Many bacteria are characterized by their ability to form highly resistant spores. The dormant spore state allows these species to survive even the harshest treatments with antimicrobial agents. Spore surface properties are particularly relevant because they influence spore dispersal in various habitats from natural to human-made environments. The spore surface in Bacillus subtilis (crust) is composed of a combination of proteins and polysaccharides. By inactivating the enzymes responsible for the synthesis of spore polysaccharides, we can assess how spore surface properties such as hydrophobicity are modulated by the addition of specific carbohydrates. Our findings indicate that several sporulation gene clusters are responsible for the assembly and allocation of surface polysaccharides. Similar mechanisms could be modulating the dispersal of infectious spore-forming bacteria.
KEYWORDS: Bacillus subtilis, cell surface, polysaccharides, spore coat, spore crust, sporulation
ABSTRACT
Polysaccharides (PS) decorate the surface of dormant endospores (spores). In the model organism for sporulation, Bacillus subtilis, the composition of the spore PS is not known in detail. Here, we have assessed how PS synthesis enzymes produced during the late stages of sporulation affect spore surface properties. Using four methods, bacterial adhesion to hydrocarbons (BATH) assays, India ink staining, transmission electron microscopy (TEM) with ruthenium red staining, and scanning electron microscopy (SEM), we characterized the contributions of four sporulation gene clusters, spsABCDEFGHIJKL, yfnHGF-yfnED, ytdA-ytcABC, and cgeAB-cgeCDE, on the morphology and properties of the crust, the outermost spore layer. Our results show that all mutations in the sps operon result in the production of spores that are more hydrophobic and lack a visible crust, presumably because of reduced PS deposition, while mutations in cgeD and the yfnH–D cluster noticeably expand the PS layer. In addition, yfnH–D mutant spores exhibit a crust with an unusual weblike morphology. The hydrophobic phenotype from sps mutant spores was partially rescued by a second mutation inactivating any gene in the yfnHGF operon. While spsI, yfnH, and ytdA are paralogous genes, all encoding glucose-1-phosphate nucleotidyltransferases, each paralog appears to contribute in a distinct manner to the spore PS. Our data are consistent with the possibility that each gene cluster is responsible for the production of its own respective deoxyhexose. In summary, we found that disruptions to the PS layer modify spore surface hydrophobicity and that there are multiple saccharide synthesis pathways involved in spore surface properties.
IMPORTANCE Many bacteria are characterized by their ability to form highly resistant spores. The dormant spore state allows these species to survive even the harshest treatments with antimicrobial agents. Spore surface properties are particularly relevant because they influence spore dispersal in various habitats from natural to human-made environments. The spore surface in Bacillus subtilis (crust) is composed of a combination of proteins and polysaccharides. By inactivating the enzymes responsible for the synthesis of spore polysaccharides, we can assess how spore surface properties such as hydrophobicity are modulated by the addition of specific carbohydrates. Our findings indicate that several sporulation gene clusters are responsible for the assembly and allocation of surface polysaccharides. Similar mechanisms could be modulating the dispersal of infectious spore-forming bacteria.
INTRODUCTION
Polysaccharides (PS) that decorate bacterial cell surfaces have been highlighted for their importance as the first point of contact in cell-to-cell, cell-to-host, or cell-to-surface interactions (1–5). The variety and intricacy of these complex sugar polymers allow for targeted recognition by other cells, for instance, through receptor specificity (e.g., with lectins). While this is often studied in the context of vegetative cells, we know that dormant bacterial endospores (spores) are also decorated with PS (6–9). In this study, we use Bacillus subtilis spores to investigate the contribution of PS synthesis enzymes to spore surface properties.
B. subtilis is a Gram-positive model organism for endospore-forming bacteria. In a nutrient-deprived environment, B. subtilis cells undergo an asymmetric division that results in one compartment forming a forespore, which will mature into a metabolically dormant spore, while the other, the mother cell, assists in forespore development and ultimately lyses (10–13). The spore is known for its ability to survive methods of sterilization, such as bleach or boiling. These resistance properties derive in large part from the complex architecture of the spore, which includes two structures built during spore formation, a thick peptidoglycan layer known as the cortex, and a protein coat made of more than 80 proteins organized in four concentric layers, the basement layer, inner coat, outer coat, and crust (14–16). Along with at least one spore membrane, these layers protect a partially dehydrated core that contains the spore chromosome. The coat layers assemble in a temporal manner and rely on the successive activation of sigma (σ) factors in the mother cell (σE in early sporulation and σK in late sporulation) (17). The outermost layer of the spore, the crust, is assembled late during sporulation by proteins under the control of σK. It is the structure most likely to influence the spore’s interactions with its environment (18, 19). The crust is composed of structural proteins that have a propensity to self-assemble (19–22) and is likely adorned by PS (9, 22, 23). The relative contributions of PS to overall spore surface properties are poorly understood, resulting in a major gap in the knowledge of how spores interact with the environment and endure harsh conditions.
Studies on the PS composition of the spore surface reveal that there is large variation between species of sporulating bacteria (6). Two pathogenic species from the Bacillus cereus group, Bacillus anthracis and Bacillus cereus, produce spores that closely resemble B. subtilis spores, although they do not have a crust; they are, however, encircled by an exosporium, a distinct, ballooning layer of proteins and glycoproteins assembled in a different manner from the coat (14, 24–26). Furthermore, B. anthracis spores have a surface deoxyhexose unique to the species called anthrose, while B. cereus has cereose and B. subtilis has quinovose (27–29). While previous studies have determined pathways for anthrose and cereose, the pathway responsible for the production of quinovose is still unclear (8, 26). At least one pathway of carbohydrate production during late sporulation is characterized in B. subtilis and is controlled by the spore polysaccharide synthesis (sps) operon. The last four genes, spsIJKL, are attributed specifically to rhamnose production (8, 9). Rhamnose is known to decorate the surface of B. subtilis and the B. cereus group spores, and σK-dependent operons responsible for rhamnose synthesis during sporulation have been found in both (8, 26). The spsIJKL gene functions have been conserved and are orthologous to the rmlACBD operon in the B. cereus group (26). Still, it is unclear in which PS synthesis pathway(s) the seven upstream genes in the sps operon, spsABCDEFG, are involved. A second spore polysaccharide synthesis locus under the control of σK, spsM, has been characterized in B. subtilis (23, 30).
Our previous research identified two additional σK-dependent gene clusters, ytdA-ytcABC and yfnHGF-yfnED, potentially involved in spore PS synthesis (31). Interestingly, ytdA and yfnH are paralogous to spsI, and they all encode glucose-1-phosphate nucleotidyltransferases. The production of glucose-1-phosphate, using nucleoside triphosphates as an energy source, is often the first step in the synthesis of deoxyhexoses from glucose (32). Since each paralog apparently utilizes a different nucleotide, they may be involved in pathways producing different deoxyhexoses. It is even possible that one of these pathways is involved in the synthesis of the B. subtilis-specific deoxyhexose quinovose. The molecular composition and the global architecture of the PS that decorate Bacillus spores are likely to dictate their interaction with the environment. Therefore, we have analyzed the contributions of these σK-dependent gene clusters to the B. subtilis spore surface properties. One of our major goals is to understand how these genes may influence spore dispersal in aqueous environments and, conversely, persistence in soils or hospital surroundings, extremely important functions of spores in human-made and natural environments (33). Studies of the B. cereus group have found that there is variability in spore properties, including hydrophobicity, between species (34, 35). The degree of hydrophobicity can dictate the level of adherence (36, 37). Since some pathogenic species might be more hydrophobic than nonpathogenic species, hydrophobicity might impact infection (28, 38). For example, the exosporium of the B. anthracis spore appears to contribute to its affinity for soil types grazed by livestock (39). Given the complexity of PS composition and their ability to dictate charge and alter other physical and chemical surface properties, the possibility that the PS present on the spore surface modulate hydrophobicity and adhesion constitutes an important area of investigation.
In this article, we continue a line of research initiated when we investigated how spore surface properties were influenced by the structural proteins responsible for proper crust assembly, focusing on cotO and cotVWXYZ (22). The deletion of these genes greatly disrupted the crust structure, resulted in perturbations of the PS distribution around the spore, and caused modifications in the hydrophobicity of the spore surface. Our data also suggested, however, that the crust might not constitute the sole anchoring point for PS and that there may be a PS layer between the outer coat and the crust. It remains unclear if the outer coat is an anchor site by default, i.e., solely used in the absence of the crust, or if wild-type spores bear more than one site for PS addition. Our previous work also looked at the cgeAB operon because of its potential role in PS anchoring in B. subtilis spores (22). Roels and Losick originally showed that cgeAB and cgeCDE are two adjacent operons expressed under the control of σK and GerE that confer specific properties to the outermost layer of the spore, possibly by glycosylation of coat proteins (40). Here, we show that genes involved in PS synthesis during sporulation influence the degree of hydrophobicity of B. subtilis spores and that these genes have distinct and sometimes antagonistic functions on the assembly of the PS layer. Our data provide insights on the mechanisms used by the spore to dictate its interactions with the environment and will guide future investigations aimed at identifying all the carbohydrates decorating the spore surface.
RESULTS
Contributions of the spsA–L operon to spore surface properties.
We utilized three methods of analysis, i.e., bacterial adherence to hydrocarbons (BATH) assays, negative staining of spore PS with India ink, and transmission electron microscopy (TEM) with ruthenium red staining, to observe the contributions of the spsA–L operon to spore surface properties and crust morphology. The BATH assay is a technique where spores are suspended in a polar aqueous phase (phosphate-buffered saline [PBS]) layered with a nonpolar organic solvent, hexadecane. The percentage of spores remaining in the aqueous layer is obtained by comparing the optical densities of the spore suspension before and after vortexing. All three methods were used recently to determine the roles played by crust structural proteins in these properties (22). Previous work has revealed that the last four genes in the spsA–L operon encode enzymes involved in rhamnose biosynthesis during the late stages of sporulation (8). The rest of the operon has not been characterized biochemically, even though detailed structural information is available for many proteins, including the crystal structure of the nucleotide-diphospho-sugar transferase SpsA (41). Every gene in the spsA–L operon shows significant homology to genes encoding either glycosyltransferases or enzymes participating in carbohydrate biosynthesis/modification reactions (Fig. 1; see also Fig. S1 in the supplemental material). All individual mutants in the operon produce spores that exhibit a strong hydrophobicity profile by BATH assay (relative to the more-hydrophilic wild-type spores), with less than 25% of spores remaining in the aqueous layer after addition of the organic solvent hexadecane and vortexing for 90 s (Fig. 2A and S2A). For genes specifically implicated in rhamnose production, BATH assays indicate that only 13% ± 2% of ΔspsI mutant spores remain in the aqueous layer (similarly, 15% ± 6% remain for ΔspsJ mutant spores, 14% ± 2% remain for ΔspsK mutant spores, and 11% ± 3% remain for ΔspsL mutant spores). Furthermore, inactivation of any one gene in the operon (as well as deletion of the entire operon) results in the production of spores devoid of any perceivable halo by negative staining with India ink, suggesting a major disruption of the PS layer (Fig. 2B and S2B). Previous research has indicated that ruthenium red staining assists in the visualization of the spore crust by TEM (7, 19). TEM analysis of sps mutant spores is consistent with the hypothesis that rhamnose synthesis is necessary for spore PS production, since ΔspsI mutant spores differ drastically from wild-type spores in the sense that the crust layer is barely detectable, whereas the rest of the coat assembles normally (Fig. 2C). Nevertheless, some features alluding to the presence of an outermost layer persist, e.g., an unstained interspace region between the outer coat and the potential localization of the crust layer. The observed defect is most likely due to a defect in PS deposition on the preassembled protein components of the crust, as it does not appear that assembly of the crust structural proteins is disrupted in the ΔspsI mutant. Specifically, green fluorescent protein (GFP) fusions to crust proteins CgeA, CotX, CotY, and CotZ localize in a similar manner in wild-type and spsI mutant spores (Fig. S2C). Taken together, our results indicate that the inability to properly synthesize rhamnose (and any carbohydrate whose biosynthesis/modification would be controlled by the spsA–L operon) results in perturbed surface properties and a severe disruption of the PS layer around the spore. This interpretation agrees with previous work highlighting the importance of the spsA–L operon or rhamnose synthesis to spore surface properties in B. subtilis and B. anthracis (6, 9, 38, 42).
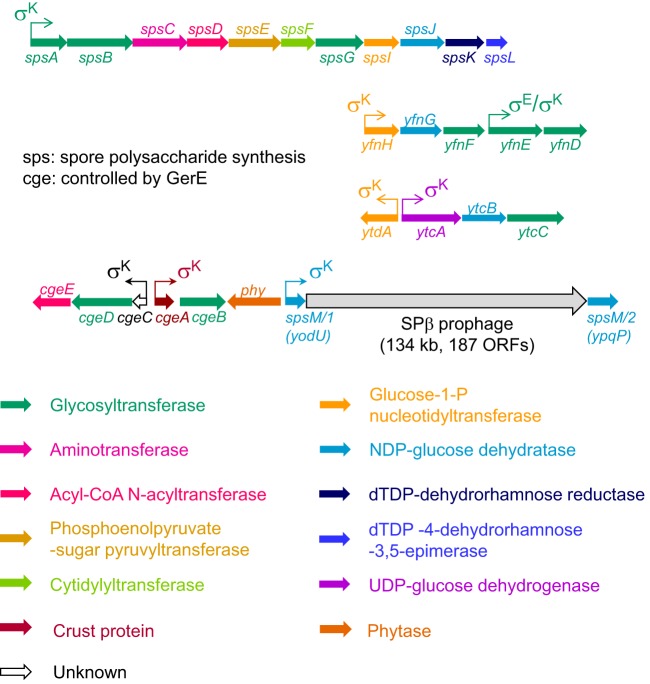
FIG 1.
Gene clusters involved in spore polysaccharide synthesis. Chromosomal regions encoding enzymes involved in spore PS synthesis in B. subtilis are shown. Arrows for each gene signal the transcriptional direction and the colors are associated with respective gene functions. Promoter locations and sporulation σ factor dependencies for each operon are also indicated with arrows (43, 45). Several of these operons have been partially characterized. The last four genes of the spsA–L operon are involved in rhamnose synthesis (8). The spsM gene (interrupted by the SPβ prophage, which is excised from the mother cell genome during sporulation) is also known to be necessary for spore PS synthesis (23). The cgeAB operon encodes a protein component of the spore crust (CgeA) and a putative glycosyltransferase (CgeB) (20, 40). The glucose-1-phosphate nucleotidyltransferase, SpsI, has two paralogs, YfnH and YtdA, both expressed during the late stages of sporulation under the control of the mother cell sigma factor σK (31). Based on sequence homology, adjacent sporulation genes also appear to be involved in PS synthesis. Putative PS synthesis pathways are detailed in Fig. S1. ORFs, open reading frames.
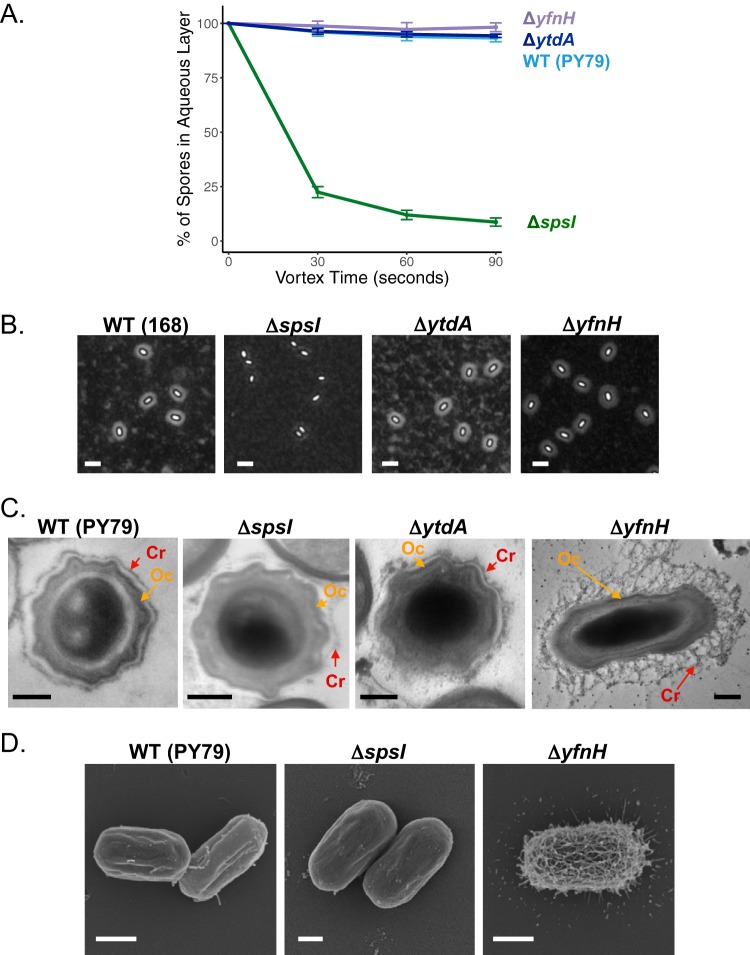
FIG 2.
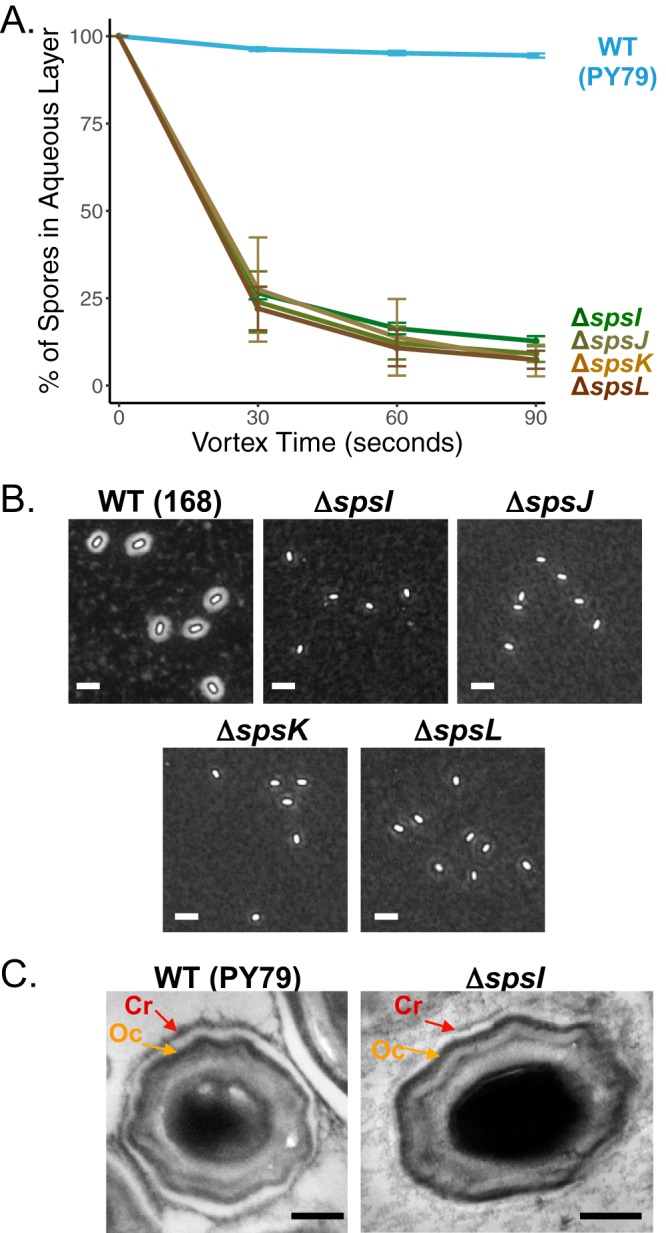
Contributions of the spsA–L operon to spore surface properties. (A) BATH assay of spores with the following gene deletions: ΔspsI (PE2763), ΔspsJ (PE3348), ΔspsK (PE3349), and ΔspsL (PE3350). An inability to synthesize rhamnose causes an increase in relative hydrophobicity profiles compared to wild-type (WT) spores (PY79). Deletions of genes upstream of spsI also result in the production of spores that are more hydrophobic (Fig. S2A). Experiments were performed in triplicate; error bars represent standard deviations. (B) India ink staining is a method revealing the presence of a PS layer by negative staining. Wild-type spores (168) are surrounded by a halo confirming the presence of a PS layer. The following sps mutant spores exhibit no halo, suggesting disruption of the PS layer: ΔspsI (BKE37840), ΔspsJ (BKE37830), ΔspsK (BKE37820), and ΔspsL (BKE37810). Similar results are obtained with deletions upstream of spsI or by deletion of the entire sps operon (Fig. S2B). Scale bars = 2.5 μm. (C) Analysis of spore crust morphology by TEM with ruthenium red staining shows that wild-type spores (PY79) are surrounded by an electron-dense outer coat layer (Oc, orange arrow) and a crust (Cr, red arrow). ΔspsI spores (PE2763) have a greatly reduced crust (red arrow), while the outer coat remains intact (orange arrow). Scale bars = 200 nm. Analysis of the localization of the crust proteins CotX-GFP, CotY-GFP, CotZ-GFP, and CgeA-GFP in ΔspsI mutant sporulating cells indicates that their localization is unaffected, suggesting that the deletion of spsI does not interfere with crust protein assembly but prevents PS deposition (Fig. S2C).
Impact of glucose-1-phosphate nucleotidyltransferases on spore surface properties.
B. subtilis contains two SpsI paralogs (YfnH and YtdA) that are also expressed during late sporulation under the control of σK (31). Based on sequence analysis, all 3 enzymes are glucose-1-phosphate nucleotidyltransferases catalyzing the addition of nucleoside diphosphates to glucose during PS synthesis (Fig. 1). Each of these enzymes, however, appears to utilize a different nucleoside triphosphate as the substrate, as SpsI is a thymidylyltransferase, YfnH is a cytidylyltransferase, and YtdA is a uridylyltransferase. These differences in energy sources suggest that these enzymes could play distinct roles in PS production, e.g., that they may be involved in the synthesis of different monosaccharides (Fig. S1). An alternative hypothesis would be that they can all participate in the same pathway and contribute different forms of activated glucose substrates. If that were the case, however, these enzymes would likely have partially redundant functions. We performed BATH assays, India ink staining, and TEM with ruthenium red staining in order to better understand the roles played by these poorly characterized enzymes in crust morphogenesis, especially how they may influence the structure of the PS layer.
As reported above (Fig. 2), ΔspsI mutant spores are more hydrophobic than are wild-type spores by BATH assays and display a severely reduced PS layer in India ink and TEM evaluations. In contrast, ΔytdA and ΔyfnH mutant spores are indistinguishable from hydrophilic wild-type spores in the BATH assays (31), settling in the aqueous phase at 94% ± 1% and 98% ± 2%, respectively (Fig. 3A). However, upon visualization by India ink staining, ΔyfnH mutant spores have an expanded PS layer (see Fig. 3B for a representative image, Fig. 4A for an additional image, and Fig. 4B and S4A for quantification of the halo areas). It is possible that ΔytdA mutant spores have a slightly expanded PS layer, but the increase in width is modest in comparison to wild-type spores (Fig. 3B and S3A for quantification of the halo area). We also investigated if genes from the σK-controlled ytcABC operon adjacent to ytdA (Fig. 1) were necessary for spore PS production but observed no significant difference between wild-type and mutant spores (Fig. S3H for BATH assays and Fig. S3I for negative staining).
FIG 3.
Putative glucose-1-phosphate nucleotidyltransferases expressed during late sporulation play different roles in spore PS synthesis. The following three σK-dependent sporulation genes encode putative glucose-1-phosphate nucleotidyltransferases: spsI, yfnH, and ytdA. (A) Analysis by BATH assays. While deletion of spsI (PE2763) causes a significant increase in spore hydrophobicity, ΔyfnH (PE2919) and ΔytdA (PE2764) mutant spores are indistinguishable from wild-type spores (PY79). Experiments were performed in triplicate; error bars represent standard deviations. (B) Analysis by India ink staining. ΔspsI mutant spores (BKE37840) show no halo, ΔytdA mutant spores (BKE30850) have halos that are similar in size to wild-type (168) spores, and ΔyfnH mutant spores (BKE07270) have expanded halos. Quantification of halo areas for ΔytdA and ΔyfnH mutant spores is reported in Fig. S3A and S4A, respectively. Scale bars = 2.5 μm. (C) Imaging of wild-type (PY79), ΔspsI mutant (PE2763), ΔytdA (PE2764), and ΔyfnH mutant (PE2919) spores by TEM with ruthenium red staining. Inactivation of yfnH results in the production of spores surrounded by a large and extensive weblike PS (red arrow) with an intact outer coat (orange arrow). Scale bars = 200 nm. Additional TEM images can be found in Fig. S3B to D. (D) Scanning electron micrographs of wild-type (PY79), ΔspsI mutant (PE2763), and ΔyfnH mutant (PE2919) spores. Scale bars = 500 nm. Additional SEM images can be found in Fig. S3E to G. BATH assays and India ink staining for deletions in the ytcABC operon are presented in Fig. S3H and I, respectively.
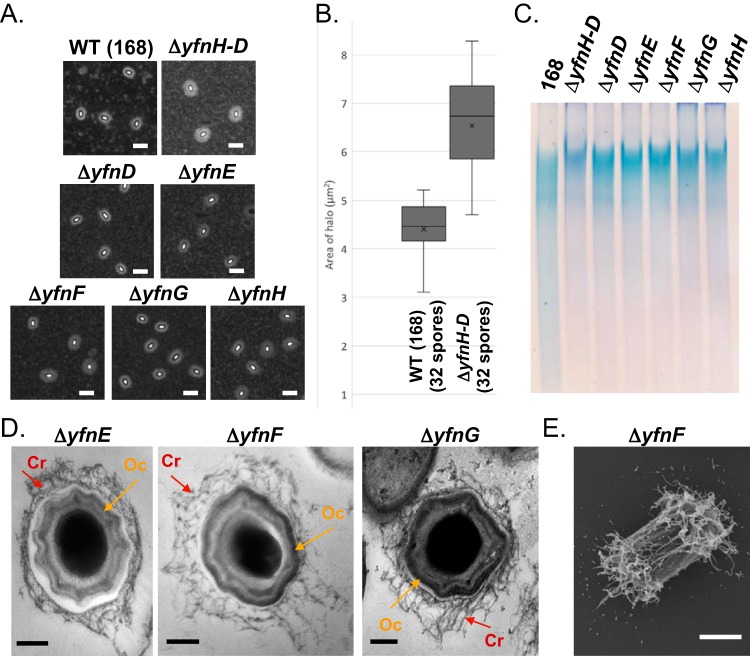
FIG 4.
Gene deletions in the yfnHGFED cluster result in expansion of the PS layer. (A) Analyses by India ink staining of a deletion mutant of the entire yfnH–D gene cluster (NY35) and individual gene deletions, as follows: ΔyfnD (BKE07310), ΔyfnE (BKE07300), ΔyfnF (BKE07290), ΔyfnG (BKE07280) and ΔyfnH (BKE07270). All deletion mutants exhibit an increase in spore halo width compared to wild-type spores (168). Scale bars = 2.5 μm. (B) Measurements of the halo areas (numbers of spores correspond to the number of spores considered for the measurements) for wild-type (168) and ΔyfnH–D (NY35) spores. Measurements for ΔyfnD (BKE07310), ΔyfnE (BKE07300), ΔyfnF (BKE07290), ΔyfnG (BKE07280) and ΔyfnH (BKE07270) mutant spores can be found in Fig. S4A. (C) Analyses of spore surface extracts by gel electrophoresis (5% polyacrylamide, Tris-borate-EDTA [TBE], stained with Stains-All). All mutants show an increase in size presumably caused by an expansion in PS content. BATH assays for each deletion mutant in the yfnH–D cluster are displayed in Fig. S4B. (D) Imaging of ΔyfnE (PE3063), ΔyfnF (PE3062), and ΔyfnG (PE2961) mutant spores by TEM with ruthenium red staining. Mutant spores exhibit extensive weblike PS (red arrow) with an intact outer coat (orange arrow). Scale bars = 200 nm. TEM images of spore fields can be found in Fig. S4C to E. (E) Analysis of ΔyfnF (PE3062) mutant spores by SEM. Scale bar = 500 nm. A larger field of spores is displayed in Fig. S4F.
Further analysis by TEM and ruthenium red staining confirms the expanded PS structure in the ΔyfnH mutant spores and reveals an unexpected weblike architecture (Fig. 3C and S3D). Wild-type, ΔspsI mutant, and ΔyfnH mutant spores were also subjected to scanning electron microscopy (SEM). Images of ΔyfnH mutant spores confirmed the presence of an intricate weblike structure. In several spores, this structure covered the entire spore surface, but in other instances, the weblike structure was found predominantly at the spore poles (Fig. S3G). Wild-type and ΔspsI mutant spores exhibited a much smoother surface appearance (Fig. 3D and S3E and F). Because the SEM protocol does not use ruthenium red staining (the traditional osmium tetroxide stain is used instead), it was unclear whether it would be possible to detect PS on the spore surface. Yet, the extra PS produced by the ΔyfnH mutant was clearly visible under these conditions. Taken together, our observations suggest that yfnH affects the formation of the PS layer and that it does so in a different manner than the spsA–L operon, possibly by changing the number or composition of the monosaccharides added to the protein components of the crust or by affecting the branching of the saccharide polymers. Importantly, the deletion of yfnH does not appear to perturb the localization of any crust proteins fused to GFP (Fig. S2C). Analysis by TEM of ΔytdA mutant spores does not reveal major differences in morphology with wild-type spores (Fig. 3C and S3B and C). The crust is present and more visible than in ΔspsI mutant spores but much less expanded than in ΔyfnH mutant spores. In summary, our data imply that the paralogs of SpsI do not have redundant activities and that YfnH especially has distinct contributions to the process of spore PS synthesis.
Systematic analysis of the yfnH–D gene cluster.
The yfnH gene belongs to a cluster composed of two operons (Fig. 1), yfnHGF, controlled by a σK-dependent promoter upstream of yfnH, and yfnED, under the control of a σE- and σK-dependent promoter upstream of yfnE (43). We confirmed that disruption of the entire cluster or individual gene deletions all resulted in expansion of the spore PS layer (see Fig. 4A for representative images; see Fig. 4B and S4A for quantification of the halo areas). The increase in size of the PS layer is also detected in electrophoresis of spore surface extracts (Fig. 4C). This technique had been previously used to confirm the implication of spsM in spore PS synthesis (23). In 5% native polyacrylamide gels stained with Stains-All (a dye that reveals PS, nucleic acids, and acidic proteins), spore surface extracts from each of the individual gene deletions in the yfnH–D cluster migrate at a higher molecular weight than do extracts from wild-type 168 spores. Consistent with this observation, evaluation of ΔyfnE, ΔyfnF, or ΔyfnG mutant spores by TEM also revealed that crusts have largely expanded PS layers with a weblike structure (Fig. 4D and S4C to E). As expected, in BATH assays, each of the mutants in the yfnH–D cluster produced spores that are as hydrophilic as wild-type spores (Fig. S4B). Analysis of ΔyfnF mutant spores by SEM (Fig. 4E and S4F) confirms the presence of an expanded weblike structure, although the expansion is more noticeable at the poles than in the center of the spore surface.
Partial complementation of the ΔspsI phenotype by inactivation of yfnF or yfnH.
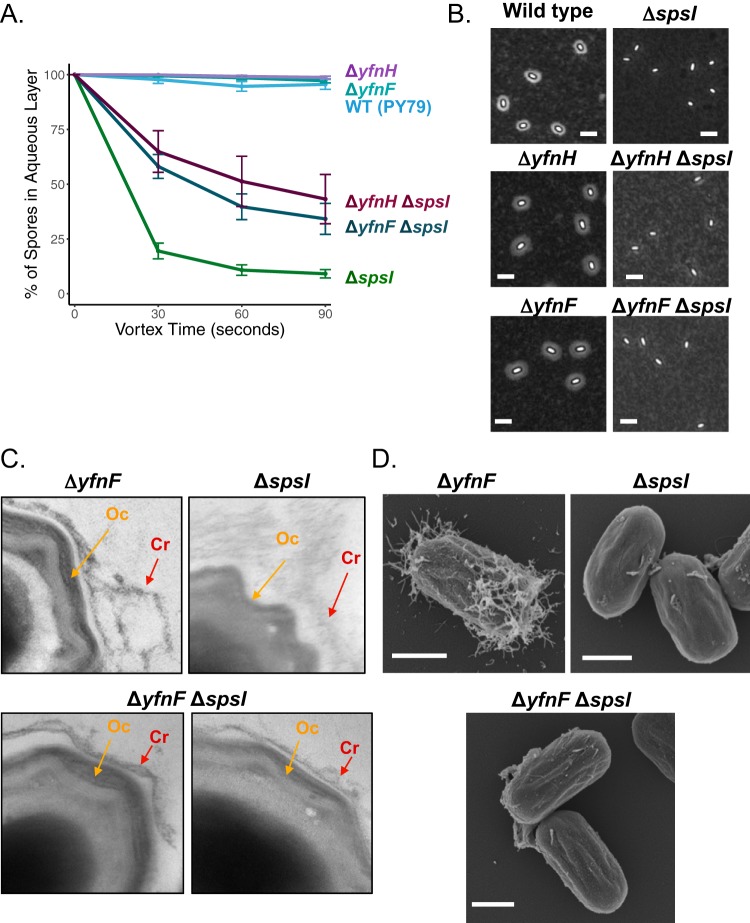
The expanded nature of the PS layer in the ΔyfnH, ΔyfnF, and ΔyfnE mutant spores is in direct contrast with the PS layer reduction in ΔspsI mutant spores, leading us to explore how these operons may interact to contribute to PS distribution and, thus, modulate spore surface properties. Specifically, we characterized the phenotypes of the ΔspsI ΔyfnH, ΔspsI ΔyfnF, and ΔspsI ΔyfnE double mutants. In BATH assays (Fig. 5A), we observe partial complementation of the spore hydrophobicity phenotype of the ΔspsI mutant with deletion of yfnH or yfnF (43% ± 11% of spores in the aqueous phase for the ΔspsI ΔyfnH mutant and 34% ± 7% for the ΔspsI ΔyfnF mutant). Yet, the recovery is incomplete since double-mutant spores remain more hydrophobic than do wild-type spores. Furthermore, not every double mutant rescues the spore hydrophobicity phenotype since inactivation of yfnD or yfnE has no effect (Fig. S5A) (7% ± 2% for the ΔspsI ΔyfnE mutant and 9% ± 3% for the ΔspsI ΔyfnD mutant). By negative staining with India ink, it does not seem that there is a significant difference between the PS layer morphology of ΔspsI mutant spores and double-mutant spores, as neither exhibits a halo (Fig. 5B). In contrast, by ruthenium red staining in TEM, a technique that might be more sensitive to certain forms of PS, a significant degree of complementation is observed in all of the double mutants (Fig. 5C and S5B for the ΔspsI ΔyfnF mutant and Fig. S5C for the ΔspsI ΔyfnE and ΔspsI ΔyfnH mutants). In ΔspsI ΔyfnF mutant spores, we detect the presence of a thin crust by TEM (Fig. 5C, bottom, red arrows), while, as reported above, little to no crust could be detected in ΔspsI mutant spores (Fig. 5C, top right), and an expanded crust is observed in ΔyfnF spores (Fig. 5C, top left, red arrow). Similarly, some restoration of the crust structure was observed in ΔspsI ΔyfnH and ΔspsI ΔyfnE mutant spores (Fig. S5C, red arrows). By SEM, however, the ΔspsI ΔyfnF mutant (Fig. 5D and S5E) and ΔspsI ΔyfnH mutant (Fig. S5D) spores are similar to the ΔspsI mutant spores. Taken together, the data collected for the double-mutant spores suggest partial complementation of the ΔspsI phenotype by inactivation of specific genes in the yfnH–D cluster. This intermediate phenotype is likely a consequence of the spsA–L operon and the yfnH–D cluster having antagonistic functions in PS assembly, since deletion of spsA–L causes reduction of the spore PS layer, whereas deletion of yfnH–D causes expansion of the spore PS layer.
FIG 5.
Partial complementation of the ΔspsI phenotype by deletion of yfnH or yfnF. (A) By BATH assay, the ΔyfnH ΔspsI (PE3119) or ΔyfnF ΔspsI (PE3118) double-deletion mutants have a greater percentage of spores remaining in the aqueous layer than did ΔspsI single-mutation spores (PE2763), and fewer spores remaining in the aqueous layer than ΔyfnH (PE2919) or ΔyfnF (PE3062) single-mutation spores, implying partial complementation. BATH assays for ΔyfnD ΔspsI (PE3357) and ΔyfnE ΔspsI (PE3117) mutants are shown in Fig. S5A. Experiments were performed in triplicate; error bars represent standard deviation. (B) Analysis of the PS layer in double mutants by India ink staining; ΔyfnH ΔspsI (PE3119) and ΔyfnF ΔspsI (PE3118) mutants do not seem to have a larger halo than do ΔspsI (BKE37840) mutant spores. Scale bars = 2.5 μm. (C) Close-ups of ΔspsI ΔyfnF mutant spores imaged by TEM and ruthenium red staining. The complete spores are shown in Fig. S5B. Top left, ΔyfnF (PE3062) mutant spores have an expanded and weblike PS layer (red arrow) in relation to the outer coat (orange arrow). Top right, ΔspsI (PE2763) mutant spores have thinned and scarcely visible crust. Bottom, ΔyfnF ΔspsI (PE3118) mutant spores have a more visible crust structure, suggesting partial rescue of the surface morphology defects observed in ΔspsI (PE2763) mutant spores. A similar analysis for ΔyfnE (PE3063), ΔyfnH (PE2919), ΔyfnE ΔspsI (PE3117), and ΔyfnH ΔspsI (PE3119) mutant spores is provided in Fig. S5C. (D) Analysis of ΔyfnF (PE3062), ΔspsI (PE2763), and ΔyfnF ΔspsI (PE3118) mutant spores by SEM. Scale bars = 500 nm. SEM images for ΔyfnH (PE2919) and ΔyfnH ΔspsI (PE3119) mutant spores can be found in Fig. S5D. An additional SEM image of a field of and ΔyfnF ΔspsI (PE3118) mutant spores is provided in Fig. S5E.
Contributions of the cgeCDE operon to spore crust structure.
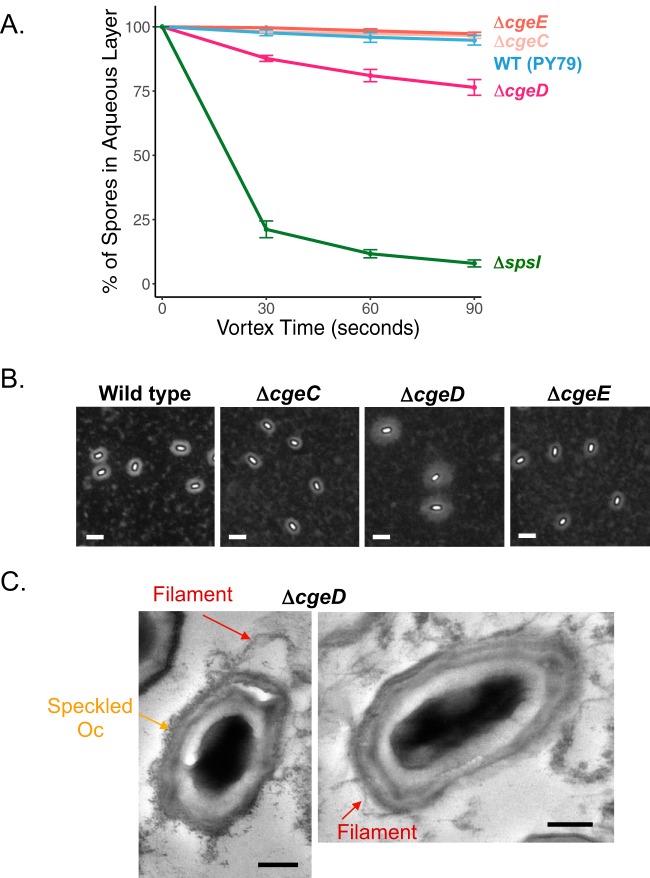
The cgeABCDE cluster is composed of two divergently oriented operons expressed during late sporulation under the control of σK and GerE (40). The spsM gene, which is known to be involved in spore PS synthesis, is in the vicinity of the cge cluster (23) (Fig. 1). Previous research has shown that both cge operons contribute to spore surface properties (40). The first operon, cgeAB, has been characterized in more detail. CgeA is a structural spore crust protein, while CgeB is a putative glycosyltransferase essential for proper crust morphology (18, 20, 22). The contributions made by the second operon, cgeCDE, are less well understood, although sequence analyses provide some clues. Notably, cgeD, which is paralogous to spsA, encodes a putative glycosyltransferase. Similarly, cgeE is homologous to spsD, and both genes encode putative acetyl-coenzyme A (acetyl-CoA) acetyltransferases. In contrast, cgeC is a small gene with no sequence similarity to genes of known function. It is currently not known if CgeC, CgeD, and CgeE belong to a specific monosaccharide synthesis pathway, but our results imply that they do play a role in proper formation of the PS layer on mature spores. By BATH assays, ΔcgeC and ΔcgeE mutant spores are as hydrophilic as wild-type spores, both settling at 97% ± 1% in the aqueous phase (Fig. 6A). There was, however, a small but consistent drop in hydrophilicity for ΔcgeD mutant spores (76% ± 3%) (Fig. 6A). By India ink staining, there is a clear increase in halo width around ΔcgeD mutant spores, suggesting an expansion of the PS layer (Fig. 6B and S6A for halo quantification and Fig. S6B for additional images). The increase is consistent with the analysis of spore surface extracts by gel electrophoresis, where extracts from ΔcgeD mutant spores show a distinctly increased size (Fig. S6C). These observations, however, do not extend to ΔcgeC or ΔcgeE mutant spores, as they appear to be indistinguishable from wild-type spores. It is interesting to contrast the phenotype of ΔcgeD mutant spores, which resembles the phenotype of ΔyfnH mutant spores, with the phenotype previously characterized for ΔspsM spores (23) that showed a drastic reduction of the PS layer, analogous to that observed for ΔspsI mutant spores. However, further inspection of spore ultrastructure by TEM with ruthenium red staining reveals clear differences between ΔcgeD and ΔyfnH mutant spores. While ΔyfnH mutant spores have normal outer coat morphology, ΔcgeD mutant spores have a speckled outer coat. Furthermore, even though ΔcgeD mutant spores have an expanded filamentous crust that balloons away from the spore surface (Fig. 6C), they do not show the weblike crust architecture observed in ΔyfnH mutant spores. In addition, while the India ink staining results are reminiscent of the expanded PS layer in ΔyfnH or ΔyfnF mutant spores, inactivation of cgeD does not rescue (even partially) the phenotype of ΔspsI mutant spores (Fig. S6D). Deletion of cgeD also fails to rescue the phenotype of ΔspsM (ΔypqP) mutant spores (Fig. S6E). Together, these data suggest that cgeD is also involved in shaping the PS layer, thus contributing to spore surface properties, but its role is different from the yfnH–D cluster since it is not acting antagonistically to the spsA–L operon.
FIG 6.
Analysis of spore surface properties in mutants with deletions in the cgeCDE operon. (A) BATH assays reveal a slight increase in hydrophobicity in ΔcgeD spores (PE2918) compared to wild-type spores (PY79), while ΔcgeC (PE2917) and ΔcgeE (PE3065) mutant spores are indistinguishable from the wild type. Experiments were performed in triplicate; error bars represent standard deviation. (B) By negative staining with India ink, ΔcgeD (NY227) mutant spores have a spread and increased width PS layer. The ΔcgeC (NY226) and ΔcgeE (NY228) mutants have no observable change to the PS layer. Scale bars = 2.5 μm. Measurements of halo areas for ΔcgeD (NY227) mutant spores are provided in Fig. S6A and additional images of spores in Fig. S6B. Analyses of spore surface extracts by gel electrophoresis can be found in Fig. S6C. Extracts from ΔcgeD (NY227) mutant spores show an increase in size presumably caused by an expansion in PS content; the size distribution for extracts from ΔcgeC (NY226) and ΔcgeE (NY228) mutant spores is similar to that observed for wild-type spores (168). (C) TEM with ruthenium red staining of ΔcgeD (PE2918) mutant spores reveals elongated crust filaments (red arrow), which can be attached or balloon away from a speckled outer coat (orange arrow). Scale bars = 200 nm.
DISCUSSION
In this study, we focused on the saccharide synthesis pathways active during spore formation in B. subtilis. The corresponding enzymes are encoded by four σK-dependent gene clusters, spsA–L, yfnHGF-yfnED, ytdA-ytcABC, and cgeCDE-cgeAB-phy-spsM (Fig. 1; also see Fig. S1 in the supplemental material). The spsA–L operon is the best characterized of the four, especially the spsIJKL cassette, which was previously shown to be involved in rhamnose synthesis (8). Our interest in yfnH and ytdA derived from the observation that these genes are paralogous to spsI, while cgeD is a paralog of spsA (and cgeE of spsD). The data we report here indicate that most of these previously uncharacterized gene regions play a role in spore surface properties. While we are still unsure of the exact molecular composition and structure of the final saccharide products, our data imply that the decoration of the spore by a PS layer significantly contributes to spore surface morphology and hydrophobicity. Based on the different phenotypes of mutants and combination of mutants uncovered by BATH assays, we believe that paralogous genes spsI, yfnH, and ytdA are likely all working toward the syntheses of their own respective monosaccharides and saccharide polymers (Fig. 1 and S1), although it is also possible that any one pathway could further modify the carbohydrates produced by a different pathway.
Deletion of the entire spsA–L operon, as well as individual disruption of the spsA–L genes, produces mature mutant spores that are all more hydrophobic than are wild-type spores, as assessed by BATH assays (Fig. 2A). While we know that spsIJKL is necessary for rhamnose synthesis, it is unclear how spsA–G contributes to monosaccharide and/or PS synthesis. The corresponding enzymes could act to further modify the composition of the PS or help their proper transfer to the crust or some other coat layer. The phenotype of spsI mutant spores is also quite characteristic when imaged by India ink and TEM with ruthenium red staining. Both methods show that the morphology of the outermost layer of the spore is severely impaired (lack of a halo by India ink, barely detectable crust by TEM [Fig. 2B and C]).
The experiments we conducted toward the characterization of the yfnH–D cluster revealed unexpected phenotypes, since mutants of either the whole cluster or individual genes displayed an opposite phenotype to the spsI mutant. Given that a BATH assay cannot uncover increases in spore hydrophilicity in comparison to wild-type spores, it did not give us clues to the contribution of yfnH–D genes to PS distribution on the crust (Fig. S4D). In contrast, yfnH mutant spores stained with India ink or ruthenium red in TEM both showed a previously unseen expansion of the PS layer. Spores with mutations in any gene in the yfnH–D cluster are surrounded by a larger halo, and a weblike expanded crust layer around the spore was observed by TEM (Fig. 4). Structurally, the PS expansion is very noticeable by TEM, but how each individual mutant achieved this phenotype is unclear. It could be that without this functional saccharide synthesis pathway, there is a build-up of the precursors to the final yfnH–D enzymatic PS output. Knowledge of the monosaccharides synthesized by the yfnH–D cluster will help to understand how the weblike phenotype is created.
A comparison of the data for the spsA–L mutant spores to the yfnH–D mutant spores suggests that the pathways of PS production by the two gene groups are somehow connected. To explore this possibility, we analyzed the phenotypes of spores carrying mutations in both gene clusters. In the BATH assays, a double mutant of yfnF and spsI (or of yfnH and spsI) shows a partial rescue of the hydrophobic phenotype (Fig. 5A). However, mutations in genes located further downstream (yfnD or yfnE) do not cause this partial rescue (Fig. S5). YfnF, YfnE, and YfnD are all annotated as putative glycosyltransferases, but the yfnH–D cluster is composed of two operons (yfnHGF and yfnED), each controlled by an individual promoter (43). It is thus possible that the functions of YfnE and YfnD are not specific to the output of the YfnHGF PS production pathway (Fig. 1). Furthermore, the promoter of the yfnED operon is also recognized by σE, implying that YfnE and YfnD are produced at an earlier stage in sporulation than all the other PS-synthesizing operons investigated here. In total, data collected for the double-mutation strains highlight the complicated interaction between the various carbohydrate synthesis pathways during sporulation. Future glycomics and biochemical analyses are required to identify the various products, the specific contributions of each enzyme, and the precise functions of the numerous transglycosylases in this process.
While we were unable to find a clear phenotypic contribution for the ytdA-ytcABC cluster (Fig. S3), future studies into these genes can help us see how they fit into the greater scheme of PS synthesis during sporulation. The precise role of the cge cluster also remains an open question (40). Previous research showed the importance of the proteins CgeA and CgeB to the structure of the spore crust (20, 22). By India ink staining, ΔcgeD mutant spores exhibit an expanded PS layer, but unlike other mutant spores with a similar phenotype, ΔcgeD mutant spores are not entirely hydrophilic by BATH, and inactivation of cgeD cannot rescue the spsI mutant (Fig. 6 and S6D) nor the spsM mutant (Fig. S6E). TEM images for ΔcgeD mutant spores show the presence of a filamentous crust, instead of the weblike structure seen with the yfnH mutant spores.
Taken together, our data indicate that at least four gene regions are integral to the proper synthesis and addition of PS to the spore surface in B. subtilis. The PS composition and organization on the outermost layer of the spore likely dictate how the spores can interact with specific environments. Furthermore, spore PS may determine recognition by other cell surfaces. Here, we have found that the previously uncharacterized yfnH–D gene cluster plays a key role in regulating proper surface PS production. Future research will be directed toward the identification of the product(s) of this enzymatic pathway. Biochemical analyses and glycomic studies will be important next steps to studying spore PS composition. While there are still many unknowns, it is evident that control of PS synthesis along with the selection of the structural proteins of the outermost layer of the spore are required to constitute a surface best adapted to the environment where spores reside. Modifications in either of the two main components of the crust (PS or proteins) can drastically alter the spore’s hydrophobicity and, therefore, its preference for aquatic or nonaquatic environments. Future work will help us understand how spores of different species evolved to adopt morphologies best suited to specific ecosystems and by which methods, if any, the cell can adjust spore surface properties to better survive a given environment.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table S1 in the supplemental material. The procedures for the deletion of the entire yfnH–D cluster (strain NY35) and the whole spsA–L operon (strain NY212) are detailed in Fig. S7A and B, respectively. Gene deletions in the cgeA–E cluster are described in Fig. S7C (strains NY226 and NY227) and Fig. S7D (strain NY228). The primers used are listed in Table S2.
Sporulation conditions for bacterial adhesion to hydrocarbon assays.
Strains were struck out onto LB plates from frozen glycerol stocks and incubated at 37°C overnight. Single colonies were then inoculated into 5 ml of Difco sporulation medium (DSM) with nutrients overnight at 37°C, and 200 μl of the liquid culture was plated onto DSM and left to sporulate for 2 days at 37°C. Spores were scraped off the agar plates and resuspended into 700 μl of cold distilled water (dH2O). Samples were washed 3 times in cold dH2O with each spin down at 16,100 relative centrifugal field for 10 min. The rest of the BATH procedure was identical to the protocol described in reference 22.
India ink staining.
Two microliters of a suspension of spores in dH2O was mixed with an equal amount of India ink (Daiso Soygo, Japan) and observed by phase-contrast microscopy, as described previously (23). The area of halo was measured using ImageJ (the bright spore area was subtracted) and plotted in a box-and-whisker plot.
Transmission electron microscopy.
Spores were fixed, stained with ruthenium red, and analyzed by thin-section TEM, as described before (19).
Scanning electron microscopy.
Spores were fixed with 1% paraformaldehyde and 2.5% glutaraldehyde in 0.05 M HEPES, adsorbed to carbon-coated and alcian blue-treated coverslips, postfixed with osmium tetroxide, dehydrated with ethanol, dried by critical-point drying, subjected to sputter coating with 3 nm gold-palladium, and examined in a field emission SEM (Leo 1530 Gemini; Carl Zeiss Microscopy, Germany) at 3 kV using the in-lens secondary electron detector.
Fluorescence microscopy.
Five milliliters of DSM was inoculated from a single colony and grown 18 h at 37°C. Five hundred microliters of sample from the sporulating culture was spun down and resuspended in 50 μl of 1× PBS, and membranes were stained with 1 μl of 1 μg/μl FM4-64. Images were collected as described before (44).
Supplementary Material
ACKNOWLEDGMENTS
This study is dedicated to the memory of coauthor Adam Driks. Adam passed away on 6 June 2019, after a long struggle with his health. He will be remembered fondly by everyone who had the privilege to interact with him. We are grateful for the opportunity to share his work one last time and add to the beautiful collection of spore images for which he was famous.
We thank Julia Bartels and Thorsten Mascher for sharing their results prior to publication. We are grateful to Felix Fuchs, Ralf Moeller, and Michael Laue for facilitating the SEM experiments. We thank Matthew Gallitto for his help in some of the pilot experiments.
This work was supported by funds from the Zegar Family Foundation and originally from NIH grant GM081571 to P.E. Additional support was provided by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (KAKENHI) (grant 15K18675 to K.A. and grant 15K07371 to T.S.).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00321-19.
REFERENCES
- 1.Power PM, Jennings MP. 2003. The genetics of glycosylation in Gram-negative bacteria. FEMS Microbiol Lett 218:211–222. doi: 10.1111/j.1574-6968.2003.tb11520.x. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt MA, Riley LW, Benz I. 2003. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol 11:554–561. doi: 10.1016/j.tim.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Tytgat HL, Lebeer S. 2014. The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol Mol Biol Rev 78:372–417. doi: 10.1128/MMBR.00007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tytgat HLP, de Vos WM. 2016. Sugar coating the envelope: glycoconjugates for microbe-host crosstalk. Trends Microbiol 24:853–861. doi: 10.1016/j.tim.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Eichler J, Koomey M. 2017. Sweet new roles for protein glycosylation in prokaryotes. Trends Microbiol 25:662–672. doi: 10.1016/j.tim.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Wunschel D, Fox KF, Black GE, Fox A. 1995. Discrimination among the B. cereus group, in comparison to B. subtilis, by structural carbohydrate profiles and ribosomal RNA spacer region PCR. System Appl Microbiol 17:625–635. doi: 10.1016/S0723-2020(11)80085-8. [DOI] [Google Scholar]
- 7.Waller LN, Fox N, Fox KF, Fox A, Price RL. 2004. Ruthenium red staining for ultrastructural visualization of a glycoprotein layer surrounding the spore of Bacillus anthracis and Bacillus subtilis. J Microbiol Methods 58:23–30. doi: 10.1016/j.mimet.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Plata G, Fuhrer T, Hsiao TL, Sauer U, Vitkup D. 2012. Global probabilistic annotation of metabolic networks enables enzyme discovery. Nat Chem Biol 8:848–854. doi: 10.1038/nchembio.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cangiano G, Sirec T, Panarella C, Isticato R, Baccigalupi L, De Felice M, Ricca E. 2014. The sps gene products affect the germination, hydrophobicity, and protein adsorption of Bacillus subtilis spores. Appl Environ Microbiol 80:7293–7302. doi: 10.1128/AEM.02893-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stragier P, Losick R. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30:297. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 11.Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 12.Higgins D, Dworkin J. 2012. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan IS, Ramamurthi KS. 2014. Spore formation in Bacillus subtilis. Environ Microbiol Rep 6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Driks A, Eichenberger P. 2016. The spore coat. Microbiol Spectr 4:TBS-0023-2016. doi: 10.1128/microbiolspec.TBS-0023-2016. [DOI] [PubMed] [Google Scholar]
- 15.McKenney PT, Driks A, Eichenberger P. 2013. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol 11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henriques AO, Moran CP Jr. 2007. Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol 61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 17.de Hoon MJ, Eichenberger P, Vitkup D. 2010. Hierarchical evolution of the bacterial sporulation network. Curr Biol 20:R735–R745. doi: 10.1016/j.cub.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imamura D, Kuwana R, Takamatsu H, Watabe K. 2010. Localization of proteins to different layers and regions of Bacillus subtilis spore coats. J Bacteriol 192:518–524. doi: 10.1128/JB.01103-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenney PT, Driks A, Eskandarian HA, Grabowski P, Guberman J, Wang KH, Gitai Z, Eichenberger P. 2010. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr Biol 20:934–938. doi: 10.1016/j.cub.2010.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamura D, Kuwana R, Takamatsu H, Watabe K. 2011. Proteins involved in formation of the outermost layer of Bacillus subtilis spores. J Bacteriol 193:4075–4080. doi: 10.1128/JB.05310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang S, Wan Q, Krajcikova D, Tang J, Tzokov SB, Barak I, Bullough PA. 2015. Diverse supramolecular structures formed by self-assembling proteins of the Bacillus subtilis spore coat. Mol Microbiol 97:347–359. doi: 10.1111/mmi.13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shuster B, Khemmani M, Abe K, Huang X, Nakaya Y, Maryn N, Buttar S, Gonzalez AN, Driks A, Sato T, Eichenberger P. 2019. Contributions of crust proteins to spore surface properties in Bacillus subtilis. Mol Microbiol 111:825–843. doi: 10.1111/mmi.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe K, Kawano Y, Iwamoto K, Arai K, Maruyama Y, Eichenberger P, Sato T. 2014. Developmentally-regulated excision of the SPbeta prophage reconstitutes a gene required for spore envelope maturation in Bacillus subtilis. PLoS Genet 10:e1004636. doi: 10.1371/journal.pgen.1004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boone TJ, Mallozzi M, Nelson A, Thompson B, Khemmani M, Lehmann D, Dunkle A, Hoeprich P, Rasley A, Stewart G, Driks A. 2018. Coordinated assembly of the Bacillus anthracis coat and exosporium during bacterial spore outer layer formation. mBio 9:e01166-18. doi: 10.1128/mBio.01166-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Driks A. 2002. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol 10:251–254. doi: 10.1016/S0966-842X(02)02373-9. [DOI] [PubMed] [Google Scholar]
- 26.Stewart GC. 2015. The exosporium layer of bacterial spores: a connection to the environment and the infected host. Microbiol Mol Biol Rev 79:437–457. doi: 10.1128/MMBR.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daubenspeck JM, Zeng H, Chen P, Dong S, Steichen CT, Krishna NR, Pritchard DG, Turnbough CL Jr. 2004. Novel oligosaccharide side chains of the collagen-like region of BclA, the major glycoprotein of the Bacillus anthracis exosporium. J Biol Chem 279:30945–30953. doi: 10.1074/jbc.M401613200. [DOI] [PubMed] [Google Scholar]
- 28.Faille C, Ronse A, Dewailly E, Slomianny C, Maes E, Krzewinski F, Guerardel Y. 2014. Presence and function of a thick mucous layer rich in polysaccharides around Bacillus subtilis spores. Biofouling 30:845–858. doi: 10.1080/08927014.2014.939073. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Mukherjee T, Bowler K, Namdari S, Snow Z, Prestridge S, Carlton A, Bar-Peled M. 2017. A four-gene operon in Bacillus cereus produces two rare spore-decorating sugars. J Biol Chem 292:7636–7650. doi: 10.1074/jbc.M117.777417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe K, Takamatsu T, Sato T. 2017. Mechanism of bacterial gene rearrangement: SprA-catalyzed precise DNA recombination and its directionality control by SprB ensure the gene rearrangement and stable expression of spsM during sporulation in Bacillus subtilis. Nucleic Acids Res 45:6669–6683. doi: 10.1093/nar/gkx466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arrieta-Ortiz ML, Hafemeister C, Bate AR, Chu T, Greenfield A, Shuster B, Barry SN, Gallitto M, Liu B, Kacmarczyk T, Santoriello F, Chen J, Rodrigues CD, Sato T, Rudner DZ, Driks A, Bonneau R, Eichenberger P. 2015. An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol Syst Biol 11:839. doi: 10.15252/msb.20156236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu HW, Thorson JS. 1994. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu Rev Microbiol 48:223–256. doi: 10.1146/annurev.mi.48.100194.001255. [DOI] [PubMed] [Google Scholar]
- 33.Joshi LT, Phillips DS, Williams CF, Alyousef A, Baillie L. 2012. Contribution of spores to the ability of Clostridium difficile to adhere to surfaces. Appl Environ Microbiol 78:7671–7679. doi: 10.1128/AEM.01862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ankolekar C, Labbe RG. 2010. Physical characteristics of spores of food-associated isolates of the Bacillus cereus group. Appl Environ Microbiol 76:982–984. doi: 10.1128/AEM.02116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koshikawa T, Yamazaki M, Yoshimi M, Ogawa S, Yamada A, Watabe K, Torii M. 1989. Surface hydrophobicity of spores of Bacillus spp. J Gen Microbiol 135:2717–2722. doi: 10.1099/00221287-135-10-2717. [DOI] [PubMed] [Google Scholar]
- 36.Rönner U, Husmark U, Henriksson A. 1990. Adhesion of bacillus spores in relation to hydrophobicity. J Appl Bacteriol 69:550–556. doi: 10.1111/j.1365-2672.1990.tb01547.x. [DOI] [PubMed] [Google Scholar]
- 37.Wiencek KM, Klapes NA, Foegeding PM. 1990. Hydrophobicity of Bacillus and Clostridium spores. Appl Environ Microbiol 56:2600–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faille C, Lequette Y, Ronse A, Slomianny C, Garenaux E, Guerardel Y. 2010. Morphology and physico-chemical properties of Bacillus spores surrounded or not with an exosporium: consequences on their ability to adhere to stainless steel. Int J Food Microbiol 143:125–135. doi: 10.1016/j.ijfoodmicro.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 39.Williams G, Linley E, Nicholas R, Baillie L. 2013. The role of the exosporium in the environmental distribution of anthrax. J Appl Microbiol 114:396–403. doi: 10.1111/jam.12034. [DOI] [PubMed] [Google Scholar]
- 40.Roels S, Losick R. 1995. Adjacent and divergently oriented operons under the control of the sporulation regulatory protein GerE in Bacillus subtilis. J Bacteriol 177:6263–6275. doi: 10.1128/jb.177.21.6263-6275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charnock SJ, Davies GJ. 1999. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38:6380–6385. doi: 10.1021/bi990270y. [DOI] [PubMed] [Google Scholar]
- 42.Bozue JA, Parthasarathy N, Phillips LR, Cote CK, Fellows PF, Mendelson I, Shafferman A, Friedlander AM. 2005. Construction of a rhamnose mutation in Bacillus anthracis affects adherence to macrophages but not virulence in guinea pigs. Microb Pathog 38:1–12. doi: 10.1016/j.micpath.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol 2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenney PT, Eichenberger P. 2012. Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol Microbiol 83:245–260. doi: 10.1111/j.1365-2958.2011.07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, Gonzalez-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, Losick R. 2003. The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol 327:945–972. doi: 10.1016/S0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.