This study addresses the microbiology of a natural ecosystem (the infant bowel) for children in a rural setting in Indonesia and in an urban environment in New Zealand. Analysis of DNA sequences generated from the microbial community (microbiota) in the feces of the infants during the first year of life showed marked differences in the composition and complexity of the bacterial collections. The differences were most likely due to differences in the prevalence and duration of breastfeeding of infants in the two countries. These kinds of studies are essential for developing concepts of microbial ecology related to the influence of nutrition and environment on the development of the gut microbiota and for determining the long-term effects of microbiological events in early life on human health and well-being.
KEYWORDS: bifidobacteria, gut, infants, microbiota
ABSTRACT
The biological succession that occurs during the first year of life in the gut of infants in Western countries is broadly predictable in terms of the increasing complexity of the composition of microbiotas. Less information is available about microbiotas in Asian countries, where environmental, nutritional, and cultural influences may differentially affect the composition and development of the microbial community. We compared the fecal microbiotas of Indonesian (n = 204) and New Zealand (NZ) (n = 74) infants 6 to 7 months and 12 months of age. Comparisons were made by analysis of 16S rRNA gene sequences and derivation of community diversity metrics, relative abundances of bacterial families, enterotypes, and cooccurrence correlation networks. Abundances of Bifidobacterium longum subsp. infantis and B. longum subsp. longum were determined by quantitative PCR. All observations supported the view that the Indonesian and NZ infant microbiotas developed in complexity over time, but the changes were much greater for NZ infants. B. longum subsp. infantis dominated the microbiotas of Indonesian children, whereas B. longum subsp. longum was dominant in NZ children. Network analysis showed that the niche model (in which trophic adaptation results in preferential colonization) of the assemblage of microbiotas was supported in Indonesian infants, whereas the neutral (stochastic) model was supported by the development of the microbiotas of NZ infants. The results of the study show that the development of the fecal microbiota is not the same for infants in all countries, and they point to the necessity of obtaining a better understanding of the factors that control the colonization of the gut in early life.
IMPORTANCE This study addresses the microbiology of a natural ecosystem (the infant bowel) for children in a rural setting in Indonesia and in an urban environment in New Zealand. Analysis of DNA sequences generated from the microbial community (microbiota) in the feces of the infants during the first year of life showed marked differences in the composition and complexity of the bacterial collections. The differences were most likely due to differences in the prevalence and duration of breastfeeding of infants in the two countries. These kinds of studies are essential for developing concepts of microbial ecology related to the influence of nutrition and environment on the development of the gut microbiota and for determining the long-term effects of microbiological events in early life on human health and well-being.
INTRODUCTION
The transition of an infant from a mainly milk diet to the introduction of solid foods (complementary feeding period) is accompanied by transformation of the composition of the gut microbiota (1–8). In Western countries, it is recommended that complementary feeding start at around 6 months of age and that infants eat mainly “family” foods by 1 year (9–11). In addition to diet, environmental (water, sanitation, hygiene, and air pollution) and lifestyle factors differ among countries and their populations. These differences may influence the gut microbiota during its development toward a microbial community more similar to that of adults (12). In general, in studies of Western infants, members of the genus Bifidobacterium that utilize lactose (and human milk oligosaccharides [HMOs] for some species) for growth are abundant in the fecal microbiota of exclusively milk-fed infants. Bacterial species capable of degrading and fermenting plant polysaccharides and their components (Bacteroidaceae, Lachnospiraceae, and Ruminococcaceae) are dominant by the end of the complementary feeding period (1–8).
Knowledge of the development of the gut microbiota in early life is important because deviations from the norm (dysbiosis) may be associated with adverse conditions later in life (13–15). Defining the normal situation for infants in different countries thus has value as concepts of the involvement of the gut microbiota in the etiology of specific diseases and conditions develop (16).
Descriptions of the fecal microbiota commonly rely on culture-independent analyses that use 16S rRNA gene sequences amplified by PCR from bulk DNA extracted from feces. Analysis of these sequences confidently provides taxonomic information about the composition of the microbiota with respect to bacterial families and genera and sometimes species (17). However, differentiation of some species, as in the case of some Bifidobacterium taxa, is difficult using 16S rRNA gene sequences. This difficulty results, in particular, in a paucity of reports of bifidobacterial subspecies distributions in human infants on a global scale (18–20). Knowledge of subspecies occurrence and abundance would be useful for developing concepts of multispecies ecology of bifidobacteria in the infant bowel during development of the microbiota. Fortunately, quantitative PCR (qPCR) provides a quick analytical method by which to accurately ascertain the abundances of bifidobacterial species and subspecies in the microbiota (21, 22).
The purpose of our study was to compare the compositions of fecal microbiotas for infants in Indonesia and New Zealand (NZ) during the complementary feeding period, using 16S rRNA gene sequences obtained by next-generation sequencing and qPCR targeting bifidobacterial species and subspecies. The Indonesian infants were from rural villages, whereas the NZ children were urban and predominantly NZ European (23–25). We found that the composition of the microbiota changed radically in NZ children during the first 12 months of life, whereas bifidobacteria, particularly Bifidobacterium longum subsp. infantis, remained at high levels in Indonesian infants during this time.
RESULTS
Comparison of the development of fecal microbiotas of Indonesian and NZ infants based on 16S rRNA gene sequences.
Alpha diversity metrics (observed amplicon sequence variants [ASVs], Faith’s phylogenetic diversity, Pielou’s evenness, and Shannon’s index) showed that the variety of bacteria in microbiotas in both cohorts increased between 6 to 7 months of age and 12 months of age, but they also indicated that community diversity was less in 12-month Indonesian microbiotas than in NZ microbiotas (observed ASVs, Pielou’s evenness, and Shannon’s index). Faith’s phylogenetic diversity showed that the microbiotas of infants at 6 to 7 months of age and 12 months of age were phylogenetically similar between cohorts, presumably due to the predominance of Bifidobacteriaceae in both groups (Fig. 1). Permutational multivariate analysis of variance (PERMANOVA) comparisons and a test for homogeneity of multivariate dispersion (PERMDISP), using four beta diversity metrics (unweighted UniFrac, weighted UniFrac, Jacard similarity matrix, and Bray-Curtis dissimilarity), supported the view that compositions of microbiotas differed between the cohorts at both 6 to 7 months and 12 months of age (Table 1; also see Fig. S1 in the supplemental material).
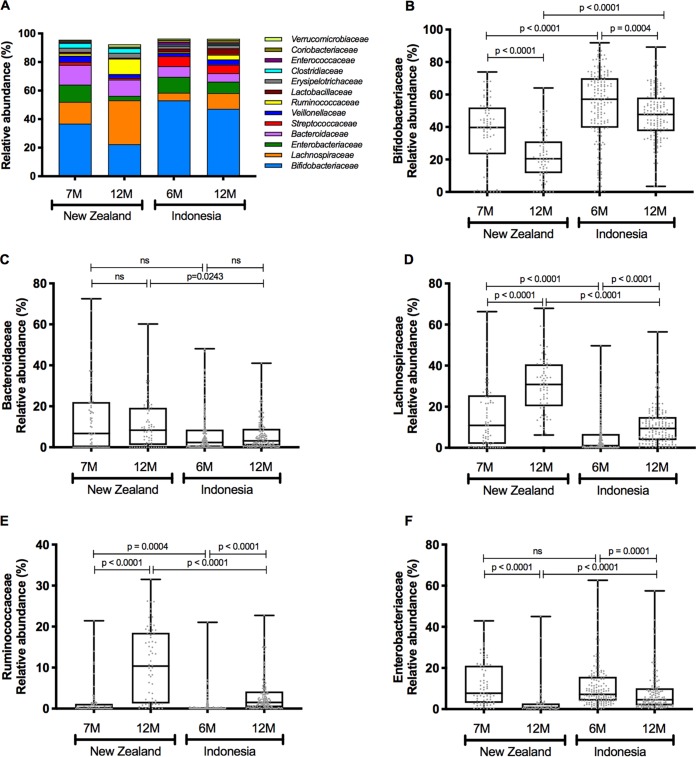
FIG 1.
Alpha diversity box plots (Tukey method; boxes are from the 25th percentile to the 75th percentile with a line at the median, whiskers are 1.5 times the interquartile range, and outliers are shown as points), with samples grouped by cohort and time point. Metrics applied are observed ASVs (A), Faith’s phylogenetic diversity (PD) (B), Pielou’s evenness (C), and the Shannon index (D). Comparisons between cohorts at 6 to 7 months and 12 months and within cohorts, comparing 6 to 7 months and 12 months, performed with the Mann-Whitney test, are shown for each metric. ns, not significant.
TABLE 1.
PERMANOVA and PERMDISP analyses comparing cohort and time groups with four beta diversity metrics
| Metric | Group 1 | Group 2 | No. of samples | PERMANOVA |
PERMDISP |
||||
|---|---|---|---|---|---|---|---|---|---|
| Pseudo-F | P | q | F | P | q | ||||
| Unweighted UniFrac | NZ, 12 mo | Indonesia, 12 mo | 209 | 16.716 | 0.001 | 0.001 | 0.017 | 0.882 | 0.938 |
| Unweighted UniFrac | NZ, 7 mo | Indonesia, 6 mo | 231 | 9.828 | 0.001 | 0.001 | 3.501 | 0.061 | 0.076 |
| Weighted UniFrac | NZ, 12 mo | Indonesia, 12 mo | 209 | 57.53 | 0.001 | 0.0011 | 27.935 | 0.001 | 0.002 |
| Weighted UniFrac | NZ, 7 mo | Indonesia, 6 mo | 231 | 17.015 | 0.001 | 0.0011 | 13.172 | 0.004 | 0.007 |
| Jaccard | NZ, 12 mo | Indonesia, 12 mo | 209 | 13.234 | 0.001 | 0.001 | 3.276 | 0.104 | 0.208 |
| Jaccard | NZ, 7 mo | Indonesia, 6 mo | 231 | 9.81 | 0.001 | 0.001 | 0.268 | 0.635 | 0.706 |
| Bray-Curtis | NZ, 12 mo | Indonesia, 12 mo | 209 | 35.777 | 0.001 | 0.0011 | 80.536 | 0.001 | 0.002 |
| Bray-Curtis | NZ, 7 mo | Indonesia, 6 mo | 231 | 13.568 | 0.001 | 0.0011 | 22.331 | 0.001 | 0.002 |
To investigate differences in taxa constituting the infant microbiotas, the relative abundances of the 13 predominant bacterial families that constituted ∼95.5% of the microbiotas were compared. Bifidobacteriaceae declined in abundance in the microbiotas of NZ and Indonesian infants between 6 to 7 months of age and 12 months of age, whereas Lachnospiraceae and Ruminococcaceae increased in abundance (Fig. 2A to F). The development of the Lachnospiraceae population was especially marked in NZ infants, with concomitant decreases in Bifidobacteriaceae and Enterobacteriaceae abundances. Levels of Bacteroidaceae remained constant between cohorts and times. Changes in Indonesian infant microbiotas were much more modest between 6 and 12 months of age.
FIG 2.
Relative abundances of the 13 predominant families, with samples grouped according to cohort and time (A), and box plots (Tukey method; boxes are from the 25th percentile to the 75th percentile with a line at the median, whiskers are 1.5 times the interquartile range, and outliers are shown as points) showing the relative abundances of Bifidobacteriaceae (B), Bacteroidaceae (C), Lachnospiraceae (D), Ruminococcaceae (E), and Enterobacteriaceae (F) in each cohort. Groups were compared using Mann-Whitney nonparametric tests. ns, not significant.
Taxonomic assignment based on the V4 region of 16S rRNA gene sequences does not allow discrimination between Bifidobacterium longum subspecies and Bifidobacterium breve. Nevertheless, the results showed that B. longum and B. breve were likely to be the most common bifidobacterial taxa in the microbiotas of both NZ and Indonesian infants (Fig. 3).
FIG 3.
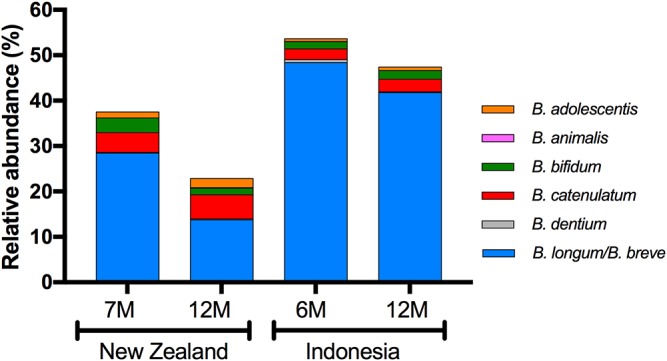
Relative abundances of bifidobacterial species, as measured by 16S rRNA gene amplicon sequencing, with samples grouped according to cohort and time. Where ASV sequences could not be assigned to a single species, all possible identities are named. Columns represent mean relative abundances.
Enterotypes detected in the fecal microbiotas of Indonesian and NZ infants.
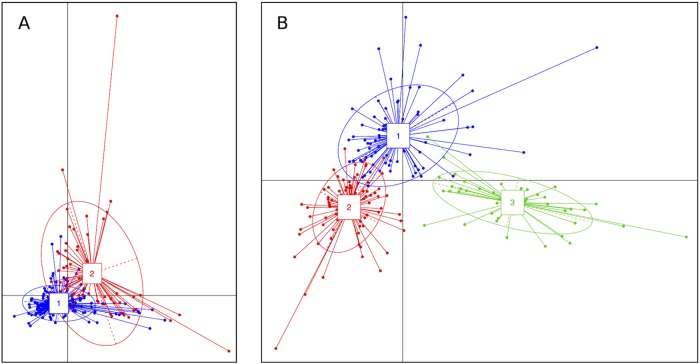
We used the Calinski-Harabasz index to search for robust clusters of taxa (enterotypes [26]) that characterized the microbiotas of the infants at 6 to 7 months of age and 12 months of age. It was predicted that two enterotypes would be optimally present in 6- to 7-month microbiotas and three enterotypes at 12 months when data were amalgamated for the respective time points. Between-class analysis (BCA) allowed the visual clustering of the enterotypes to be shown (Fig. 4). Using Statistical Analysis of Taxonomic and Functional Profiles (STAMP) software (27), it was determined that, with clustering into two enterotypes at 6 to 7 months, samples were dominated by an enterotype defined by B. longum/B. breve regardless of the country of origin. A second enterotype, defined by Ruminococcus gnavus, Bacteroides fragilis, and Clostridium ramosum, was observed with greater prevalence in the NZ cohort (Fig. 5A).
FIG 4.
BCA plots, showing clustering of samples from combined cohort data into two enterotypes at 6 to 7 months of age (A) and three enterotypes at 12 months of age (B).
FIG 5.
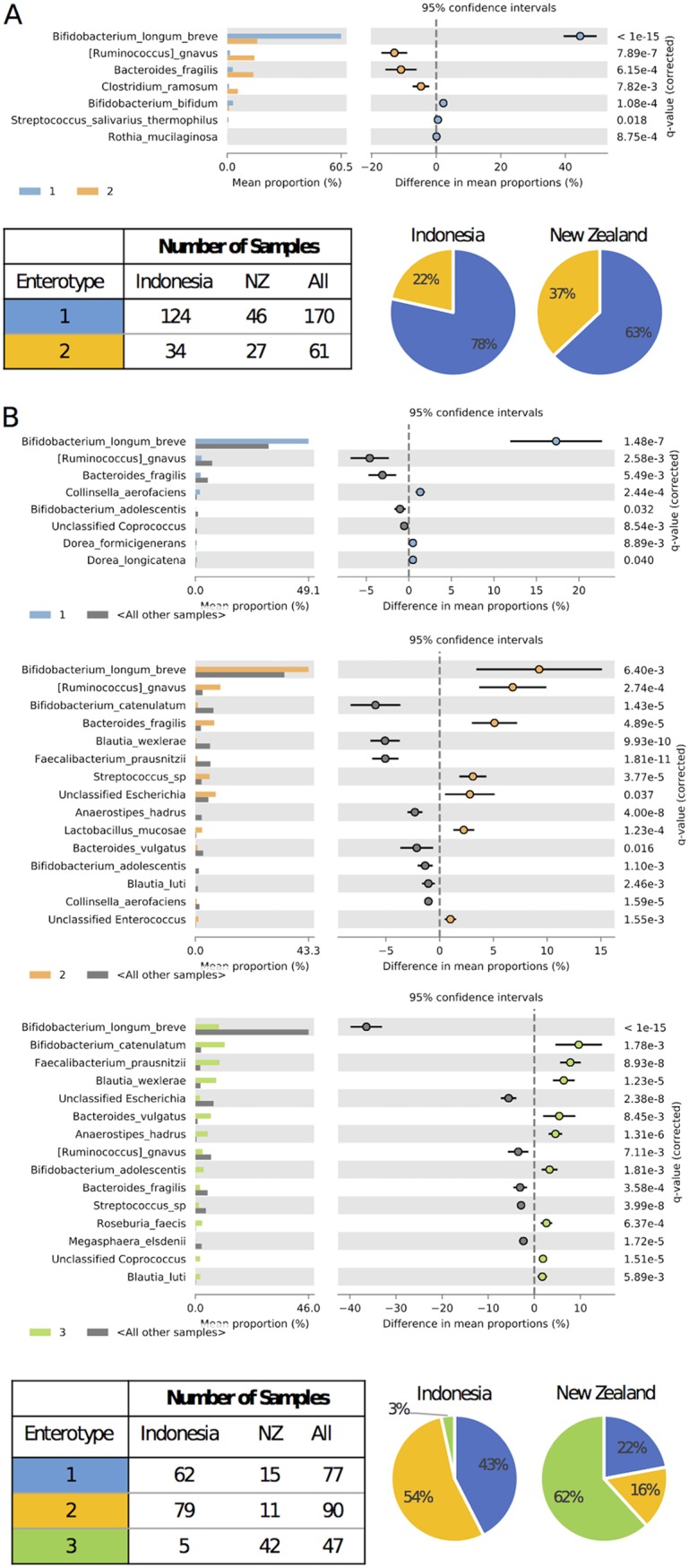
Description of enterotypes predicted for infants at 6 to 7 months of age (A) and 12 months of age (B). Each panel contains the STAMP output (extended error bar plot) depicting species-level features within each enterotype with significant differential abundance (Welch’s t test with Benjamini-Hochberg false discovery rate), compared to all other enterotypes, a table reporting the number of individuals from each cohort within each enterotype, and pie charts showing the proportion of individuals from each cohort associated with each enterotype.
Microbiotas at 12 months showed one enterotype dominated by B. longum/B. breve, a second enterotype defined by B. longum/B. breve, R. gnavus, and B. fragilis, and a third enterotype defined by Bifidobacterium catenulatum, Faecalibacterium prausnitzii, and Blautia wexlerae. In this case, the Indonesian cohort microbiotas were dominated by the B. longum/B. breve-containing enterotypes, whereas the NZ infant microbiotas were dominated by the third enterotype, defined by B. catenulatum, F. prausnitzii, and B. wexlerae (Fig. 5B).
In summary, both Indonesian and NZ microbiotas were characterized by B. longum/B. breve populations at 6 to 7 months. Indonesian microbiotas remained characterized by B. longum/B. breve populations at 12 months but the NZ microbiotas at this age were especially characterized by an enterotype in which these bacterial species did not predominate (Fig. 5A and B).
Cooccurrence networks in the fecal microbiotas of Indonesian and NZ infants.
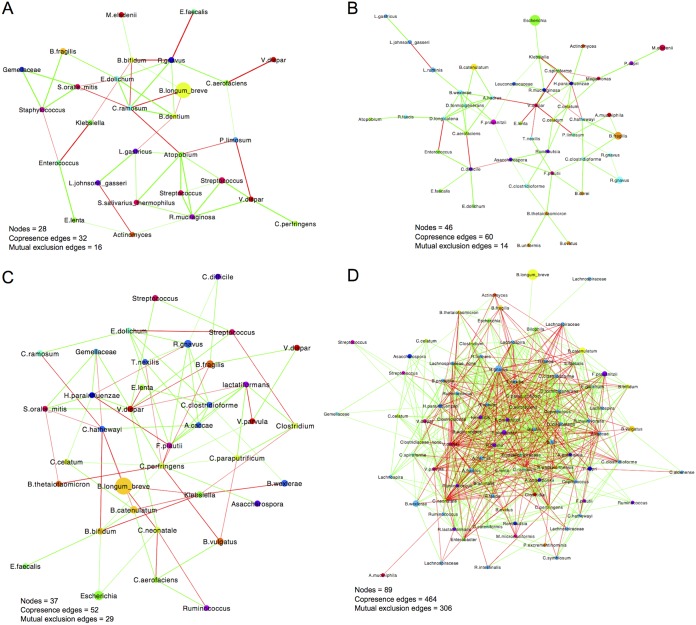
Construction of microbial (correlation) networks from sequencing data may facilitate understanding community structures. The networks show individual microbes (ASVs or features) as nodes (hub species) and species cooccurrence or mutual exclusion as feature-feature pairs (edges); an edge may imply a biological or biochemical relationship between features. Microbes that benefit each another may be positively correlated, whereas microbes that compete for the same niche may be negatively correlated (28, 29).
Correlation networks constructed from our data underlined the different development of microbiotas of Indonesian infants, relative to those of NZ infants. Indonesian networks displayed an increase in node numbers between 6 and 12 months of age (28 versus 46 nodes) and a 1.5-fold increase in edge numbers (48 versus 74 edges). There was also an increase in the proportion of positive correlations at 12 months (67% at 6 months and 81% at 12 months) (Fig. 6A and B). In contrast, the NZ 12-month network was much more complex, with a greater number of nodes being observed at 12 months than at 7 months (89 and 37 nodes, respectively). There was a 9.5-fold increase in edge numbers (81 edges at 6 months versus 770 edges at 12 months). The two time points showed similar proportions of positive correlations (64% at 7 months and 60% at 12 months). However, 40% of correlations were negative (mutual exclusion edges) at 12 months, in contrast to the Indonesian network, in which 19% were negative. In summary, Indonesian microbiotas had similar numbers of negative correlations between nodes at 6 and 12 months (16 and 14 mutual exclusion edges, respectively), whereas nodes in 12-month NZ microbiotas were more negatively correlated than those at 7 months (306 and 29 mutual exclusion edges, respectively).
FIG 6.
CoNet-derived networks for Indonesian infants at 6 months of age (A), Indonesian infants at 12 months of age (B), NZ infants at 7 months of age (C), and NZ infants at 12 months of age (D). Cooccurrence edges (positive correlations) are shown in green, while mutual exclusion edges (negative correlations) are shown in red. Edge weight is determined by the q value assigned to the relationship between nodes. Nodes are colored by family and weighted by abundance.
Bifidobacterial subspecies abundances measured by qPCR in the fecal microbiotas of Indonesian and NZ infants.
We determined the abundances of B. breve, B. bifidum, B. longum subsp. infantis, and B. longum subsp. longum because they are common members of fecal microbiotas in early life (18, 30–37). They have distinctly different biochemical capacities with regard to the utilization of HMOs and other carbohydrates (22, 38, 39). Differentiation of the B. longum subspecies and B. longum/B. breve is difficult with standard 16S RNA gene sequence comparisons, necessitating the use of a qPCR assay (21, 22) to determine their proportions in the microbiota. While the four taxa that we enumerated were present in all microbiotas, the Indonesian bifidobacterial populations at both 6 and 12 months were dominated by B. longum subsp. infantis. The NZ bifidobacterial populations at 7 and 12 months were composed mainly of B. longum subsp. longum. These striking results further differentiated the fecal microbiotas of Indonesian and NZ children (Fig. 7).
FIG 7.
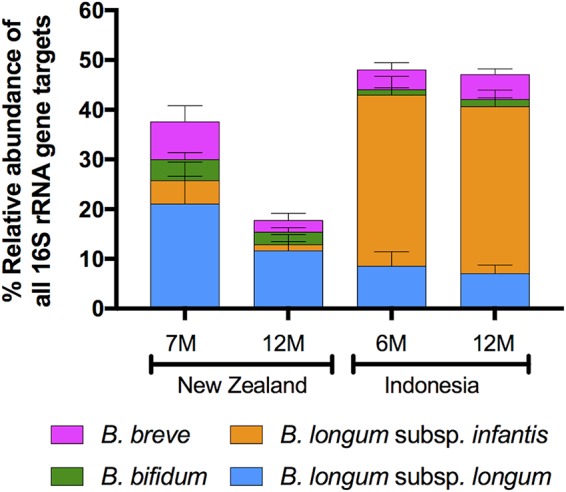
Relative abundance of four bifidobacterial species/subspecies measured by qPCR. Values are the mean percentage of total community 16S rRNA gene target. Error bars show 95% confidence intervals.
DISCUSSION
Comparison of the development of the fecal microbiota of Indonesian and NZ infants during the first year of life showed clear differences in the composition and complexity of the bacterial communities. Enterotypes dominated by B. longum/B. breve (in fact, B. longum subsp. infantis, as shown by qPCR) were common in Indonesian infants at both 6 and 12 months of age, whereas a third enterotype was common in the NZ infant microbiotas at 12 months. This enterotype was not dominated by B. longum/B. breve.
The processes that underpin the assembly of communities in natural systems are of much interest to ecologists. Two contrasting models to explain the development of diversity in communities have been proposed, namely, the niche selection and neutral theories of ecology (40–43). The dynamics of the niche selection model, it is proposed, are dominated by resource selection, with species of particular trophic adaptations being preferential colonizers of the habitat. Sharing of the habitat’s resources might result in interactions of mutual benefit that would result in the development of a stable community (44–46). In contrast, the dynamics of the neutral model are stochastic, implying that species that assemble are functionally equivalent; therefore, competition will be a feature of community assembly.
The gut microbiotas of Indonesian children favor the niche model, because cooccurrence networks at 6 and 12 months have similar numbers of nodes and most of the correlations between nodes are positive (copresence edges). These findings suggest a relatively stable environment with direct reliance on environmental resources by bacteria with special trophic adaptations that permit cohabitation (for example, differential utilization of HMOs for growth [22, 38, 44]). In contrast, NZ microbiotas at 12 months were competitive (large proportions of mutual exclusion edges), with much more complexity (more nodes). These findings suggest that acquisition of bacterial species was more random (neutral theory of assemblage) and involved an assortment of competing bacteria with similar trophic requirements (for example, utilization of dietary fiber).
Niche selection with HMOs is highly likely to explain the situation in Indonesian infants. HMOs and their components are used by B. longum subsp. infantis, following transport into the cell, for growth. B. longum subsp. infantis has a genome that contains a large gene cluster that is specifically involved in HMO utilization (39). In contrast, B. longum subsp. longum has limited capacity to utilize HMO components (22). The extent of breast milk feeding is different in Indonesia and NZ. While initial breast milk feeding uptake is high in both countries (∼100%), rates fall to national averages of 65% of infants by 6 months after birth in NZ and 36% by 12 months of age, whereas rates remain high (national average of 84% at 12 months of age) in Indonesia (47, 48). Cohort-specific values were greater for both groups in our study, but breast milk feeding was still much more common among Indonesian infants 12 months of age (NZ, 64%; Indonesia, 98%). Breastfeeding after 6 months of age refers to continued breastfeeding and not exclusive breastfeeding, rates of which are substantially lower by 6 months of age in both countries because complementary feeding needs to start by this age.
The composition of breast milk has been reported to differ between mothers in different countries and even between mothers in different locations in the same country (49–55). Comparisons of HMO contents and other biochemical features of Indonesian and NZ breast milks have not been made, which may be a topic worth investigating further. Additionally, volumes of milk consumed daily by infants in different settings would be useful information in considering the relationship between microbiotas and infant nutrition.
Evidence from studies of the fecal microbiotas of twins and of cohabiting adults and children indicate the potential importance of shared environments and similar diets in the assembly and maintenance of the microbiota of the human body (56–58). Shared environments provided by communal living, such as in rural Indonesia, might result in greater ease of dispersal (horizontal transmission) of gut commensals such as B. longum subsp. infantis among the infant population. Dispersal theory (59–62) has been advocated to explain the differing diversity, as well as greater similarity, of microbiotas in nonindustrialized populations (for example, rural Papua-New Guinea [12]), relative to Western microbiotas. Easier dispersal of gut microbes in a rural village setting might favor the acquisition of B. longum subsp. infantis, whereas Western methods of sanitation, water treatment, and hygiene might be inimical to this organism. Although B. longum subsp. infantis was detectable in the microbiotas of some NZ infants, it did not dominate the bacterial community, as was the case for Indonesian children. Thus, B. longum subsp. infantis is not an extinct lineage in NZ microbiotas but is most likely to have been influenced by lack of HMO enrichment in the infant gut due to breastfeeding practices. At least in the case of adults, studies of fecal microbiotas in industrialized countries have not shown major differences between nationalities (such as in Japan, Italy, Spain, Denmark, France, and the United States), indicating that genetic factors probably have only minor influences on the composition of microbiotas (26).
Overall, the transition from infant to more adult-like microbiotas during the first year of life was more pronounced in NZ infants than in Indonesian infants. Bifidobacteria are common in infants from both countries at 6 to 7 months of age. However, NZ infants have complex microbiotas, dominated by an enterotype different than that predominating in Indonesian infants at 12 months of age. The outcomes of the study show that the development of the fecal microbiota is not the same for infants in all countries and point to the necessity of obtaining a better understanding of the factors that control the colonization of the gut in early life. Such knowledge is important because early life influences due to the microbiota may have lifelong consequences; this concept was encapsulated by Rene Dubos and colleagues as “biological Freudianism” (63) and is supported by more recent studies of influences of the microbiota on infant health and well-being (64–66).
MATERIALS AND METHODS
Indonesian and NZ infants.
The members of the Indonesian cohort were from the Tanjungsari, Sukasari, and Pamulihan subdistricts of the Sumedang district, West Java, Indonesia. The majority of the infants were of Sundanese ethnicity. A cohort of breast-milk-fed infants was enrolled at 6 months of age after random selection from 30 villages in the three subdistricts, using local birth registry data. Metadata related to these infants and their parents have been published previously (24, 25). Fecal samples were obtained from 158 infants at 6 months of age and 147 infants at 12 months of age. Fecal samples for both time points were available for 101 individuals. Fifty-seven individuals provided samples at 6 months only, while 46 individuals provided samples at 12 months only.
The NZ cohort has been described previously (8) and comprised a subset of infants enrolled in the Baby-Led Introduction to SolidS (BLISS) randomized controlled trial. The design of this trial and metadata for the infants and mothers have been published previously (23). Fecal microbiota data from BLISS and non-BLISS infants were amalgamated because there were no differences in the relative abundances of the most abundant families (>1% relative abundance [8]). Fecal samples were obtained from these infants at 7 months of age (73 infants) and at 12 months of age (68 infants). Fecal samples for both time points were available for 67 individuals. Six individuals provided samples at 7 months only, while 1 individual provided a sample at 12 months only. Complementary feeding practices follow WHO recommendations in NZ and Indonesia (some solid foods are included in the diet from 6 months of age).
Ethics approval for these studies was obtained from the Human Ethics Committees of Padjadjaran University, Indonesia, and the University of Otago, New Zealand (Indonesian study), and from the Lower South Regional Ethics Committee of New Zealand and the University of Otago Human Ethics Committee (NZ study). Written informed consent to participate in the studies was given by the parents or primary guardians of the infants. Participants were free to withdraw from the study at any time.
Fecal DNA extraction, 16S rRNA gene sequencing, and sequence analysis.
DNA was extracted from 250 mg feces according to the kit protocol provided by the manufacturer (PowerSoil DNA isolation kit, product no. 12855-100; Mo Bio). Amplification of the 16S rRNA gene V4 region, library preparation, and sequencing were carried out at Argonne National Laboratories (University of Chicago), using paired-end reads (2 by 250 bp), on an Illumina MiSeq instrument. Initial quality control and read pairing was carried out using QIIME2 v2018.8 (17). Sequence error correction and generation of ASVs were achieved using DADA2 (67). Taxonomic classifications were made using the q2-feature-classifier plugin (68) and the SILVA v123 database (69). A summary of sequence outputs is available in Table S1 in the supplemental material.
The composition of microbiotas was described by using four alpha diversity measures, namely, the number of ASVs (a proxy for observed species), phylogenetic diversity, Pielou’s evenness, and the Shannon index. Therefore, two indices (observed species and phylogenetic diversity) described microbial richness alone (i.e., number of species), one index (Pielou’s evenness) described community evenness (i.e., the equality of distribution of the species’ frequencies), and one index (the Shannon index) described richness and evenness. The feature table was rarefied to the minimum sample count (10,500 sequences) for calculation of the alpha diversity measures. Relative abundance at the family level was calculated by collapsing the raw ASV feature table based on seven-level taxonomic strings obtained from the SILVA v123 database.
Beta diversity metrics (Bray-Curtis index, unweighted UniFrac, weighted UniFrac, and Jaccard distance) were applied by using the QIIME2 v2018.8 command line interface and the core-metrics-phylogenetic plugin, with a sampling depth of 10,500 sequences. Group significance for each metric was measured with PERMANOVA (70), and group dispersion was measured with PERMDISP (71).
Enterotypes were predicted in R by using the approach described by Arumugam et al. (26) and following the tutorial provided by EMBL (https://enterotype.embl.de/). Input tables were filtered to contain data from either the 6- to 7-month samples or the 12-months samples, in order to concentrate on enterotype clusters at distinct time points. Differential abundance testing to determine which species were driving enterotypes was carried out on the full ASV feature table, modified for input to STAMP (27) and filtered within STAMP to focus on time points. Each enterotype was compared to all other samples using Welch’s t test with the Benjamini-Hochberg false discovery rate multiple-test correction.
Bacterial interaction networks were generated by using the CoNet v1.1.1-beta plugin (72) and following recommendations provided in the CoNet tutorial (http://psbweb05.psb.ugent.be/conet/microbialnetworks/conet_new.php), including the following methods: Pearson correlation, Spearman correlation, Bray-Curtis dissimilarity, Kullback-Leibler dissimilarity, and mutual information. Networks were visualized using Cytoscape v3.6.1. Input feature tables were filtered to contain cohort- and time-specific samples, and features found in <25% of samples were removed.
Quantitative PCR of bifidobacterial species and subspecies.
A previously described method was used for 16S rRNA gene-based quantitative differentiation of B. longum subsp. longum and B. longum subsp. infantis (22). For quantitation of B. breve and Bifidobacterium bifidum, primers described by Matsuki et al. (73) were used at a final concentration of 400 nM. Primers for universal detection of 16S rRNA gene targets (74) were used at a final concentration of 300 nM. All reactions were carried out on a Life Technologies ViiA 7 real-time PCR system, in MicroAmp Fast Optical 384-well plates with optical adhesive film (Applied Biosystems, Carlsbad, CA), using 10-μl volumes containing 1× PerfeCTa SYBR FastMix (Quantabio, Beverly, MA, USA), primers, and approximately 0.2 ng of template DNA. The thermocycling profile consisted of initial activation of the polymerase at 95°C for 30 s, followed by 40 cycles of 95°C for 1 s and 62°C for 20 s. Fluorescence levels were measured after the 62°C annealing/extension step. A melting curve was generated to analyze product specificity. Genomic DNA from B. longum subsp. longum (ATCC 15707T), B. longum subsp. infantis (DSM 20088T), B. breve (ATCC 15700T), and B. bifidum (DSM 20456T) was used for control reactions and for generation of standard curves. The standard DNA was quantified spectrophotometrically using a NanoDrop 1000 spectrophotometer (Thermo Scientific) and was diluted in 5-fold steps from 5 × 106 to 3.2 × 102 genomes/reaction, calculated using target gene copies per genome obtained from genome sequence information (NCBI). All reactions were carried out in duplicate and were run twice on separate plates. No-template controls were included on each plate. Statistical analyses of data used GraphPad Prism 7 (GraphPad Software Inc., La Jolla, CA).
Accession number(s).
The DNA sequence data have been deposited in the NCBI database under BioProject accession number PRJNA419227 (for the NZ infants) and BioProject accession number PRJNA528344 (for the Indonesian infants).
Supplementary Material
ACKNOWLEDGMENTS
The Indonesian study was supported by a grant from Meat and Livestock Australia and the University of Otago; the BLISS study was supported by Lottery Health Research, Meat and Livestock Australia, the Karitane Products Society, Perpetual Trustees, the New Zealand Women’s Institute, and the University of Otago. Supplementary support was provided by the Riddet CORE. K.M.-G. was supported by a summer student scholarship provided by Microbiome Otago.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01105-19.
REFERENCES
- 1.Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, Gil A, Vieites JM, Norin E, Young D, Scott JA, Dore J, Edwards CA. 2011. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology 157:1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 2.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergstrom A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, Molgaard C, Michaelsen KF, Licht TR. 2014. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol 80:2889–2900. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Laursen MF, Andersen LBB, Michaelsen KF, Mølgaard C, Trolle E, Bahl MI, Licht TR. 2016. Infant gut microbiota development is driven by transition to family foods independent of maternal obesity. mSphere 1:e00069-15. doi: 10.1128/mSphere.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laursen MF, Bahl MI, Michaelsen KF, Licht TR. 2017. First foods and gut microbes. Front Microbiol 8:356. doi: 10.3389/fmicb.2017.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qasem W, Azad MB, Hossain Z, Azad E, Jorgensen S, Castillo San Juan S, Cai C, Khafipour E, Beta T, Roberts LJ, Friel J. 2017. Assessment of complementary feeding of Canadian infants: effects on microbiome & oxidative stress, a randomized controlled trial. BMC Pediatr 17:54. doi: 10.1186/s12887-017-0805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leong C, Haszard JJ, Lawley B, Otal A, Taylor RW, Szymlek-Gay EA, Fleming EA, Daniels L, Fangupo LJ, Tannock GW, Heath AM. 2018. Mediation analysis as a means of identifying dietary components that differentially affect the fecal microbiota of infants weaned by modified baby-led and traditional approaches. Appl Environ Microbiol 84:e00914-18. doi: 10.1128/AEM.00914-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.New Zealand Ministry of Health. 2008. Food and nutrition guidelines for healthy infants and toddlers (aged 0–2): a background paper, 4th ed New Zealand Ministry of Health, Wellington, New Zealand. [Google Scholar]
- 10.National Health and Medical Research Council. 2012. Infant feeding guidelines: information for health workers. National Health and Medical Research Council, Canberra, Australia. [Google Scholar]
- 11.Health Canada. 2014. Nutrition for healthy term infants: recommendations from 6 to 24 months. http://www.hc-sc.gc.ca/fn-an/nutrition/infant-nourisson/recom/recom-6-24-months-6-24-mois-eng.php. [DOI] [PubMed]
- 12.Martínez I, Stegen JC, Maldonado-Gómez MX, Eren AM, Siba PM, Greenhill AR, Walter J. 2015. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep 11:527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 13.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, CHILD Study Investigators, Mohn WW, Turvey SE, Finlay BB. 2015. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 14.Arrieta MC, Arévalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, Vaca M, Boutin RCT, Morien E, Jin M, Turvey SE, Walter J, Parfrey LW, Cooper PJ, Finlay B. 2018. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol 142:424–434.e10. doi: 10.1016/j.jaci.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker WA. 2017. The importance of appropriate initial bacterial colonization of the intestine in newborn, child, and adult health. Pediatr Res 82:387–395. doi: 10.1038/pr.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker WA, Lawley TD. 2013. Therapeutic modulation of intestinal dysbiosis. Pharmacol Res 69:75–86. doi: 10.1016/j.phrs.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet C, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope E, Da Silva R, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley G, Janssen S, Jarmusch AK, Jiang L, Kaehler B, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MG, Lee J, Ley R, Liu Y, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton J, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson M II, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CH, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. 2018. QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Preprints 6:e27295v2 10.7287/peerj.preprints.27295v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, Kubota H, Swinkels S, Sakai T, Oishi K, Kushiro A, Knol J. 2016. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 11:e0158498. doi: 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youn SY, Seo JM, Ji GE. 2007. Evaluation of the PCR method for identification of Bifidobacterium species. Lett Appl Microbiol 46:7–13. doi: 10.1111/j.1472-765X.2007.02263.x. [DOI] [PubMed] [Google Scholar]
- 20.Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB. 2014. Stool microbiota and vaccine responses of infants. Pediatrics 134:e362–e372. doi: 10.1542/peds.2013-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawley B, Munro K, Hughes A, Hodgkinson AJ, Prosser CG, Lowry D, Zhou SJ, Makrides M, Gibson RA, Lay C, Chew C, Lee PS, Wong KH, Tannock GW. 2017. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. PeerJ 5:e3375. doi: 10.7717/peerj.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawley B, Centanni M, Watanabe J, Sims I, Carnachan S, Broadbent R, Lee PS, Wong KH, Tannock GW. 2018. tuf gene sequence variation in Bifidobacterium longum subsp. infantis detected in the fecal microbiota of Chinese infants. Appl Environ Microbiol 84:e00336-18. doi: 10.1128/AEM.00336-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daniels L, Heath A-L, Williams SM, Cameron SL, Fleming EA, Taylor BJ, Wheeler BJ, Gibson RS, Taylor RW. 2015. Baby-Led Introduction to SolidS (BLISS) study: a randomised controlled trial of a baby-led approach to complementary feeding. BMC Pediatr 15:179. doi: 10.1186/s12887-015-0491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diana A, Haszard JJ, Purnamasari DM, Nurulazmi I, Luftimas DE, Rahmania S, Nugraha GI, Erhardt J, Gibson RS, Houghton L. 2017. Iron, zinc, vitamin A and selenium status in a cohort of Indonesian infants after adjusting for inflammation using several different approaches. Br J Nutr 118:830–839. doi: 10.1017/S0007114517002860. [DOI] [PubMed] [Google Scholar]
- 25.Diana A, Mallard SR, Haszard JJ, Purnamasari DM, Nurulazmi I, Herliani PD, Nugraha GI, Gibson RS, Houghton L. 2017. Consumption of fortified infant foods reduces dietary diversity but has a positive effect on subsequent growth in infants from Sumedang district, Indonesia. PLoS One 12:e0175952. doi: 10.1371/journal.pone.0175952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. 2014. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer E, Thiele I. 2018. From network analysis to functional metabolic modeling of the human gut microbiota. mSystems 3:e00209-17. doi: 10.1128/mSystems.00209-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Röttjers L, Faust K. 2018. From hairballs to hypotheses: biological insights from microbial networks. FEMS Microbiol Rev 42:761–780. doi: 10.1093/femsre/fuy030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biavati B, Castagnoli P, Crociani F, Trovatelli LD. 1984. Species of the genus Bifidobacterium in the feces of infants. Microbiologica 7:341–345. [PubMed] [Google Scholar]
- 31.Favier CF, Vaughan EE, De Vos WM, Akkermans A. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol 68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young SL, Simon MA, Baird MA, Tannock GW, Bibiloni R, Spencely K, Lane JM, Fitzharris P, Crane J, Town I, Addo-Yobo E, Murray CS, Woodcock A. 2004. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested from cord blood. Clin Diagn Lab Immunol 11:686–690. doi: 10.1128/CDLI.11.4.686-690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. 2010. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156:3329–3341. doi: 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- 34.Grönlund M-M, Grześkowiak Ł, Isolauri E, Salminen S. 2011. Influence of mother’s intestinal microbiota on gut colonization in the infant. Gut Microbes 2:227–233. doi: 10.4161/gmic.2.4.16799. [DOI] [PubMed] [Google Scholar]
- 35.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O’Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makino H, Kushiro A, Ishikawa E, Kubota H, Gawad A, Sakai T, Oishi K, Martin R, Ben-Amor K, Knol J, Tanaka R. 2013. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS One 8:e78331. doi: 10.1371/journal.pone.0078331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, Tett A, Segata N, van Sinderen D, Ventura M. 2015. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. 2014. Breast milk oligosaccharides: structure-function relationships in the neonate. Annu Rev Nutr 34:143–169. doi: 10.1146/annurev-nutr-071813-105721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A 105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fargione J, Brown CS, Tilman D. 2003. Community assembly and invasion: an experimental test of neutral versus niche processes. Proc Natl Acad Sci U S A 100:8916–8920. doi: 10.1073/pnas.1033107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tilman D. 2004. Niche tradeoffs, neutrality, and community structure: a stochastic theory of resource competition, invasion, and community assembly. Proc Natl Acad Sci U S A 101:10854–10861. doi: 10.1073/pnas.0403458101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraft NJB, Valencia R, Ackerly DD. 2008. Functional traits and niche-based tree community assembly in an Amazonian forest. Science 322:580–582. doi: 10.1126/science.1160662. [DOI] [PubMed] [Google Scholar]
- 43.Fisher CK, Mehta P. 2014. The transition between niche and neutral regimes in ecology. Proc Natl Acad Sci U S A 111:13111–13116. doi: 10.1073/pnas.1405637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Centanni M, Ferguson SA, Sims IM, Biswas A, Tannock GW. 2019. Bifidobacterium bifidum ATCC 15696 and Bifidobacterium breve 24b metabolic interaction based on 2ʹ-O-fucosyl-lactose studied in steady-state cultures in a Freter-style chemostat. Appl Environ Microbiol 85:e02783-18. doi: 10.1128/AEM.02783-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tannock GW, Wilson CM, Loach D, Cook GM, Eason J, O'Toole PW, Holtrop G, Lawley B. 2012. Resource partitioning in relation to cohabitation of Lactobacillus species in the mouse forestomach. ISME J 6:927–938. doi: 10.1038/ismej.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tannock GW. 2017. Understanding the gut microbiota. John Wiley and Sons, Hoboken, NJ. [Google Scholar]
- 47.Null C, Beatty A, Ingwersen N, Leith W, Borkum E, Brecher-Haimson J, Gage A, Peckarsky M, Rangarajan A. 2016. MCC Indonesia Nutrition Project impact evaluation baseline report. Mathematica Policy Research, Princeton, NJ: https://www.mathematica-mpr.com/our-publications-and-findings/publications/mcc-indonesia-nutrition-project-impact-evaluation-baseline-report. [Google Scholar]
- 48.Castro T, Grant C, Wall C, Welch M, Marks E, Fleming C, Teixeira J, Bandara D, Berry S, Morton S. 2017. Breastfeeding indicators among a nationally representative multi-ethnic sample of New Zealand children. N Z Med J 130:34–44. [PubMed] [Google Scholar]
- 49.Castanys-Muñoz E, Martin MJ, Prieto PA. 2013. 2ʹ-Fucosyllactose: an abundant, genetically determined soluble glycan present in human milk. Nutr Rev 71:773–789. doi: 10.1111/nure.12079. [DOI] [PubMed] [Google Scholar]
- 50.Marx C, Bridge R, Wolf AK, Rich W, Kim JH, Bode L. 2014. Human milk oligosaccharide composition differs between donor milk and mother’s own milk in the NICU. J Hum Lact 30:54–61. doi: 10.1177/0890334413513923. [DOI] [PubMed] [Google Scholar]
- 51.McGuire MK, Meehan CL, McGuire MA, Williams JE, Foster J, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SE, Prentice AM, Kvist LJ, Otoo GE, Brooker SL, Price WJ, Shafii B, Placek C, Lackey KA, Robertson B, Manzano S, Ruíz L, Rodríguez JM, Pareja RG, Bode L. 2017. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr 105:1086–1100. doi: 10.3945/ajcn.116.139980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Autran CA, Kellman BP, Kim JH, Asztalos E, Blood AB, Spence ECH, Patel AL, Hou J, Lewis NE, Bode L. 2018. Human milk oligosaccharide composition predicts risk of necrotising enterocolitis in preterm infants. Gut 67:1064–1070. doi: 10.1136/gutjnl-2016-312819. [DOI] [PubMed] [Google Scholar]
- 53.Gay MCL, Koleva PT, Slupsky CM, Toit ED, Eggesbo M, Johnson CC, Wegienka G, Shimojo N, Campbell DE, Prescott SL, Munblit D, Geddes DT, Kozyrskyj AL. 2018. Worldwide variation in human milk metabolome: indicators of breast physiology and maternal lifestyle? Nutrients 10:1151. doi: 10.3390/nu10091151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gómez-Gallego C, Morales JM, Monleón D, Du Toit E, Kumar H, Linderborg KM, Zhang Y, Yang B, Isolauri E, Salminen S, Collado MC. 2018. Human breast milk NMR metabolomic profile across specific geographical locations and its association with the milk microbiota. Nutrients 10:1355. doi: 10.3390/nu10101355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Leeuwen SS, Stoutjesdijk E, Ten Kate GA, Schaafsma A, Dijck-Brouwer J, Muskiet FAJ, Dijkhuizen L. 2018. Regional variations in human milk oligosaccharides in Vietnam suggest FucTx activity besides FucT2 and FucT3. Sci Rep 8:16790. doi: 10.1038/s41598-018-34882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Knights D, Clemente JC, Nakielny S, Gordon JI, Fierer N, Knight R. 2013. Cohabiting family members share microbiota with one another and with their dogs. Elife 2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, Lucas SK, Beura LK, Thompson EA, Till LM, Batres R, Paw B, Pergament SL, Saenyakul P, Xiong M, Kim AD, Kim G, Masopust D, Martens EC, Angkurawaranon C, McGready R, Kashyap PC, Culhane-Pera KA, Knights D. 2018. US immigration westernizes the human gut microbiome. Cell 175:962–972.e10. doi: 10.1016/j.cell.2018.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cadotte MW. 2006. Dispersal and species diversity: a meta-analysis. Am Nat 167:913–924. doi: 10.1086/504850. [DOI] [PubMed] [Google Scholar]
- 60.Vellend M. 2010. Conceptual synthesis in community ecology. Q Rev Biol 85:183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- 61.Chase JM, Myers JA. 2011. Disentangling the importance of ecological niches from stochastic processes across scales. Philos Trans R Soc B 366:2351–2363. doi: 10.1098/rstb.2011.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dubos R, Savage D, Schaedler R. 1966. Biological Freudianism: lasting effects of early environmental influences. Pediatrics 38:789–800. [PubMed] [Google Scholar]
- 64.Gordon JI, Dewey KG, Mills DA, Medzhitov RM. 2012. The human gut microbiota and undernutrition. Sci Transl Med 4:137ps12. doi: 10.1126/scitranslmed.3004347. [DOI] [PubMed] [Google Scholar]
- 65.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA Jr, Ahmed T, Gordon JI. 2014. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, Liu J, Houpt E, Li JV, Holmes E, Nicholson J, Knights D, Ursell LK, Knight R, Gordon JI. 2013. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Caporaso JG. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. [Google Scholar]
- 71.Anderson MJ. 2006. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62:245–253. doi: 10.1111/j.1541-0420.2005.00440.x. [DOI] [PubMed] [Google Scholar]
- 72.Faust K, Raes J. 2016. CoNet app: inference of biological association networks using Cytoscape. F1000Res 5:1519. doi: 10.12688/f1000research.9050.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, Tanaka R. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol 70:167–173. doi: 10.1128/aem.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, Eisen JA. 2009. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci U S A 106:17187–17192. doi: 10.1073/pnas.0904847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.