The glucan phosphatase family member LIKE SEX4 1 does not act as a phosphatase during starch degradation, instead functioning as a protein scaffold on the starch granule surface to bind β-amylases.
Abstract
In Arabidopsis (Arabidopsis thaliana) leaves, starch is synthesized during the day and degraded at night to fuel growth and metabolism. Starch is degraded primarily by β-amylases, liberating maltose, but this activity is preceded by glucan phosphorylation and is accompanied by dephosphorylation. A glucan phosphatase family member, LIKE SEX4 1 (LSF1), binds starch and is required for normal starch degradation, but its exact role is unclear. Here, we show that LSF1 does not dephosphorylate glucans. The recombinant dual specificity phosphatase (DSP) domain of LSF1 had no detectable phosphatase activity. Furthermore, a variant of LSF1 mutated in the catalytic cysteine of the DSP domain complemented the starch-excess phenotype of the lsf1 mutant. By contrast, a variant of LSF1 with mutations in the carbohydrate binding module did not complement lsf1. Thus, glucan binding, but not phosphatase activity, is required for the function of LSF1 in starch degradation. LSF1 interacts with the β-amylases BAM1 and BAM3, and the BAM1-LSF1 complex shows amylolytic but not glucan phosphatase activity. Nighttime maltose levels are reduced in lsf1, and genetic analysis indicated that the starch-excess phenotype of lsf1 is dependent on bam1 and bam3. We propose that LSF1 binds β-amylases at the starch granule surface, thereby promoting starch degradation.
INTRODUCTION
Starch, the major storage carbohydrate in plants, accumulates in leaves, as well as the storage tissues of seeds, tubers, and roots. In leaves, starch is synthesized in chloroplasts during the day from photo-assimilated carbon and is degraded at night to fuel metabolism in the dark. Starch consists of two glucan polymers, amylopectin and amylose. In amylopectin (the major polymer), α-1,4-linked glucan chains are connected to each other by α-1,6 linkages (branch points) to form a racemose, tree-like structure. Adjacent glucan chains in amylopectin form double helices and adopt a stable, semicrystalline lamellar structure. By contrast, amylose is essentially linear and is thought to occupy the spaces between the crystalline regions of amylopectin (Buléon et al., 1998; Zeeman et al., 2010; Pfister and Zeeman, 2016).
A number of different enzymes are required for starch degradation in Arabidopsis (Arabidopsis thaliana) leaves (Zeeman et al., 2010). The first step is the phosphorylation of a small proportion of the glucosyl residues at the granule surface by the enzymes GLUCAN, WATER DIKINASE (GWD) and PHOSPHOGLUCAN, WATER DIKINASE (PWD), which phosphorylate glucan chains in the C6 and C3 positions, respectively (Baunsgaard et al., 2005; Kötting et al., 2005; Ritte et al., 2006). Phosphorylation is believed to disrupt the crystalline packing of the amylopectin, allowing hydrolytic enzymes access to the glucan chains (Edner et al., 2007; Hejazi et al., 2008). The major class of glucan hydrolase that participates in leaf starch degradation at night is β-amylase, an exoamylase that removes maltose from the non-reducing ends of the α-1,4-linked glucan chains (Scheidig et al., 2002; Kaplan and Guy, 2004; Weise et al., 2004, 2005). Nine genes in the Arabidopsis genome encode β-amylases and β-amylase-like proteins (BAMs; Fulton et al., 2008). At least four proteins (BAM1, BAM2, BAM3, and BAM4) localize to the chloroplasts. Two proteins (BAM7 and BAM8) localize to the nucleus and act as transcription factors rather than glucosyl hydrolases (Reinhold et al., 2011; Soyk et al., 2014). Of the four plastidial β-amylases, BAM1 and BAM3 are the major, enzymatically active isoforms. The bam3 knockout mutant is unable to fully degrade its starch during the night and thus has a starch excess (sex) phenotype (Fulton et al., 2008). The bam1 mutant does not have a sex phenotype, but the bam1 bam3 double mutant lacking both isoforms accumulates more starch than the bam3 single mutant, indicating some redundancy in BAM1 and BAM3 function. BAM4 possesses an atypical glucosyl hydrolase domain and is thought to act as a regulator of starch degradation, whereas no role has been attributed to BAM2 (Fulton et al., 2008).
Other enzyme activities participate in the complete degradation of starch, since β-amylases cannot hydrolyze branch points or phosphorylated glucose residues nor cleave past them (French and Summer, 1956; Takeda and Hizukuri, 1981). Branch points are hydrolyzed by the debranching enzymes ISOAMYLASE3 (ISA3) and LIMIT DEXTRINASE (LDA), which preferentially remove external chains of amylopectin that have been shortened by β-amylolysis (Hussain et al., 2003; Wattebled et al., 2005; Delatte et al., 2006; Takashima et al., 2007). The chloroplastic α-AMYLASE3 (AMY3) is an endoamylase that can cleave internally of the branch points, releasing a range of linear and branched malto-oligosaccharides (Streb et al., 2012; Seung et al., 2013).
Phosphate groups, having served the purpose of disrupting the semicrystalline structure of amylopectin, are removed again to allow complete hydrolysis of starch. This process is mediated by the phosphoglucan phosphatases STARCH EXCESS4 (SEX4; Zeeman et al., 1998; Niittylä et al., 2006; Kötting et al., 2009; Hejazi et al., 2010; Vander Kooi et al., 2010; Meekins et al., 2014) and LIKE SEX4 2 (LSF2; Santelia et al., 2011; Meekins et al., 2013). SEX4 preferentially dephosphorylates the C6 position of the glucosyl residues, while LSF2 preferentially dephosphorylates the C3 position (Santelia et al., 2011; Meekins et al., 2013, 2014). The sex4 mutant has a strong sex phenotype, while the lsf2 mutant has only slightly more starch than the wild type (Santelia et al., 2011). However, the lsf2 sex4 double mutant has a much greater starch excess than the sex4 single mutant alone. The enzymes involved in glucan phosphorylation and dephosphorylation work synergistically with the glucan hydrolases. In vitro experiments showed that starch degradation by ISA3 and BAM3 is stimulated by GWD activity, but also that GWD activity is stimulated by β-amylolysis (Ritte et al., 2004; Edner et al., 2007). The inclusion of SEX4 in such in vitro experiments to create a cycle of glucan phosphorylation and dephosphorylation further increases starch degradation by ISA3 and BAM3 (Hejazi et al., 2009; Kötting et al., 2009).
Vascular plants contain another chloroplastic protein with sequence similarity to SEX4 and LSF2, called LSF1 (for LIKE SEX FOUR 1). These three proteins share homology in their dual specificity phosphatase (DSP) domains as part of the protein tyrosine phosphatase (PTP) family (Silver et al., 2014; White-Gloria et al., 2018). LSF1 also possesses a carbohydrate binding module (CBM; Fordham-Skelton et al., 2002; Kerk et al., 2006), similar to SEX4, and binds to starch granules in vivo (Comparot-Moss et al., 2010). However, unlike the other two proteins, LSF1 contains a region toward its N terminus with homology to the protein-protein interaction domain of the PDZ-like type (Silver et al., 2014). PDZ stands for postsynaptic density protein, drosophila disc large tumor suppressor, zonula occludens-1 protein (Ponting, 1997; Jeleń et al., 2003; Lee and Zheng, 2010). LSF1 is required for proper starch degradation, as the lsf1 mutant has a sex phenotype (Comparot-Moss et al., 2010), but there is currently no evidence that it is a glucan phosphatase. In sex4 mutants, phosphorylated oligosaccharides accumulate during starch degradation (Kötting et al., 2009) and in lsf2, the starch contains elevated levels of phosphate (Santelia et al., 2011). By contrast, lsf1 mutants neither accumulate phospho-oligosaccharides nor display elevated levels of starch-bound phosphate (Comparot-Moss et al., 2010).
In this study, to elucidate the role of LSF1 in starch degradation, we investigated the LSF1 protein in vitro and found that unlike SEX4 and LSF2, LSF1 is not an active glucan phosphatase. Using site-directed mutagenesis, we confirmed in vivo that the loss of putative LSF1 phosphatase activity is not the cause of the sex phenotype in the lsf1 mutant. Using a combination of techniques, we showed that LSF1 interacts with the chloroplastic β-amylases, BAM1 and BAM3. Based on these findings, we propose a non-enzymatic role for LSF1 in starch degradation.
RESULTS
LSF1 Is a Starch Binding Protein with No Detectable Phosphatase Activity
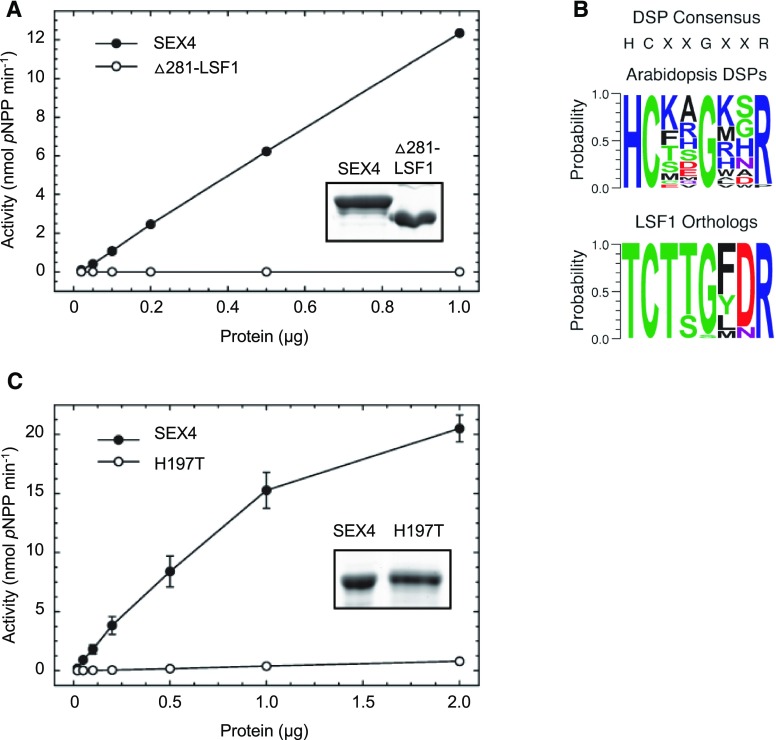
The LSF1 protein consists of a putative protein-protein interaction domain (PDZ-like; Supplemental Figure 1), a DSP domain, and a carbohydrate binding module 48 (CBM48) domain, which is typically involved in starch or glycogen binding. Despite the sequence similarities between the DSP and CBM domains of LSF1 and the known glucan phosphatase SEX4, whether LSF1 is itself a glucan phosphatase has been unclear. To further investigate whether LSF1 has phosphatase activity, we conducted phosphatase assays with LSF1 recombinant protein expressed in and purified from Escherichia coli. We produced the Δ61-LSF1 protein, which is the full-length mature polypeptide lacking the predicted 61-amino acid chloroplast transit peptide (cTP), as well as a further truncated Δ281-LSF1 protein that lacks the first 281 amino acids at the N terminus, including the cTP and the PDZ-like domain but still retaining the DSP and CBM domains. The Δ61-LSF1 protein was unstable and rapidly degraded in E. coli, but the Δ281-LSF1 was stable. However, the Δ281-LSF1 protein showed no activity against the generic phosphatase substrate p-nitrophenyl phosphate (pNPP), even when up to 1 µg of protein was used in the assay (Figure 1A). By contrast, phosphatase activity was detected with the recombinant SEX4 protein even when using 20-fold less protein in the assay (50 ng).
Figure 1.
LSF1 Has No Detectable Phosphatase Activity In Vitro.
(A) Phosphatase assays using SEX4 and the Δ281-LSF1 protein. Proteins were preincubated with 10 mM DTT, the incubation medium exchanged (see “Methods”), and the proteins incubated with pNPP for 30 min at 30°C. Data points represent mean activity of replicate incubations (n = 4, ±se). Inset, Coomassie-stained SDS gel of purified proteins (2 µg).
(B) Catalytic signature motifs of plant DSPs. The sizes of the amino acid symbols represent residue probabilities within the HCX5R sequence of LSF1 orthologs and other Arabidopsis DSPs. Sequence logos were generated using WebLogo (Crooks et al., 2004). Accession numbers of proteins used to generate the logos are given in the Supplemental Table.
(C) Phosphatase assays using SEX4 and the H197T mutant protein, as in (A).
The DSP domain is characterized by the signature motif HCX5R within the active site. This highly conserved motif contains a Cys residue involved in the formation of the phospho-enzyme reaction intermediate (Kaneko et al., 2012; Silver et al., 2013). Aligning the sequences of various Arabidopsis DSP domains (not including that of LSF1), we found that the catalytic site of LSF1 has an additional conserved Gly, forming a HCX2GX2R motif (Figure 1B). Although the Cys was conserved in all LSF1 orthologs examined, the surrounding motif in LSF1 was atypical. The highly conserved His preceding the conserved Cys in the DSP motif was replaced with a Thr in the LSF1 orthologs (Figure 1B). This His lowers the pKa of the conserved Cys via electrostatic interactions during catalysis, preventing protonation of the thiolate anion at physiological pH (Zhang and Dixon, 1993; Meekins et al., 2015). This His-to-Thr substitution could explain why we were unable to detect phosphatase activity for LSF1. To test this, we introduced the same substitution into the SEX4 recombinant protein (H197T). This resulted in a ∼25-fold reduction in the specific activity of the SEX4 protein (Figure 1C). We attempted the reverse substitution (T389H) in LSF1 to determine whether this would restore measurable phosphatase activity. However, the T389H variant had low expression and was degraded in E. coli, preventing us from testing this possibility.
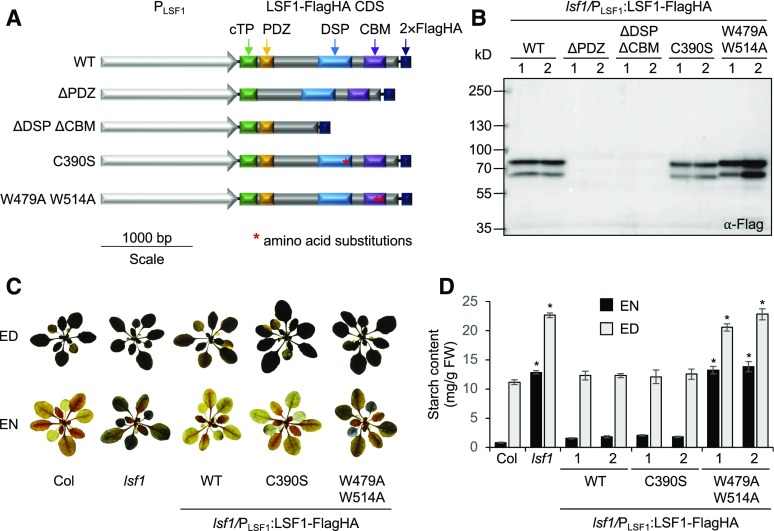
Although these experiments suggested that LSF1 cannot dephosphorylate pNPP, there was still the possibility that the protein could dephosphorylate specific substrates in vivo. Potential targets of the DSP domain could include phosphorylated proteins as well as glucans. Alternatively, LSF1 could be an inactive phosphatase that plays its primary role in starch degradation using its other domains, such as its PDZ-like domain to interact with other proteins and/or its CBM48 domain to interact with starch. We therefore designed a set of constructs to test which features of the LSF1 protein are important for its role in starch degradation (Figure 2A). Two of the constructs encoded LSF1 variants with a deletion of either the PDZ-like domain (LSF1 ∆PDZ, lacking amino acids 76–129) or the DSP and CBM domains (LSF1 ∆DSP ∆CBM, lacking amino acids 291–545). The borders of the DSP and CBM domains were defined based on a previous report (Comparot-Moss et al., 2010). For the PDZ-like domain, we aligned the LSF1 sequence with the consensus PDZ sequence (Supplemental Figure 1), which revealed that amino acids 76–129 shared the most homology with the consensus sequence. A second region, amino acids 163–187, also shared some homology with the consensus sequence, but this region was not contiguous with the first region. Thus, this second region was not included in the deletion. Another construct (LSF1 C390S) encoded a mutated LSF1 protein where the conserved Cys (Cys390 within the HCX5R motif) that is present in all DSPs was substituted with a serine. This mutation abolishes the phosphatase activity of SEX4 and LSF2 (Kötting et al., 2009; Santelia et al., 2011). The other construct, LSF1 W479A W514A, had the two conserved glucan binding tryptophan residues (Trp479 and Trp514) of the CBM48 changed to an alanine. These mutations abolish glucan binding in other CBM48s (McBride et al., 2009; Seung et al., 2015, 2017). All constructs contained a C-terminal fusion to a dual Flag and human influenza hemagglutinin (HA) tag (referred to as FlagHA) and were driven by the native LSF1 promoter. The glufosinate (BASTA) resistance marker was used to select transformants.
Figure 2.
Transformation of lsf1 Plants with LSF1 Variants.
(A) Schematic diagram of the LSF1 constructs with their domain structures indicated. Point mutations in the DSP and CBM48 domains are marked with a red asterisk, and changes in nucleotide sequences are shown. The constructs were transformed into lsf1 plants, with expression driven by the native LSF1 promoter (PLSF1).
(B) Immunoblot analysis of total protein extracts of transformed lsf1 plants. SDS-PAGE gels were loaded on an equal leaf area basis.
(C) Representative images of 4-week-old rosettes of the wild type (Col), untransformed lsf1, and complemented lsf1 plants with the wild-type LSF1, LSF1 C390S, and LSF1 W479A W514A expressed under the control of the native promoter. The rosettes were stained with iodine to detect starch.
(D) Starch content at the end of day (ED) and end of night (EN) measured in 3-week-old plant rosettes. Values are the mean ± se from six individual plants. Values marked with asterisks (*) are significantly different from the corresponding wild type (Col) using a two-tailed t test at P < 0.05.
The numbers 1 and 2 in (B) and (D) refer to two different homozygous lines for each transgene from independent transformation events.
We transformed the constructs into lsf1 plants and selected BASTA-resistant T1 plants. The segregation of the BASTA resistance marker in subsequent generations was used to identify multiple independent transgenic lines for each construct, two of which were shown to harbor the transgene insertion at a single genetic locus. We selected homozygous plants and used them for subsequent experiments. To compare the transgene expression levels among the selected lines, we extracted total protein from leaf tissue and performed immunoblot analysis using an anti-Flag antibody. The LSF1 wild type, C390S, and W479A W514A proteins were expressed to comparable levels in all of the selected lines (Figure 2B). Interestingly, two bands were detected for all of the LSF1-FlagHA proteins, although the endogenous LSF1 from wild-type extracts runs as a single band (Comparot-Moss et al., 2010). The lower band ran at the expected molecular weight of the tagged protein, while the upper band ran higher than expected. The reason for this is unknown, but it could be due to a posttranslational modification or the presence of the immature peptide still containing the N-terminal cTP. The proteins of the constructs carrying entire domain deletions (LSF1 ∆PDZ and LSF1 ∆DSP ∆CBM) were undetectable in plant extracts, despite screening 15–30 independent transformants in the T1 generation. These results suggest that these domains are required for stable protein accumulation in vivo.
We then investigated whether the expression of the mutated LSF1 proteins could complement the sex phenotype of lsf1 plants. The transgenic plants were grown under a 12-h light/12-h dark regime, and entire rosettes were harvested at the end of the day and end of the night. First, we observed starch in the rosette by iodine staining (Figure 2C). As expected, lsf1 had a sex phenotype, as it stained positive for starch at the end of the night. This phenotype was complemented by the expression of the wild-type protein (LSF1 WT-FlagHA). Interestingly, the expression of the LSF1 C390S-FlagHA protein also complemented the starch excess phenotype of lsf1. By contrast, the staining pattern of the lines expressing LSF1 W479A/W514A-FlagHA was identical to that of untransformed lsf1 plants. Starch quantification confirmed the iodine staining results: lines expressing LSF1 WT-FlagHA or LSF1 C390S-FlagHA had identical starch content to untransformed wild-type plants at both the end of day and end of night, while the starch content of lines expressing LSF1 W479A W514A-FlagHA was identical to that of untransformed lsf1 plants (Figure 2D). These data suggest that any putative phosphatase activity of LSF1 is not required for proper starch degradation under normal growth conditions. However, a functional CBM48 domain of LSF1 is essential for this process.
Since no protein accumulation was detected in lines harboring the LSF1 variants with entire domain deletions (LSF1 ∆PDZ, LSF1 ∆DSP ∆CBM), we tested whether we could observe protein expression when driven under the strong, constitutive Cauliflower mosaic virus (CaMV) 35S promoter. Indeed, expression driven by the CaMV 35S promoter resulted in detectable protein accumulation (Supplemental Figure 2A). However, iodine staining and starch quantification showed that neither protein could complement the sex phenotype of lsf1 (Supplemental Figures 2B and 2C).
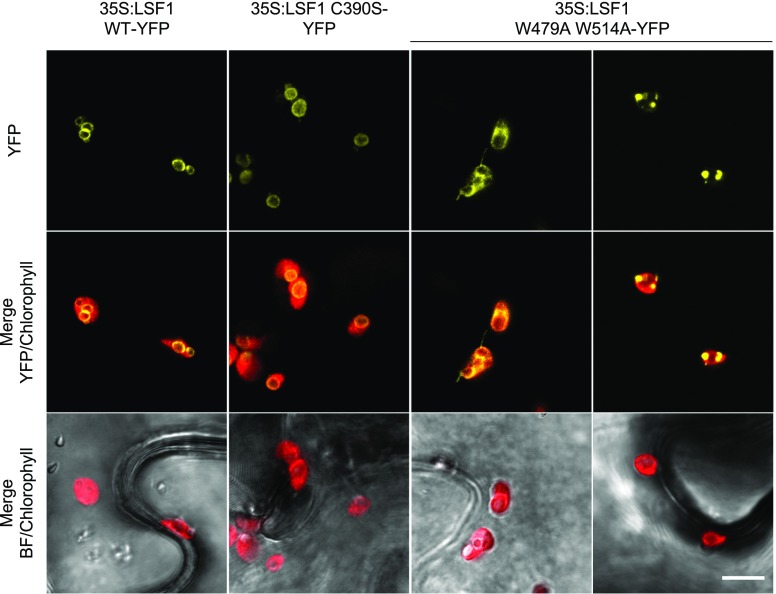
We verified that the W479A W514A mutations abolished starch binding of the LSF1 protein in vivo. For this, we used constructs encoding the wild-type or mutated proteins fused at their C termini to the yellow fluorescent protein (YFP) and driven by the CaMV 35S promoter. We infiltrated cultures of Agrobacterium cells harboring each construct into Nicotiana benthamiana leaves and visualized the protein using confocal microscopy 3 d after transformation. The wild-type protein localized to the starch granules (Figure 3), as previously observed (Comparot-Moss et al., 2010). However, the W479A W514A variant showed a different localization, usually appearing as a diffuse signal spread throughout the chloroplast stroma. However, in cells highly expressing the protein, signals were sometimes observed as bright spots around the starch granules, possibly due to protein aggregation caused by overexpression. However, in no chloroplasts did the W479A W514A variant adopt the same starch-bound localization as the wild-type protein, suggesting that the CBM domain is critical for starch binding in the LSF1 protein. The LSF1 C390S protein had the same starch-bound localization as the wild type, indicating that the mutation of this residue does not affect starch binding (Figure 3).
Figure 3.
The Subcellular Localization of LSF1 Wild-Type, C390S, and W479A W514A Proteins Transiently Expressed in Nicotiana benthamiana Leaves.
Confocal microscopy of N. benthamiana leaf epidermal cells transiently overexpressing LSF1 constructs tagged with YFP. All panels are shown to the same scale. Scale bar, 10 µm.
We also assessed the localization of the LSF1 ∆PDZ and LSF1 ∆DSP ∆CBM proteins. However, these proteins showed a stromal localization, similar to that observed for the LSF1 W479A W514A protein (Supplemental Figure 2D). This localization was expected for the LSF1 ∆DSP ∆CBM protein, as it lacks the CBM domain. However, the finding that the LSF1 ∆PDZ protein also showed this localization suggests that the PDZ-like domain is also required for starch binding.
LSF1 Interacts with the β-Amylases BAM1 and BAM3
Since the LSF1 C390S protein mediated proper starch degradation whereas the W479A W514A variant did not, we hypothesized that LSF1 might function as an enzymatically inactive protein scaffold during starch degradation, rather than acting as a phosphatase. We therefore searched for interaction partners of LSF1 using tandem-affinity purification (TAP). We created a construct encoding LSF1 with a C-terminal TAP-tag (LSF1-TAP), driven by the CaMV 35S promoter, and transformed it into the lsf1 mutant. Like the LSF1-FlagHA protein mentioned above, the LSF1-TAP protein also complemented the starch-excess phenotype of lsf1, showing that it is functional (Supplemental Figure 3). LSF1-TAP was purified from extracts of transformed plants using TAP. Each TAP experiment was performed in duplicate, and extracts from wild-type plants were used as negative controls. Proteins that copurified with LSF1-TAP were identified using mass spectrometry of peptides after trypsin digestion. For confident identification of proteins, a minimum of two unique peptides for each protein was required, and proteins identified in the negative-control purifications were removed from the list of potential LSF1 interactors. The remaining proteins (125 proteins in total) that were identified as potential interactors of LSF1 are shown in the Supplemental Data Set. While many of these interactions are likely unspecific, we found three proteins known to be involved in starch degradation. Indeed, both of the major chloroplastic β-amylases, BAM1 and BAM3, copurified with the LSF1-TAP, with particularly large numbers of peptides for BAM1 (Table 1). We also consistently found a small number of peptides matching the chloroplastic α-amylase AMY3. Interaction with these amylases could indicate a role for LSF1 in starch degradation that is distinct from glucan dephosphorylation, and it could explain why lsf1 mutants have a starch-excess phenotype.
Table 1. Interactions between LSF1 and Amylases Detected by TAP Purifications and MS.
| Protein | LSF1-TAP | BAM1-TAP | BAM3-TAP | BAM2-TAP | ||||
|---|---|---|---|---|---|---|---|---|
| Rep. 1 | Rep. 2 | Rep. 1 | Rep. 2 | Rep. 1 | Rep. 2 | Rep. 1 | Rep. 2 | |
| LSF1 | 37 | 40 | 16 | 9 | 14 | 42 | 0 | 0 |
| BAM1 | 21 | 10 | 29 | 14 | 0 | 0 | 0 | 0 |
| BAM3 | 5 | 6 | 0 | 0 | 145 | 168 | 0 | 0 |
| BAM2 | 0 | 0 | 0 | 0 | 0 | 0 | 19 | 2 |
| AMY3 | 2 | 4 | 0 | 4 | 8 | 5 | 0 | 0 |
Two replicate purifications of TAP-tagged BAM1, BAM2, BAM3, and LSF1 were analyzed by mass spectrometry. Values represent the total spectral counts of peptides matching each protein indicated. The full list of matching proteins can be found in the Supplemental Data Set. The numbers in bold represent the total spectral counts of peptides matching the TAP-tagged protein within each purification.
To confirm the interaction with the β-amylases, we conducted reciprocal TAP-tagging experiments. The coding sequence of either BAM1 or BAM3 was cloned downstream of the CaMV 35S promoter and in-frame with the C-terminal TAP tag. As a control, another chloroplast-localized β-amylase (BAM2) that did not copurify with LSF1-TAP was also TAP tagged. Each BAM-TAP construct was transformed into the knockout mutant lacking the respective endogenous BAM isoform. The TAP-tagging experiment was performed as described above, with the full list of potential interaction partners shown in the Supplemental Data Set. LSF1 peptides were found consistently in both BAM1-TAP and BAM3-TAP samples, but not in the BAM2-TAP samples (Table 1). AMY3 peptides were found in both replicates of the BAM3-TAP, but in only one replicate of the BAM1-TAP. Interestingly, no peptides matching BAM3 were found in the BAM1-TAP samples, and no peptides matching BAM1 were identified in the BAM3-TAP samples, suggesting that LSF1 resides in individual complexes with either BAM1 or BAM3, but not with both simultaneously.
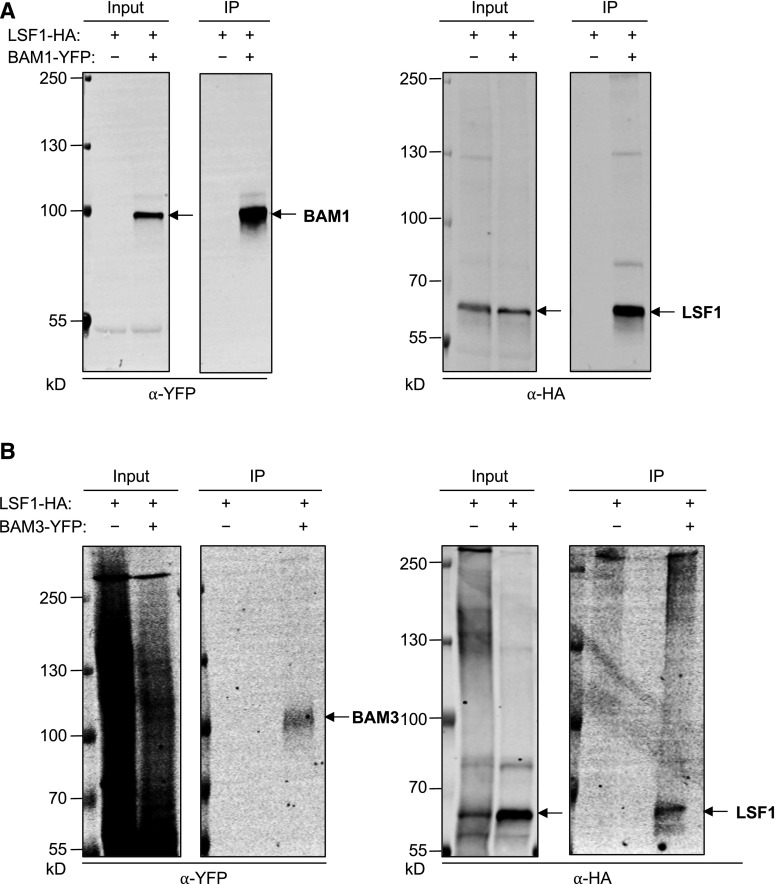
To further validate the protein-protein interactions between LSF1 and the BAMs, we used an immunoprecipitation-based technique with transiently expressed tagged proteins in N. benthamiana leaves. LSF1 with a C-terminal HA tag (LSF1-HA) was coexpressed with either BAM1-YFP or BAM3-YFP using leaf-infiltration of Agrobacterium cultures, as done for the experiment shown in Figure 3. Immunoprecipitations were conducted using extracts of transformed leaves using anti-YFP beads. The extracts (inputs) as well as the immunoprecipitates were analyzed by immunoblotting using HA and YFP antibodies. We found that LSF1-HA copurified with BAM1-YFP (Figure 4A) and with BAM3-YFP (Figure 4B), confirming interactions between LSF1 and each BAM isoform.
Figure 4.
Pairwise Interaction Tests Using Immunoprecipitation of LSF1 and BAMs Coexpressed in Nicotiana Benthamiana.
(A) BAM1-YFP and LSF1-HA were coexpressed in N. benthamiana leaves. BAM1-YFP was purified from extracts (Input) using anti-GFP beads. Proteins were detected in the input and immunoprecipitate (IP) fractions by immunoblotting with anti-GFP and anti-HA antibodies.
(B) As for (A), but with BAM3-YFP and LSF1-HA.
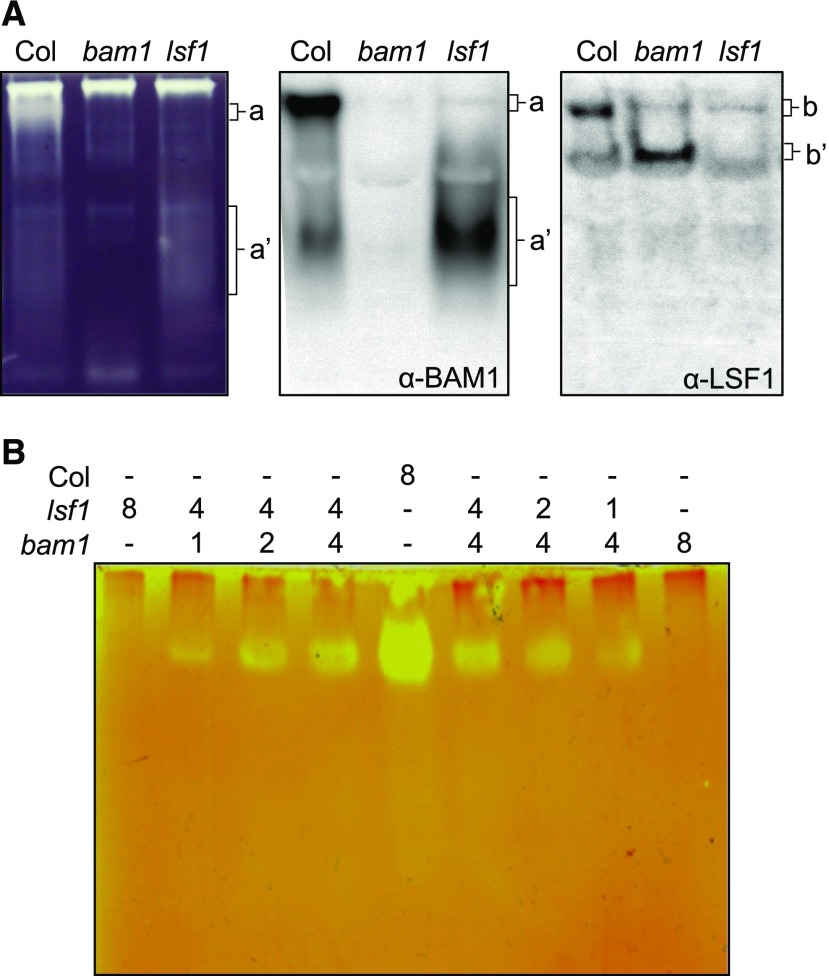
We also observed the interaction between LSF1 and BAM1 using several additional techniques. First, we performed zymography on native PAGE gels containing amylopectin to separate and visualize different amylolytic activities in crude extracts. Unfortunately, no activities corresponding to either BAM3 or AMY3 were observed in the gels, as verified with bam3 and amy3 mutants (Supplemental Figure 4). However, two bands of activity could be attributed to BAM1: a consistent, well-focused band of low electrophoretic mobility at the top of the gel and a more variable diffuse region of activity with higher electrophoretic mobility (Figure 5A). Both these activity bands were missing in the bam1 mutant. Interestingly, in the lsf1 mutant, the low-mobility BAM1 activity band was missing, while the intensity of the faster migrating activity band was increased relative to that observed in wild-type extracts. Probing blots of amylopectin-containing native gels with BAM1-specific antisera revealed that the BAM1 protein colocalizes with the two bands of BAM1 activity, as expected. In wild-type extracts, the majority of the BAM1 protein was present in the low-mobility form. However, in lsf1 extracts, all of the detectable BAM1 protein was present as the faster migrating form. Probing blots with LSF1-specific antisera revealed that the LSF1 protein colocalized with the low-mobility BAM1 activity (Figure 5A). In the absence of BAM1, the LSF1 protein also migrated more rapidly in the gel, further supporting the idea that BAM1 and LSF1 are present in the same high-molecular-weight complex. We investigated whether the putative BAM1-LSF1 complex could form in vitro by mixing bam1 and lsf1 extracts and incubating them prior to native PAGE. For this, we used gels containing glycogen, since the low-mobility BAM1 activity was particularly clearly resolved using this method. In the mixed extracts, the low-mobility BAM1 activity was visible; indeed, mixing different amounts of bam1 and lsf1 extracts restored the low-mobility activity in a ratiometric manner (Figure 5B). Thus, we observed a complex between BAM1 and LSF1 on native PAGE gels, and the complex formed in vitro.
Figure 5.
Two β-Amylase Activities of Low and High Electrophoretic Mobility (a and a′, Respectively) Are Missing in bam1.
(A) Extracts of total soluble protein from leaves of the wild type, bam1, and lsf1 were analyzed on native gels containing 0.1% (w/v) amylopectin (left). Two amylolytic activities (a and a′) were undetectable in the bam1 mutant. Immunoblots of amylopectin-containing native gels probed with the BAM1 antibody (middle) reveals two BAM1 bands (a and a′) that coincide with the β-amylase activities. In lsf1, only as the high mobility form of BAM1 (a′) is present. Protein gel blots probed with the LSF1 antibody (right) reveals two LSF1 bands (b and b′). In bam1, LSF1 is present only as the high-mobility form (b′). The low-mobility form (B) comigrates with the low-mobility β-amylase activity (A).
(B) Different amounts of bam1 and lsf1 extracts were mixed in the indicated relative amounts/ratios, separated on a native gel containing 0.3% (w/v) glycogen, and subjected to ratiometric analysis. Note the ratiometric restoration of the low-mobility β-amylase activity.
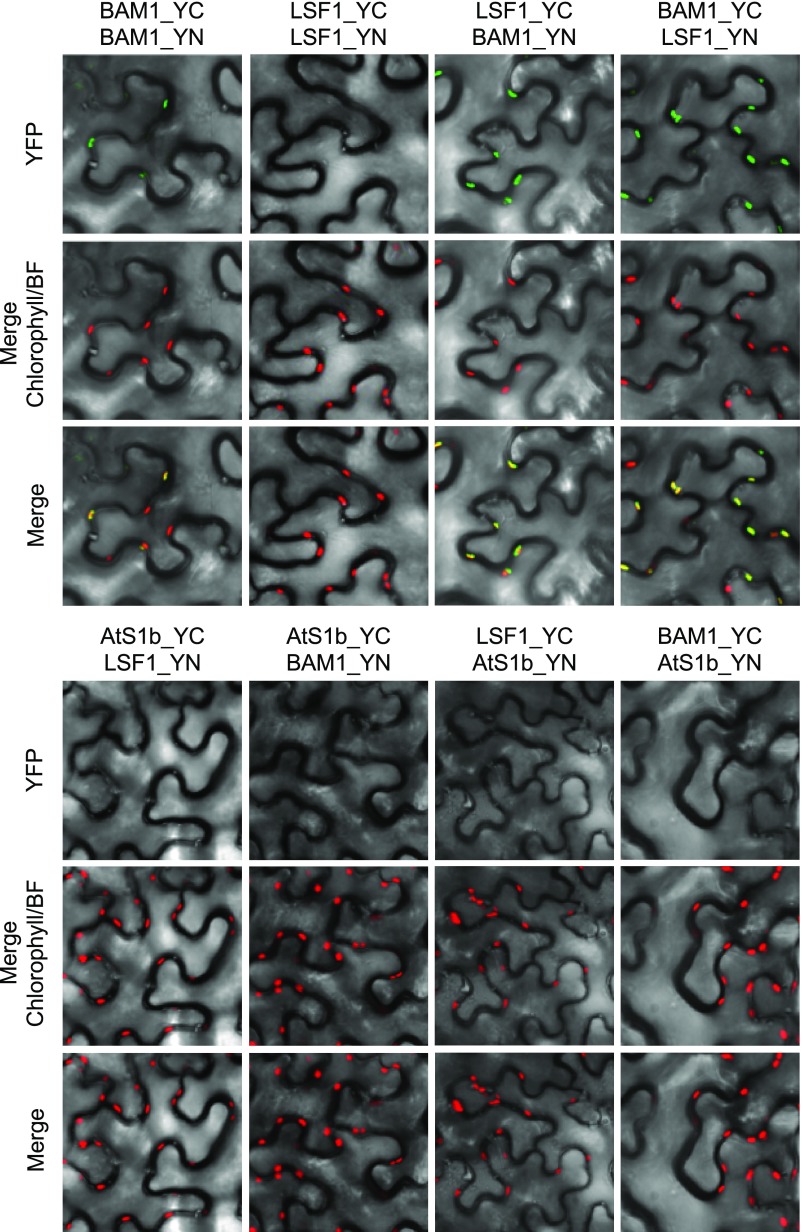
Finally, bimolecular fluorescence complementation (BiFC) further confirmed the BAM1-LSF1 interaction in planta. The N- and C-terminal parts of YFP were fused in-frame to the C-terminal ends of LSF1, BAM1, and BAM3 and expressed in N. benthamiana leaf cells. Fluorescence from the reconstituted YFP protein was visible when BAM1 and LSF1 constructs were coexpressed, and it colocalized with chlorophyll autofluorescence, suggesting that the two proteins are in close contact within the chloroplast (Figure 6). Controls for oligomerization of LSF1 and for self-assembly of the YFP protein did not result in YFP fluorescence. However, low levels of fluorescence were observed when BAM1 fused to the N-terminal part of YFP and BAM1 fused with the C-terminal part of YFP were coexpressed, suggesting possible oligomerization of BAM1.
Figure 6.
Bimolecular Fluorescence Complementation Assay Showing That BAM1 and LSF1 Interact in the Chloroplast.
C-terminal fusion constructs of BAM1 and LSF1 with N-terminal (YN) and C-terminal (YC) parts of YFP were cotransformed into N. benthamiana leaf cells using A. tumefaciens. Fusion constructs of the RbcS transit peptide (AtS1b) were used as negative controls for YFP self-assembly. YFP and chlorophyll fluorescence were assayed 72 h after infiltration using a confocal microscope. Fluorescence signals were overlaid onto a bright field image (YFP and Chlorophyll) and an image merging both YFP and chlorophyll fluorescence (Merge) was obtained using the Leica Application Suite software.
Our investigations of the BAM3-LSF1 complex were hindered by the finding that BAM3 activity was not detectable on native gels (Supplemental Figure 3). It is possible that the running conditions of the native PAGE (e.g. alkaline pH) are not suited for the preservation of BAM3 activity. Also, the BiFC constructs for the transient expression of BAM3 fused to the N- or C-terminal parts of YFP did not result in reliable protein expression, even though the full-length BAM3-YFP protein could be expressed (Figure 4). The reason for these difficulties in analyzing BAM3 may be related to its short half-life in vivo (see "Discussion" section; Li et al., 2017).
Phosphoglucan Degradation by LSF1-BAMs Is Dependent on External Phosphatase Activity
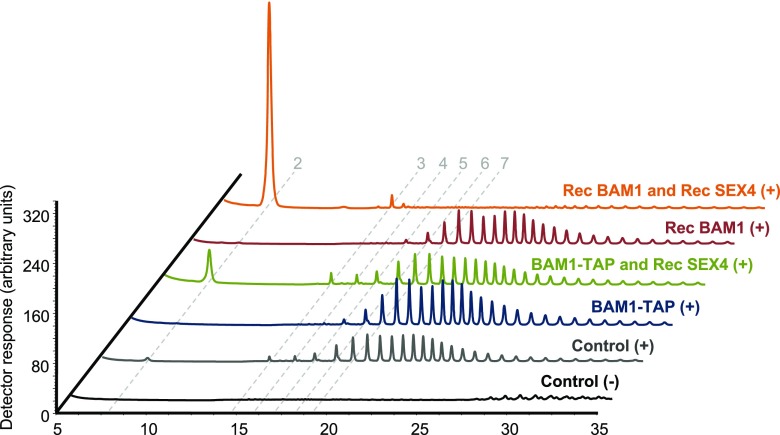
The finding that LSF1 interacts with BAMs allowed us to further substantiate our initial observation that LSF1 lacks phosphatase activity. We used the TAP method to purify the LSF1-containing BAM1-TAP complex (as described above) and tested whether it could degrade phospho-oligosaccharides. The phospho-oligosaccharides were pretreated with commercial β-amylase so that the phosphate groups were close to the nonreducing end and to obstruct further β-amylolysis (see “Methods”). As expected, further treatment of these purified phospho-oligosaccharides with recombinant BAM1 protein did not result in any release of maltose. The purified BAM1-TAP complex was also unable to release maltose, despite the presence of LSF1 (Figure 7). These results suggest that LSF1 cannot dephosphorylate the phospho-oligosaccharides, preventing the action of the associated BAM1. Coincubation of recombinant SEX4 with the recombinant BAM1 or with the BAM1-TAP complex allowed maltose to be released from the phospho-oligosaccharides. These results indicate that in the presence of an active glucan phosphatase (SEX4), the BAM1-TAP complex was able to degrade the phospho-oligosaccharides, supporting our finding that LSF1 itself is not an active glucan phosphatase.
Figure 7.
Phosphatase-Dependent Degradation of Phospho-Oligosaccharides to Maltose by BAM1.
Phospho-oligosaccharides prepared from potato amylopectin (see “Methods”) were incubated alone (controls) or with the enzyme combinations shown (BAM1-TAP, the TAP-purified LSF1-containing complex; Rec BAM1, recombinant BAM1; Rec SEX4, recombinant SEX4). After incubation, the reactions were stopped by boiling (5 min), and the remaining phospho-oligosaccharides dephosphorylated by treatment with excess recombinant SEX4 (+) or left untreated (−). Free oligosaccharides were analyzed by HPAEC-PAD. No maltose was released from phospho-oligosaccharides by recombinant BAM1 or by the LSF1-containing BAM1-TAP complex. However, coincubation with SEX4 allowed the release of maltose by recombinant BAM1 or by the BAM1-TAP complex. The dashed lines indicate the retention times of linear malto-oligosaccharides with the degree of polymerization indicated.
LSF1 Deficiency Reduces BAM Function In Vivo
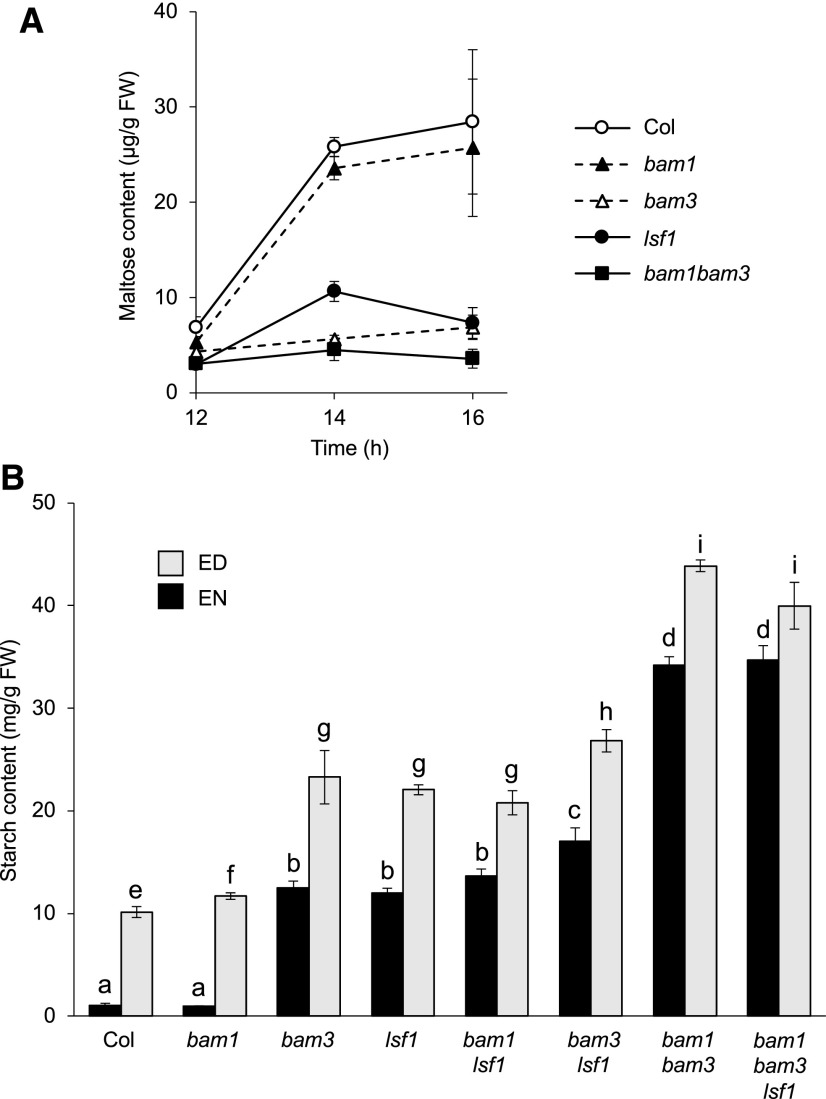
Our results suggest that LSF1 contributes to starch degradation in vivo through its interaction with BAM1 and/or BAM3 and not via phosphatase activity. To investigate this idea further, we analyzed the maltose content of the lsf1 mutant during the first 4 h of the night. Maltose levels were markedly reduced in lsf1 relative to the wild type, as would be expected from reduced BAM activity. However, the maltose levels were not as low as those observed in the bam3 and bam1 bam3 mutants (Figure 8A). This result suggests that the loss of LSF1 reduces BAM1 and BAM3 function but does not abolish it completely. The bam1 mutant had similar maltose levels to the wild type, which is consistent with the observation that it does not have a sex phenotype (Figure 8B).
Figure 8.
Maltose and Starch Levels in bam1 and lsf1.
(A) Maltose levels in the leaves of wild type and in bam1 and lsf1 harvested during the first 4 h of the night (ZT12–ZT16). Mean values are given (n = 5, ±se).
(B) Starch levels in the leaves of wild type (WT) and in bam1 and lsf1 measured at the end of the night (EN) and at the end of the day (ED). Mean values are given (n = 5, ±se). Statistical significance under a two-tailed t test are denoted with letters (a–d for EN values; e–i for ED values), whereby values with the same letter are not significantly different and values with different letters are significantly different at P < 0.05.
Next, we generated double mutants lacking LSF1 and either BAM1 or BAM3, as well as the triple mutant. We measured starch levels in plants harvested at the end of the night and at the end of the day (Figure 8B). As shown previously, the starch content of bam1 was similar to that of the wild type, whereas lsf1 and bam3 had elevated levels of starch (Kaplan and Guy, 2005; Fulton et al., 2008; Comparot-Moss et al., 2010). The bam1 bam3 double mutant had greatly elevated starch levels (Fulton et al., 2008). The starch levels of bam1 lsf1 resembled those of lsf1, whereas the bam3 lsf1 double mutant had slightly elevated starch levels relative to either single mutants. Notably, the starch content of the triple mutant was similar to that of the bam1 bam3 double mutant. These findings suggest that in the absence of both BAM proteins, the additional loss of LSF1 has no further impact on starch metabolism.
DISCUSSION
The Putative Phosphatase Activity of LSF1 Is Not Important for Starch Degradation
Despite the similarity of LSF1 to the phosphoglucan phosphatase SEX4, our in vitro and in planta experiments do not support the idea that LSF1 itself is enzymatically active as a phosphatase. No phosphatase activity of the Δ281-LSF1 recombinant protein was detected against the generic phosphatase substrate pNPP despite the presence of the entire DSP domain (Figure 1), and the LSF1- and BAM1-containing complex purified from plants had no activity against phospho-oligosaccharides (Figure 7). This adds to the previous observation that there was no change in the total phosphoglucan phosphatase activity in crude extracts of lsf1 compared with the wild type and that unlike sex4, no phospho-oligosaccharide intermediates accumulate in lsf1 during starch degradation (Comparot-Moss et al., 2010). Also, unlike lsf2 starch, the levels of starch-bound phosphate in lsf1 starch are similar to those of wild-type starch (Comparot-Moss et al., 2010; Santelia et al., 2011). Here, we also demonstrated that the starch excess phenotype of lsf1 is not caused by the loss of its putative phosphatase activity, since the expression of the LSF1 C390S protein, which lacked the conserved catalytic Cys in the DSP domain, fully complemented the starch excess phenotype of the lsf1 mutant (Figure 2). However, the LSF1 W479A W514A protein, which could not bind to starch, was unable to complement the lsf1 starch excess phenotype (Figure 2). These results suggest that the lsf1 mutant phenotype is specifically caused by the absence of the LSF1 protein itself at the starch granule.
Nevertheless, it is important to note that LSF1 has a strictly conserved catalytic Cys residue within the DSP domain (Figure 1). It is possible that the Cys is conserved to maintain proper protein structure and stability rather than to mediate catalysis, perhaps by forming a disulfide bond. In SEX4, the same catalytic Cys can form a disulfide bond, which regulates enzyme activity according to redox potential (Silver et al., 2013). Also, it remains possible that LSF1 has phosphatase activity toward an as-yet-unidentified substrate in vivo, although this activity is clearly not required for normal starch degradation.
Complex Formation between LSF1 and Chloroplastic BAMs
Several lines of evidence support the idea of a complex containing both LSF1 and BAMs. First, the reciprocal TAP-tag purification and mass spectrometry approach unambiguously identified that BAM1 and BAM3 copurified with LSF1 (Table 1). LSF1 was also consistently identified among proteins copurified with BAM1-TAP and with BAM3-TAP. However, the two BAMs did not copurify with each other, suggesting that LSF1 interacts in a complex with one or the other BAM, but not with both. It is worth noting that BAM1 and BAM3 fall into the same subfamily of β-amylases (designated as subfamily 2; Fulton et al., 2008) and that the proteins share a high degree of sequence similarity. No evidence for an LSF1-containing complex was obtained by tagging the BAM2 protein, which falls into a different β-amylase subfamily (subfamily 4; Fulton et al., 2008). Similarly, no other BAM isoforms were present in the purifications of LSF1-TAP. Therefore, we propose that BAM1 and BAM3 specifically interact with the LSF1-containing complex via a shared amino acid motif or surface structure.
The interaction between LSF1 and either BAM1 or BAM3 was also observed when the proteins were coexpressed in N. benthamiana leaves and the complex purified by immunoprecipitation (Figure 4). Our ability to detect two forms of BAM1 on native gels provided a further line of evidence for its interaction with LSF1: LSF1 comigrated with the low-mobility form of BAM1, while in the absence of LSF1, BAM1 migrated as a diffuse high-mobility band (Figure 5). Our data suggest that most but not all BAM1 is bound to LSF1 in leaf extracts. Furthermore, the observation that the BAM1-LSF1 low-mobility band was restored when bam1 and lsf1 extracts were mixed suggests that the interaction forms readily in solution. Finally, our BiFC analyses showed that the BAM1 and LSF1 proteins must either interact directly or be in very close proximity to one another (Figure 6).
Unfortunately, similar native PAGE and BiFC analyses were not successful for investigating the BAM3-LSF1 interaction because BAM3 activity could not be detected on our native gels and because BAM3 could not be reliably expressed in N. benthamiana when fused to the split YFP halves leaves. BAM3 is one of the most rapidly degraded proteins in Arabidopsis leaves, with a half-life of 0.43 days (Li et al., 2017). It is possible that BAM3 in extracts is degraded rapidly and that BAM3 is stabilized by the large YFP tag but not the smaller split YFP. Thus, the relative abundance and functional comparison between the BAM1- and BAM3-containing complexes also require further investigation.
Further work is also required to test other potential members of the complex. Relatively few AMY3 peptides were found in the LSF1-TAP samples, but more peptides were identified in the BAM3-TAP samples. Thus, it is possible that AMY3 interacts closely with BAM3 rather than directly through LSF1. However, it is unknown whether the association of AMY3 with the complex plays a significant role, since the bam1 bam3 lsf1 triple mutant had the same level of starch as the bam1 bam3 double mutant.
A survey of all proteins identified in the TAP experiments revealed that only one other protein was consistently found among LSF1, BAM1, and BAM3 interactors. This protein is a plastid-localized NAD-dependent malate dehydrogenase (At3g47520; pdNAD-MDH; Backhausen et al., 1998; Berkemeyer et al., 1998; Beeler et al., 2014; Selinski et al., 2014; Schreier et al., 2018), and the numbers of matching peptides were high (LSF1-TAP, 36/40; BAM1-TAP, 16/9; BAM3-TAP, 10/20; Supplemental Table), suggesting that a complex forms consisting of pdNAD-MDH, LSF1 and either BAM1 or BAM3. Immunoprecipitation of pdNAD-MDH-YFP expressed in 4-week-old rosettes also resulted in the copurification of BAM1, BAM3, and LSF1, further substantiating this interaction (Schreier et al., 2018). The functional significance of pdNAD-MDH in this complex is unclear. This malate dehydrogenase interconverts oxaloacetate and malate using NADH as a cofactor. However, it was recently shown that pdNAD-MDH has a distinct and essential moonlighting role in stabilizing the FtsH12 protein complex at the chloroplast inner envelope (Schreier et al., 2018). Therefore, perhaps pdNAD-MDH also stabilizes other plastidial protein complexes, such as those containing BAMs and LSF1.
LSF1 Could Function as a BAM Binding Scaffold at the Starch Granule
We propose that LSF1 plays a critical role in facilitating starch breakdown via its association with BAM1 and BAM3. As with SEX4, LSF1 possesses a CBM and associates with starch granules (Figure 2; Comparot-Moss et al., 2010). The LSF1 W479A W514A protein that could not bind to starch via this domain was also unable to function in starch degradation (Figure 2). Thus, the role of LSF1 may be to recruit and bind BAMs at the surface of the starch granule. We showed previously that GRANULE-BOUND STARCH SYNTHASE (GBSS), which is tightly associated with granules yet lacks a specialized CBM domain, is assisted in its interaction with the granule by the CBM-containing protein PTST1 (PROTEIN TARGETING TO STARCH; Seung et al., 2015). Similarly, all plant BAMs lack recognizable CBM domains and may be assisted in interacting with starch by virtue of the CBM domain of LSF1. Interestingly, bacterial β-amylases do possess a distinct CBM, supporting the idea that such a domain can assist in β-amylase function (Miyake et al., 2003).
We propose that the starch-excess phenotype of the lsf1 mutant is caused by reduced BAM1 and BAM3 activity at the granule surface. The marked reduction in maltose levels in lsf1 at the onset of the night is consistent with a deficiency in β-amylolysis. Also, the starch content of the bam1 bam3 lsf1 triple mutant is comparable to that of the bam1 bam3 double mutant, suggesting that the loss of LSF1 in the absence of both BAMs has no further effect on starch degradation. However, while LSF1 may enhance BAM1 and BAM3 activity on starch granules, it may not be absolutely essential for their activity, as lsf1 does not have the same phenotype as the bam1 bam3 double mutant. Indeed, bam1 bam3 accumulates much more starch than lsf1, suggesting that the BAM isoforms retain some activity in the absence of LSF1. Perhaps the BAMs have some starch binding capacity that is independent of LSF1. Indeed, both BAM1 and BAM3 can bind to amylopectin in vitro in the absence of LSF1 (Li et al., 2009). Also, according to our model, LSF1 would affect BAM1 and BAM3 activity on the starch granule surface, but it is likely that the activity of the BAMs on soluble oligosaccharides is unaffected by the loss of LSF1. Consistent with this idea, total BAM activity against the short soluble glucan substrate, PNP-G5, in crude extracts of lsf1 was not significantly different from that measured in wild-type extracts (Supplemental Figure 5). BAM1 and BAM3 may therefore actively degrade soluble oligosaccharides released from starch by AMY3 and ISA3 in the lsf1 mutant. The complete loss of this activity (i.e., in the bam1 bam3 or bam1 bam3 lsf1 mutant) results in greater starch excess. Likewise, the residual activity of the BAMs in the absence of LSF1 may also explain why the bam1 lsf1 and bam3 lsf1 double mutants did not have the same starch contents as bam1 bam3.
Our work not only provides evidence for physical interactions between proteins involved in starch degradation, but it also reveals distinct ways in which the phosphoglucan phosphatase family facilitates starch degradation in Arabidopsis leaves. On one hand, the nonenzymatic LSF1 recruits BAM proteins to the starch granule surface, ensuring their availability to participate in glucan degradation. On the other hand, after the initial phosphorylation of glucan chains by GWD or PWD to disrupt the crystalline packing, SEX4 and LSF2 actively dephosphorylate the glucans, clearing the way for BAM action. Both aspects of phosphoglucan phosphatase family activity at the granule surface appear to be required for effective starch granule degradation, as demonstrated by the sex phenotypes of sex4 and lsf1.
Further research will be needed to determine the dynamics of the BAM-LSF1 complex formation and dissociation and whether it is modulated to control starch degradation rates. It appears that LSF1 is not required for adjusting the starch degradation rate in response to an early night (Scialdone et al., 2013), but it could control the starch degradation rate in response to stress. Our analysis has focused on the starch content of the leaf mesophyll tissue under standard conditions, but there is growing evidence that starch and β-amylases play key roles during various abiotic stresses (e.g., osmotic stress) or in specific cell types (e.g., guard cell, nectary; Kaplan and Guy, 2004, 2005; Valerio et al., 2011; Maruyama et al., 2014; Monroe et al., 2014; Horrer et al., 2016; Thalmann et al., 2016; Zanella et al., 2016; Solhaug et al., 2019). The effect of the posttranslational modifications of both BAM isoforms on the interaction with LSF1 should also be investigated, such as the redox-regulation of BAM1 by thioredoxins (Sparla et al., 2006), the glutathionylation of BAM3 (Storm et al., 2018), and the phosphorylation of both proteins (Kötting et al., 2010). Also, we do not yet know the binding site of LSF1 on the starch granule. In SEX4, the CBM48 domain and the catalytic site form a continuous binding pocket for phospho-oligosaccharides (Meekins et al., 2014). Since LSF1 also has a CBM48 domain, it is possible that LSF1 retains the ability to bind phosphorylated glucan substrates, despite not being able to hydrolyze them, and that it directs BAM activity to phosphorylated regions of the granule surface that are amenable to degradation. Finally, complex formation with LSF1 may have a direct influence on the turnover rates of BAM1 and BAM3. The measured turnover rate of BAM1 is not as rapid as that of BAM3 (Li et al., 2017). It would be interesting to use a similar approach to measure the turnover rates of both proteins in the lsf1 mutant.
METHODS
Plant Material
All Arabidopsis (Arabidopsis thaliana) plants were grown in a climate-controlled Percival AR-95 growth chamber (CLF Plant Climatics) fitted with fluorescent lamps and supplemented with red LED panels. The diurnal cycle was set to 12-h light/12-h dark with a uniform light intensity of 150 μmol photons m−2 s−1. Relative humidity was 70%, and the temperature was 20°C. Seeds were sown onto potting compost, covered with plastic propagator tops, and stratified for 48 h at 4°C. The propagator tops were removed after cotyledon emergence. Two weeks after germination, individual seedlings were transplanted to 5-cm pots. The T-DNA insertion mutant lines were obtained from the Salk Institute (San Diego, California) and the GABI-Kat project (Max Plank Institute for Plant Breeding Research; Cologne, Germany). The following accessions characterized by Fulton et al. (2008) and Comparot-Moss et al. (2010) were used in this work: bam1, SALK_039895; bam2, GABI_132E06; bam3, CS92461; lsf1, SALK_053285. The bam1 lsf1 and bam3 lsf1 double mutants were produced by crossing the respective single mutants, while the bam1 bam3 lsf1 triple mutant was isolated from a cross between bam1 bam3 and lsf1. PCR-based genotyping was performed as described (Fulton et al., 2008; Comparot-Moss et al., 2010).
Sequence Alignment of the Catalytic Motifs of DSPs
The protein sequences of LSF1 orthologs were retrieved by searching the phylum Viridiplantae data set of the Phytozome 7 database (http://www.phytozome.net) using the LSF1 sequence as a query. No orthologs were found in green algae species, as noted previously (Comparot-Moss et al., 2010). All other Arabidopsis DSPs were as defined previously (Kerk et al., 2002, 2008). These sequences were aligned and trimmed to the DSP catalytic signature with JalView (Waterhouse et al., 2009). Graphical representations of the residue probabilities for these motifs (i.e., sequence logos) were generated using WebLogo (Crooks et al., 2004). Accession numbers for all proteins are given in the Supplemental Table.
Sequence Alignment of the PDZ Domain
The PDZ consensus protein sequence was retrieved from the NCBI Conserved Domain Database (CDD; https://www.ncbi.nlm.nih.gov/cdd/; accession cd00992). The PDZ consensus sequence was aligned to the Arabidopsis LSF1 amino acid sequence using the ClustalO program. The alignment is presented in Supplemental Figure 1.
Recombinant Protein Expression and Phosphatase Assays
The SEX4 cDNA encoding the mature protein (lacking the N-terminal cTP, residues 54–379; Niittylä et al., 2006; Kerk et al., 2008) including its stop codon was cloned into pET101/D/TOPO using Gateway cloning (Thermo Fisher Scientific). The H197T mutant version of SEX4 was generated using a QuikChange II Site-Directed Mutagenesis kit (Agilent) according to the manufacturer’s instructions. Recombinant proteins were expressed in Escherichia coli BL21 Star (DE3) cells (Thermo Fisher Scientific) that were induced for 4 h at 22°C with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The cells were harvested by centrifugation (4000g; 10 min at 4°C) and disrupted using a cell press (1000 psi; three passes) in lysis buffer containing 50 mM Tris-HCl (pH 7.4), 1 mM EDTA, 1 mM EGTA, 5% (v/v) glycerol, 0.5 mM phenylmethanesulfonylfluoride (PMSF), and 0.5 mM benzamidine. The lysate was cleared by centrifugation (100,000g; 35 min at 4°C), adjusted to 2 mM DTT, and loaded onto a Q-Sepharose anion-exchange column (GE Healthcare). Proteins were eluted with a 0–200 mM NaCl gradient over 15-column volumes at a flow rate of 3 mL min−1 using an ÄKTA FPLC system (GE Healthcare). Fractions containing SEX4/SEX4 H197T were pooled and loaded on a prepacked Superdex 75 size-exclusion column (GE Healthcare) run with a flow rate of 1 mL min−1 with 25 mM Tris-HCl (pH 7.4), 200 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 5% (v/v) glycerol. Pure SEX4/SEX4 H197T fractions were pooled, concentrated, flash-frozen in liquid N2, and stored at −80°C.
The LSF1 cDNA encoding the mature protein (lacking the N-terminal cTP; residues 62–591) was cloned and expressed as described for the SEX4 constructs, except that no stop codon was inserted, resulting in a HIS-tagged protein. An N-terminal truncated version, Δ281-LSF1 (residues 282–591), was also cloned. Expressed proteins were purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography. Clarified lysates were incubated with 0.5 mL Ni-NTA agarose resin (Qiagen) for 1 h at 4°C on an end-over-end mixer. The resin was washed with 200 volumes of a high-stringency wash buffer containing 25 mM Tris-HCl (pH 7.5), 1 M NaCl, 30 mM imidazole, 0.05% (v/v) Triton X-100, 0.5 mM PMSF, 0.5 mM benzamidine, followed by 50 volumes of low-stringency wash buffer containing 25 mM Tris-HCl (pH 7.5), 1 M NaCl, and 30 mM imidazole. Proteins were eluted with 5 volumes of wash buffer containing 250 mM imidazole, concentrated, frozen in liquid N2, and stored at −80°C.
Phosphatase assays were performed to measure in vitro enzyme activity by monitoring the hydrolysis of pNPP. Purified proteins were incubated with 10 mM DTT for 10 min at 20°C and buffer exchanged into pNPP buffer (100 mM HEPES-NaOH, pH 7.0; 150 mM NaCl, 1 mM EDTA) using an Amicon Ultra Centrifugal Filter (Millipore). Various amounts of SEX4 and H197T (0–2 µg) or Δ281-LSF1 (0–1 µg) were incubated with 4 mM pNPP for 30 min at 30°C and neutralized with an equal volume of 2 M NaOH. Absorbance at 405 nm was measured. Four independent experiments were performed, each with measurements in triplicate.
Cloning of Expression Vectors for Plant Transformation
Expression constructs of LSF1 driven by the native LSF1 promoter were generated using the Multi-site Gateway cloning system (Thermo Fisher Scientific). The LSF1 promoter (1560 bp) was amplified from Arabidopsis genomic DNA, flanked with attB4 and attB1R recombination sites, and recombined into pDONR P4-P1r using BP Clonase. The full-length coding sequence of LSF1 was amplified from Arabidopsis cDNA with attB1 and attB2 sites and recombined into the pDONR221 using BP Clonase. The point mutations in the DSP and CBM domains, as well as the entire deletions of the PDZ and DSP CBM domains, were generated in the LSF1:pDONR221 vector using a QuikChange Site-Directed Mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. The Flag-HA tag in pDONR P2R-P3 was acquired from Tschopp et al. (2017). The promoter, appropriate coding sequence of LSF1, and in-frame C-terminal FlagHA tag were recombined into the multisite Gateway binary vector pB7m34GW,0 via an LR Clonase II plus reaction.
To visualize subcellular localization, the appropriate LSF1 coding sequence (wild type, C390S, W479A W514A, ∆PDZ, ∆DSP ∆CBM) in pDONR221 was recombined into the pB7YWG2 vector via an LR reaction, in-frame with the C-terminal YFP tag. For the overexpression of LSF1 ∆PDZ and LSF1 ∆DSP ∆CBM, the appropriate coding sequence in the pDONR221 vector was recombined into the pC-TAP (pYL436) vector (Rubio et al., 2005) via an LR Clonase reaction. The pC-TAPa (pYL436) vector contains a C-terminal immunoglobulin G (IgG) binding domain, preceded by 9Xmyc epitopes and a His tag, with a protease cleavage site between them. Expression in planta is driven by the CaMV 35S promoter.
To construct vectors used to generate plants for TAP, full-length cDNAs of BAM1, BAM2, BAM3, and LSF1 were cloned into the pC-TAPa vector via the pDONR221 intermediate using procedures similar to those described above. For the immunoprecipitation experiments, the BAM1 coding sequence in pDONR221 was recombined into the pB7YWG2,0 vector (with a CaMV 35S promoter and in-frame C-terminal YFP tag; Karimi et al., 2002); the BAM3 coding sequence in pDONR221 was recombined into pUBC-YFP (with a UBIQUITIN10 promoter from Arabidopsis and in-frame C-terminal YFP tag; Grefen et al., 2010); the LSF1 coding sequence in pDONR221 was recombined into pJCV52 (with a CaMV 35S promoter and in-frame C-terminal HA tag; Department of Plant Systems Biology, VIB-Ghent University).
For the BiFC analysis, the coding sequences for BAM1 and LSF1 were cloned using a pCR8/GW/TOPO TA kit (Thermo Fisher Scientific). The initial 84 bp encoding the transit peptide of the ribulose bisphosphate carboxylase small chain 3B (RbcS) were amplified and cloned as a negative control (Hiltbrunner et al., 2004). All constructs were recombined into the Gateway destination vectors encoding C-terminal fusions to the N-terminal half (YN) or C-terminal half (YC) of the yellow fluorescent protein (YFP); pGPTVII.Bar.YN-GW, pGPTVII.Bar.GW-YN, pGPTVII.Hyg.YC-GW, and pGPTVII.Hyg.GW-YC using LR Clonase (Stolpe et al., 2005).
Plant Transformation and Complementation
Plasmids were transformed into Agrobacterium tumefaciens (strain GV3101 or C58C1, carrying the helper plasmid pCH32). Arabidopsis plants were transformed using the floral-dip method, and at least three different plants (T0) were dipped per construct (Clough and Bent, 1998). Transformed seedlings were either selected on soil by spraying with Basta (for constructs in pB7m34Gw,0) or on Murashige and Skoog plates containing gentamycin (35 μg mL−1; for constructs in pC-TAPa). Approximately 5–10 independent transformants from each T0 plant were screened for protein expression in the T1 generation. For starch measurements, two independent lines (i.e., from different T0 plants) homozygous for a single insertion (as determined by analyzing the segregation of the selection marker) were selected, and the measurements were performed in the T3 generation. For complementation analysis and TAP, several lines from independent transformation events were grown on selective plates alongside the respective wild-type and mutant controls on nonselective plates. Gentamycin-resistant seedlings and the relevant controls were transplanted to soil and grown for at least 2 more weeks.
For the BiFC experiments, each transformed A. tumefaciens strain and C58C1 carrying the P19 silencing suppressor plasmid were grown at 28°C in Luria-Bertani medium supplemented with the appropriate antibiotics for 22–24 h. Bacteria were sedimented by centrifugation at 5000g for 15 min at 20°C and resuspended in 10 mM MgCl2 and 150 μM acetosyringone (Sigma-Aldrich). Cells were adjusted to an optical density (OD) at 600 nm of 0.22–0.5, mixed, and incubated at 20°C for 2 h. The A. tumefaciens mixture was infiltrated into Nicotiana benthamiana leaves as described previously (Witte et al., 2004). For immunoprecipitation experiments in N. benthamiana, A. tumefaciens cells (strain GV3101) carrying the appropriate vectors were resuspended in water to OD600 = 1 and infiltrated into leaves of 4-week-old plants. Transformed leaves were analyzed after 2 to 3 d after infiltration.
Confocal Microscopy
To image YFP in leaf tissue, confocal laser-scanning microscopy was conducted as previously described (Beeler et al., 2014). For BiFC analysis, fluorescence was assayed 72 h after infiltration using a 63× oil-immersion objective (1.32 numerical aperture) on an inverted Leica DM IRB confocal microscope. YFP signals were monitored with a 488-nm excitation wavelength and an emission wavelength window of 505–565 nm.
TAP
TAP was performed according to Rubio et al. (2005) with minor modifications. Plant material (up to 10 g) harvested at the end of the night was ground in liquid N2 with a mortar and pestle. Protein was extracted from the leaf powder by further grinding in all-glass homogenizers in ice-cold extraction buffer containing 100 mM Tris-HCl (pH 7.5), 10% (v/v) glycerol, 150 mM NaCl, 0.1% (v/v) Triton X-100, with Complete Protease Inhibitor Cocktail (Roche), at a tissue:medium ratio of 1:2 (w/v). Homogenates were filtered through two layers of Miracloth and insoluble material removed by centrifugation (13,000g, 4°C for 15 min). Supernatants were incubated with IgG beads (GE Healthcare; 300 µL per 5 g starting material) on a rotating incubator for 4 h at 4°C. After incubation, the beads were washed thrice with 10 mL extraction buffer and once with 10 mL extraction medium supplemented with 1 mM DTT. Cleavage from the IgG beads was performed by incubation with 25 units of 3C PreScission protease (GE Healthcare) in 5 mL extraction medium with 1 mM DTT on a rotating incubator (2.5 h at 4°C). Eluted proteins were incubated for an additional 2 h at 4°C with 1 mL of Ni-Sepharose 6 Fast Flow beads (GE Healthcare). The beads were washed with thrice with 10 mL extraction medium and bound proteins eluted in 5 mL medium supplemented with 150 mM imidazole. Eluates were concentrated 40-fold using Amicon Ultra-4 10 kD Centrifugal Filter Devices (Millipore). This purification procedure was repeated at least twice for each tagged protein. As a negative control, extracts of wild-type plants were processed the same way.
Sample Preparation and Mass Spectrometry
Protein samples obtained from TAP-tagging purifications were subjected to SDS-PAGE on 10% (w/v) gels, which were sliced into several fractions after silver staining, as described previously (Shevchenko et al., 2002). Each gel slice was diced into small pieces. In-gel digestion was performed essentially as described previously (Shevchenko et al., 1996). After digestion, dried peptides were dissolved in 3% (v/v) acetonitrile, 0.2% (v/v) trifluoroacetic acid and cleaned up using C18 ZipTips (Millipore). Clean samples were dried and dissolved in 5% (v/v) acetonitrile, 0.1% (v/v) formic acid for mass spectrometry.
Peptides were analyzed on an LTQ Orbitrap mass spectrometer (Thermo Fischer Scientific) coupled to an Eksigent-Nano-HPLC system (Eksigent Technologies). Peptide mixtures were loaded onto laboratory-made capillary columns (75 μm inner diameter (BGB Analytik), 8 cm long, packed with Magic C18 AQ beads, 3 μm, 100 Å (Michrom BioResources). Peptides were eluted from the column with an increasing concentration of acetonitrile (from 5% [v/v] acetonitrile, 0.2% [v/v] formic acid to 40% [v/v] acetonitrile, 0.2% [v/v] formic acid over 74 min, followed by a 10-min wash step at 5% [v/v] acetonitrile, 0.2% [v/v] formic acid). Full-scan mass spectrometry (MS) spectra (300–2000 m/z) were acquired with a resolution of 60,000 at 400 m/z after accumulation to a target value of 500,000. Collision induced dissociation tandem MS (MS/MS) spectra were recorded in a data-dependent manner in the ion trap from the six most intense signals above a threshold of 500, using a normalized collision energy of 35% and an activation time of 30 ms. Charge state screening was enabled and single charge states were rejected. Precursor masses already selected for MS/MS were excluded for further selection for 120 s, and the exclusion window was set to 20 ppm. The size of the exclusion list was set to a maximum of 500 entries.
MS/MS spectra were searched with Mascot (Matrix Science) version 2.2.04 against the Arabidopsis TAIR9 protein database with a concatenated decoy database supplemented with contaminants (67,079 entries). The search parameters were as follows: requirement for tryptic ends, one missed cleavage allowed, mass tolerance of ± 5 ppm. Beside carbamidomethylation of cysteines as fixed modification, oxidation of methionine was included as variable modification. Individual ion scores higher than 22 indicated identity or extensive homology. Peptide identification was accepted with a minimal Mascot ion score of 23 and a Mascot expectation value below 0.05. To increase protein identification confidence, a minimum of two unique peptides for each identified protein was required. The spectrum false discovery rate was calculated by dividing the number of decoy database spectrum assignments by the number of spectrum assignments in the final data set. The false positive rate was below 1% for all measured biological replicates of the TAP-tagging experiments.
Immunoprecipitation Experiments
Proteins were extracted from N. benthamiana leaves transiently expressing epitope-tagged proteins by homogenizing in immunoprecipitation medium (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% [v/v] Triton X-100, 1 mM DTT, and Complete Protease Inhibitor cocktail [Roche]). Insoluble material was removed by centrifugation. The supernatant was incubated for 1 h at 4°C with μMACS magnetic beads conjugated to α-YFP (Miltenyi Biotec). After incubation, the beads were recovered using a μColumn (Miltenyi Biotec) on a magnetic stand. The beads were washed five times with immunoprecipitation medium before eluting the bound proteins with SDS-PAGE loading buffer. Input and elution samples were loaded onto a SDS-PAGE and immunoblotted with anti-green fluorescent protein (GFP; Abcam ab290; 1:1000) and anti-HA (Abcam ab9110; 1:7000) antibodies.
Native PAGE and Native PAGE Blotting
Soluble proteins were extracted from plant leaf tissue in ice-cold medium containing 100 mM Mops (pH 7.2), 1 mM EDTA, 1 mM DTT, and 10% (v/v) ethanediol at a tissue:medium ratio of 1:6 (w/v). The tissue was ground in all-glass homogenizers and insoluble material removed by centrifugation (10 min, 16,000g, 4°C). Total soluble protein levels were determined using a Bradford kit (Bio-Rad). Native PAGE was performed using 20 µg of total protein, as described previously (Zeeman et al., 1998). In brief, the gels contained 6% (w/v) polyacrylamide and either 1% (w/v) oyster-glycogen (Sigma-Aldrich) or 0.2% (w/v) potato amylopectin (Sigma-Aldrich). After 3 h of electrophoresis at 4°C, the gels were incubated for either 1–2 h at 37°C or overnight at 25°C in medium containing 100 mM Tris-HCl (pH 7.2), 1 mM MgCl2, 1 mM CaCl2, and 2.5 mM DTT. The gels were stained with Lugol’s iodine solution (Sigma-Aldrich). For immunoblot detection of proteins after native PAGE, the gels were incubated twice for 5 min in 20 mM Tris-HCl (pH 8.3), 150 mM Gly, 1% (w/v) SDS, at 75°C. The gels were rinsed in the same medium and blotted using standard electroblotting procedures. Antibodies and immunodetection conditions were as described in Fulton et al. (2008) and Comparot-Moss et al. (2010).
Carbohydrate Measurements
Whole rosettes of 3-to 4-week-old plants were harvested at the indicated times and frozen directly in liquid N2. The frozen plant material was pulverized using a Mixer Mill (Retsch). The frozen powder was extracted in ice-cold 0.7 M perchloric acid for 30 min with intermittent mixing. Subsequent steps were as described in Delatte et al. (2005). Starch levels in the insoluble fraction were determined by measuring the amount of glucose released by treatment with α-amylase and amyloglucosidase, as described previously (Smith and Zeeman, 2006).
Maltose levels were determined using high performance anion exchange chromatography with pulsed amperometric detection. Samples of the neutralized soluble fraction (100 µL) were sequentially applied to 1.5-mL columns of Dowex 50 W and Dowex 1 (Sigma-Aldrich). Neutral compounds were eluted with water, lyophilized, and redissolved in 100 μL of water. Maltose was separated on a Dionex PA-100 column (Thermo Fisher Scientific) according to the following conditions: eluent A, 100 mM NaOH; eluent B, 100 mM NaOH and 50 mM sodium acetate; eluent C, 150 mM NaOH and 500 mM sodium acetate. The gradient was as follows: 0 to 5 min, 50% A and 50% B; 5 to 25 min, a concave gradient to 50% A, 10% B, and 40% C; 25 to 32 min, step to 10% B and 90% C; 32 to 36 min, step to 50% A and 50% B. Peaks were identified by coelution with known standards. Peak areas were determined using Chromeleon software. To account for unequal losses between samples during the Dowex steps and the lyophilization, samples were spiked with a known concentration of cellobiose and peak areas were corrected accordingly.
For iodine staining, plant rosettes were harvested at the end of the day and the end of the night and were incubated in 80% (v/v) ethanol to remove the chlorophyll. Excess ethanol was washed away with water before staining with Lugol’s solution (KI/I2 solution; Sigma-Aldrich). Excess iodine was washed away with water.
Preparation of Phospho-oligosaccharides
Ten grams of amylopectin from potato (Solanum tuberosum) starch (Sigma-Aldrich) were dissolved in 200 mL of 5 mM sodium acetate (pH 4.8) at 60°C. Partial enzymatic degradation was achieved by incubation with isoamylase (25,000 U; from Pseudomonas amyloderamosa, Sigma-Aldrich), pullulanase (3.6 U; from Klebsiella planticola, Megazyme), and β-amylase (1000 U; from Hordeum vulgare; Megazyme) at 37°C for 19 h, followed by the addition of α-amylase (3000 U; from pig pancreas, Roche) and an additional 3-h incubation. The reaction was stopped by heating to 95°C for 15 min. Insoluble material was removed by centrifugation (30 min, 5000g, 4°C). The supernatant was filtered through a 0.45-μm filter and diluted to 1 L with water, the pH was adjusted to 7.0 with NaOH, and the sample applied to a 50 mL anion-exchange chromatography column (Q-Sepharose FF, GE Healthcare; flow rate of 2 mL min−1, 4°C). The column was washed with 2 volumes of water and phospho-oligosaccharides were eluted into fractions with 200 mM NaCl, 10 mM HCl (flow rate of 5 mL min−1). Positive fractions, as determined by a reducing-end assay (Anthon and Barrett, 2002), were pooled and the phospho-oligosaccharides precipitated in 75% (v/v) ethanol for 30 min on ice and pelleted by centrifugation (30 min, 12,000g, 4°C). The supernatant was removed and the pellet dried and redissolved in 2 mL 2 mM HEPES-NaOH (pH 7.0) to a concentration of 45.5 µmol Glc equivalents mL−1.
After the specified treatments, the phospho-oligosaccharides were dephosphorylated with recombinant SEX4 protein in 0.1 M sodium acetate, 50 mM bis-Tris, 50 mM Tris-HCl, pH 6.0, 2 mM DTT for 2 h at 37°C (Kötting et al., 2009). The dephosphorylated phospho-oligosaccharides were analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) as described previously (Kötting et al., 2009).
Accession Numbers
Sequence data from this article can be found in TAIR (www.arabidopsis.org) under the following accession numbers: LSF1 (AT3G01510), BAM1 (AT3G23920), BAM3 (AT4G17090), SEX4 (AT3G52180), BAM2 (AT4G00490), AMY3 (AT1G69830), and pdNAD-MDH (AT3G47520). The TAIR accession numbers of LSF1 orthologs and other Arabidopsis DSPs are provided in the Supplemental Table, and the TAIR accession numbers of potential interaction partners of LSF1, BAM1, BAM3, and BAM2 are provided in the Supplemental Data Set.
Supplemental Data
Supplemental Figure 1. Amino acid sequence alignment of the putative LSF1 PDZ domain with the PDZ consensus sequence.
Supplemental Figure 2. Overexpression of the LSF1 ∆PDZ and LSF1 ∆DSP ∆CBM constructs in lsf1 plants does not complement the starch excess phenotype.
Supplemental Figure 3. LSF1-TAP expression restores near-normal levels of starch in lsf1.
Supplemental Figure 4. No activities corresponding to BAM3 or AMY3 are observed with native PAGE.
Supplemental Figure 5. Total β-amylase activity is not altered in extracts of lsf1.
Supplemental Table. Accession and/or phytozome numbers of protein sequences used to generate logos in Figure 1B.
Supplemental Data Set. Proteins identified by tandem mass spectrometry in TAP-tagging experiments.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Paolo Nanni (Functional Genomics Centre Zurich) for help with proteomic analysis, David Brändli (Institute of Molecular Plant Biology, ETH Zurich) for technical help, and Matthias Hirsch-Hoffmann (Institute of Molecular Plant Biology, ETH Zurich) for bioinformatic help. We thank the Roche Research Foundation, the ETH Foundation (for a Heinz Imhof fellowship sponsored by Syngenta), and the Swiss-South African Joint Research Programme (grant 08 IZ LS Z3122916). This work was also funded in part by the National Science Council (grant NSC 96-2311-B-001-004 to J.C.), the Natural Sciences and Engineering Research Council of Canada (to G.B.G.M.), the Korea Research Foundation, funded by the Korean Government (MOEHRD, Basic Research Promotion Fund; grant KRF-2008-C00143to S.-K.L.), a Zurich-Basel Plant Science Centre Syngenta postdoctoral fellowship (to S.B.), and the Swiss National Foundation (SNF) (grant 31003A_156987 to O.K. and S.C.Z. and grant PP00P3_176957 to S.B.).
AUTHOR CONTRIBUTIONS
T.B.S., M.U., J.C., G.B.G.M., and S.C.Z. conceived and directed the research; T.B.S., M.U., S.-K.L., D.Si., A.G., and D.Se. designed the experiments; T.B.S., M.U., S.-K.L., W.-L.L., Z.S., D.Si., A.G., A.M., S.E., M.S.-W., S.B., and O.K. performed research and analyzed data; T.B.S., M.U., and S.C.Z. wrote the article with input from all of the authors.
Footnotes
Articles can be viewed without a subscription.
References
- Anthon G.E., Barrett D.M. (2002). Determination of reducing sugars with 3-methyl-2-benzothiazolinonehydrazone. Anal. Biochem. 305: 287–289. [DOI] [PubMed] [Google Scholar]
- Backhausen J.E., Emmerlich A., Holtgrefe S., Horton P., Nast G., Rogers J.J.M., Müller-Röber B., Scheibe R. (1998). Transgenic potato plants with altered expression levels of chloroplast NADP-malate dehydrogenase: Interactions between photosynthetic electron transport and malate metabolism in leaves and in isolated intact chloroplasts. Planta 207: 105–114. [Google Scholar]
- Baunsgaard L., Lütken H., Mikkelsen R., Glaring M.A., Pham T.T., Blennow A. (2005). A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated α-glucans and is involved in starch degradation in Arabidopsis. Plant J. 41: 595–605. [DOI] [PubMed] [Google Scholar]
- Beeler S., Liu H.-C., Stadler M., Schreier T., Eicke S., Lue W.-L., Truernit E., Zeeman S.C., Chen J., Kötting O. (2014). Plastidial NAD-dependent malate dehydrogenase is critical for embryo development and heterotrophic metabolism in Arabidopsis. Plant Physiol. 164: 1175–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkemeyer M., Scheibe R., Ocheretina O. (1998). A novel, non-redox-regulated NAD-dependent malate dehydrogenase from chloroplasts of Arabidopsis thaliana L. J. Biol. Chem. 273: 27927–27933. [DOI] [PubMed] [Google Scholar]
- Buléon A., Colonna P., Planchot V., Ball S. (1998). Starch granules: Structure and biosynthesis. Int. J. Biol. Macromol. 23: 85–112. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Comparot-Moss S., et al. (2010). A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol. 152: 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte T., Trevisan M., Parker M.L., Zeeman S.C. (2005). Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J. 41: 815–830. [DOI] [PubMed] [Google Scholar]
- Delatte T., Umhang M., Trevisan M., Eicke S., Thorneycroft D., Smith S.M., Zeeman S.C. (2006). Evidence for distinct mechanisms of starch granule breakdown in plants. J. Biol. Chem. 281: 12050–12059. [DOI] [PubMed] [Google Scholar]
- Edner C., Li J., Albrecht T., Mahlow S., Hejazi M., Hussain H., Kaplan F., Guy C., Smith S.M., Steup M., Ritte G. (2007). Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial beta-amylases. Plant Physiol. 145: 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordham-Skelton A.P., Chilley P., Lumbreras V., Reignoux S., Fenton T.R., Dahm C.C., Pages M., Gatehouse J.A. (2002). A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J. 29: 705–715. [DOI] [PubMed] [Google Scholar]
- French D., Summer R. (1956). Action of beta-amylase on branched oligosaccharides. J. Biol. Chem. 222: 469–477. [PubMed] [Google Scholar]
- Fulton D.C., et al. (2008). Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Hejazi M., Fettke J., Haebel S., Edner C., Paris O., Frohberg C., Steup M., Ritte G. (2008). Glucan, water dikinase phosphorylates crystalline maltodextrins and thereby initiates solubilization. Plant J. 55: 323–334. [DOI] [PubMed] [Google Scholar]
- Hejazi M., Fettke J., Paris O., Steup M. (2009). The two plastidial starch-related dikinases sequentially phosphorylate glucosyl residues at the surface of both the A- and B-type allomorphs of crystallized maltodextrins but the mode of action differs. Plant Physiol. 150: 962–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejazi M., Fettke J., Kötting O., Zeeman S.C., Steup M. (2010). The Laforin-like dual-specificity phosphatase SEX4 from Arabidopsis hydrolyzes both C6- and C3-phosphate esters introduced by starch-related dikinases and thereby affects phase transition of alpha-glucans. Plant Physiol. 152: 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A., Grünig K., Alvarez-Huerta M., Infanger S., Bauer J., Kessler F. (2004). AtToc90, a new GTP-binding component of the Arabidopsis chloroplast protein import machinery. Plant Mol. Biol. 54: 427–440. [DOI] [PubMed] [Google Scholar]
- Horrer D., Flütsch S., Pazmino D., Matthews J.S.A., Thalmann M., Nigro A., Leonhardt N., Lawson T., Santelia D. (2016). Blue light induces a distinct starch degradation pathway in guard cells for stomatal opening. Curr. Biol. 26: 362–370. [DOI] [PubMed] [Google Scholar]
- Hussain H., Mant A., Seale R., Zeeman S., Hinchliffe E., Edwards A., Hylton C., Bornemann S., Smith A.M., Martin C., Bustos R. (2003). Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. Plant Cell 15: 133–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeleń F., Oleksy A., Śmietana K., Otlewski J. (2003). PDZ domains—common players in the cell signaling. Acta Biochim. Pol. 50: 985–1017. [PubMed] [Google Scholar]
- Kaneko T., Joshi R., Feller S.M., Li S.S.C. (2012). Phosphotyrosine recognition domains: The typical, the atypical and the versatile. Cell Commun. Signal. 10: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F., Guy C.L. (2004). beta-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 135: 1674–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F., Guy C.L. (2005). RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J. 44: 730–743. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kerk D., Bulgrien J., Smith D.W., Barsam B., Veretnik S., Gribskov M. (2002). The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 129: 908–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk D., Conley T.R., Rodriguez F.A., Tran H.T., Nimick M., Muench D.G., Moorhead G.B.G. (2006). A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J. 46: 400–413. [DOI] [PubMed] [Google Scholar]
- Kerk D., Templeton G., Moorhead G.B.G. (2008). Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 146: 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O., Pusch K., Tiessen A., Geigenberger P., Steup M., Ritte G. (2005). Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 137: 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O., Santelia D., Edner C., Eicke S., Marthaler T., Gentry M.S., Comparot-Moss S., Chen J., Smith A.M., Steup M., Ritte G., Zeeman S.C. (2009). STARCH-EXCESS4 is a laforin-like Phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell 21: 334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O., Kossmann J., Zeeman S.C., Lloyd J.R. (2010). Regulation of starch metabolism: The age of enlightenment? Curr. Opin. Plant Biol. 13: 321–329. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Zheng J.J. (2010). PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun. Signal. 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Francisco P., Zhou W., Edner C., Steup M., Ritte G., Bond C.S., Smith S.M. (2009). Catalytically-inactive β-amylase BAM4 required for starch breakdown in Arabidopsis leaves is a starch-binding-protein. Arch. Biochem. Biophys. 489: 92–98. [DOI] [PubMed] [Google Scholar]
- Li L., Nelson C.J., Trösch J., Castleden I., Huang S., Millar A.H. (2017). Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell 29: 207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Urano K., Yoshiwara K., Morishita Y., Sakurai N., Suzuki H., Kojima M., Sakakibara H., Shibata D., Saito K., Shinozaki K., Yamaguchi-Shinozaki K. (2014). Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 164: 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A., Ghilagaber S., Nikolaev A., Hardie D.G. (2009). The glycogen-binding domain on the AMPK beta subunit allows the kinase to act as a glycogen sensor. Cell Metab. 9: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekins D.A., Guo H.-F., Husodo S., Paasch B.C., Bridges T.M., Santelia D., Kötting O., Vander Kooi C.W., Gentry M.S. (2013). Structure of the Arabidopsis glucan phosphatase like sex four2 reveals a unique mechanism for starch dephosphorylation. Plant Cell 25: 2302–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekins D.A., Raththagala M., Husodo S., White C.J., Guo H.-F., Kötting O., Vander Kooi C.W., Gentry M.S. (2014). Phosphoglucan-bound structure of starch phosphatase Starch Excess4 reveals the mechanism for C6 specificity. Proc. Natl. Acad. Sci. USA 111: 7272–7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekins D.A., Raththagala M., Auger K.D., Turner B.D., Santelia D., Kötting O., Gentry M.S., Vander Kooi C.W. (2015). Mechanistic insights into glucan phosphatase activity against polyglucan substrates. J. Biol. Chem. 290: 23361–23370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake H., Kurisu G., Kusunoki M., Nishimura S., Kitamura S., Nitta Y. (2003). Crystal structure of a catalytic site mutant of beta-amylase from Bacillus cereus var. mycoides cocrystallized with maltopentaose. Biochemistry 42: 5574–5581. [DOI] [PubMed] [Google Scholar]
- Monroe J.D., Storm A.R., Badley E.M., Lehman M.D., Platt S.M., Saunders L.K., Schmitz J.M., Torres C.E. (2014). β-Amylase1 and β-amylase3 are plastidic starch hydrolases in Arabidopsis that seem to be adapted for different thermal, pH, and stress conditions. Plant Physiol. 166: 1748–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T., Comparot-Moss S., Lue W.L., Messerli G., Trevisan M., Seymour M.D.J., Gatehouse J.A., Villadsen D., Smith S.M., Chen J., Zeeman S.C., Smith A.M. (2006). Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J. Biol. Chem. 281: 11815–11818. [DOI] [PubMed] [Google Scholar]
- Pfister B., Zeeman S.C. (2016). Formation of starch in plant cells. Cell. Mol. Life Sci. 73: 2781–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P. (1997). Evidence for PDZ domains in bacteria, yeast, and plants. Protein Sci. 6: 464–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold H., Soyk S., Simková K., Hostettler C., Marafino J., Mainiero S., Vaughan C.K., Monroe J.D., Zeeman S.C. (2011). β-amylase-like proteins function as transcription factors in Arabidopsis, controlling shoot growth and development. Plant Cell 23: 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G., Scharf A., Eckermann N., Haebel S., Steup M. (2004). Phosphorylation of transitory starch is increased during degradation. Plant Physiol. 135: 2068–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G., Heydenreich M., Mahlow S., Haebel S., Kötting O., Steup M. (2006). Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 580: 4872–4876. [DOI] [PubMed] [Google Scholar]
- Rubio V., Shen Y., Saijo Y., Liu Y., Gusmaroli G., Dinesh-Kumar S.P., Deng X.W. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41: 767–778. [DOI] [PubMed] [Google Scholar]
- Santelia D., Kötting O., Seung D., Schubert M., Thalmann M., Bischof S., Meekins D.A., Lutz A., Patron N., Gentry M.S., Allain F.H.-T., Zeeman S.C. (2011). The phosphoglucan phosphatase like sex Four2 dephosphorylates starch at the C3-position in Arabidopsis. Plant Cell 23: 4096–4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidig A., Fröhlich A., Schulze S., Lloyd J.R., Kossmann J. (2002). Downregulation of a chloroplast-targeted β-amylase leads to a starch-excess phenotype in leaves. Plant J. 30: 581–591. [DOI] [PubMed] [Google Scholar]
- Schreier T.B., Cléry A., Schläfli M., Galbier F., Stadler M., Demarsy E., Albertini D., Maier B.A., Kessler F., Hörtensteiner S., Zeeman S.C., Kötting O. (2018). Plastidial NAD-dependent malate dehydrogenase: A moonlighting protein involved in early chloroplast development through its interaction with an FtsH12-FtsHi protease complex. Plant Cell 30: 1745–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialdone A., Mugford S.T., Feike D., Skeffington A., Borrill P., Graf A., Smith A.M., Howard M. (2013). Arabidopsis plants perform arithmetic division to prevent starvation at night. eLife 2: e00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinski J., König N., Wellmeyer B., Hanke G.T., Linke V., Neuhaus H.E., Scheibe R. (2014). The plastid-localized NAD-dependent malate dehydrogenase is crucial for energy homeostasis in developing Arabidopsis thaliana seeds. Mol. Plant 7: 170–186. [DOI] [PubMed] [Google Scholar]
- Seung D., Thalmann M., Sparla F., Abou Hachem M., Lee S.K., Issakidis-Bourguet E., Svensson B., Zeeman S.C., Santelia D. (2013). Arabidopsis thaliana AMY3 is a unique redox-regulated chloroplastic α-amylase. J. Biol. Chem. 288: 33620–33633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D., Soyk S., Coiro M., Maier B.A., Eicke S., Zeeman S.C. (2015). PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol. 13: e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D., Boudet J., Monroe J., Schreier T.B., David L.C., Abt M., Lu K.J., Zanella M., Zeeman S.C. (2017). Homologs of PROTEIN TARGETING TO STARCH Control Starch Granule Initiation in Arabidopsis Leaves. Plant Cell 29: 1657–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Jensen O.N., Podtelejnikov A.V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Shevchenko A., Boucherie H., Mann M. (1996). Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 93: 14440–14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Schaft D., Roguev A., Pijnappel W.W.M.P., Stewart A.F., Shevchenko A. (2002). Deciphering protein complexes and protein interaction networks by tandem affinity purification and mass spectrometry: Analytical perspective. Mol. Cell. Proteomics 1: 204–212. [DOI] [PubMed] [Google Scholar]
- Silver D.M., Silva L.P., Issakidis-Bourguet E., Glaring M.A., Schriemer D.C., Moorhead G.B.G. (2013). Insight into the redox regulation of the phosphoglucan phosphatase SEX4 involved in starch degradation. FEBS J. 280: 538–548. [DOI] [PubMed] [Google Scholar]
- Silver D.M., Kötting O., Moorhead G.B.G. (2014). Phosphoglucan phosphatase function sheds light on starch degradation. Trends Plant Sci. 19: 471–478. [DOI] [PubMed] [Google Scholar]
- Smith A.M., Zeeman S.C. (2006). Quantification of starch in plant tissues. Nat. Protoc. 1: 1342–1345. [DOI] [PubMed] [Google Scholar]
- Solhaug E.M., Roy R., Chatt E.C., Klinkenberg P.M., Mohd-Fadzil N.A., Hampton M., Nikolau B.J., Carter C.J. (2019). An integrated transcriptomics and metabolomics analysis of the Cucurbita pepo nectary implicates key modules of primary metabolism involved in nectar synthesis and secretion. Plant Direct 3: e00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyk S., Simková K., Zürcher E., Luginbühl L., Brand L.H., Vaughan C.K., Wanke D., Zeeman S.C. (2014). The enzyme-like domain of Arabidopsis nuclear β-amylases is critical for DNA sequence recognition and transcriptional activation. Plant Cell 26: 1746–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla F., Costa A., Lo Schiavo F., Pupillo P., Trost P. (2006). Redox regulation of a novel plastid-targeted β-amylase of Arabidopsis. Plant Physiol. 141: 840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolpe T., Süsslin C., Marrocco K., Nick P., Kretsch T., Kircher S. (2005). In planta analysis of protein-protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma 226: 137–146. [DOI] [PubMed] [Google Scholar]
- Storm A.R., Kohler M.R., Berndsen C.E., Monroe J.D. (2018). Glutathionylation inhibits the catalytic activity of Arabidopsis β-amylase3 but not that of paralog β-amylase1. Biochemistry 57: 711–721. [DOI] [PubMed] [Google Scholar]
- Streb S., Eicke S., Zeeman S.C. (2012). The simultaneous abolition of three starch hydrolases blocks transient starch breakdown in Arabidopsis. J. Biol. Chem. 287: 41745–41756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y., Senoura T., Yoshizaki T., Hamada S., Ito H., Matsui H. (2007). Differential chain-length specificities of two isoamylase-type starch-debranching enzymes from developing seeds of kidney bean. Biosci. Biotechnol. Biochem. 71: 2308–2312. [DOI] [PubMed] [Google Scholar]
- Takeda Y., Hizukuri S. (1981). Re-examination of the action of sweet-potato beta-amylase on phosphorylated (1→4)-χ-d-glucan. Carbohydr. Res. 89: 174–178. [Google Scholar]
- Thalmann M., Pazmino D., Seung D., Horrer D., Nigro A., Meier T., Kölling K., Pfeifhofer H.W., Zeeman S.C., Santelia D. (2016). Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 28: 1860–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp M.-A., Iki T., Brosnan C.A., Jullien P.E., Pumplin N. (2017). A complex of Arabidopsis DRB proteins can impair dsRNA processing. RNA 23: 782–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio C., Costa A., Marri L., Issakidis-Bourguet E., Pupillo P., Trost P., Sparla F. (2011). Thioredoxin-regulated β-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. J. Exp. Bot. 62: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Kooi C.W., Taylor A.O., Pace R.M., Meekins D.A., Guo H.-F., Kim Y., Gentry M.S. (2010). Structural basis for the glucan phosphatase activity of Starch Excess4. Proc. Natl. Acad. Sci. USA 107: 15379–15384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. (2009). Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattebled F., Dong Y., Dumez S., Delvallé D., Planchot V., Berbezy P., Vyas D., Colonna P., Chatterjee M., Ball S., D’Hulst C. (2005). Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol. 138: 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise S.E., Weber A.P.M., Sharkey T.D. (2004). Maltose is the major form of carbon exported from the chloroplast at night. Planta 218: 474–482. [DOI] [PubMed] [Google Scholar]
- Weise S.E., Kim K.S., Stewart R.P., Sharkey T.D. (2005). beta-Maltose is the metabolically active anomer of maltose during transitory starch degradation. Plant Physiol. 137: 756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Gloria C., Johnson J.J., Marritt K., Kataya A., Vahab A., Moorhead G.B. (2018). Protein kinases and phosphatases of the plastid and their potential role in starch metabolism. Front. Plant Sci. 9: 1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte C.P., Noël L.D., Gielbert J., Parker J.E., Romeis T. (2004). Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol. Biol. 55: 135–147. [DOI] [PubMed] [Google Scholar]
- Zanella M., Borghi G.L., Pirone C., Thalmann M., Pazmino D., Costa A., Santelia D., Trost P., Sparla F. (2016). β-amylase 1 (BAM1) degrades transitory starch to sustain proline biosynthesis during drought stress. J. Exp. Bot. 67: 1819–1826. [DOI] [PubMed] [Google Scholar]
- Zeeman S.C., Northrop F., Smith A.M., Rees T. (1998). A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J. 15: 357–365. [DOI] [PubMed] [Google Scholar]
- Zeeman S.C., Kossmann J., Smith A.M. (2010). Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61: 209–234. [DOI] [PubMed] [Google Scholar]
- Zhang Z.Y., Dixon J.E. (1993). Active site labeling of the Yersinia protein tyrosine phosphatase: The determination of the pKa of the active site cysteine and the function of the conserved histidine 402. Biochemistry 32: 9340–9345. [DOI] [PubMed] [Google Scholar]