Different bacterial strains compete to occupy the same niche. The outcome of such competition can be affected by the type VI secretion system (T6SS), an intercellular killing mechanism of bacteria. Here an animal-bacterial symbiosis is used as a platform for study of the genetic factors that promote the T6SS-mediated killing of one strain by another. Identification of the molecular determinants of T6SS function in vivo contributes to the understanding of how different strains interact within a host.
KEYWORDS: Vibrio fischeri, cell-cell interaction, invertebrate-microbe interactions, secretion systems, symbiosis
ABSTRACT
Bacteria that have the capacity to fill the same niche will compete with one another for the space and resources available within an ecosystem. Such competition is heightened among different strains of the same bacterial species. Nevertheless, different strains often inhabit the same host. The molecular mechanisms that impact competition between different strains within the same host are poorly understood. To address this knowledge gap, the type VI secretion system (T6SS), which is a mechanism for bacteria to kill neighboring cells, was examined in the marine bacterium Vibrio fischeri. Different strains of V. fischeri naturally colonize the light organ of the bobtail squid Euprymna scolopes. The genome of FQ-A001, a T6SS-positive strain, features two hcp genes that are predicted to encode identical subunits of the T6SS. Coincubation assays showed that either hcp gene is sufficient for FQ-A001 to kill another strain via the T6SS in vitro. Additionally, induction of hcp expression is sufficient to induce killing activity in an FQ-A001 mutant lacking both hcp genes. Squid colonization assays involving inocula of FQ-A001-derived strains mixed with ES114 revealed that both hcp genes must be deleted for FQ-A001 and ES114 to occupy the same space within the light organ. These experimental results provide insight into the genetic factors necessary for the T6SS of V. fischeri to function in vivo, thereby increasing understanding of the molecular mechanisms that impact strain diversity within a host.
IMPORTANCE Different bacterial strains compete to occupy the same niche. The outcome of such competition can be affected by the type VI secretion system (T6SS), an intercellular killing mechanism of bacteria. Here an animal-bacterial symbiosis is used as a platform for study of the genetic factors that promote the T6SS-mediated killing of one strain by another. Identification of the molecular determinants of T6SS function in vivo contributes to the understanding of how different strains interact within a host.
INTRODUCTION
Epithelial surfaces are typically colonized by microbes that impact the physiology of their hosts (1). The variability in physical and chemical composition associated with these surfaces results in a vast array of habitats that can support microbial growth, which has contributed to the tremendous genetic diversity of animal microbiomes (2, 3). Such habitats select for specific microbial species that can grow under the prevailing conditions (3–5). However, microbes also exhibit genetic diversity at the level of individual strains, and the molecular mechanisms that promote occupancy by certain strains remain poorly understood (6, 7). In theory, competition for resources between two strains within the same ecological niche will result in competitive exclusion, i.e., the extinction of the strain with lower fitness (8). However, alternative outcomes to competitive exclusion are possible. For example, new adaptations in one strain that redefine its niche could promote cooccupancy of the habitat (9). Conversely, intercellular interactions between different strain types could promote the occupancy of a habitat by a single strain even if that strain has lower fitness than the other (10). Bacteria exhibit various mechanisms that impact the outcome of intercellular interactions, including systems that can kill neighboring cells (11). Determining the extent to which contact-dependent killing mechanisms function in vivo is important for understanding how specific strains become established within the microbial ecosystems associated with epithelial surfaces.
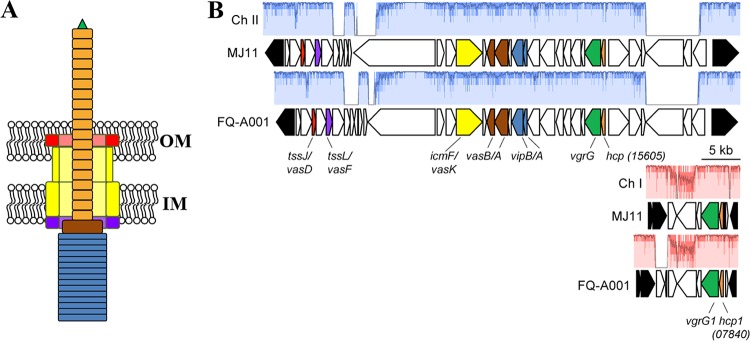
One contact-dependent killing mechanism is mediated by the type VI secretion system (T6SS), which features a secretion apparatus that is structurally homologous to the contractile tail of a bacteriophage (12). The baseplate of the T6SS anchors the secretion system to the inner membrane, thereby promoting the assembly of a complex within the cytoplasm containing a tube with a spiked tip that is housed within an outer tube (13, 14). The spiked tip is formed by a trimer of VgrG (15), to which other proteins are attached. The inner tube consists of stacked rings, with each ring containing a hexamer of Hcp, resulting in a hollow interior that is hypothesized to facilitate the translocation of macromolecules (13, 16). The inner tube and spiked tip also function as a needle: upon contraction of the outer tube, the inner tube is fired through the cell wall (17) (Fig. 1A), resulting in the translocation of Hcp, VgrG, and any other proteins associated with the inner tube to the exterior of the cell (13). If another cell is in close proximity, the needle can puncture its exterior to deliver effectors that kill the neighboring cell (12). In this manner, T6SSs can introduce antagonism into the cellular interactions that take place within a microbial community, which has the potential to alter community composition and, consequently, the way in which the community functions within the ecosystem. For example, T6SS-mediated antagonism enables certain Bacteroides spp. to protect their niche within the polymicrobial environment of the mammalian gastrointestinal tract (18).
FIG 1.
Genetic model for the T6SS in FQ-A001. (A) Cartoon of the T6SS in V. fischeri based on the homologous system in V. cholerae. The contracted state of the needle is shown. OM, outer membrane; IM, inner membrane. (B) Main and auxiliary gene clusters of the T6SS in the FQ-A001 genome. Several genes are colored in accordance with the model shown in panel A. Genes adjoining one another in ES114 are shown in black. Similarity was determined by progressiveMauve alignment; regions without color (red for chromosome [Ch] I; blue for Ch II) represent loci at which there is not syntenic DNA in the other strain. The hcp ORF on FQ-A001 chromosome II (VFFQA001_15605) is syntenic with MJ11 VFMJ11_A0831; the hcp1 ORF on FQ-A001 chromosome I (VFFQA001_07840) is syntenic with MJ11 VFMJ11_1495.
T6SSs are ubiquitous; roughly 25% of all bacterial genomes are predicted to contain T6SS-related genes (19). A functional T6SS gene cluster was recently identified in certain strains of Vibrio fischeri (20), a marine bacterium that establishes a symbiotic association with the Hawaiian bobtail squid Euprymna scolopes (21). This symbiosis is based on the production of bioluminescence by populations of V. fischeri that are housed within epithelium-lined crypt spaces in the host’s light organ (22, 23). The symbiosis is established shortly after the animals hatch from their eggs, when the juvenile squid are exposed to seawater containing various strains of V. fischeri (24). To establish symbiosis, typically 1 or 2 V. fischeri cells initially colonize each crypt space, and the subsequent period of growth results in the light-emitting populations (25). Thus, the initial colonization of the light organ presents an opportunity for different strains to interact physically. In addition, no other microbial species is able to occupy the crypt spaces (22). This scenario provides a simplified platform for studying intraspecies interactions in vivo.
Previous work has shown that two symbiotic strains of V. fischeri, ES114 and FQ-A001, fail to coexist within the same crypt space (26). This incompatibility in vivo depends on the T6SS of FQ-A001, which can kill ES114 in vitro (20). Here we report the identification of two putative hcp genes in FQ-A001 that are necessary for T6SS-dependent activities, including the prevention of other strains of V. fischeri from colonizing the same crypt space. These findings contribute to knowledge of how the T6SS functions in V. fischeri during symbiosis establishment, providing insight into the molecular mechanisms that impact bacterial interactions in vivo.
RESULTS
The large T6SS gene cluster of FQ-A001 features an hcp gene.
FQ-A001 is a V. fischeri strain that was isolated from the light organ of an adult E. scolopes squid and exhibits T6SS-dependent killing activity toward other symbiotic strains of V. fischeri (20, 26). To gain more insight into this system at a molecular level, the genome of FQ-A001 was sequenced. The de novo draft genome (NCBI genome accession number SJSX00000000) comprises three contigs with synteny to MJ11 chromosome I and one contig with synteny to MJ11 chromosome II (27).
The killing activity of FQ-A001 toward ES114 in vitro depends on the vasA_2 gene (20), which is predicted to encode a baseplate component of the T6SS (Fig. 1A). The vasA_2 gene was identified on the second largest contig (FQ-A001_contig4) generated during genome assembly. This contig resembles the second chromosome of MJ11 and contains a cluster of 35 open reading frames (ORFs) predicted to encode factors that assemble a functional T6SS, including vasA_2 (Fig. 1B and Table 1). Of relevance to this study was the identification of a gene within this cluster that is predicted to encode Hcp (locus tag VFFQA001_15605) (Fig. 1B; see also Fig. S1 in the supplemental material). In the T6SSs of other microbes, Hcp both comprises the inner tube of the apparatus and is a member of the T6SS secretome (28). Therefore, the hcp gene represented an ideal target for investigating the molecular basis of the T6SS in FQ-A001.
TABLE 1.
Protein identities for FQ-A001 T6SS loci
| Chr.a | Locus |
% aa identity (no. of identical aa/total no.)b | Genec | Predicted function in FQ-A001 | |
|---|---|---|---|---|---|
| FQ-A001 | MJ11 | ||||
| II | VFFQA001_15465 | VFMJ11_A0804 | 99 (198/200) | Hypothetical protein | |
| II | VFFQA001_15470 | VFMJ11_A0805 | 86 (419/486) | tagH | Type VI secretion system-associated FHA domain protein TagH |
| II | VFFQA001_15475 | VFMJ11_A0806 | 100 (166/166) | tssJ (vasD) | Type VI secretion system lipoprotein TssJ |
| II | VFFQA001_15480 | VFMJ11_A0807 | 99 (439/441) | tssK | Type VI secretion system baseplate subunit TssK |
| II | VFFQA001_15485 | VFMJ11_A0808 | 99 (262/263) | tssL (vasF) | DotU family type IV/VI secretion system protein |
| II | VFFQA001_15490 | VFMJ11_A0809 | 95 (477/504) | DUF4150 domain-containing protein | |
| II | VFFQA001_15495 | VFMJ11_A0810 | 21 (52/253) | Hypothetical protein | |
| II | VFFQA001_15500 | No hits on chromosome II | Hypothetical protein | ||
| II | VFFQA001_15505 | No hits on chromosome II | Hypothetical protein | ||
| II | VFFQA001_15510 | VFMJ11_A0812 | 91 (148/163) | Hypothetical protein | |
| II | VFFQA001_15515 | VFMJ11_A0813 | 98 (139/142) | Hypothetical protein | |
| II | VFFQA001_15520 | VFMJ11_A0814 | 88 (2,560/2,912) | Hypothetical protein | |
| II | VFFQA001_15525 | VFMJ11_A0815 | 96 (298/309) | Hypothetical protein | |
| II | VFFQA001_15530 | VFMJ11_A0816 | 93 (403/435) | Hypothetical protein | |
| II | VFFQA001_15535 | VFMJ11_A0817 | 99 (1,116/1,132) | tssM | Type VI secretion system membrane subunit TssM |
| II | VFFQA001_15540 | VFMJ11_A0818 | 100 (149/149) | Lrp/AsnC family transcriptional regulator | |
| II | VFFQA001_15545 | VFMJ11_A0819 | 98 (325/332) | tssG | Type VI secretion system baseplate subunit TssG |
| II | VFFQA001_15550 | VFMJ11_A0820 | 99 (575/582) | vasB (tssF) | Type VI secretion system baseplate subunit TssF |
| II | VFFQA001_15555 | VFMJ11_A0821 | 99 (134/136) | vasA_2 (tssE) | Type VI secretion system baseplate subunit TssE |
| II | VFFQA001_15560 | VFMJ11_A0822 | 99 (490/492) | tssC | Type VI secretion system contractile sheath large subunit |
| II | VFFQA001_15565 | VFMJ11_A0823 | 100 (169/169) | vipB (tssB) | Type VI secretion system contractile sheath small subunit |
| II | VFFQA001_15570 | VFMJ11_A0824 | 92 (478/519) | vipA (tssA) | Type VI secretion system protein TssA |
| II | VFFQA001_15575 | VFMJ11_A0825 | 98 (645/661) | pknA | Protein kinase family protein |
| II | VFFQA001_15580 | VFMJ11_A0826 | 98 (303/313) | Hypothetical protein | |
| II | VFFQA001_15585 | VFMJ11_A0827 | 99 (321/325) | DUF2169 domain-containing protein | |
| II | VFFQA001_15590 | VFMJ11_A0828 | 97 (428/441) | Hypothetical protein | |
| II | VFFQA001_15595 | VFMJ11_A0829 | 99 (162/164) | Hypothetical protein | |
| II | VFFQA001_15600 | VFMJ11_A0830 | 99 (688/692) | vrgG (tssI) | Type VI secretion system tip protein VgrG |
| II | VFFQA001_15605 | VFMJ11_A0831 | 98 (168/172) | hcp | Hcp family type VI secretion system effector |
| II | VFFQA001_15610 | VFMJ11_A0832 | 99 (882/888) | tssH (clpV) | Type VI secretion system ATPase TssH |
| II | VFFQA001_15615 | VFMJ11_A0833 | 95 (509/534) | vasH | Sigma 54-dependent Fis family transcriptional regulator |
| II | VFFQA001_15620 | VFMJ11_A0834 | 97 (161/166) | Hypothetical protein | |
| II | VFFQA001_15625 | VFMJ11_A0835 | 27 (431/1,615) | OmpA family protein | |
| II | VFFQA001_15630 | VFMJ11_A0842 | 37 (136/364) | WD40 repeat domain-containing protein | |
| II | VFFQA001_15635 | VFMJ11_A0837 | 45 (252/564) | Hypothetical protein | |
| I | VFFQA001_07810 | No hits on chromosome I | Pseudogene; frameshifted in FQ-A001 | ||
| I | VFFQA001_07815 | *d | * | Hypothetical protein | |
| I | VFFQA001_07820 | VFMJ11_1491 | 74 (264/359) | Hypothetical protein | |
| I | VFFQA001_07825 | VFMJ11_1492 | 52 (426/817) | LysM peptidoglycan-binding domain-containing protein | |
| I | VFFQA001_07830 | VFMJ11_1493 | 95 (187/197) | DUF4123 domain-containing protein | |
| I | VFFQA001_07835 | VFMJ11_1494 | 98 (741/754) | vrgG1 (tssI1) | Type VI secretion system tip protein VgrG |
| I | VFFQA001_07840 | VFMJ11_1495 | 100 (172/172) | hcp1 | Hcp family type VI secretion system effector |
| I | VFFQA001_07845 | VFMJ11_1497 | 100 (110/110) | Zinc chelation protein SecC | |
Chr., chromosome.
aa, amino acid.
Gene names in parentheses are alternative gene names that have been reported.
*, the top hit for VFFQA001_07815 (59 aa) was VFMJ11_1492 (844 aa).
Symbiosis establishment by FQ-A001 is independent of hcp.
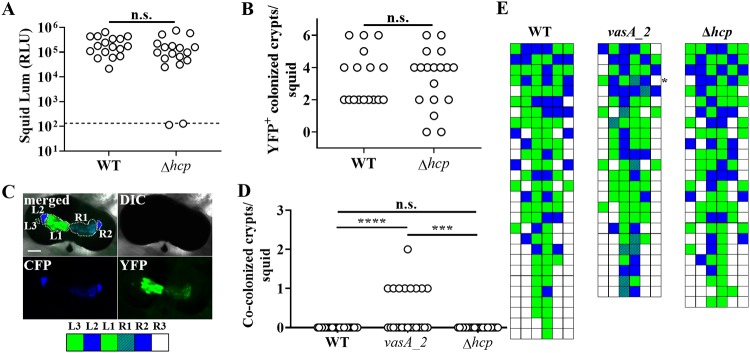
The inability of FQ-A001 and ES114 cells to occupy the same crypt space depends on the T6SS of FQ-A001 (20). To begin investigating the impact of Hcp on the T6SS-mediated activities of FQ-A001 in vivo, we constructed a mutant of FQ-A001 containing an in-frame deletion allele of the hcp gene within the main T6SS gene cluster, resulting in genotype FQ-A001 Δhcp. To assess the impact of this gene on symbiosis establishment, the Δhcp mutant was used in squid colonization assays. The majority (16/18) of juvenile squid exposed to an inoculum with the Δhcp mutant were luminescent by 48 h (Fig. 2A), and the luminescence levels were comparable to those of animals exposed to FQ-A001. In addition, the numbers of colonized crypt spaces within squid were comparable between animal groups (Fig. 2B), suggesting that the ability of the mutant to colonize the host was unimpaired. Together, these results for animals exhibiting luminescence and possessing crypt spaces colonized by bacteria suggest that symbiosis establishment by FQ-A001 is independent of the hcp gene.
FIG 2.
Impact of hcp on symbiosis establishment by FQ-A001. (A) Luminescence (Lum) of animals at 48 h after initial exposure to an inoculum containing either FQ-A001 (WT) or TIM416 (Δhcp) harboring the YFP expression plasmid pSCV38. The dashed line indicates the 95% tail of luminescence associated with animals within an aposymbiotic group, above which animals are scored as luminescent. Seventeen or 18 animals were used in each group. A Mann-Whitney test determined that the medians for luminescent animals within each group are not significantly different from those for other groups (α = 0.05). n.s., not significant (P > 0.05). The experiment was performed twice, with similar results. RLU, relative light units. (B) Number of crypt spaces per animal in panel A exhibiting YFP fluorescence. A Mann-Whitney test determined that the median for animals within each group was not significantly different from those for other groups (α = 0.05). (C) (Top) Images of a light organ containing fluorescently labeled strains of V. fischeri. In the merged image, crypt spaces that harbor V. fischeri populations are outlined by dotted lines and labeled according to crypt type. (Bottom) Strategy for visualizing the colonization state of the light organ by scoring crypt spaces according to strain type (white, empty; blue, CFP; green, YFP; striped pattern, CFP plus YFP). (D) Number of crypts exhibiting CFP and YFP fluorescence per squid. Squid were exposed to an inoculum in which CFP-labeled ES114 and the indicated FQ-A001-derived strain labeled with YFP (WT, FQ-A001; vasA_2, ANS2098; Δhcp, TIM416) were mixed evenly. A Kruskal-Wallis test determined significant differences among groups (P < 0.0001; H = 22.18), and Dunn’s multiple-comparison test was performed to test for significant differences between specific groups (****, P < 0.0001; ***, P < 0.001). The experiment was performed twice, with similar results. (E) Colonization states of light organs of the squid for which data are shown in panel D. Each row represents an individual animal. The asterisk indicates the light organ imaged in panel C.
To determine whether hcp affects strain compatibility in vivo, squid cocolonization assays were performed using mixed inocula of ES114 and FQ-A001-derived strains. To distinguish between strain types in vivo, ES114 was labeled with cyan fluorescent protein (CFP) and FQ-A001-derived strains were labeled with yellow fluorescent protein (YFP) (Fig. 2C). As a negative control for a functional T6SS, squid colonization assays were performed using FQ-A001 vasA_2, a mutant that does not exhibit strain incompatibility in vivo (20). None of the squid (n = 28) exposed to the mixed inoculum containing ES114 and wild-type (WT) FQ-A001 exhibited crypts colonized by both strain types (Fig. 2D and E). In contrast, 9/24 squid exposed to a mixed inoculum of ES114 and the vasA_2 mutant had at least one crypt space cocolonized by both strains (Fig. 2D and E). These results are consistent with previous findings suggesting that a T6SS is necessary for ES114 and FQ-A001 incompatibility in vivo (20). However, none of the 25 squid exposed to the mixed inoculum of ES114 and the Δhcp mutant harbored any cocolonized crypt spaces (Fig. 2D and E), suggesting that the T6SS of FQ-A001 functions in vivo without the hcp gene product.
Assessment of impact of the hcp gene on interactions between FQ-A001 and ES114 in vitro.
The strain incompatibility observed between ES114 and the Δhcp mutant in vivo was unexpected, because disruption of a different T6SS structural gene within the main T6SS gene cluster, i.e., vasA_2, was sufficient to permit occupancy of the same crypt space as ES114 (20). To determine whether other T6SS-dependent phenotypes are affected in the Δhcp mutant, we turned to in vitro coincubation assays, which can be used to qualitatively assess the ability of one strain to inhibit the growth of another (20). Briefly, cultures of two strain types differentially labeled with CFP and YFP are combined in equal amounts and are then introduced onto the surface of a solid medium as a concentrated spot. Following 24 h of incubation, the patterns of CFP and YFP fluorescence of each spot are acquired in order to determine the extent to which either strain type grew.
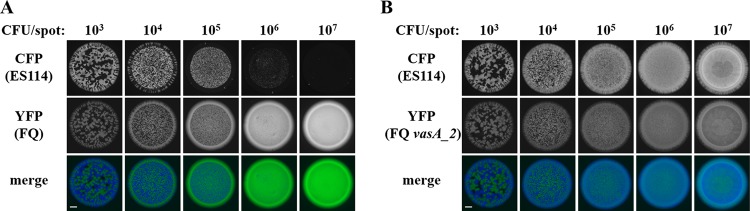
Regardless of which strain types were used in the assay, when ∼103 total CFU were spotted, the resulting fluorescence patterns within the spot revealed some regions without growth and other regions containing either CFP or YFP (Fig. 3), suggesting that the cells that had been within the spot when the assay was initiated were spatially separated and grew by clonal expansion into structures resembling adjacent CFU. Increasing the initial total CFU up to ∼105 resulted in the loss of regions without growth and the presence of smaller regions of CFP and YFP fluorescence (Fig. 3), findings consistent with the notion that the cells initially within the spot were closer to one another. However, as the total initial CFU increased to ∼106 or 107 for spots initiated with ES114 and FQ-A001, the resulting spot featured only YFP fluorescence (Fig. 3A), suggesting that the high initial cell density led to inhibition of ES114 growth. In contrast, spots that contained the vasA_2 mutant and were initiated with high cell densities resulted in growth exhibiting YFP and CFP fluorescence signals that colocalized throughout (Fig. 3B). This result suggests that the T6SS of FQ-A001 inhibits the growth of ES114, and without the functional T6SS, colocalization of the two strains could be observed. This assay was then used to assess the impact of hcp on the growth-inhibitory effect of FQ-A001 on ES114 observed with a high initial cell density. Coincubation of ES114 and the FQ-A001 Δhcp mutant resulted in a spot exhibiting only YFP fluorescence (Fig. 4A), suggesting that the growth of ES114 was inhibited and that the FQ-A001 T6SS is still functional without the hcp gene.
FIG 3.
Impact of initial cell density on the growth of ES114 during coincubation with FQ-A001. Shown are images of CFP fluorescence (top) or YFP fluorescence (center) and merged images (bottom) of spots derived from 10-μl cell suspensions grown on LBS agar for 24 h. ES114 harboring pYS112 (CFP) was mixed with the indicated strain harboring pSCV38 (YFP), and the mixture was serially diluted using 10-fold dilution steps. The YFP-labeled strains were FQ-A001 (FQ) (A) and ANS2098 (FQ vasA_2) (B). Bars, 1 mm.
FIG 4.
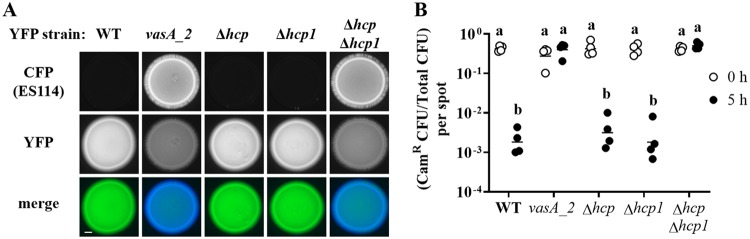
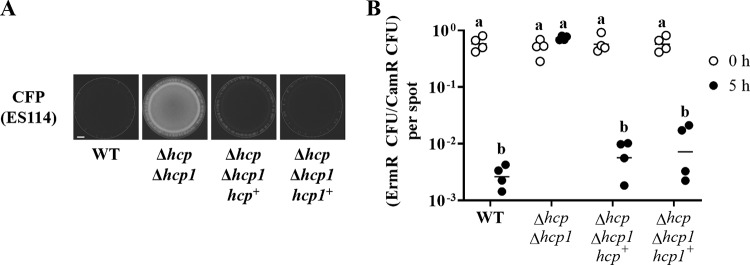
Impact of hcp genes on the inhibition of ES114 growth by FQ-A001. (A) Coincubation assay with ES114 and the indicated FQ-A001-derived strains, harboring pYS112 and pSCV38, respectively. FQ-A001-derived strains are FQ-A001 (WT), ANS2098 (vasA_2), TIM416 (Δhcp), NPW57 (Δhcp1), and NPW58 (Δhcp Δhcp1). Shown are images of CFP fluorescence (top) or YFP fluorescence (center) and merged images of the bacterial growth resulting from the 24-h incubation of cellular mixtures spotted onto agar (bottom). Approximately 5 × 107 total CFU were introduced into each spot. Bar, 1 mm. (B) Ratio of Camr CFU to total CFU resulting from a 5-h coincubation of Camr-labeled ES114 with each of the indicated FQ-A001-derived strains. Each point represents the ratio associated with an individual spot at 0 h (open symbols) or 5 h (filled symbols), and each horizontal line represents the geometric mean (n = 4). Approximately 5 × 107 total CFU were introduced into each spot. Two-way analysis of variance revealed significant differences among the means of log-transformed ratios due to time (F1,30 = 54.78; P < 0.0001), genotype (F4,30 = 47.20; P < 0.0001), and their interaction (F4,30 = 54.78; P < 0.0001). A Sidak post hoc test was performed to compare the means statistically, with P values adjusted for multiple comparisons. Groups with different letters have significantly different medians (P < 0.0001), and groups with the same letter do not have significantly different medians (P ≥ 0.05).
To quantify the impact of FQ-A001-derived strains on ES114 during coincubation, the assay described above was modified to determine the abundances of the strains within each spot. To generate each spot, a strain of ES114 harboring a plasmid with chloramphenicol resistance (Camr) was combined with a FQ-A001-derived strain that was unlabeled, i.e., chloramphenicol sensitive. At 0 and 5 h, the spots were harvested to determine total and Camr CFU counts (see Fig. S2 in the supplemental material), which were used to calculate the proportion of ES114 cells within each spot (Fig. 4B). After 5 h, the total CFU associated with each spot increased (Fig. S2A), indicating that growth had occurred. For the ES114–FQ-A001 spots, the proportion of Camr CFU decreased 230-fold (Fig. 4B). In addition, the abundance of ES114 had decreased within these spots (Fig. S2B), suggesting that ES114 cells died during the coincubation. When grown separately on a solid medium, ES114 and FQ-A001 exhibit generation times of 71 min and 66 min, respectively (data not shown), suggesting that differences in growth rates cannot account for the effects observed from their coincubation. For the spots comprising ES114 and the vasA_2 mutant, the proportion of Camr CFU remained constant during growth (Fig. 4B), suggesting that ES114 is killed via the T6SS of FQ-A001. When the Δhcp mutant was used in the coincubation assay, the spots showed a decrease in the proportion of Camr CFU similar to that observed with the FQ-A001 control (Fig. 4B), suggesting that the T6SS of FQ-A001 can kill ES114 without the hcp gene.
Either hcp gene is sufficient for T6SS-mediated killing.
The T6SS of Vibrio cholerae depends on two hcp genes that encode nearly identical Hcp subunits (13, 29). These hcp genes are not genetically linked to the main T6SS gene cluster; instead, two distinct auxiliary gene clusters each contain one of the hcp genes. Examination of the largest contig assembled during the sequencing of the FQ-A001 genome (FQ-A001-contig2) revealed another hcp gene (hcp1; locus tag VFFQA001_07840 [Fig. 1]). Notably, hcp1 is predicted to encode an Hcp subunit that is 100% identical to the Hcp subunit encoded by the hcp gene within the main cluster (Fig. S1 in the supplemental material). In MJ11, the homologue of hcp1, VFMJ11_1495, resides on chromosome I. The Hcp subunits encoded by VFMJ11_1495 and VFMJ11_A0831 (the hcp gene located within the large gene cluster of MJ11) share 100% and 98% identity, respectively, with the Hcp homologues in FQ-A001.
To determine the impact of hcp1 on the function of the T6SS in FQ-A001, an in-frame deletion allele of hcp1 was constructed and then introduced into FQ-A001 and the Δhcp mutant to generate the Δhcp1 and Δhcp Δhcp1 mutants, respectively. The growth of ES114 was inhibited when it was coincubated with the Δhcp1 mutant (Fig. 4A), suggesting that the T6SS is functional in this strain. However, when coincubated with the Δhcp Δhcp1 mutant, ES114 could grow to a level comparable to that observed with the vasA_2 mutant (Fig. 4A), suggesting that the Hcp proteins encoded in the main and auxiliary T6SS gene clusters contribute to the growth inhibition function of the T6SS in FQ-A001. Determination of CFU abundance after 5 h of coincubation revealed that the proportion of ES114 remained constant when it was exposed to the Δhcp Δhcp1 mutant (Fig. 4B), suggesting that the hcp genes are functionally redundant and that either is sufficient for the killing function of the T6SS in FQ-A001. The generation time of the Δhcp Δhcp1 mutant when grown alone was determined to be 75 min, comparable to that of the WT.
We also tested for genetic complementation by introducing a plasmid containing either hcp gene with the corresponding promoter region into the Δhcp Δhcp1 mutant. During 24-h coincubation assays, the growth of ES114 was impaired when the Δhcp Δhcp1 mutant harboring either hcp gene was used (Fig. 5A). The impact of expressing either hcp gene in trans on the killing function of the T6SS was assessed by the 5-h coincubation assay. Because the plasmids containing the hcp genes require chloramphenicol for maintenance, chloramphenicol selection could not be used to determine the abundance of the ES114 strain within the spots. Therefore, the assay was modified to use TIM313, an erythromycin-resistant (Ermr) derivative of ES114 that can be enumerated using erythromycin selection in place of ES114. As observed in the coincubation assays described above, the proportion of Camr CFU that were Ermr was reduced by 5 h in spots initiated with TIM313 and FQ-A001 (Fig. 5B). Proportions similar to that with this positive control were observed with the Δhcp Δhcp1 mutant harboring either an hcp+ or an hcp1+ plasmid (Fig. 5B). Together, these results suggest that either hcp gene can promote the T6SS-mediated activities of FQ-A001 in vitro.
FIG 5.
Impact of expression of hcp genes in trans on T6SS function in vitro. (A) Coincubation assay with ES114 and the indicated FQ-A001-derived strains. ES114 is labeled with CFP via pYS112. The FQ-A001-derived strains are FQ-A001 (WT) and NPW58 (Δhcp Δhcp1), which harbors the vector control pVSV105 (unlabeled), pNPW16 (hcp1+), or pNPW17 (hcp+). Images show the CFP fluorescence of the bacterial growth resulting from the 24-h incubation of cellular mixtures spotted onto agar. Approximately 5 × 107 total CFU were introduced into each spot. Dotted lines indicate the border of bacterial growth within each spot. Bar, 1 mm. (B) Ratio of Ermr CFU to Camr CFU resulting from a 5-h coincubation of the Ermr ES114-derived strain TIM313 labeled with Camr by plasmid pYS112 with the FQ-A001-derived strains described in the legend to panel A. Each point represents the ratio associated with an individual spot at 0 h (open symbols) or 5 h (filled symbols), and each horizontal line represents the geometric mean (n = 4). Approximately 5 × 107 total CFU were introduced into each spot. Two-way analysis of variance revealed significant differences among the means of log-transformed ratios due to time (F1,24 = 299.5; P < 0.0001), genotype (F3,24 = 37.46; P < 0.0001), and their interaction (F3,24 = 42.95; P < 0.0001). A Sidak post hoc test was performed to compare the means statistically, with P values adjusted for multiple comparisons. Groups with different letters have significantly different medians (P < 0.0001), and groups with the same letter do not have significantly different medians (P ≥ 0.05).
Crypt incompatibility requires at least one hcp gene in FQ-A001.
The results described above suggest that at least one hcp gene is necessary for T6SS-mediated activities in vitro. To test whether the function of the T6SS in vivo has a similar dependency, the Δhcp1 and Δhcp Δhcp1 mutants were used in squid colonization assays. Examination of the luminescence and crypt occupancy of squid exposed to either mutant revealed no effect (see Fig.S3 and S4 in the supplemental material), suggesting that the ability of FQ-A001 to establish symbiosis is independent of Hcp. Cocolonization assays using an inoculum in which ES114 and the Δhcp1 mutant were mixed evenly yielded animals containing crypt spaces with only one strain type (Fig. S3C), suggesting that the Δhcp1 mutant could prevent ES114 from occupying the same crypt space. However, exposure of squid to an inoculum in which ES114 and the Δhcp Δhcp1 mutant were mixed evenly yielded animals containing some crypt spaces colonized with both strain types (Fig. 6), suggesting that the ability of FQ-A001 to prevent ES114 from occupying the same crypt space becomes impaired without Hcp production.
FIG 6.
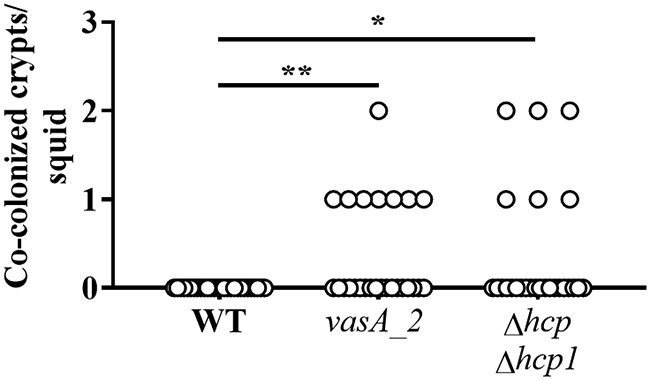
Impact of hcp and hcp1 on crypt cooccupancy. Shown are the numbers of crypts per squid that exhibit both CFP and YFP fluorescence. Squid were exposed to an inoculum in which CFP-labeled ES114 and the indicated FQ-A001-derived strain labeled with YFP (WT, FQ-A001; vasA2, ANS2098; Δhcp Δhcp1, NPW58) were mixed evenly. Twenty-six to 28 animals were used in each group. A Kruskal-Wallis test determined significant differences among group medians (P = 0.0097; H = 9.269). Dunn’s post hoc test was performed for statistical comparison of the median of each group to that of the WT group, with P values adjusted for multiple comparisons (**, P < 0.01; *, P < 0.05).
Induction of hcp expression is sufficient for T6SS-mediated activities.
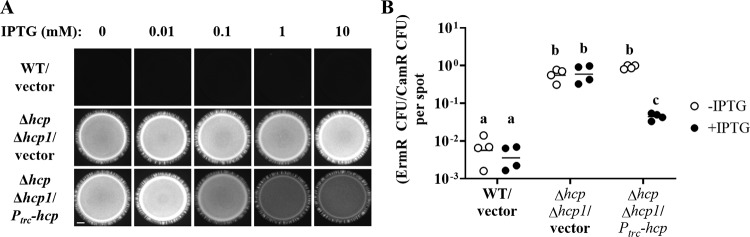
The results described above show that the expression of either hcp gene from its native promoter is sufficient to restore normal killing activity in the Δhcp Δhcp1 mutant (Fig. 5B), suggesting that Hcp levels may be a limiting factor for T6SS-mediated activities under the conditions used in the coincubation assays. To test this hypothesis, the hcp gene was cloned downstream of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter Ptrc, which has been successfully used to induce the expression of other genes in V. fischeri (30). The resulting construct (Ptrc-hcp) was introduced into the Δhcp Δhcp1 mutant to generate a strain in which the Hcp levels can be controlled exogenously by supplementing the growth medium with IPTG.
The coincubation assays described above were modified by including an IPTG treatment when the cell mixtures were spotted onto a solid medium. This IPTG treatment affected neither the ability of FQ-A001 to inhibit the growth of ES114 nor the way ES114 grew when it was coincubated with the Δhcp Δhcp1 mutant harboring a vector control (Fig. 7A). Without the IPTG treatment, coincubation of ES114 with the Δhcp Δhcp1 mutant harboring Ptrc-hcp for 24 h resulted in spots that exhibited CFP fluorescence comparable to that with the vector control (Fig. 7A). In contrast, the same pair of strains grown in the presence of at least 0.1 mM IPTG resulted in growth exhibiting decreased CFP fluorescence (Fig. 7A), suggesting that the growth of ES114 was inhibited. Because the CFP fluorescence in these spots was higher than that in spots containing FQ-A001 (Fig. 7A), the extent of growth inhibition due to the IPTG induction of hcp appeared to be less than that with the WT.
FIG 7.
Impact of induction of hcp expression on T6SS-mediated activities. (A) Coincubation assay with ES114 and the indicated FQ-A001-derived strains. ES114 is labeled with CFP via pYS112. FQ-A001-derived strains are FQ-A001 (WT) and NPW58 (Δhcp Δhcp1), which harbors pTM214 (vector) or pKRG018 (Ptrc-hcp). Images show the CFP fluorescence of the bacterial growth resulting from a 24-h incubation of cellular mixtures spotted onto agar containing the indicated concentration of IPTG. Approximately 5 × 107 total CFU were introduced into each spot. Bar, 1 mm. (B) Ratio of Ermr CFU to Camr CFU resulting from a 5-h coincubation of the Ermr ES114-derived strain TIM313 labeled with Camr by the plasmid pYS112 with the FQ-A001-derived strains described in the legend to panel A. Each point represents the ratio associated with an individual spot at 5 h without IPTG (open symbols) or with 1 mM IPTG (filled symbols), and each horizontal line represents the geometric mean (n = 4). Approximately 5 × 107 total CFU were introduced into each spot. Two-way analysis of variance revealed significant differences among the means of log-transformed ratios due to genotype (F2,18 = 165.7; P < 0.0001), IPTG treatment (F1,18 = 24.88; P < 0.0001), and their interaction (F2,18 = 17.45; P < 0.0001). Tukey’s post hoc test was performed to compare the means statistically, with P values adjusted for multiple comparisons. Groups with different letters have significantly different medians (P < 0.001), and groups with the same letter do not have significantly different medians (P ≥ 0.05).
To determine the extent to which induction of hcp could promote killing by FQ-A001, 5-h coincubation assays were performed using the Ermr ES114-derived strain TIM313 described above. In addition, 1 mM IPTG was used as the treatment due to the saturated effect observed in the 24-h coincubation assay (Fig. 7A). Treatment with 1 mM IPTG had no effect on the relative abundance of TIM313 for either the WT or the Δhcp Δhcp1 mutant control group (Fig. 7B). Also, in the absence of IPTG treatment, the proportion of TIM313 when it was coincubated with the Δhcp Δhcp1 mutant harboring the Ptrc-hcp construct was comparable to that with the vector control (Fig. 7B), in agreement with the results of the 24-h fluorescence-based assay (Fig. 7A). In contrast, the relative abundance of TIM313 coincubated with the Δhcp Δhcp1 mutant harboring the Ptrc-hcp construct was 8.6-fold lower with IPTG treatment (Fig. 7B), suggesting that the corresponding induction of hcp expression is sufficient to promote killing by the Δhcp Δhcp1 mutant. Taken together, these data suggest that the functions of the T6SS in FQ-A001 can be controlled through the regulation of hcp expression.
DISCUSSION
Along host epithelial surfaces, bacterial cells are often in direct contact with each other, permitting certain intercellular interaction systems to alter the physiology of neighboring cells (31). In this study, the T6SS of V. fischeri strain FQ-A001 was investigated in order to gain insight into the molecular determinants underlying the killing activity that this strain exhibits toward ES114. Two genes, hcp and hcp1, are predicted to encode identical Hcp subunits and were shown to contribute to the ability of FQ-A001 to kill ES114 (Fig. 5B; also see Fig. S1 in the supplemental material). While hcp is located within the main T6SS gene cluster, hcp1 is located on a different chromosome (Fig. 1B). Deletion of both the hcp and the hcp1 gene is necessary to disrupt killing activity (Fig. 4B), and expression of hcp in trans in a Δhcp Δhcp1 mutant is sufficient to restore killing activity (Fig. 5B and 7B). Finally, the ability of FQ-A001 to prevent the cocolonization of crypt spaces with ES114 was shown to require at least one hcp gene (Fig. 6).
Taken together, the results of this study contribute to the model of how the T6SS functions during establishment of the squid-Vibrio symbiosis (Fig. 8). When a T6SS-positive cell colonizes the same crypt space as a T6SS-susceptible cell, the initial growth of each strain type results in subpopulations that come into direct contact with one another. The expression of the hcp genes by the T6SS-positive strain in vivo is sufficiently high for the killing of T6SS-susceptible cells. Elimination of the T6SS-susceptible subpopulation results in the dominance of the T6SS-positive strain within the crypt space by the time the light-producing symbiosis is established. Thus, the T6SS has the potential to limit V. fischeri strain diversity within the squid light organ during symbiosis establishment.
FIG 8.
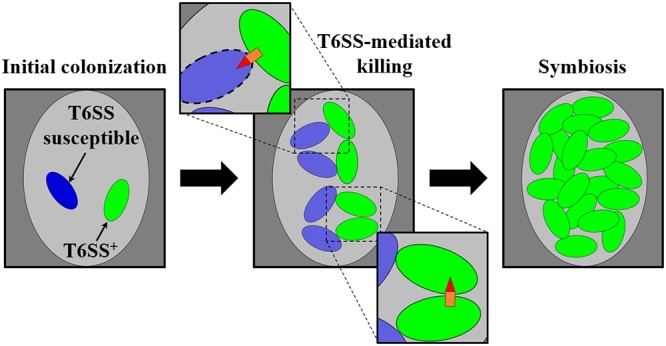
Model of T6SS-mediated killing during symbiosis establishment. (Left) A single crypt space (light gray oval) initially colonized by a cell of a T6SS-positive V. fischeri strain type (green) and a cell of a V. fischeri strain type susceptible to the T6SS (blue). (Center) Clonal expansion of each strain type results in contact between cells, which promotes T6SS-mediated interactions. (Top inset) T6SS-mediated killing of T6SS-susceptible cells. (Bottom inset) Survival of T6SS-positive cells after a T6SS-mediated attack. (Right) As a result of continued clonal expansion of the population, the crypt space harbors only the T6SS-positive strain type during symbiosis.
The killing activity by the T6SS of FQ-A001 depends on the extent to which hcp is expressed. In V. cholerae, the transcriptional activation of hcp genes depends on the σ54-dependent transcription factor VasH (32). The expression of vasH, which resides within the large gene cluster in V. cholerae, is controlled by two transcription factors associated with the natural competence pathway that promotes the uptake and chromosomal incorporation of exogenous DNA (32–34). This pathway is activated when V. cholerae is exposed to high levels of chitin, the polymer that constitutes the exoskeleton of the copepods that serve as the natural reservoir for V. cholerae (35, 36). Furthermore, HapR, a transcription factor associated with intercellular quorum sensing (37), positively regulates hcp expression (38). The consequence of these regulatory inputs is that the T6SS functions when V. cholerae is present at high abundance on the chitinaceous surfaces, which has been hypothesized to contribute to the diversification of V. cholerae strains within the environment (39, 40). Our finding that induction of hcp expression promotes T6SS-mediated activities in FQ-A001 is significant because it suggests that transcriptional regulation of the hcp genes is a potential mechanism for V. fischeri to alter T6SS function in response to environmental conditions. Therefore, future studies will be devoted to examining the extent to which hcp and hcp1 are transcribed and to identifying the factors that impact their transcription. Because homologues of transcription factors that regulate hcp transcription in V. cholerae, such as vasH, exist in FQ-A001, future studies will investigate the extent to which these homologues impact hcp expression in this strain so as to determine how the T6SS-dependent activities of V. fischeri respond to environmental conditions.
Elimination of the T6SS-mediated killing activity through the deletion of hcp and hcp1 appears to have no impact on the ability of FQ-A001 to establish symbiosis (Fig. S3 in the supplemental material). This finding is significant because it suggests that the functions of this T6SS during symbiosis establishment are specific to intercellular interactions involving other bacterial cells rather than host cells. This is in contrast to the T6SS of V. cholerae, which promotes the displacement of a natural symbiont of the zebrafish gut by strengthening intestinal contractions rather than by directly killing the symbiont (41). The effector was shown to be VgrG-1, with the actin cross-linking domain (ACD) located at the C terminus of VgrG (41). Neither vgrG nor vgrG1, the VgrG homologues in FQ-A001 that are located downstream of hcp and hcp1, respectively, is predicted to encode this 514-residue ACD. Instead, the VgrG homologues associated with the T6SS gene clusters in V. fischeri have small C-terminal domains. Our results suggest that the V. fischeri T6SS mediates only interactions between bacterial cells, as opposed to host-microbe interactions, a conclusion consistent with the lack of the ACD in the V. fischeri proteins.
In summary, two hcp genes exhibit functional redundancy for T6SS-dependent activities in V. fischeri. The ability of V. fischeri to establish symbiosis appears to be independent of a functional T6SS, further supporting the role of the T6SS in mediating the direct interactions that can occur between strains during host colonization. This study has revealed knowledge of how the T6SS of V. fischeri functions during symbiosis establishment, thereby increasing understanding of the molecular mechanisms on which bacterial symbionts depend in order to associate with a host.
MATERIALS AND METHODS
Media and growth conditions.
V. fischeri strains were grown aerobically in LBS medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 2% [wt/vol] NaCl, 50 mM Tris–HCl [pH 7.5]) (42). When appropriate, LBS was supplemented with chloramphenicol at a final concentration of 2.5 μg ml−1 and/or erythromycin at a final concentration of 5 μg ml−1.
Strains.
The V. fischeri strains used in this study are listed in Table 2.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| ES114 | Wild-type V. fischeri | 46 |
| FQ-A001 | Wild-type T6SS+ V. fischeri | 26 |
| ANS2098 | FQ-A001 vasA_2::erm | 20 |
| TIM416 | FQ-A001 Δhcp | This study |
| NPW57 | FQ-A001 Δhcp1 | This study |
| NPW58 | FQ-A001 Δhcp Δhcp1 | This study |
| TIM313 | ES114 Tn7::erm | 47 |
| Plasmids | ||
| pSCV38 | pVSV105 PtetA-yfp PtetA-mCherry | 48 |
| pYS112 | pVSV105 PproD-cfp PtetA-mCherry | 26 |
| pVSV105 | R6Kori ori(pES213) RP4 oriT cat | 49 |
| pEVS79 | pBC SK (+) oriT cat | 50 |
| pTM430 | pEVS79 Δhcp | This study |
| pAGC007 | pEVS79 Δhcp1 | This study |
| pNPW16 | pVSV105 hcp1+ | This study |
| pNPW17 | pVSV105 hcp+ | This study |
| pTM214 | pVSV105 Ptrc-mCherry | 51 |
| pKRG018 | Ptrc-hcp | This study |
| pEVS104 | R6Kori RP4 oriT trb tra kan | 50 |
(i) Construction of deletion alleles.
The deletion alleles Δhcp and Δhcp1 were each generated from FQ-A001 genomic DNA by PCR amplification of 1.3- to 1.5-kb regions on either side of the corresponding genes and cloning of the products into pEVS79 to yield plasmids pTM430 and pAGC007, respectively. The primers and restriction sites used in the cloning process are listed in Table 3. To generate a mutant with a particular deletion allele, the corresponding plasmid was introduced into the parental strain by conjugation using pEVS104 and screening by PCR for the double-crossover event, as described previously (42).
TABLE 3.
Primers used in this study
| Primer name | Sequence (5′ → 3′) |
|---|---|
| Deletion alleles | |
| Δhcp | |
| Δhcp-5-KpnI-u | GGGGTACCTCCAGTTTAACTAATTGGAATCGAC |
| Δhcp-5-XbaI-l | GGTCTAGACATATATGCTGGAGTTGGCATGCT |
| Δhcp-3-XbaI-u | GGTCTAGAACACCACGCGAAGCATAATCGTTT |
| Δhcp-3-SacI-l | GGGAGCTCTTATCAGTATAAATTTCTTCACC |
| Δhcp1 | |
| Δhcp1-5-KpnI-u | GGGGTACCGTCGGTTTGGGTATTCCGGGAATG |
| Δhcp1-5-XbaI-l | GGTCTAGACATATACGCTGGAGTTGGCATGGC |
| Δhcp1-3-XbaI-u | GGTCTAGAACACCGCGCGAAGCGTAATGATTA |
| Δhcp1-3-SacI-l | GGGAGCTCGTCACTAGAGGTCTGTCTGGGTCC |
| Complementation vectors | |
| hcp+ | |
| hcp-XbaI-u | GCTCTAGAAGTTAGTTTCAATCATTAGCCAGAG |
| hcp-KpnI-l | GTGGTACCATCTTTTACCTGATACTAATATGCA |
| hcp1+ | |
| hcp1-XbaI-u | GCTCTAGACTTGGAGCCTTATAATTTTAGAACAC |
| hcp1-KpnI-l | GTGGTACCTCATAAACCATAAGCAATGCAATAG |
| IPTG-inducible vectors | |
| Ptrc-hcp-u | CACGTCGACGTTGACACTCTATCATTGATAG |
| Ptrc-hcp-l | GAAGATTTTCTTATTTGTATAGTTCATCC |
(ii) Construction of complementation vectors.
Plasmids pNPW16 and pNPW17 contain the hcp1+ and hcp+ alleles, respectively. To generate each construct, these genes and their corresponding promoter regions were first amplified by PCR from genomic DNA of FQ-A001 and then cloned into pVSV105. The primers and the restriction sites used in cloning are shown in Table 3.
(iii) Construction of Ptrc-hcp.
Plasmid pKRG018, which contains the hcp gene downstream of the IPTG-inducible trc promoter, was constructed by PCR amplification of hcp from FQ-A001 genomic DNA and cloning of the product into pTM214 upstream of the mCherry gene.
Comparative genomics.
The FQ-A001 genome sequence (NCBI genome accession number SJSX00000000 [version SJSX01000000]) was analyzed using Mauve (progressiveMauve algorithm; snapshot 2015_02_25 macOS) and NCBI BLAST (blastp, version 2.9.0+), with comparisons against the V. fischeri MJ11 genome (NCBI accession numbers CP001139.1 [chromosome I] and CP001133.1 [chromosome II]) (27, 43–45).
Squid colonization assays.
Squid colonization assays were performed as described previously (20). Briefly, LBS cultures grown overnight at 28°C with shaking were first normalized to an optical density at 600 nm (OD600) of 1.0 and then diluted 1:100 into LBS medium. For strains harboring plasmids for labeling with fluorescence, the medium contained 2.5 μg/ml chloramphenicol. At an OD600 of 1.0, cultures were diluted into filter-sterilized seawater (FSSW) and were combined as indicated in the figure legends to generate each inoculum. Freshly hatched juvenile squid were exposed to the inoculum for 3.5 h, after which the inoculum was washed off the animals by transfer to fresh FSSW. At 24 h postinoculation (p.i.), another wash step was performed. At 48 h p.i., animals were cooled on ice and were then fixed and imaged by fluorescence microscopy as described previously (20).
Coincubation assays.
Coincubation assays were performed as described previously (20), with the modifications described here. LBS cultures grown overnight at 28°C with shaking were first diluted into LBS to achieve an OD600 of 1.0. To initiate the assay, 50-μl cell suspensions of each strain were combined in a microcentrifuge tube, and then a 10-μl volume was spotted onto the surface of LBS medium containing 1.5% agar, which was incubated at 24°C. As described below, the spots were either harvested for CFU counts or visualized by fluorescence microscopy for qualitative structural features of the strains within the spots.
To determine cellular abundance within a spot, flame-sterilized forceps were used to extract the spot and the corresponding agar fragment, which were then introduced into a microcentrifuge tube containing 1 ml LBS. Cells were released from the agar by vortexing for 10 s, after which serial dilutions were performed, and cells were plated onto LBS agar with or without an antibiotic for CFU counts.
For fluorescence measurements, spots were incubated for 24 h and were then examined at ×2.5 magnification using an SZX16 dissecting microscope (Olympus, Waltham, MA) equipped with an SDF PLFL 0.3× objective and filter sets for CFP and YFP fluorescence. Images of blue (CFP) and yellow (YFP) fluorescence signals of each spot were obtained using an EOS Rebel T5 camera (Canon, Melville, NY) attached to the camera port of the microscope and set for the RAW image format. Images were converted to the RGB TIFF format using ImageJ (NIH, Bethesda, MD) with the DCRaw reader plug-in, and only the blue (or green) channel was retained from the blue (or yellow) fluorescence images. To emphasize the structural features of the spots associated with a particular fluorophore, the lookup table of the images was scaled uniformly across the samples in the same experiment.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01 GM129133 (to T.M.), R35 GM119627 (to M.J.M.), and R21 AI117262 (to M.J.M.) and by National Science Foundation grant IOS-1757297 (to M.J.M.). Trainee support for K.M.B. was provided on NIH grant T32 GM008349.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00221-19.
REFERENCES
- 1.Esser D, Lange J, Marinos G, Sieber M, Best L, Prasse D, Bathia J, Ruhlemann MC, Boersch K, Jaspers C, Sommer F. 2018. Functions of the microbiota for the physiology of animal metaorganisms. J Innate Immun 11:393–404. doi: 10.1159/000495115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi T, Voisin B, Kim DY, Kennedy EA, Jo JH, Shih HY, Truong A, Doebel T, Sakamoto K, Cui CY, Schlessinger D, Moro K, Nakae S, Horiuchi K, Zhu J, Leonard WJ, Kong HH, Nagao K. 2019. Homeostatic control of sebaceous glands by innate lymphoid cells regulates commensal bacteria equilibrium. Cell 176:982–997.e16. doi: 10.1016/j.cell.2018.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim M, Ashida H, Ogawa M, Yoshikawa Y, Mimuro H, Sasakawa C. 2010. Bacterial interactions with the host epithelium. Cell Host Microbe 8:20–35. doi: 10.1016/j.chom.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Leser TD, Molbak L. 2009. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol 11:2194–2206. doi: 10.1111/j.1462-2920.2009.01941.x. [DOI] [PubMed] [Google Scholar]
- 5.Wessler S, Backert S. 2008. Molecular mechanisms of epithelial-barrier disruption by Helicobacter pylori. Trends Microbiol 16:397–405. doi: 10.1016/j.tim.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. 2017. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27:626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joyce EA, Chan K, Salama NR, Falkow S. 2002. Redefining bacterial populations: a post-genomic reformation. Nat Rev Genet 3:462–473. doi: 10.1038/nrg820. [DOI] [PubMed] [Google Scholar]
- 8.Nowak MA, Sigmund K. 2002. Bacterial game dynamics. Nature 418:138–139. doi: 10.1038/418138a. [DOI] [PubMed] [Google Scholar]
- 9.Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, Kupiec M, Gophna U, Sharan R, Ruppin E. 2011. Competitive and cooperative metabolic interactions in bacterial communities. Nat Commun 2:589. doi: 10.1038/ncomms1597. [DOI] [PubMed] [Google Scholar]
- 10.Majeed H, Gillor O, Kerr B, Riley MA. 2011. Competitive interactions in Escherichia coli populations: the role of bacteriocins. ISME J 5:71–81. doi: 10.1038/ismej.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Bayona L, Comstock LE. 2018. Bacterial antagonism in host-associated microbial communities. Science 361:eaat2456. doi: 10.1126/science.aat2456. [DOI] [PubMed] [Google Scholar]
- 12.Ho BT, Dong TG, Mekalanos JJ. 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A 104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A 106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD. 2014. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. 2016. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci U S A 113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speare L, Cecere AG, Guckes KR, Smith S, Wollenberg MS, Mandel MJ, Miyashiro T, Septer AN. 2018. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci U S A 115:E8528–E8537. doi: 10.1073/pnas.1808302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McFall-Ngai M. 2014. Divining the essence of symbiosis: insights from the squid-vibrio model. PLoS Biol 12:e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyholm SV, McFall-Ngai MJ. 2004. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol 2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 23.Jones BW, Nishiguchi MK. 2004. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Marine Biol 144:1151–1155. doi: 10.1007/s00227-003-1285-3. [DOI] [Google Scholar]
- 24.Koch EJ, Miyashiro T, McFall-Ngai MJ, Ruby EG. 2014. Features governing symbiont persistence in the squid-vibrio association. Mol Ecol 23:1624–1634. doi: 10.1111/mec.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wollenberg MS, Ruby EG. 2009. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol 75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, LaSota ED, Cecere AG, LaPenna KB, Larios-Valencia J, Wollenberg MS, Miyashiro T. 2016. Intraspecific competition impacts Vibrio fischeri strain diversity during initial colonization of the squid light organ. Appl Environ Microbiol 82:3082–3091. doi: 10.1128/AEM.04143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bultman KM, Cecere AG, Miyashiro T, Septer AN, Mandel MJ. 2019. Draft genome sequences of type VI secretion system-encoding Vibrio fischeri strains FQ-A001 and ES401. Microbiol Resour Announc 8:e00385-19. doi: 10.1128/MRA.00385-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altindis E, Dong T, Catalano C, Mekalanos J. 2015. Secretome analysis of Vibrio cholerae type VI secretion system reveals a new effector-immunity pair. MBio 6:e00075. doi: 10.1128/mBio.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joshi A, Kostiuk B, Rogers A, Teschler J, Pukatzki S, Yildiz FH. 2017. Rules of engagement: the type VI secretion system in Vibrio cholerae. Trends Microbiol 25:267–279. doi: 10.1016/j.tim.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao X, Studer SV, Wassarman K, Zhang Y, Ruby EG, Miyashiro T. 2012. The novel sigma factor-like regulator RpoQ controls luminescence, chitinase activity, and motility in Vibrio fischeri. MBio 3:e00285-11. doi: 10.1128/mBio.00285-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stubbendieck RM, Straight PD. 2016. Multifaceted interfaces of bacterial competition. J Bacteriol 198:2145–2155. doi: 10.1128/JB.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. 2011. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol 193:6471–6482. doi: 10.1128/JB.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto S, Morita M, Izumiya H, Watanabe H. 2010. Chitin disaccharide (GlcNAc)2 induces natural competence in Vibrio cholerae through transcriptional and translational activation of a positive regulatory gene tfoXVC. Gene 457:42–49. doi: 10.1016/j.gene.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Lo Scrudato M, Borgeaud S, Blokesch M. 2014. Regulatory elements involved in the expression of competence genes in naturally transformable Vibrio cholerae. BMC Microbiol 14:327. doi: 10.1186/s12866-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol 45:275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 37.Kovacikova G, Skorupski K. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol Microbiol 46:1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- 38.Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. 2009. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One 4:e6734. doi: 10.1371/journal.pone.0006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostiuk B, Unterweger D, Provenzano D, Pukatzki S. 2017. T6SS intraspecific competition orchestrates Vibrio cholerae genotypic diversity. Int Microbiol 20:130–137. doi: 10.2436/20.1501.01.294. [DOI] [PubMed] [Google Scholar]
- 40.Bernardy EE, Turnsek MA, Wilson SK, Tarr CL, Hammer BK. 2016. Diversity of clinical and environmental isolates of Vibrio cholerae in natural transformation and contact-dependent bacterial killing indicative of type VI secretion system activity. Appl Environ Microbiol 82:2833–2842. doi: 10.1128/AEM.00351-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Logan SL, Thomas J, Yan J, Baker RP, Shields DS, Xavier JB, Hammer BK, Parthasarathy R. 2018. The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc Natl Acad Sci U S A 115:E3779–E3787. doi: 10.1073/pnas.1720133115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wasilko NP, Larios-Valencia J, Steingard CH, Nunez BM, Verma SC, Miyashiro T. 2019. Sulfur availability for Vibrio fischeri growth during symbiosis establishment depends on biogeography within the squid light organ. Mol Microbiol 111:621–636. doi: 10.1111/mmi.14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. 2009. A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. 2010. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol 77:1556–1567. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verma SC, Miyashiro T. 2016. Niche-specific impact of a symbiotic function on the persistence of microbial symbionts within a natural host. Appl Environ Microbiol 82:5990–5996. doi: 10.1128/AEM.01770-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunne C, Dolan B, Clyne M. 2014. Factors that mediate colonization of the human stomach by Helicobacter pylori. World J Gastroenterol 20:5610–5624. doi: 10.3748/wjg.v20.i19.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stabb EV, Ruby EG. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol 358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- 51.Miyashiro T, Klein W, Oehlert D, Cao X, Schwartzman J, Ruby EG. 2011. The N-acetyl-d-glucosamine repressor NagC of Vibrio fischeri facilitates colonization of Euprymna scolopes. Mol Microbiol 82:894–903. doi: 10.1111/j.1365-2958.2011.07858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.