To the Editor,
For nearly 3 decades, the human MLL (KMT2A) gene and its rearrangements have been investigated in many different laboratories around the world. At our diagnostic center (DCAL Frankfurt), our standard strategy for the identification of MLL-r is based on two independent approaches, namely “Multiplex” (MP)-polymerase chain reaction (PCR) and “Long distance inverse” (LDI)-PCR approach [1]. The MP-PCR approach is used to rapidly identify the eight most frequent MLL fusions (AF4, AF6, AF9, AF10, ENL, ELL, EPS15, and PTDs) which encompass ~90% of all diagnosed MLL-r leukemia patients, while LDI-PCR is used for all other patients (~10%). By applying both technologies, we have accumulated 94 direct MLL-gene fusions and 247 reciprocal fusion partner genes [2]. Nearly, all breakpoints have been identified in the major breakpoint cluster region (BCR) of the MLL gene (MLL exons 8–14). However, some of the patients remained negative, although they were positively prescreened by various methods.
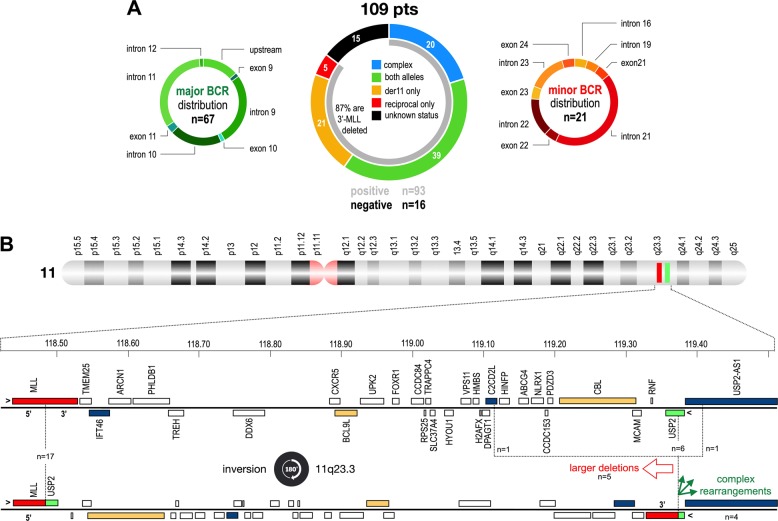
In order to diagnose MLL breakpoints in every patient, a total of 2688 overlapping Illumina capture probes covering the whole-MLL gene were designed and used to analyze a cohort of AL patients (n = 109) where we had either limited (n = 4; PCR positive but not sequenced) or no information (n = 105) on their molecular status. As depicted in Fig. 1a, we identified chromosomal rearrangements in 93 out of 109 patient cases. Sixteen patients remained MLL-r negative and were therefore assigned as patients with “unknown status”. The data analyses of the remaining 93 patients revealed the following distribution: for 67 patients (72%) a breakpoint could be analyzed in the major BCR; 5 patients (5%) displayed only the reciprocal der(TP) with breakpoints in exon 11 (putative CEP83-MLL spliced fusion), intron 11 (n = 3; putative FKBP8-MLL spliced fusion, AF9-MLL, RELA-MLL) and intron 27 (IFT46-MLL), respectively. Surprisingly, an additional 21 patients (23%) had their breakpoints outside of the major BCR, but inside a novel, minor BCR. This novel BCR is localizing between MLL intron 19 and exon 24 (with a clear preference for MLL intron 21–23).
Fig. 1.
Overview about all analyzed patients, their molecular information and breakpoint distribution. a Data from 109 patients which were analyzed with NGS in percentages. Their breakpoint distribution is displayed left (major BCR; n = 67) and right (minor BCR; n = 21). Five patients displayed only a reciprocal fusion, while 16 cases displayed no MLL rearrangement. b Top: chromosome 11 is depicted with highlighting of the MLL (red) and USP2 (green) genes. Below: all the genes between MLL and USP; blue marked genes: additional genes found in this study to be rearranged with MLL; orange marked genes: genes that have been earlier described to be rearranged with MLL. Recombinations between MLL and USP2 are caused by an inversion, with reciprocal alleles that carry additional deletions or complex rearrangements
Most of the new BCR cases represented MLL–USP2 gene fusions (n = 17). USP2 is localized about 1 Mbp telomer to MLL at 11q23.3 and transcriptionally orientated in direction of the centromere of chromosome 11, classifying all these fusions as intrachromosomal inversions (see Fig. 1b). In addition, we identified four balanced translocations in the minor BCR: one patient with an USP8 fusion (see Fig. 2 and Suppl. Figure S1), two with AF4 and one with AF9.
Fig. 2.
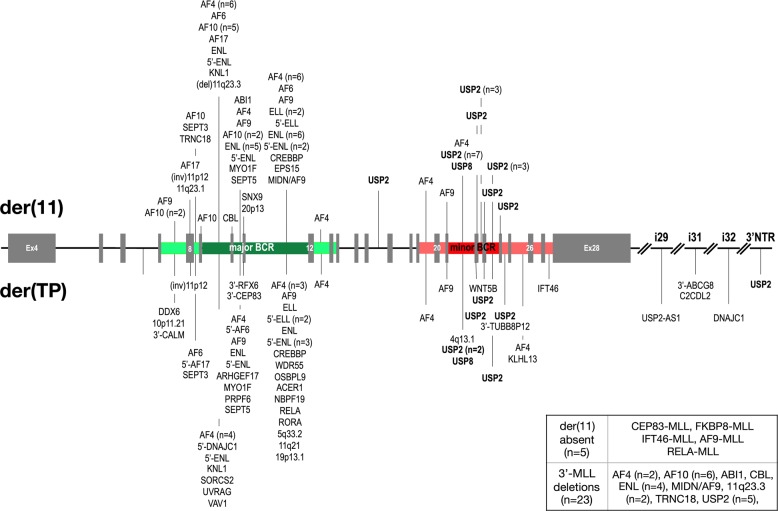
Detailed distribution of all breakpoints in both BCRs of MLL. The MLL gene is depicted from exon 4 to the end. The major BCR is marked in green, the minor BCR in red. Main breakpoint regions are depicted in dark green/red while regions with fewer breakpoints are depicted in light green/red. The fusions sites and the fusion partners are shown. Information about the 5 cases with no der(11) or the 23 cases with 3′-MLL deletions are given in the box at the right bottom
MLL–USP2 and MLL–USP8 alleles seem to be restricted to the minor BCR (see Fig. 2), because they were never diagnosed in association with the major BCR. Most of the reciprocal USP2–MLL fusions were scattered over a larger region at 11q23.3 (see Fig. 2), involving also upstream (C2CD2L) and downstream genes (USP2-AS1). Our analysis revealed also five patients with 3′-MLL deletions that were caused by microdeletions (<200 bp), larger deletions (up to 34 kbp), or complex rearrangements including other chromosomes as well (n = 4; chromosome regions 2p21, 4q13.1, 12p13.33, and 18p11.32). A detailed picture of the investigated MLL–USP2 and MLL–USP8 and their reciprocal fusions is shown in the Suppl. Fig. S2A–D.
All patients with a rearrangement of USP2 or USP8 fused the conserved “UCH-domain” to an extended 5′-MLL portion (see Suppl. Fig. S1A). This may indicate that the UCH domain has a functional importance for the resulting MLL fusion protein. USP genes belong to a large group of deubiquitinating proteins binding to specific target proteins [3–5]. The USP family exhibits a ubiquitin-specific protease (UCH domain) that is characterized by several conserved amino acids that are summarized as CYS- and ASP-box (see Suppl. Fig. 1B). USP2 protein deubiquinates and stabilizes MDM2, leading to an enhanced degradation of p53 [6]. This in turn activates MYC, because active p53 induces the transcription of several microRNAs that target MYC mRNA.
MLL fusions with the conserved 3′-UCH domain of USP2 and USP8 may change profoundly the functions of these novel MLL fusion proteins. It has already been shown that PHD2 [7] and PHD3/BD [8] both bind to proteins (CDC34 and ESCASB2) that mediate the destruction of MLL by poly-ubiquitination and proteasomal degradation. Fusing single or all PHD domains to a der(11) product (MLL-AF9 and MLL-ENL) caused even a strong drop of their transforming potential [9, 10]. This well-described degradation mechanism of MLL may now be counteracted by the UCH domain of MLL–USP2 or MLL–USP8, and thus, restoring their oncogenic transformation capacity.
In our cohort, we also identified new MLL fusion partner genes (n = 3). These novel fusion genes were SNX9 (6q25.3), USP8 (15q21.2), and SEPT3 (22q13.2). SNX9 encodes a protein known to be a member of the sorting nexin family which contain a phosphoinositide binding domain and are involved in intracellular trafficking. The SNX9 protein has a variety of interaction partners, including an adapter protein 2, dynamin, tyrosine kinase nonreceptor 2, Wiskott–Aldrich syndrome-like, and ARP3 actin-related protein 3. USP8 has diverse functions, being required for the internalization of liganded receptor tyrosine kinases and stabilization of ESCRT components. The USP8 protein is thought to regulate the morphology of the endosome by ubiquitination of proteins on this organelle and is involved in cargo sorting and membrane trafficking at the early endosome stage. SEPT3 is the seventh member of the septin family of GTPases that is fused to MLL. Members of this family are required for cytokinesis.
A few cases of MLL–USP2 fusions have already been described. However, these were single patient cases and they were classified as exceptional rearrangements [11–13]. Our NGS approach allowed for the first time the recurrent characterization of breakpoints in this novel minor BCR region of the MLL gene. Moreover, our targeted NGS approach enabled us to overcome the technical limitations associated with LDI-PCR and MP-PCR approaches.
Another advantage of the targeted NGS approach is the simultaneous identification of 3′ MLL deletions or copy number variations. In the current study, 23 of the patients (out of 88: 26%) had a 3′ MLL deletion. According to our data, 3′-MLL deletions were present in both breakpoint groups (major and minor) to a similar extent with 26.9% and 23.8%, respectively. This seems to be much higher than previously described (Andersson et al. [12]: 13%; Peterson et al. [14]: 7%).
In diagnostic fluorescence in situ hybridization analyses, these MLL–USP2 cases revealed two major patterns: (1) loss of the 3′-MLL probe signal, and (2) a normal pattern typical for MLL wild-type (Suppl. Table S1b, Suppl. Fig. S3). Considering the clinical data (Suppl. Table S1a–c), our 17 patients with MLL–USP2 were divided into 8 males and 9 females. All of them were children, and the median age at diagnosis was 17 months (range: 3–120 months). The median leukocyte count was 30.4 × 109/L (range: 3.4–324.0 × 109/L), and the disease phenotype was predominantly B-ALL (n = 12), followed by mixed-phenotype acute leukemia (MPAL) (n = 4) and acute myeloid leukemia (n = 1). The MPAL cases all had mixed myeloid and B-cell phenotype. The patients were treated with diverse therapy protocols. Five patients (29%) presented with central nervous system disease, and 13 patients (76%) had positive-minimal residual disease (MRD) levels at day 33. Prednisone response was measured in 12 patients with a poor response in 5 patients (42%). The median follow-up of the patients was 1.2 years (range: 0.1–11.1 years), and 2 cases died after 5 and 9 months following diagnosis. The remaining patients are still at first clinical remission.
In conclusion, we have identified a minor BCR within the human MLL gene that is recurrently associated in acute leukemia patients with MLL–USP2 fusion alleles as well as MLL fusion partnerships with USP8, AF4, and AF9. However, with 17 cases out of ~2500 analyzed patients the incidence is less than 1% while still ranking fourteenth of our updated fusion gene list (see Table 1 of reference 1). The discovery of a second, minor BCR extends our knowledge of the MLL-recombinome and MLL-r oncogenesis. Moreover, these findings will enable many labs to make changes in their diagnostic set-up for MLL-MRD diagnostics to ensure the best medical treatment for a group of patients that is still very hard to cure.
Supplementary information
Acknowledgements
BAL received a fellowship provided by CAPES and the Alexander von Humboldt Foundation (#88881.136091/2017-01). ME is supported by CNPq (PQ-2017#305529/2017-0) and FAPERJ-JCNE (#26/203.214/2017) research scholarships, and ZZ by grant RVO-VFN64165. GC is supported by the AIRC Investigator grant IG2015 grant no. 17593 and RS by Cancer Australia grant PdCCRS1128727. This work was supported by grants to RM from the “Georg und Franziska Speyer’sche Hochsschulstiftung”, the “Wilhelm Sander foundation” (grant 2018.070.1) and DFG grant MA 1876/12-1.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Claus Meyer, Bruno A. Lopes
Supplementary information
The online version of this article (10.1038/s41375-019-0451-7) contains supplementary material, which is available to authorized users.
References
- 1.Meyer C, Schneider B, Reichel M, Angermueller S, Strehl S, Schnittger S, et al. Diagnostic tool for the identification of MLL rearrangements including unknown partner genes. Proc Natl Acad Sci USA. 2005;102:449–54. doi: 10.1073/pnas.0406994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer C, Burmeister T, Gröger D, Tsaur G, Fechina L, Renneville A, et al. The MLL recombinome of acute leukemias in 2017. Leukemia. 2018;32:273–84. doi: 10.1038/leu.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W, Sulea T, Tao L, Cui Q, Purisima EO, Vongsamphanh R, et al. Contribution of active site residues to substrate hydrolysis by USP2: insights into catalysis by ubiquitin specific proteases. Biochemistry. 2011;50:4775–85. doi: 10.1021/bi101958h. [DOI] [PubMed] [Google Scholar]
- 4.Nishi R, Wijnhoven P, le Sage C, Tjeertes J, Galanty Y, Forment JV, et al. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat Cell Biol. 2014;16:1016–26. doi: 10.1038/ncb3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbé S. Deubiquitylases from genes to organism. Physiol Rev. 2013;93:1289–315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 6.Sacco JJ, Coulson JM, Clague MJ, Urbé S. Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB Life. 2010;62:140–57. doi: 10.1002/iub.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Muntean AG, Hess JL. ECSASB2 mediates MLL degradation during hematopoietic differentiation. Blood. 2012;119:1151–61. doi: 10.1182/blood-2011-06-362079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Muntean AG, Wu L, Hess JL. A subset of mixed lineage leukemia proteins has plant homeodomain (PHD)-mediated E3 ligase activity. J Biol Chem. 2012;287:43410–6. doi: 10.1074/jbc.M112.423855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muntean AG, Giannola D, Udager AM, Hess JL. The PHD fingers of MLL block MLL fusion protein-mediated transformation. Blood. 2008;112:4690–3. doi: 10.1182/blood-2008-01-134056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Santillan DA, Koonce M, Wei W, Luo R, Thirman MJ, et al. Loss of MLL PHD finger 3 is necessary for MLL-ENL-induced hematopoietic stem cell immortalization. Cancer Res. 2008;68:6199–207. doi: 10.1158/0008-5472.CAN-07-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–15. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, et al. St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project. The genetic basis and cell of origin of mixed phenotype acute leukaemia. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:330–7. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander TB, Gu Z, Iacobucci I, Dickerson K, Choi JK, Xu B, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature. 2018;562:373–9. doi: 10.1038/s41586-018-0436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson JF, Baughn LB, Pearce KE, Williamson CM, Benevides Demasi JC, Olson RM, et al. KMT2A (MLL) rearrangements observed in pediatric/young adult T-lymphoblastic leukemia/lymphoma: A 10-year review from a single cytogenetic laboratory. Genes Chromosomes Cancer. 2018;57:541–6. doi: 10.1002/gcc.22666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.