Endosidin2-14 shows potential as a small molecule chemical inhibitor as it targets EXO70, a subunit of the exocyst complex, resulting in inhibition of exocytosis in plants and fungal pathogens.
Abstract
The evolutionarily conserved octameric exocyst complex tethers secretory vesicles to the site of membrane fusion during exocytosis. The plant exocyst complex functions in cell wall biosynthesis, polarized growth, stress responses, and hormone signaling. In fungal pathogens, the exocyst complex is required for growth, development, and pathogenesis. Endosidin2 (ES2) is known to inhibit exocytosis in plant and mammalian cells by targeting the EXO70 subunit of the exocyst complex. Here we show that an analog of ES2, ES2-14, targets plant and two fungal EXO70s. A lower dosage of ES2-14 than of ES2 is required to inhibit plant growth, plant exocytic trafficking, and fungal growth. ES2-14 treatments inhibit appressorium formation and reduce lesion sizes caused by Magnaporthe oryzae. Inhibition of EXO70 by ES2-14 in Botrytis cinerea also reduces its virulence in Arabidopsis (Arabidopsis thaliana). Interestingly, ES2-14 did not affect EXO70 localization or transferrin recycling in mammalian cells. Overall, our results indicate that a minor change in ES2 affects its specificity in targeting EXO70s in different organisms and they demonstrate the potential of using ES2-14 to study the mechanisms of plant and fungal exocytosis and the roles of exocytosis in fungus-plant interactions.
Plant growth and development require dynamic regulation of membrane trafficking in a spatiotemporal manner for material delivery and signaling purposes. Exocytosis is an important step of membrane trafficking that delivers materials such as proteins and lipids to the plasma membrane (PM) and extracellular space. Cargo proteins of exocytosis include enzymes for cell wall synthesis, transporters or receptors for hormone signaling, proteins that facilitate nutrient uptake, and proteins involved in stress responses. For examples, cellulose synthase complex (Zhu et al., 2018), Brassinosteroid-Insensitive 1 (BRI1) brassinosteroid receptor (Geldner et al., 2007), and some PIN-FORMED (PIN) auxin transporters (Drdová et al., 2013) are constitutively delivered to the PM through exocytosis during normal plant growth (Kleine-Vehn et al., 2011; Luschnig and Vert, 2014).
The conserved octameric exocyst complex is an essential component in exocytosis that tethers secretory vesicles to the site of membrane fusion (Heider and Munson, 2012; Wu and Guo, 2015). Each exocyst complex contains one molecule of EXO70, EXO84, SEC3, SEC5, SEC6, SEC8, SEC10, and SEC15 proteins (TerBush et al., 1996; Guo et al., 1999). In plants, by analyzing genetic mutants, it has been found that the exocyst complex functions in cell plate formation (Fendrych et al., 2010), cell wall deposition (Kulich et al., 2010), immune responses (Stegmann et al., 2012), root development, polarized growth (Synek et al., 2006), embryo development (Zhang et al., 2013), xylem development (Vukašinović et al., 2017), and hormone signaling (Drdová et al., 2013). In rice (Oryza sativa), the exocyst complex is involved in effector recognition during invasion of the rice blast fungus Magnaporthe oryzae and is essential for rice defense against insect invasion (Fujisaki et al., 2015; Guo et al., 2018). The cellular mechanisms behind the functioning of plant exocyst complex with other cellular components, such as GTPase signaling, lipid signaling, and cytoskeletons, need further investigation. Plant EXO70s exist as large families (Synek et al., 2006) and it is expected that small-molecule inhibitors will be useful in overcoming genetic redundancy to study the functions of these genes.
The exocyst complex also has been shown to be essential for growth and pathogenesis in a number of filamentous fungi. In M. oryzae, the causal agent of rice blast and a model for studying fungus-plant interactions, deletion of exocyst components not only inhibits growth but also affects the formation of highly specialized infection structures known as appressoria and effector delivery during plant infection (Giraldo et al., 2013; Chen et al., 2015; Gupta et al., 2015). Unlike the hemibiotrophic pathogen M. oryzae, the gray mold fungus Botrytis cinerea is a necrotrophic pathogen. The function of exocyst in B. cinerea is not well characterized, but BcEXO84 appears to be required for growth and pathogenicity (Giesbert et al., 2012).
Previously, a small molecule endosidin2 (ES2) was found to target the AtEXO70A1 in Arabidopsis (Arabidopsis thaliana) and EXO70 in mammalian cells to inhibit exocytosis (Zhang et al., 2016). Cellular localization of proteins that undergo constitutive exocytosis was affected by short-term ES2 treatments. Upon treatment with 40 μm ES2 for 2 h, the abundance of the PIN2 auxin transporter was reduced at the PM but increased at the prevacuolar compartments (PVCs) and the vacuole. ES2 also inhibits the recovery trafficking of PIN2 from brefeldin A (BFA)-induced large cellular compartments. The equilibrium dissociation constant (Kd) between AtEXO70A1 and ES2 was between 250 and 400 μm, depending on the biochemical assays used (Zhang et al., 2016).
Recently, ES2 has been used as a tool not only in understanding plant exocytosis regulation but also in mammalian cell membrane trafficking and cancer biology (Gómez-Escudero et al., 2017; Mayers et al., 2017; Wang et al., 2017; Cole et al., 2018; O'Neill et al., 2018). Previously, two analogs of ES2 were found to inhibit PIN2 trafficking in plants, but their activities in other organisms were not characterized. In this study, we characterized the activities of ES2-14 in targeting plant and fungal EXO70s using growth assays, biochemical binding assays, live cell imaging, and pathogenicity tests. We show that ES2-14 directly interacted with AtEXO70A1 and inhibited plant exocytosis at a lower dosage than that required for ES2. ES2-14 also directly interacted with fungal EXO70 proteins and inhibited the growth and pathogenicity of M. oryzae and B. cinerea. These results indicate that ES2 and ES2-14 can be used to inhibit plant and fungal exocytosis by targeting EXO70s in these organisms.
RESULTS
ES2-14 Is a More Potent Growth Inhibitor Than ES2
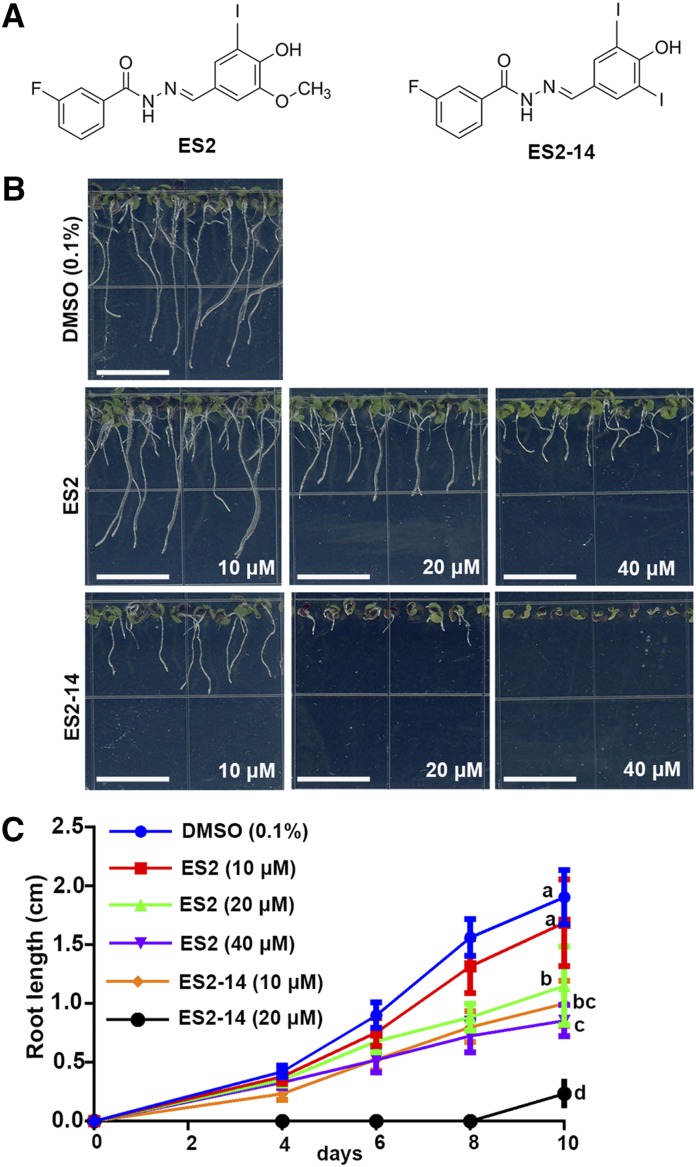
In a previous structure-activity relationship analysis on ES2, two analogs were found to be active in inhibiting the trafficking of PIN2 to the PM (Zhang et al., 2016). ES2-14 is one of these active analogs that have a minor structural difference from that of ES2 by replacing the methoxy group in one of the benzene rings with an iodine (Fig. 1A). To assay the effect of this modification on plant growth inhibition, Arabidopsis seedlings were grown on growth media supplemented with different concentrations of ES2, ES2-14, or the dimethyl sulfoxide (DMSO) solvent control. No obvious differences in growth were observed between 10-d-old seedlings grown on media with 10 μm ES2 or DMSO. However, Arabidopsis seedlings had significantly shorter roots when grown on growth media with 10 μm ES2-14 than on media with DMSO. At concentrations of 20 or 40 μm, seedlings grown on media with ES2-14 were significantly smaller and had shorter roots than those grown on media with the same concentration of ES2 (Fig. 1B). In fact, on growth media supplemented with 40 μm ES2-14, the roots of Arabidopsis seedlings failed to elongate at all. Statistical analysis on the root length of seedlings grown on media with different concentrations of ES2 and ES2-14 at 10 d confirmed that ES2-14 is a more potent growth inhibitor than ES2 (Fig. 1C). The half maximal inhibitory concentration (IC50) for ES2 and ES2-14 for Arabidopsis are 32 and 15 μm, respectively.
Figure 1.
Inhibition of Arabidopsis root growth by ES2 and ES2-14. A, Molecular structures of ES2 and ES2-14. B, Representative images of 10-d-old Arabidopsis seedlings grown on one-half strength Murashige and Skoog (MS) media supplemented with 0.1% DMSO or different concentrations of ES2 and ES2-14. Bars = 1 cm. C, A time-course assay for root growth in Arabidopsis seedlings grown on one-half strength MS media with different concentrations of ES2 and ES2-14 at the indicated time points. Data represent the mean ± sd from three independent replicates (n = 15). Statistically significant differences were determined by one-way ANOVA test followed by Tukey’s multiple comparisons test. Lowercase letters indicate significant differences between groups (P < 0.05) with regard to root length of Arabidopsis seedlings at 10 d.
ES2-14 Is Also a More Potent Inhibitor of Exocytosis Than ES2 in Arabidopsis
To test whether ES2-14 has effects similar to those of ES2 on exocytic trafficking (Zhang et al., 2016), we first examined the cellular localization of different organelle markers upon ES2-14 treatment. Treatments with 40 μm ES2-14 for 2 h had no obvious effects on localization of the fluorescence-tagged endoplasmic reticulum resident protein His-Asp-Glu-Leu (HDEL), Golgi-localized Golgi transport protein 1 (GOT1p), trans-Golgi network protein vacuolar proton ATPase a1 (VHA-a1), and PM-localized proteins Rho of Plant 6 (ROP6), plasma membrane intrinsic protein 2a (PIP2a), and P-glycoprotein 4 (PGP4; Supplemental Fig. S1). These data indicate that, like ES2, ES2-14 does not disturb the general membrane system in Arabidopsis.
We then compared the effects of ES2-14 and ES2 on cellular localization of PIN2, which goes through exocytic and endocytic trafficking constitutively during normal growth (Kleine-Vehn et al., 2011; Drdová et al., 2013). Whereas it is predominantly localized to the PM when treated with DMSO, PIN2-GFP was found to accumulate at the PVCs after treatment with 40 μm ES2 for 2 h (Zhang et al., 2016). We first tested whether ES2-14 caused the same PIN2 trafficking phenotypes as ES2. We treated PIN2:GFP; RFP:ARA7 plants with DMSO (0.1%), 40 μm ES2, or 40 μm ES2-14 for 2 h. Treatment with ES2-14, similar to that with ES2, caused PIN2:GFP to localize to intracellular compartments that also contain RFP:ARA7, a PVC marker protein (Supplemental Fig. S2), which indicates that ES2-14 accelerates PIN2 trafficking to the vacuole.
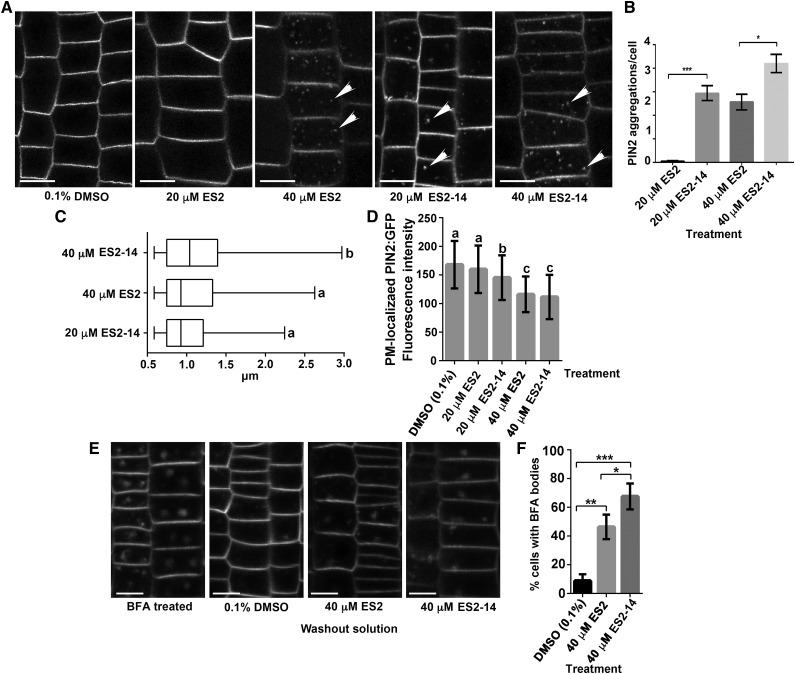
In order to compare the activity of ES2 and ES2-14, we examined PIN2:GFP localization after treatment with different concentrations of ES2 or ES2-14. When treated with 20 μm ES2 for 2 h, we found only a few PVCs that contained PIN2:GFP (Fig. 2, A and B). However, treatment with 20 μm ES2-14 for 2 h significantly increased the number of PVCs that contained PIN2:GFP in comparison with that from 20 μm ES2 treatment (Fig. 2, A and B). The number of PVCs containing PIN2:GFP was similar between the treatment group with 20 μm ES2-14 and that with 40 μm ES2 for 2 h. Nevertheless, treatments with 40 μm ES2-14 for 2 h or longer further increased the number of PVCs with GFP fluorescence (Fig. 2, A and B). We next compared the size of PVCs labeled by PIN2:GFP after treatment with different concentrations of ES2 and ES2-14. The Feret diameter of PIN2:GFP-labeled PVCs caused by 20 μm ES2-14 treatment was 1.01 ± 0.37 μm (mean ± sd, n = 170, from 90 cells of eight seedlings), which was comparable to the diameter of 1.08 ± 0.47 μm caused by 40 μm ES2 treatment for 2 h (mean ± sd, n = 190, from 100 cells of 8 seedlings), and 40 μm ES2-14 treatment further increased the diameter of PIN2-GFP-labeled PVCs to 1.15 ± 0.54 μm (mean ± sd, n = 185, from 95 cells of eight seedlings; Fig. 2C). The fluorescence intensity of PM-localized PIN2-GFP was not significantly reduced after 20 μm ES2 treatment for 2 h; however, 2 h of 20 μm ES2-14 treatment significantly reduced the fluorescence intensity of PIN2:GFP at the PM (Fig. 2D). PM-localized PIN2:GFP was further reduced by 40 μm ES2 and ES2-14 treatments (Fig. 2D). These quantitative data indicate that ES2-14 is more potent than ES2 in affecting PIN2 trafficking.
Figure 2.
Inhibition of PIN2 trafficking by treatment with ES2 and ES2-14. A, PIN2:GFP localization after treatment with DMSO or different concentrations of ES2 and ES2-14 for 2 h. Scale bars = 10 μm. B, Numbers of PVCs containing PIN2:GFP in Arabidopsis root epidermal cells treated with ES2 or ES2-14 for 2 h. ES2-14 is more effective in promoting PIN2 localization to the PVC. Data represent the means ± sd (n = 90 cells from eight seedlings). C, Boxplot showing the size distribution (Feret diameter) of PVCs containing PIN2:GFP, as shown in B. D, Quantification of the fluorescence intensity of PM-localized PIN2 on root epidermal cells treated with DMSO, ES2, or ES2-14. Data represent means ± sd (n = 120 cells from eight seedlings). E, Representative images of PIN2 localization after treatment with 40 μm of BFA for 1 h followed by 80 min recovery in one-half strength MS liquid media with 0.1% DMSO, 40 μm ES2, or 40 μm ES2-14. F, Numbers of BFA-induced compartments with PIN2:GFP in root epidermal cells after 80 min recovery in one-half strength liquid MS. ES2-14 is more effective than ES2 in reducing PIN2:GFP exocytic trafficking. Data represent the means ± sd (n = 110 cells from four seedlings). Asterisks indicate significant difference as determined by paired t test: *P < 0.05; **P < 0.01; ***P < 0.0001. Lowercase letters in C indicate significant differences between groups (P < 0.05) as determined by paired t test, and those in D indicate significant differences between groups (P < 0.05) as determined by one-way ANOVA test followed by Tukey’s multiple comparisons test.
BFA is a fungal lactone that inhibits exocytic trafficking of proteins such as PIN2 (Jásik et al., 2016). To assay the inhibitory effects of ES2-14 on exocytic transport, 7-d-old PIN2::PIN2:GFP seedlings were pretreated with 40 μm BFA for 60 min (Fig. 2E) and then recovered in one-half strength MS liquid media containing 0.1% DMSO, 40 μm ES2, or 40 μm ES2-14. After 80 min of recovery, Arabidopsis root epidermal cells were examined by confocal microscopy for PIN2 localization. In comparison with the DMSO control, ES2 or ES2-14 treatment samples showed significantly reduced recovery of cells from BFA treatment. The percentage of cells containing BFA compartments was ∼67% in seedlings recovered in media with ES2-14. Under the same conditions, only ∼45% of cell contained BFA-induced compartments in seedlings recovered in media with ES2 (Fig. 2, E and F). Taken together, these results indicate that ES2-14 is effective in inhibiting PIN2 exocytic trafficking from BFA-induced compartments in Arabidopsis.
ES2-14 Directly Interacts with AtEXO70A1
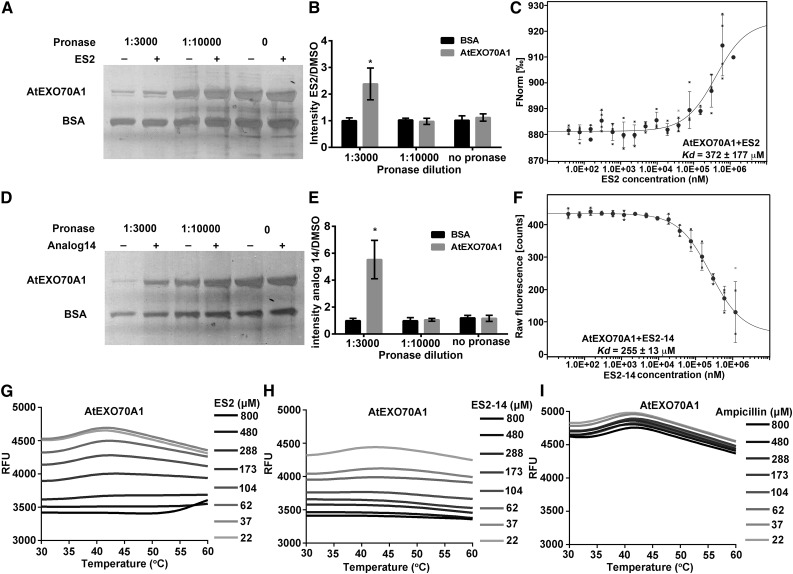
Because ES2 directly interacts with AtEXO70A1, a subunit of the exocyst complex (Zhang et al., 2016), we predicted that ES2-14 had activity similar to that of ES2 and thus assayed the interaction between ES2-14 and AtEXO70A1. The full-length AtEXO70A1 protein fused with the Histidine and small ubiquitin-like modifier (His-SUMO) tag was purified (Supplemental Fig. S3A, lane 2) and tested for its interaction with ES2-14 using the drug affinity responsive target stability (DARTS) assay, which is based on ligand protection of receptor proteins from degradation by proteases (Lomenick et al., 2009). Consistent with a previous report (Zhang et al., 2016), ES2 protected AtEXO70A1 from degradation by pronase, a mixture of different types of proteases, at 1:3,000 dilution (Fig. 3, A and B). As an internal control for the DARTS assay, bovine serum albumin (BSA) was not protected by ES2. We also performed a microscale thermophoresis (MST) assay to confirm the interaction between ES2 and AtEXO70A1. The MST assay detects the interaction of biomolecules by quantifying the temperature-induced fluorescence change of the target protein in response to different dosages of the ligand (Jerabek-Willemsen et al., 2011; Seidel et al., 2013). AtEXO70A1 protein labeled with amine-reactive red dye NT-647 (Zhang et al., 2016) was titrated with different concentrations of ES2 or ES2-14 in MST assays. Consistent with previous reports (Zhang et al., 2016), ES2 interacted with AtEXO70A1 at a Kd of 372 ± 177 μm (Fig. 3C).
Figure 3.
Biochemical assays for direct interaction of AtEXO70A1 with ES2 and ES2-14. A and B, DARTS assay for interaction between ES2 and recombinant AtEXO70A1 with different concentrations of pronase. A, Silver staining of proteins. B, Quantification of ratios of AtEXO70A1 and BSA intensities in samples treated with ES2 and DMSO, as shown in A. Data represent means ± sd (n = 3). C, MST assay for the interaction between AtEXO70A1 and ES2. The thermophoresis binding curve of the interaction of NT-647-labeled recombinant AtEXO70A1 with different concentrations of ES2 is shown. Data represent the means ± sd (n = 4). D to F, The same assays as in A to C, respectively, but with ES2-14 used instead of ES2. Data in E and F represent the means ± sd (n = 3). The raw fluorescence of NT-647-labeled recombinant AtEXO70A1 with different concentrations of ES2-14 is shown in F, because ES2-14 quenches NT-647 fluorescence at higher concentrations tested, which may be due to the interaction with the fluorophore at the binding region. For B and E, silver-staining gels from three independent DARTS assays were used for quantification. *P < 0.05 by paired t test. G to I, DSF assay showing the thermal stability of AtEXO70A1 in the presence of increasing concentrations of ES2 (G), ES2-14 (H), and Ampicillin (I; negative control). RFU, relative fluorescence units. Data shown are representative of three independent repeats.
When we performed the DARTS assay using recombinant AtEXO70A1 and ES2-14, ES2-14 protected AtEXO70A1, but not BSA, from degradation at 1:3,000 pronase dilution (Fig. 3, D and E). These results showed that, like ES2, ES2-14 was able to interact with AtEXO70A1 and protect it from degradation by proteases. We next used the MST assay to further test the direct interaction between ES2-14 and AtEXO70A1. AtEXO70A1 protein labeled with NT-647 (Zhang et al., 2016) was titrated with different concentrations of ES2-14 in MST assays. From the dosage response curve, ES2-14 interacted with AtEXO70A1 at a Kd of 255 ± 13 μm (Fig. 3F). Results from these two biochemical binding assays confirm that ES2-14 directly interacts with AtEXO70A1.
To further confirm the interaction of AtEXO70A1 with ES2 and ES2-14, we used another biochemical assay called differential scanning fluorimetry (DSF; Niesen et al., 2007), which has been commonly used to detect the interaction between small molecules and protein ligands (Ablinger et al., 2013; Baud et al., 2014; Athuluri-Divakar et al., 2016). The DSF assay is based on the theory that when a small-molecule ligand binds to the target protein, it could stabilize the protein and make the protein more tolerant to denaturation caused by high temperature. A fluorescence dye that binds to the hydrophobic parts of the protein is used to measure the unfolding process of the protein at increased temperature. Purified recombinant protein (Supplemental Fig. S3B, lane2) was used to test the interaction of AtEXO70A1 with ES2 and ES2-14 by DSF assay. As shown in Figure 3, G and H, the unfolding of AtEXO70A1 was protected by increased concentrations of both ES2 and ES2-14, as shown by the reduced fluorescence intensity when ES2 or ES2-14 concentration was increased. By contrast, ampicillin, as a negative control, did not show protection of AtEXO70A1 unfolding (Fig. 3I). We also tested for the interaction of AtEXO70A1 with ES2 and ES2-14 using the NMR chemical shift assay (Reibarkh et al., 2006; Shortridge et al., 2008; Arai et al., 2012; Furukawa et al., 2016). In this experiment, we added different concentrations of purified protein to ES2 or ES2-14 solution and observed the spectra of protons in ES2 and ES2-14. We found that increased concentrations of AtEXO70A1 led to a chemical shift and line broadening of protons in ES2 and ES2-14, indicating direct interaction of AtEXO70A1 with ES2 and ES2-14 (Supplemental Fig. S4).
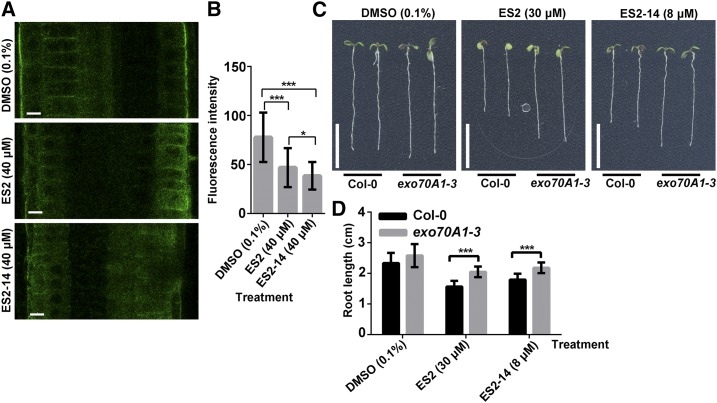
In order to further confirm that ES2-14 has activity similar to that of ES2 in inhibiting the exocyst complex, we performed further localization analyses and plant growth assays. Previously, it was found that ES2 reduced the polar localization of AtEXO70A1 in the outer lateral side of root epidermal cells and the heterozygous plants of exo70A1-3 showed reduced sensitivity to ES2 inhibition (Zhang et al., 2016). We treated plants expressing GFP-AtEXO70A1 with ES2-14 and found that like ES2 treatment, ES2-14 treatment reduced the localization of GFP-AtEXO70A1 at the outer lateral side of epidermal cells in the root transition zone (Fig. 4A). We quantified the maximum fluorescence intensity of GFP-EXO70A1 at the outer lateral PM and found that both ES2 and ES2-14 treatment reduced PM-localized GFP-EXO70A1 compared to treatment with the DMSO control (Fig. 4B). We also grew seeds from heterozygous exo70A1-3 plants and found that these seedlings showed reduced sensitivity to both ES2 and ES2-14 (Fig. 4, C and D). Our results indicate that ES2-14 targets the exocyst complex in plants in a way similar to that observed for ES2.
Figure 4.
ES2-14 affects polar localization of AtEXO70A1 and has a genetic interaction with AtEXO70A1. A, Representative images of GFP-AtEXO70A1 in cells at the root elongation zone in plants treated with DMSO, ES2, or ES2-14 for 2 h. Polar localization of GFP-AtEXO70A1 at the outer lateral side of root epidermal cells was altered by ES2 and ES2-14 treatment. Scale bars = 10 μm. B, Quantification of the fluorescence intensity of GFP-AtEXO70A1 at the outer lateral sides of root epidermal cells as shown in A. Data represent means ± sd (n = 50 from nine seedlings). C, Representative 5-d-old heterozygous exo70A1-3 plants grown on one-half strength MS media supplemented with DMSO, ES2, or ES2-14. Bars = 1 cm. D, Quantification of the root length of seedlings grown on DMSO, ES2, or ES2-14 at the indicated concentration. Data represent means ± sd (n = 13). Asterisks indicate a significant difference as determined by paired t test: *P < 0.05; ***P < 0.0001.
ES2 and ES2-14 Differ in Their Inhibitory Activities on Exocytosis in Mammalian Cells
ES2 is active in targeting mammalian EXO70s and it can inhibit the recycling of transferrin and the localization of Rattus norvegicus EXO70 (RnEXO70) to the PM (Zhang et al., 2016). Because ES2-14 is an active exocytosis inhibitor in plants, we then assayed whether it could be used as an exocytosis inhibitor in mammalian cells. We first tested the effect of ES2-14 on the localization of GFP-RnEXO70 in Hela cells. Consistent with the earlier report (Zhang et al., 2016), ES2 reduced the localization of RnEXO70 to the PM and caused its accumulation in intracellular compartments containing Rab family small GTPase Rab8 (Supplemental Fig. S5). However, treatment with ES2-14 did not affect cellular localization of RnEXO70 in Hela cells (Supplemental Fig. S5). We also performed transferrin recycling assays and found that ES2-14 also did not significantly inhibit the recycling of fluorescence-labeled transferrin in Hela cells (Supplemental Fig. S6). Our results indicate that ES2-14 is not an effective inhibitor of exocytosis in mammalian cells. Therefore, minor changes in the ES2 structure could affect its activity in different organisms.
Both M. oryzae and B. cinerea Are More Sensitive to ES2-14 Than to ES2
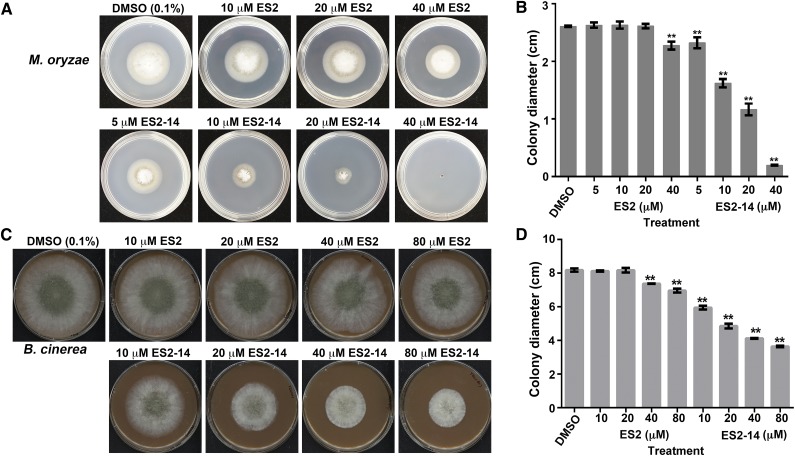
Because an inhibitor of the pathogen exocytosis will be a valuable tool in studying fungus-plant interactions, we then tested the effects of ES2 and ES2-14 on M. oryzae and B. cinerea, two fungal pathogens that employ different infection mechanisms (Dean et al., 2012). When assayed for growth on media with different concentrations of ES2 and ES2-14, we found that M. oryzae was more sensitive to ES2-14 than to ES2 (Fig. 5, A and B). Whereas 5 μm ES2-14 was sufficient to cause significant reduction in growth, 40 μm ES2 or higher was necessary to significantly reduce the growth of M. oryzae (Fig. 5, A and B). The IC50 values for ES2 and ES2-14 for M. oryzae are 562 and 16 μm, respectively. In B. cinerea, concentrations of 40 μm of ES2 and 10 μm or higher of ES2-14 significantly inhibited the growth rate of B. cinerea (Fig. 5, C and D). The IC50 of ES2-14 for B. cinerea was 47 μm, whereas the IC50 of ES2 for B. cinerea could not be calculated due to its abnormal growth curve in the presence of ES2. In both M. oryzae and B. cinerea, a lower concentration of ES2-14 than of ES2 was required for reducing the growth rate (Fig. 5), indicating that ES2-14 is a more potent fungal growth inhibitor.
Figure 5.
Inhibition of B. cinerea and M. oryzae growth by ES2 and ES2-14. A, Representative cultures of M. oryzae grown on complete minimal medium supplemented with 0.1% DMSO or different concentrations of ES2 or ES2-14 for 4 d. B, Diameter of 4-d-old complete minimal medium cultures of M. oryzae in the presence of ES2 or ES2-14. Data represent means ± sd (n = 6). C, Four-day-old V8 cultures of B. cinerea colonies with 0.1% DMSO or different concentrations of ES2 or ES2-14. D, Diameter of 4-d-old V8 B. cinerea colonies in the presence of different concentrations of ES2 or ES2-14. Data represent means ± sd (n = 6). The diameters of the petri dishes are 6 cm for M. oryzae and 10 cm for B. cinerea. **P < 0.01 (B and D) as compared with the DMSO control, determined by paired t test.
ES2-14 Directly Interacts with MoEXO70 and BcEXO70
In phylogeny, AtEXO70A1 and RnEXO70 are grouped together, whereas MoEXO70 and BcEXO70 are in a separate clade (Supplemental Fig. S7). Two amino acids in AtEXO70A1 (L596 and I613) that were previously predicted to be important for the interaction of AtEXO70A1 with ES2 were not conserved in fungal protein as well (Zhang et al., 2016; Supplemental Fig. S7). However, since ES2 and ES2-14 show inhibitory effects on fungal growth, we wanted to test whether there is a direct interaction between ES2 and ES2-14 and fungal EXO70 proteins. We expressed and purified MoEXO70 and BcEXO70 proteins fused with the His-SUMO tag (Supplemental Fig. S3A, lanes 3 and 4, respectively) for DARTS assays. Because MST assays detect protein and ligand interactions by fluorescence, we expressed and purified GFP-MoEXO70 and GFP-BcEXO70 using the His-SUMO tag for these assays (Supplemental Fig. S3A, lanes 5 and 6, respectively). We expressed GFP with His-SUMO tag as the negative control for the MST assays (Supplemental Fig. S3A, lane 7). We used MoEXO70 and BcEXO70 without any tag (Supplemental Fig. S3B, lanes 3 and 4, respectively) for DSF assays and NMR chemical shift assays.
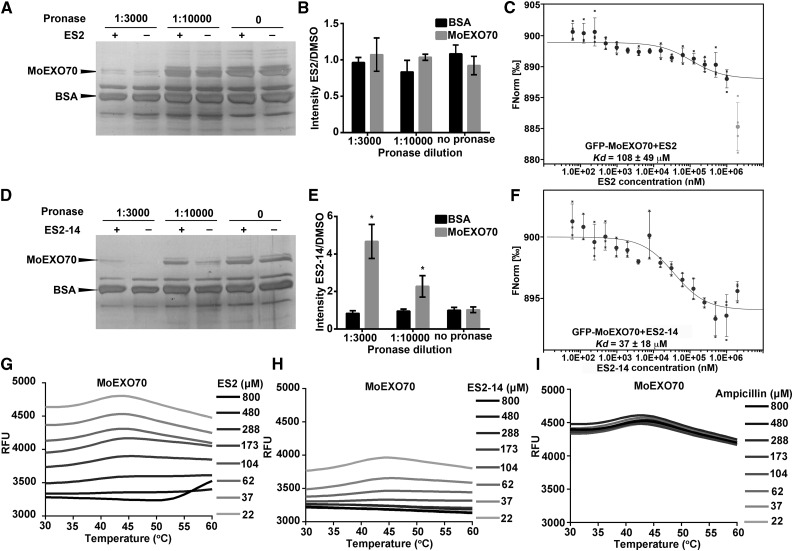
We first tested whether ES2 and ES2-14 directly interact with MoEXO70. In DARTS assays with ES2 and MoEXO70, ES2 did not significantly protect MoEXO70 from degradation after pronase digestion at 1:3,000 and 1:10,000 dilutions (Fig. 6, A and B). However, MST dosage response curves showed that ES2 interacted with GFP-MoEXO70 with a calculated Kd of 108 ± 49 μm (Fig. 6C). When we tested the interaction between ES2-14 and MoEXO70 using DARTS assays, ES2-14 protected MoEXO70, but not BSA, from degradation by pronase at 1:3,000 and 1:10,000 dilutions (Fig. 6, D and E). After protease digestion, the abundance of MoEXO70 was significantly higher in reactions containing ES2-14 than in those with the DMSO control. ES2-14 interacted with GFP-MoEXO70 with a calculated Kd of 37 ± 18 μm in MST assays (Fig. 6F). As controls for MST assays, GFP with His-SUMO tag did not interact with ES2 or ES2-14 (Supplemental Fig. S8). In DSF assays, both ES2 and ES2-14 protected MoEXO70 from unfolding during thermal denaturation, whereas ampicillin as a negative control showed no protective effect (Fig. 6, G–I). When we performed the NMR chemical shift assay, increased concentrations of MoEXO70 induced chemical shift and line broadening of both ES2 and ES2-14 (Supplemental Fig. S9). We found inconsistent results for the interaction between MoEXO70 and ES2. We detected interaction between MoEXO70 and ES2 using MST, DSF, and NMR chemical shift assays but we did not detect direct interaction between MoEXO70 and ES2 using the DARTS assay. However, results from DARTS, MST, DSF, and NMR chemical shift assays indicated that ES2-14 directly interacts with MoEXO70. Considering the lower efficiency of ES2 in inhibiting M. oryzae growth compared with that of ES2-14, we conclude that ES2-14 is a more efficient inhibitor of MoEXO70 than ES2.
Figure 6.
Biochemical assays for the interaction of ES2 and ES2-14 with MoEXO70. A and B, DARTS assays for the interaction between ES2 and recombinant MoEXO70 with different concentrations of pronase. A, Silver staining of proteins in the DARTS assay of MoEXO70 with ES2. B, Quantification of ratios of MoEXO70 and BSA intensities in silver staining gels between samples treated with ES2 and DMSO. Data represent means ± sd (n = 3). C, MST assay for the interaction between MoEXO70 and ES2. Data represent means ± sd (n = 4). The thermophoresis binding curve of the interaction of GFP-MoEXO70 with different concentrations of ES2 is shown. The gray outlier data point at the highest ES2 concentration was not used for curve fitting and Kd estimation. D to F, The same assays as shown in A to C, respectively, except that ES2-14 was used instead of ES2. Data in E and F represent means ± sd (n = 3). Silver staining gels from three independent DARTS experiments were used to generate each chart in B and E. ES2 did not protect MoEXO70 from degradation by the dilutions of pronase that were tested. ES2-14 protected MoEXO70 from degradation by pronase. *P < 0.05, paired t test. G to I, DSF assay showing the thermal stability of MoEXO70 in the presence of increasing concentrations of ES2 (G), ES2-14 (H) and Ampicillin (I; negative control). RFU, relative fluorescence units. Data shown are representative of three independent repeats.
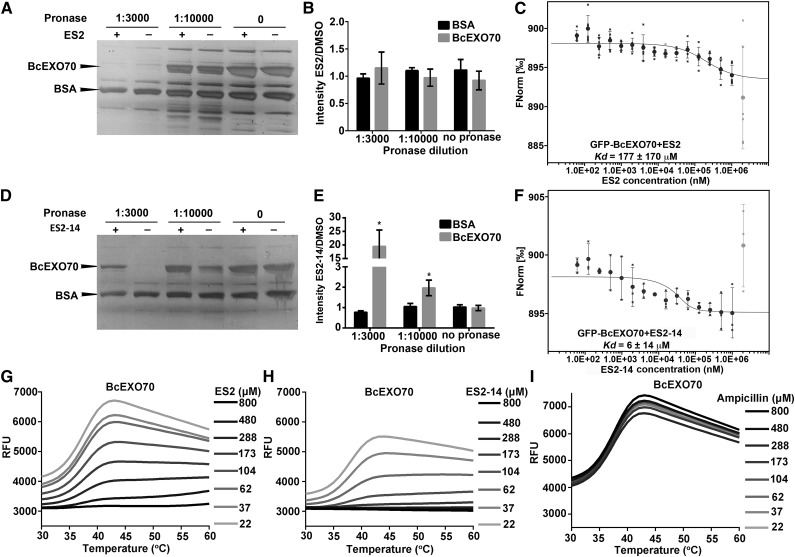
We next tested whether ES2 and ES2-14 directly interact with BcEXO70. In DARTS assays with ES2 and BcEXO70, ES2 did not significantly protect BcEXO70 from degradation after pronase digestion at 1:3,000 and 1:10,000 dilutions (Fig. 7, A and B). However, MST assays showed that ES2 interacted with GFP-BcEXO70 at a Kd of 177 ± 170 μm (Fig. 7C). In DARTS assays, ES2-14 protected BcEXO70, but not BSA, from degradation by pronase at 1:3,000 and 1:10,000 dilutions (Fig. 7, D and E). In MST assays, ES2-14 interacted with GFP-BcEXO70 with a calculated Kd of 6 ± 14 μm (Fig. 7F). In DSF assays, both ES2 and ES2-14 protected BcEXO70 from unfolding during thermal denaturation, whereas ampicillin as a negative control showed no protective effect (Fig. 7, G–I). In NMR chemical shift assays, increased concentrations of BcEXO70 induced chemical shifts and line broadening of both ES2 and ES2-14 (Supplemental Fig. S10). We detected some inconsistent results for the interaction between ES2 and BcEXO70 in different biochemical assays. We did not detect the protection of BcEXO70 degradation by ES2 in DARTS assays, although we detected direct interaction between ES2 and BcEXO70 in MST, DSF, and NMR chemical shift assays. However, all biochemical assays we used consistently confirmed that ES2-14 directly interacts with BcEXO70. Due to the weak inhibition of ES2 on B. cinerea growth, we conclude that ES2-14 is a more efficient inhibitor of BcEXO70 than ES2.
Figure 7.
Biochemical assays for the interaction of ES2 and ES2-14 with BcEXO70. A and B, DARTS assays for the interaction between ES2 and recombinant BcEXO70 with different concentrations of pronase. A, Silver staining of proteins in the DARTS assay of BcEXO70 with ES2. B, Quantification of ratios of BcEXO70 and BSA intensities in the silver staining gels shown in A, between samples treated with ES2 and DMSO. Data represent means ± sd (n = 3). C, MST assay for the interaction between BcEXO70 and ES2. Data represent means ± sd (n = 6). The thermophoresis binding curve for the interaction of GFP-BcEXO70 with different concentrations of ES2 is shown. D to F, The same assays as A to C, respectively, except that ES2-14 was used instead of ES2. Data in E represent means ± sd (n = 3). Data in F represent means ± sd (n = 3). Silver staining gels from three independent DARTS experiments were used to generate each chart in B and E. ES2 did not protect BcEXO70 from degradation by the dilutions of pronase that were tested. ES2-14 protected BcEXO70 from degradation by pronase. *P < 0.05, paired t test. In C and F, the gray outlier data points at the highest ES2 (C) and ES2-14 (F) concentrations was not used for curve fitting and Kd estimation. G to I, DSF assay showing the thermal stability of BcEXO70 in the presence of increasing concentrations of ES2 (G), ES2-14 (H), and Ampicillin (I; negative control). RFU, relative fluorescence units. Data shown are representative of three independent repeats.
ES2 and ES2-14 Affect Cellular Localization of MoEXO70
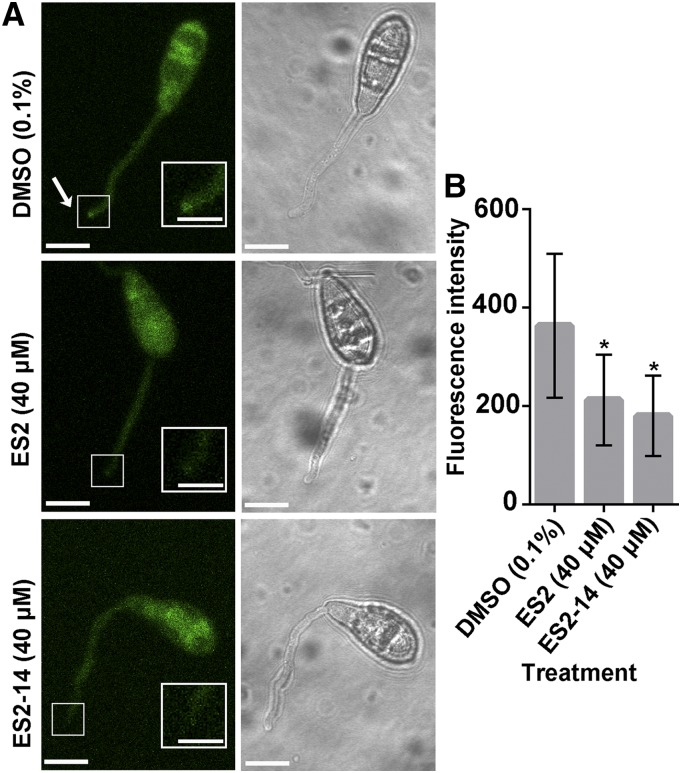
The exocyst complex has been found to be an important component for the establishment of rice blast fungal infection in host plants (Gupta et al., 2015). As both ES2 and ES2-14 affect M. oryzae growth and directly interact with MoEXO70, we wonder whether the two compounds interfere with MoEXO70 localization. MoEXO70-GFP was found to localize to the tips of germinating conidia during appressoria formation in the presence of DMSO (Fig. 8A). However, in samples treated with ES2 (40 μm) or ES2-14 (40 μm), conidia could germinate, but the fluorescence of MoEXO70-GFP was not enriched at the tip region (Fig. 8). Our results show that ES2 and ES2-14 affect MoEXO70 localization at the tip region of the conidia during appressoria formation.
Figure 8.
ES2 and ES2-14 affect cellular localization of MoEXO70. A, Representative images of MoEXO70-GFP in conidia treated with DMSO, ES2, or ES2-14 for 3 h. ES2 and ES2-14 reduce the localization of MoEXO70-GFP at the tips of germinated conidia. The arrow points to the tip of a germinated conidium. Scale bars = 5 μm outside the insets and 2.5 μm in the insets. B, Quantification of the intensity of MoEXO70-GFP at the tips of germinated conidia, as shown in A. Data represent means ± sd (n = 10). *P < 0.05, paired t test.
ES2-14 Inhibits Appressorium Formation and Plant Infection in M. oryzae
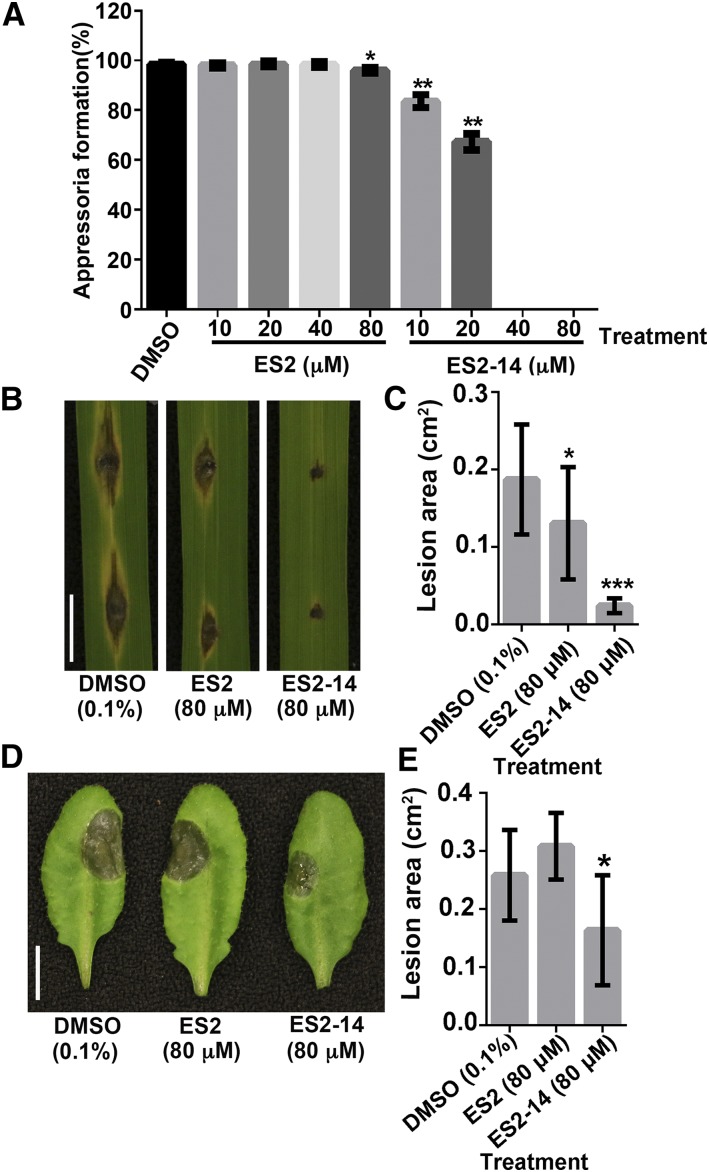
In M. oryzae, the formation of appressoria is essential for the penetration of plant cells. We first tested the effects of ES2 and ES2-14 on the formation of appressoria on artificial hydrophobic surfaces. Different concentrations of ES2 or ES2-14 were added to the spore suspensions. Formation of appressoria was observed after 24 h of incubation under moist conditions. Whereas ES2 appeared to have limited effects, ES2-14 inhibited appressorium formation (Fig. 9A). Treatment with 10 μm ES2-14 was sufficient to significantly reduce appressorium formation. Appressorium formation was almost completely blocked in the presence of 40 or 80 μm ES2-14 (Fig. 9A).
Figure 9.
Inhibitory effects of ES2 and ES2-14 on the pathogenicity of M. oryzae and B. cinerea. A, Inhibition of appressorium formation in M. oryzae by ES2 and ES2-14. The percentage of germ tubes that formed appressoria was examined after incubating spore suspensions with the indicated concentrations of ES2 or ES2-14 on plastic coverslips for 24 h. Whereas ES2 only slightly inhibited appressorium formation at 80 μm, ES2-14 completely blocked the formation of appressoria at 40 μm. Data represent means ± sd (n = 300). B, Rice leaves inoculated with M. oryzae spores mixed with DMSO, ES2, or ES2-14 were photographed at 6 dpi. C, Average size of lesions on rice leaves inoculated with M. oryzae spores mixed with the indicated concentrations of DMSO, ES2, or ES2-14 at 6 dpi. Data represent means ± sd (n = 20 from 10 plants). D, Arabidopsis leaves inoculated with B. cinerea spores mixed with DMSO, ES2, or ES2-14 were examined at 3 dpi. E, Average size of lesions caused by B. cinerea in treatments with DMSO, ES2, or ES2-14. Data represent means ± sd (n = 10 from 10 plants). Scale bars = 1 cm (B and D). *P < 0.05; **P < 0.01, ***P < 0.001, as determined by paired t test.
We then mixed spores of M. oryzae with ES2 or ES2-14 for infection assays with rice seedlings. On leaves drop-inoculated with 4-μL spore suspensions with 80 μm ES2-14, only limited necrosis was observed right below the spore drops at 6 d postinoculation (dpi). No extensive spreading of typical blast lesions was observed in the presence of 80 μm ES2-14 (Fig. 9, B and C). Leaves inoculated with spore suspensions with 80 μm ES2 still developed blast lesions that were smaller than those observed after inoculation with the DMSO control (Fig. 7, B and C). These results indicate that ES2 and ES2-14, particularly the latter, reduce the virulence of M. oryzae on rice leaves.
Virulence of B. cinerea Is Also Reduced by ES2-14
B. cinerea is a pathogen that can infect different plant species, including Arabidopsis. To test their effects on the virulence of B. cinerea, we mixed its spores with 80 μm of ES2 or ES2-14. On the leaves of 3-week-old Arabidopsis plants inoculated with spore suspensions of B. cinerea, ES2 did not affect the development of lesions compared with the DMSO control. However, treatment with ES2-14 significantly reduced the lesion size compared with control treatment (Fig. 9, D and E). These results show that ES2-14 is an inhibitor of B. cinerea fungal virulence.
DISCUSSION
Small molecule chemical inhibitors are valuable tools in studying dynamic cellular processes, such as exocytosis. These inhibitors bind to endogenous proteins to affect their functions without using genetic mutants. Previously, ES2 was found to inhibit exocytosis in plant and mammalian cells by targeting the EXO70 subunit of the conserved exocyst complex. Although AtEXO70A1 and RnEXO70 only share 23% sequence identity and 57% sequence similarity, their three-dimensional structures are quite similar and ES2 can directly interact with both. Due to the divergence of EXO70s in different organisms, minor modification of ES2 structure could affect its specificity and efficiency in targeting different EXO70s. Indeed, we found that ES2-14 had different specificity for EXO70s in different organisms we have tested. In comparison with ES2, ES2-14 is more potent in inhibiting Arabidopsis root growth and PIN2:GFP exocytosis, given that a lower dosage of ES2-14 than of ES2 was required to effect inhibition. Like ES2, ES2-14 directly interacted with AtEXO70A1 in DARTS, MST, DSF, and NMR chemical shift assays. These results show that ES2-14 can be used as an efficient exocyst inhibitor in Arabidopsis. ES2-14 did not inhibit the localization of other PM proteins tested, such as PIP2a, ROP6, and PGP4. There are a few possible explanations for this result: (1) these cargo proteins do not require the exocyst complex to localize to the PM; (2) ES2-14 only targets a subset of the 23 EXO70s in Arabidopsis and these cargos use those non-ES2-14 targets for PM localization; or (3) those proteins have a slower delivery rate and we could not detect the difference at the PM within 2 h of treatment. These are interesting and important questions that are worth future investigation.
It has been shown that ES2 interacts with mammalian EXO70 and inhibits exocytosis in animal cells. We found that ES2-14 was efficient in inhibiting plant exocytosis but it was not an efficient inhibitor of mammalian exocyst. Unlike ES2, ES2-14 treatment did not cause the mislocalization of RnEXO70 in Hela cells and did not inhibit the recycling of fluorescence-labeled transferrin. Although fungi are phylogenetically closer to animals than to plants, we found that both ES2 and ES2-14 inhibited the growth of the two plant pathogenic fungi M. oryzae and B. cinerea. In fact, a lower dosage of ES2-14 than of ES2 was required for growth inhibition of both fungi, indicating that ES2-14 is a more effective fungal growth inhibitor. In DARTS, MST, DSF, and NMR chemical shift assays, direct interactions between ES2-14 and fungal EXO70s were reliably detected. However, direct interactions between ES2 and fungal EXO70s were detected in MST, DSF, and NMR chemical shift assays, but not in DARTS assays. Taken together, our results from growth inhibition and biochemical binding assays showed that ES2-14 is an efficient exocyst inhibitor in M. oryzae and B. cinerea.
Exocytosis is essential for fungal growth, but it also plays critical roles in plant penetration and infectious growth in host tissues (Giraldo et al., 2013; Chen et al., 2015; Gupta et al., 2015). In M. oryzae, ES2-14 efficiently inhibited the formation of appressorium on artificial hydrophobic surfaces. In the presence of 10 μm ES2-14, conidium germination was not affected, but appressorium formation was blocked after incubation for 24 h. Under the same conditions, ES2 did not inhibit appressorium formation. Therefore, ES2-14 is also a more effective appressorium formation inhibitor than ES2. In infection assays with both M. oryzae and B. cinerea, treatments with ES2-14 significantly reduced the severity of disease development. On rice leaves inoculated with M. oryzae, ES2-14 treatment resulted in the formation of limited black spots under the inoculum (without extensive necrosis), which was likely due to its inhibitory effects on the development and growth of invasive hyphae in plant tissues. Due to the importance of active exocytosis, we expect that ES2-14 can be a useful inhibitor in understanding the regulation of exocytosis in fungal growth, development, and pathogenesis.
CONCLUSION
In this study, we characterized an analog of ES2, a reported inhibitor of plant and mammalian EXO70. In plant and fungal growth assays, ES2-14 was found to be more potent than ES2 in its inhibitory activity in Arabidopsis and the two important fungal pathogens M. oryzae and B. cinerea. At the cellular level, ES2-14 is more efficient in inhibiting PIN2:GFP exocytic trafficking in Arabidopsis. ES2-14 directly interacts with AtEXO70A1, MoEXO70, and BcEXO70. Consistent with earlier genetic studies with exocytosis mutants, we showed that binding of ES2-14 to MoEXO70 inhibits appressorium formation and reduces virulence of M. oryzae. Treatment with ES2-14 or ES2 also reduces hyphal growth of B. cinerea and its virulence during Arabidopsis infection. Interestingly, ES2-14 is not effective in inhibiting RnEXO70 localization and transferrin recycling in mammalian cells at the dosage tested. Therefore, minor changes in the ES2 structure could affect its specificity in targeting EXO70s in different organisms. ES2-14 can be used as an effective inhibitor in studying the mechanisms of plant and fungal exocytosis and the roles of exocytosis in fungus-plant interactions.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
To test the inhibitory effect of ES2-14 on plant growth, Arabidopsis (Arabidopsis thaliana) wild-type Col-0 plants were used. To test the effect of ES2-14 on cellular localization of proteins in different organelles, transgenic plants expressing fluorescence-tagged PIP2a, HDEL, VHA-a1, PGP4, ROP6, and GOT1p were used (Cutler et al., 2000; Matsushima et al., 2003; Dettmer et al., 2006; Cho et al., 2007; Fu et al., 2009; Geldner et al., 2009). To test the effect of ES2-14 on exocytic transport, the PIN2::PIN2:GFP line was used (Xu and Scheres, 2005). To test the localization of PIN2 at PVCs, the PIN2::PIN2:GFP;RFP:ARA7 line was used (Zhang et al., 2016). Seeds for plants that were used for live cell imaging or growth assay were sequentially sterilized with 50% (v/v) bleach (5.25% [w/v] sodium hypochlorite) and 75% (v/v) ethanol. After washing with sterilized water, the seeds were sowed on one-half strength MS growth media with 1% (w/v) Suc and 0.8% (w/v) agar at pH 5.8. The plants were grown under continuous light of 130 μmol m−2 s−1 intensity at 22°C. To test the effect of ES2 and ES2-14 on the pathogenicity of Botrytis cinerea on Arabidopsis, wild-type Col-0 plants were grown in soil at 22°C under a 16-h light/8-h dark cycle. To test the effect of ES2 and ES2-14 on the pathogenicity of Magnaporthe oryzae on rice (Oryza sativa), the rice cultivar Nipponbare was used and the plants were grown at 26°C under a 12-h light/12-h dark cycle.
Plant Growth Assay
In order to quantify the inhibitory effect of ES2 and ES2-14 on Arabidopsis root growth, sterilized wild-type Col-0 seeds were sowed on one-half strength MS medium supplemented with different concentrations of ES2 or ES2-14 on 10 × 10 cm square petri dishes with grids. The plates were placed in vertical orientation in the growth chamber for root measurement. Starting from 4 d after the plates were placed in the growth chamber, the plates were scanned using the Epson Perfection V550 scanner every 2 d. The root length of plants was measured using ImageJ. About 100 seedlings were measured from each treatment.
Live Cell Imaging of Fluorescence-Tagged Proteins and Image Analysis
To test the effect of ES2 and ES2-14 on cellular localization of fluorescence-tagged proteins, transgenic plants expressing different fluorescence-tagged proteins were grown on one-half strength MS agar plates for 5 d. The seedlings were incubated in one-half strength MS liquid medium supplemented with different concentrations of ES2 or ES2-14 for 2 h. The images were collected using a Zeiss 710 laser scanning confocal microscope equipped with a 40× water objective with NA 1.2. For imaging GFP-tagged proteins, a 488 nm laser line was used as the excitation source and the emission light of 493–598 nm was collected. For imaging yellow fluorescent protein-tagged proteins, a 514 nm laser line was used as the excitation source and the emission light of 519–621 nm was collected. The detailed procedure for ES2 and ES2-14 treatment and the BFA washout experiment can be found in a previously published protocol (Huang and Zhang, 2018).
To quantify the intracellular localized PIN2 after ES2 and ES2-14 treatment, Z-stack images from treated cells were thresholded and the cell outline was drawn using the polygon selection tool in ImageJ. The intracellular PVCs that contain PIN2:GFP were quantified using the Analyze Particles tool in ImageJ in selected cells. The compartments that are <0.1 μm2 were considered as background and were discarded. A total of eight images from about 90 cells were quantified for each drug treatment. The Feret diameter of PIN2:GFP compartments were measured using the Analyze Particles tool in ImageJ. The fluorescence intensity of PIN2:GFP at the PM was quantified using the Plot Profile tool in ImageJ in selected cells. Fluorescence intensity was calculated by subtracting background fluorescence from peak fluorescence. For each chemical treatment, 120 cells from eight seedlings were quantified. The fluorescence intensity of GFP-AtEXO70A1 was quantified using the Plot Profile tool in ImageJ in selected outer lateral cells. Fluorescence intensity was calculated by subtracting background fluorescence from peak fluorescence. For each drug treatment, 50 cells from nine seedlings were quantified.
To quantify MoEXO70-GFP fluorescence at the tip of germinated conidia, conidia from the M. oryzae MoEXO70-GFP strain harvested from complete medium agar cultures were resuspended to 5 × 105 conidia/mL in sterile water containing 0.1% (v/v) DMSO, 40 μm ES2, or 40 μm ES2-14. Of each conidia suspension, 20 μL was dropped on a hydrophobic coverslip in a moist petri dish at 28°C for 3 h in dark condition. MoEXO70-GFP was examined using a Zeiss LSM710 confocal with 100×/1.3 oil objective under a system-recommended GFP image setting. The fluorescence intensity of MoEXO70-GFP was quantified using the Plot Profile tool in ImageJ in selected hyphal tips. Fluorescence intensity was calculated by subtracting background fluorescence from peak fluorescence. For each drug treatment, 10 hyphal tips were quantified.
Protein Expression and Purification
To obtain full-length AtEXO70A1, MoEXO70, and BcEXO70 for the DARTS assay, the coding sequences of Arabidopsis EXO70A1, B. cinerea EXO70, and M. oryzae EXO70 were cloned from complementary DNA into a modified pRSF-Duet-1 vector. To obtain GFP-labeled full-length MoEXO70 and BcEXO70 protein for the MST assay, the pRSF-Duet-1 vector was further modified by inserting the GFP coding sequence into the vector using SacI/PstI restriction sites. Full-length complementary DNA of MoEXO70 and BcEXO70 was cloned in frame to the C-terminal region of GFP to express the fusion protein using the NotI and PstI/NotI sites. Primers used for cloning are listed in Supplemental Table S1. Verified recombinant clones were transformed into BL21(DE3) competent cells for protein expression. The cells carrying expression plasmids were grown at 37°C until the optical density at 600 nm reached 0.6 and then were induced for protein expression using 0.1 mm isopropyl β-D-1-thiogalactopyranoside at 16°C. After overnight incubation, the cells were lysed using sonication and the fusion protein was purified using a HisTrap HP His-tagged protein purification column in an ÄKTA pure FPLC system (GE Healthcare).
To remove the His-SUMO tag on purified EXO70 proteins, His-ULP1 was incubated together with His-SUMO-EXO70 proteins at 4°C for 12 h and further purified with a HisTrap HP His-tagged protein purification column. Purified protein without the tag was dialyzed overnight for further biochemical experiments. The protein concentration was quantified using UV absorption at 280 nm and calculated with corresponding extinction coefficient. All the purified proteins have a purity of >95%.
Testing the Effect of ES2 and ES2-14 on Fungal Growth
To test the effect of ES2 and ES2-14 on the growth of B. cinerea, different concentrations of the compounds were added to V8 medium (36% [v/v] V8 juice, 0.2% [w/v] CaCO3, and 2% [w/v] agar). To test the effect of ES2 and ES2-14 on the growth of M. oryzae, complete medium (10 g/L D-Glc, 2 g/L peptone, 1 g/L yeast extract, 1 g/L casamino acid, 1× nitrate salts, 1× vitamin, and 15 g/L agar, pH 6.5) was used with different concentrations of the compounds. For M. oryzae, a 1.5-mm-diameter block of culture from a culture plate was used to inoculate the plates with equal volumes of growth media and different concentrations of compounds. For B. cinerea, a 3-mm-diameter block of culture from a culture plate without compound was used to inoculate. The inoculated cultures were grown at 22°C under continuous fluorescence light. The culture plates were scanned with an Epson Perfection V550 scanner and the diameters of colonies were measured 4 d after inoculation for B. cinerea and M. oryzae.
Plant Infection Assays
For M. oryzae infection assays, conidia harvested from complete medium agar cultures were resuspended to 5 × 105 conidia/mL in sterile water containing 0.1% (w/v) DMSO, 80 μm ES2, or 80 μm ES2-14. The second leaf from 3-week-old rice plants was detached and inoculated with two drops (4 μL each) of conidium suspensions. The inoculated leaves were kept in a culture dish containing 0.1% 6-Benzylaminopurine in darkness for 24 h and then transferred to the growth chamber under 12-h/12-h (light/dark) conditions. The inoculated leaves were scanned at 6 dpi and the size of the lesions was measured using ImageJ. To test the effects of ES2 and ES2-14 on M. oryzae appressorium formation, we mixed different concentrations of ES2 or ES2-14 with M. oryzae spores. The spore suspension was applied to the plastic coverslips in petri dishes containing wet paper towels. The formation of appressoria was observed after 24 h of incubation. For B. cinerea infection assays, conidia of strain B05.10 from V8 medium agar cultures were resuspended in 1% Sabouraud Maltose Broth containing 0.1% (w/v) DMSO, 80 μm ES2, or 80 μm ES2-14 to 1.25 × 105 conidia/mL. Of the conidial suspension, 5 μL was applied to the surface of third, fourth, and fifth leaves of 4-week-old Arabidopsis plants. The inoculated plants were kept under a transparent cover under continuous light at 22°C. The size of the lesions was measured 3 d after inoculation.
DARTS Assay
To test the interaction between ES2 or ES2-14 and EXO70 proteins using DARTS assays (Lomenick et al., 2009), purified AtEXO70A1, MoEXO7O, or BcEXO70 was used. We mixed 2.5 μg of purified protein with 2.5 μg of BSA in 200-μL reactions. The protein mixture was incubated with 2% (w/v) DMSO, 400 μm ES2, or 400 μm ES2-14 for 1 h at room temperature with rotating. After incubation, each protein and chemical mixture was divided into three tubes, each with 60 μL of mixture. To the different aliquots was added 1 μL pronase at 1:3,000 or 1:10,000 dilutions from 10 mg/mL stock or 1 μL water. After 30 min of digestion, the reaction was terminated by adding SDS loading buffer and denaturing at 100°C for 5 min. The samples were loaded to SDS-PAGE and the protein was detected using silver staining. The silver-stained gel was scanned and the intensity of the protein band was quantified using ImageJ.
MST Assays
MST assays were carried out using a Monolith NT.115 (NanoTemper) at the Chemical Genomics Facility at Purdue University. To test the interaction between small molecules and AtEXO70, purified AtEXO70A1 with a 74 amino acid deletion at the N-terminal region was labeled with NT-647 via amine conjugation (NanoTemper), as described previously in Zhang et al. (2016). To test the interaction between small molecules and MoEXO70 and BcEXO70, purified recombinant full-length MoEXO70 and BcEXO70 with GFP-tag were used. GFP was used as a negative control for MST experiments. Increasing concentrations of ES2 or ES2-14 were titrated against 50 nm of the protein in a standard MST buffer (50 mm Tris·HCl, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 0.05% [w/v] Tween 20). The small molecules were dissolved in DMSO and the final concentration of DMSO was 5% (v/v) with an equal volume of solution with target protein in all reactions. MST standard capillaries were used to load the samples to the MST instrument. At least three repeated reactions were performed for each test. The MST data were processed using MO.Affinity Analysis Version 2.3 software (NanoTemper). We noticed that the interaction between ES2-14 and NT-647 labeled AtEXO70A1 reduced the intensity of the fluorescence. The reaction curve was thus plotted using raw fluorescence and the concentration of ES2-14, per recommendation of the manual.
Detecting the Effect of ES2-14 on the Exocytosis in Mammalian Cells
To test the effect of ES2 and ES2-14 on the secretory vesicles in Hela cells, plasmids of RnEXO70 and Rab8 were cotransfected to Hela cells. The cells were treated with 40 µm ES2 or ES2-14 for 4 h and the localization of RnEXO70 and Rab8 was detected using a Leica DMI6000 microscope. The transferrin recycling assay was performed as described previously in Zhang et al. (2016).
DSF Assays
Purified tag-free full-length AtEXO70A1, MoEXO70, and BcEXO70 protein were used for the DSF experiment. The experiment was performed with the CFX connect qPCR instrument (BioRad), following published protocols (Niesen et al., 2007; Vivoli et al., 2014). Briefly, purified protein was diluted and mixed with ES2, ES2-14, and ampicillin in a 96-well, white, low-profile qPCR plate (USA Scientific). The final incubation buffer contains 1 μm protein, 50 mm Tris·HCl (pH 8.0), 150 mm NaCl, 2% (v/v) DMSO, 5× SYPRO orange (Invitrogen). Serial gradient dilution of ES2, ES2-14, and ampicillin was established from 800 μm to 22 μm in a 3/5 manner (800 μm, 480 μm, 288 μm, etc.). Each well contains 30 μL mixture. The plate was placed in the qPCR instrument to run a melting curve from 25°C to 95°C at 1°C per 0.5 min increment and read SYBR-green fluorescence every 0.5 min. Three repeats were performed for each protein-ligand combination. The data were exported from CFX manager 3.1 software and analyzed in Excel and GraphPad.
NMR Chemical Shift Assays
To detect the effect of different proteins on the chemical shifts of protons in ES2 and ES2-14 molecules, purified full-length EXO70 proteins without any tag were added to 200 μm of ES2 or ES2-14. All one-dimensional proton spectra were acquired on a Bruker Avance-III 800 MHz spectrometer equipped with a cryo QCI probe. The buffer for each NMR sample contained 50 mm Tris·HCl (pH 8), 0.5 m NaCl, 5% (v/v) D2O, and 0.5% (v/v) DMSO. All free induction decays were collected at 25°C with WATERGATE, line broadened by 1 Hz, Fourier-transformed, phased, and baseline corrected. For clarity, only the aromatic regions of ES2 and ES2-14 are shown.
Statistical Analysis
Statistical analyses were carried out using Excel 2010 for the two-tailed student’s t test and GraphPad Prism 6 for Tukey’s one-way ANOVA multiple comparisons. The type of statistical test and the numbers of samples used in the test have been described in the corresponding figure legends.
Protein Sequence Analysis
Protein sequence alignment of AtEXO70A1, RnEXO70, BcEXO70 and MoEXO70 was performed using MAFFT Version 7 with G-INS-i method (Katoh et al., 2017). Alignment was visualized by JalView software and colored by Clustal format. Phylogenetic analysis of AtEXO70A1, RnEXO70, BcEXO70 and MoEXO70 was constructed using unroot Neighbor Joining (NJ) tree with MEGA7 software (Kumar et al., 2016).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AtEXO70A1 (Q9LZD3), RnEXO70 (O54922), BcEXO70 (A0A384JDU6), and MoEXO70 (A0A060CW34).
SUPPLEMENTAL DATA
The following supplemental information is available.
Supplemental Figure S1. ES2-14 does not disturb general membrane system in Arabidopsis.
Supplemental Figure S2. ES2-14 causes PIN2:GFP to localize to PVC.
Supplemental Figure S3. Coomassie staining of proteins used in biochemical binding assays.
Supplemental Figure S4. NMR chemical shift of protons on ES2 and ES2-14 in response to AtEXO70A1.
Supplemental Figure S5. ES2-14 does not affect the cellular localization of RnEXO70 in Hela cells.
Supplemental Figure S6. ES2-14 does not inhibit exocytosis in Hela cells.
Supplemental Figure S7. Comparison of AtEXO70A1, RnEXO70, MoEXO70 and BcEXO70.
Supplemental Figure S8. Thermophoresis binding curve of purified GFP with different concentrations of ES2 or ES2-14.
Supplemental Figure S9. NMR chemical shift of protons on ES2 and ES2-14 in response to MoEXO70.
Supplemental Figure S10. NMR chemical shift of protons on ES2 and ES2-14 in response to BcEXO70.
Supplemental Table S1. Primers used for cloning of EXO70s.
Acknowledgments
We thank Dr. Jin-Rong Xu at Purdue University for helping us with substantial editing of the manuscript and advice in M. oryzae experiments. We thank Dr. Cankui Zhang at Purdue University for providing us with rice seeds and sharing the CFX connect qPCR instrument. We thank Dr. Nick Talbot at The Sainsbury Laboratory for providing the MoEXO70-GFP fungal strain. We thank Dr. M. Catherine Aime at Purdue University for helping with fungal strain maintenance. We thank the Chemical Genomics Facility at the Purdue Institute for Drug Discovery for providing us access to the MST equipment and Dr. Lan Chen and Li Wu at Purdue University for discussions on performing MST assays.
Footnotes
This project was funded by Purdue University, as part of AgSEED Crossroads funding to support Indiana's Agriculture and Rural Development.
Articles can be viewed without a subscription.
References
- Ablinger E, Leitgeb S, Zimmer A (2013) Differential scanning fluorescence approach using a fluorescent molecular rotor to detect thermostability of proteins in surfactant-containing formulations. Int J Pharm 441: 255–260 [DOI] [PubMed] [Google Scholar]
- Arai M, Ferreon JC, Wright PE (2012) Quantitative analysis of multisite protein-ligand interactions by NMR: Binding of intrinsically disordered p53 transactivation subdomains with the TAZ2 domain of CBP. J Am Chem Soc 134: 3792–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athuluri-Divakar SK, Vasquez-Del Carpio R, Dutta K, Baker SJ, Cosenza SC, Basu I, Gupta YK, Reddy MV, Ueno L, Hart JR, et al. (2016) A small molecule RAS-mimetic disrupts RAS association with effector proteins to block signaling. Cell 165: 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud MGJ, Lin-Shiao E, Cardote T, Tallant C, Pschibul A, Chan KH, Zengerle M, Garcia JR, Kwan TT, Ferguson FM, et al. (2014) Chemical biology. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes. Science 346: 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ebbole DJ, Wang Z (2015) The exocyst complex: Delivery hub for morphogenesis and pathogenesis in filamentous fungi. Curr Opin Plant Biol 28: 48–54 [DOI] [PubMed] [Google Scholar]
- Cho M, Lee SH, Cho HT (2007) P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19: 3930–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RA, Peremyslov VV, Van Why S, Moussaoui I, Ketter A, Cool R, Moreno MA, Vejlupkova Z, Dolja VV, Fowler JE (2018) A broadly conserved NERD genetically interacts with the exocyst to affect root growth and cell expansion. J Exp Bot 69: 3625–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, et al. (2012) The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13: 414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drdová EJ, Synek L, Pečenková T, Hála M, Kulich I, Fowler JE, Murphy AS, Zárský V (2013) The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. Plant J 73: 709–719 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Synek L, Pecenková T, Toupalová H, Cole R, Drdová E, Nebesárová J, Sedinová M, Hála M, Fowler JE, et al. (2010) The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. Plant Cell 22: 3053–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Xu T, Zhu L, Wen M, Yang Z (2009) A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr Biol 19: 1827–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki K, Abe Y, Ito A, Saitoh H, Yoshida K, Kanzaki H, Kanzaki E, Utsushi H, Yamashita T, Kamoun S, et al. (2015) Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. Plant J 83: 875–887 [DOI] [PubMed] [Google Scholar]
- Furukawa A, Konuma T, Yanaka S, Sugase K (2016) Quantitative analysis of protein-ligand interactions by NMR. Prog Nucl Magn Reson Spectrosc 96: 47–57 [DOI] [PubMed] [Google Scholar]
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J (2007) Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesbert S, Schumacher J, Kupas V, Espino J, Segmüller N, Haeuser-Hahn I, Schreier PH, Tudzynski P (2012) Identification of pathogenesis-associated genes by T-DNA-mediated insertional mutagenesis in Botrytis cinerea: A type 2A phosphoprotein phosphatase and an SPT3 transcription factor have significant impact on virulence. Mol Plant Microbe Interact 25: 481–495 [DOI] [PubMed] [Google Scholar]
- Giraldo MC, Dagdas YF, Gupta YK, Mentlak TA, Yi M, Martinez-Rocha AL, Saitoh H, Terauchi R, Talbot NJ, Valent B (2013) Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat Commun 4: 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Escudero J, Moreno V, Martín-Alonso M, Hernández-Riquer MV, Feinberg T, Colmenar Á, Calvo E, Camafeita E, Martínez F, Oudhoff MJ, et al. (2017) E-cadherin cleavage by MT2-MMP regulates apical junctional signaling and epithelial homeostasis in the intestine. J Cell Sci 130: 4013–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Xu C, Wu D, Zhao Y, Qiu Y, Wang X, Ouyang Y, Cai B, Liu X, Jing S, et al. (2018) Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat Genet 50: 297–306 [DOI] [PubMed] [Google Scholar]
- Guo W, Grant A, Novick P (1999) Exo84p is an exocyst protein essential for secretion. J Biol Chem 274: 23558–23564 [DOI] [PubMed] [Google Scholar]
- Gupta YK, Dagdas YF, Martinez-Rocha AL, Kershaw MJ, Littlejohn GR, Ryder LS, Sklenar J, Menke F, Talbot NJ (2015) Septin-dependent assembly of the exocyst is essential for plant infection by Magnaporthe oryzae. Plant Cell 27: 3277–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider MR, Munson M (2012) Exorcising the exocyst complex. Traffic 13: 898–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Zhang C (2018) Use endosidin2 to study plant exocytosis and vacuolar trafficking. Methods Mol Biol 1789: 167–175 [DOI] [PubMed] [Google Scholar]
- Jásik J, Bokor B, Stuchlík S, Mičieta K, Turňa J, Schmelzer E (2016) Effects of auxins on PIN-FORMED2 (PIN2) dynamics are not mediated by inhibiting PIN2 endocytosis. Plant Physiol 172: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol 9: 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Wabnik K, Martinière A, Łangowski Ł, Willig K, Naramoto S, Leitner J, Tanaka H, Jakobs S, Robert S, Luschnig C, Govaerts W, et al. (2011) Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol Syst Biol 7: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulich I, Cole R, Drdová E, Cvrcková F, Soukup A, Fowler J, Zárský V (2010) Arabidopsis exocyst subunits SEC8 and EXO70A1 and exocyst interactor ROH1 are involved in the localized deposition of seed coat pectin. New Phytol 188: 615–625 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, et al. (2009) Target identification using drug affinity responsive target stability (DARTS). Proc Natl Acad Sci USA 106: 21984–21989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Vert G (2014) The dynamics of plant plasma membrane proteins: PINs and beyond. Development 141: 2924–2938 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Kondo M, Nishimura M, Hara-Nishimura I (2003) A novel ER-derived compartment, the ER body, selectively accumulates a beta-glucosidase with an ER-retention signal in Arabidopsis. Plant J 33: 493–502 [DOI] [PubMed] [Google Scholar]
- Mayers JR, Hu T, Wang C, Cárdenas JJ, Tan Y, Pan J, Bednarek SY (2017) SCD1 and SCD2 form a complex that functions with the exocyst and RabE1 in exocytosis and cytokinesis. Plant Cell 29: 2610–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2: 2212–2221 [DOI] [PubMed] [Google Scholar]
- O'Neill P. R., Castillo-Badillo J. A., Meshik X., Kalyanaraman V., Melgarejo K. Gautam N.. 2018. Membrane flow drives an adhesion-independent amoeboid cell migration mode. Dev Cell 46: 9–22.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reibarkh M, Malia TJ, Wagner G (2006) NMR distinction of single- and multiple-mode binding of small-molecule protein ligands. J Am Chem Soc 128: 2160–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel SA, Dijkman PM, Lea WA, van den Bogaart G, Jerabek-Willemsen M, Lazic A, Joseph JS, Srinivasan P, Baaske P, Simeonov A, et al. (2013) Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59: 301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge MD, Hage DS, Harbison GS, Powers R (2008) Estimating protein-ligand binding affinity using high-throughput screening by NMR. J Comb Chem 10: 948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Anderson RG, Ichimura K, Pecenkova T, Reuter P, Žársky V, McDowell JM, Shirasu K, Trujillo M (2012) The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24: 4703–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synek L, Schlager N, Eliás M, Quentin M, Hauser MT, Zárský V (2006) AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J 48: 54–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P (1996) The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J 15: 6483–6494 [PMC free article] [PubMed] [Google Scholar]
- Vivoli M, Novak HR, Littlechild JA, Harmer NJ (2014) Determination of protein-ligand interactions using differential scanning fluorimetry. J Vis Exp 91: 51809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukašinović N, Oda Y, Pejchar P, Synek L, Pečenková T, Rawat A, Sekereš J, Potocký M, Žárský V (2017) Microtubule-dependent targeting of the exocyst complex is necessary for xylem development in Arabidopsis. New Phytol 213: 1052–1067 [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang L, Tang Y, Tang R, Jing Y, Zhang C, Zhang B, Li X, Cui Y, Zhang C, et al. (2017) Arabidopsis choline transporter-like 1 (CTL1) regulates secretory trafficking of auxin transporters to control seedling growth. PLoS Biol 15: e2004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Guo W (2015) The exocyst at a glance. J Cell Sci 128: 2957–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Scheres B (2005) Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Brown MQ, van de Ven W, Zhang ZM, Wu B, Young MC, Synek L, Borchardt D, Harrison R, Pan S, et al. (2016) Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc Natl Acad Sci USA 113: E41–E50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Immink R, Liu CM, Emons AM, Ketelaar T (2013) The Arabidopsis exocyst subunit SEC3A is essential for embryo development and accumulates in transient puncta at the plasma membrane. New Phytol 199: 74–88 [DOI] [PubMed] [Google Scholar]
- Zhu X, Li S, Pan S, Xin X, Gu Y (2018) CSI1, PATROL1, and exocyst complex cooperate in delivery of cellulose synthase complexes to the plasma membrane. Proc Natl Acad Sci USA 115: E3578–E3587 [DOI] [PMC free article] [PubMed] [Google Scholar]