Abstract
A new approach to single-molecule DNA sequencing in which dNTPs, released by pyrophosphorolysis from the strand to be sequenced, are captured in microdroplets and read directly could have substantial advantages over current sequence-by-synthesis methods; however, there is no existing method sensitive enough to detect a single nucleotide in a microdroplet. We have developed a method for dNTP detection based on an enzymatic two-stage reaction which produces a robust fluorescent signal that is easy to detect and process. By taking advantage of the inherent specificity of DNA polymerases and ligases, coupled with volume restriction in microdroplets, this method allows us to simultaneously detect the presence of and distinguish between, the four natural dNTPs at the single-molecule level, with negligible cross-talk.
INTRODUCTION
It is well documented (1,2) that, under the correct conditions, the DNA polymerization reaction can proceed in reverse, releasing deoxyribonucleoside triphosphates (dNTPs) sequentially from double-stranded DNA. Coupling this pyrophosphorolysis reaction with the ability to accurately detect and identify single dNTPs offers the potential for a novel approach to direct single molecule sequencing. If the DNA to be sequenced is immobilized, and microdroplets containing an enzyme capable of pyrophosphorolysis are passed over it at a suitably high rate, the dNTPs liberated will be individually captured in microdroplets. Subsequent merging of these droplets with reagents for dNTP identification would permit the microdroplets to serve as distinct reaction vessels (3–8) while simultaneously allowing for the order of dNTPs, and therefore the sequence, to be maintained.
Herein, we describe a method for dNTP detection that is suitable for identifying single dNTP molecules in microdroplets, is linear over a sufficient range of concentrations, can simultaneously detect and distinguish between each of the four natural dNTPs with negligible amounts of crosstalk and results in a robust fluorescence signal that is easy to read and process. Moreover, it requires only simple instrumentation (thermocycler and fluorescence plate reader for bulk samples, standard fluorescence microscopy for droplets) and small quantities of reagent. By incorporating a signal-amplifying reaction in our detection method, we overcome the inherent difficulty of obtaining sufficient signal strength from a single analyte molecule (9,10), while maintaining a level of specificity that is inherent to enzymatic reactions. As well as being the key component in the realization of a single molecule sequencing system, such a chemistry also offers the potential for quantification of dNTPs from very low numbers of cells, opening the door to new diagnostic methods.
Currently, detection and quantification of dNTPs is a vital tool in the study of such diverse diseases as cancer (11–13) and HIV infection (14,15) as well as therapeutic approaches toward them (16,17). DNA replication and repair require that the four canonical dNTPs be present in the correct concentrations and ratios, and any deviation could result in increased mutation rates or incorrect DNA repair (18–20). dNTP imbalances can also be indicative of a variety of pathologies involving nucleotide metabolism (21–25) and may even trigger apoptosis (26). The most sensitive nucleotide assay in general use—a photometric luciferin/luciferase assay—has picomolar sensitivity (27), but is limited to the detection of adenosine triphosphate (ATP). For other triphosphates—including dNTPs—‘classical’ analytical techniques, such as liquid chromatography (with high nanomolar sensitivity) (28–32), often coupled to mass spectrometry (increasing sensitivity to the low nanomolar range) (28,33–37), as well as biochemical assays (38–40), often relying on radioactive isotopes (41–44), are required. Even the most sensitive of these methods require about 75 femtomoles or more (40) of dNTPs, or concentrations in the nanomolar range for typical analyte volumes. With none of these methods is it possible to identify single dNTPs or quantify the dNTP concentration from very low numbers of cells.
MATERIALS AND METHODS
In a typical detection reaction, the following ingredients were combined in our buffer (see Supplementary Table S8 for buffer composition) for a total volume of 135 μl: Probe oligos in four colors (4–10 nM, depending on color), ligation oligos in four colors (10–40 nM, depending on color), capture oligos in four colors (500 pM), spermine (1 mM), spermidine (1 mM), Tetraethylenepentamine (0.015%), Bst L. F. DNA polymerase (28.6 U/ml), Escherichia coli DNA ligase (17.85 U/ml), PfuUltra II Fusion HS DNA polymerase (1.43 Reactions/ml), and Thermostable Inorganic Pyrophosphatase (TIPP, 66.6 U/ml). From this mixture, two samples of 63 μl were taken and 7 μl of dNTP solution (250 pM) or water were added. The samples were split into aliquots of 15 μl each and incubated (10 min at 37°C/90 min at 69.5°C). If desired, some samples were removed during this incubation to provide data points at intermediate times. After incubation, samples were transferred to a 384-well microplate (10.5 μl per well) and read on a BMG Labtech Clariostar microplate reader. See Supplementary Tables S1, 2, 3 and 7 for full details.
Microdroplet samples were produced by combining, in ratio 1:4, the detection reaction mix described above with 1% ABIL EM 90 (a non-ionic, polymeric silicone based surfactant, Evonik) in paraffin oil (Sigma-Aldrich) followed by 15 min on a vortex mixer (Grant Instruments). For incubation and measurement a portion of the resulting droplets was pipetted into the bottom of a well formed from a PMMA coated fused silica substrate (175 μm thickness, JGS2 grade, University Wafer) bonded to an aluminum annulus. This well is filled with 1% ABIL EM90 in paraffin oil then capped with a circular quartz coverslip (UQG Ltd).
Incubation was performed on a series of calibrated hotplates (10 min at 37°C/90 min at 69.5°C). Droplet shrinkage was prevented by sealing the well in an aluminum chamber partially filled with water to maintain high humidity.
Fluorescent images of microdroplets were acquired using a custom fluorescent microscope consisting of an Andor Zyla 5.5 sCMOS camera, four laser excitation lines (532, 594, 640, 701 nm), a filter cube changer to select the appropriate excitation, dichroic and emission filters (see Supplementary Table S10), and a galvanometer scanning mirror system to raster the laser over the area of interest. The exposure time per laser for each 300 × 300 μm field of view was 30 s.
Further experimental details can be found in the Supplementary Data section.
RESULTS AND DISCUSSION
Detection reaction
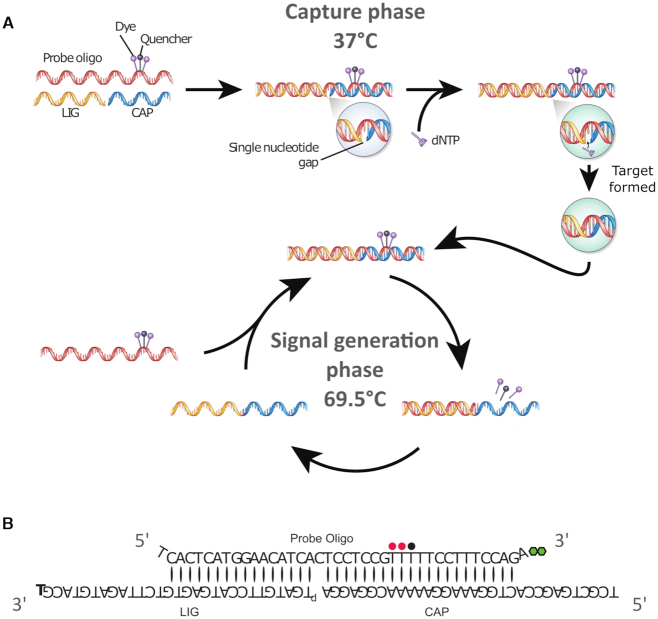
Our detection reaction is based on a molecular scaffold of DNA oligomers (Figure 1A) allowing us to take advantage of the specificity of DNA polymerases and DNA ligases to keep error rates to a minimum. The reaction consists of two phases—a capture phase and a signal generation phase. In the capture phase, at 37°C, a labeled DNA oligomer (‘Probe Oligo’) anneals to two shorter oligomers (‘Capture Oligo’ or ‘CAP’ and ‘Ligation Oligo’ or ‘LIG’, respectively) such that a 1-nt gap is created. The Probe Oligo is decorated with 2–4 fluorophores and one fluorescence quencher (see Supplementary Data); the fluorophores and quencher are in close contact with one another, resulting in >99% quenching of fluorescence emission. A complementary dNTP is incorporated into the gap by a DNA polymerase, and a DNA ligase then ligates the extended CAP to LIG, forming a complete dsDNA construct consisting of Probe Oligo and ‘Target’ (the ligated CAP and LIG). For the signal generation phase, the temperature is raised to 69.5°C at which point unligated CAP and LIG oligos dissociate from the Probe Oligo, but ligated Target remains annealed. A second DNA polymerase with strong double-strand-dependent 3′→5′ exonuclease activity digests the dye-labeled Probe Oligo annealed to the Target, thereby releasing the dye-labeled deoxyribonucleotides, which restores their fluorescence emission. A new Probe Oligo can now anneal to the Target, allowing the cycle to repeat multiple times, isothermally. The use of four Probe Oligos, each with a different sequence and capturing one of the four natural dNTPs, labeled with dyes fluorescing at different wavelengths (ATTO 532, ATTO 594, ATTO 655, ATTO 700), allows the different types of dNTP to be identified.
Figure 1.
(A) Mechanism of dNTP detection. In the first (capture) phase, three oligonucleotides—LIG, CAP and Probe Oligo—anneal to form a gapped double strand. The Probe Oligo carries fluorophores and a quencher which suppresses their fluorescence. A dNTP molecule, complementary to the base in the gap, is incorporated by a DNA polymerase and the nick is ligated, forming a fully double-stranded molecule consisting of Probe Oligo and Target. In the second (signal generation) phase, the Probe Oligo is digested by the exonuclease activity of a polymerase, releasing fluorophores which can now fluoresce. A new Probe Oligo can now anneal to the Target strand, allowing the cycle to repeat. (B) Schematic representation of the oligos used to capture dGTP, showing mismatches at both ends of the Probe Oligo (top, bearing fluorophores (red) and quencher (black)), as well as a 3′-hexanediol protection group (green). The protection group on LIG (T, bottom left) is a 5′-3′-inverted dT. Note the overhangs on the 5′ and 3′ ends of the Probe Oligo that help ensure that the Probe Oligo is the target for exonuclease digestion.
Oligo design
The DNA oligomers comprising our detection scaffold required careful design (see Figure 1B). In addition to the need to have CAP and LIG annealed at the capture temperature, protecting groups are needed to prevent unintended digestion reactions. Unintended digestion may result in false-signal due to release of fluorescent dye from Probe Oligo or an inability to generate signal due to digestion of Target. To this end, the LIG oligo features a 5′-3′ inverted deoxythymidine at the 3′ terminus as this group was found to offer the best protection (see Supplementary Figure S11). CAP and LIG were designed to provide a 5′ overhang at the 3′-end of Probe Oligo and a 3′-overhang at the 5′ end of the Probe Oligo, thus directing the exonuclease toward digestion of the Probe Oligo as well as preventing unintended dNTP capture at the 3′ end of LIG. For the latter reason, a mismatch between the 3′-end of the Probe Oligo and CAP is also required. The lengths of the annealed regions in the dsDNA construct were chosen with attention to their melting behavior. The three oligos must hybridize stably during the capture phase at 37°C, but non-ligated CAP and LIG oligos must dissociate from the Probe Oligo during the signal generation phase at 69.5°C in order to differentiate between constructs that captured a dNTP and those that did not.
The design of the Probe Oligo in particular is crucial to efficient signal generation. As the exonuclease activity is not entirely limited to dsDNA constructs, there is always some residual single-strand activity that results in false signal. In order to minimize this, it is crucial to place the dye modifications away from the 3′-end of the Probe Oligo; a distance of 12 bases was optimal for the 40 base Probe Oligos used here. In addition a 3′-hexanediol group was added to further reduce single-strand exonuclease activity. At the same time, the dye modifications must not interfere with capture and ligation. We found that a distance of 5–10 bases from the capture site was sufficient to ensure that the capture polymerase and ligase are not impeded. Moving the dye modifications too close to the 5′-end of the Probe oligo led to the dsDNA construct melting before digestion had run as far as the dyes, thereby reducing signal. For DNA sequencing, the ability to simultaneously detect and distinguish all four dNTPs is required, meaning that it is necessary to design four orthogonal sets of oligos with four different fluorophores (colors) and sequences that need to work optimally at the same temperatures. It is essential to ensure identical melting behavior, which was achieved by matching oligomer length and GC content (see Supplementary Table S6 for oligomer sequences). Where required, intentional mismatches in the sequences of the Probe Oligo and CAP or LIG were added to lower the melting temperature.
Choice of enzymes
The capture polymerase needs to be able to rapidly and efficiently incorporate all four dNTPs into single base gaps with high fidelity and have no exonuclease activity. The ligase should be efficient and also possess high fidelity, while the exonuclease should be able to differentiate between dsDNA and ssDNA and be tolerant of the dyes and quenchers used. Using a combination of several DNA-binding enzymes in the same reaction is inherently difficult, as competitive binding can lead to inhibition of required enzymatic activity. By performing the capture and signal generation phases at two different temperatures, we minimize these unfavorable interactions. A large number of polymerases, exonucleases and ligases were screened, using multiple buffers (see Supplementary Figures S1–10 and Tables S8–9). The capture phase is performed at 37°C by Bst L. F. DNA polymerase and E. coli ligase. For the signal generation phase, the 3′→5′ exo-active hot-start DNA polymerase PfuUltra II Fusion HS is used, which is active above 50°C and was found to have optimal activity in our reaction at 69.5°C. At this temperature, E. coli ligase is heat-inactivated within minutes, making any subsequent ligation impossible. The two phases of the reaction are, therefore, decoupled, allowing the signal generation phase to remain isothermal.
Typical results in four colors
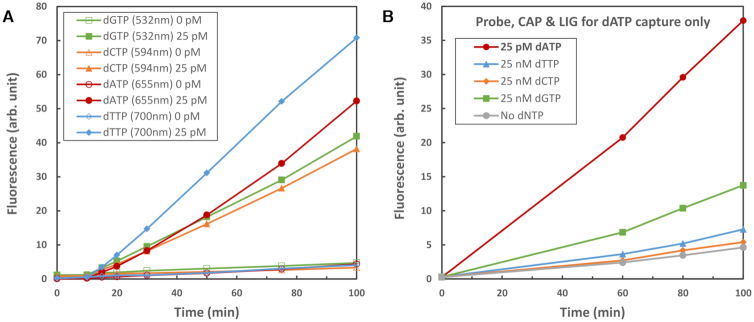
Figure 2A shows typical results for dNTP detection in four colors, performed simultaneously. To this end, four sets of detecting oligos (4× Probe Oligo, 4× CAP and 4× LIG) were combined and a mixture containing dATP, dCTP, dGTP and dTTP (final concentration of each: 25 pM) or water (0 pM), was added. Incubation time was typically 10 min at 37°C for capture and ligation, during which no signal is generated, followed by 90 min at 69.5°C for the signal generation phase, during which a linear increase in signal with time is observed. Signal detection was carried out in a standard fluorescence plate reader and ratios of signal:false-signal (the signal from dNTP-free reactions) of 10–16 were observed, depending on color. Comparing the signal intensity with that obtained by digesting Probe Oligo to completion with a combination of efficient single-strand exonucleases we estimate that, depending on the oligos used, we digest approximately 22–42% of the available Probe Oligos. This implies that each target molecule (hence each dNTP molecule) leads to the digestion of about 100 Probe Oligos (see Supplementary Tables S4–5), releasing 200–400 fluorophores for detection. If desired, the signal generation phase can be shortened (e.g. for higher dNTP concentrations) or extended (for samples closer to the detection limit).
Figure 2.
(A) Typical results of 25 pM dNTP detection, compared to 0 pM (water) controls, performed in four colors (dGTP detection in 532 nm, dCTP detection in 594 nm, dATP detection in 655 nm, dTTP detection in 700 nm) in a single tube. Incubation was 10 min at 37°C (no signal increase), followed by 90 min at 69.5°C (linear signal increase). (B) Testing for chemical cross-talk by adding either intended or ‘incorrect’ dNTP to the reaction. The individual capture channels were tested separately and the 655 nm channel is shown. Note that in each case the intended dNTP is present at 1000-fold lower concentration than the three others.
Interestingly, even in absence of the corresponding dNTP, we observe a small amount of signal being generated. This false-signal can be caused by a number of undesired reaction pathways or by dNTP contamination. For example, the exonuclease activity employed to generate signal is not completely limited to dsDNA constructs. It is therefore not surprising that small amounts of the Probe Oligo are digested as ssDNA in the absence of dNTP capture and oligo ligation. Alternatively, a small percentage of CAP and/or LIG oligo might transiently anneal to Probe Oligo even at the elevated temperature of the signal generation phase, or, stabilized by enzymes bound to them, might not melt off quickly enough upon heating, thereby presenting an easily digestible dsDNA construct to the exonuclease. It is also possible that the enzymes used in our experiments could be contaminated with dNTPs as they are isolated from live cell cultures.
Specificity of detection—cross-talk
Owing to the high natural fidelity of the polymerase (45) and ligase (46), our detection reaction shows high specificity for the intended dNTPs. In an experiment where 25 nM ‘incorrect’ dNTPs were added to the Probe, CAP and LIG oligos designed to capture a particular dNTP (655 nm channel shown in Figure 2B, others in Supplementary Figure S12), the signal detected is less than that from 25pM of the ‘correct’ dNTP. This is despite the fact that 25 nM of ‘incorrect’ dNTP is a concentration 1000 times higher than that of the dNTP for which the oligos are intended to be specific, which implies a specificity better than 99.9%. Given that the dNTP stocks used in these experiments are ≥99% pure, the small amounts of false signal could be due to trace amounts (<0.1%) of the incorrect dNTPs in our stocks.
Detection range
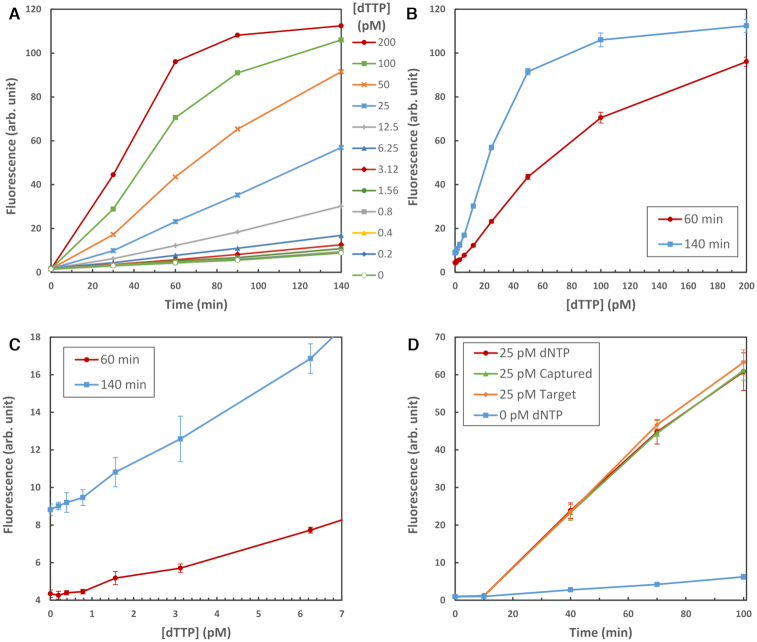
Our reaction conditions were optimized for 25 pM dNTP concentration to emphasize maximum sensitivity; Figures 3A and B show that a range of concentrations are detectable. Depending on incubation time, the signal is approximately linear up to a concentration of 50–100 pM. At higher concentrations, quantitative detection requires a dilution of the dNTP sample prior to detection. More interestingly, we can readily detect concentrations of dNTP as low as 1 pM, as is evident from Figure 3C. This is the approximate equivalent of the extract obtained from a few 10’s of eukaryotic cells and this limit is only due to our relatively large typical working volume of 15 μl. For an unknown sample a standard calibration curve would be measured and compared against for determination of dNTP concentration.
Figure 3.
(A) Signal intensity at [dTTP] from 0.2 to 200 pM; incubation was 10 min at 37°C, followed by 130 min at 69.5°C. (B) Plot of fluorescence intensity versus concentration of dTTP at 60 min and at 140 min. (C) Detail of B) showing the linearity at lower concentrations of dTTP. (D) Comparison of dNTP detection with signal obtained from CAP oligos with a pre-captured dNTP added during oligo synthesis, and a completed target oligo (i.e. CAP, captured dNTP and LIG) synthesized as a whole. This allows detection efficiency to be estimated. In all cases, the data points are the mean of six measurements and the error bars are ±1 standard deviation.
Capture and ligation efficiency
For this chemistry to be useful in a single molecule sequencing system capture and ligation of dNTPs should proceed close to completion. If not, missed bases will result in deletion errors. To estimate the efficiency with which our chemistry can detect dNTPs, we compared the signal levels obtained from 25 pM dNTPs with those from reactions where instead of dNTPs we added 25 pM of either CAP oligo with the corresponding base synthesized in place as if already captured, or synthesized Target oligo to simulate that capture and ligation had already occurred. This is shown in Figure 3D and as can be seen the signal traces lie close to each other, all within one standard deviation of the dNTP measurements, indicating a high efficiency of capture and ligation.
Detection of single dNTPs
Microdroplets are a convenient platform for fluidic manipulation and their use as reaction vessels in biology and chemistry is well documented (3–8). Building upon the reaction detailed above, one can see that appropriate reduction of the reaction volume should allow detection of a single dNTP. The central concept is that individual aqueous droplets, separated by a continuous oil phase, are independent. This, and their small scale, allows simple parallel testing for the presence of each particular dNTP type in a large number of droplets.
To demonstrate the detection of single dNTPs we use vortex mixing to emulsify a sample containing 12.5 pM of each of dCTP, dGTP, dTTP and 37.5 pM of dATP. The additional 25 pM dATP was added as an internal control to highlight changing droplet occupancy with dNTP concentration. The continuous phase is a hydrocarbon oil containing a polymeric surfactant. A portion of the resulting emulsion is added to a well with a fused silica base for incubation and subsequent measurement. The emulsion contains a wide distribution of droplet sizes from which we analyse only the size range of interest. While control of droplet size is better with microfluidics we use this method here as we want to examine a range of droplet sizes which allows us to better illustrate that we truly are detecting single dNTP molecules.
The probability of a droplet containing k dNTPs of a particular type is expected to follow a Poisson distribution, P(k) = λk e−λ / k!, with the expectation value (mean number of dNTPs per droplet) given by λ = NA c V, where NA is the Avogadro constant, c is molar dNTP concentration and V is droplet volume. Occupancy probabilities are given for 12.5 pM dNTP concentration and a range of spherical droplet diameters in Table 1. The concentrations of all other reagents are chosen to be high (>500 pM for all reagents sold with known molarity), such that many copies are present in each droplet and we do not see stochastic effects arising from them.
Table 1.
Droplet volume, mean bases per droplet and occupation probabilities for a 12.5 pM dNTP concentration at a selection of spherical droplet diameters
| Droplet diameter (μm) | Droplet volume, V (fL) | Mean dNTP number per droplet, λ | P(0) | P(1) | P(2) | P(≥3) |
|---|---|---|---|---|---|---|
| 3.0 | 14.14 | 0.11 | 0.899 | 0.096 | 0.005 | 0.000 |
| 4.0 | 33.51 | 0.25 | 0.777 | 0.196 | 0.025 | 0.002 |
| 5.0 | 65.45 | 0.49 | 0.611 | 0.301 | 0.074 | 0.014 |
| 6.0 | 113.10 | 0.85 | 0.427 | 0.363 | 0.155 | 0.055 |
| 7.0 | 179.59 | 1.35 | 0.259 | 0.350 | 0.236 | 0.155 |
The droplet diameter range of 3–7 μm is chosen as a balance between having ≤1 dNTP per droplet with a good signal:false-signal ratio, shown to be true for 12.5 pM in Figure 3A, and having droplets large enough to image using standard optical instrumentation. Moving to larger droplets would require reduction of the dNTP concentration in order to maintain ≤1 dNTP per droplet but this would reduce the signal:false-signal ratio.
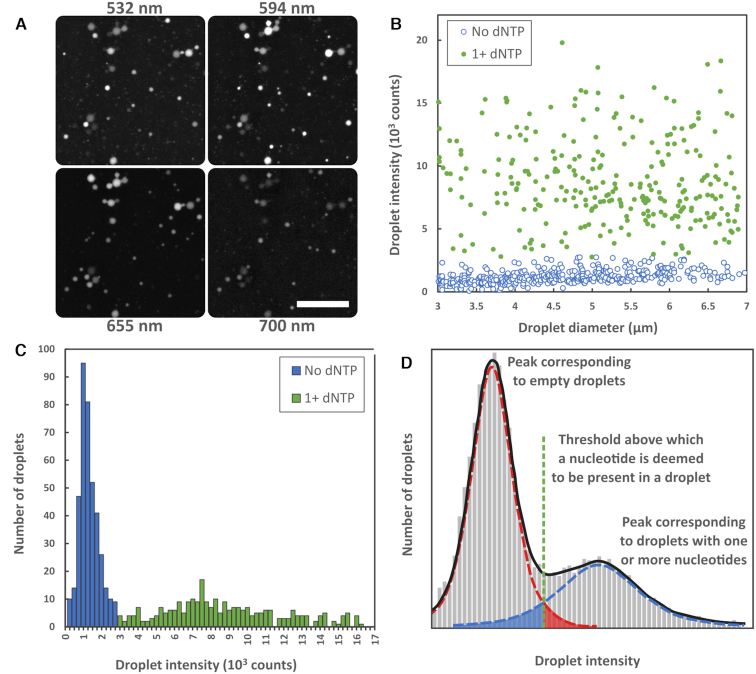
After incubation, for every field of view of droplets, a fluorescence image in each color channel is measured. An example is shown in Figure 4A. A brightfield image is also acquired and used to identify droplet positions and diameters via a droplet finding algorithm developed in-house (see Supplementary Data for more detail). For all droplets found within the desired size range we calculate, in each fluorescence channel, the average counts per pixel from the central portion (defined by a circle of 0.35 droplet diameter) which we call the ‘droplet intensity’. This is plotted against droplet diameter for the 594 nm channel (dCTP) in Figure 4B, with the other channels shown in Supplementary Figure S14. As the fluorescence detection is not confocal we expect the droplet intensity for a given concentration of fluorophores to scale linearly with droplet diameter (as total fluorophore number is proportional to volume, while the number of pixels a droplet covers is proportional to droplet area). Thus the false-signal scales linearly with droplet diameter. For droplets containing dNTPs there will be the additional fluorescence from the detection reaction which will depend upon the local dNTP concentration in the droplet (determined by number of dNTPs in the droplet and inversely proportional to the droplet volume), but which is limited by the point at which the reaction saturates (cf. Figure 3A).
Figure 4.
(A) Typical fluorescence images of microdroplets. Scale bar is 50 μm. (B) Droplet intensity (average counts per pixel from central portion of droplet) versus diameter extracted from the 594 nm fluorescence channel. (C) Histogram showing frequency of occurrence versus droplet intensity for the 594 nm channel in the 3–7 μm diameter range. Two peaks are visible, one for droplets which contain no nucleotide (blue) and one for droplets which contain one (or more) nucleotides (green). D) An illustrative example of the droplet intensity versus frequency histogram including the threshold above which a droplet is deemed to contain a nucleotide and the graphical representation of the false negative (solid blue region) and false positive errors (solid red region). The data for all dNTP channels is included in the Supplementary Data (Supplementary Figures S13–15).
For a particular diameter range the histogram of frequency of occurrence versus droplet intensity can be extracted, as shown in Figure 4C for the 594 nm channel in the 3–7 μm diameter range (see Supplementary Figure S15 for other color channels). The vertical axis records the number of droplets with intensity within a certain range. Figure 4D depicts an illustrative, annotated version of this histogram. These histograms show two distinct peaks, the lower intensity peak corresponds to droplets with no dNTP and the higher intensity peak corresponds to droplets with one or more nucleotides. This is strong evidence for the ability to detect single dNTPs.
Were the intrinsic spread in droplet intensity small enough, it might be expected that distinct peaks for 0, 1, 2 … dNTPs per droplet would be apparent. However, we see only a narrow peak of droplets with no dNTP and a single broad peak corresponding to droplets with one or more dNTPs. The broadening of the ‘with dNTP’ peaks into a single wide peak has a number of sources, including variation in enzymatic activity, focus drift between images, errors in assigning diameters and uneven illumination. Heavy-tailed distributions can be fitted to the peaks (to reduce the impact of errors that severely affect a few droplets (47)) and their crossing point acts as an intensity threshold (dashed line, Figure 4D). If the intensity of a droplet is above the threshold in a particular color, a dNTP corresponding to that color is deemed to be present in the droplet. False positives are given by the section of the ‘no dNTP’ peak above the threshold (solid red region, Figure 4D) and false negatives by the section of the ‘1+ dNTP’ peak below the threshold (solid blue region, Figure 4D).
There are a number of additional features in this data set which reinforce the conclusion that we are observing stochastic incorporation of dNTPs into droplets, and so have the ability to detect a single dNTP.
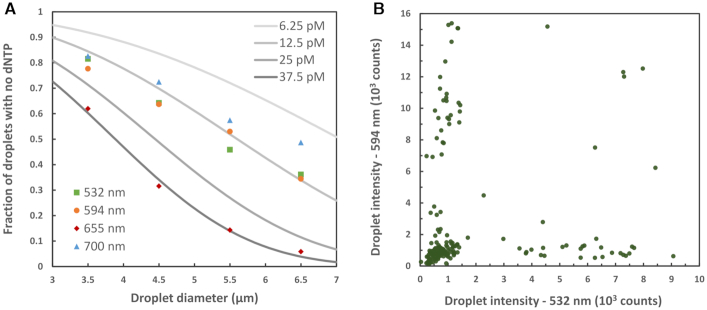
First, the fraction of droplets containing no dNTP, which is plotted against droplet diameter in Figure 5A, should vary with droplet diameter according to the Poisson distribution described previously (Table 1). The expected ‘no dNTP’ fraction, for a range of dNTP concentrations, is included for comparison. The best fit to the data allows estimation of the dNTP concentrations as dGTP (532 nm) 14 pM, dCTP (594 nm) 14 pM, dATP (655 nm) 37 pM, and dTTP (700 nm) 10 pM. This is in close agreement with the nominal concentrations.
Figure 5.
(A) Measured fraction of droplets with no dNTP plotted against droplet diameter for all four color channels (filled dots). The expected fractions calculated from the Poisson distribution, for a range of dNTP concentrations, are included for comparison (gray lines). (B) Droplet intensity in 532 nm channel versus 594 nm channel for the 3–4 μm subset of droplet diameters. The data for all dNTP channels and the 3–7 μm diameter range is included in the Supplementary Data (Supplementary Figures S16 and 17).
Additionally, if incorporation of dNTPs into droplets is random then there should be no correlation of droplet intensity in different colors. Plotting intensity in one color against that in another color allows this to be verified. This is shown for 532 nm (dGTP) versus 594 nm (dCTP), at droplet diameters of 3–4 μm, in Figure 5B. The Pearson correlation coefficient for these data is 0.07 (r2 = 0.005) indicating no significant linear correlation. Due to the low double occupancy probability at this small droplet size and dNTP concentration (Table 1), we expect to see few droplets along the diagonal, rather there would be a cluster close to the origin corresponding to droplets with no dNTP and two bands extending horizontally and vertically from this corresponding to droplets with one or the other of the dNTPs respectively. For higher dNTP concentration or larger droplets a fraction of the droplets would begin to appear on the diagonal as the probability of a droplet containing more than one type of dNTP becomes larger. This behavior can clearly be observed in Figure 5B and Supplementary Data Figures S16 and 17.
Potential problems such as runaway enzymatic activity or broadband fluorescent contamination would affect all droplets equally so would be easily spotted in plots such as Figure 5. The nature of the reaction, which releases many fluorophores per dNTP, ensures that contamination from fluorescent molecules is negligible in comparison or else is not stochastic. Indeed, the main contamination issue faced is with dNTP molecules themselves. Significant deviation from the expected concentration in Figure 5A would signify this.
All this provides strong evidence that we are observing stochastic dNTP presence in droplets and that single dNTPs can be detected. Indeed, for 12.5 pM dNTP in 3–4 μm droplets >90% of positive detections are single dNTP events.
CONCLUSION
We have developed a novel method to detect dNTPs with a detection limit that is orders of magnitude more sensitive than the best methods currently in use. The four dNTPs can be detected simultaneously and with high specificity, with cross-talk being 3–4 orders of magnitude lower than the intended signal. The fluorescence signal generated is robust and readily detectable, thereby making expensive instrumentation unnecessary. However, it is the high sensitivity of this method that makes it particularly attractive. It allows us to detect a few attomoles of dNTPs in bulk solution, the equivalent amount found in very low numbers of cells. This will open the door to new diagnostic methods which require drastically reduced sample sizes, and potentially single cell dNTP quantification. More significantly, the high sensitivity enables us to detect, and distinguish between, single dNTP molecules in microdroplets. This result supports the use of individual dNTP detection at the core of a new approach to single-molecule DNA sequencing.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Prof. Tom Brown for his advice on the design, and synthesis of, the Probe Oligos.
Author contributions’: B.B., K.J., Ma. St., A.-L.S., A.N., N.M.B., T.H.I., M.D., J.C., L.A.I., R.N.P., J.B., A.J.D., D.M.L., O.P., D.J.K., A.S., M.W., Ma. So., P.H.D, F.T., E.P., M.A., L.R.S., M.S.S. and E.S planned and performed experiments. J.K., G.J.P. and B.W.B. designed experiments, C.A.F. supervised research. All authors analyzed experimental data. B.B. and K.J. wrote the paper with help from Ma. St., A.-L.S., A.N., N.M.B., P.H.D and B.W.B.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Base4 Innovation, Ltd. (Base4). Funding for open access charge: Base4 Innovation, Ltd.
Conflict of interest statement. The authors are or have been employed by Base4 Innovation,Ltd. (Base4), a privately held company and may hold stock or stock options. Base4 has filed provisional applications with the US Patent and Trademark Office on aspects of this research (PCT/GB2015/052119).
REFERENCES
- 1. Bessman M.J., Lehman I.R., Simms E.S., Kornberg A.. Enzymatic synthesis of deoxyribonucleic Acid. II. General properties of the reaction. J. Biol. Chem. 1958; 233:171–177. [PubMed] [Google Scholar]
- 2. Deutscher M.P., Kornberg A.. Enzymatic synthesis of deoxyribonucleic Acid. XXVIII. The pyrophosphate exchange and pyrophosphorolysis reactions of deoxyribonucleic acid polymerase. J. Biol. Chem. 1969; 244:3019–3028. [PubMed] [Google Scholar]
- 3. Song H., Chen D.L., Ismagilov R.F.. Reactions in droplets in microfluidic channels. Angew. Chem. Int. Ed. 2006; 45:7336–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Theberge A.B., Courtois F., Schaerli Y., Fischlechner M., Abell C., Hollfelder F., Huck W.T.S.. Microdroplets in microfluidics: an evolving platform for discoveries in chemistry and biology. Angew. Chem. Int. Ed. 2010; 49:5846–5868. [DOI] [PubMed] [Google Scholar]
- 5. Huebner A., Sharma S., Srisa-Art M., Hollfelder F., Edel J.B., DeMello A.J.. Microdroplets: a sea of applications. Lab. Chip. 2008; 8:1244–1254. [DOI] [PubMed] [Google Scholar]
- 6. Rotem A., Ram O., Shoresh N., Sperling R.A., Goren A., Weitz D.A., Bernstein B.E.. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 2015; 33:1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baroud C.N., de Saint Vincent M.R., Delville J.-P.. An optical toolbox for total control of droplet microfluidics. Lab. Chip. 2007; 7:1029–1033. [DOI] [PubMed] [Google Scholar]
- 8. Mashaghi S., Abbaspourrad A., Weitz D.A., van Oijen A.M.. Droplet microfluidics: a tool for biology, chemistry and nanotechnology. Trends Anal. Chem. 2016; 82:118–125. [Google Scholar]
- 9. Holzmeister P., Acuna G.P., Grohmann D., Tinnefeld P.. Breaking the concentration limit of optical single-molecule detection. Chem. Soc. Rev. 2014; 43:1014–1028. [DOI] [PubMed] [Google Scholar]
- 10. Ma F., Li Y., Tang B., Zhang C.Y.. Fluorescent biosensors based on single-molecule counting. Acc. Chem. Res. 2016; 49:1722–1730. [DOI] [PubMed] [Google Scholar]
- 11. Gad H., Koolmeister T., Jemth A.S., Eshtad S., Jacques S.A., Ström C.E., Svensson L.M., Schultz N., Lundbäck T., Einarsdottir B.O. et al.. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014; 508:215–221. [DOI] [PubMed] [Google Scholar]
- 12. Qu J., Sun W., Zhong J., Lv H., Zhu M., Xu J., Jin N., Xie Z., Tan M., Lin S.H. et al.. Phosphoglycerate mutase 1 regulates dNTP pool and promotes homologous recombination repair in cancer cells. J. Cell Biol. 2017; 216:409–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pfister S.X., Markkanen E., Jiang Y., Sarkar S., Woodcock M., Orlando G., Mavrommati I., Pai C.C., Zalmas L.P., Drobnitzky N. et al.. Inhibiting WEE1 selectively kills histone H3K36me3-Deficient cancers by dNTP starvation. Cancer Cell. 2015; 28:557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amie S.M., Daly M.B., Noble E., Schinazi R.F., Bambara R.A., Kim B.. Anti-HIV host factor SAMHD1 regulates viral sensitivity to nucleoside reverse transcriptase inhibitors via modulation of cellular deoxyribonucleoside triphosphate (dNTP) levels. J. Biol. Chem. 2013; 288:20683–20691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van Laethem K., Witvrouw M., Pannecouque C., Van Remoortel B., Schmit J.C., Esnouf R., Kleim J.P., Balzarini J., Desmyter J., De Clercq E. et al.. Mutations in the non-nucleoside binding-pocket interfere with the multi-nucleoside resistance phenotype. Aids. 2001; 15:553–561. [DOI] [PubMed] [Google Scholar]
- 16. Chen X., Seifert S.M., Castillo-Mancilla J.R., Bushman L.R., Zheng J.H., Kiser J.J., MaWhinney S., Anderson P.L.. Model linking plasma and intracellular tenofovir/emtricitabine with deoxynucleoside triphosphates. PLoS One. 2016; 11:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Puigvert J.C., Sanjiv K., Helleday T.. Targeting DNA repair, DNA metabolism and replication stress as anti-cancer strategies. FEBS J. 2016; 283:232–245. [DOI] [PubMed] [Google Scholar]
- 18. Weinberg G., Ullman B., Martin D.W.. Mutator phenotypes in mammalian cell mutants with distinct biochemical defects and abnormal deoxyribonucleoside triphosphate pools. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:2447–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp. Cell Res. 1989; 181:305–316. [DOI] [PubMed] [Google Scholar]
- 20. Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 1988; 57:349–374. [DOI] [PubMed] [Google Scholar]
- 21. Aird K.M., Zhang G., Li H., Tu Z., Bitler B.G., Garipov A., Wu H., Wei Z., Wagner S.N., Herlyn M. et al.. Suppression of nucleotide metabolism underlies the establishment and maintenance of Oncogene-Induced senescence. Cell Rep. 2013; 3:1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gandhi V.V., Samuels D.C.. A review comparing deoxyribonucleoside triphosphate (dNTP) concentrations in the mitochondrial and cytoplasmic compartments of normal and transformed cells. Nucleosides Nucleotides Nucleic Acids. 2011; 30:317–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schaaper R.M., Mathews C.K.. Mutational consequences of dNTP pool imbalances in E. coli. DNA Repair (Amst). 2013; 12:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guan Z., Wang X., Dong Y., Xu L., Zhu Z., Wang J., Zhang T., Niu B.. dNTP deficiency induced by HU via inhibiting ribonucleotide reductase affects neural tube development. Toxicology. 2015; 328:142–151. [DOI] [PubMed] [Google Scholar]
- 25. Wheeler L.J., Mathews C.K.. Nucleoside triphosphate pool asymmetry in mammalian mitochondria. J. Biol. Chem. 2011; 286:16992–16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oliver F.J., Collins M.K.L., López-Rivas A.. dNTP pools imbalance as a signal to initiate apoptosis. Experientia. 1996; 52:995–1000. [DOI] [PubMed] [Google Scholar]
- 27. Lyman G.E., DeVincenzo J.P.. Determination of picogram amounts of ATP using the luciferin-luciferase enzyme system. Anal. Biochem. 1967; 21:435–443. [DOI] [PubMed] [Google Scholar]
- 28. Coulier L., Gerritsen H., van Kampen J.J.A., Reedijk M.L., Luider T.M., Osterhaus A.D.M.E., Gruters R.A., Brüll L.. Comprehensive analysis of the intracellular metabolism of antiretroviral nucleosides and nucleotides using liquid chromatography-tandem mass spectrometry and method improvement by using ultra performance liquid chromatography. J. Chromatogr. B. 2011; 879:2772–2782. [DOI] [PubMed] [Google Scholar]
- 29. Cross D.R., Miller B.J., James S.J.. A simplified HPLC method for simultaneously quantifying ribonucleotides and deoxyribonucleotides in cell extracts or frozen tissues. Cell Prolif. 1993; 26:327–336. [DOI] [PubMed] [Google Scholar]
- 30. Decosterd L.A., Cottin E., Chen X., Lejeune F., Mirimanoff R.O., Biollaz J., Coucke P.A.. Simultaneous determination of deoxyribonucleoside in the presence of ribonucleoside triphosphates in human carcinoma cells by high-performance liquid chromatography. Anal. Biochem. 1999; 270:59–68. [DOI] [PubMed] [Google Scholar]
- 31. Nakamura M., Kakutani T., Samejima K., Wataya Y., Hayatsu H.. Deoxyribonucleoside triphosphates in mouse fetal liver cells assayed by High-Pressure liquid chromatography. Toxicol. Methods. 1998; 8:11–16. [Google Scholar]
- 32. Rimerman R.A., Prorok G.D., Cordel K.L., Shahwan A.M., Vaughan W.P.. Improved high-performance liquid chromatographic analysis of intracellular deoxyribonucleoside triphosphate levels. J. Chromatogr. B Biomed. Sci. Appl. 1993; 619:29–35. [DOI] [PubMed] [Google Scholar]
- 33. Chen P., Liu Z., Liu S., Xie Z., Aimiuwu J., Pang J., Klisovic R., Blum W., Grever M.R., Marcucci G. et al.. A LC-MS/MS method for the analysis of intracellular nucleoside triphosphate levels. Pharm. Res. 2009; 26:1504–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fromentin E., Gavegnano C., Obikhod A., Schinazi R.F.. Simultaneous quantification of intracellular natural and antiretroviral nucleosides and nucleotides by liquid chromatography-tandem mass spectrometry. Anal. Chem. 2010; 82:1982–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Henneré G., Becher F., Pruvost A., Goujard C., Grassi J., Benech H.. Liquid chromatography-tandem mass spectrometry assays for intracellular deoxyribonucleotide triphosphate competitors of nucleoside antiretrovirals. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003; 789:273–281. [DOI] [PubMed] [Google Scholar]
- 36. Kamčeva T., Bjånes T., Svardal A., Riedel B., Schjøtt J., Eide T.. Liquid chromatography/tandem mass spectrometry method for simultaneous quantification of eight endogenous nucleotides and the intracellular gemcitabine metabolite dFdCTP in human peripheral blood mononuclear cells. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015; 1001:212–220. [DOI] [PubMed] [Google Scholar]
- 37. Thomas D., Herold N., Keppler O.T., Geisslinger G., Ferreirós N.. Quantitation of endogenous nucleoside triphosphates and nucleosides in human cells by liquid chromatography tandem mass spectrometry. Anal. Bioanal. Chem. 2015; 407:3693–3704. [DOI] [PubMed] [Google Scholar]
- 38. Dong J., Wu T., Xiao Y., Xu L., Fang S., Zhao M.. A fuel-limited isothermal DNA machine for the sensitive detection of cellular deoxyribonucleoside triphosphates. Chem. Commun. 2016; 52:11923–11926. [DOI] [PubMed] [Google Scholar]
- 39. Gao W.Y., Johns D.G., Mitsuya H.. Enzymatic assay for quantification of deoxynucleoside triphosphates in human cells exposed to antiretroviral 2′,3′-Dideoxynucleosides. Anal. Biochem. 1994; 222:116–122. [DOI] [PubMed] [Google Scholar]
- 40. Wilson P.M., Labonte M.J., Russell J., Louie S., Ghobrial A.A., Ladner R.D.. A novel fluorescence-based assay for the rapid detection and quantification of cellular deoxyribonucleoside triphosphates. Nucleic Acids Res. 2011; 39:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ferraro P., Franzolin E., Pontarin G., Reichard P., Bianchi V.. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 2010; 38:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hollenbaugh J.A., Kim B.. Prasad V, Kalpana G. HIV-1 reverse transcriptase-based assay to determine cellular dNTP concentrations. HIV Protocols. Methods in Molecular Biology. 2016; 1354:NY: Humana Press; 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Roy B., Beuneu C., Roux P., Buc H., Lemaire G., Lepoivre M.. Simultaneous determination of pyrimidine or purine deoxyribonucleoside triphosphates using a polymerase assay. Anal. Biochem. 1999; 269:403–409. [DOI] [PubMed] [Google Scholar]
- 44. Smid K., Van Moorsel C., Noordhuis P., Voorn D., Peters G.. Interference of gemcitabine triphosphate with the measurements of deoxynucleotides using an optimized DNA polymerase elongation assay. Int. J. Oncol. 2001; 19:157–162. [PubMed] [Google Scholar]
- 45. Rittié L., Perbal B.. Enzymes used in molecular biology: A useful guide. J. Cell Commun. Signal. 2008; 2:25–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chauleau M., Shuman S.. Kinetic mechanism and fidelity of nick sealing by Escherichia coli NAD+ -dependent DNA ligase (LigA). Nucleic Acids Res. 2016; 44:2298–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lange K.L., Little R.J.A., Taylor J.M.G.. Robust statistical modeling using the t distribution. J. Am. Stat. Assoc. 1989; 84:881–896. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.