Abstract
The peptidoglycan (PG) cell wall is an essential structure for the growth of most bacteria. However, many are capable of switching into a wall-deficient L-form state, which is resistant to antibiotics that target cell wall synthesis, under osmoprotective conditions, including host environments. L-form cells might have an important role in chronic or recurrent infections. Crucially, the cellular pathways involved in switching to and from the L-form state are still poorly understood. This work shows that the lack of cell wall or blocking its synthesis by β-lactam antibiotics, results in an increased flux through glycolysis. This leads to the production of reactive oxygen species (ROS) from the respiratory chain (RC), which prevents L-form growth. Compensation for the metabolic imbalance by slowing down glycolysis, activating gluconeogenesis, or depleting oxygen, enables L-form growth in Bacillus subtilis, Listeria monocytogenes and Staphylococcus aureus. These effects do not occur in Enterococcus faecium, which lacks the RC pathway. Our results collectively show that when cell wall synthesis is blocked under aerobic and glycolytic conditions the perturbation of cellular metabolism causes cell death. We provide a mechanistic framework for many anecdotal descriptions of the optimal conditions for L-form growth and non-lytic killing by β-lactam antibiotics.
Many bacteria retain the ability to switch into a wall-deficient state called the L-form 1, which is completely resistant to antibiotics working specifically on cell wall synthesis, including β-lactams. L-forms have been identified as antibiotic resistant organisms in samples from humans, and thus they might contribute to chronic or recurrent infections 2–4. Crucially, in both a free living Bacillus subtilis and a pathogenic Staphylococcus aureus, host lytic enzymes, especially lysozyme, can promote an L-form switch that enables the organism to evade β-lactam action and continue proliferating 5.
B. subtilis has been used as a tractable model system for studying the molecular biology of L-forms 4,6. Inhibition of de novo PG synthesis traps the membrane-bound L-form inside the original PG wall. Autolytic enzymes or exogenous PG hydrolases, such as lysozyme, can degrade the wall, enabling L-form to escape, and survival in isotonic environments 5,7 (Supplementary Fig. 1a). L-form proliferation seems to involve a simple biophysical effect based on an increased rate of surface area to volume synthesis, driving cell shape deformations that lead to spontaneous scission 8. However, because ROS originating from the RC pathway are abnormally increased in the cell-wall-deficient cells, reduction of RC activity or high levels of ROS scavengers are normally required to support robust L-form growth 9 (Supplementary Fig. 1a). L-form growth of the Gram-negative bacterium Escherichia coli was also stimulated under anaerobic conditions or by adding a ROS scavenger, suggesting that oxidative damage might be an important impediment to L-form growth in a wide range of bacteria 9–11. Nevertheless, it remained unclear why the L-form transition should result in increased ROS.
ROS is generated as a natural side effect of cellular metabolism and aerobic respiration. The PG wall synthesis pathway is one of the major drains on carbon in the central metabolic pathway (Fig. 1a), so we favor a model in which a block in PG synthesis results in re-routing of phospho-sugars towards catabolic rather anabolic pathways. This could, in turn, result in stimulating flux into the TCA cycle, leading to increased generation of ROS as a by-product of the metabolism of molecular oxygen in the RC pathway.
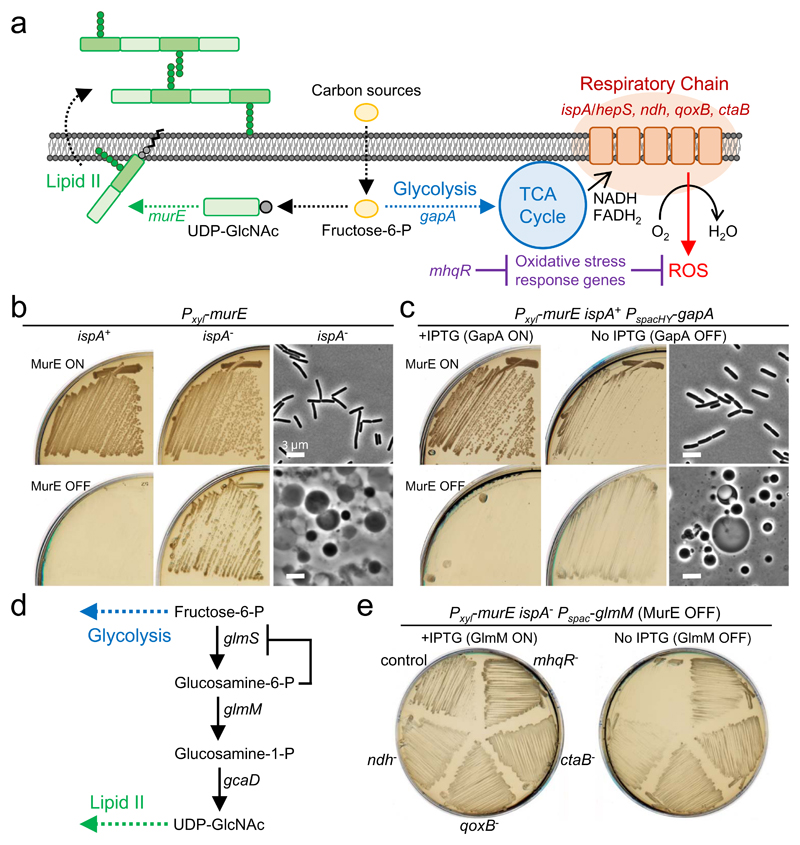
Figure 1. Blocking PG synthesis causes glycolysis-mediated cell death.
a, Schematic representation of the drains on carbon in the central metabolic pathway in B. subtilis.
b, Effects of repression of ispA on cell growth in the walled and L-form states. B. subtilis strains BS115 (Pxyl-murE ispA+) and LR2 (Pxyl-murE ispA-) were streaked on NA/MSM (sucrose) plates with (MurE ON) or without (MurE OFF) 1% xylose and incubated at 30°C (left and middle panels). Phase contrast micrographs of LR2 (right panels) were taken from the plates with (MurE ON) or without (MurE OFF) xylose shown in middle.
c, Effects of repression of gapA on cell growth in the walled and L-form states. B. subtilis strain YK1621 (Pxyl-murE PspacHY-gapA) was streaked on NA/MSM (sucrose) plates with (GapA ON) or without (GapA OFF) 1 mM IPTG, and also in the presence (MurE ON) or absence (MurE OFF) of 1% xylose and incubated at 30°C. Phase contrast micrographs of YK1621 (right panels) were taken from the plates without IPTG (GapA OFF), and in the presence (MurE ON) or absence (MurE OFF) of xylose shown in middle.
d, Schematic representation of UDP-GlcNAc pathway in B. subtilis.
e, Effects of repression of glmM on L-form growth. B. subtilis strains YK1563 (Pxyl-murE ispA- Pspac-glmM), YK1763 (YK1563 ndh-), YK1764 (YK1563 qoxB-), YK1765 (YK1563 ctaB-) and YK1995 (YK1563 mhqR-) were streaked on NA/MSM plates with (GlmM ON) or without (GlmM OFF) 1 mM IPTG in the absence of xylose (MurE OFF) and incubated at 30°C.
The figures are representative of at least three independent experiments (b, c and e).
Here, we show that the abnormal generation of ROS in the L-form transition correlates with an increase in glycolytic activity in B. subtilis and likely other Gram-positive bacteria, including pathogens. Moreover, we show that a reduction of glycolytic activity or activation of gluconeogenesis, which acts as the reverse of glycolysis, can compensate for the metabolic imbalance, reducing the oxidative damage and thus promoting L-form growth, thereby evading β-lactam killing. Consistent with this model, the glycolysis-mediated killing did not occur during the L-form transition in the pathogen Enterococcus faecium, which lacks the RC pathway 12,13. Our results show how a specific metabolic diversion induced by β-lactam antibiotics contributes to cell killing in Gram-positive bacteria, and provide an understanding of the conditions influencing the ability of cells to grow in the L-form state.
Results
Repression of glycolytic activity promotes L-form growth
Figure 1b and supplementary figures 1b-e recapitulate our previous findings that repression of the murE operon (containing genes for several steps in the PG precursor, lipid II, pathway) induces an efficient L-form switch, when the cells have an ispA mutation (or other mutations which affect the RC pathway) but not in ispA+ cells 6,8. The ispA product is thought to contribute to the synthesis of menaquinone, an electron carrier in the RC pathway 9,14 (Fig. 1a). To test whether the requirement for the ispA mutation was due to glycolytic activity, we put the gapA gene, which encodes glyceraldehyde 3-phosphate dehydrogenase, an essential component of the glycolytic pathway 15 (Fig. 1a), under IPTG control in the ispA+ background. In the normal walled state (MurE ON), repression of gapA partially impaired growth on isotonic NA/sucrose plates (nutrient agar containing 0.5 M sucrose) (Fig. 1c, MurE ON / GapA OFF), consistent with expectation that glycolysis is required for efficient growth of the walled cells under these culture conditions. When the precursor pathway was blocked (MurE OFF), no L-form growth was observed in the presence of IPTG (Fig. 1c, MurE OFF/GapA ON). However, growth did occur in the absence of IPTG, and phase contrast microscopy showed the presence of heterogenous spheroidal L-forms on the plates (Fig. 1c, MurE OFF / GapA OFF), similar to those of the ispA- strain (Fig. 1b). Thus, reduction of glycolytic activity can substitute for ispA mutation in promoting L-form growth.
Metabolic re-routing prevents L-form growth
Uridine 5’-diphospho-N-acetylglucosamine (UDP-GlcNAc), an essential precursor of lipid II, is generated from fructose-6-phosphate, a glycolytic intermediate (Fig. 1a), through the action of the glmS, glmM and gcaD gene products (Fig. 1d). The glmS riboswitch is a ribozyme that self-cleaves upon binding glucosamine-6-phosphate, the product of the enzyme encoded by glmS 16 (Fig. 1d). We expected that blockage of the UDP-GlcNAc pathway by repressing glmM and indirectly also glmS should result in a re-routing of glucose metabolism towards glycolysis. To test the effects of this metabolic switch on L-form growth, we put the glmM gene under IPTG control in the LR2 strain (Pxyl-murE ispA-). In the presence of IPTG (GlmM ON) but no xylose (MurE OFF), efficient L-form growth was observed (Fig. 1e, left, control). However, growth did not occur when the expression of glmM was repressed (Fig. 1e, right, control). If the growth defect was caused by increased ROS production from the RC pathway through increased glycolytic activity, it should be suppressed by reducing RC activity. As shown in Figure 1a, ndh, qoxB and ctaB gene products are involved in the RC system: ndh encodes a major NADH dehydrogenease 17; qoxB encodes cytochrome aa3 quinol oxidase subunit I 18; and ctaB encodes heme O synthase 19. We examined the effects of mutations in these genes on L-form growth with glmM repression and found that they all rescued the growth defect (Fig. 1e). Similar results were obtained by introducing a mutation in mhqR (Fig. 1e), which encodes a transcriptional repressor for genes induced by the thiol-specific oxidative and/or electrophile stress response 20 (Fig. 1a). Thus, carbon flux through glycolysis seems to be a serious impediment to L-form growth and it can be bypassed either by downregulating RC activity, or upregulating oxidative stress response genes.
Inhibition of the sugar uptake system promotes L-form growth
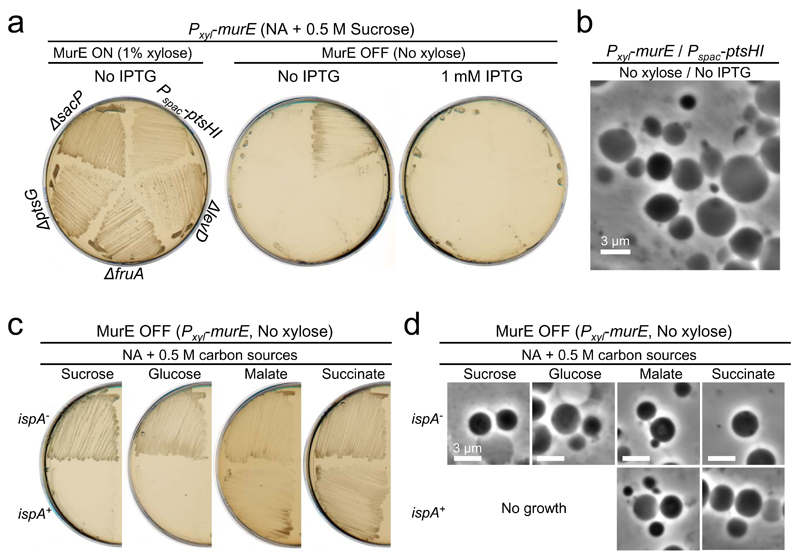
Sucrose is a disaccharide sugar consisting of glucose and fructose that has frequently been used in L-form culture media to provide an isotonic environment. Many bacteria, including B. subtilis, take up sucrose by the phosphotransferase system (PTS) and consume this through glycolysis to provide cellular energy 21. In B. subtilis, levansucrase and endolevanase can cleave sucrose externally to glucose and fructose 22, which are then also utilised via the PTS pathway and glycolysis. According to our model, limiting the uptake of those glycolytic sugars should promote L-form growth by reducing glycolytic flux in the MurE OFF background. The PTS consists of two general components, enzyme I (EI) and phosphocarrier protein (HPr), and a suite of sugar-specific transporters (EII) 23. We constructed mutants for the sucrose (ΔsacP), glucose (ΔptsG) and fructose (ΔfruA and ΔlevD) transporters, respectively, and an IPTG-inducible mutant for the co-regulated universal EI and Hpr proteins (Pspac-ptsH-ptsI operon) in Pxyl-murE background. In the walled state (MurE ON), all of the strains grew well, except the ptsHI mutant, which showed slightly slower growth on NA/sucrose plates (Fig. 2a and Supplementary Fig. 2a), suggesting that not only sucrose but also other sugars in the plate are utilized as carbon sources under these conditions. When PG synthesis was blocked (MurE OFF), none of the single deletion mutants showed significant growth (Fig. 2a, middle and right). However, the ptsHI mutant showed efficient L-form growth (Fig. 2b) when repressed (Fig. 2a, middle) but not when induced (right panel). Thus, inhibition of uptake for glycolytic sugars by repressing the PTS system promotes L-form growth without the need for the ispA mutation, likely by reducing glycolytic flux.
Figure 2. The glycolysis-mediated cell death is suppressed under gluconeogenic conditions.
a, Effects of repression of sugar uptake system, PTS, on L-form growth. B. subtilis strains YK1601 (Pxyl-murE Pspac-ptsHI), YK1602 (Pxyl-murE ΔptsG), YK2124 (Pxyl-murE ΔsacP), YK2125 (Pxyl-murE ΔfruA) and YK2126 (Pxyl-murE ΔlevD) were streaked on NA/MSM (sucrose) plates with or without 1 mM IPTG, and in the presence (MurE ON) or absence (MurE OFF) of 1% xylose, and incubated at 30°C. No effects were observed in the walled cell growth in the presence of IPTG and xylose.
b, Phase contrast micrographs of YK1601 were taken from the plates without IPTG and xylose shown in panel a.
c, Effects of additional carbon sources on L-form growth on a complex rich medium (NA). B. subtilis strains BS115 (Pxyl-murE ispA+) and LR2 (Pxyl-murE ispA-) were streaked on NA plates with 0.5 M various carbon sources (sucrose, glucose, malate or succinate) in the absence of xylose (MurE OFF), and incubated at 30°C.
d, Phase contrast micrographs of BS115 or LR2 were taken from the plates shown in panel c.
The figures are representative of at least three independent experiments (a-d).
Since the PTS is also crucial for the signal-transduction pathways in carbon catabolite repression (CCR) 23, we tested the effects of deleting ccpA, which encodes a key transcriptional factor in the CCR, but no significant stimulation of L-form growth was observed (Supplementary Fig. 2a).
Contrasting effects of gluconeogenic carbon sources on L-form growth
Figures 2c and d show that glucose (NA + 0.5M glucose), which is a highly favoured glycolytic carbon source in many bacteria, could substitute for sucrose in providing osmotic support for B. subtilis L-form growth, but again this required a mutation such as ispA. These findings were largely independent of the growth medium because similar results were obtained when the cells were cultured on minimal medium (MM) containing 0.5 M sucrose or glucose as the sole carbon source (Supplementary Fig. 2b and c). Again L-form growth was readily observed in the ispA mutant, but not in the ispA+ background, confirming that the reduction of RC activity by the ispA mutation is crucial for B. subtilis L-form growth under glycolytic conditions.
Gluconeogenesis is a metabolic pathway that results in the generation of glucose from certain non-carbohydrate carbon substrates, including malate and succinate, and the reactions occur much like the reverse of glycolysis. Imported malate enters the TCA cycle, but only 10% is respired via the TCA cycle in B. subtilis 24. The majority is converted to phosphoenolpyruvate and pyruvate, resulting in an increased gluconeogenic flux and overflow metabolism. Similarly, it has been shown that flux through the TCA cycle is also reduced in the presence of succinate 25. To test whether increased flux through gluconeogenesis (and/or overflow metabolism) could counteract the inhibitory effect of glycolysis on L-form growth, we streaked Pxyl-murE strains with or without an ispA mutation on NA plates containing 0.5 M malate or succinate: L-form growth occurred efficiently even without the ispA mutation (Fig. 2c and d). Similar results were obtained on MM plates (Supplementary Fig. 2b and c). Thus, a reduction of RC activity by the ispA mutation is not required for L-form growth under gluconeogenic conditions.
L-form growth does not require mutational changes under gluconeogenic conditions
β-lactams, including penicillin G (PenG), remain widely used antibiotics, which target the penicillin-binding proteins that assemble the PG wall 26. We recently discovered that innate immune PG hydrolases, such as lysozyme, can rescue various Gram-positive bacteria from β-lactam killing by converting them to L-forms5. Supplementary Figures 1b-e and 2d show that the promotion of L-form growth by combined treatment with PenG and lysozyme (NB these cells were murE+) still required an ispA mutation or other mutations in genes acting on the RC pathway under aerobic glycolytic conditions. However, under gluconeogenic conditions, a completely wild type (i.e. murE+, ispA+) B. subtilis strain could be switched into the L-form state by PenG and lysozyme. As shown in Figure 3a, the presence of PenG blocked B. subtilis growth on NA/succinate plates (because the L-form protoplast remains trapped inside the cell 5), but a lawn of small colonies was readily detectable on plates containing both PenG and lysozyme after 2 days of incubation (because the lysozyme enables the protoplast to escape from the cell and initiate L-form growth 5). Phase contrast microscopy of the smaller colonies showed the presence of heterogeneous spheroidal L-forms (Fig. 3b, ii). The longer incubation also promoted the emergence of several larger colonies on plates (Fig. 3a, i), which appeared to contain walled cells with typical rod-shaped morphology (Fig. 3b, i). These presumably arose by reversion from L-forms when the antibiotic selection decayed, as described previously 5,27. Similar results were observed on succinate plates containing a cephalosporin β-lactam, cephalexin, except that fewer revertant colonies were evident (Supplementary Fig. 2e). Thus, under gluconeogenic conditions in the presence of lysozyme, wild type B. subtilis cells can escape from β-lactam killing by switching to an L-form state without the need for any mutational change.
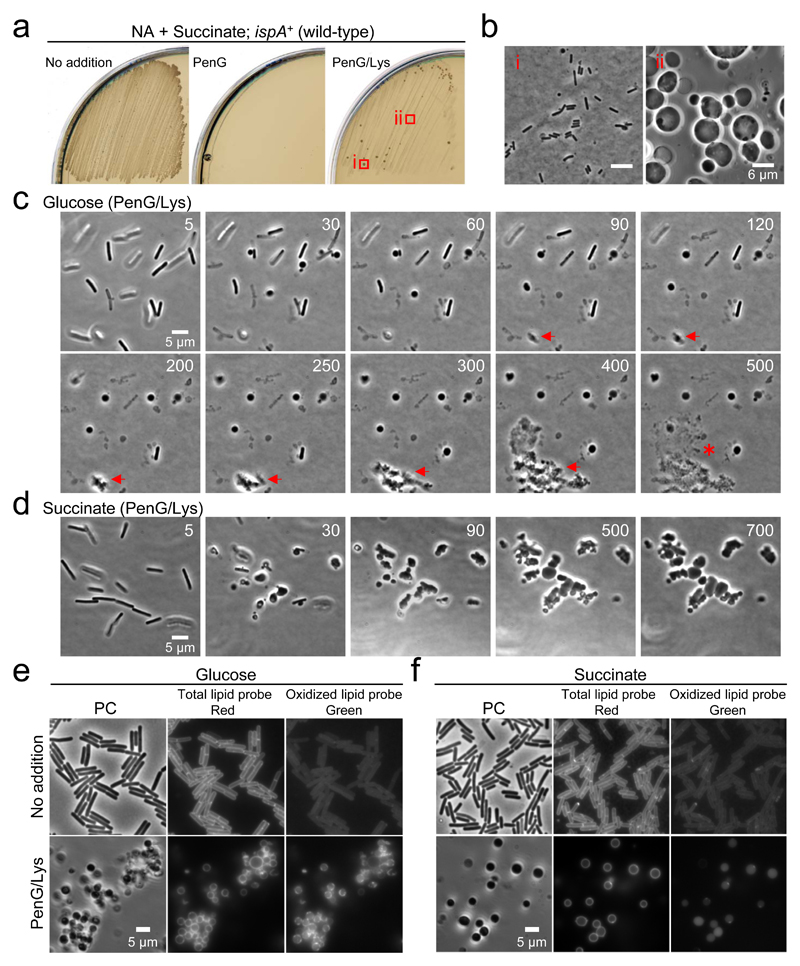
Figure 3. L-form growth under gluconeogenic conditions.
a, B. subtilis wild-type L-form growth on succinate plates containing PenG and lysozyme. B. subtilis wild-type strain (168CA; murE+ ispA+) was streaked on NA/succinate plates with or without 200 μg/ml PenG, or PenG and 100 μg/ml chicken egg white lysozyme (Lys), and incubated at 30°C for 2-3 days.
b, Phase contrast micrographs of walled cells (i) and L-forms (ii) were taken from the plates shown in panel a (PenG/Lys).
c, Effects of glucose on L-form transition of B. subtilis wild-type strain. Exponentially growing B. subtilis wild-type cells were placed on NA/glucose with PenG and lysozyme for time-lapse microscopy. Individual frames are extracted from Supplementary Video 1. Numbers in the top right corner of each frame represent time (min) elapsed in the movie.
d, Effects of succinate on L-form transition of B. subtilis wild-type strain. Exponentially growing B. subtilis wild-type cells were placed on NA/succinate with PenG and lysozyme for time-lapse microscopy. Individual frames are extracted from Supplementary Video 1. Numbers in the top right corner of each frame represent time (min) elapsed in the movie.
e, f, Effects of L-form transition on oxidative damage in B. subtilis wild-type strain. Exponentially growing B. subtilis wild-type cells in NB/glucose (e) or NB/succinate (f) were treated with PenG and lysozyme. The fluorescent probe C11-BODIPY581/591 was used as an indicator of lipid peroxidation. The probe undergoes a shift from red to green fluorescence emission upon peroxidation.
The figures are representative of at least two independent experiments (a-f).
Time-lapse microscopy was used to view the L-form switch in a wild-type strain. Under glycolytic conditions in the presence of PenG, addition of lysozyme promoted the emergence of L-forms within 30 min, but they did not grow and frequently lysed (Fig. 3c and Supplementary Video 1). Although we occasionally observed successful L-form growth (Fig. 3c, red arrows), they eventually lysed (asterisk at 500 min). In contrast, under gluconeogenic conditions, L-forms emerged from the walled cells and propagated in a typical manner (Fig. 3d and Supplementary Video 1).
To test whether the failure of L-form growth under glycolytic conditions was due to oxidative damage, we took advantage of a fluorescent fatty acid analogue, C11-BODYPY581/591, which has been used as an indicator of oxidative damage to lipids 9,28. The probe is incorporated into membranes, and the fluorescent properties shift from red to green upon free radical-induced oxidation. In wild-type B. subtilis walled cells exponentially growing in NB/glucose medium, the cells exhibited a regular and smooth red fluorescence at the cell surface, but no clear green fluorescence was detectable (Fig. 3e, No addition). In contrast, green fluorescence was readily detectable in cells treated with PenG and lysozyme (Fig. 3e, PenG/Lys), suggesting oxidative damage to lipids in L-forms under glycolytic conditions. Crucially, such a fluorescent shift was not observed in L-forms cultured under gluconeogenic conditions (Fig. 3f).
Increased carbon flux through glycolysis generates ROS toxicity
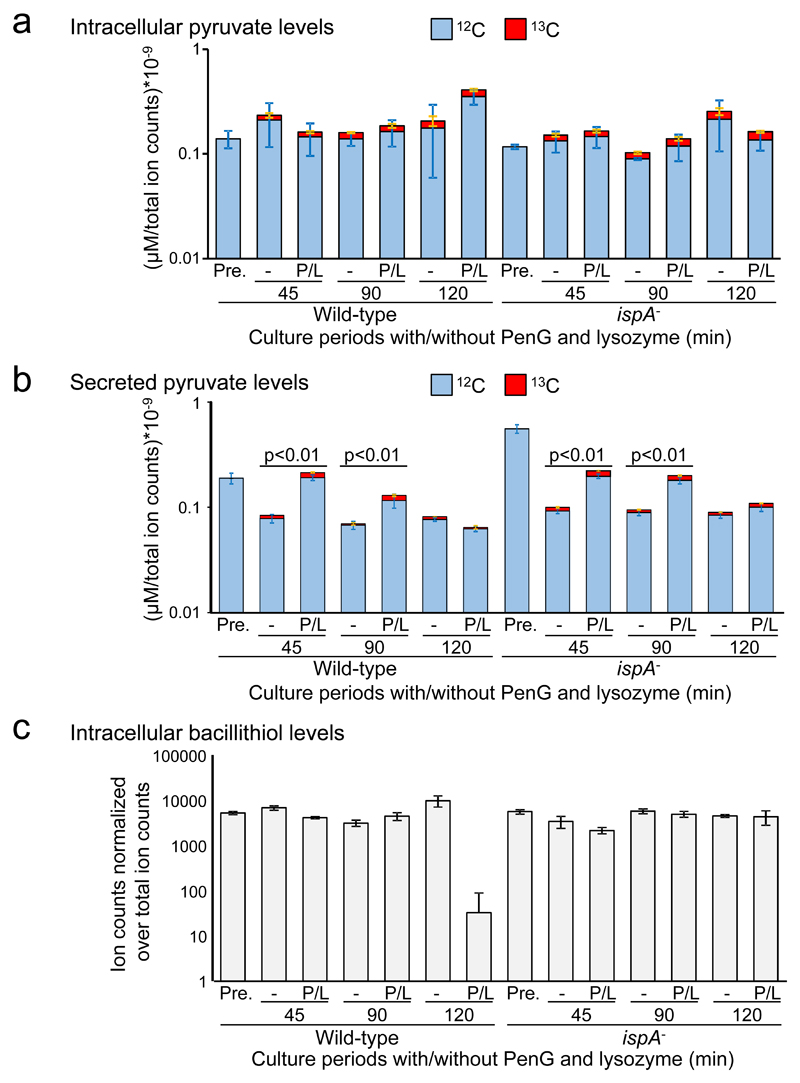
The above results suggested that the generation of L-forms from walled cells and/or L-form growth causes a perturbation in cellular metabolism under glycolytic conditions, leading to ROS-mediated cell death (Supplementary Fig. 3a). To test this, we carried out 13C-based metabolic analysis. B. subtilis wild-type or ispA- cells were pre-cultured in NB containing 0.4 M glucose medium to obtain sufficient biomass and then transferred to fresh 0.4 M glucose medium containing 20 % universally 13C-labelled (U-13C) glucose in the presence or absence of PenG and lysozyme. Pyruvate is the end product of glycolysis, and previous work has shown that B. subtilis excretes excess pyruvate during growth on glycolytic substrates 29. Although no significant changes occurred in the pool size of intracellular pyruvate (Fig. 4a), we observed increased extracellular pyruvate in the presence of PenG and lysozyme (Fig. 4b). This finding provided direct support for the idea that flux through glycolysis is increased during L-form growth.
Figure 4. Increase in glycolytic flux and ROS toxicity during L-form transitions.
a, b, Effects of the L-form transition on glycolysis in B. subtilis. The charts show the pool size of labelled (13C, red) and unlabelled (12C, blue) intracellular pyruvate (a), or secreted extracellular pyruvate (b) from ~108 cells of B. subtilis wild-type (168CA) and ispA- (RM81) cultures in NB/glucose with or without PenG (P) and lysozyme (L). Pre-cultures of B. subtilis in the presence 0.4 M unlabelled glucose were also analysed as a control for the pyruvate levels before incubation in the presence of labelled glucose (Pre.). The concentrations were normalised over total ion counts. The bars represent the averages and the standard deviation from four biological replicates. The p value (two-sided) was calculated using a student t test. To ensure that cell lysis did not contribute to the extracellular values we also measured succinate and found diminished (rather than increased) extracellular levels in the presence of PenG and lysozyme (Supplementary Table 1).
c, Effects of the L-form transition on intracellular bacillithiol. Samples from the above experiment were analysed for total pool size of intracellular bacillithiol from ~108 cells of B. subtilis cultures. The identification of bacillithiol was based on m/z ion counts, which were normalised over total ion counts. The bars represent the averages and the standard deviation from four biological replicates.
Bacillithiol (BSH) plays an important role in detoxification of ROS 30 and promotion of L-form growth under aerobic glycolytic conditions (Supplementary Fig. 3b-d) 9. We analysed the samples used for the above experiments and found that intracellular BSH levels were significantly reduced in the wild-type cells treated with PenG and lysozyme (Fig. 4c and Supplementary Fig. 4), suggesting that BSH is consumed due to the accumulation of ROS. However, the reduction did not occur in the ispA mutant cells (Fig. 4 and Supplementary Fig. 4), again consistent with the idea that increased flux through glycolysis due to the inhibition of PG synthesis causes increased ROS generation from the RC pathway, leading to cell death.
Gluconeogenic growth protects S. aureus and L. monocytogenes from β-lactam killing
We examined whether these observations held also for a pathogenic Gram-positive bacterium S. aureus. Here, we replaced lysozyme with lysostaphin, a more effective hydrolytic enzyme for S. aureus PG 31. As shown in Figures 5a and b, no growth was seen on either glucose or succinate plates containing PenG. However, L-form growth was readily detectable on succinate plates with PenG and lysostaphin but not on glucose plates. Note that although we and others have reported that S. aureus L-form growth does occur on complex rich medium containing sucrose as osmoprotectant 5,27,32,33, that L-form growth requires uptake and metabolism of glycerol, which is a substrate for gluconeogenesis 32, suggesting that sucrose may not be the primary carbon source under those conditions.
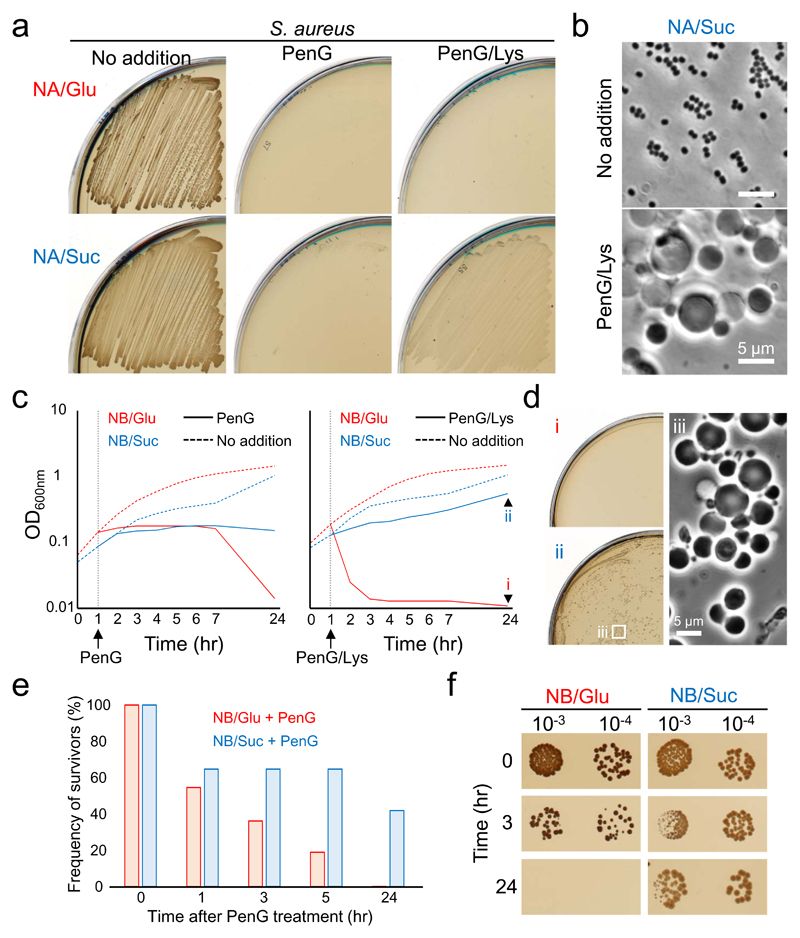
Figure 5. S. aureus escapes from β-lactam killing under gluconeogenic conditions.
a, Effects of carbon sources on S. aureus L-form growth. S. aureus strain RN4220 was streaked on NA/glucose (Glu) or succinate (Suc) plates with or without 200 μg/ml PenG and/or 10 μg/ml lysostaphin, and incubated at 30°C for 2-3 days.
b, Phase contrast micrographs of S. aureus cells on NA/succinate plates with or without PenG and lysostaphin were taken from the plates shown in panel a.
c, Growth profiles of S. aureus cells in NB/glucose (Glu) or succinate (Suc) with or without 100 μg/ml PenG (left panel), or PenG and 10 μg/ml lysostaphin (right panel). S. aureus cells were incubated at 30°C without shaking.
d, The overnight cultures of S. aureus cells in NB/glucose (panel c, i) or succinate (panel c, ii) with PenG and lysostaphin were placed on NA/glucose (i) or succinate (ii), and incubated at 30°C for 3 days. Phase contrast micrograph of S. aureus L-forms was taken from the NA/succinate plates (iii).
e, Relative PenG sensitivity of S. aureus in NB/glucose (Glu) compared to NB/succinate (Suc). Time course measurements of CFU (ml-1) in S. aureus cultures (panel c, left) after addition of PenG were shown as a percentage of the survivors for the cells before PenG treatment (time 0) in the same cultures.
f, S. aureus cells in NB/glucose (Glu) or succinate (Suc) after addition of PenG (panel c, left) were diluted (10-fold series) and 10 μl spots were placed on NA/Glu or Suc plates, respectively, and incubated at 30°C for 2-3 days.
The figures are representative of at least three independent experiments (a-f).
PenG prevents PG assembly and is generally thought to induce explosive cell lysis under classical microbiological culture conditions, which are mainly hypotonic. Although, under osmoprotective conditions, the explosive death is largely suppressed, bacteria still die 5, by mechanisms that have not yet been resolved. Given the above results, we wondered whether gluconeogenic growth could also protect walled cells from PenG killing in isotonic medium. We cultured S. aureus walled cells in nutrient broth (NB) containing 0.5 M glucose (Glu) or succinate (Suc). The growth arrest was induced in both cultures shortly after addition of PenG (Fig. 5c, left). Although no significant changes in optical density (OD) occurred in the succinate culture following PenG treatment, prolonged incubation of the glucose culture resulted in a severe decrease in OD. In parallel with this, the colony forming units (CFU) of the glucose culture decreased over time in the presence of PenG and had dropped at least 1,000-fold after overnight incubation (Fig. 5e and f, and Supplementary Table 2). In contrast, in the succinate culture CFU fell no more than 10 fold after overnight incubation with PenG (Fig. 5e and f, and Supplementary Table 2). It thus appears that the efficiency of PenG killing in walled cells is greatly reduced under gluconeogenic conditions. This suggests that oxidative damage through glycolysis contributes greatly to PenG killing in walled S. aureus.
Consistent with a key role for lysozyme in the switch to L-form growth upon β-lactam treatment, the OD of the succinate culture was significantly increased by combined treatment with PenG and lysostaphin (Fig. 5c, right, and d). However, in the glucose culture, rapid cell death occurred (Fig. 5c, right). We do not fully understand the mechanism of the rapid death but the glycolysis-mediated killing via oxidative damage or other mechanisms, such as futile cycling of PG synthesis and turnover, as occurs in E. coli 34, might be enhanced by stripping the PG wall. Nevertheless, those killing effects are largely counteracted under gluconeogenic conditions, leading to robust L-form growth that can evade β-lactam action.
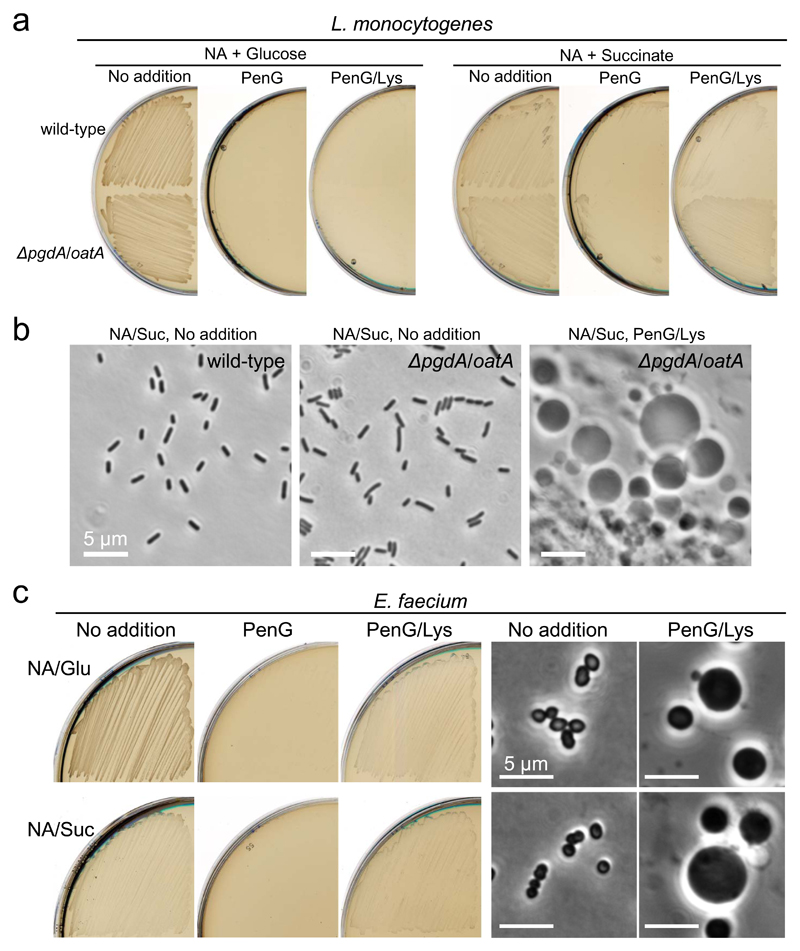
We then examined another pathogenic Firmicute, Listeria monocytogenes that is naturally resistant to lysozyme due to N-deacetylation or O-acetylation on the PG wall: elimination of the PG modification by use of a pgdA/oatA double mutant results in sensitivity to lysozyme 35,36. As shown in Figures 6a and b, formation and significant growth of L. monocytogenes L-forms did not occur on glucose plates containing PenG and lysozyme. In contrast, L-form colonies appeared on succinate plates after 2 days, showing that Listeria behaves similarly to B. subtilis and S. aureus in the effects of glycolytic vs gluconeogenic substrates. Note, also, that L-form growth was much more abundant for the pgdA/oat double mutant than for the wild type (Fig. 6a, Succinate, PenG/Lys), providing further evidence for the importance of cell wall degradation in L-form switching and protection from β-lactam killing 5.
Figure 6. Bacteria lacking the RC pathway evades glycolysis-mediated β-lactam killing.
a, Effects of carbon sources on L. monocytogenes L-form growth. L. monocytogenes wild-type EDGe and EDGe ΔpgdA Δoat double mutant were streaked on NA/glucose (Glu) or succinate (Suc) plates with or without 200 μg/ml PenG and/or 100 μg/ml lysozyme (Lys), and incubated at 30°C for 2-3 days.
b, Phase contrast micrographs were taken from the plates shown in panel a.
c, Effects of carbon sources on E. faecium L-form growth. E. faecium (ATCC19434) was streaked on NA/glucose (Glu) or succinate (Suc) plates with or without 200 μg/ml PenG and/or 100 μg/ml lysozyme, and incubated at 30°C for 2-3 days. Phase contrast micrographs were taken from the plates shown in left.
The figures are representative of at least three independent experiments (a-c).
Lack of the RC pathway allows escape from glycolysis-mediated β-lactam killing
Finally, we examined Enterococcus faecium, which is also a pathogenic Firmicute but is strictly fermentative and completely lacks the RC pathway 12,13. Crucially, significant E. faecium L-form growth was seen not only on succinate plates but also on glucose plates containing PenG and lysozyme within 2 days of incubation (Fig. 6c). Presumably, lack of the RC pathway enables E. faecium to escape from β-lactam killing by switching to the L-form state even under glycolytic conditions.
Discussion
β-lactam antibiotics inhibit bacterial cell wall assembly and it is generally assumed that the main killing effect arises from loss of cell wall integrity, leading to explosive lysis. However, this kind of lysis does not seem generally to occur for many Gram-positive bacteria, even under non-osmoprotective (hypotonic) conditions: the bacteria are still prevented from growing and die by other mechanisms 5,37. Kohanski et al. have proposed a crucial role for ROS 38, which are generated as a natural side effect of aerobic respiration, in antibiotic-mediated killing of bacteria, including that arising from β-lactam treatment, although the extent to which this is responsible for cell killing remains controversial 39,40. Nevertheless, recent studies that reveal an important role for cellular metabolism in β-lactam killing further support the involvement of ROS as a metabolic by-product 41–43. Our results now provide a possible mechanism for this effect. We show that blocking the cell wall pathway stimulates glycolytic flux, leading to ROS production via aerobic respiration (Fig. 1a). It is probably not surprising that the inhibition of cell wall synthesis results in dramatic changes in cell physiology because wall synthesis is a major drain on cellular resources, especially in Gram-positive bacteria where it comprises 10-20% of total cell mass. Crucially, our results appear to show that physiological compensation for the metabolic imbalance, by reducing glycolytic activity or activating gluconeogenesis, moderates the β-lactam toxicity, leading to the proliferation of wall-free L-form bacteria in the presence of lysozyme. The persistent growth during β-lactam treatment under glycolytic conditions of E. faecium, which naturally lacks a respiratory chain 12,13, provides strong support for this view (Fig. 6c). The physiological effects we have uncovered, especially in relation to the sensitivity of cells to intrinsically produced ROS, will be important for efforts to engineer simplified or artificial cells. The work suggests that the utilization of aerobic respiration comes at a potentially serious cost, and that in many organisms cellular metabolism is finely balanced to provide maximum energetic benefit. Perturbations of key central pathways, or in the future, incorrect design of metabolic pathways, may have seriously damaging consequences for the cell.
In many diverse bacteria, the L-form state can be induced experimentally by treatment of cells with antibiotics and/or lytic enzymes that inhibit cell wall synthesis 2–5. A variety of protocols for generating L-forms to specific lineages have been described through decades of research, and those culture methods often involve anaerobic conditions10,44, consistent with a role for oxygen toxicity in L-form generation. Our genetic experiments have now provided some clarity on the physiological changes responsible for this effect; thus, inhibition of the uptake of glycolytic sugars, reduction of glycolytic activity, or reduction of RC activity can all work in promoting L-form growth under aerobic glycolytic conditions. It seems likely that utilization of sugar-phosphate intermediates through glycolysis results in an increased generation of ROS from the RC pathway, which then prevents L-form growth. Stimulation of L-form growth by gluconeogenic growth or oxygen depletion, without the need for any mutational changes provides further support for this view (Fig. 3 and Supplementary Fig. 3).
Since blocking cell wall synthesis eliminates the wall, its precursors and turnover products, which could normally act as ROS scavengers, it is formally possible that loss of these molecules increases sensitivity to normal levels of ROS. However, we do not favour this idea at present because cells treated with β-lactam but no lysozyme, and which therefore still possess both wall and precursors, nevertheless suffer from ROS toxicity 38. Also, growth of L-forms lacking a wall and blocked for precursor synthesis is strongly influenced by modulation of the glycolytic pathway (Fig. 1). It remains to be determined how the increased flux through glycolysis affects the RC pathway thereby impacting on L-form growth.
Our results also reveal that the avoidance of β-lactam killing by switching to the L-form state depends on two key factors; i) escape from the cell wall sacculus – which can be effected by exogenous lytic enzymes, and ii) avoidance of ROS toxicity – which, via reduced glycolytic flux, is suppressed by growth on gluconeogenic carbon substrates (or lack of an RC). These findings show that the L-form transition can be supported, for a wide range of bacteria, perhaps including both free living and pathogenic organisms, by conditions found in the natural environment. Indeed, historically, L-form isolation has been reported from a wide range of sites including animal, human or plant hosts, as well as the natural environment 2–5. Although the possible role of L-forms in infectious diseases is presently unclear, β-lactam treatment could effectively facilitate L-form generation in certain niches within the human body because immune PG hydrolases and gluconeogenic substrates, such as lactate, glycerol and amino acids, are abundantly present. Finally, many antibiotics, including penicillin, work by attacking the bacterial cell wall. Our understanding of the physiological changes that are generated by the inhibition of cell wall synthesis, together with their implications for L-form generation and survival should prompt renewed interest into the possible role of L-forms in persistent or recurrent infections.
Methods
Bacterial Strains and Growth Conditions
The bacterial strains in this study are listed in Supplementary Table 3. Nutrient broth (NB, Oxoid) and agar (NA, Oxoid) was used for bacterial growth at 30°C. Bacterial L-forms were induced and grown on osmoprotective medium composed of 2x magnesium-sucrose-maleic acid (MSM) pH7 (40 mM magnesium chloride, 1 M sucrose, and 40 mM maleic acid) mixed 1:1 with 2x NA, or 2x minimal 45 (0.4% ammonium sulfate, 2.8% dipotassium phosphate, 1.2% potassium dihydrogen phosphate, 0.2% sodium citrate dehydrate, 0.04% magnesium sulphate) medium supplemented with 20 μg/ml tryptophane, 0.1% yeast extract, 1 μg/ml ferric ammonium citrare, 0.1% glutamic acid, 0.02% casamino acids and 1% agar. 1 M sucrose in 2x MSM was replaced by 1 M glucose, malate or succinate as necessary. Supplements, 1 mM IPTG or 1% xylose were added to media when required, and also antibiotics were added at the following concentrations: 1 μg/ml erythromycin, 200 μg/ml PenG, 100 μg/ml cephalexin, 100 μg/ml lysozyme and/or 10 μg/ml lysostaphin. 1 μg/ml 8J (FtsZ inhibitor) 46 was used to prevent the growth of walled cells when required 8. Anaerobic growth condition was maintained using anaerobic atmosphere generation bags (AnaeroGenTM, Oxoid) in an anaerobic jar for growth on plates. 10 mM Nitrate and 0.1% glutamic acid were added for B. subtilis growth under anaerobic conditions. For growth curve and time course CFU measurement, overnight cell cultures in NB were diluted at 10-2 into fresh NB/glucose or succinate and incubated at 30°C with shaking for several hours, and the exponential cell cultures (optical density 600 nm of about 0.1) were prepared for the experiments. The exponential cell cultures were incubated at 30°C without shaking.
Construction of IPTG-Inducible gapA gene, ptsHI operon and glmM gene
To construct the IPTG-inducible gapA mutant, the first 200-300 bp of the gapA gene containing Shine-Dalgarno sequence was amplified by PCR from genomic DNA of the B. subtilis strain 168CA using the primers pM4SD-gapA-F and pM4-gapA-R (Supplementary Table 2), then cloned between the EcoRI and BamHI sites of plasmid pMutin4 47, creating pM4-Pspac-gapA. The resulting plasmid was introduced into the strain BS115 to generate YK1571 (Supplementary Table 2). In this strain, the full-length gapA gene is expressed from the IPTG-inducible promoter Pspac, but the growth is not fully complemented, suggesting insufficient expression to compensate for the native levels of GapA. We therefore introduced a second copy of PspacHY-gapA fusion in the strain YK1571. The gapA gene containing the Shine–Dalgano sequence was amplified by PCR from genomic DNA of B. subtilis strain 168CA, then cloned between the XbaI and SphI sites of plasmid pPL82 48, creating pPL82–gapA. The resulting plasmid was used to generate YK1621 in which the full-length gapA are expressed from the IPTG-inducible promoter PspacHY at the amyE locus on the chromosome (Supplementary Table 2).
To construct the IPTG-inducible ptsHI mutant, the first 200~300 bp of the ptsH gene containing Shine-Dalgarno sequence was amplified by PCR from genomic DNA of the B. subtilis strain 168CA using the primers pM4SD-ptsHI-F and pM4-ptsHI-R (Supplementary Table 2), then cloned between the EcoRI and BamHI sites of plasmid pMutin4 47, creating pM4-Pspac-ptsHI. The resulting plasmid was introduced into the strain BS115 to generate YK1601 (Supplementary Table 2). In this strain, the full-length ptsHI operon is expressed from the IPTG-inducible promoter Pspac.
To construct the IPTG-inducible glmM mutant, the first 200~300 bp of the glmM gene containing Shine-Dalgarno sequence was amplified by PCR from genomic DNA of the B. subtilis strain 168CA using the primers pM4SD-glmM-F and pM4-glmM-R (Supplementary Table 2), then cloned between the EcoRI and BamHI sites of plasmid pMutin4 47, creating pM4-Pspac-glmM. The resulting plasmid was introduced into the strain LR2 to generate YK1563 (Supplementary Table 2). In this strain, the full-length glmM is expressed from the IPTG-inducible promoter Pspac. DNA manipulations and transformations were carried out using standard methods.
Construction of disruption mutants
To construct the ptsG, sacP, fruA and levD disruption mutant, an internal segment of those genes were amplified by PCR from the B. subtilis strain 168CA genomic DNA using the primers in each case Supplementary Table 2), then cloned between the EcoRI and BamHI sites of plasmid pMutin4 47, creating pMutin4-ptsG, sacP, fruA and levD. The resulting plasmids were introduced into the strain BS115 to generate disruption mutants (Supplementary Table 2). DNA manipulations and transformations were carried out using standard methods.
Microscopic Imaging
For snapshot live-cell imaging, cells were mounted on microscope slides covered with a thin film of 1 % agarose in water, or NB/MSM for L-forms. Images were acquired with a Sony Cool-Snap HQ2 cooled CCD camera (Roper Scientific) attached to a Zeiss Axiovert 200M, or a Rolera EM-C2 (Q-imaging) camera attached to a Nikon TiE microscope, and analysed using Metamorph (Molecular Devices). For time lapse imaging, 300 μL of exponentially growing B. subtilis walled cells in NB/glucose or succinate media were placed on 35 mm sterile glass bottom microwell dishes (ibidi GmbH, Munich, Germany), which were pre-coated with 2 mg/ml bovine serum albumin (BSA), and incubated for 5 min. The dishes were centrifuged at 100 g for 2 min using a Beckman Allegra X-12R centrifuge. Non-adherent cells were removed, and a thin layer of NB/g with 0.2% agar bacteriological (Oxoid), 100 μg/ml PenG and lysozyme was placed on the top of the dishes. The dishes were placed on the microscope stage at 30°C. Images were acquired with a Sony Cool-Snap HQ2 cooled CCD camera (Roper Scientific) attached to a Zeiss Axiovert 200M, and analyzed using Metamorph (Molecular Devices). Pictures and videos were prepared for publication using ImageJ (https://imagej.nih.gov/ij) and Adobe Photoshop.
Lipid peroxidation
Lipid oxidation was detected using a fluorescent probe (C11-BODIPY581/591; Life Technologies) as described previously 9. Exponentially growing B. subtilis walled cells were cultured in NB/glucose or succinate media at 37°C, and then 5 μM C11-BODIPY581/591 was added to the culture and incubated for 1 hr at 30°C before being used for microscopic analysis. For L-forms, 100 μg/ml PenG and lysozyme were added to the cultures with exponentially growing B. subtilis walled cells and incubated for 1 hr at 37°C. The cultures were further incubated for several hours at 30°C without shaking, and then 5 μM C11-BODIPY581/591 was added to the culture.
Metabolome analysis
For metabolome analysis nutrient broth (Difco) supplemented with 5 g/L NaCl and 0.4M glucose was used (NBG). The NBG was supplemented with 15 g/L agar (NAG) to generate solid plates.
For intracellular metabolome analysis, B. subtilis cells were grown in NBG until mid-log phase. 108 cells were collected on 0.22 μm filters and the transferred in NAG and incubated overnight at 30°C to allow replication and producing biomass. Then, the cells-laden filters were used as an unlabelled pre-culture control or placed in fresh NAG plates containing 20% U-13C-glucose (Sigma-Aldrich), with or without 200 μg/ml PenG and lysozyme, and incubated at 30 °C. The cells were collected at 0 (pre-culture control), 45, 90 and 120 mins and polar metabolites were extracted in pre-chilled (-40 °C) methanol:acetonitrile:water solution as previously described 49.
For exo-metabolome analysis, B. subtilis cultures were grown in NBG until mid-log phase and the culture filtrates were collected by 0.2 μm filters as an unlabelled pre-culture control. 108 cells were collected from the log phase cultures and re-suspended in fresh 4 ml NBG containing 20% U-13C-glucose (Sigma-Aldrich), with or without 400 μg/ml PenG and lysozyme, and incubated at 30 °C. The culture filtrates were collected by 0.2 μm filters at 45, 90 and 120 mins and then diluted 1:3 with methanol:acetonitrile:water solution.
The liquid chromatography-mass spectrometry was performed with a modified protocol described previously 50. Briefly, aqueous normal phase liquid chromatography was performed using an Agilent 1200 LC system at controlled temperature (4 °C). Flow rate of 0.4 mL/min was used. Elution polar were performed using a gradient of two solvents, A (mQ water and 0.1 % of formic acid) and B (acetonitrile and 0.1 % of formic acid). The accurate mass spectrometry was performed using an Agilent Accurate Mass 6230 TOF apparatus equipped with Dual Agilent Jet Stream ESI source. The data were analysed by Profinder B.08.00 software and Mass Hunter Qualitative Analysis B07.00. The metabolites were identified comparing the accurate m/z (error <10 ppm) and the retention time with the accurate m/z and the retention time of standard solutions for the specific metabolite. The metabolites were quantified using standards calibration curves and then were normalised over total ion counts present in each extract. Since the bacillithiol standard is not commercially available, the identification of this molecule (the reduced form, monomer) was based on m/z only. We could not detect the oxidised form (dimer).
Supplementary Material
Acknowledgments
We thank Pascale Cossart for L. monocytogenes strains. All work in the Errington lab was funded by a European Research Council award (grant number 670980). Work in the LPSC lab was supported by a Wellcome Trust New Investigator Award (104785/B/14/Z). The LPSC lab is also funded by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001060), the UK Medical Research Council (FC001060), and the Wellcome Trust (FC001060).
Footnotes
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Author Contributions
Y.K. performed and analysed most of the experiments. R.M. and K.M. contributed strain constructions and preliminary genetic and microscopic experiments. A.S. and L.P.S.C. performed and analysed the metabolome and mass spectrometry experiments. All authors contributed to experimental design and concepts. Y.K. and J.E. wrote the main text with contributions from all other authors.
Competing Interests
The authors declare no competing interests.
References
- 1.Klieneberger E. The natural occurrence of pleuropneumonia-like organisms in apparent symbiosis with Streptobacillus moniliformis and other bacteria. J Pathol Bacteriol. 1935;40:93–105. [Google Scholar]
- 2.Allan EJ, Hoischen C, Gumpert J. Bacterial L-forms. Adv Appl Microbiol. 2009;68:1–39. doi: 10.1016/S0065-2164(09)01201-5. [DOI] [PubMed] [Google Scholar]
- 3.Domingue GJ, Sr, Woody HB. Bacterial persistence and expression of disease. Clin Microbiol Rev. 1997;10:320–344. doi: 10.1128/cmr.10.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Errington J, Mickiewicz K, Kawai Y, Wu LJ. L-form bacteria, chronic diseases and the origins of life. Philos Trans R Soc Lond B Biol Sci. 2016;371 doi: 10.1098/rstb.2015.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawai Y, Mickiewicz K, Errington J. Lysozyme Counteracts beta-Lactam Antibiotics by Promoting the Emergence of L-Form Bacteria. Cell. 2018;172:1038–1049 e1010. doi: 10.1016/j.cell.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leaver M, Dominguez-Cuevas P, Coxhead JM, Daniel RA, Errington J. Life without a wall or division machine in Bacillus subtilis. Nature. 2009;457:849–853. doi: 10.1038/nature07742. [DOI] [PubMed] [Google Scholar]
- 7.Domínguez-Cuevas P, Mercier R, Leaver M, Kawai Y, Errington J. The rod to L-form transition of Bacillus subtilis is limited by a requirement for the protoplast to escape from the cell wall sacculus. Mol Microbiol. 2012;83:52–66. doi: 10.1111/j.1365-2958.2011.07920.x. [DOI] [PubMed] [Google Scholar]
- 8.Mercier R, Kawai Y, Errington J. Excess membrane synthesis drives a primitive mode of cell proliferation. Cell. 2013;152:997–1007. doi: 10.1016/j.cell.2013.01.043. [DOI] [PubMed] [Google Scholar]
- 9.Kawai Y, et al. Cell growth of wall-free L-form bacteria is limited by oxidative damage. Curr Biol. 2015;25:1613–1618. doi: 10.1016/j.cub.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber TW, Brinkley AW. Growth of cell wall-defective variants of Escherichia coli: comparison of aerobic and anaerobic induction frequencies. J Clin Microbiol. 1977;6:166–171. doi: 10.1128/jcm.6.2.166-171.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jutras BL, Jacobs-Wagner C. Bacterial evolution: what goes around comes around. Curr Biol. 2015;25:R496–498. doi: 10.1016/j.cub.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Willett HP. In: Zinsser's Microbiology. 20th ed. Joklik Wolfgang K, Willett Hilda P, Amos Dennis B., editors. Appleton: Lange; 1992. pp. 53–75. [Google Scholar]
- 13.van Schaik W, et al. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics. 2010;11:239. doi: 10.1186/1471-2164-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julsing MK, Rijpkema M, Woerdenbag HJ, Quax WJ, Kayser O. Functional analysis of genes involved in the biosynthesis of isoprene in Bacillus subtilis. Appl Microbiol Biotechnol. 2007;75:1377–1384. doi: 10.1007/s00253-007-0953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blencke HM, et al. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab Eng. 2003;5:133–149. doi: 10.1016/s1096-7176(03)00009-0. [DOI] [PubMed] [Google Scholar]
- 16.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 17.Gyan S, Shiohira Y, Sato I, Takeuchi M, Sato T. Regulatory loop between redox sensing of the NADH/NAD(+) ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J Bacteriol. 2006;188:7062–7071. doi: 10.1128/JB.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santana M, et al. Molecular cloning, sequencing, and physiological characterization of the qox operon from Bacillus subtilis encoding the aa3-600 quinol oxidase. J Biol Chem. 1992;267:10225–10231. [PubMed] [Google Scholar]
- 19.Mogi T. Over-expression and characterization of Bacillus subtilis heme O synthase. J Biochem. 2009;145:669–675. doi: 10.1093/jb/mvp024. [DOI] [PubMed] [Google Scholar]
- 20.Töwe S, et al. The MarR-type repressor MhqR (YkvE) regulates multiple dioxygenases/glyoxalases and an azoreductase which confer resistance to 2-methylhydroquinone and catechol in Bacillus subtilis. Mol Microbiol. 2007;66:40–54. doi: 10.1111/j.1365-2958.2007.05891.x. [DOI] [PubMed] [Google Scholar]
- 21.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira Y, Petit-Glatron MF, Chambert R. yveB, Encoding endolevanase LevB, is part of the sacB-yveB-yveA levansucrase tricistronic operon in Bacillus subtilis. Microbiology. 2001;147:3413–3419. doi: 10.1099/00221287-147-12-3413. [DOI] [PubMed] [Google Scholar]
- 23.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 24.Kleijn RJ, et al. Metabolic fluxes during strong carbon catabolite repression by malate in Bacillus subtilis. J Biol Chem. 2010;285:1587–1596. doi: 10.1074/jbc.M109.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schilling O, et al. Transcriptional and metabolic responses of Bacillus subtilis to the availability of organic acids: transcription regulation is important but not sufficient to account for metabolic adaptation. Appl Environ Microbiol. 2007;73:499–507. doi: 10.1128/AEM.02084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovering AL, Safadi SS, Strynadka NC. Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem. 2012;81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 27.Kawai Y, Mercier R, Errington J. Bacterial cell morphogenesis does not require a preexisting template structure. Curr Biol. 2014;24:863–867. doi: 10.1016/j.cub.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drummen GP, van Liebergen LC, Op den Kamp JA, Post JA. C11-BODIPY(581/591), an oxidation-sensitive fluorescent lipid peroxidation probe: (micro)spectroscopic characterization and validation of methodology. Free Radic Biol Med. 2002;33:473–490. doi: 10.1016/s0891-5849(02)00848-1. [DOI] [PubMed] [Google Scholar]
- 29.Chubukov V, et al. Transcriptional regulation is insufficient to explain substrate-induced flux changes in Bacillus subtilis. Mol Syst Biol. 2013;9:709. doi: 10.1038/msb.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandrangsu P, Loi VV, Antelmann H, Helmann JD. The Role of Bacillithiol in Gram-Positive Firmicutes. Antioxid Redox Signal. 2018;28:445–462. doi: 10.1089/ars.2017.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu JA, Kusuma C, Mond JJ, Kokai-Kun JF. Lysostaphin disrupts Staphylococcus aureus and Staphylococcus epidermidis biofilms on artificial surfaces. Antimicrob Agents Chemother. 2003;47:3407–3414. doi: 10.1128/AAC.47.11.3407-3414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han J, et al. Glycerol uptake is important for L-form formation and persistence in Staphylococcus aureus. PLoS One. 2014;9:e108325. doi: 10.1371/journal.pone.0108325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mercier R, Kawai Y, Errington J. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. Elife. 2014;3 doi: 10.7554/eLife.04629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho H, Uehara T, Bernhardt TG. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell. 2014;159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rae CS, Geissler A, Adamson PC, Portnoy DA. Mutations of the Listeria monocytogenes peptidoglycan N-deacetylase and O-acetylase result in enhanced lysozyme sensitivity, bacteriolysis, and hyperinduction of innate immune pathways. Infect Immun. 2011;79:3596–3606. doi: 10.1128/IAI.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burke TP, et al. Listeria monocytogenes is resistant to lysozyme through the regulation, not the acquisition, of cell wall-modifying enzymes. J Bacteriol. 2014;196:3756–3767. doi: 10.1128/JB.02053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez-Tébar A, Rojo F, Vazquez D. Interaction of beta-lactam antibiotics with penicillin-binding proteins from Bacillus megaterium. Eur J Biochem. 1982;126:161–166. doi: 10.1111/j.1432-1033.1982.tb06761.x. [DOI] [PubMed] [Google Scholar]
- 38.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 39.Dwyer DJ, Collins JJ, Walker GC. Unraveling the physiological complexities of antibiotic lethality. Annu Rev Pharmacol Toxicol. 2015;55:313–332. doi: 10.1146/annurev-pharmtox-010814-124712. [DOI] [PubMed] [Google Scholar]
- 40.Van Acker H, Coenye T. The Role of Reactive Oxygen Species in Antibiotic-Mediated Killing of Bacteria. Trends Microbiol. 2017;25:456–466. doi: 10.1016/j.tim.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Lobritz MA, et al. Antibiotic efficacy is linked to bacterial cellular respiration. Proc Natl Acad Sci USA. 2015;112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belenky P, et al. Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage. Cell Rep. 2015;13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ladjouzi R, et al. Loss of Antibiotic Tolerance in Sod-Deficient Mutants Is Dependent on the Energy Source and Arginine Catabolism in Enterococci. J Bacteriol. 2015;197:3283–3293. doi: 10.1128/JB.00389-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dienes L, Weinberger HJ. The L forms of bacteria. Bacteriol Rev. 1951;15:245–288. doi: 10.1128/br.15.4.245-288.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spizizen J. Transformation of Biochemically Deficient Strains of Bacillus subtilis by Deoxyribonucleate. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams DW, Wu LJ, Czaplewski LG, Errington J. Multiple effects of benzamide antibiotics on FtsZ function. Mol Microbiol. 2011;80:68–84. doi: 10.1111/j.1365-2958.2011.07559.x. [DOI] [PubMed] [Google Scholar]
- 47.Vagner V, Dervyn E, Ehrlich SD. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144(Pt 11):3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 48.Quisel JD, Burkholder WF, Grossman AD. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J Bacteriol. 2001;183:6573–6578. doi: 10.1128/JB.183.22.6573-6578.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agapova A, et al. Flexible nitrogen utilisation by the metabolic generalist pathogen Mycobacterium tuberculosis. Elife. 2019;8 doi: 10.7554/eLife.41129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larrouy-Maumus G, et al. Cell-Envelope Remodeling as a Determinant of Phenotypic Antibacterial Tolerance in Mycobacterium tuberculosis. ACS Infect Dis. 2016;2:352–360. doi: 10.1021/acsinfecdis.5b00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.