Abstract
The cryoEM method Microcrystal Electron Diffraction (MicroED) involves transmission electron microscope (TEM) and electron detector working in synchrony to collect electron diffraction data by continuous rotation. We previously reported several protein, peptide, and small molecule structures by MicroED using manual control of the microscope and detector to collect data. Here we present a procedure to automate this process using a script developed for the popular open-source software package SerialEM. With this approach, SerialEM coordinates stage rotation, microscope operation, and camera functions for automated continuous-rotation MicroED data collection. Depending on crystal and substrate geometry, more than 300 datasets can be collected overnight in this way, facilitating high-throughput MicroED data collection for large-scale data analyses.
Keywords: MicroED, microcrystal, electron diffraction, cryoEM, transmission electron microscopy, SerialEM
1. Introduction
The cryoEM method Microcrystal Electron Diffraction (MicroED) is gaining momentum and is at the stage at which automation could further expand its use. In MicroED, a nanocrystal that is continuously rotated is exposed to the electron beam in diffraction mode while the diffraction data is collected on a fast camera as a movie [1,2]. This mode of MicroED data collection is called continuous rotation and is analogous to the rotation method in X-ray crystallography [3]. Combined with a camera operating in shutterless mode, continuous-rotation MicroED avoids partially-measured reflections by sampling all reciprocal space throughout the rotation range of the crystal, and provides both faster data collection and simpler data processing [1]. Continuous rotation has also improved the quality of the measurements [1] and allowed MicroED to tackle more complex structure determination projects [4]; it has therefore become the standard method of data collection in MicroED.
MicroED data are collected from a grid which might contain thousands of nanocrystals. On most modern electron microscopes, a maximum of 140° of data can be collected per crystal (+/− 70° tilt) so that, depending on symmetry, a single nanocrystal may yield an almost complete dataset [5]. Data from several crystals can be merged to increase completeness if each crystal is oriented differently on the grid [6]. MicroED data is then processed using standard X-ray crystallography software where structures are determined and refined as described before [7,8].
MicroED could benefit from automation, where multiple targets are selected for acquisition in a single run, as is commonly done with other cryoEM modalities such as electron tomography (ET) [9] and single particle analysis (SPA) [10]. These methodologies utilize software that controls the electron microscope and camera in order to perform the operations necessary during long-running data acquisition sessions with minimum human intervention. Automation is appealing because it increases the throughput of the instrument, which is particularly relevant to busy facilities where microscopes are operated as shared resources. It also reduces error arising from manual execution of repetitive tasks and allows for data collection 24/7.
SerialEM [11] is a freely-available and open-source Microsoft Windows-based program used to coordinate microscope tasks and acquire digital images on TEMs. As of September 2018, SerialEM has been installed on over 500 electron microscopes worldwide. It is compatible with modern electron microscopes and imaging detectors from several major manufacturers, and provides a consistent user interface across different hardware platforms. The software is highly extensible and allows microscope and camera acquisition tasks to be automated through its scripting command processor. Updates to the software and user-created scripts have enabled increasingly automated SPA [12] and ET [13] data collection. Other TEM automation software programs exist [10], but SerialEM is currently the only one with the ability to control the microscope in diffraction mode in conjunction with a continuously-rotating stage.
2. SerialEM data collection procedure
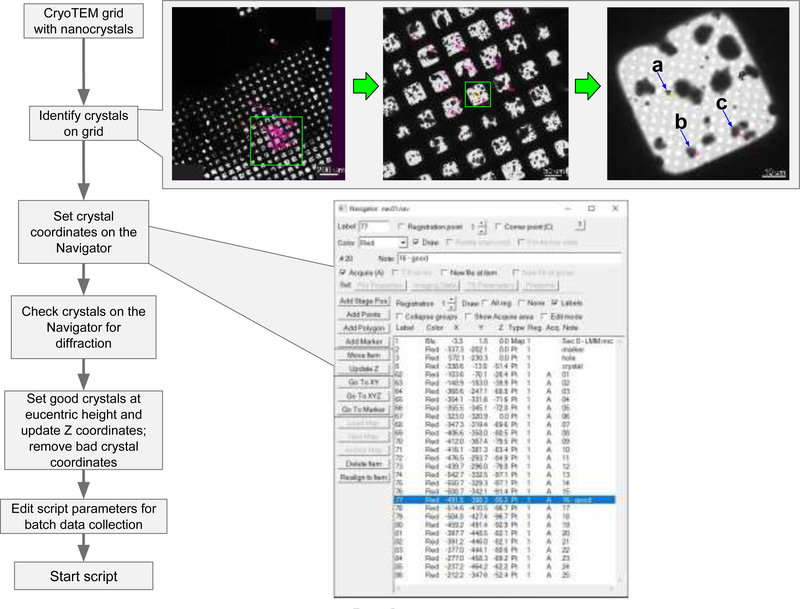
We developed a procedure using a script for SerialEM (http://bio3d.colorado.edu/SerialEM) which enables large-scale MicroED data collection on TEMs by Thermo Fisher Scientific (formerly FEI Company, Philips Electron Optics) coupled to electron detectors from various manufacturers. An overview of this procedure is presented in Figure 1 and generally follows similar protocols commonly used for collecting SPA data. Once a grid containing nanocrystals is loaded into the microscope, the operator identifies potential crystals either by inspecting the grid manually using low-dose imaging [14], or by montaging a whole-grid atlas. An atlas provides a fast way to visually screen an entire grid for crystals offline and keeps the exposure of the sample at a minimum. The location of each crystal is then marked in a similar way to selecting regions of holes/gridsquares for SPA data collection. The operator can then collect a sample diffraction pattern from each of the marked crystals to narrow down the selection of crystals for complete data collection (Figure 2). This step can be done either manually or automatically in SerialEM using its Navigator tools.
Fig. 1.
SerialEM automation workflow for MicroED. A cryoTEM grid containing protein nanocrystals is examined via a whole-grid atlas collected at low magnification. Crystal areas are then identified from this atlas and added into the SerialEM Navigator queue for subsequent diffraction screening. Diffracting crystals on this list are selected for subsequent MicroED data collection in batch. (a), (b), and (c) denote crystals selected for diffraction screening in Fig. 2.
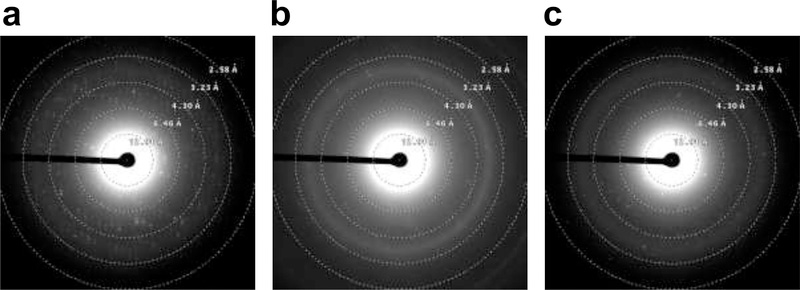
Fig. 2.
Electron diffraction screening of selected crystals. (a), (b), and (c) are the resulting diffraction patterns of crystals (a), (b), and (c) in Fig. 1.
Before data collection can begin on a selected set of crystals, the eucentric height needs to be determined at each data collection point on the grid. Akin to crystal centering in X-ray diffraction, this ensures that the crystal does not move out of the confines of the measurement during the continuous-rotation experiment [2]. Several reviews and papers have been published on crystal identification, troubleshooting, and microscope setup for diffraction [2,7,15–17]. The stage coordinates of the selected, well-diffracting crystals are then loaded to the automated data collection pipeline.
Our pipeline coordinates all components to automate data collection. The operator inputs the desired rotation angle span (within the range from −70° to +70°), rotation speed (entered as a multiplier to the intended degrees per second), exposure time per frame (seconds), and destination directory for the data output; these are applied to all crystals on a job list. Data collection then proceeds in batch, where the SerialEM script directs the microscope sequentially through the list of selected crystals, one crystal at a time.
SerialEM synchronizes crystal rotation and the start of data collection, such that the crystal is rotating at the desired, constant rate when the first frame is recorded. The entire rotation range is recorded as set, then camera recording stops, the stage rotation stops, and the data is saved. SerialEM then continues to the next crystal location in the job list. The relevant metadata, such as rotation rate and camera configuration, are automatically stored with the diffraction images in directories on the filesystem of the data collection host. These output files need to be converted to formats readable by typical crystallography integration packages such as DIALS [18], iMosflm/MOSFLM [19], or XDS [20] using our conversion software [7], which is freely available from https://cryoem.ucla.edu/pages/MicroED. A step-by-step protocol of SerialEM setup and instructions for MicroED data collection is presented in Supplementary Material.
3. Methods
Proteinase K was used a test sample for SerialEM-enabled MicroED data collection.
3.1. Crystal growth.
The fungal serine protease Proteinase K (E. album) from Sigma-Aldrich (St. Louis, MO, USA) was prepared by combining 3 mL of protein solution (50 mg mL−1) with 3 mL of precipitant solution (1.0–1.3 M ammonium sulfate, 0.1 M Tris @ pH 8.0) [15] and set in a 24-well tray as 4-μL hanging drops.
3.2. CryoEM sample preparation.
3 μL of proteinase K solution was pipetted onto a Quantifoil R 2/2 Cu 300-mesh grid (Quantifoil Micro Tools GmbH, Großlöbichau, Germany) which had been pretreated with glow-discharge plasma at 15 mA for 30 s (PELCO easiGlow, Ted Pella Inc., Redding, CA, USA). The grid was blotted onto 595 filter paper (Ted Pella Inc.) at a blot force setting of 5 for 10 s and plunge-frozen into liquid ethane using a Thermo Fisher Vitrobot Mark IV (Thermo Fisher Scientific, Hillsboro, OR) with a chamber environment of 22°C and 100% humidity.
3.3. TEM configuration.
Vitrified grids were examined with a Thermo Fisher Talos Arctica TEM operated at 200 kV and fitted with a Thermo Fisher CetaD scintillated CMOS 4k × 4k camera. The specimen was kept at ~100 K in the microscope. Low-magnification imaging for whole-grid crystal searching/montaging was done at a microscope magnification of 155×. TEM diffraction beam settings were saved into the SerialEM Low Dose Control for low-dose diffraction imaging [14] and coherent-beam diffraction for data collection. In this Low Dose Control, the diffraction beam used for data collection was set to an electron dose of 0.01 e− Å−2 s−1 and saved as the “Record mode” beam. The “View mode” beam setting was used for local (gridsquare) area imaging of crystals for screening on the FluCam (phosphor screen) of the Arctica TEM, and was set to an electron dose rate of ~1 × 10−6 e− Å−2 s−1. A diffraction-defocus offset was applied to view the crystal using the SerialEM Low Dose Control’s defocus “offset for View mode” setting.
3.4. Microcrystal screening.
Grids were searched for thin ice and microcrystals first using the low magnification setting (155×). This initial search either took place manually, by observing the FluCam and moving the stage, or via the SerialEM Full Montage function, where a series of images are collected at the low magnification setting during a rasterization of the stage across the entire grid surface. Crystal stage coordinates were identified and saved to the SerialEM Navigator for later diffraction testing. To identify crystals suitable for data collection in an automated way, we set the Record mode camera parameters to collect a 4-second exposure. This uses the “Record Mode” diffraction beam setting as mentioned above for crystal exposure. The selected area (SA) aperture was inserted, the size of which was set to accommodate the dimensions of the largest crystal chosen for diffraction. We then used the “Acquire at Points” function to acquire diffraction pattern exposures at each previously-saved crystal coordinate. For those crystals which showed protein diffraction, the position was revisited to set the crystal at eucentric height using the microscope controls, then queued in the Navigator for batch MicroED data collection.
3.5. MicroED data collection.
Microcrystal coordinates prepared for data collection were set to “Acquire” in the SerialEM Navigator in the previous section. Before starting, the parameters for data collection, particularly output directory, rotation speed, angular range, frame exposure time, and frame binning, were configured in the CRmov script for this batch run. A detailed protocol for SerialEM-automated MicroED data collection and the CRmov script are available in the Supplementary Material.
4. Discussion and Conclusion
This work lays the foundation for automating MicroED data collection by using software to control the repetitive and intricate tasks involved, allowing the experimenter to spend more time on those aspects of structure determination where computer-controlled procedures are not yet feasible, such as crystal optimization and TEM grid preparation. Our procedures may be further developed to automatically detect crystals on a grid and determine optimal data collection parameters as is often done at synchrotrons for X-ray crystallography (e.g., BEST [21], STRATEGY [22]). Currently SerialEM only has access to the necessary microscope commands on Thermo Fisher systems; work on compatibility with JEOL microscopes is ongoing and support for other microscopes and cameras is anticipated.
Once MicroED data is recorded, the CRmov script directs SerialEM to save the images as TIF files which are then converted to an image format that is readable by downstream X-ray crystallography processing software [7]. The data can be processed by several different standard crystallographic packages including DIALS [18], iMosflm/MOSFLM [19], and XDS [20]. Diffraction intensities are then integrated, scaled, and merged (as necessary). In order to generate high-resolution structures, these intensities are combined with phases calculated either from similar models by molecular replacement, or determined experimentally via ab initio direct methods [23], isomorphous replacement by heavy-atom derivatives, or by extracting phases from images of crystals.
Interest in cryoEM methods for biological investigation has recently surged as developments in hardware (increasingly stable electron optics, in-microscope robotic multiple-specimen holders allowing longer contamination-free storage times, and fast cameras allowing new ways of data collection) and software (automation for data collection and analysis) have opened cryoEM to the wider community. We show here that MicroED data can be collected in a consistent and automated way using the open-source software application SerialEM with minimal user intervention. This level of automation for MicroED simplifies the data collection experience for the end-user and makes MicroED accessible to laboratories and facilities without prior extensive experience. Depending on the type of camera used and the speed of rotation, data collection can take as little as 1 minute per crystal for a full 140° dataset that is often sufficient for structure determination. Under such a regime, data from more than 300 crystals can be collected autonomously overnight, bringing MicroED to the X-ray crystallography timescale and enabling analyses of big datasets that could be used for time-resolved studies, drug discovery, and protein dynamics. Thus, large-scale automation represents the next phase of MicroED studies.
Supplementary Material
Highlights.
A procedure for automating TEM and electron detector to collect MicroED data is described.
The widely-used open-source software program SerialEM is used to coordinate TEM goniometer movement and detector image recording.
A SerialEM script and usage protocol for Thermo Fisher TEMs is provided.
Acknowledgments
We are grateful to David Mastronarde for feature support and technical assistance with SerialEM. This research was supported by the NIH/NCI Cancer Center Support Grant P30 CA008748 to Memorial Sloan Kettering Cancer Center. The Gonen laboratory is supported by funds from the Howard Hughes Medical Institute.
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Nannenga BL, Shi D, Leslie AGW, Gonen T, High-resolution structure determination by continuous-rotation data collection in MicroED., Nat. Methods 11 (2014) 927–930. doi: 10.1038/nmeth.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shi D, Nannenga BL, de la Cruz MJ, Liu J, Sawtelle S, Calero G, Reyes FE, Hattne J, Gonen T, The collection of MicroED data for macromolecular crystallography., Nat. Protoc 11 (2016) 895–904. doi: 10.1038/nprot.2016.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Arndt UW, Wonacott AJ, The Rotation method in crystallography: data collection from macromolecular crystals, North-Holland, Amsterdam, 1977. [Google Scholar]
- [4].Nannenga BL, Shi D, Hattne J, Reyes FE, Gonen T, Structure of catalase determined by MicroED, Elife. 3 (2014) e03600. doi: 10.7554/eLife.03600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Glaser RM, Tong L, Kim SH, Three-dimensional reconstructions from incomplete data: Interpretability of density maps at “atomic” resolution, Ultramicroscopy. 27 (1989) 307–318. doi: 10.1016/0304-3991(89)90021-1. [DOI] [PubMed] [Google Scholar]
- [6].Gallagher-Jones M, Glynn C, Boyer DR, Martynowycz MW, Hernandez E, Miao J, Zee C-T, Novikova IV, Goldschmidt L, McFarlane HT, Helguera GF, Evans JE, Sawaya MR, Cascio D, Eisenberg DS, Gonen T, Rodriguez JA, Sub-ångström cryo-EM structure of a prion protofibril reveals a polar clasp, Nat. Struct. Mol. Biol (2018). doi: 10.1038/s41594-017-0018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hattne J, Reyes FE, Nannenga BL, Shi D, De La Cruz MJ, Leslie AGWW, Gonen T, MicroED data collection and processing, Acta Crystallogr. Sect. A Found. Adv 71 (2015) 353–360. doi: 10.1107/S2053273315010669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Weiss SC, Kim SK, Hinck CS, Hinck AP, Calero G, Eisenberg D, Gonen T, Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED, Nat. Methods 14 (2017) 399–402. doi: 10.1038/nmeth.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Koning RI, Koster AJ, Sharp TH, Advances in cryo-electron tomography for biology and medicine, Ann. Anat 217 (2018) 82–96. doi: 10.1016/j.aanat.2018.02.004. [DOI] [PubMed] [Google Scholar]
- [10].Tan YZ, Cheng A, Potter CS, Carragher B, Automated data collection in single particle electron microscopy, Reprod. Syst. Sex. Disord 65 (2016) 43–56. doi: 10.1093/jmicro/dfv369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements, J. Struct. Biol 152 (2005) 36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- [12].Stroupe ME, Xu C, Goode BL, Stroupe ME, Xu C, Goode BL, Grigorieff N,Actin filament labels for localizing protein components in large complexes viewed by electron microscopy Actin filament labels for localizing protein components in large complexes viewed by electron microscopy, (2009) 244–248. doi: 10.1261/rna.1313609. [DOI] [PMC free article] [PubMed]
- [13].Hagen WJH, Wan W, Briggs JAG, Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging, J. Struct. Biol (2016). doi: 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nakamura N, Shimizu Y, Shinkawa T, Nakata M, Bammes B, Zhang J, Chiu W, Automated specimen search in cryo-TEM observation with DIFF-defocus imaging, J. Electron Microsc. (Tokyo). 59 (2010) 299–310. doi: 10.1093/jmicro/dfq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hattne J, Shi D, De La Cruz MJ, Reyes FE, Gonen T, Modeling truncated pixel values of faint reflections in MicroED images, J. Appl. Crystallogr 49 (2016) 1029–1034. doi: 10.1107/S1600576716007196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hattne J, Shi D, Glynn C, Martynowycz MW, Rodriguez JA, Gonen T, Hattne J, Shi D, Glynn C, Zee C, Gallagher-jones M, Martynowycz MW, Analysis of Global and Site-Specific Radiation Damage in Cryo-EM, Structure. 26 (2018) 759−−766.e4. doi: 10.1016/j.str.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gonen T, The Collection of High-Resolution Electron Diffraction Data, in: Schmidt-Krey I, Cheng Y (Eds.), Electron Crystallogr. Soluble Membr. Proteins Methods Protoc. Methods Mol. Biol, Springer Science+Business Media LLC, 2013: pp. 153–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Clabbers MTB, Gruene T, Parkhurst JM, Abrahams JP, Waterman DG,Electron diffraction data processing with DIALS, Acta Crystallogr. Sect. D Struct. Biol 74 (2018) 506–518. doi: 10.1107/S2059798318007726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Powell HR, Battye TGG, Kontogiannis L, Johnson O, Leslie AGW, Integrating macromolecular X-ray diffraction data with the graphical user interface iMosflm, Nat. Protoc 12 (2017) 1310–1325. doi: 10.1038/nprot.2017.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kabsch W, Xds, Acta Crystallogr. Sect. D Biol. Crystallogr 66 (2010) 125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Popov AN, Bourenkov GP, Choice of data-collection parameters based on statistic modelling, Acta Crystallogr. - Sect. D Biol. Crystallogr 59 (2003) 1145–1153. doi: 10.1107/S0907444903008163. [DOI] [PubMed] [Google Scholar]
- [22].Ravelli RBG, Sweet RM, Skinner JM, Duisenberg AJM, Kroon J,STRATEGY: A program to optimize the starting spindle angle and scan range for X-ray data collection, J. Appl. Crystallogr 30 (1997) 551–554. doi: 10.1107/S0021889897003543. [DOI] [Google Scholar]
- [23].Usón I, Sheldrick GM, An introduction to experimental phasing of macromolecules illustrated by SHELX ; new autotracing features, Acta Crystallogr.Sect. D Struct. Biol 74 (2018) 106–116. doi: 10.1107/S2059798317015121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.