Abstract
Background:
DNA methylation (DNAm) may contribute to processes that underlie associations between air pollution and poor health. Therefore, our objective was to evaluate associations between DNAm and ambient concentrations of particulate matter (PM) ≤2.5, ≤10, and 2.5–10 μm in diameter (PM2.5; PM10; PM2.5–10).
Methods:
We conducted a methylome-wide association study among twelve cohort- and race/ethnicity-stratified subpopulations from the Women’s Health Initiative and the Atherosclerosis Risk in Communities study (n = 8397; mean age: 61.5 years; 83% female; 45% African American; 9% Hispanic/Latino American). We averaged geocoded address-specific estimates of daily and monthly mean PM concentrations over 2, 7, 28, and 365 days and 1 and 12 months before exams at which we measured leukocyte DNAm in whole blood. We estimated subpopulation-specific, DNAm-PM associations at approximately 485,000 Cytosine-phosphate-Guanine (CpG) sites in multi-level, linear, mixed-effects models. We combined subpopulation- and site-specific estimates in fixed-effects, inverse variance-weighted meta-analyses, then for associations that exceeded methylome-wide significance and were not heterogeneous across subpopulations (P < 1.0 × 10−7; PCochran’s Q > 0.10), we characterized associations using publicly accessible genomic databases and attempted replication in the Cooperative Health Research in the Region of Augsburg (KORA) study.
Results:
Analyses identified significant DNAm-PM associations at three CpG sites. Twenty-eight-day mean PM10 was positively associated with DNAm at cg19004594 (chromosome 20; MATN4; P = 3.33 × 10−8). One-month mean PM10 and PM2.5–10 were positively associated with DNAm at cg24102420 (chromosome 10; ARPP21; P = 5.84 × 10−8) and inversely associated with DNAm at cg12124767 (chromosome 7; CFTR; P = 9.86 × 10−8). The PM-sensitive CpG sites mapped to neurological, pulmonary, endocrine, and cardiovascular disease-related genes, but DNAm at those sites was not associated with gene expression in blood cells and did not replicate in KORA.
Conclusions:
Ambient PM concentrations were associated with DNAm at genomic regions potentially related to poor health among racially, ethnically and environmentally diverse populations of U.S. women and men. Further investigation is warranted to uncover mechanisms through which PM-induced epigenomic changes may cause disease.
Keywords: Particulate matter, DNA methylation, Epigenetics, Air pollution, Epigenome-wide association study
1. Introduction
Ambient particulate matter (PM) air pollution is a modifiable exposure that has been consistently associated with morbidity and mortality (Cohen et al., 2017; Di et al., 2017; Miller et al., 2007) attributed to cardiovascular disease (Brook et al., 2004, 2010), respiratory disease (Dominici et al., 2006; Gan et al., 2013; Laumbach and Kipen, 2012), and lung cancer (Pope et al., 2002; Raaschou-Nielsen et al., 2013). Despite the ubiquity of air pollution exposure and the continued population burden of PM (Cohen et al., 2017), the causal mechanisms underlying PM associations with poor health have not been adequately investigated.
One such mechanism could involve methylation of deoxyribonucleic acids (DNAm), conventionally measured at Cytosine-phosphate-Guanine (CpG) sites. DNAm is a heritable, but dynamic epigenetic modification that can influence gene expression without altering the DNA sequence (Clouaire and Stancheva, 2008; Neidhart, 2016) and may be central to mediation of PM-associated disease risk (Baccarelli et al., 2010; Bollati and Baccarelli, 2010; Zhong et al., 2016). Indeed, PM exposure has been implicated in whole blood DNAm near candidate genes involved in inflammation, oxidative stress, coagulation and vasoconstriction (Bellavia et al., 2013; Chen et al., 2015, 2016; Tarantini et al., 2009, 2013), abnormalities of which have established associations with cardiovascular and respiratory disease. A few studies have agnostically evaluated DNAm associations with PM on a methylome-wide scale (de F.C. Lichtenfels et al., 2018; Panni et al., 2016; Plusquin et al., 2017), but none have done so in large, sociodemographically and environmentally diverse, well-characterized populations of adult women and men.
The present study therefore examined methylome-wide associations between DNAm and ambient concentrations of PM ≤ 2.5, ≤10, and 2.5–10 μm in diameter (PM2.5, PM10, and PM2.5–10) within the Women’s Health Initiative (WHI) and the Atherosclerosis Risk in Communities study (ARIC) cohorts, and their replication in subpopulations of the Cooperative Health Research in the Region Augsburg (KORA) study.
2. Methods
2.1. Study design and populations
The study included 8397 consenting participants from subpopulations within the WHI and ARIC cohorts who had available peripheral blood leukocyte DNA.
The WHI is a multicenter prospective study of risk factors for cardiovascular disease (CVD), cancer, osteoporotic fractures, and other causes of morbidity and mortality among postmenopausal women (Anderson et al., 2003; NIH, 1998). Between 1993 and 1998, women aged 50–79 years from forty WHI clinical centers throughout the United States (US) were enrolled in the Clinical Trials (CT) (n = 68,132) or Observational Study (OS) (n = 93,676). All WHI participants completed a screening visit (SV). CT participants also completed an annual visit (AV) at one, three, six, and nine years after randomization (AV1, AV3, AV6, AV9), and OS participants three years after enrollment (AV3). An additional visit of CT and OS participant subsets occurred between 2011 and 2012 (ranging from 14 to 19 years after enrollment) as part of the WHI Long Life Study (LLS) (Anderson and LaCroix, n.d.).
For the current study, WHI participants were drawn from three ancillary studies: Epigenetic Mechanisms of PM-Mediated CVD Risk (WHI-EMPC) (Whitsel, n.d.), Broad Agency Announcement 23 (WHI-BAA23) (Assimes et al., n.d.) and Ancillary Study 311 (WHI-AS311) (Jordahl et al., 2018). WHI-EMPC is a study of epigenetic mechanisms underlying associations between ambient PM air pollution and CVD within the WHI CT. From this population, DNAm was measured in 2200 randomly selected participants (stage 1: SV, AV3, or AV6), remeasured in 200 participants at a second visit (stage 2: AV3 or AV6), and remeasured again in 43 participants at a third visit among those who participated in the WHI Long Life Study (stage 3: LLS), yielding 2443 total observations. WHI-BAA23, also known as Integrative Genomics and Risk of CHD and Related Phenotypes in the Women’s Health Initiative, is a case-control study of coronary heart disease within the WHI CT (n = 1546) and OS (n = 442). By design, WHI-BAA23 oversampled African Americans and Hispanic/Latino Americans and required all participants to have undergone genome-wide genotyping and profiling of seven cardiovascular disease biomarkers. DNAm was measured in blood collected at the SV, before the incidence of coronary heart disease. WHI-AS311 is a matched case-control study of bladder cancer among women within the WHI CT (n = 405) and OS (n = 455). Bladder cancer cases were matched to controls based on enrollment year, age at enrollment, follow-up time, and DNAm extraction method. DNAm was measured in blood collected at the SV, before the incidence of bladder cancer.
ARIC is a community-based prospective study of atherosclerosis and its clinical outcomes in four US communities: Washington County, Maryland; Forsyth County, North Carolina; selected suburbs of Minneapolis, Minnesota; and Jackson, Mississippi (ARIC Investigators, 1989). Enrollment in 1987–1989 (Visit 1) was followed by five sub-sequent visits (Visits 2–6) between 1990 and 2017. The present study included all 2796 African Americans from Forsyth County or Jackson (ARIC-AA) with DNA and 1139 European Americans from Forsyth County or Minneapolis (ARIC-EA) with cerebral magnetic resonance imaging data (Mosley et al., 2005), all at Visits 2 (1990–1992) or 3 (1993–1995).
Replication involved up to 2176 participants from two studies of the population-based KORA cohort: F3 (n = 464) and F4 (n = 1712). KORA F3 (2004–2005) and F4 (2006–2008) are follow-up studies of the KORA S3 and S4 cohort participants, including German nationals aged 25–74 years from Augsburg, Germany (Holle et al., 2005; Wichmann et al., 2005).
2.2. Particulate matter exposure estimation
The study focuses on three ambient particulate matter (PM) air pollutants, including two (PM2.5 and PM10) that are regulated under the Clean Air Act by the US Environmental Protection Agency (EPA) according to its National Ambient Air Quality Standards (NAAQS) (EPA, 2017).
PM exposures were estimated at all geocoded WHI and ARIC participant addresses (Whitsel et al., 2004, 2006) in the contiguous US since the baseline examinations using two exposure modeling approaches, both based on US EPA Air Quality System (AQS) monitoring data for PM10 (since 1987) and PM2.5 (since 1999). In the WHI, the median distance from geocoded participant addresses to PM10 and PM2.5 EPA monitors was 7.8 and 7.6 km. In ARIC, it was 4.8 and 7.2 km. Geocoded address-specific daily mean PM10 concentrations (μg/m3) were spatially estimated using national-scale, log-normal ordinary kriging. Exposure measurement error using kriging methods may yield misclassification and increase variance or bias associations (Alexeeff et al., 2014; Lee et al., 2012), therefore validity of the estimation was assessed, using standard cross-validation statistics: average prediction error (PE), standardized prediction error (SPE), root mean square standardized (RMSS), and standard error (SE). Observed values of PE and SPE near zero, RMSS near one, and RMS near SE have provided evidence of model validity (Liao et al., 2006, 2007).
Also, geocoded address-specific monthly mean concentrations (μg/m3) were spatiotemporally estimated using generalized additive mixed models and geographic information system-based predictors. Because EPA AQS monitoring data for PM2.5 were not widely available until 1999, spatiotemporal estimation also involved the log-transformed ratio of PM2.5 to predicted PM10 between 1987 and 1999. A five- or ten-fold, out-of-sample cross-validation of the estimates in which the squared Pearson correlation between excluded monthly observations and model predictions (R2 = 0.68–0.77) indicated that estimation models performed well (Yanosky et al., 2014).
Daily mean concentrations of PM10 were averaged over the 2-, 7-, 28-, and 365-day periods ending on (including) the examination day. Monthly mean concentrations of PM2.5 and PM10 were averaged over the 12-month period ending on (including) the calendar month of ex-amination. Finally, coarse PM (PM2.5–10) concentrations for each averaging duration were calculated as differences between PM10 and PM2.5 concentrations.
2.3. DNA methylation
Peripheral blood leukocytes were isolated from visit-specific, fasting blood drawn from study participants. DNA was extracted from the peripheral blood leukocytes and then DNAm was measured on a methylome-wide scale at 485,577 CpG sites using the Illumina 450K Infinium Methylation BeadChip (Illumina Inc.; San Diego, CA, USA). Methylation was quantitatively represented by beta, the proportion of methylated cytosines over the sum of methylated and unmethylated cytosines across the same loci. The data from all studies were quality controlled (Table S1), Beta Mixture Quantile (BMIQ)-normalized to adjust for probe bias (Teschendorff et al., 2013), and in WHI-EMPC, ComBat-adjusted for stage and plate using empirical Bayes methods (Johnson et al., 2007). Otherwise, technical covariates (assay plate, chip, and row) were available to control for batch effects; and leukocyte proportions (CD8+ T cell, CD4+ T cell, B cell, natural killer cell, monocyte, and granulocyte) to account for leukocyte composition (Houseman et al., 2012). Among ARIC-AA participants, missing lymphocyte, monocyte, neutrophil, eosinophil, and basophil proportions were imputed based on measured proportions. Analyses excluded CpG sites at which DNAm distributions were multi-modal (Andrews et al., 2016) in at least one study.
2.4. Multiple imputation
To avoid potential for selection bias in complete-data analysis when data are missing at random (Hernan et al., 2004), multivariate imputation by chained equations (MICE) (Azur et al., 2011; Stuart et al., 2009) as implemented in SAS 9.3 (Cary, NC) was used to impute in-frequently missing PM2.5, PM10, and PM2.5–10 concentrations (missing range: 3.3%, 3.5%) and other covariates (missing range: 0%, 10.4%), excluding methylome-wide DNAm. Binary and categorical data were imputed using the logistic and discriminant functions whereas interval-scale data were imputed using predictive means matching with a k-nearest neighbor (k = 5) approach.
2.5. Statistical analysis
All analyses were stratified by cohort and race/ethnicity (African-, European-, and Hispanic/Latino-American) and adjusted for age (years) at blood draw, education (high school education or lower, more than high school), smoking status (current, former, never), alcohol use (current, former, never), physical activity (metabolic equivalent of task [MET-hours/week]), body mass index (BMI, kg/m2), neighborhood socioeconomic status (Roux et al., 2001), mean temperature (°C), mean dew point (°C), mean barometric pressure (kPa), season, and methylation-related variables, which included ten principal components (PCs) for genetic ancestry (when available), leukocyte proportions, and technical covariates. Analyses additionally controlled for cohort-specific covariates, including binary sex (male, female) in ARIC; randomly assigned treatment group (CT subpopulations of WHI-AS311, WHI-BAA23, WHI-EMPC); case-control status (WHI-AS311, WHI-BAA23); and control matching criteria (WHI-AS311).
In each subpopulation, covariate-adjusted, multi-level, linear, mixed-effects models (LMMs) were used to estimate DNAm-PM associations. In WHI-EMPC, three-level, longitudinal models had a random intercept for examination at the participant level, a random intercept and slope for PM at the WHI center level, and a random intercept for chip, as given by
| (1) |
In WHI-BAA23 CT & OS, and WHI-AS311 CT & OS, two-level cross-sectional models had a random intercept and slope for PM at the WHI center level and a random intercept for plate and chip, as given by
| (2) |
In ARIC-AA and ARIC-EA, one-level cross-sectional models had a random intercept for plate and chip, as given by
| (3) |
Above, i, j and k denote the ith examination of the jth participant in the kth center; DNAm is the CpG site-specific beta value; β0 is the intercept; PM is the 2-, 7-, 28-, 365-day, or 1- or 12-month mean of PM2.5, PM10, or PM2.5–10; and Z is a vector of covariates. The terms are a random intercept and a random slope for PM at the center level, is a random intercept for examination at the participant level, are random intercepts for technical covariates, and is the random error at the examination level. Measures of association (β1) and their 95% confidence intervals (β1 ± 1.96 × standard error) were reported as an absolute percentage change in DNAm per 10 μg/m3 increase in PM.
Given the focus on fixed effects, LMMs were fit with maximum likelihood using the MixedModels package (Bates, 2017) in Julia v0.6 (Bezanson et al., 2017). Stratum-specific results were combined using fixed-effects, inverse-variance weighted meta-analysis. Homogeneity of associations was assessed using Cochran’s Q test statistic (Cochran, 1954). A PCochran’s Q < 0.10 and Bonferroni-corrected threshold of P < 1 × 10−7 (i.e. assuming 500,000 independent CpG tests) were used to identify significant CpG associations. The threshold of suggestive significance was P < 1 × 10−5.
Examination of stratified and meta-analyzed results included reviewing quantile-quantile (QQ) plots of the observed −log10-transformed P values for each CpG site against the expected values from a theoretical χ2 distribution and estimating the associated genomic inflation factor (λ), where λ is defined as the ratio of the observed to expected median −log10P values (Devlin et al., 2001).
2.6. Technical validation
In a random subset of 200 WHI-EMPC participants, bisulfite pyrosequencing was used to validate the Illumina 450K measures of DNAm at ten PM10- or PM2.5-sensitive CpG sites (P < 1 × 10−5). CpG sites with poor next generation sequencing data or situated in CpG-rich, repetitive element, or low sequence complexity regions of the genome were not candidates for pyrosequencing. Site-specific comparisons of DNAm measures were based on mean Illumina 450K minus bisulfite pyrosequencing differences (Δ), Pearson correlation coefficients (r), and Deming regression estimates of their intercepts (α) and slopes (β) (Cornbleet and Gochman, 1979). When the two measures are nearly identical, Δ, r, α, and β approach values of 0, 1, 0, and 1, respectively.
2.7. Functional annotation
Published genotype-phenotype associations for variants annotated to or within 100 kilobases of genes containing statistically significant PM-sensitive CpG sites were identified in the National Human Genome Research Institute (NHGRI) Genome-Wide Association Study (GWAS) Catalog (Welter et al., 2014). Tissue-specific gene expression was assessed using the Genotype-Tissue Expression (GTEx) database (Lonsdale et al., 2013) and associations between DNAm and gene expression in human blood cells were obtained from a study of approximately 400,000 CpG sites and > 13,000 transcripts in the Multi-Ethnic Study of Atherosclerosis (MESA) and Grady Trauma Project (GTP) cohorts (Kennedy et al., 2018). PM-sensitive CpG sites (P < 1 × 10−5) were functionally characterized using experimentally derived Functional element Overlap analysis of ReGions from EWAS (eFORGE) v2.0 (Breeze et al., 2016) with data from the Encyclopedia of DNA elements (ENCODE) (Consortium, 2012), Roadmap Epigenomics Project (Bernstein et al., 2010), and BLUEPRINT (Stunnenberg et al., 2016). Overlap of CpG site-specific PM sensitivity, histone modification, and DNase I hypersensitivity were evaluated in eFORGE with a false discovery rate (FDR) threshold of 0.05.
2.8. Replication
Significant CpG sites that were not heterogeneous across sub-populations (P < 1.0 × 10−7; PCochran’s Q > 0.10) underwent replication and meta-analyses in KORA F3 and F4. Pollutant-and averaging duration-specific replication thresholds were Bonferroni-corrected by dividing the conventional alpha level (0.05) by the number of CpG sites carried into replication.
3. Results
The study consisted of twelve ARIC and WHI subpopulations, collectively representing 8397 participants, of whom 45.8% were African American, 8.4% were Hispanic/Latino American, and 83.0% were female (Table 1). Participants were on average 61.3 years of age and contributed methylation data at ≥461,014 CpG sites. One-month mean concentrations of PM10, PM2.5, and PM2.5–10 were 20.9, 13.2, and 7.7 μg/m3; varied by subpopulation and race/ethnicity (Tables 1 and S2); and did not exceed NAAQS in place at the time of data collection. Between-pollutant Pearson correlation coefficients depended on size fraction and averaging duration (Table 2). Overall, the median (range) was 0.35 (−0.14, 0.79) and among 2-, 7-, 28, and 365-day mean PM10 concentrations, it was 0.64 (0.43, 0.79). Correlations between PM10 and PM2.5 concentrations were 0.73 and 0.64 when they were averaged over 1 and 12 months.
Table 1.
Characteristics of the study participants, by subpopulation.
| Subpopulation | Race/ethnicity | n | % female | Age, yrs |
Maximum CpGs | PM (μg/m3), 1 mo (SD) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| (SD) | PM10 | PM2.5 | PM2.5–10 | |||||||
| ARIC | AA | 2664 | 63% | 56.6 (5.9) | 463,431 | 20.5 (4.6) | 13.2 (3.1) | 7.3 (2.1) | ||
| EA | 1100 | 58% | 59.9 (5.4) | 462,543 | 23.2 (5.3) | 15.4 (4.3) | 7.8 (3.5) | |||
| WHI | AS311 | CT | EA | 351 | 100% | 64.7 (7.1) | 461,136 | 19.8 (6.6) | 11.9 (3.82) | 7.9 (4.6) |
| OS | EA | 395 | 100% | 66.2 (6.9) | 461,136 | 19.9 (5.7) | 12.0 (3.9) | 7.9 (4.1) | ||
| BAA23 | CT | AA | 371 | 100% | 61.8 (6.3) | 461,014 | 22.6 (6.2) | 14.3 (4.2) | 8.3 (3.8) | |
| EA | 926 | 100% | 67.8 (6.2) | 461,014 | 19.7 (5.7) | 11.7 (3.7) | 8.0 (4.4) | |||
| HLA | 220 | 100% | 60.7 (6.4) | 461,014 | 21.4 (8.1) | 10.3 (4.1) | 11.1 (5.7) | |||
| OS | AA | 259 | 100% | 62.8 (6.8) | 461,014 | 22.3 (5.9) | 14.0 (4.0) | 8.3 (4.2) | ||
| HLA | 174 | 100% | 62.8 (7.3) | 461,014 | 23.0 (8.1) | 11.0 (4.2) | 11.9 (6.4) | |||
| EMPCa | AA | 553 | 100% | 62.7 (6.9) | 463,916 | 22.2 (6.2) | 15.2 (5.1) | 7.0 (4.7) | ||
| EA | 1072 | 100% | 64.6 (7.1) | 463,916 | 19.4 (6.0) | 13.0 (5.0) | 6.4 (5.2) | |||
| HLA | 312 | 100% | 61.5 (6.1) | 463,916 | 21.9 (7.1) | 12.8 (6.3) | 9.1 (5.3) | |||
| All | AA (45.8%) | |||||||||
| HLA (8.4%) | 8397 | 83% | 61.3 (7.4) | 463,916 | 20.9 (5.8) | 13.2 (4.3) | 7.7 (4.0) | |||
| EA (45.8%) | ||||||||||
Abbreviations: AA, African American; ARIC, Atherosclerosis Risk in Communities; AS311, Ancillary Study 311; BAA23, Broad Agency Award 23; CpG, Cytosine-phosphate-Guanine; CT, Clinical Trial; EA, European American; EMPC, Epigenetic Mechanisms of PM-Mediated CVD Risk; HLA, Hispanic/Latino American; mo, month; OS, Observational Study; PM10, PM < 10 μm in diameter; PM2.5, PM < 2.5 μm in diameter; PM2.5–10, PM > 2.5 and < 10 μm in diameter; SD, standard deviation; WHI, Women’s Health Initiative; , mean.
At the 1st visit. Methylation data also were available among 185 & 43 WHI-EMPC participants @ the 2nd & 3rd visits
Table 2.
Particulate matter concentration (μg/m3) means and Pearson correlations in the total population (n = 8397).
| PM10 | PM10 | PM10 | PM10 | PM10 | PM10 | PM2.5 | PM2.5 | PM2.5–10 | PM2.5–10 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| 2 d | 7 d | 28 d | 365 d | 1 mo | 12 mo | 1 mo | 12 mo | 1 mo | 12 mo | ||
|
| |||||||||||
| (SD) | 31.9 (12.1) | 31.1 (9.2) | 30.9 (7.1) | 31.2 (5.1) | 20.9 (5.8) | 20.9 (4.0) | 13.2 (4.3) | 13.2 (3.0) | 7.7 (4.0) | 7.8 (3.1) | |
| PM10 | 2 d | 1.00 | |||||||||
| PM10 | 7 d | 0.74 | 1.00 | ||||||||
| PM10 | 28 d | 0.58 | 0.79 | 1.00 | |||||||
| PM10 | 365 d | 0.43 | 0.56 | 0.70 | 1.00 | ||||||
| PM10 | 1 mo | 0.39 | 0.48 | 0.54 | 0.27 | 1.00 | |||||
| PM10 | 12 mo | 0.15 | 0.18 | 0.24 | 0.35 | 0.62 | 1.00 | ||||
| PM2.5 | 1 mo | 0.29 | 0.36 | 0.41 | 0.17 | 0.73 | 0.39 | 1.00 | |||
| PM2.5 | 12 mo | 0.11 | 0.12 | 0.15 | 0.23 | 0.40 | 0.64 | 0.66 | 1.00 | ||
| PM2.5–10 | 1 mo | 0.25 | 0.31 | 0.35 | 0.21 | 0.67 | 0.48 | −0.02 | −0.13 | 1.00 | |
| PM2.5–10 | 12 mo | 0.08 | 0.12 | 0.17 | 0.23 | 0.41 | 0.67 | −0.14 | −0.14 | 0.74 | 1.00 |
Abbreviations: d, day; mo, month; PM, particulate matter; PM10, PM < 10 μm in diameter; PM2.5, PM < 2.5 μm in diameter; PM2.5–10, PM > 2.5 and < 10 μm in diameter; SD, standard deviation; , mean.
QQ plots (Fig. 1) based on the trans-ethnic, fixed-effects, inverse variance-weighted meta-analyses provided little evidence of inflation across pollutants and averaging durations: median (range) λ = 1.01, (0.89–1.07). Manhattan plots (Fig. 2) show three significant (P < 1 × 10−7) and 55 suggestively significant (1 × 10−5 < P < 1 × 10−7) PM-sensitive CpG sites (Tables 3 and S3). The three significant CpG sites (cg19004594; cg24102420; cg12124767) were neither within ten base pairs of single nucleotide polymorphisms (minor allele frequency > 1%) nor previously identified as cross-reactive probes (Chen et al., 2013).
Fig. 1.
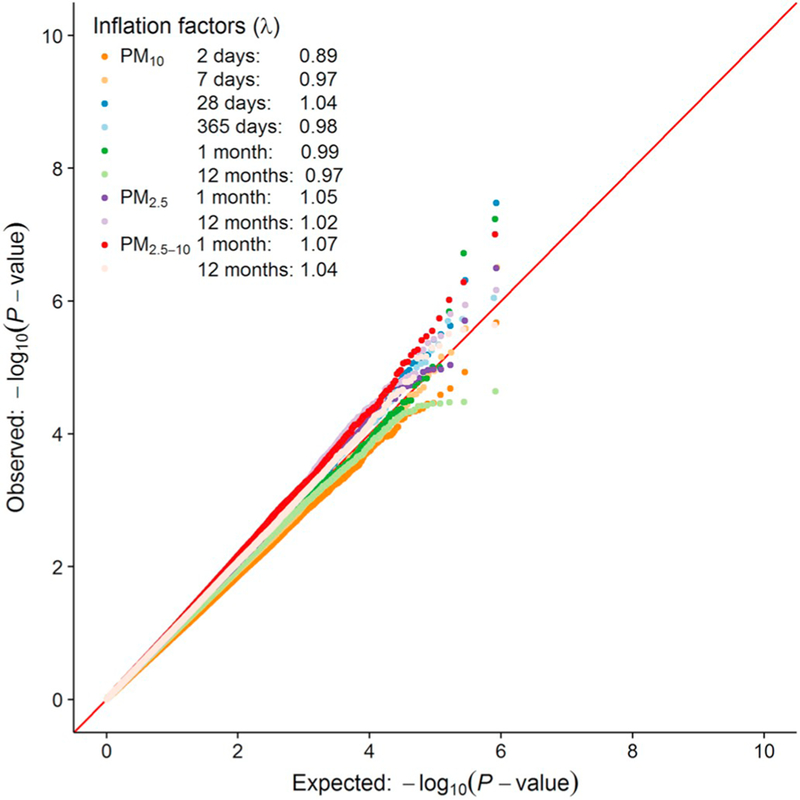
Quantile-quantile (QQ) plot of observed vs. expected −log10 P-value of each CpG site from trans-ethnic, fixed-effects meta-analyses of 2-, 7-, 28-, and 365-day PM10 and 1- and 12-month PM10 and PM2.5. The red diagonal line references the methylome-wide significance threshold (P < 1.0 × 10−7). Lambda (λ) is the inflation factor. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Manhattan plot of −log10 P-value vs. chromosomal position of each CpG site from trans-ethnic, fixed-effects meta-analyses of 2-, 7-, 28-, and 365-day PM10 and 1- and 12-month PM10 and PM2.5. The red line references the methylome-wide significance threshold (P < 1.0 × 10−7). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 3.
Findings from trans-ethnic, fixed-effects meta-analyses (P < 1 × 10−7, PCochran’s Q > 0.10).
| Chr | Positiona | CpG | Exposure | %Δ (95% CI)b | P | nobs | Gene |
|---|---|---|---|---|---|---|---|
| 20 | 43,926,884 | cg19004594 | PM10, 28 d | 0.3 (0.2, 0.4) | 3.33 × 10−8 | 8622 | MATN4 |
| 3 | 35,785,890 | cg24102420 | PM10, 1 mo | −0.5 (−0.7, −0.3) | 5.84 × 10−8 | 8575 | ARPP21/miR128–2 |
| 7 | 117,299,297 | cg12124767 | PM2.5–10, 1 mo | −0.5 (−0.7, −0.3) | 9.96 × 10−8 | 8577 | CFTR |
Abbreviations: Δ, change; Chr, chromosome; CI, confidence interval; CpG, Cytosine-phosphate-Guanine; d, days; mo, month; PM10, PM < 10 μm in diameter; PM2.5, PM < 2.5 μm in diameter; PM2.5–10, PM > 2.5 and < 10 μm in diameter.
Build 37.
Absolute percentage point per 10 μg/m3 increase in PM10.
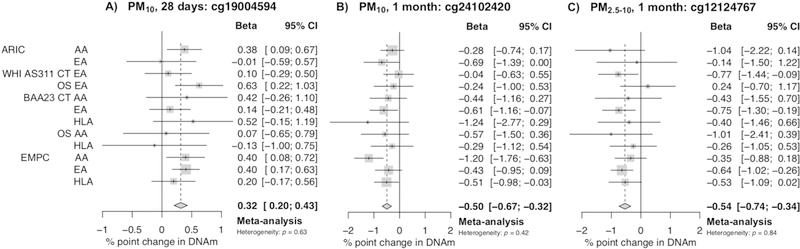
On chromosome 20 within an exonic CpG island of MATN4, a 10 μg/m3 increase in 28-day mean PM10 was associated with a 0.3% (95% confidence interval [CI]: 0.2, 0.4) higher DNAm at cg19004594 (P = 3.33 × 10−8; Fig. 3A). On chromosome 3 intronic to ARPP21, a 10 μg/m3 increase in 1-month mean PM10 was associated with a 0.5% (95% CI: 0.3, 0.7) lower DNAm at cg24102420 (P = 5.84 × 10−8; Fig. 3B). Cg24102420 is approximately 200 base pairs upstream from the transcriptional start site for microRNA 128–2 (miR128–2). On chromosome 7 intronic to CFTR, a 10 μg/m3 increase in 1-month mean PM2.5–10 was associated with a 0.5% (95% CI: 0.3, 0.7) lower DNAm at cg12124767 (P = 9.86 × 10−8; Fig. 3C). Furthermore, PM associations with cg19004594, cg24102420, and cg12124767 were similar across race/ethnic strata (Fig. S1). Complete annotations for all PM-sensitive CpG sites (P < 1 × 10−7) are available in Excel Table S1.
Fig. 3.
Forest plots of PM-CpG associations (95% confidence intervals) for A) cg19004594, B) cg2410240, and C) cg12124767 with a 10 μg/m3 increase in PM by subpopulation and overall after fixed-effects meta-analysis.
3.1. Technical validation
Overall, bisulfite pyrosequencing and Illumina 450K-based DNAm measures were similar (Table S4). The medians (interdecile ranges) of Δ, r, α and β were: 0.01 (−0.06, 0.07), 0.73 (0.20, 0.83), 0.04 (−0.27, 0.24), and 0.98 (0.09, 1.62). Corresponding estimates (95% CIs) for cg24102420 were −0.04 (−0.04, −0.03), 0.79 (0.73, 0.83), −0.16 (−0.38, 0.07) and 1.13 (0.88, 1.39). Cg19004594 and cg12124767 were not pyrosequenced.
3.2. Functional annotation
MATN4 is highly expressed in the pancreas, reproductive tract, and skin (Fig. S2), but variants of this gene have not been significantly associated (P < 5 × 10−8) with any phenotypes in prior GWAS. ARPP21 is primarily expressed in the brain (Fig. S3), is significantly associated with neuroticism and severe H1N1 influenza, and suggestively associated (5 × 10−8 < P < 5 × 10−6) with entorhinal cortical thickness and childhood-onset asthma in prior GWAS. CFTR is expressed in various tissues, including the pancreas, colon, minor salivary gland, digestive tract, and lung (Fig. S4). CFTR polymorphisms are associated with cystic fibrosis (CF), Barrett’s esophagus/esophageal carcinoma, and coronary artery disease.
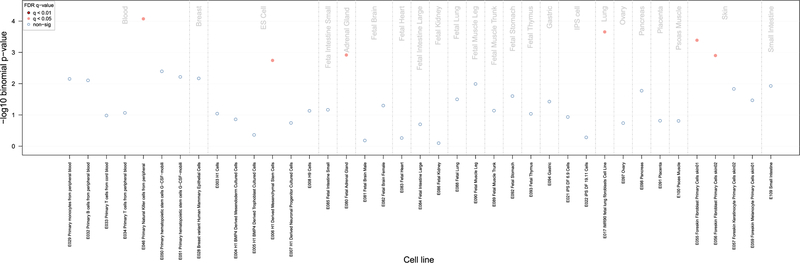
Differential methylation at cg19004594, cg24102420, or cg12124767 was not associated with gene expression in blood cells at any of the > 13,000 transcripts evaluated (P > 10−5) in the MESA/GTP cohorts. Although genomic regions around PM-sensitive CpG sites were associated with tri-methylation of histone 3 at lysine 9 (H3K9me3) in natural killer cells, derived mesenchymal stem cells, the fetal adrenal gland, fetal lung fibroblasts, and foreskin fibroblasts (FDR < 0.05; Fig. 4), they were not associated with mono- or tri-methylation of histone 3 at lysine 4, 27, or 36 (H3K4me1, H3K4me3, H3K27me3, or H3K36me3) or DNase I hypersensitivity in any tissues catalogued by eFORGE.
Fig. 4.
Enrichment of PM-sensitive CpG sites in regions overlapping H3K9me3 using Roadmap data.
3.3. Replication
The three statistically significant, non-heterogeneous PM-sensitive CpG sites (cg19004594; cg24102420; cg12124767) did not replicate in KORA F3/F4 (Table S5).
4. Discussion
This methylome-wide association study (MWAS) discovered three CpG sites at which higher levels of monthly mean ambient particulate matter air pollution concentrations were associated with DNAm. The DNAm-PM associations at all three CpG sites were homogeneous across the twelve subpopulations and each site was annotated to a neurological, pulmonary, endocrine, or cardiovascular disease-related gene (MATN4, ARPP21 or CFTR). Although a recent MWAS also implicated cigarette smoking in DNA methylation at ARPP21 and CFTR (Joehanes et al., 2016)—two genes that may underlie epigenetically mediated responses to inhalable environmental exposures—the CpG sites discovered herein are in different regions of ARPP21 and CFTR, suggesting varied responses to particulate exposures, and none of them were associated with gene expression of blood cells in MESA/GTP.
Methylation of cg19004594 (exon of MATN4) was positively associated with 28-day mean PM10 concentrations. MATN4 encodes Matrilin 4, a von Willebrand factor A domain-containing protein, which contributes to cardiac remodeling (Barallobre-Barreiro et al., 2012) and inhibits the proliferation of hematopoietic stem cells at rest. Additionally, environmental stressors trigger expression of the CXCL12- encoded chemokine (SDF1) (Liberda et al., 2010) and activation of its G protein-coupled receptor (CXCR4), leading to inhibition of Matrilin 4 and subsequent expansion of hematopoietic stem cell pools (Uckelmann et al., 2016). SDF1-activated CXCR4 also inhibits beta-adrenergically activated calcium influx through myocardial L-type calcium ion channels (Pyo et al., 2006), a process that may affect PM10-associated ventricular action potential and electrocardiographic QT interval duration (Gondalia et al., 2017). Methylation of MATN4 may therefore underlie commonly observed hematological and electrocardiographic of effects of PM10.
Methylation at cg24102420 (intron of ARPP21) was positively associated with 1-month mean PM10 concentrations. ARPP21 encodes a neuronal cAMP-regulated phosphoprotein, a regulator of calmodulin signaling (RCS) that is highly enriched in medium spiny neurons within the basal ganglia, cerebral cortex, and other regions of the brain (Rakhilin et al., 2004), with dual evidence of expression in cardiac tissues (Kahr et al., 2011; Kirchhof et al., 2011; Mathar et al., 2013). Variants of ARPP21 have been associated with entorhinal cortical thickness (Furney et al., 2010). Calmodulin signaling (O’Day et al., 2015), entorhinal cortical thickness (Velayudhan et al., 2013), and PM air pollution (Cacciottolo et al., 2017) are all associated with Alzheimer’s disease progression, suggesting a potential epigenetic mechanism of PM10-related neuropathology.
Indeed, ARPP21 and miR128–2, a microRNA within ARPP21, are both regulators of dendritic growth (Rehfeld et al., 2018). In a study of rats, exposure to ammonium sulfate, a major component of PM2.5, was associated with diminished dendritic complexity in hippocampal neurons (Cheng et al., 2017). Additionally, miR128 expression in peripheral blood of steel plant workers increased with increases in PM exposure, as was confirmed by an in vitro study of PM-treated pulmonary tissue (Bollati et al., 2015). Additional roles of miR128 include the inhibition of ABCA1 and ABCG1, adenosine triphosphate-binding cassette (ABC) transporter genes also involved in homeostasis of cholesterol (Adlakha et al., 2013), an established risk factor for stroke, myocardial infarction, and other common forms of cardiovascular disease.
Methylation at cg12124767 (intron of CFTR) was inversely associated with 1-month mean PM2.5–10 concentrations. CFTR encodes a transmembrane conductance regulator; specifically, an ABC transporter of chloride and thiocyanate ions. The CFTR-encoded ABC transporter controls fluid secretion and absorption in epithelial tissues (Saint-Criq and Gray, 2017). Its most common mutation impairs folding and trafficking of the encoded protein in pulmonary and pancreatic epithelia, causing CF and CF-related diabetes (Brennan et al., 2004). However, cigarette smoke and chronic inflammation also reduce CFTR chloride channel function (Rasmussen et al., 2014), a hypothesized molecular pathway underlying the development of chronic obstructive pulmonary disease (Rab et al., 2013). Furthermore, CFTR chloride channel currents in the myocardium shorten action potential and QT interval duration (Duan, 2013). Their activation by cAMP protein kinase A (PKA), protein kinase C (PKC), or extracellular adenosine triphosphate (ATP) through purinergic receptors (al-Awqati, 1995; Duan, 2013) can be arrhythmogenic (Cacciapuoti et al., 1991; Engler and Yellon, 1996; Leonard et al., 2017; Najeed et al., 2002; Yamazaki and Hume, 1997). Hypomethylation of CFTR at this site therefore highlights another epigenetic mechanism that may underlie PM10-related pulmonary and electrocardiographic manifestations of disease.
While the putative mechanisms described above are biologically plausible, analyses on which they are based are limited by their reliance on DNAm derived from leukocytes. Although other (e.g. heart, lung, nervous) tissues may be more appropriate for studying the role of DNAm on human disease, their collection is highly invasive (McCullough et al., 2017; Zhong et al., 2016); as such, leukocytes extracted from peripheral blood are widely used surrogate tissues (Zhong et al., 2016) with demonstrated consistency of DNAm patterns across relevant tissues types (Byun et al., 2009; Fan and Zhang, 2009; Ma et al., 2014). Still, DNAm at cg19004594, cg24102420, cg12124767 was not associated with gene expression of blood cells in GTP/MESA (Kennedy et al., 2018). Unlike DNAm patterns though, gene expression is highly variable by tissue type (Aguet et al., 2017), and MATN4, ARPP21 and CFTR are primarily expressed in other tissues.
The inability to replicate associations in KORA F3 and F4 participants is noteworthy. Although independent from the discovery populations, KORA represents a population of white, European men and women living in Augsburg, Germany, one distinct from that of the environmentally diverse, multi-racial/ethnic U.S. populations in the discovery. In addition, PM composition in ARIC and WHI (1990–2012) may differ from that in Augsburg during KORA F3 and F4 (2004–2006). Furthermore, PM concentrations in KORA were measured at community monitors, while those in WHI and ARIC were spatially or spatiotemporally estimated at participant geocoded addresses from monitoring networks in the 48 contiguous US states.
DNAm associations with PM2.5 – potentially the driver for PM-associated disease (Brook et al., 2010) – were not detected in this study. Inability to do so may be due to lower power to detect PM2.5 versus PM10 associations with DNAm given lower-variance PM2.5 exposure estimates, lack of short-duration PM2.5 data before 1999 when EPA AQS started monitoring it, and/or induction of PM2.5 health effects that are not epigenetically mediated.
The analyses also were limited by predominantly cross-sectional data, high multiple testing burden, small effect sizes, and residual need for functional characterization. However, repeated measures of PM and DNAm over time were leveraged in WHI-EMPC to increase statistical power. Among-pollutant correlations also were moderate in this context, so the multiple comparisons made were not strictly independent. Similarly, the Bonferroni-corrected threshold used herein (P < 1 × 10−7) is conservative because of methylome-wide correlations among CpG sites (Saffari et al., 2018; Tsai et al., 2012), decreasing the likelihood of false positives. Moreover, observed effect sizes were consistent with those seen in other epigenetic studies of particulate matter exposure (de F.C. Lichtenfels et al., 2018; Panni et al., 2016; Plusquin et al., 2017) and smoking (Joehanes et al., 2016). Further investigation is nonetheless needed to determine the clinical impact of CpG-specific changes in methylation although functional validation of epigenetic associations was outside the scope of presently funded work. Still, this is a well-powered study of geographically diverse, multi-racial/ethnic populations of women and men with methylome-wide DNAm and geocoded address-specific PM data, that leveraged multi-variate imputation to minimize selection-related biases otherwise known to affect epidemiologic associations in complete data analyses.
5. Conclusions
Findings from this large, racially/ethnically and environmentally diverse methylome-wide association study of women and men in EPA regions 1–10 suggest that ambient particulate matter air pollution affects DNAm at regions of the genome potentially related to neurological, pulmonary, endocrine, and cardiovascular disease. Although the discovered associations are biologically plausible, functional characterization in relevant tissues or animal models remains necessary to validate associations and elucidate putative epigenetic mechanisms of PM-associated disease.
Supplementary Material
Acknowledgements
The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HHSN268201700005I). The authors thank the staff and participants of the ARIC study for their important contributions. Funding was also supported by 5RC2HL102419 and R01NS087541. Data from the ARIC study are available on request at https://www2.cscc.unc.edu/aric/distribution-agreements.
1The WHI program is funded by the NHLBI, U.S. Department of Health and Human Services, through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C. WHI-AS311 was supported by American Cancer Society award 125299-RSG-13–100-01-CCE. All contributors to WHI science are listed at https://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf. Data from the WHI are available on request at https://www.whi.org/researchers/SitePages/Write%20a%20Paper.aspx.
The KORA study was initiated and financed by the Helmholtz Zentrum München – German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ.
This work was supported by NIEHS grant R01-ES020836 (LH, AB, EAW), NHLBI contract HHSN268201100046C (KC), NIEHS grant R01-ES017794 (EAW), NHLBI National Research Service Award T32-HL007055 (RG), NIEHS National Research Service Award T32-ES007018 (KH), and NCI grant R25-CA094880 (KJ).
Abbreviations:
- AA
African American
- AV
annual visit
- ARIC
Atherosclerosis Risk in Communities
- AS311
Ancillary Study 311
- AQS
United States Environmental Protection Agency Air Quality System
- BAA23
Broad Agency Award 23
- CI
confidence interval
- CpG
Cytosine-phosphate-Guanine
- CT
Clinical Trial
- DNAm
deoxyribonucleic acid methylation
- CVD
cardiovascular disease
- EA
European American
- eFORGE
Functional element Overlap analysis of Regions
- EMPC
Epigenetic Mechanisms of PM-Mediated CVD Risk
- FDR
false discovery rate
- GTP
Grady Trauma Project
- GWAS
genome-wide association study
- HLA
Hispanic/Latino American
- KORA
Cooperative Health Research in the Region Augsburg study
- LLS
Long Life Study
- LMM
linear mixed models
- MESA
Multi-Ethnic Study of Atherosclerosis
- MICE
multiple imputation by chained equations
- MWAS
methylome-wide association study
- NAAQS
National Ambient Air Quality Standards
- OS
Observational Study
- PE
prediction error
- PM10
PM < 10 μm in diameter
- PM2.5
PM < 2.5 μm in diameter
- PM2.5–10
PM > 2.5 and < 10 μm in diameter
quantile-quantine
- RMSS
root mean square standardized
- SD
standard deviation
- SE
standard error
- SPE
standardized prediction error
- WHI
Women’s Health Initiative
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2019.03.071.
Conflicts of interest
No authors have declared a potential conflicts of interest.
References
- Adlakha YK, Khanna S, Singh R, Singh VP, Agrawal A, Saini N, 2013. Pro-apoptotic miRNA-128–2 modulates ABCA1, ABCG1 and RXRα expression and cholesterol homeostasis. Cell Death Dis 4, e780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguet F; Brown AA; Castel SE; Davis JR; He Y; Jo B; Mohammadi P; Park Y; Parsana P; Segrè AV; Strober BJ; Zappala Z; Cummings BB; Gelfand ET; Hadley K; Huang KH; Lek M; Li X; Nedzel JL; Nguyen DY; Noble MS; Sullivan TJ; Tukiainen T; MacArthur DG; Getz G; Addington A; Guan P; Koester S; Little AR; Lockhart NC; Moore HM; Rao A; Struewing JP; Volpi S; Brigham LE; Hasz R; Hunter M; Johns C; Johnson M; Kopen G; Leinweber WF; Lonsdale JT; McDonald A; Mestichelli B; Myer K; Roe B; Salvatore M; Shad S; Thomas JA; Walters G; Washington M; Wheeler J; Bridge J; Foster BA; Gillard BM; Karasik E; Kumar R; Miklos M; Moser MT; Jewell SD; Montroy RG; Rohrer DC; Valley D; Mash DC; Davis DA; Sobin L; Barcus ME; Branton PA; Abell NS; Balliu B; Delaneau O; Frésard L; Gamazon ER; Garrido-Martín D; Gewirtz ADH; Gliner G; Gloudemans MJ; Han B; He AZ; Hormozdiari F; Li X; Liu B; Kang EY; McDowell IC; Ongen H; Palowitch JJ; Peterson CB; Quon G; Ripke S; Saha A; Shabalin AA; Shimko TC; Sul JH; Teran NA; Tsang EK; Zhang H; Zhou Y-H; Bustamante CD; Cox NJ; Guigó R; Kellis M; McCarthy MI; Conrad DF; Eskin E; Li G; Nobel AB; Sabatti C; Stranger BE; Wen X; Wright FA; Ardlie KG; Dermitzakis ET; Lappalainen T; Aguet F; Ardlie KG; Cummings BB; Gelfand ET; Getz G; Hadley K; Handsaker RE; Huang KH; Kashin S; Karczewski KJ; Lek M; Li X; MacArthur DG; Nedzel JL; Nguyen DT; Noble MS; Segrè AV; Trowbridge CA; Tukiainen T; Abell NS; Balliu B; Barshir R; Basha O; Battle A; Bogu GK; Brown A; Brown CD; Castel SE; Chen LS; Chiang C; Conrad DF; Cox NJ; Damani FN; Davis JR; Delaneau O; Dermitzakis ET; Engelhardt BE; Eskin E; Ferreira PG; Frésard L; Gamazon ER; Garrido-Martín D; Gewirtz ADH; Gliner G; Gloudemans MJ; Guigo R; Hall IM; Han B; He Y; Hormozdiari F; Howald C; Kyung Im H; Jo B; Yong Kang E; Kim Y; Kim-Hellmuth S; Lappalainen T; Li G; Li X; Liu B; Mangul S; McCarthy MI; McDowell IC; Mohammadi P; Monlong J; Montgomery SB; Muñoz-Aguirre M; Ndungu AW; Nicolae DL; Nobel AB; Oliva M; Ongen H; Palowitch JJ; Panousis N; Papasaikas P; Park Y; Parsana P; Payne AJ; Peterson CB; Quan J; Reverter F; Sabatti C; Saha A; Sammeth M; Scott AJ; Shabalin AA; Sodaei R; Stephens M; Stranger BE; Strober BJ; Sul JH; Tsang EK; Urbut S; van de Bunt M; Wang G; Wen X; Wright FA; Xi HS; Yeger-Lotem E; Zappala Z; Zaugg JB; Zhou Y-H; Akey JM; Bates D; Chan J; Chen LS; Claussnitzer M; Demanelis K; Diegel M; Doherty JA; Feinberg AP; Fernando MS; Halow J; Hansen KD; Haugen E; Hickey PF; Hou L; Jasmine F; Jian R; Jiang L; Johnson A; Kaul R; Kellis M; Kibriya MG; Lee K; Billy Li J; Li Q; Li X; Lin J; Lin S; Linder S; Linke C; Liu Y; Maurano MT; Molinie B; Montgomery SB; Nelson J; Neri FJ; Oliva M; Park Y; Pierce BL; Rinaldi NJ; Rizzardi LF; Sandstrom R; Skol A; Smith KS; Snyder MP; Stamatoyannopoulos J; Stranger BE; Tang H; Tsang EK; Wang L; Wang M; Van Wittenberghe N; Wu F; Zhang R; Nierras CR; Branton PA; Carithers LJ; Guan P; Moore HM; Rao A; Vaught JB; Gould SE; Lockart NC; Martin C; Struewing JP; Volpi S; Addington AM; Koester SE; Little AR; Brigham LE; Hasz R; Hunter M; Johns C; Johnson M; Kopen G; Leinweber WF; Lonsdale JT; McDonald A; Mestichelli B; Myer K; Roe B; Salvatore M; Shad S; Thomas JA; Walters G; Washington M; Wheeler J; Bridge J; Foster BA; Gillard BM; Karasik E; Kumar R; Miklos M; Moser MT; Jewell SD; Montroy RG; Rohrer DC; Valley DR; Davis DA; Mash DC; Undale AH; Smith AM; Tabor DE; Roche NV; McLean JA; Vatanian N; Robinson KL; Sobin L; Barcus ME; Valentino KM; Qi L; Hunter S; Hariharan P; Singh S; Um KS; Matose T; Tomaszewski MM; Barker LK; Mosavel M; Siminoff LA; Traino HM; Flicek P; Juettemann T; Ruffier M; Sheppard D; Taylor K; Trevanion SJ; Zerbino DR; Craft B; Goldman M; Haeussler M; Kent WJ; Lee CM; Paten B; Rosenbloom KR; Vivian J; Zhu J Genetic effects on gene expression across human tissues. Nature 2017;550:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al-Awqati Q, 1995. Regulation of ion channels by ABC transporters that secrete ATP. Science 269, 805–806. [DOI] [PubMed] [Google Scholar]
- Alexeeff SE, Schwartz J, Kloog I, Chudnovsky A, Koutrakis P, Coull BA, 2014. Consequences of kriging and land use regression for PM2.5 predictions in epidemiologic analyses: insights into spatial variability using high-resolution satellite data. Journal of Exposure Science and Environmental Epidemiology 25, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GL; LaCroix A, W64 - Long Life Study (Long Life Study)
- Anderson GL, Manson J, Wallace R, Lund B, Hall D, Davis S, Shumaker S, Wang C-Y, Stein E, Prentice RL, 2003. Implementation of the Women’s Health Initiative study design. Ann. Epidemiol 13, S5–S17. [DOI] [PubMed] [Google Scholar]
- Andrews SV, Ladd-Acosta C, Feinberg AP, Hansen KD, Fallin MD, 2016. “Gap hunting” to characterize clustered probe signals in Illumina methylation array data. Epig netics Chromatin 9 (56). [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARIC, 1989. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am. J. Epidemiol 129, 687–702. [PubMed] [Google Scholar]
- Assimes T; Tsao P; Absher D; Horvath S, BA23 - Integrative Genomics and Risk of CHD and Related Phenotypes in the Women’s Health Initiactive
- Azur MJ, Stuart EA, Frangakis C, Leaf PJ, 2011. Multiple imputation by chained equations: what is it and how does it work? Int. J. Methods Psychiatr. Res 20, 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Rienstra M, Benjamin EJ, 2010. Cardiovascular epigenetics. Circ. Cardiovasc. Genet 3, 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barallobre-Barreiro J, Didangelos A, Schoendube FA, Drozdov I, Yin X, Fernández-Caggiano M, Willeit P, Puntmann VO, Aldama-López G, Shah AM, Doménech N, Mayr M, 2012. Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation 125, 789–802. [DOI] [PubMed] [Google Scholar]
- Bates D, 2017. Mixed-Effects Models in Julia. GitHub
- Bellavia A, Urch B, Speck M, Brook RD, Scott JA, Albetti B, Behbod B, North M, Valeri L, Bertazzi PA, Silverman F, Gold D, Baccarelli A,A, 2013. DNA hypomethylation, ambient particulate matter, and increased blood pressure: findings from controlled human exposure experiments. J. Am. Heart Assoc 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA, 2010. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol 28, 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanson J, Edelman A, Karpinski S, Shah VB, 2017. Julia: A fresh approach to numerical computing. SIAM Rev 59, 65–98. [Google Scholar]
- Bollati V, Baccarelli A, 2010. Environmental epigenetics. Heredity 105, 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Angelici L, Rizzo G, Pergoli L, Rota F, Hoxha M, Nordio F, Bonzini M, Tarantini L, Cantone L, Pesatori AC, Apostoli P, Baccarelli AA, Bertazzi PA, 2015. Microvesicle-associated microRNA expression is altered upon particulate matter exposure in healthy workers and in A549 cells. J. Appl. Toxicol 35, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze Charles E., Paul Dirk S., van Dongen J, Butcher Lee M., Ambrose John C., Barrett James E., Lowe R, Rakyan Vardhman K., Iotchkova V, Frontini M, Downes K, Ouwehand Willem H., Laperle J, Jacques P-É, Bourque G, Bergmann Anke K., Siebert R, Vellenga E Saeed S, Matarese F, Martens Joost H.A., Stunnenberg Hendrik G., Teschendorff Andrew E., Herrero J, Birney E, Dunham I, Beck S, 2016. eFORGE: a tool for identifying cell type-specific signal in epigenomic data. Cell Rep 17, 2137–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan AL, Geddes DM, Gyi KM, Baker EH, 2004. Clinical importance of cystic fibrosis-related diabetes. J. Cyst. Fibros 3, 209–222. [DOI] [PubMed] [Google Scholar]
- Brook RD, Franklin B, Cascio W, Hong Y, Howard G, Lipsett M, Luepker R, Mittleman M, Samet J, Smith SC, Tager I, 2004. Air pollution and cardiovascular disease. Circulation 109, 2655. [DOI] [PubMed] [Google Scholar]
- Brook RD, Rajagopalan S, Pope CA, Brook JR, Bhatnagar A, Diez-Roux AV, Holguin F, Hong Y, Luepker RV, Mittleman MA, Peters A, Siscovick D, Smith SC, Whitsel L, Kaufman JD, 2010. Particulate matter air pollution and cardio-vascular disease. Circulation 121, 2331. [DOI] [PubMed] [Google Scholar]
- Byun H-M, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, Yang AS, 2009. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum. Mol. Genet 18, 4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciapuoti F, Spiezia R, Bianchi U, Lama D, D’Avino M, Varricchio M, 1991. Effectiveness of glibenclamide on myocardial ischemic ventricular arrhythmias in non-insulin-dependent diabetes mellitus. Am. J. Cardiol 67, 843–847. [DOI] [PubMed] [Google Scholar]
- Cacciottolo M, Wang X, Driscoll I, Woodward N, Saffari A, Reyes J, Serre ML, Vizuete W, Sioutas C, Morgan TE, Gatz M, Chui HC, Shumaker SA, Resnick SM, Espeland MA, Finch CE, Chen JC, 2017. Particulate air pollutants, APOE alleles and their contributions to cognitive impairment in older women and to amyloidogenesis in experimental models. Transl. Psychiatry 7, e1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R, 2013. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Qiao L, Li H, Zhao Y, Zhang Y, Xu W, Wang C, Wang H, Zhao Z, Xu X, Hu H, Kan H, 2015. Fine particulate matter constituents, nitric oxide synthase DNA methylation and exhaled nitric oxide. Environmental Science & Technology 49, 11859–11865. [DOI] [PubMed] [Google Scholar]
- Chen R, Meng X, Zhao A, Wang C, Yang C, Li H, Cai J, Zhao Z, Kan H, 2016. DNA hypomethylation and its mediation in the effects of fine particulate air pollution on cardiovascular biomarkers: a randomized crossover trial. Environ. Int 94, 614–619. [DOI] [PubMed] [Google Scholar]
- Cheng L, Lau WKW, Fung TKH, Lau BWM, Chau BKH, Liang Y, Wang Z, So KF, Wang T, Chan CCH, Lee TMC, 2017. PM2.5 exposure suppresses dendritic maturation in subgranular zone in aged rats. Neurotox. Res 32, 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, Stancheva I, 2008. Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin? Cell. Mol. Life Sci 65, 1509–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran WG, 1954. The combination of estimates from different experiments. Biometrics 10, 101–129. [Google Scholar]
- Cohen AJ, Brauer M, Burnett R, Anderson HR, Frostad J, Estep K, Balakrishnan K, Brunekreef B, Dandona L, Dandona R, Feigin V, Freedman G, Hubbell B, Jobling A, Kan H, Knibbs L, Liu Y, Martin R, Morawska L, Pope CA III, Shin H, Straif K, Shaddick G, Thomas M, Dingenen R, van Donkelaar A, Vos T, Murray CJL, Forouzanfar MH, 2017. Estimates and 25-year trends of the global burden of disease attributable to ambient air pollution: an analysis of data from the Global Burden of Diseases Study 2015. Lancet 389, 1907–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP, 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornbleet PJ, Gochman N, 1979. Incorrect least-squares regression coefficients in method-comparison analysis. Clin. Chem 25, 432–438. [PubMed] [Google Scholar]
- de FC Lichtenfels AJ, van der Plaat DA, de Jong K, van Diemen CC, Postma DS, Nedeljkovic I, van Duijn CM, Amin N, la Bastide-van Gemert S, de Vries M, Ward-Caviness CK, Wolf K, Waldenberger M, Peters A, Stolk RP, Brunekreef B, Boezen HM, Vonk JM, 2018. Long-term air pollution exposure, genome-wide DNA methylation and lung function in the LifeLines cohort study. Environ. Health Perspect 027004, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Wasserman L, 2001. Genomic control, a new approach to genetic-based association studies. Theor. Popul. Biol 60, 155–166. [DOI] [PubMed] [Google Scholar]
- Di Q, Wang Y, Zanobetti A, Wang Y, Koutrakis P, Choirat C, Dominici F, Schwartz JD, 2017. Air pollution and mortality in the Medicare population. N. Engl. J. Med 376, 2513–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici F, Peng RD, Bell ML, Pham L, McDermott A, Zeger SL, 2006. Fine particulate air pollution and hospital admission for cardiovascular and respiratory diseases. JAMA 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan DD, 2013. Phenomics of cardiac chloride channels. Comprehensive Physiology [DOI] [PMC free article] [PubMed]
- Engler RL, Yellon DM, 1996. Sulfonylurea KATP blockade in type II diabetes and preconditioning in cardiovascular disease. Time for Reconsideration 94, 2297–2301. [DOI] [PubMed] [Google Scholar]
- EPA, 2017. What Are the Air Quality Standards for PM? U.S. Environmental Protection Agency. [Google Scholar]
- Fan S, Zhang X, 2009. CpG island methylation pattern in different human tissues and its correlation with gene expression. Biochem. Biophys. Res. Commun 383, 421–425. [DOI] [PubMed] [Google Scholar]
- Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P, Hodges A, Powell J, Wahlund LO, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Spenger C, Lathrop M, Shen L, Kim S, Saykin AJ, Weiner MW, Lovestone S, 2010. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Mol. Psychiatry 16, 1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan WQ, FitzGerald JM, Carlsten C, Sadatsafavi M, Brauer M, 2013. Associations of ambient air pollution with chronic obstructive pulmonary disease hospitalization and mortality. Am. J. Respir. Crit. Care Med 187, 721–727. [DOI] [PubMed] [Google Scholar]
- Gondalia R, Avery CL, Napier MD, Mendez-Giraldez R, Stewart JD, Sitlani CM, Li Y, Wilhelmsen KC, Duan Q, Roach J, North KE, Reiner AP, Zhang ZM, Tinker LF, Yanosky JD, Liao D, Whitsel EA, 2017. Genome-wide association study of susceptibility to particulate matter-associated QT prolongation. Environ. Health Perspect 125, 067002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernan MA, Hernandez-Diaz S, Robins JM, 2004. A structural approach to selection bias. Epidemiology 15, 615–625. [DOI] [PubMed] [Google Scholar]
- Holle R, Happich M, Löwel H, Wichmann H-E, Group n.f.t.M.K.S, 2005. KORA-a research platform for population based health research. Das Gesundheitswesen 67, 19–25. [DOI] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT, 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joehanes R; Just AC; Marioni RE; Pilling LC; Reynolds LM; Mandaviya PR; Guan W; Xu T; Elks CE; Aslibekyan S; Moreno-Macias H; Smith JA; Brody JA; Dhingra R; Yousefi P; Pankow JS; Kunze S; Shah S; McRae AF; Lohman K; Sha J; Absher DM; Ferrucci L; Zhao W; Demerath EW; Bressler J; Grove ML; Huan T; Liu C; Mendelson MM; Yao C; Kiel DP; Peters A; Wang-Sattler R; Visscher PM; Wray NR; Starr JM; Ding J; Rodriguez CJ; Wareham NJ; Irvin MR; Zhi D; Barrdahl M; Vineis P; Ambatipudi S; Uitterlinden AG; Hofman A; Schwartz J; Colicino E; Hou L; Vokonas PS; Hernandez DG; Singleton AB; Bandinelli S; Turner ST; Ware EB; Smith AK; Klengel T; Binder EB; Psaty BM; Taylor KD; Gharib SA; Swenson BR; Liang L; DeMeo DL; Connor GT; Herceg Z; Ressler KJ; Conneely KN; Sotoodehnia N; Kardia SLR; Melzer D; Baccarelli AA; van Meurs JBJ; Romieu I; Arnett DK; Ong KK; Liu Y; Waldenberger M; Deary IJ; Fornage M; Levy D; London SJ Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet 2016; [DOI] [PMC free article] [PubMed]
- Johnson WE, Li C, Rabinovic A, 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127. [DOI] [PubMed] [Google Scholar]
- Jordahl KM, Randolph TW, Song X, Sather CL, Tinker LF, Phipps AI, Kelsey KT, White E, Bhatti P, 2018. Genome-wide DNA methylation in prediagnostic blood and bladder cancer risk in the Women’s Health Initiative. Cancer Epidemiol. Biomark. Prev 27, 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahr PC, Piccini I, Fabritz L, Greber B, Schöler H, Scheld HH, Hoffmeier A, Brown NA, Kirchhof P, 2011. Systematic analysis of gene expression differences between left and right atria in different mouse strains and in human atrial tissue. PLoS One 6, e26389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Goehring GN, Nichols MH, Robins C, Mehta D, Klengel T, Eskin E, Smith AK, Conneely KN, 2018. An integrated-omics analysis of the epigenetic landscape of gene expression in human blood cells. BMC Genomics 19, 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld H-H, Rotering H, Fortmueller L, Laakmann S, Verheule S, Schotten U, Fabritz L, Brown NA, 2011. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ. Cardiovasc. Genet 4, 123–133. [DOI] [PubMed] [Google Scholar]
- Laumbach RJ, Kipen HM, 2012. Respiratory health effects of air pollution: update on biomass smoke and traffic pollution. J. Allergy Clin. Immunol 129, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Serre Marc L, van Donkelaar A, Martin Randall V, Burnett Richard T, Jerrett M, 2012. Comparison of geostatistical interpolation and remote sensing techniques for estimating long-term exposure to ambient PM2.5 concentrations across the continental United States. Environ. Health Perspect 120, 1727–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CE, Hennessy S, Han X, Siscovick DS, Flory JH, Deo R, 2017. Pro- and antiarrhythmic actions of sulfonylureas: mechanistic and clinical evidence. Trends in Endocrinology & Metabolism 28, 561–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Peuquet DJ, Duan Y, Whitsel EA, Dou J, Smith RL, Lin H-M, Chen J-C, Heiss G, 2006. GIS approaches for the estimation of residential-level ambient PM concentrations. Environ. Health Perspect 1374–1380. [DOI] [PMC free article] [PubMed]
- Liao D, Peuquet DJ, Lin H-M, Duan Y, Whitsel EA, Smith RL, Heiss G, 2007. National kriging exposure estimation: Liao et al. respond. Environ. Health Perspect 115, A338–A339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberda EN, Cuevas AK, Gillespie PA, Grunig G, Qu Q, Chen LC, 2010. Exposure to inhaled nickel nanoparticles causes a reduction in number and function of bone marrow endothelial progenitor cells. Inhal. Toxicol 22, 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, Foster B, Moser M, Karasik E, Gillard B, Ramsey K, Sullivan S, Bridge J, Magazine H, Syron J, Fleming J, Siminoff L, Traino H, Mosavel M, Barker L, Jewell S, Rohrer D, Maxim D, Filkins D, Harbach P, Cortadillo E, Berghuis B, Turner L, Hudson E, Feenstra K, Sobin L, Robb J, Branton P, Korzeniewski G, Shive C, Tabor D, Qi L, Groch K, Nampally S, Buia S, Zimmerman A, Smith A, Burges R, Robinson K, Valentino K, Bradbury D, Cosentino M, Diaz-Mayoral N, Kennedy M, Engel T, Williams P, Erickson K, Ardlie K, Winckler W, Getz G, DeLuca D, MacArthur D, Kellis M, Thomson A, Young T, Gelfand E, Donovan M, Meng Y, Grant G, Mash D, Marcus Y, Basile M, Liu J, Zhu J, Tu Z, Cox NJ, Nicolae DL, Gamazon ER, Im HK, Konkashbaev A, Pritchard J, Stevens M, Flutre T, Wen X, Dermitzakis ET, Lappalainen T, Guigo R, Monlong J, Sammeth M, Koller D, Battle A, Mostafavi S, McCarthy M, Rivas M, Maller J, Rusyn I, Nobel A, Wright F, Shabalin A, Feolo M, Sharopova N, Sturcke A, Paschal J, Anderson JM, Wilder EL, Derr LK, Green ED, Struewing JP, Temple G, Volpi S, Boyer JT, Thomson EJ, Guyer MS, Ng C, Abdallah A, Colantuoni D, Insel TR, Koester SE, Little AR, Bender PK, Lehner T, Yao Y, Compton CC, Vaught JB, Sawyer S, Lockhart NC, Demchok J, Moore HF, 2013. The genotype-tissue expression (GTEx) project. Nat. Genet 45, 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Wilker EH, Willis-Owen SAG, Byun H-M, Wong KCC, Motta V, Baccarelli AA, Schwartz J, Cookson WOCM, Khabbaz K, Mittleman MA, Moffatt MF, Liang L, 2014. Predicting DNA methylation level across human tissues. Nucleic Acids Res 42, 3515–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathar I, Kecskes M, Van Der Mieren G, Jacobs G, Uhl S, Camacho Londoño JE, Flockerzi V, Voets T, Freichel M, Nilius B, Herijgers P, Vennekens R, 2013. Increased ß-adrenergic inotropy in ventricular myocardium from Trpm4−/− mice. Circ. Res 114, 283–294. [DOI] [PubMed] [Google Scholar]
- McCullough SD, Dhingra R, Fortin MC, Diaz-Sanchez D, 2017. Air pollution and the epigenome: a model relationship for the exploration of toxicoepigenetics. Current Opinion in Toxicology 6, 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KA, Siscovick DS, Sheppard L, Shepherd K, Sullivan JH, Anderson GL, Kaufman JD, 2007. Long-term exposure to air pollution and incidence of cardiovascular events in women. N. Engl. J. Med 356, 447–458. [DOI] [PubMed] [Google Scholar]
- Mosley TH, Knopman DS, Catellier DJ, Bryan N, Hutchinson RG, Grothues CA, Folsom AR, Cooper LS, Burke GL, Liao D, Szklo M, 2005. Cerebral MRI findings and cognitive functioning. Neurology 64, 2056. [DOI] [PubMed] [Google Scholar]
- Najeed SA, Khan IA, Molnar J, Somberg JC, 2002. Differential effect of glyburide glibenclamide and metformin on qt dispersion: a potential adenosine triphosphate sensitive k+ channel effect. Am. J. Cardiol 90, 1103–1106. [DOI] [PubMed] [Google Scholar]
- Neidhart M, 2016. DNA Methylation and Complex Human Disease EdÊds Elsevier, Amsterdam. [Google Scholar]
- NIH, 1998. Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Control. Clin. Trials 19, 61–109. [DOI] [PubMed] [Google Scholar]
- O’Day DH, Eshak K, Myre MA, 2015. Calmodulin binding proteins and Alzheimer’s disease. J. Alzheimers Dis 46, 553–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panni T, Mehta AJ, Schwartz JD, Baccarelli AA, Just AC, Wolf KA, 2016. Genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the normative aging study. Environ. Health Perspect 124, 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusquin M, Guida F, Polidoro S, Vermeulen R, Raaschou-Nielsen O, Campanella G, Hoek G, Kyrtopoulos SA, Georgiadis P, Naccarati A, Sacerdote C, Krogh V, Bas Bueno-de-Mesquita H, Monique Verschuren WM, Sayols-Baixeras S, Panni T, Peters A, Hebels DGAJ, Kleinjans J, Vineis P, Chadeau-Hyam M, 2017. DNA methylation and exposure to ambient air pollution in two prospective cohorts. Environ. Int 108, 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA III, Burnett RT, Thun MJ, Calle EE, Krewski D, Ito K, Thurston GD, 2002. Lung cancer, cardiopulmonary mortality, and long-term exposure to fine particulate air pollution. Jama 287, 1132–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo RT, Sui J, Dhume A, Palomeque J, Blaxall BC, Diaz G, Tunstead J, Logothetis DE, Hajjar RJ, Schecter AD, 2006. CXCR4 modulates contractility in adult cardiac myocytes. J. Mol. Cell. Cardiol 41, 834–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaschou-Nielsen O, Andersen ZJ, Beelen R, Samoli E, Stafoggia M, Weinmayr G, Hoffmann B, Fischer P, Nieuwenhuijsen MJ, Brunekreef B, Xun WW, Katsouyanni K, Dimakopoulou K, Sommar J, Forsberg B, Modig L, Oudin A, Oftedal B, Schwarze PE, Nafstad P, De Faire U, Pedersen NL, Östenson C-G, Fratiglioni L, Penell J, Korek M, Pershagen G, Eriksen KT, Sørensen M, Tjønneland A, Ellermann T, Eeftens M, Peeters PH, Meliefste K, Wang M, Bueno-de-Mesquita B, Key TJ, de Hoogh K, Concin H, Nagel G, Vilier A, Grioni S, Krogh V, Tsai M-Y, Ricceri F, Sacerdote C, Galassi C, Migliore E, Ranzi A, Cesaroni G, Badaloni C, Forastiere F, Tamayo I, Amiano P, Dorronsoro M, Trichopoulou A, Bamia C, Vineis P, Hoek G, 2013. Air pollution and lung cancer incidence in 17 European cohorts: prospective analyses from the European Study of Cohorts for Air Pollution Effects (ESCAPE). The Lancet Oncology 14, 813–822. [DOI] [PubMed] [Google Scholar]
- Rab A, Rowe SM, Raju SV, Bebok Z, Matalon S, Collawn JF, 2013. Cigarette smoke and CFTR: implications in the pathogenesis of COPD. Am. J. Phys. Lung Cell. Mol. Phys 305, L530–L541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhilin SV, Olson PA, Nishi A, Starkova NN, Fienberg AA, Nairn AC, Surmeier DJ, Greengard PA, 2004. Network of control mediated by regulator of calcium/calmodulin-dependent signaling. Science 306, 698–701. [DOI] [PubMed] [Google Scholar]
- Rasmussen JE, Sheridan JT, Polk W, Davies CM, Tarran R, 2014. Cigarette smoke-induced Ca2+ release leads to cystic fibrosis transmembrane conductance regulator (CFTR) dysfunction. J. Biol. Chem 289, 7671–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeld F, Maticzka D, Grosser S, Knauff P, Eravci M, Vida I, Backofen R, Wulczyn FG, 2018. The RNA-binding protein ARPP21 controls dendritic branching by functionally opposing the miRNA it hosts. Nat. Commun 9, 1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux AVD, Merkin SS, Arnett D, Chambless L, Massing M, Nieto FJ, Sorlie P, Szklo M, Tyroler HA, Watson RL, 2001. Neighborhood of residence and incidence of coronary heart disease. N. Engl. J. Med 345, 99–106. [DOI] [PubMed] [Google Scholar]
- Saffari A, Silver MJ, Zavattari P, Moi L, Columbano A, Meaburn EL, Dudbridge F, 2018. Estimation of a significance threshold for epigenome-wide association studies. Genet. Epidemiol 42, 20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Criq V, Gray MA, 2017. Role of CFTR in epithelial physiology. Cell. Mol. Life Sci 74, 93–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart EA, Azur M, Frangakis C, Leaf P, 2009. Multiple imputation with large data sets: a case study of the Children’s Mental Health Initiative. Am. J. Epidemiol 169, 1133–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunnenberg HG, Abrignani S, Adams D, de Almeida M, Altucci L, Amin V, Amit I, Antonarakis SE, Aparicio S, Arima T, Arrigoni L, Arts R, Asnafi V, Esteller M, Bae J-B, Bassler K, Beck S, Berkman B, Bernstein BE, Bilenky M, Bird A, Bock C, Boehm B, Bourque G, Breeze CE, Brors B, Bujold D, Burren O, Bussemakers MJ, Butterworth A, Campo E, Carrillo-de-Santa-Pau E, Chadwick L, Chan KM, Chen W, Cheung TH, Chiapperino L, Choi NH, Chung H-R, Clarke L, Connors JM, Cronet P, Danesh J, Dermitzakis M, Drewes G, Durek P, Dyke S, Dylag T, Eaves CJ, Ebert P, Eils R, Eils J, Ennis CA, Enver T, Feingold EA, Felder B, Ferguson-Smith A, Fitzgibbon J, Flicek P, Foo RSY, Fraser P, Frontini M, Furlong E, Gakkhar S, Gasparoni N, Gasparoni G, Geschwind DH, Glažar P, Graf T, Grosveld F, Guan X-Y, Guigo R, Gut IG, Hamann A, Han B-G, Harris RA, Heath S, Helin K, Hengstler JG, Heravi-Moussavi A, Herrup K, Hill S, Hilton JA, Hitz BC, Horsthemke B, Hu M, Hwang J-Y, Ip NY, Ito T, Javierre B-M, Jenko S, Jenuwein T, Joly Y, Jones SJM, Kanai Y, Kang HG, Karsan A, Kiemer AK, Kim SC, Kim B-J, Kim H-H, Kimura H, Kinkley S, Klironomos F, Koh I-U, Kostadima M, Kressler C, Kreuzhuber R, Kundaje A, Küppers R, Larabell C, Lasko P, Lathrop M, Lee DHS, Lee S, Lehrach H, Leitão E, Lengauer T, Lernmark Å, Leslie RD, Leung GKK, Leung D, Loeffler M, Ma Y, Mai A, Manke T, Marcotte ER, Marra MA, Martens JHA, Martin-Subero JI, Maschke K, Merten C, Milosavljevic A, Minucci S, Mitsuyama T, Moore RA, Müller F, Mungall AJ, Netea MG, Nordström K, Norstedt I, Okae H, Onuchic V, Ouellette F, Ouwehand W, Pagani M, Pancaldi V, Pap T, Pastinen T, Patel R, Paul DS, Pazin MJ, Pelicci PG, Phillips AG, Polansky J, Porse B, Pospisilik JA, Prabhakar S, Procaccini DC, Radbruch A, Rajewsky N, Rakyan V, Reik W, Ren B, Richardson D, Richter A, Rico D, Roberts DJ, Rosenstiel P, Rothstein M, Salhab A, Sasaki H, Satterlee JS, Sauer S, Schacht C, Schmidt F, Schmitz G, Schreiber S, Schröder C, Schübeler D, Schultze JL, Schulyer RP, Schulz M, Seifert M, Shirahige K, Siebert R, Sierocinski T, Siminoff L, Sinha A, Soranzo N, Spicuglia S, Spivakov M, Steidl C, Strattan JS, Stratton M, Südbeck P, Sun H, Suzuki N, Suzuki Y, Tanay A, Torrents D, Tyson FL, Ulas T, Ullrich S, Ushijima T, Valencia A, Vellenga E, Vingron M, Wallace C, Wallner S, Walter J, Wang H, Weber S, Weiler N, Weller A, Weng A, Wilder S, Wiseman SM, Wu AR, Wu Z, Xiong J, Yamashita Y, Yang X, Yap DY, Yip KY, Yip S, Yoo J-I, Zerbino D, Zipprich G, Hirst M, 2016. The international human epigenome consortium: a blueprint for scientific collaboration and discovery. Cell 167, 1145–1149. [DOI] [PubMed] [Google Scholar]
- Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, Cantone L, Rizzo G, Hou L, Schwartz J, Bertazzi PA, Baccarelli A, 2009. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ. Health Perspect 117, 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantini L, Bonzini M, Tripodi A, Angelici L, Nordio F, Cantone L, Apostoli P, Bertazzi PA, Baccarelli AA, 2013. Blood hypomethylation of inflammatory genes mediates the effects of metal-rich airborne pollutants on blood coagulation. Occup. Environ. Med 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S, 2013. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P-C, Spector TD, Bell JT, 2012. Using epigenome-wide association scans of DNA methylation in age-related complex human traits. Epigenomics 4, 511–526. [DOI] [PubMed] [Google Scholar]
- Uckelmann H, Blaszkiewicz S, Nicolae C, Haas S, Schnell A, Wurzer S, Wagener R, Aszodi A, Essers MAG, 2016. Extracellular matrix protein Matrilin-4 regulates stress-induced HSC proliferation via CXCR4. J. Exp. Med 213, 1961–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayudhan L, Proitsi P, Westman E, Muehlboeck JS, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Soininen H, Spenger C, Hodges A, Powell J, Lovestone S, Simmons A, 2013. Entorhinal cortex thickness predicts cognitive decline in Alzheimer’s disease. Journal of Alzheimer’s disease: JAD 33, 755–766. [DOI] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H, 2014. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42, D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsel EA, AS315 - Epigenetic Mechanisms of PM-mediated CVD Risk
- Whitsel EA, Rose KM, Wood JL, Henley AC, Liao D, Heiss G, 2004. Accuracy and repeatability of commercial geocoding. Am. J. Epidemiol 160, 1023–1029. [DOI] [PubMed] [Google Scholar]
- Whitsel EA, Quibrera PM, Smith RL, Catellier DJ, Liao D, Henley AC, Heiss G, 2006. Accuracy of commercial geocoding: assessment and implications. Epidemiologic Perspectives & Innovations: EP + I 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann H, Gieger C, Illig T, Group MKS, 2005. KORA-gen—resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen (Bundesverband der Arzte des Offentlichen Gesundheitsdienstes (Germany)) 67, S26. [DOI] [PubMed] [Google Scholar]
- Yamazaki J, Hume JR, 1997. Inhibitory effects of glibenclamide on cystic fibrosis transmembrane regulator, swelling-activated, and Ca2+-activated Cl− channels in mammalian cardiac myocytes. Circ. Res 81, 101–109. [DOI] [PubMed] [Google Scholar]
- Yanosky JD, Paciorek CJ, Laden F, Hart JE, Puett RC, Liao D, Suh HH, 2014. Spatio-temporal modeling of particulate air pollution in the conterminous United States using geographic and meteorological predictors. Environmental Health: A Global Access Science Source 13 (63). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Agha G, Baccarelli AA, 2016. The role of DNA methylation in cardiovascular risk and disease. Methodological Aspects, Study Design, and Data Analysis for Epidemiological Studies 118, 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.