Salmonella is one of the most important food-borne pathogens, infecting over one million people in the United States every year. These bacteria use a needle-like device to interact with intestinal epithelial cells, leading to invasion of the cells and induction of inflammatory diarrhea. A complex regulatory network controls expression of the invasion system in response to numerous environmental signals. Here we explore the molecular mechanisms by which the small RNA PinT contributes to this regulation, facilitating inactivation of the system after invasion. PinT controls several important virulence systems in Salmonella, tuning the transition between different stages of infection.
KEYWORDS: PhoPQ, SPI1, Salmonella, sRNA
ABSTRACT
Salmonella enterica serovar Typhimurium induces inflammatory diarrhea and bacterial uptake into intestinal epithelial cells using the Salmonella pathogenicity island 1 (SPI1) type III secretion system (T3SS). HilA activates transcription of the SPI1 structural components and effector proteins. Expression of hilA is activated by HilD, HilC, and RtsA, which act in a complex feed-forward regulatory loop. Many environmental signals and other regulators are integrated into this regulatory loop, primarily via HilD. After the invasion of Salmonella into host intestinal epithelial cells or during systemic replication in macrophages, the SPI T3SS is no longer required or expressed. We have shown that the two-component regulatory system PhoPQ, required for intracellular survival, represses the SPI1 T3SS mostly by controlling the transcription of hilA and hilD. Here we show that PinT, one of the PhoPQ-regulated small RNAs (sRNAs), contributes to this regulation by repressing hilA and rtsA translation. PinT base pairs with both the hilA and rtsA mRNAs, resulting in translational inhibition of hilA, but also induces degradation of the rts transcript. PinT also indirectly represses expression of FliZ, a posttranslational regulator of HilD, and directly represses translation of ssrB, encoding the primary regulator of the SPI2 T3SS. Our in vivo mouse competition assays support the concept that PinT controls a series of virulence genes at the posttranscriptional level in order to adapt Salmonella from the invasion stage to intracellular survival.
IMPORTANCE Salmonella is one of the most important food-borne pathogens, infecting over one million people in the United States every year. These bacteria use a needle-like device to interact with intestinal epithelial cells, leading to invasion of the cells and induction of inflammatory diarrhea. A complex regulatory network controls expression of the invasion system in response to numerous environmental signals. Here we explore the molecular mechanisms by which the small RNA PinT contributes to this regulation, facilitating inactivation of the system after invasion. PinT controls several important virulence systems in Salmonella, tuning the transition between different stages of infection.
INTRODUCTION
Salmonella enterica uses a type III secretion system (T3SS) encoded on the Salmonella pathogenicity island 1 (SPI1) to both invade the intestinal epithelium of the host and induce inflammatory diarrhea (1–4). The SPI1 T3SS injects bacterial effector proteins directly into the host cell cytosol to manipulate host cytoskeletal rearrangement and signal transduction (5–9). The bacteria that invade are subsequently capable of replicating in macrophages to cause life-threatening disease. In the phagosome of either invaded epithelial cells or macrophages, SPI1 gene expression is turned off, and neither SPI1 nor the coordinately regulated flagellar system is expressed or required during systemic stages of diseases (10).
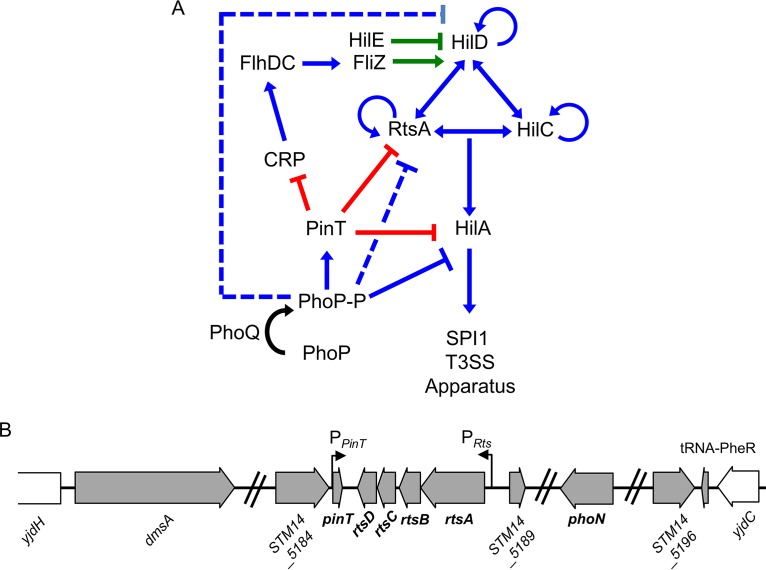
The bacteria integrate environmental signals and input from a variety of global regulatory systems to precisely control transcription and translation of the SPI1 T3SS genes (5, 9, 11). HilA (hyperinvasion locus A), encoded in the SPI1 locus, directly activates transcription of the SPI1 T3SS structural and primary effector genes (8, 12–14). Three AraC-like proteins, HilD, HilC, and RtsA, activate transcription of hilA by binding the promoter (15). In addition, HilD, HilC, and RtsA regulate their own transcription and the transcription of each other, forming a complex feed-forward regulatory loop (15–18) (Fig. 1). Environmental signals and regulatory systems are integrated into the SPI1 regulatory circuit primarily at the level of HilD activity or hilD translation (11), while HilC and RtsA amplify these inducing signals for efficient system activation (11, 15, 19) (Fig. 1). Several regulatory systems have been implicated in controlling hilD mRNA stability or translation, acting at either the 5′ untranslated region (UTR) or 3′ UTR of the hilD mRNA (20–25). Other systems directly control hilA expression or more globally affect the entire system (12, 26–28). We previously showed that small regulatory RNAs (sRNAs) play a pivotal role in the regulation of SPI1 T3SS in response to oxygen, a key environmental signal controlling Salmonella virulence (21).
FIG 1.
SPI1 T3SS regulatory circuit and the Rts-PinT locus. (A) Simplified regulatory model of the SPI1 T3SS and related regulators. Blue lines indicate transcriptional regulation, green lines indicate regulation at the protein level, and red lines indicate regulation at posttranscriptional level. Dotted lines indicate that the exact mechanism of regulation is not known and is likely indirect. (B) The pinT gene and rts operon are located within a 15-kb insert located near tRNA-PheR. Gray arrows represent the genes within the genomic island, and white arrows are the surrounding chromosomal genes. Not all genes are shown. The illustration was created from the strain 14028 genome sequence (100).
sRNAs are abundant posttranscriptional regulators of gene expression in bacteria. Induction of a given sRNA is often affected by environmental or stress conditions, and sRNAs play roles in many aspects of bacterial physiology, including metabolism, metal homeostasis, stress responses and pathogenesis (29–33). sRNAs typically base pair with target mRNAs to negatively or positively control translation and/or stability of the mRNA (30, 31, 34–37). Hfq and ProQ are involved as RNA chaperones that mediate stability of the sRNAs and/or facilitate sRNA-mRNA interaction (35, 38–42). In Salmonella, both experimental and bioinformatic approaches have been used to identify more than 325 sRNAs, and the functional roles of several sRNAs in regulating physiology and pathogenesis have been studied (21, 43–47).
The PhoPQ two-component system is a key virulence regulator in Salmonella, controlling myriad factors required for intracellular survival and growth (48–55). PhoQ is the sensor kinase and phosphorylates its cognate response regulator, PhoP, in response to antimicrobial peptides, low levels of divalent cations, and low pH (56–58). Either impairing or constitutively activating the PhoPQ two-component system confers defects in Salmonella virulence, suggesting that precise control of the two-component system is crucial for adaptation to various niches in the host (1, 52, 59). The PhoPQ two-component system regulates SPI1 gene expression by repressing hilA transcription (11), but the mechanism of action was not clear. We have shown that the PhoP represses hilA transcription by blocking activation of the promoter and also indirectly affects hilD and rtsA transcription, providing mechanistic insight into how the SPI1 system is shut off after invasion (60). PhoPQ also induces two sRNAs: MgrR and PinT (33, 61, 62). MgrR regulates genes involved in lipopolysaccharide modification and phosphate import in response to intracellular magnesium levels (61–63). The 80-nucleotide (nt) sRNA PinT is produced as a primary transcript, is induced in intracellular Salmonella, and regulates virulence-associated genes (33, 44). PinT was suggested to function as a modulator of SPI1 and SPI2 gene expression in response to the intracellular environment, but little of the molecular mechanisms was revealed (33).
Here we show that PinT posttranscriptionally affects SPI1 gene expression by directly regulating hilA and rtsA translation and indirectly affecting fliZ expression. PinT inhibits translation of hilA and promotes degradation of the rts transcript. Thus, PinT contributes additional posttranscriptional regulation of the SPI1 T3SS to the transcriptional repression governed by PhoPQ, ensuring efficient downregulation in the niches where the SPI1 T3SS is no longer required. The data support a model in which PinT directly and indirectly controls SPI1, flagellar biosynthesis, and SPI2 gene expression to facilitate intracellular survival within the host.
RESULTS
PinT, a PhoPQ-activated sRNA, regulates hilA and rtsA translation.
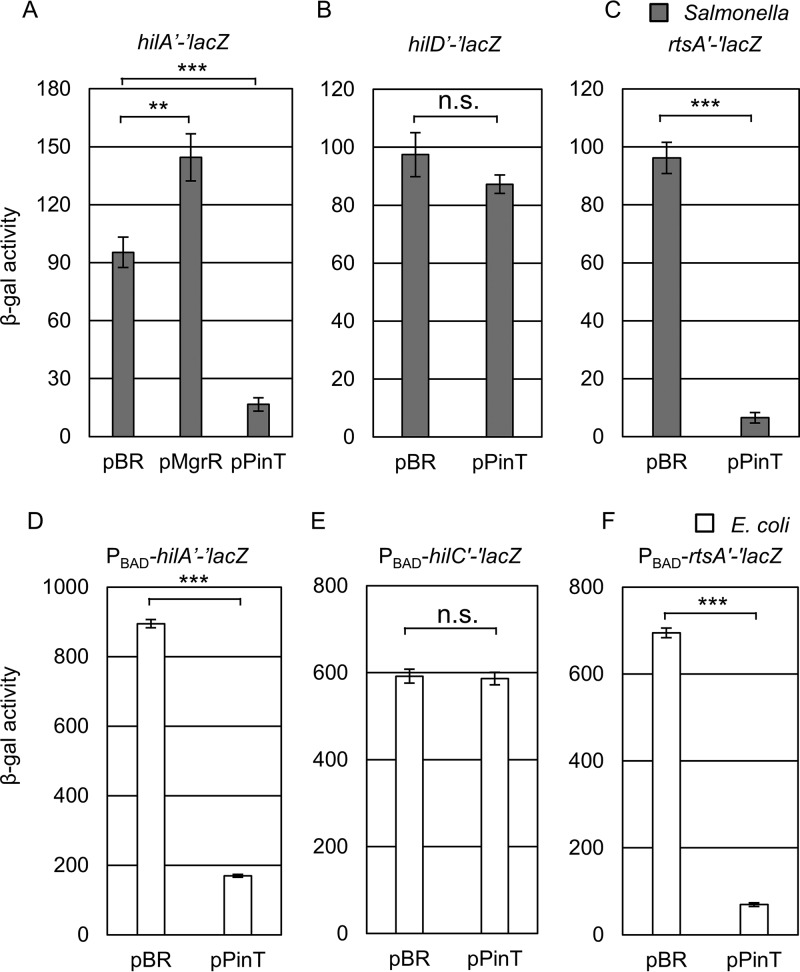
The PhoPQ two-component system regulates expression of the SPI1 genes primarily by repressing hilA and hilD transcription (11, 60) (Fig. 1A). The PhoPQ two-component system also controls expression of two sRNAs, MgrR and PinT (33, 61). We hypothesized that these sRNAs could contribute to the PhoPQ-mediated regulation of the SPI1 T3SS at the posttranscriptional level. To test this hypothesis, both MgrR and PinT from Salmonella enterica serovar Typhimurium strain 14028 were cloned into the pBRpLac vector (64). We then tested the effect of overproduction of each sRNA on expression of an in-locus hilA′-′lacZ translational fusion, which contains the 350-nt 5′ UTR and the first 31 codons of hilA fused in frame with LacZ under the control of the native hilA promoter. When MgrR was overexpressed from the plasmid, expression increased ∼1.5-fold. However, overexpression of PinT caused significant repression of the hilA′-′lacZ fusion activity (∼5-fold) (Fig. 2A). We focused our analysis on PinT.
FIG 2.
PinT downregulates SPI1 expression by repressing hilA and rtsA translation. (A to C) β-Galactosidase activity in Salmonella strains containing an hilA′-′lacZ (A), hilD′-′lacZ (B), or rtsA′-′lacZ (C) translational fusion and plasmids overexpressing PinT grown under SPI1-inducing conditions. (D to F) β-Galactosidase activity in E. coli strains containing a PBAD-hilA′-′lacZ (D), PBAD-hilC′-′lacZ (E), or PBAD-rtsA′-′lacZ (F) translational fusion and plasmids overexpressing PinT grown in the presence of 100 μM IPTG and 0.001% arabinose to induce sRNA expression and fusion expression, respectively. β-Galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation, where n = 3. P values (unpaired t test) are indicated as follows: **, P < 0.01; ***, P < 0.001; n.s., not significant. The strains used were JS892, JS2333, JS2334, JMS6503, JMS6504, and JMS6505, each with plasmid pBRplac or pPinT.
Based on the feed-forward loop model of hilA activation, it is possible that PinT represses the hilA′-′lacZ fusion directly and/or by controlling the upstream regulators HilD, HilC, and/or RtsA (15). To elucidate the regulatory mechanism of PinT in hilA expression, we tested the effect of PinT on a PBAD-hilA′-′lacZ fusion in Escherichia coli. Using the E. coli fusions reduces the complications imposed by the complex feed-forward loop controlling SPI1 gene expression in Salmonella (11, 15). Overexpression of PinT significantly repressed expression of the PBAD-hilA′-′lacZ fusion (Fig. 2D), showing that PinT directly represses hilA translation. To test whether PinT also regulates expression of the upstream regulators, we tested the effect of overexpression of PinT on an hilD′-′lacZ fusion in Salmonella (Fig. 2B). There was no effect on expression of this fusion. To test the possible involvement of PinT in regulation of rtsA or hilC expression, we tested the overexpression of PinT in a PBAD-rtsA′-′lacZ or PBAD-hilC′-′lacZ translational fusion in E. coli. Overexpression of PinT did not affect the hilC′-′lacZ fusion (Fig. 2E) but caused significant repression of the rtsA′-′lacZ fusion (Fig. 2F). This repression is also reproduced in an rtsA′-′lacZ fusion in Salmonella (Fig. 2C), suggesting that PinT also directly represses rtsA translation in Salmonella. Lastly, we addressed whether overexpression of PinT affects hilD expression via the 3′ UTR, which is known to affect the stability of the hilD mRNA (24, 25). It is possible that PinT binding to the 3′ UTR could lead to degradation of the mRNA, contributing to decreased expression of hilA. We created an hilD′-lacZ+ fusion where transcriptional lacZ is fused at the end of hilD 3′ UTR (see Fig. S1A in the supplemental material). In the absence of rtsA, the hilD′-lacZ+ 3′ UTR fusion is not regulated by overproducing PinT, showing that PinT does not regulate hilD at posttranscriptional level (Fig. S1B). Thus, we identified that PinT represses hilA expression by regulating both hilA and rtsA translation.
PinT base pairs directly with the hilA and rtsA mRNAs.
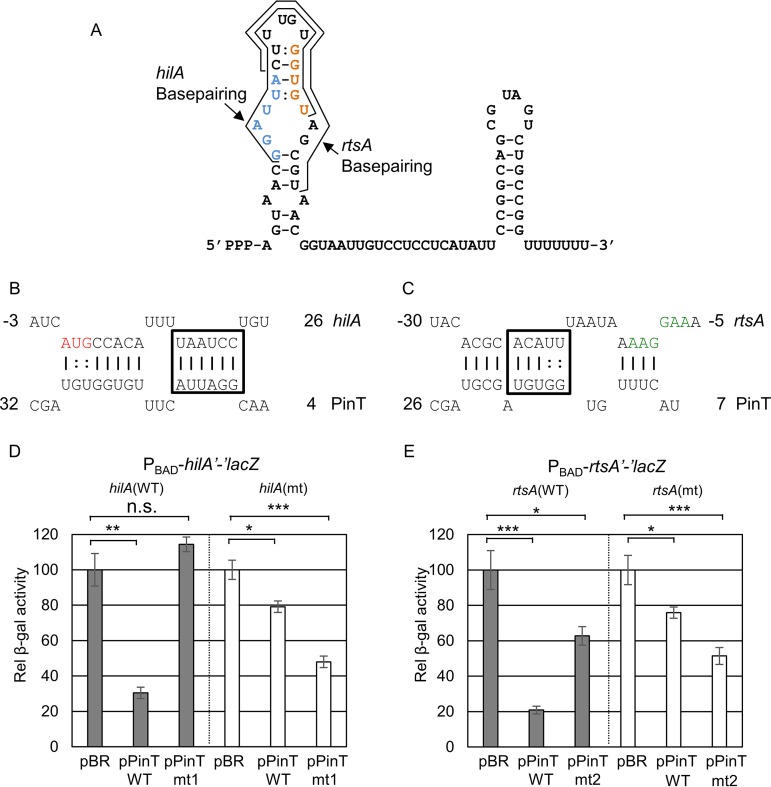
We hypothesized that PinT regulates SPI1 via base-pairing interactions with the hilA and rtsA mRNAs. Bioinformatic predictions (65) suggested that PinT binds near the AUG start codon of the hilA mRNA and near the ribosome binding site (RBS) of the rtsA mRNA (Fig. 3B and C). To test these predictions, we performed mutational analyses using the PBAD-hilA′-′lacZ or PBAD-rtsA′-′lacZ fusion in E. coli. PinT nt 7 to 29 are predicted to base pair with the hilA mRNA from nt +1 to +23 relative to the initiation AUG (Fig. 3A and B). Consistent with this prediction, the PinT-mt1, in which the boxed PinT nuncleotides were mutated, was reduced in its ability to regulate the wild-type hilA′-′lacZ fusion (Fig. 3D). Introducing compensatory mutations at positions 18 to 23 in the hilA fusion largely restored the regulation by the mutant PinT (Fig. 3D). These genetic data support the model that PinT requires this base-pairing interaction to regulate hilA translation.
FIG 3.
PinT regulates hilA and rtsA translation by direct base-pairing interaction. (A) The structure of PinT based on the study by Westermann et al. (33). The regions of base pairing with hilA and rtsA are indicated. Mutated base pairs are indicated in blue and orange. (B and C) Predicted base-pairing interactions between PinT and hilA (B) or rtsA (C) mRNA are shown. For hilA and rtsA, nucleotides are numbered from the translational start site. The ribosome binding site of rtsA mRNA is highlighted in green, and the translational start site of hilA is highlighted in red. Boxes mark nucleotides for which complementary mutations were created in hilA or rtsA and PinT. (D and E) Relative β-galactosidase activity in E. coli strains containing the wild-type or mutated hilA′-′lacZ (D) or rtsA′-′lacZ (E) translational fusion and plasmids overexpressing either the wild-type (pPinT) or mutated (pPinT-mts) sRNA grown as indicated in Fig. 2D to F. β-Galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation, where n = 3. P values (unpaired t test) are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant. The strains used are JMS6505, JMS6506, JMS6504, and JMS6507, each with plasmid pBRplac, pPinT, pPinT-mt1, or pPinT-mt2.
PinT nt 9 to 23 are predicted to interact with rtsA nt −9 to −27 relative to the AUG (Fig. 3A and C). Mutating the boxed PinT nucleotides (PinT-mt2) largely disrupted the interaction with the rtsA mRNA (Fig. 3E). Introduction of compensatory mutations in the rtsA fusion restored the regulation by the mutant PinT, suggesting that PinT requires this base-pairing interaction to regulate rtsA translation. Altogether, we concluded that PinT directly base pairs with both hilA and rtsA mRNAs to regulate expression.
Mechanism of PinT-mediated regulation of hilA and rtsA translation.
The above data suggest that PinT independently regulates translation of rtsA and hilA. RtsA, however, contributes to the transcription of hilA. Overexpression of PinT led to an ∼3-fold repression of the hilA′-′lacZ fusion in Salmonella (Fig. 4A). This regulation is a sum of the regulation of PinT on rtsA translation and hilA translation. When we tested the overexpression of PinT on hilA′-′lacZ in an rtsA null background, PinT caused only a 2-fold repression of hilA expression (Fig. 4A), consistent with the contribution of RtsA in activation of hilA transcription (15). When we introduced an hilC deletion mutation into the hilA′-′lacZ fusion strain, the overexpression of PinT still repressed hilA translation ∼3-fold, similar to the level of repression that we observed in the wild-type background, showing that the effect is specific to RtsA (see Fig. S2 in the supplemental material).
FIG 4.
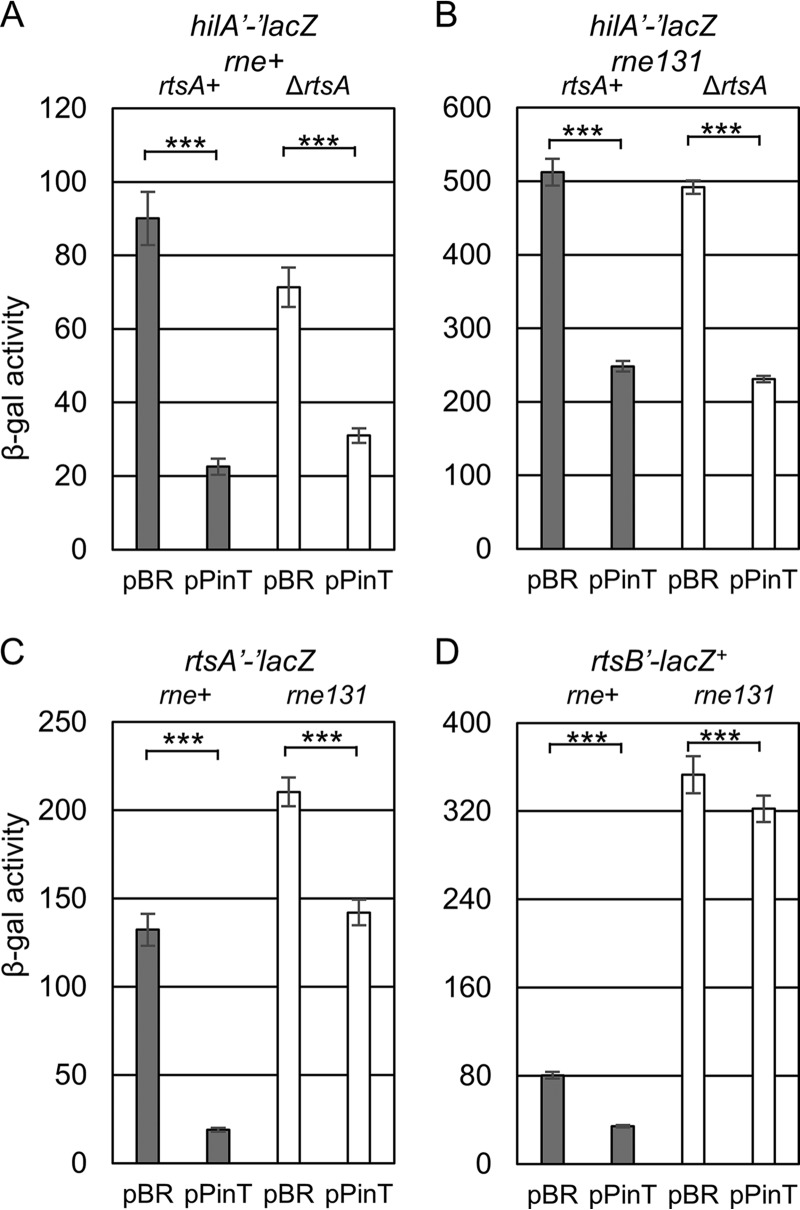
Mechanism of PinT regulation of hilA or rtsA mRNA translation. Shown is β-galactosidase activity in Salmonella strains containing the hilA′-′lacZ translational fusion (A and B), rtsA′-′lacZ translational fusion (C), or rtsB′-lacZ+ transcriptional fusion (D) in the indicated genetic backgrounds with either the empty vector or plasmids overexpressing PinT. β-Galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation, where n = 3. P values (unpaired t test) are indicated as follows: ***, P < 0.001. The strains used are JS2333, JS2337, JS2336, JS2338, JS2334, JS2336, JS325, and JS2339, each with plasmid pBRplac or pPinT.
When an sRNA base pairs with its target mRNA near the ribosome binding site (RBS) and AUG start codon, the base-pairing interaction either can induce simple translational inhibition or, additionally, can initiate RNA degradosome-dependent mRNA degradation (37). The rne131 mutation truncates RNase E, maintaining enzymatic activity, but preventing assembly of the degradosome, which in many cases also eliminates sRNA-dependent mRNA turnover (21, 37, 66, 67). To elucidate the mechanism of PinT-mediated regulation of hilA and rtsA translation, we introduced the rne131 mutation of RNase E into either the hilA′-′lacZ or rtsA′-′lacZ Salmonella strains. Introduction of the rne131 mutation partially relieved repression of hilA expression from 3-fold to ∼2-fold (Fig. 4A and B). However, in the absence of RtsA, overexpression of PinT decreased hilA translation to the same level in both the wild-type and rne131 backgrounds. These data suggest that PinT directly blocks translation of the hilA mRNA and this repression does not require mRNA degradation.
We also compared the PinT effects on the rtsA′-′lacZ fusion in either the wild-type or rne131 background. In the wild-type background, the overexpression of PinT caused approximately a 10-fold repression of rtsA expression (Fig. 4C). However, the overexpression of PinT repressed less than 2-fold in the rne131 background (Fig. 4C). Thus, initiation of mRNA degradation is apparently important for PinT to regulate rtsA. In the absence of this degradation, regulation of hilA results from only the direct effect of PinT on hilA translation, explaining the fact that RtsA plays no role in hilA expression in the strain lacking a functional degradosome (Fig. 4B). The rts operon contains four genes (Fig. 1B). We tested the effects on expression of the downstream genes by monitoring an rtsB′-lacZ+ transcriptional fusion. Overexpression of PinT decreased expression of the rtsB′-lacZ+ fusion ∼3-fold (Fig. 4D). Introduction of the rne131 allele into this background resulted in an almost 4-fold increase in lac activity, and in this background, overexpression of PinT had barely any effect on rtsB expression. These data suggest that the major consequence of the PinT-mediated regulation of rtsA translation is the degradation of the polycistronic mRNA by the RNA degradosome, regulating expression of all the genes in the operon. Thus, PinT represses both hilA and rtsA translation, but the mechanisms of regulation are distinct.
PhoPQ-mediated regulation of SPI1 T3SS involves PinT acting posttranscriptionally.
PinT expression is activated by the PhoPQ two-component system and is highly induced in infected host cells (33). To confirm that PinT is controlled by PhoPQ, we created a transcriptional fusion to pinT and measured expression after growth in N-minimal medium with either a low (10 μM) or high (10 mM) concentration of Mg2+. As shown in Fig. S3, the transcription of pinT is induced under low-Mg2+ conditions. Deletion of phoPQ led to strongly reduced expression, whereas introduction of the phoQ24 allele (68, 69) resulted in high-level constitutive expression of pinT (Fig. S3). These data confirm that PinT is regulated by the PhoPQ two-component system.
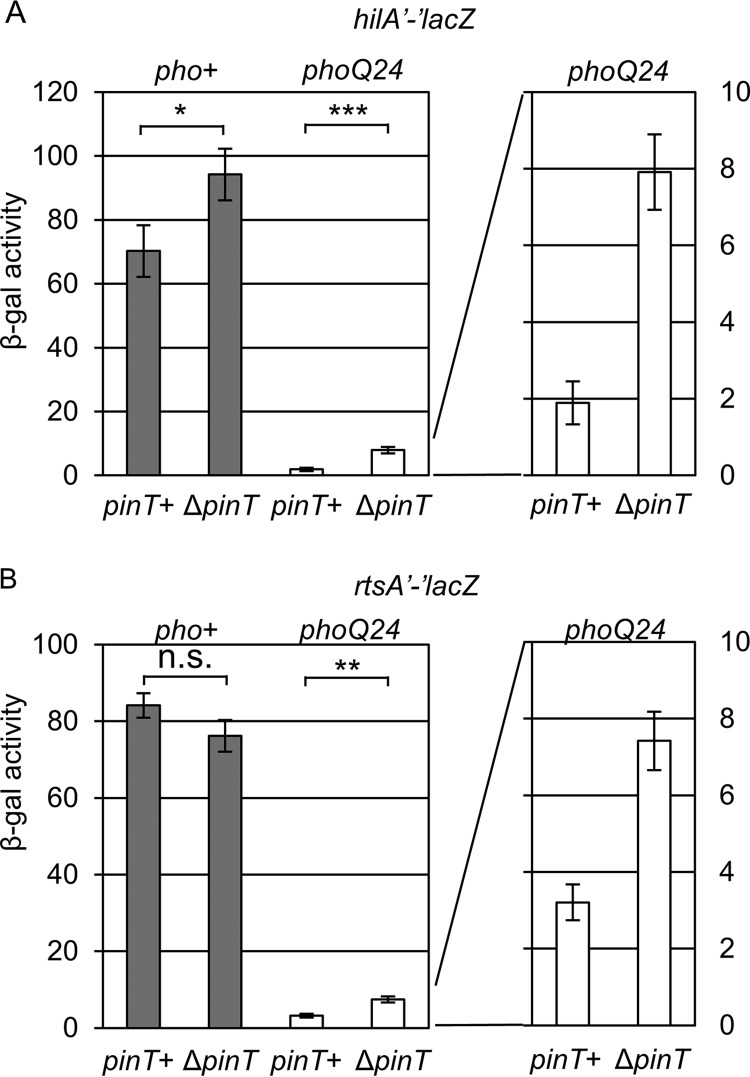
PhoPQ represses SPI1 expression primarily by blocking activation of the hilA promoter and indirectly repressing hilD and rtsA transcription (60). Given the effects of PinT on hilA and rtsA translation, we tested how PinT contributes to the PhoPQ-mediated regulation of the SPI1 T3SS. We deleted pinT in the wild-type and phoQ24 backgrounds and measured hilA and rtsA expression. As shown in Fig. 5, deletion of pinT in the wild-type background slightly increased hilA expression, but had no effect on rtsA expression when these strains were cultured under SPI1-inducing conditions. As expected, constitutive activation of PhoP in the phoQ24 background led to significant repression of both hilA and rtsA (and hilD) (60). Deletion of pinT in the phoQ24 background conferred significant increases in both hilA (∼4-fold) and rtsA (∼2-fold) expression. However, the major effect of phoQ24 on the expression of SPI1 is via transcriptional regulation of hilD, rtsA, and hilA (60). Thus, regulation of hilA and rtsA by PinT is an additional layer on top of the transcriptional repression of the SPI1 T3SS mediated by the PhoPQ two-component system to effect complete and rapid shutoff of the system.
FIG 5.
PinT contributes posttranscriptional regulation to PhoP-mediated repression of SPI1 T3SS expression by regulating hilA and rtsA translation. Shown is β-galactosidase activity in Salmonella strains containing an hilA′-′lacZ (A) or rtsA′-′lacZ (B) translational fusion in the indicated mutant background grown under SPI1-inducing conditions. β-Galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation, where n = 3. P values (unpaired t test) are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant. The strains used are JS2333, JS2341, JS2342, JS2343, JS2334, JS2344, JS2345, and JS2346.
PinT represses motility and fliZ expression in Salmonella.
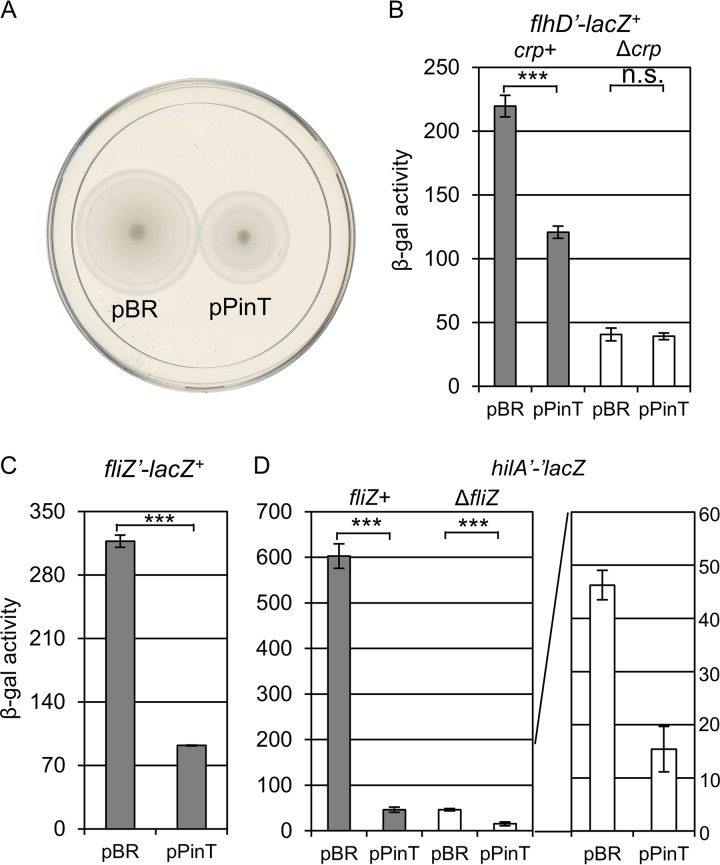
As shown in Fig. 4D, overexpression of PinT represses rtsB expression as part of the rts operon. We have previously shown that RtsB regulates flagellar gene expression by repressing the transcription of flhDC, encoding the master regulator of the flagellar regulon (19, 70, 71). We hypothesized that overexpression of PinT could lead to increased flagellar gene expression by alleviating the repression by RtsB. To test this possibility, we monitored motility in a strain overexpressing PinT compared to the control strain containing the pBRpLac empty vector. In contrast to our initial hypothesis, overexpression of PinT caused an observable motility defect compared to the empty vector control (Fig. 6A). To further investigate the effects of PinT on flagellar genes, we measured expression of flhDC, encoding a master regulator of flagellar genes (72, 73). As shown in Fig. 6B, overexpression of PinT led to decreased flhDC expression. It was shown that PinT represses crp expression presumably via direct base-pairing interaction (33). cAMP receptor protein (CRP) also directly activates flhDC transcription (74–76). We tested whether PinT represses flhDC expression via CRP. Deletion of crp significantly decreased flhDC expression, as expected. Moreover, overexpression of PinT no longer had an effect in this background (Fig. 6B). Thus, we conclude that PinT represses flagellar gene expression through regulation of crp expression.
FIG 6.
PinT regulates motility and flagellar genes in Salmonella. (A) The strains containing pBRpLac or pPinT were applied as spots in motility plates with 0.3% agar. (B to D) β-Galactosidase activity in Salmonella strains containing the flhD′-lacZ+ transcriptional fusion (B), fliZ′-lacZ+ transcriptional fusion (C), or hilA′-′lacZ translational fusion (D) in the indicated background and either the empty vector or plasmids overexpressing PinT. Cells were subcultured 1:100 into 2 ml of HSLB in a 13-mm tube and grown on a roller drum for 4 h. β-Galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation, where n = 3. P values (unpaired t test) are indicated as follows: ***, P < 0.001; n.s., not significant. The strains used are 14028, JS2347, JS2349, JS696, JS2333, and JS2350, each with plasmid pBRplac or pPinT.
Given the effect on flagellar gene expression, we then hypothesized that PinT could also control hilA by affecting expression of the FlhDC-activated flagellar gene fliZ, encoding a major regulator of HilD protein activity (72, 73, 77). We measured the effect of PinT on an fliZ′-lacZ+ fusion in Salmonella (Fig. 6D). As expected, PinT repressed fliZ expression (Fig. 6D). We then compared expression of the hilA′-lacZ+ fusion in wild-type and ΔfliZ backgrounds (Fig. 6E). As shown previously, loss of fliZ resulted in decreased expression of hilA (77). Moreover, the effect of PinT overproduction was reduced from 15-fold in the wild-type background to 3-fold in the ΔfliZ background (Fig. 6E). Thus, PinT-mediated regulation of fliZ expression has a significant role in regulating hilA expression under these conditions.
Impact of PinT on Salmonella virulence in mice.
Westermann et al. suggested that PinT plays a role in timing the transition between SPI1 and SPI2 expression based on global transcriptomic data (33). They noted that the biggest regulatory role for PinT was to decrease SPI2 gene expression and suggested that PinT acts upstream of the primary transcriptional regulator SsrB. PinT regulates the expression of CRP, which could have an indirect regulatory effect on SPI2 T3SS expression (33). However, we observed that the overexpression of PinT also represses a PBAD-ssrB′-′lacZ fusion in E. coli, suggesting that PinT directly regulates SPI2 T3SS expression (see Fig. S4 in the supplemental material). Westermann et al. (33) provided evidence that PinT negatively regulates expression of the SPI1 effectors SopE and SopE2. (Strain 14028 does not encode SopE [78].) However, they did not detect PinT-mediated regulation of the SPI1 apparatus per se. PinT was also previously identified in a large-scale TraDIS screen as being important for intestinal infection in pigs and cattle, but there was no apparent effect of insertions in the sRNA from the same mutational library during systemic infection in mice (79).
To more carefully examine the potential role of PinT during infection, we used oral mouse competition assays (dependent on the SPI1 T3SS) and intraperitoneal (i.p.) infection (bypassing the need for SPI1). To determine whether any observed effects were due to changes in SPI1 expression, we also performed competition assays in an spi1 null background (Δspi1 ΔrtsA). In oral infection, the ΔpinT strain competed equally with the wild-type strain in both the spi1+ and spi1 null backgrounds in the intestine, suggesting that any effects on SPI1 expression are too subtle to be detected in this assay (Table 1). However, the ΔpinT mutant significantly outcompeted the wild type during systemic infections based on competitive index (CI) values in spleen samples after either oral or i.p. infections. This effect was SPI1 independent, as expected. This is perhaps due to increased expression of SPI2 genes in the mutant background (33).
TABLE 1.
PinT affects systemic infection of Salmonella in mice
| Strain Aa | Strain Ba | Infection routeb |
Organb | CIc | P valued | No. of mice |
|---|---|---|---|---|---|---|
| ΔpinT mutant | WT | i.p. | Sp | 2.93 | 0.02 | 6 |
| Oral | SI | 1.28 | NS | 5 | ||
| Sp | 2.34 | 0.03 | 5 | |||
| ΔpinT Δspi1 mutant | Δspi1 | i.p. | Sp | 7.29 | 0.01 | 5 |
| Oral | SI | 1.32 | NS | 5 | ||
| Sp | 4.19 | 0.09 | 5 | |||
The strains used were JS749, JS2358, JS2359, and JS2360.
Bacteria were recovered from the spleen (Sp) after intraperitoneal (i.p.) infections or from the small intestine (SI) and spleen after oral infection.
The competitive index (CI) was calculated as described in Materials and Methods.
Student’s t test was used to compare the CIs to the inocula. NS, not significant (P ≫ 0.05).
DISCUSSION
Salmonella strains sense various environmental niches in the host and adjust their gene expression accordingly during the course of infection. The SPI1 T3SS is induced to initiate intestinal invasion and inflammatory diarrhea (1–4), but once the bacteria are inside the phagosome of epithelial cells or in macrophages during systemic stages of disease, the SPI1 T3SS is no longer required or expressed (80–82). The PhoPQ two-component system, induced and required for survival in the phagosome, appears to be primarily responsible for turning off SPI1 (11, 60, 83). Here we show that the sRNA PinT, transcriptionally induced by PhoP, contributes to this regulatory control of SPI1 and the coordinate regulation of both the flagellar regulon and the SPI2 T3SS.
The PinT sRNA affects SPI1 T3SS expression through three mechanisms. First, PinT directly downregulates hilA translation. Second, PinT represses rtsA translation and induces degradation of the rts mRNA. Third, PinT regulates the expression of flagellar genes via CRP. The cross talk between the SPI1 T3SS and the flagellar regulon affects the dynamics and the expression level of each system (19, 71). The result is decreased expression of FliZ, which leads to decreased HilD activity. PinT also directly blocks translation of SopE (not found in strain 14028 [78]) and SopE2, primary SPI1 T3SS effectors that promote invasion.
The 5′ region of PinT is predicted to fold into a stem-loop with two bulges; the 3′ end also forms a terminator stem-loop structure (33) (Fig. 3). Nucleotides in the 5′ stem-loop of PinT are crucial for base pairing with its target mRNAs (33) (Fig. 3). PinT downregulates both hilA and rtsA, but the mechanisms of regulation are distinct. PinT base pairs near the initiation AUG codon of hilA, leading to direct inhibition of ribosome binding; RNase E degradosome activity is not required for the regulation of hilA translation. On the other hand, PinT base pairs near the ribosome binding site of the rtsA mRNA, causing both translational inhibition and degradation of the entire rts mRNA in an RNase E degradosome-dependent manner. This results in coordinated downregulation of all genes in the rts operon.
Both flagella and the SPI T3SS are expressed in the intestine and then shut off after invasion (15, 80–82, 84–86). The two systems are coordinately regulated via several factors (5, 11, 72, 73). The second gene in the rts operon, rtsB, encodes a negative regulator of flhDC transcription (70). Thus, both PinT and RtsB negatively regulate flagellar gene expression, but PinT negatively regulates RtsB. Loss of RtsB affects the relative timing of flagellar and SPI1 gene expression, but the effect is subtle (71). Certainly, under the conditions used in this study, the negative effect of PinT on flhDC transcription via CRP seems to outweigh the repression of RtsB.
Loss of PinT did not confer any phenotype in the intestine after oral infection. This is presumably due to the subtle posttranscriptional regulatory effect of ΔpinT on hilA expression (Fig. 5A). Moreover, we think that the PhoPQ-mediated transcriptional regulation and PinT-mediated posttranscriptional regulation on the SPI1 T3SS are primarily exerted after the invasion of Salmonella into the host intestinal epithelium, where the PhoPQ becomes active (48, 49, 51, 52). The ΔpinT mutant did outcompete the wild-type strain during systemic stages of infection (Table 1). The advantage in systemic infection is presumably due to increased SPI2 expression in ΔpinT background. Westermann et al. (33) showed that PinT regulates CRP and provided data suggesting that PinT affects the expression of SPI2 in a CRP-dependent manner. In addition, we have shown that PinT directly affects ssrB translation (Fig. S4). Therefore, PinT also regulates the expression of SPI2 via multiple mechanisms.
PinT is encoded immediately downstream of the rtsABCD operon in Salmonella Typhimurium. These genes are located on a horizontally acquired locus near tRNA-PheR (annotated tRNA-PheU in E. coli). This locus in many Salmonella serovars also encodes the acid phosphatase PhoN, which is regulated by PhoPQ, explaining the “Pho” nomenclature (87). The gene repertoire between ydiH (dcuS) and tRNA-PheR is highly variable in various serovars of Salmonella and other Enterobacteriaceae (88–90). Table S1 shows the relative conservation of various genes in some representative Salmonella strains. RtsA and RtsB are highly conserved in all S. enterica serovars as well as the Salmonella bongori species. The genes coding for RtsC and RtsD, for which we do not know the function, are pseudogenes in many serovars. PinT is not found in S. enterica serovar Gallinarum or S. bongori but is 100% conserved in the other serovars.
In summary, the sRNA PinT is a posttranscriptional regulator of three major systems critical for Salmonella virulence: the SPI1 and SPI2 type III secretion systems and the flagellar regulon. PhoPQ is a primary regulator of all of these systems, with PinT adding an additional layer of control to the timing of expression of these factors, allowing for efficient adaptation between niches in the host.
MATERIALS AND METHODS
Strain construction.
Bacterial strains and plasmids are listed in Table S2 in the supplemental material. All Salmonella enterica serovar Typhimurium strains created for this study are isogenic derivatives of strain 14028 (American Type Culture Collection) and were constructed using P22 HT105/1 int-201 (P22)-mediated transduction (91). Deletion of various genes and concomitant insertion of an antibiotic resistance cassette were performed using lambda Red-mediated recombination (92–94). The endpoints of each deletion are indicated in Table S2. In all cases, the appropriate insertion of the antibiotic resistance marker was confirmed by PCR analysis. In each case, the constructs resulting from this procedure were moved into an unmutagenized background by P22 transduction. When appropriate, antibiotic resistance cassettes were removed using the temperature-sensitive plasmid pCP20 carrying the FLP recombinase (95). To create transcriptional lacZ fusions to pinT, the insertion mutation in pinT was converted to a transcriptional lac fusion using FLP/FLP recombination target (FRT)-mediated site-specific recombination, as previously described (94). The translational lacZ reporter fusions were constructed using lambda Red-mediated recombination in E. coli strain PM1205, as described previously (21, 64). To create translational fusions with mutations in the hilA and rtsA 5′ UTR, the nucleotide changes were introduced in the amplifying primers (see Table S3 in the supplemental material). After electroporation of the PCR fragment into PM1205, the recombinants were selected on sucrose minimal plates (M63 salts, 0.2% glycerol, 5% sucrose) containing 40 μg ml−1 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
Plasmid construction and site-directed mutagenesis of sRNA constructs.
The pPinT plasmid was constructed by PCR amplifying pinT (STnc440) from strain 14028 using primers F-AatII-PinT and R-EcoRI-PinT (Table S3). The PCR product was subsequently cloned into the pBR-plac vector after digestion with AatII and EcoRI (64). Various bioinformatics tools (65, 96, 97) were used to predict the region of PinT base pairing with hilA or rtsA mRNA. The QuikChange Lightning site-directed mutagenesis kit (Stratagene) was used to create the corresponding mutant PinT constructs with the primers listed in Table S3.
Media, reagents, and enzymatic assays.
Lysogeny broth medium containing 10 g tryptone, 5 g yeast extract, and 0 g NaCl per liter (designated no-salt LB [NSLB]), 5 g NaCl per liter (low-salt LB [LSLB]), or 10 g NaCl per liter (high-salt LB [HSLB]) was used as indicated. Modified N-minimal medium (pH 5.8) was supplemented with either 10 mM or 10 μM MgCl2 (98). Superoptimal broth with catabolite repression (SOC) was used for the recovery of transformants (91). Bacterial strains were normally grown at 37°C, except for the strains containing the temperature-sensitive plasmid pCP20 or pKD46, which were grown at 30°C. When required, antibiotics were used at the following concentrations: 100 μg ml−1 ampicillin, 20 μg ml−1 chloramphenicol, 50 μg ml−1 kanamycin, 25 μg ml−1 tetracycline, and 50 μg ml−1 apramycin. Primers were purchased from IDT. Enzymes were purchased from New England Biolabs or Invitrogen.
β-Galactosidase assays were performed using a microtiter plate assay as previously described (99) on strains grown under the indicated conditions. β-Galactosidase activity units are defined as (μmol of ortho-nitrophenol [ONP] formed min−1) × 106/(optical density at 600 nm [OD600] × ml of cell suspension) and are reported as mean ± standard deviation with three biological replicates (n = 3) and analyzed statistically using an unpaired t test. Cultures used to measure β-galactosidase activity in Salmonella were grown overnight in NSLB and subcultured 1/100 in 3 ml of HSLB in a 13- by 100-mm tube and grown statically overnight to induce expression of SPI1. Cultures used to measure β-galactosidase activity in E. coli were initially inoculated into LSLB, grown overnight, and subcultured 1/100 in LSLB and grown to an OD600 of 0.5 with 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside) and 0.001% arabinose.
Virulence assays.
Bacteria were initially grown overnight in NSLB and then subcultured 1/35 in 4 ml HSLB in 125-ml flasks and grown for 4 h with aeration at 200 rpm. BALB/c mice (Harlan) (10 to 13 weeks old) were inoculated either orally or intraperitoneally (i.p.) with 0.2 ml of a bacterial suspension. For oral infections, the bacteria were washed and suspended at 5 × 108 CFU (wild-type background) or 109 CFU (spi1 background) per 0.2 ml in sterile 0.1 M sodium phosphate buffer, pH 8.0. Before infection, food and water were withheld for 4 h, and mice were orally inoculated with the indicated number of bacteria, after which the food and water were provided immediately. For intraperitoneal infections, the cells were diluted to 103 CFU per 0.2 ml in sterile phosphate-buffered saline (PBS). For oral infections, mice were sacrificed by CO2 asphyxiation at 3.5 days after infection, and the spleens and small intestines were harvested. For i.p. infection, the mice were sacrificed by CO2 asphyxiation 4.5 days after infection, and spleens were harvested. These organs were homogenized, and serial dilutions of the homogenates were plated on the HSLB plates with appropriate antibiotics to determine the number of CFU per organ. The relative percentage of each strain recovered was determined by replica plating to the appropriate antibiotic-containing HSLB plates. In all competition assays, the inoculum consisted of a 1:1 mixture of two bacterial strains. The actual CFU and relative percentage represented by each strain were determined by direct plating of the inoculum. The competitive index (CI) was calculated as (% strain A recovered/% strain B recovered)/(% strain A inoculated/% strain B inoculated), and Student's t test was used to determine whether the output ratio was significantly different from the input ratio (21).
Ethics statement.
All animal work was reviewed and approved by the University of Illinois Institutional Animal Care and Use Committee (IACUC). Procedures were performed in our AAALAC-accredited facility in accordance with University and PHS guidelines under protocol 18204. All efforts were made to minimize animal suffering.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH grant GM120182 to J.M.S. and C.K.V.
We thank Gary Olsen and Danielle Campbell for assistance with bioinformatic analyses and the Vanderpool lab and Slauch lab members for helpful discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00312-19.
REFERENCES
- 1.Galán JE, Curtiss R. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A 86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou D, Mooseker MS, Galán JE. 1999. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc Natl Acad Sci U S A 96:10176–10181. doi: 10.1073/pnas.96.18.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finlay BB, Heffron F, Falkow S. 1989. Epithelial cell surfaces induce Salmonella proteins required for bacterial adherence and invasion. Science 243:940–943. doi: 10.1126/science.2919285. [DOI] [PubMed] [Google Scholar]
- 4.Wallis TS, Galyov EE. 2000. Molecular basis of Salmonella-induced enteritis. Mol Microbiol 36:997–1005. doi: 10.1046/j.1365-2958.2000.01892.x. [DOI] [PubMed] [Google Scholar]
- 5.Ellermeier JR, Slauch JM. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol 10:24–29. doi: 10.1016/j.mib.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Galan JE. 2001. Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol 17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 7.Galán JE, Collmer A. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 8.Bajaj V, Hwang C, Lee CA. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol 18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 9.Jones BD. 2005. Salmonella invasion gene regulation: a story of environmental awareness. J Microbiol 43:110–117. [PubMed] [Google Scholar]
- 10.Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, Dormann J, Hardt WD. 2011. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog 7:e1002143. doi: 10.1371/journal.ppat.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. 2012. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190:79–90. doi: 10.1534/genetics.111.132779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajaj V, Lucas RL, Hwang C, Lee CA. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol 22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 13.Darwin KH, Miller VL. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J Bacteriol 181:4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichelberg K, Galan JE. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect Immun 67:4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellermeier CD, Ellermeier JR, Slauch JM. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol 57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- 16.Olekhnovich IN, Kadner RJ. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J Bacteriol 184:4148–4160. doi: 10.1128/jb.184.15.4148-4160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olekhnovich IN, Kadner RJ. 2006. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J Mol Biol 357:373–386. doi: 10.1016/j.jmb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Olekhnovich IN, Kadner RJ. 2007. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J Bacteriol 189:6882–6890. doi: 10.1128/JB.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saini S, Ellermeier JR, Slauch JM, Rao CV. 2010. The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella. PLoS Pathog 6:e1001025. doi: 10.1371/journal.ppat.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang L, Feng L, Yang B, Zhang W, Wang P, Jiang X, Wang L. 2017. Signal transduction pathway mediated by the novel regulator LoiA for low oxygen tension induced Salmonella Typhimurium invasion. PLoS Pathog 13:e1006429. doi: 10.1371/journal.ppat.1006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim K, Golubeva YA, Vanderpool CK, Slauch JM. 2018. Oxygen-dependent regulation of SPI1 type three secretion system by small RNAs in Salmonella enterica serovar Typhimurium. Mol Microbiol 111:570–587. doi: 10.1111/mmi.14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez LC, Yakhnin H, Camacho MI, Georgellis D, Babitzke P, Puente JL, Bustamante VH. 2011. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol 80:1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Mouali Y, Gaviria-Cantin T, Sanchez-Romero MA, Gibert M, Westermann AJ, Vogel J, Balsalobre C. 2018. CRP-cAMP mediates silencing of Salmonella virulence at the post-transcriptional level. PLoS Genet 14:e1007401. doi: 10.1371/journal.pgen.1007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaviria-Cantin T, El Mouali Y, Le Guyon S, Römling U, Balsalobre C. 2017. Gre factors-mediated control of hilD transcription is essential for the invasion of epithelial cells by Salmonella enterica serovar Typhimurium. PLoS Pathog 13:e1006312. doi: 10.1371/journal.ppat.1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez-Garrido J, Puerta-Fernandez E, Casadesus J. 2014. A eukaryotic-like 3' untranslated region in Salmonella enterica hilD mRNA. Nucleic Acids Res 42:5894–5906. doi: 10.1093/nar/gku222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schechter LM, Jain S, Akbar S, Lee CA. 2003. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect Immun 71:5432–5435. doi: 10.1128/IAI.71.9.5432-5435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Troxell B, Sikes ML, Fink RC, Vazquez-Torres A, Jones-Carson J, Hassan HM. 2011. Fur negatively regulates hns and is required for the expression of HilA and virulence in Salmonella enterica serovar Typhimurium. J Bacteriol 193:497–505. doi: 10.1128/JB.00942-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song M, Kim HJ, Kim EY, Shin M, Lee HC, Hong Y, Rhee JH, Yoon H, Ryu S, Lim S, Choy HE. 2004. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J Biol Chem 279:34183–34190. doi: 10.1074/jbc.M313491200. [DOI] [PubMed] [Google Scholar]
- 29.Jagodnik J, Brosse A, Le Lam TN, Chiaruttini C, Guillier M. 2017. Mechanistic study of base-pairing small regulatory RNAs in bacteria. Methods 117:67–76. doi: 10.1016/j.ymeth.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Desnoyers G, Bouchard M-P, Massé E. 2013. New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet 29:92–98. doi: 10.1016/j.tig.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 31.De Lay N, Schu DJ, Gottesman S. 2013. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem 288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fröhlich KS, Vogel J. 2009. Activation of gene expression by small RNA. Curr Opin Microbiol 12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 33.Westermann AJ, Forstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Muller L, Reinhardt R, Stadler PF, Vogel J. 2016. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529:496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- 34.Darfeuille F, Unoson C, Vogel J, Wagner E. 2007. An antisense RNA inhibits translation by competing with standby ribosomes. Mol Cell 26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Desnoyers G, Massé E. 2012. Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev 26:726–739. doi: 10.1101/gad.182493.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gottesman S, Storz G. 2011. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massé E, Escorcia FE, Gottesman S. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geissmann TA, Touati D. 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J 23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell 9:23–30. doi: 10.1016/S1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 40.Soper TJ, Doxzen K, Woodson SA. 2011. Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA 17:1544–1550. doi: 10.1261/rna.2767211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vecerek B, Moll I, Afonyushkin T, Kaberdin V, Blasi U. 2003. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol Microbiol 50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 42.Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colgan AM, Kroger C, Diard M, Hardt WD, Puente JL, Sivasankaran SK, Hokamp K, Hinton JC. 2016. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet 12:e1006258. doi: 10.1371/journal.pgen.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srikumar S, Kroger C, Hebrard M, Colgan A, Owen SV, Sivasankaran SK, Cameron AD, Hokamp K, Hinton JC. 2015. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog 11:e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 46.Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, Hinton JC. 2012. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westermann AJ, Venturini E, Sellin ME, Forstner KU, Hardt WD, Vogel J. 2019. The major RNA-binding protein ProQ impacts virulence gene expression in Salmonella enterica serovar Typhimurium. mBio 10:e02504-18. doi: 10.1128/mBio.02504-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalebroux ZD, Miller SI. 2014. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr Opin Microbiol 17:106–113. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia Vescovi E, Soncini FC, Groisman EA. 1994. The role of the PhoP/PhoQ regulon in Salmonella virulence. Res Microbiol 145:473–480. doi: 10.1016/0923-2508(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 50.Gunn JS. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol 16:284–290. doi: 10.1016/j.tim.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 51.Miller SI. 1991. PhoP/PhoQ: macrophage-specific modulators of Salmonella virulence? Mol Microbiol 5:2073–2078. doi: 10.1111/j.1365-2958.1991.tb02135.x. [DOI] [PubMed] [Google Scholar]
- 52.Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A 86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol 50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- 54.Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol 31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 55.Fass E, Groisman EA. 2009. Control of Salmonella pathogenicity island-2 gene expression. Curr Opin Microbiol 12:199–204. doi: 10.1016/j.mib.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcia Vescovi E, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 57.Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol 178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kato A, Tanabe H, Utsumi R. 1999. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J Bacteriol 181:5516–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fields PI, Groisman EA, Heffron F. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 60.Palmer AD, Kim K, Slauch JM. 2019. PhoP-mediated repression of the SPI1 T3SS in Salmonella enterica serovar Typhimurium. J Bacteriol doi: 10.1128/jb.00264-19:JB.00264-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moon K, Gottesman S. 2009. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol Microbiol 74:1314–1330. doi: 10.1111/j.1365-2958.2009.06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin X, Wu Orr M, Wang H, Hobbs EC, Shabalina SA, Storz G. 2018. The small protein MgtS and small RNA MgrR modulate the PitA phosphate symporter to boost intracellular magnesium levels. Mol Microbiol 111:131–144. doi: 10.1111/mmi.14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moon K, Six DA, Lee HJ, Raetz CR, Gottesman S. 2013. Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol Microbiol 89:52–64. doi: 10.1111/mmi.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mandin P, Gottesman S. 2009. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol 72:551–565. doi: 10.1111/j.1365-2958.2009.06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Busch A, Richter AS, Backofen R. 2008. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24:2849–2856. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lopez PJ, Marchand I, Joyce SA, Dreyfus M. 1999. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol Microbiol 33:188–199. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- 67.Rice JB, Vanderpool CK. 2011. The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res 39:3806–3819. doi: 10.1093/nar/gkq1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller SI, Mekalanos JJ. 1990. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol 172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Véscovi EG, Ayala YM, Di Cera E, Groisman EA. 1997. Characterization of the bacterial sensor protein PhoQ: evidence for distinct binding sites for Mg2+ and Ca2+. J Biol Chem 272:1440–1443. doi: 10.1074/jbc.272.3.1440. [DOI] [PubMed] [Google Scholar]
- 70.Ellermeier CD, Slauch JM. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J Bacteriol 185:5096–5108. doi: 10.1128/jb.185.17.5096-5108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saini S, Slauch JM, Aldridge PD, Rao CV. 2010. Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes. J Bacteriol 192:5767–5777. doi: 10.1128/JB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aldridge P, Hughes KT. 2002. Regulation of flagellar assembly. Curr Opin Microbiol 5:160–165. doi: 10.1016/S1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- 73.Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev 64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Komeda Y, Suzuki H, Ishidsu JI, Iino T. 1976. The role of cAMP in flagellation of Salmonella typhimurium. Mol Gen Genet 142:289–298. [DOI] [PubMed] [Google Scholar]
- 75.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181:7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yokota T, Gots JS. 1970. Requirement of adenosine 3′, 5′-cyclic phosphate for flagella formation in Escherichia coli and Salmonella typhimurium. J Bacteriol 103:513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chubiz JE, Golubeva YA, Lin D, Miller LD, Slauch JM. 2010. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar Typhimurium. J Bacteriol 192:6261–6270. doi: 10.1128/JB.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang S, Santos RL, Tsolis RM, Stender S, Hardt WD, Bäumler AJ, Adams LG. 2002. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect Immun 70:3843–3855. doi: 10.1128/iai.70.7.3843-3855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chaudhuri RR, Peters SE, Pleasance SJ, Northen H, Willers C, Paterson GK, Cone DB, Allen AG, Owen PJ, Shalom G, Stekel DJ, Charles IG, Maskell DJ. 2009. Comprehensive identification of Salmonella enterica serovar Typhimurium genes required for infection of BALB/c mice. PLoS Pathog 5:e1000529. doi: 10.1371/journal.ppat.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ibarra JA, Knodler LA, Sturdevant DE, Virtaneva K, Carmody AB, Fischer ER, Porcella SF, Steele-Mortimer O. 2010. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology 156:1120–1133. doi: 10.1099/mic.0.032896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. 2010. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci U S A 107:17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Laughlin RC, Knodler LA, Barhoumi R, Payne HR, Wu J, Gomez G, Pugh R, Lawhon SD, Baumler AJ, Steele-Mortimer O, Adams LG. 2014. Spatial segregation of virulence gene expression during acute enteric infection with Salmonella enterica serovar Typhimurium. mBio 5:e00946. doi: 10.1128/mBio.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bijlsma JJ, Groisman EA. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol Microbiol 57:85–96. doi: 10.1111/j.1365-2958.2005.04668.x. [DOI] [PubMed] [Google Scholar]
- 84.Adams P, Fowler R, Kinsella N, Howell G, Farris M, Coote P, O'Connor CD. 2001. Proteomic detection of PhoPQ- and acid-mediated repression of Salmonella motility. Proteomics 1:597–607. doi:. [DOI] [PubMed] [Google Scholar]
- 85.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47:103–118. [DOI] [PubMed] [Google Scholar]
- 86.Sano G-i, Takada Y, Goto S, Maruyama K, Shindo Y, Oka K, Matsui H, Matsuo K. 2007. Flagella facilitate escape of Salmonella from oncotic macrophages. J Bacteriol 189:8224–8232. doi: 10.1128/JB.00898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kier LD, Weppelman RM, Ames BN. 1979. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J Bacteriol 138:155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hansen-Wester I, Hensel M. 2002. Genome-based identification of chromosomal regions specific for Salmonella spp. Infect Immun 70:2351–2360. doi: 10.1128/iai.70.5.2351-2360.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hensel M. 2004. Evolution of pathogenicity islands of Salmonella enterica. Int J Med Microbiol 294:95–102. doi: 10.1016/j.ijmm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 90.Nieto PA, Pardo-Roa C, Salazar-Echegarai FJ, Tobar HE, Coronado-Arrázola I, Riedel CA, Kalergis AM, Bueno SM. 2016. New insights about excisable pathogenicity islands in Salmonella and their contribution to virulence. Microbes Infect 18:302–309. doi: 10.1016/j.micinf.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 91.Maloy SR, Stewart VJ, Taylor RK. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainville, NY. [Google Scholar]
- 92.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ellermeier CD, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161. doi: 10.1016/S0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 95.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 96.Kruger J, Rehmsmeier M. 2006. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Groisman EA, Kayser J, Soncini FC. 1997. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J Bacteriol 179:7040–7045. doi: 10.1128/jb.179.22.7040-7045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Slauch JM, Silhavy TJ. 1991. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol 173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jarvik T, Smillie C, Groisman EA, Ochman H. 2010. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J Bacteriol 192:560–567. doi: 10.1128/JB.01233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.