Significance
Mosquito-transmitted flaviviruses such as Zika virus (ZIKV) are responsible for over 400 million human infections each year. Unfortunately, the molecular mechanisms that facilitate flavivirus transmission by mosquitoes remain unclear. Here, we demonstrate that noncoding subgenomic flavivirus RNA (sfRNA), that is produced by all flaviviruses, plays a critical role in ZIKV transmission by Aedes aegypti mosquitoes. ZIKV requires sfRNA to overcome the mosquito midgut barrier and efficiently accumulate in the mosquito saliva. We reveal that the mosquito protein ME31B has antiviral activity and specifically binds to sfRNA. These results establish sfRNA as a determinant of ZIKV transmission by mosquitoes and provide mechanistic insights into the functions of this noncoding RNA.
Keywords: Zika virus, subgenomic flavivirus RNA, Aedes aegypti, RNA-affinity purification, transmission
Abstract
Zika virus (ZIKV) is an arthropod-borne flavivirus predominantly transmitted by Aedes aegypti mosquitoes and poses a global human health threat. All flaviviruses, including those that exclusively replicate in mosquitoes, produce a highly abundant, noncoding subgenomic flavivirus RNA (sfRNA) in infected cells, which implies an important function of sfRNA during mosquito infection. Currently, the role of sfRNA in flavivirus transmission by mosquitoes is not well understood. Here, we demonstrate that an sfRNA-deficient ZIKV (ZIKVΔSF1) replicates similar to wild-type ZIKV in mosquito cell culture but is severely attenuated in transmission by Ae. aegypti after an infectious blood meal, with 5% saliva-positive mosquitoes for ZIKVΔSF1 vs. 31% for ZIKV. Furthermore, viral titers in the mosquito saliva were lower for ZIKVΔSF1 as compared to ZIKV. Comparison of mosquito infection via infectious blood meals and intrathoracic injections showed that sfRNA is important for ZIKV to overcome the mosquito midgut barrier and to promote virus accumulation in the saliva. Next-generation sequencing of infected mosquitoes showed that viral small-interfering RNAs were elevated upon ZIKVΔSF1 as compared to ZIKV infection. RNA-affinity purification followed by mass spectrometry analysis uncovered that sfRNA specifically interacts with a specific set of Ae. aegypti proteins that are normally associated with RNA turnover and protein translation. The DEAD/H-box helicase ME31B showed the highest affinity for sfRNA and displayed antiviral activity against ZIKV in Ae. aegypti cells. Based on these results, we present a mechanistic model in which sfRNA sequesters ME31B to promote flavivirus replication and virion production to facilitate transmission by mosquitoes.
Flaviviruses (family Flaviviridae), such as Zika virus (ZIKV), dengue virus (DENV), and West Nile virus (WNV) are arthropod-borne viruses (arboviruses) of serious concern for human health (1, 2). Annually, arboviruses are responsible for over 400 million cases of human infection, and, with rising global temperatures and increased trade and travel, this number is expected to further increase (3–8). Flaviviruses have a single-stranded positive-sense RNA genome that contains 1 open reading frame (ORF) which is flanked by highly structured 5′ and 3′ untranslated regions (UTRs), that are essential for virus replication (9, 10). During flavivirus infection, the viral genomic RNA (vgRNA) is degraded by host 5′ → 3′ exoribonucleases (XRNs), including XRN1 in mammalian hosts and its homolog Pacman in insects (11). However, when these XRNs encounter the highly structured 3′ UTR, they stall on XRN-resistant RNA structures. In vitro XRN1 digestion assays and X-ray crystallography have revealed that XRNs stall on tightly folded RNA structures, that form a so-called XRN-resistant “molecular knot” (12, 13). The stalling produces a residual RNA molecule of 0.4 to 0.6 kb referred to as subgenomic flavivirus RNA (sfRNA), that is very abundant in infected cells of mosquitoes and vertebrate hosts (11, 13–15). The 3′ UTR of ZIKV contains several secondary and tertiary RNA structures: duplicated stem-loop (SL) structures SL-I and SL-II, 2 dumbbell (DB)-like structures Ѱ-DB and DB-1, and a terminal 3′ SL (16) (SI Appendix, Fig. S1). ZIKV produces at least 2 species of sfRNA (sfRNA1 and sfRNA2) in infected cells through stalling of XRNs on SL-I and SL-II, respectively (13, 16, 17).
Significant functions of sfRNA have been implicated in mammalian hosts, such as the induction of pathogenicity in mice and cytopathicity in cell culture (11, 18), evasion of the type I/II IFN response (19–21), decreased mRNA turnover (22), and enhanced virus replication in mammalian cells (11, 23). Lately, the importance of sfRNA formation for the infection and subsequent transmission by the mosquito vector has been suggested. We showed that sfRNA is important for WNV transmission by Culex pipiens mosquitoes (24). Moreover, it was recently demonstrated in Aedes aegypti that DENV-2 strains which produce lower amounts of sfRNA display decreased transmission efficiencies, suggesting that sfRNA may also have important functions during infection of Ae. aegypti (25). Currently, evidence for sfRNA-mediated flavivirus infection of and transmission by Ae. aegypti mosquitoes is lacking, and a molecular mechanism by which sfRNA may mediate the infection of mosquitoes remains enigmatic.
Here, we investigated the importance of ZIKV sfRNA for infection of and transmission by Ae. aegypti using an sfRNA-deficient ZIKV mutant (ZIKVΔSF1). Through comparison of mosquito infection via blood feeding and intrathoracic injections, we investigated the relative contribution of ZIKV sfRNA in overcoming the mosquito midgut barrier. We applied a small RNA sequencing approach to assess the role of sfRNA as an RNA interference (RNAi) suppressor in vivo in Ae. aegypti. By RNA-affinity purification, we identified mosquito proteins that interact with sfRNA of both ZIKV and WNV, and we mapped the RNA structures in sfRNA required for protein binding. Through RNAi-based silencing of these sfRNA-interacting mosquito proteins, we investigated the significance of these interactions for ZIKV and WNV replication in Ae. aegypti cells. Our findings establish sfRNA as a key determinant of flavivirus transmission by mosquitoes and lead to a mechanistic model that describes how this noncoding viral RNA mediates flavivirus transmission.
Results
ZIKV sfRNA Is Required to Overcome the Mosquito Midgut and Salivary Gland Barriers and Determines ZIKV Transmission by Ae. aegypti.
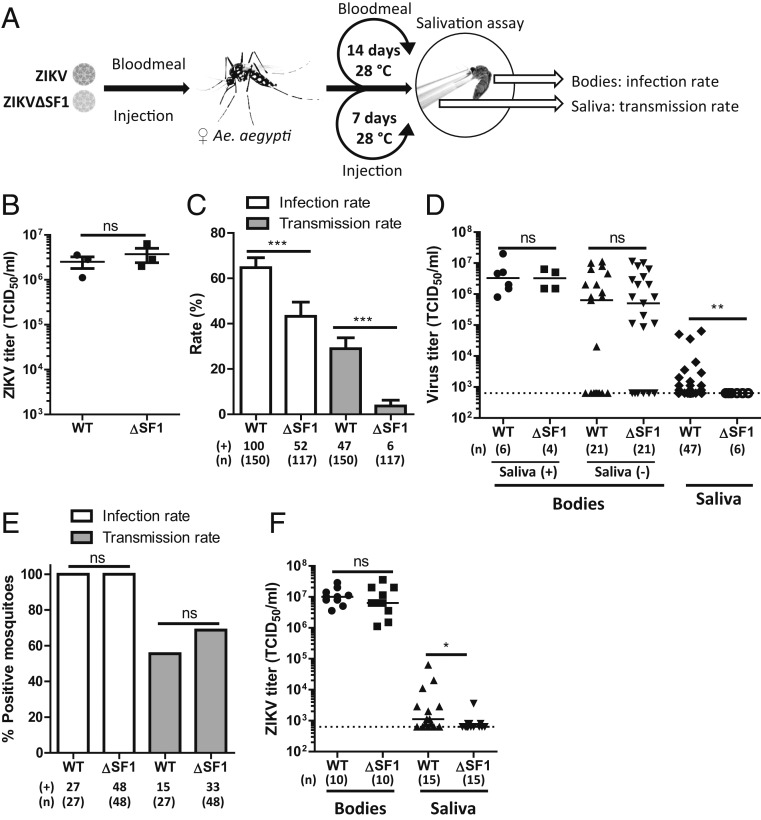
Arbovirus transmission by mosquitoes largely depends on the ability of the virus to overcome the midgut and salivary gland barriers of the mosquito (2, 26). To investigate the role of ZIKV sfRNA in virus transmission by Ae. aegypti, an sfRNA1-deficient ZIKV-mutant (ZIKVΔSF1) was generated using a ZIKV infectious clone (27) (SI Appendix, Fig. S1A). This mutant is deficient in sfRNA1 formation in mosquito cells (SI Appendix, Fig. S1B) but replicates similarly to wild-type ZIKV in Ae. aegypti Aag2 cells (SI Appendix, Fig. S1C). Female Ae. aegypti were fed with an infectious blood meal containing ZIKV or ZIKV∆SF1 (Fig. 1A). Titration of the prepared infectious blood meals confirmed that equal titers of ZIKV and ZIKV∆SF1 were administered (Fig. 1B). At 14 d postexposure, ZIKV reached an infection rate of 66% while a significantly lower infection rate of 44% (P < 0.001) was observed for ZIKV∆SF1 (Fig. 1C), demonstrating that sfRNA1 is important for efficient ZIKV infection in Ae. aegypti. Furthermore, ZIKV reached a transmission rate of 31%, versus only 5% for the ZIKV∆SF1-exposed mosquitoes (P < 0.001). Viral titers of ZIKV and ZIKV∆SF1 were similar in the bodies of both saliva-positive (Fig. 1D) (3.6 to 4.6 × 106 tissue culture infectious dose 50% [TCID50]/mL; P = 1.00) and saliva-negative mosquitoes (Fig. 1D) (1.8–2.2 × 106 TCID50/mL; P = 0.63), but the mean titer in the saliva of ZIKV∆SF1-infected mosquitoes was lower than in the ZIKV-infected mosquitoes (Fig. 1D) (4.1 × 104 vs. 6.3 × 102 TCID50/mL; P = 0.002). It should also be noted that none of the virus-positive saliva samples from ZIKV∆SF1-infected mosquitoes had saliva titers exceeding the end-point dilution assay (EPDA) detection limit.
Fig. 1.
Infectious blood meals and intrathoracic injections of Ae. aegypti with ZIKV or ZIKV∆SF1 indicate that sfRNA1 determines virus transmission. (A) Schematic overview of the experimental setups. (B) ZIKV titers of the infectious blood meals used in the 3 replicate experiments were determined by end-point dilution assay (EPDA). Shown is the mean titer ± SEM. Statistics were performed by unpaired t test. (C) Female Ae. aegypti were fed with an infectious blood meal containing 3.0 × 106 TCID50/mL ZIKV or ZIKV∆SF1. Engorged females were incubated for 14 d at 28 °C, and infection and transmission rates were determined by infectivity assay on Vero cells. Shown are the mean infection and transmission rates ± SEM. Statistics were performed by Fisher’s exact test on cumulative data. (D) Viral titers in the bodies of mosquitoes with ZIKV-positive [Saliva (+)] and ZIKV-negative [Saliva (−)] saliva and titers of ZIKV-positive saliva samples were determined by EPDA. Shown are the median titers, and statistics were performed by Mann–Whitney U test. (E) Female Ae. aegypti were intrathoracically injected with ∼400 TCID50 of ZIKV or ZIKV∆SF1. At 7 d postinjection, infection and transmission rates were determined. Statistics were performed by Fisher’s exact test on cumulative data. (F) Viral titers in the bodies of virus-positive bodies and saliva samples were determined by EPDA. Shown are the median titers, and statistics were performed by Mann–Whitney U test. Dotted lines indicate the EPDA detection limit. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ns, not significant; WT, wild type.
To investigate whether full-length sfRNA is important for ZIKV to cross the midgut barrier, we intrathoracically injected female Ae. aegypti with ZIKV or ZIKV∆SF1, which bypasses the midgut barrier (Fig. 1E). All injected mosquitoes (100%) were infected, and similar transmission rates were observed for ZIKV and ZIKV∆SF1 (55% vs. 68%; P = 0.32). Furthermore, these results confirm our earlier observations that Ae. aegypti has a salivary gland barrier for ZIKV (28) as both ZIKV and ZIKVΔSF1 do not reach transmission rates of >70%. Titration of the bodies and saliva samples that were virus-positive in the infectivity assay demonstrated that ZIKV and ZIKV∆SF1 reached similar titers in the bodies of Ae. aegypti (Fig. 1F) (1.1 to 1.2 × 107 TCID50/mL; P = 0.49). This demonstrates that sfRNA1 is not essential for replication of ZIKV in the mosquito body after passing the Ae. aegypti midgut barrier. However, the titers of the mosquito salivas were significantly lower for ZIKV∆SF1 compared to ZIKV (3.6 × 103 vs. 6.3 × 104 TCID50/mL; P = 0.01), indicating that sfRNA1 promotes efficient virus dissemination through the salivary gland barrier and subsequent virus accumulation in the saliva of Ae. aegypti. Since the Ae. aegypti Rockefeller strain is highly laboratory adapted, we have confirmed our observations in 2 recently colonized Ae. aegypti strains from French Guiana and Kenya (SI Appendix, Fig. S2).
ZIKV sfRNA as Modulator of Mosquito Innate Immune Responses.
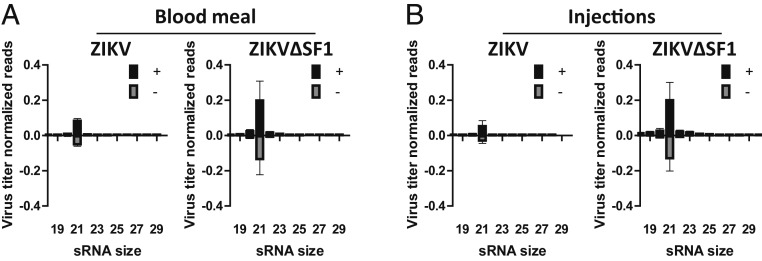
For DENV in Ae. aegypti mosquitoes, it was recently demonstrated that sfRNA slightly modulates the expression of genes involved in the Toll pathway (29), which correlated with enhanced DENV transmission (25). However, analysis of gene expression for Toll and Janus kinase/signal transducers and activators of transcription (JAK/STAT)-regulated genes after ZIKV, ZIKV∆SF1, WNV, or WNV∆SF1+2 infection of Ae. aegypti Aag2 cells did not indicate that sfRNA suppresses the induction of a Toll or JAK/STAT signaling response (SI Appendix, Fig. S3). Previous research has also implicated sfRNA as a putative suppressor of the RNAi response (24, 30, 31). The antiviral RNAi response in mosquitoes is mediated by 21-nucleotide (nt) small-interfering RNAs (siRNAs) that are produced through cleavage of viral double-strand RNA (dsRNA) intermediates by the endonuclease Dicer (32). We sequenced the small RNAs from female Ae. aegypti with fully disseminated ZIKV and ZIKV∆SF1 infections, which were established by infectious blood meal or by intrathoracic injection (3 pools of 5 to 6 mosquitoes for each treatment). After normalizing the number of reads to the number of reads per library and the viral titer of the mosquito pools that were used for next-generation sequencing of small RNAs, the results demonstrated an elevated siRNA response for ZIKV∆SF1 infections compared to infections with wild-type ZIKV (Fig. 2 A and B) although this difference was not apparent before normalizing for viral titer (SI Appendix, Fig. S4 A and B). The correlation between sfRNA production and a decreased number of viral siRNAs suggests that ZIKV sfRNA may suppress the Ae. aegypti RNAi response. However, there was no noticeable effect of sfRNA on the genome distribution of 21-nt small RNAs on the (+) or (−) strand after infection via infectious blood meal or intrathoracic injection (SI Appendix, Fig. S4 C–F). Furthermore, no clear 25- to 30-nt PIWI-interacting (pi) RNAs were observed in our data from fully disseminated ZIKV-infected Ae. aegypti mosquitoes (SI Appendix, Fig. S4 A and B), and the limited population of 25- to 30-nt small RNA reads did not display a ping-pong piRNA signature (SI Appendix, Fig. S5).
Fig. 2.
Small RNA responses in ZIKV and ZIKV∆SF1-infected Ae. aegypti. Small RNAs were sequenced at 14 d postexposure from 3 individual pools of 5 to 6 fully-disseminated Ae. aegypti mosquitoes infected with ZIKV or ZIKV∆SF1 via either a blood meal (A) or intrathoracic injections (B). Shown are size distributions of small RNAs that mapped to the ZIKV genome. Small RNA reads were normalized as percentage of reads from the total number of reads in the library and additionally normalized to the ZIKV titer in the pool of sequenced mosquitoes.
Identification of ZIKV and WNV sfRNA Binding Proteins in Ae. aegypti Cells Using Streptavidin Aptamer-Based sfRNA-Affinity Purification.
We performed sfRNA-affinity purification using the 4XS1m aptamer, which has high affinity for streptavidin (SA) (33), to identify ZIKV and WNV sfRNA-binding proteins (SI Appendix, Fig. S6A). We included WNV sfRNA in this experiment since we previously demonstrated its importance for WNV transmission by C. pipiens mosquitoes (24). Incorporation of 4 sequential (4XS1m) aptamers led to the most efficient RNA-to-bead coupling (SI Appendix, Fig. S6B). We therefore produced in vitro transcribed RNA corresponding to the sequences of 4XS1m-ZIKV-sfRNA, 4XS1m-WNV-sfRNA, and 4XS1m-Control and verified the folding of ZIKV and WNV sfRNA in complex with the aptamers by in vitro XRN1-stalling assay (SI Appendix, Fig. S6C). As anticipated, addition of in vitro XRN1 resulted in the accumulation of ZIKV-sfRNA1 for 4XS1m-ZIKV-sfRNA, WNV-sfRNA1 for 4XS1m-WNV-sfRNA, and no RNA product for 4XS1m-Control (SI Appendix, Fig. S6C). This indicates that ZIKV and WNV sfRNA have retained their functionality to stall XRN1 in vitro in the presence of the 4XS1m sequence.
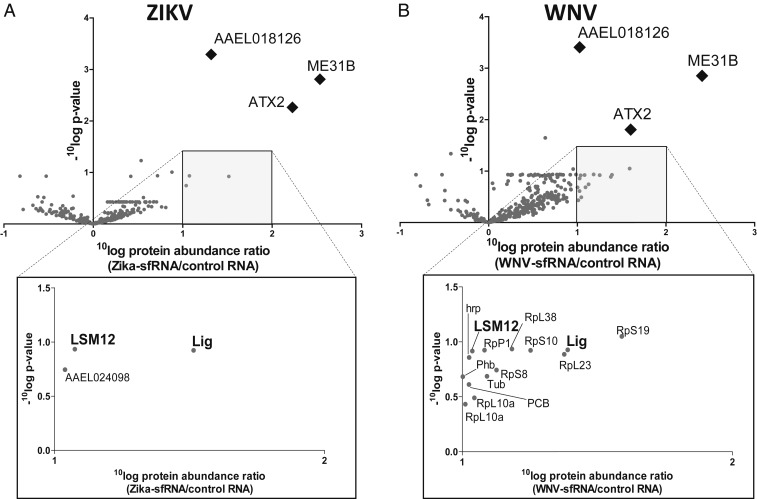
To determine which mosquito proteins interact with ZIKV and WNV sfRNA, we employed the sfRNA-affinity purification in combination with a mass spectrometry approach in 3 independent biological replicates (see SI Appendix, Fig. S6A for a schematic overview). Volcano plots were generated for both ZIKV and WNV sfRNA where proteins with a ≥10-fold relative enrichment in the sfRNA samples and a P value of ≤0.05 were considered as significant (Fig. 3 A and B). A complete list of detected proteins is included in SI Appendix, Table S2 (ZIKV) and SI Appendix, Table S3 (WNV). The ZIKV (Fig. 3A) and WNV (Fig. 3B) sfRNA samples were significantly enriched for ME31B (ortholog of human DDX6), Ataxin-2 (ATX2), and AAEL018126 (∼17% sequence conservation with Drosophila protein twenty-four [TYF]). Additionally, 3 and 14 proteins were ≥10-fold enriched with lower significance in the ZIKV and WNV samples, respectively (Fig. 3 A and B, Insets), of which Lingerer (Lig) and LSM12 were ≥10-fold enriched in both datasets.
Fig. 3.
Identification of ZIKV and WNV sfRNA-binding proteins in Ae. aegypti cells using a streptavidin-binding aptamer approach and nano LC-MS/MS. 4XS1m-aptamer bound RNAs were bound to streptavidin beads and used to purify Ae. aegypti proteins with affinity for the bait RNA. Streptavidin beads were incubated with in vitro transcribed RNA of 4XS1m-ZIKV-sfRNA, 4XS1m-WNV-sfRNA, or 4XS1m-control. RNA-bound proteins were eluted by RNase A digestion of the bait RNA and identified and quantified by mass spectrometry. (A and B) Volcano plots of eluted proteins detected by mass spectrometry. The x axis shows the mean 10Log difference in protein abundance between the ZIKV sfRNA (A) or WNV sfRNA (B) and control samples from 3 independent biological replicates. The y axis shows the 10Log of the P value by Student’s t test from comparison of the protein abundance in the ZIKV sfRNA (A) or WNV sfRNA (B) with the control samples. Significantly enriched proteins are shown as diamonds. Insets below show proteins that are ≥10-fold enriched in the sfRNA samples but not observed significantly different from the control.
Those proteins that were ≥3-fold up-regulated for both ZIKV and WNV sfRNA were further analyzed using the STRING database with high confidence level (≥0.7) based on their Drosophila orthologs (SI Appendix, Fig. S7). These sfRNA-binding proteins were significantly enriched in protein–protein interactions among each other (P = 2 × 10−15), indicating that these proteins are likely biologically connected and involved in similar processes. Moreover, 2 main interaction networks were implicated (SI Appendix, Fig. S7), and protein function analysis using the Panther Gene Ontology server implicated their involvement in posttranscriptional regulation of gene expression (P = 6.4 × 10−6) and cytoplasmic translation (P = 3.9 × 10−2).
sfRNA Has Specific Affinity for the DEAD-Box Helicase ME31B.
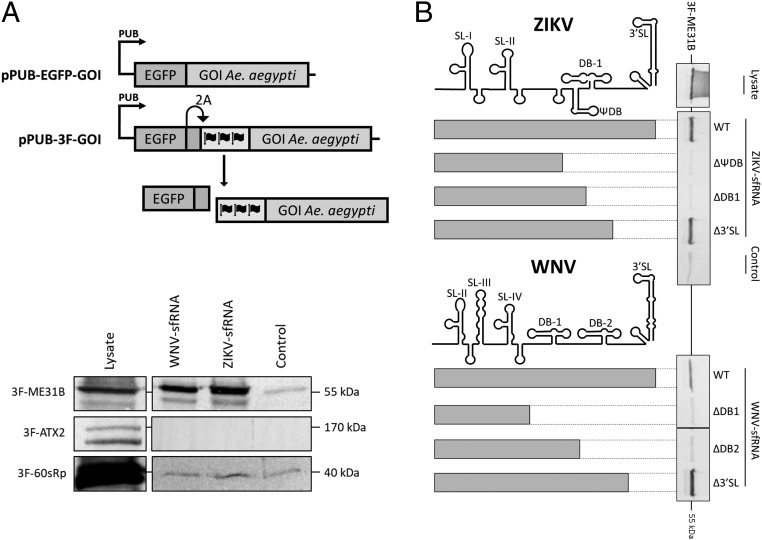
To confirm the observed interaction of the 3 most significant sfRNA-interacting proteins (ME31B, ATX2, and AAEL018126), we attempted to clone their complementary DNAs (cDNAs) from Ae. aegypti Aag2 cells into a mosquito polyubiquitin (PUB) promoter-driven plasmid and N-terminally fused to EGFP-2A-3XFLAG (3F) or EGFP (Fig. 4 A, Top). We also included the 60s ribosomal protein (60sRp) as a positive control that binds equally to ZIKV sfRNA, WNV sfRNA, and the control RNA (SI Appendix, Tables S2 and S3). Expression vectors for ME31B and ATX2 were successfully constructed, but we were unable to clone the entire AAEL018126 ORF. Next, Aag2 cells were transfected with pPUB-3F-ME31B, pPUB-3F-ATX2, or pPUB-3F-60sRp, and lysates were subjected to RNA-affinity purification and input and eluted proteins were analyzed by Western blot with α-FLAG antibodies (Fig. 4 A, Bottom). As expected, the 60sRp control bound equally to both sfRNAs and the control RNA while ME31B was highly enriched in the RNA-affinity purification for WNV and ZIKV sfRNA, but not for the control RNA (Fig. 4A), confirming that sfRNA has specific affinity for ME31B. Surprisingly, ATX2 was not enriched for WNV nor ZIKV sfRNA (Fig. 4A). As ATX2 is predicted to interact directly with ME31B (SI Appendix, Fig. S7), we hypothesized that ATX2 could indirectly interact with sfRNA via ME31B. We therefore performed a similar RNA-affinity purification using cell lysates of Aag2 cells cotransfected with both pPUB-EGFP-ME31B and pPUB-3F-ATX2 (SI Appendix, Fig. S8). Interestingly, for cotransfected cell lysates, both 3F-ATX-2 and EGFP-ME31B were enriched for WNV and ZIKV sfRNA, but not the control RNA, confirming that, indeed, sfRNA indirectly interacts with ATX2 via ME31B.
Fig. 4.
Ae. aegypti ME31B binds to the second dumbbell RNA structure of ZIKV and WNV sfRNA. (A, Top) Schematic overview of the used plasmid constructs. The genes of interest (GOI) were expressed fused N-terminally to EGFP followed by a foot-and-mouth disease virus 2A ribosome skipping sequence and a triple FLAG (3F)-tag which allows the expression of separate EGFP-2A and 3F-GOI. (A, Bottom) RNA-affinity purification to confirm ZIKV and WNV sfRNA binding proteins. Lysates of Aag2 cells transfected with pPUB-3F-ME31B, pPUB-3F-ATX2, pPUB-3F-LSM12, or pPUB-3F-60sRp were prepared. Lysates were subjected to RNA-affinity purification using streptavidin beads coated with in vitro transcribed RNA of 4XS1m-ZIKV-sfRNA, 4XS1m-WNV-sfRNA, or 4XS1m-control. RNA-bound proteins were eluted with RNase A and detected by Western blot with α-Flag antibodies. (B) Lysates of Aag2 cells transfected with pPUB-3F-ME31B were subjected to RNA-affinity purification with 4XS1m-aptamer fused 3′-truncated sfRNAs of ZIKV and WNV. A schematic overview of the used truncations is included in the figure. RNA-bound proteins were eluted with RNase A and detected by Western blot with α-Flag antibodies.
We then investigated which structure(s) of WNV and ZIKV sfRNA are essential for ME31B binding via an RNA-affinity purification using 3′ truncated sfRNAs (Fig. 4B). As expected, ME31B was enriched for full-length sfRNA of ZIKV and WNV, but not the control RNA. When the highly conserved 3′ SL was deleted (sfRNA∆3′SL), the interaction with ME31B was maintained. However, when the second DB was deleted (ZIKV: sfRNA∆DB1; WNV: sfRNA∆DB2) this enrichment was lost, implicating that ME31B binds at least to the second DB structure of ZIKV and WNV sfRNA.
ME31B Is an Antiviral Gene Inhibiting ZIKV and WNV Replication in Ae. aegypti Cells.
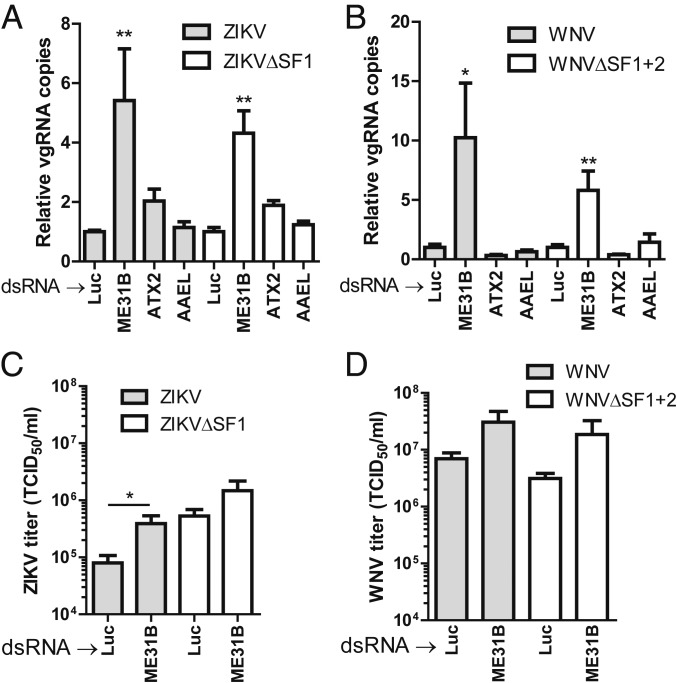
As the interaction of sfRNA with ME31B, indirectly with ATX2 and possibly with AAEL018126, implicates a putative role of these mosquito proteins during flavivirus infection, we investigated the role of these proteins for ZIKV and WNV replication in Ae. aegypti Aag2 cells. Gene silencing of 75% to 90% was achieved by dsRNA transfections targeting ME31B, ATX2, AAEL018126, or luciferase (Luc) (SI Appendix, Fig. S9). The silenced cells were infected with ZIKV, ZIKV∆SF1, WNV, or WNV∆SF1+2 at a multiplicity of infection (MOI) of 0.1. At 48 h postinfection (hpi), total RNA was extracted from the cell monolayers and subjected to qRT-PCR to quantify the viral genomic RNA (vg RNA) copies. Silencing of ME31B resulted in significantly higher vgRNA levels for ZIKV, ZIKV∆SF1 (Fig. 5A), WNV, and WNV∆SF1+2 (Fig. 5B) while silencing of ATX2 and AAEL018126 did not affect ZIKV or WNV replication (Fig. 5 A and B). To further investigate the effect of ME31B silencing on virus production, the viral titers were determined in ME31B-silenced and control cells (Fig. 5 C and D). For ZIKV, silencing of ME31B resulted in a significant increase in viral titer (∼1 log, similar to the increase in vgRNA copies), suggesting that ME31B acts antiviral during ZIKV infection. For ZIKV∆SF1, WNV, and WNV∆SF1+2, silencing of ME31B slightly, but not significantly, increased the viral titers. Together, these results suggest that ME31B inhibits ZIKV and WNV viral RNA replication and virion production in Aag2 cells.
Fig. 5.
Depletion of ME31B increases ZIKV and WNV replication in Ae. aegypti cells. (A and B) Effect of ME31B, ATX2, and AAEL018126 (AAEL) silencing on viral RNA replication of ZIKV and WNV. At 6 h post-second dsRNA transfection, cells were infected with (A) ZIKV, ZIKV∆SF1, (B) WNV, or WNV∆SF1+2 at a multiplicity of infection (MOI) of 0.1. Viral genomic RNA (vgRNA) copies were determined at 48 h postinfection (hpi) by qRT-PCR normalized to the rpS7 reference gene and computed relative to the dsLuc transfected control. Statistics were performed by one-way ANOVA with Tukey post hoc test. (C and D) Effect of ME31B silencing on the virus growth kinetics of ZIKV and WNV. At 4 h post-second dsRNA transfection, cells were infected with (C) ZIKV, ZIKV∆SF1, (D) WNV, or WNV∆SF1+2 at an MOI of 0.1. Viral titers were determined at 48 hpi by end-point dilution assay on Vero cells. Mann–Whitney U tests were used to compare viral titers and relative vgRNA copies. *P ≤ 0.05; **P ≤ 0.01.
Discussion
The molecular determinants of arbovirus transmission by mosquitoes are largely unknown, and few interactions between arboviruses and their vectors have been described in molecular detail. Here, we demonstrate that the noncoding, viral sfRNA is crucial for ZIKV to overcome the mosquito midgut barrier and efficiently accumulate in the mosquito saliva. Moreover, we uncover that sfRNA interacts with specific mosquito proteins and indicate that ZIKV sfRNA expression correlates with a weakened RNAi response in Ae. aegypti. Furthermore, we show that the most potent sfRNA-interacting protein of Ae. aegypti, ME31B, has an antiviral function during flavivirus replication in mosquito cells. The combined effect of sequestering the antiviral mosquito protein ME31B and the suppression of the RNAi response may enable ZIKV to overcome the midgut and salivary gland barriers to ultimately become transmissible to the vertebrate host. Together, these results establish sfRNA as a key determinant of ZIKV transmission by Ae. aegypti and provide leads to understand the underlying molecular mechanism.
While several molecular mechanisms have been proposed of how sfRNA mediates flavivirus transmission by mosquitoes have been proposed (16, 24, 25, 31, 34), none of these had previously been confirmed convincingly. SfRNA is not efficiently packaged into virions and is only formed after sufficient vgRNA replication has occurred (11). Therefore, functions of sfRNA are inherently executed at a later stage of replication in infected cells, and in case of a mosquito infection after entry into the midgut epithelial cells and the establishment of early virus replication. SfRNA could act as a viral suppressor of mosquito innate immune responses: e.g., the RNAi response, Toll pathway, or JAK/STAT pathway (32, 35), thereby facilitating virus transmission. In vitro cell culture studies and in vivo experiments with Culex mosquitoes showed that sfRNA suppresses Dicer-2 activity, possibly by acting as an RNA decoy (30, 31). Here, we demonstrate that Ae. aegypti produces higher siRNA levels relative to the viral titer during infection with ZIKVΔSF1 compared to wild-type ZIKV infection. However, as considerable numbers of siRNAs (>45,000) were still produced in response to wild-type ZIKV infection, it remains to be investigated to which extent the observed RNAi suppression contributes directly to increased virus transmission. Furthermore, in our previous study with WNV-infected Culex mosquitoes, no significant difference in the abundance of viral small interfering RNAs was observed between wild-type and sfRNA1-deficient WNV (24). Of note, a recent study indicated that the midgut of Ae. aegypti mosquitoes does not express the protein Loquacious 2, which is essential for siRNA-mediated silencing (36). This finding questions the role of the RNAi response in infection of the Ae. aegypti midgut and might indicate that RNAi suppression by sfRNA is likely more important to facilitate systemic ZIKV infection. Indeed, we did observe functional activity of sfRNA beyond the midgut, since ZIKV accumulated to higher titers than ZIKVΔSF1 in the mosquito saliva, even after intrathoracic injections.
Besides small RNA responses, the JAK/STAT and Toll signaling pathways (35) were previously implicated as antiviral during flavivirus infection (29, 37–40). In a study on DENV-2–infected Ae. aegypti, higher levels of sfRNA correlated with suppression of Toll-regulated genes while JAK/STAT-regulated genes were not suppressed (25). However, the differences in mRNA expression in these studies were not large (<2 fold), which makes it uncertain whether sfRNA may sufficiently down-regulate the protein levels of these immune regulatory pathways to affect virus transmission. In our present study, we did not find an induction of Toll- or JAK/STAT-regulated gene expression after ZIKV or WNV infection of Ae. aegypti cells; therefore, this does not logically invite further research on sfRNA as an antagonist of these innate immune pathways to mediate ZIKV transmission by Ae. aegypti mosquitoes.
In addition to suppression of innate immune responses, sfRNA could act as a “molecular sponge” that interacts with mosquito RNA-binding proteins to antagonize or redirect their functionality, as previously hypothesized (41). This sponging effect is often observed for host and viral noncoding RNAs due to their high abundance and ability to sequester proteins, small RNAs, and mRNA transcripts (reviewed in ref. 42). In mammalian cells, sfRNA has been demonstrated to sequester G3BP1, G3BP2, and Caprin, which prevents induction of the IFN response (43). By RNA-affinity purification and mass spectrometry analysis, we uncovered that the Ae. aegypti proteins ME31B, ATX2, and AAEL018126 (a possible TYF homolog) significantly bind to ZIKV and WNV sfRNA, and additionally LSM12 and Lig were enriched in both sfRNA samples, albeit not significantly.
ME31B is involved in mRNA decapping, translation inhibition, mRNA storage, and miRNA-mediated mRNA decay (44, 45). ME31B localizes to processing bodies (PBs), which are nucleoprotein granules that are constitutively present, contain several proteins involved in mRNA turnover, and function as a storage hub for mRNAs that are translationally repressed (46). Many viruses interfere with PB assembly (47), including members of the Flaviviridae family (48–50). In our study, silencing of ME31B resulted in increased ZIKV and WNV RNA replication, suggesting that ME31B acts antiviral during flavivirus infection. Given that silencing of ME31B is known to result in the disassembly of PBs (50), it is also possible that PBs, and not ME31B per se, display antiviral activity in Ae. aegypti cells. Attempts to silence ME31B via dsRNA injection in vivo resulted in high mortality rates in the first 2 d postinjection, combined with poor silencing efficiency in the surviving mosquitoes at day 5, which may suggest that ME31B is important for mosquito homeostasis.
ATX2, which we demonstrated to indirectly bind to sfRNA via ME31B, is known to interact with TYF, ME31B, and poly(A)-binding protein (PABP) in Drosophila and acts as a regulator of protein translation (51–53) while a complex of ATX2, LSM12, and TYF regulates cap-dependent translation in Drosophila neurons (53). Thus, sequestration of (a complex of) these proteins by sfRNA may remodel the host cell mRNA and protein translation landscape to favor virus replication and translation. Indeed, sfRNA expression correlates with decreased mRNA turnover in mammalian cells (22), supporting a role of sfRNA in the regulation of host mRNAs.
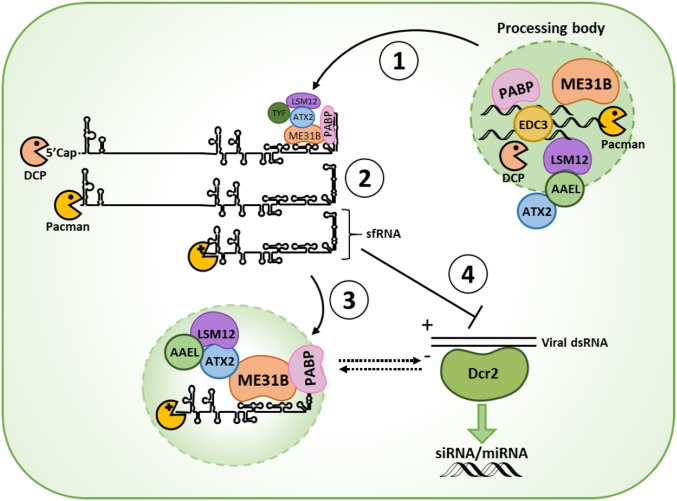
We now present a mechanistic working model in which sfRNA enhances flavivirus replication in mosquitoes to ultimately increase the efficiency of virus transmission, by sequestering a complex of PB components, consisting at least of ME31B, ATX2, LSM12, and AAEL018126 (Fig. 6, step 1). It is important to realize that, as the 3′ UTR of the vgRNA is identical in sequence and highly similar in RNA structure to sfRNA, the sfRNA binding proteins likely also interact with the 3′ UTR of the vgRNA (Fig. 6, step 1). As PB components are heavily involved in mRNA degradation, their recruitment by the 3′ UTR of the vgRNA likely induces the degradation of vgRNA, including the 5′ → 3′ degradation by XRN1. Stalling of XRN1 on specific RNA stem loop structures in the viral 3′ UTR results in the formation of sfRNA (Fig. 6, step 2).
Fig. 6.
Mechanistic model of sfRNA during flavivirus infection of mosquitoes. (Step 1) In the early stage of replication, (+) strand vgRNA molecules bind ME31B (ortholog of human DDX6) via the 3′ untranslated region (UTR) to recruit processing-body (PB) components to viral replication complexes. (Step 2) Recruitment of PB components involved in decapping and mRNA degradation, including decapping enzymes (DCPs) and the exoribonuclease Pacman (ortholog of human XRN1), induces decapping of free (+) viral genomic RNA followed by degradation by Pacman. Pacman stalls on resistant stem-loop RNA structures in the 3′ untranslated region (UTR), leading to the formation of sfRNA. (Step 3) SfRNA competes with the 3′ UTR for binding of PB-associated proteins and sequesters these proteins into cytoplasmic foci. (Step 4) SfRNA suppresses the RNAi response either directly by acting as a decoy substrate for Dcr2, or indirectly by remodeling PBs and associated RNA silencing complexes. PBs, processing bodies; Dcr2, Dicer2; PABP, poly-(A) binding protein; DCP, decapping enzyme; SL, stem-loop; DB, dumbbell; AAEL, AAEL018126.
The initial recruitment of such PB components to replication complexes can be beneficial for flavivirus replication since flaviviruses lack a poly-(A) tail and rely on the recruitment of viral and host proteins to their structured 5′ and 3′ UTRs for the assembly of translation initiation complexes (9, 54–56). For example, PABP binds to the DB region in the DENV 3′ UTR (54), and eukaryotic elongation factor (eEF)-1α binds to the 3′ SL of WNV and DENV (57, 58). Further, the mammalian ME31B ortholog DDX6 is recruited to WNV and hepatitis C virus (HCV) replication sites (48, 49) and promotes efficient DENV and ZIKV replication in mammalian cells (59, 60).
When flavivirus infection progresses, the accumulating sfRNA molecules will start to compete with protein binding to the 3′ UTR of the vgRNA and will sequester PB components and translation factors (Fig. 6, step 3). Since ME31B and ATX2 have been implicated in small RNA-mediated silencing (44, 45, 61), it is possible that the sfRNA–ME31B interaction modulates the RNAi response through remodeling of PBs and associated small RNA silencing complexes (Fig. 6, step 4). While uncovering additional details of how these virus–mosquito interactions promote flavivirus transmission by mosquitoes remains the focus of ongoing research, the presented findings have revealed important molecular function(s) of this intriguing noncoding RNA during flavivirus infection of the mosquito vector.
Materials and Methods
Cell Lines and Viruses.
African green monkey kidney Vero E6 cells (ATCC CRL-1586), Ae. aegypti Aag2, Aedes pseudoscutellaris AP-61 cells, and Aedes albopictus C6/36 (ATCC CRL-1660) and U4.4 cells were cultured as described previously (28, 62) (details in SI Appendix). To generate infectious clone-derived ZIKV, a pCC1-BAC–based and SP6 promoter-driven infectious clone of the BeH819015 ZIKV isolate from Brazil (GenBank KU365778.1) was retrieved from Andres Merits, University of Tartu, Tartu, Estonia (27), hereafter named pZIKVIC. Site-directed mutagenesis was used to introduce 4-nucleotide substitution in the pseudoknot of SL-I, to generate the sfRNA1-deficient mutant ZIKVΔSF1 and an additional 3-nucleotide substitution in the pseudoknot of SL-II to generate ZIKVΔSF1+2 (SI Appendix, Fig. S1A) (details in SI Appendix). pZIKVIC, pZIKV∆SF1, or pZIKV∆SF1+2-derived, in vitro transcribed, capped RNA was transfected into a preseeded monolayer of Vero cells in 6-well plates using Lipofectamine 2000 (Invitrogen). Six days posttransfection, the supernatant was harvested and passaged once on Vero cells before P2 and P3 stocks were generated on Ap-61 cells and stored at −80 °C until further use. Infectious clone-derived WNV GR10 strain (WNVIC) (GenBank KC496015.1) and sfRNA-deficient mutants WNV∆SF1 and WNV∆SF1+2 were reported previously (24) and were used as a P2 from C6/36 cells. Virus titers of stocks were determined after 1 freeze–thaw cycle by end-point dilution assay (EPDA) on Vero cells (24).
Infectious Blood Meals and Intrathoracic Injections.
Infectious blood meals were performed with Ae. aegypti (Rockefeller strain, obtained from Bayer AG) that were reared, maintained, and processed as described previously (28). Mosquitoes were fed for 1 h under light conditions at 24 °C and 70% relative humidity (RH) and anesthetized with 100% CO2, and fully engorged females were selected and maintained at 28 °C in a 12:12 light:dark cycle, 70% RH, and provided with 6% glucose solution. After 14 d, mosquitoes were anesthetized and salivated for ∼45 min as described previously (28). Briefly, mosquitoes were anesthetized with 100% CO2 and immobilized by removal of their legs and wings, and their proboscis was inserted into a 200-µL pipet tip containing 5 µL of a 50% sugar water, 50% FBS mixture. Intrathoracic injections were performed as described previously (28) on anesthetized Ae. aegypti by injecting 69 nL (∼400 tissue culture infectious dose 50% [TCID50]) of ZIKV or ZIKV∆SF1 using a Drummond Nanoject II injector (Drummond Scientific) with glass capillaries (3.5-inch Drummond no. 3-000-203-G/X; Drummond Scientific). Injected mosquitoes were maintained at 28 °C for 7 d in a 12:12 light:dark cycle, 70% relative humidity, and provided with 6% glucose solution. Presence of ZIKV in the mosquito bodies and salivas was determined by infectivity assay based on ZIKV-induced cytopathic effect (CPE) in Vero cells at 3 to 4 d postinfection (dpi), as described before (28). For virus titrations, the TCID50/mL was determined by EPDA on Vero cells as described previously (28). The TCID50/mL was scored at 3 to 4 dpi by virus-induced CPE.
Transfections and dsRNA-Mediated Gene Silencing.
Transfections of mosquito cells with DNA plasmids (cloning details in SI Appendix) were performed with Fugene HD (Promega) in serum-free media following the manufacturer’s protocol. Transfections of Vero cells with in vitro transcribed RNA (details in SI Appendix) were performed using Lipofectamine 2000 (Invitrogen) in Opti-MEM (Gibco). At 4 h posttransfection, the transfection mix was aspirated and replaced with fresh culture media. For silencing experiments, dsRNA was transfected into preseeded Aag2 cell monolayers in 24-well plates using Fugene HD. After 48 h, the cell monolayers were transfected a second time with dsRNA to ensure proper silencing efficiency (63). At 5 to 6 h after second transfection, cells were split into 96-well plates, and a fraction was stored for RNA isolation and quantification of gene silencing by quantitative real-time (qRT)-PCR. The 96-well plates of freshly seeded silenced cells were inoculated with WNV, WNV∆SF1+2, ZIKV, or ZIKV∆SF1 at the indicated multiplicity of infection (MOI). Viral titers in the supernatant were determined by EPDA on Vero cells at the indicate time point postinfection.
qRT-PCR.
Total RNA was extracted from cell monolayers with TRIzol reagent, and RNA yield was quantified by NanoDrop (Thermo). Equal amounts of total RNA were treated with RQ1-RNase-free DNase (Promega) to remove residual DNA contamination and subjected to first-strand synthesis using SuperScript II reverse transcriptase (Invitrogen) and random hexamers (Roche) according to the manufacturer’s protocols. The resulting cDNA was diluted 1:5 with MilliQ-H2O and subjected to relative quantification by qRT-PCR using SYBR Select Master mix (Invitrogen) and primer nos. 45 to 62 on a CFX96 Real-Time PCR system (Bio-Rad). Cycling conditions were 95 °C (2 min), (95 °C [15 s], 55 °C [30 s], 72 °C [30 s]) ×40, 72 °C (30 s), followed by a 55 to 95 °C melt curve (0.5 °C/5 s) to verify amplicon homogeneity. Data analysis was performed using Bio-Rad CFX Maestro software with relative quantification to a standard curve, consisting of 10-fold serial dilutions of Aag2 cells total RNA-derived cDNA and normalized to the rpS7 reference gene.
RNA-Affinity Purification.
RNA-affinity purification was based on the 4× S1m optimized streptavidin-binding RNA-aptamer system which has high affinity for streptavidin (33) (details in SI Appendix). Briefly, 50% Streptavidin Sepharose High Performance bead-slurry (SA-beads; GE Healthcare) was equilibrated by washing thrice with SA-RNP lysis buffer supplemented with protease inhibitors. Per 100 µL of equilibrated SA-beads, 20 µL of DNase-treated renatured in vitro transcribed 4× S1m aptamer-containing RNA was added and coupled to the SA-beads by incubation at 4 °C for 2.5 h with overhead rotation. RNA-bound beads were incubated with precleared Aag2 cell lysates (2.7 to 3.0 mg of protein) at 4 °C for 3.5 h under overhead rotation. Beads were collected and washed 5 times with SA-wash buffer, and RNA-bound proteins were eluted from the final pellet by addition of 30 µL of SA-elution buffer containing 0.6 µg of RNase A (Invitrogen) followed by 10-min incubation at 4 °C. Beads were removed, and ∼30 µL of eluate was collected and supplemented with 10 µL of 4× SDS loading buffer. Samples were stored at −20 °C before protein identification by mass spectrometry or Western blot (details in SI Appendix).
Next-Generation Small RNA Sequencing.
Total RNA was isolated from triplicate pools of 5 to 6 virus-positive mosquitoes (blood-fed or injected with ZIKV or ZIKVΔSF1) using TRIzol reagent (Invitrogen) following an adjusted version of the manufacturer’s protocol. Mosquitoes were incubated for 4 h in TRIzol while shaking, isopropanol precipitation was performed for 1 h at −80 °C, and an additional 75% ethanol wash step was incorporated. Small RNAs of 18 to 30 nt were sequenced from 2.5 µg of total RNA on a BGISEQ-500 sequencer (BGI Genomics), as described previously (62). Small RNA sequencing reads were uploaded to the NCBI Sequence Read Archive (SRA) under BioProject PRJNA525617. Single-end FASTQ reads were generated with an in-house filtering protocol of BGI. Small RNA libraries were analyzed on the Galaxy webserver (64). Quality of the small RNA libraries was confirmed by FastQC version 0.11.7 (65). Reads were mapped with Bowtie2 version 2.3.4.2 (66) to the genome of ZIKV or ZIKV∆SF1, allowing 1-nt mismatch in a seed length of 28 nt. Read counts were normalized against the total number of reads in each small RNA library and to the viral titers of mosquitoes within the different pools. Small-interfering RNA genome distributions were determined by filtering reads with a length of 21 nt and mapping the 5′ end of each read. Analysis of Piwi-interacting RNAs was performed on the https://mississippi.snv.jussieu.fr/ Galaxy server, as described in ref. 67.
Statistics.
For a detailed description of statistics, please see SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Andres Merits for providing the ZIKV infectious clone; Dr. Ronald van Rij for providing Aag2 cells; Dr. Byron Martina for providing AP-61 cells; Marleen Henkens for help in sample collection and mosquito titrations; Corinne Geertsema for maintenance of cell culture and assistance during mosquito experiments; Dr. Martijn van Hemert and Dr. Peter Bredenbeek for fruitful scientific discussions; Julian Bakker for help in constructing the pDONR-EGFP vector; and Dr. Mark Sterken for statistical analysis. We acknowledge Dr. Pascal Miesen for providing an efficient silencing protocol in Aag2 cells and for rearing Ae. aegypti Kenya mosquitoes, which were kindly contributed to our study by Dr. Yaw Afrane and Dr. Mariangela Bonizzoni. Ae. aegypti ir0115 mosquito eggs were obtained via the European Union Horizon 2020 Research Infrastructure Program Infravec2.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Small RNA sequencing reads were uploaded to the National Center for Biotechnology Information Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra (BioProject PRJNA525617).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905617116/-/DCSupplemental.
References
- 1.Weaver S. C., Charlier C., Vasilakis N., Lecuit M., Zika, chikungunya, and other emerging vector-borne viral diseases. Annu. Rev. Med. 69, 395–408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogels C. B., Göertz G. P., Pijlman G. P., Koenraadt C. J., Vector competence of European mosquitoes for West Nile virus. Emerg. Microbes Infect. 6, e96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatt S., et al. , The global distribution and burden of dengue. Nature 496, 504–507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer S. V., Tesh R. B., Vasilakis N., The emergence of arthropod-borne viral diseases: A global prospective on dengue, chikungunya and zika fevers. Acta Trop. 166, 155–163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO “Global burden of major vector-borne diseases, as of March 2017” in Global vector control response 2017–2030 (World Health Organization, 2017), pp. 41–42. [Google Scholar]
- 6.Brown J. E., et al. , Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution 68, 514–525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraemer M. U. G., et al. , The global distribution of the arbovirus vectors Aedes aegypti and Ae. Albopictus. Elife 4, e08347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatem A. J., Hay S. I., Rogers D. J., Global traffic and disease vector dispersal. Proc. Natl. Acad. Sci. U.S.A. 103, 6242–6247 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinton M. A., Basu M., Functions of the 3′ and 5′ genome RNA regions of members of the genus Flavivirus. Virus Res. 206, 108–119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinton M. A., Replication cycle and molecular biology of the West Nile virus. Viruses 6, 13–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pijlman G. P., et al. , A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4, 579–591 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Chapman E. G., et al. , The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 344, 307–310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akiyama B. M., et al. , Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 354, 1148–1152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva P. A. G. C., Pereira C. F., Dalebout T. J., Spaan W. J. M., Bredenbeek P. J., An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J. Virol. 84, 11395–11406 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y. S., Fan Y. H., Tien C. F., Yueh A., Chang R. Y., The conserved stem-loop II structure at the 3′ untranslated region of Japanese encephalitis virus genome is required for the formation of subgenomic flaviviral RNA. PLoS One 13, e0201250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Göertz G. P., Abbo S. R., Fros J. J., Pijlman G. P., Functional RNA during Zika virus infection. Virus Res. 254, 41–53 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Filomatori C. V., et al. , Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells. PLoS Pathog. 13, e1006265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan Y.-H., et al. , Small noncoding RNA modulates Japanese encephalitis virus replication and translation in trans. Virol. J. 8, 492 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donald C. L., et al. , Full genome sequence and sfRNA interferon antagonist activity of Zika virus from Recife, Brazil. PLoS Negl. Trop. Dis. 10, e0005048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuessler A., et al. , West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J. Virol. 86, 5708–5718 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manokaran G., et al. , Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350, 217–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moon S. L., et al. , A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA 18, 2029–2040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Funk A., et al. , RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 84, 11407–11417 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Göertz G. P., et al. , Noncoding subgenomic Flavivirus RNA is processed by the mosquito RNA interference machinery and determines West Nile virus transmission by Culex pipiens mosquitoes. J. Virol. 90, 10145–10159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pompon J., et al. , Dengue subgenomic flaviviral RNA disrupts immunity in mosquito salivary glands to increase virus transmission. PLoS Pathog. 13, e1006535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franz A. W., Kantor A. M., Passarelli A. L., Clem R. J., Tissue barriers to arbovirus infection in mosquitoes. Viruses 7, 3741–3767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mutso M., et al. , Reverse genetic system, genetically stable reporter viruses and packaged subgenomic replicon based on a Brazilian Zika virus isolate. J. Gen. Virol. 98, 2712–2724 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Göertz G. P., Vogels C. B. F., Geertsema C., Koenraadt C. J. M., Pijlman G. P., Mosquito co-infection with Zika and chikungunya virus allows simultaneous transmission without affecting vector competence of Aedes aegypti. PLoS Negl. Trop. Dis. 11, e0005654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xi Z., Ramirez J. L., Dimopoulos G., The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 4, e1000098 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnettler E., et al. , Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J. Virol. 86, 13486–13500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon S. L., et al. , Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology 485, 322–329 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blair C. D., Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 6, 265–277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leppek K., Stoecklin G., An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins. Nucleic Acids Res. 42, e13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeh S.-C., Pompon J., Flaviviruses produce a subgenomic Flaviviral RNA that enhances mosquito transmission. DNA Cell Biol. 37, 154–159 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Sim S., Jupatanakul N., Dimopoulos G., Mosquito immunity against arboviruses. Viruses 6, 4479–4504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olmo R. P., et al. , Control of dengue virus in the midgut of Aedes aegypti by ectopic expression of the dsRNA-binding protein Loqs2. Nat. Microbiol. 3, 1385–1393 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Colpitts T. M., et al. , Alterations in the Aedes aegypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PLoS Pathog. 7, e1002189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Souza-Neto J. A., Sim S., Dimopoulos G., An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. U.S.A. 106, 17841–17846 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartholomay L. C., et al. , Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science 330, 88–90 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez J. L., Dimopoulos G., The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev. Comp. Immunol. 34, 625–629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roby J. A., Pijlman G. P., Wilusz J., Khromykh A. A., Noncoding subgenomic flavivirus RNA: Multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses 6, 404–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charley P. A., Wilusz J., Sponging of cellular proteins by viral RNAs. Curr. Opin. Virol. 9, 14–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bidet K., Dadlani D., Garcia-Blanco M. A., G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 10, e1004242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostareck D. H., Naarmann-de Vries I. S., Ostareck-Lederer A., DDX6 and its orthologs as modulators of cellular and viral RNA expression. Wiley Interdiscip. Rev. RNA 5, 659–678 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Chu C. Y., Rana T. M., Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 4, e210 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Standart N., Weil D., P-bodies: Cytosolic droplets for coordinated mRNA storage. Trends Genet. 34, 612–626 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Poblete-Durán N., Prades-Pérez Y., Vera-Otarola J., Soto-Rifo R., Valiente-Echeverría F., Who regulates whom? An overview of RNA granules and viral infections. Viruses 8, 1–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ariumi Y., et al. , Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 85, 6882–6892 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chahar H. S., Chen S., Manjunath N., P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology 436, 1–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emara M. M., Brinton M. A., Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U.S.A. 104, 9041–9046 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Ling J., Yuan C., Dubruille R., Emery P., A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science 340, 879–882 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim C., Allada R., ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science 340, 875–879 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Lee J., et al. , LSM12 and ME31B/DDX6 define distinct modes of posttranscriptional regulation by ATAXIN-2 protein complex in Drosophila circadian pacemaker neurons. Mol. Cell 66, 129–140.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Polacek C., Friebe P., Harris E., Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J. Gen. Virol. 90, 687–692 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Paranjape S. M., Harris E., Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J. Biol. Chem. 282, 30497–30508 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Holden K. L., Harris E., Enhancement of dengue virus translation: Role of the 3′ untranslated region and the terminal 3′ stem-loop domain. Virology 329, 119–133 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Blackwell J. L., Brinton M. A., Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 71, 6433–6444 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Nova-Ocampo M., Villegas-Sepúlveda N., del Angel R. M., Translation elongation factor-1α, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 295, 337–347 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Ward A. M., et al. , Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 8, 1173–1186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McIntyre W., et al. , Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 46, 5776–5791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCann C., et al. , The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc. Natl. Acad. Sci. U.S.A. 108, E655–E662 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Göertz G. P., et al. , Mosquito small RNA responses to West Nile and insect-specific virus infections in Aedes and Culex mosquito cells. Viruses 11, E271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miesen P., Ivens A., Buck A. H., van Rij R. P., Small RNA profiling in dengue virus 2-infected Aedes mosquito cells reveals viral piRNAs and novel host miRNAs. PLoS Negl. Trop. Dis. 10, e0004452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goecks J., Nekrutenko A., Taylor J.; Galaxy Team , Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andrews S., FastQC: A Quality Control Tool for High Throughput Sequence Data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 1 May 2019.
- 66.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miesen P., Girardi E., van Rij R. P., Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res. 43, 6545–6556 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.