48.Chevalier C; Stojanovic O; Colin DJ; Suarez-Zamorano N; Tarallo V; Veyrat-Durebex C; Rigo D; Fabbiano S; Stevanovic A; Hagemann S; Montet X; Seimbille Y; Zamboni N; Hapfelmeier S; Trajkovski M, Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell 2015, 163, 1360–1374. [DOI] [PubMed] [Google Scholar]
- 49.Robinson CJ; Bohannan BJ; Young VB, From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev 2010, 74, 453–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spor A; Koren O; Ley R, Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 2011, 9, 279–90. [DOI] [PubMed] [Google Scholar]
- 51.Ley RE; Turnbaugh PJ; Klein S; Gordon JI, Microbial ecology: human gut microbes associated with obesity. Nature 2006, 444, 1022–3. [DOI] [PubMed] [Google Scholar]
- 52.Hildebrandt MA; Hoffmann C; Sherrill-Mix SA; Keilbaugh SA; Hamady M; Chen YY; Knight R; Ahima RS; Bushman F; Wu GD, High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 2009, 137, 1716–24 e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bloom SM; Bijanki VN; Nava GM; Sun LL; Malvin NP; Donermeyer DL; Dunne WM; Allen PM; Stappenbeck TS, Commensal Bacteroides Species Induce Colitis in Host-Genotype-Specific Fashion in a Mouse Model of Inflammatory Bowel Disease. Cell Host & Microbe 2011, 9, 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shang QS; Shi JJ; Song G; Zhang MF; Cai C; Hao JJ; Li GY; Yu GL, Structural modulation of gut microbiota by chondroitin sulfate and its oligosaccharide. International Journal of Biological Macromolecules 2016, 89, 489–498. [DOI] [PubMed] [Google Scholar]
- 55.Verdam FJ; Fuentes S; de Jonge C; Zoetendal EG; Erbil R; Greve JW; Buurman WA; de Vos WM; Rensen SS, Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity 2013, 21, E607–E615. [DOI] [PubMed] [Google Scholar]
- 56.Cann I; Bernardi RC; Mackie RI, Cellulose degradation in the human gut: Ruminococcus champanellensis expands the cellulosome paradigm. Environmental Microbiology 2016, 18, 307–310. [DOI] [PubMed] [Google Scholar]
- 57.Derrien M; Van Baarlen P; Hooiveld G; Norin E; Muller M; de Vos WM, Modulation of Mucosal Immune Response, Tolerance, and Proliferation in Mice Colonized by the Mucin-Degrader Akkermansia muciniphila. Front Microbiol 2011, 2, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Everard A; Belzer C; Geurts L; Ouwerkerk JP; Druart C; Bindels LB; Guiot Y; Derrien M; Muccioli GG; Delzenne NM; de Vos WM; Cani PD, Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. P Natl Acad Sci USA 2013, 110, 9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin R; Liu WT; Piao MY; Zhu H, A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids 2017, 49, 2083–2090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.