Abstract
Long-lasting changes in synaptic functions after an appropriate stimulus require altered protein expression at the synapse. To restrict changes in protein composition to activated synapses, proteins may be synthesized locally as a result of transmitter receptor-triggered signaling pathways. Second messenger-controlled mechanisms that affect mRNA translation are essentially unknown. Here we report that a receptor for activated C kinase, RACK1, is a component of messenger ribonucleoprotein (mRNP) complexes. RACK1 is predominantly associated with polysome-bound, polyA-mRNAs that are being actively translated. We find it to be present in a complex with β-tubulin and at least two mRNA-binding proteins, polyA-binding protein 1 and a 130 kDa polyA-mRNA binding protein (KIAA0217). Activation of PKCβ2 in vitro by phosphatidylserine/diacylglycerol or in hippocampal slices by metabotropic glutamate receptor stimulation increased the amount of RACK1/PKCβ2 associated with polysome-bound polyA-mRNAs.In vitro, PKCβ2 can phosphorylate a subset of polyA-mRNA-associated proteins that are also phosphorylated underin vivo conditions. On the basis of these findings plus the somatodendritic localization of RACK1, we hypothesize that metabotropic glutamate receptor-triggered binding of activated PKCβ2 to mRNP complexes bound to polyA-mRNAs is involved in activity-triggered control of protein synthesis.
Keywords: translational control, messenger ribonucleoproteins, metabotropic glutamate receptor, protein kinase C, protein phosphorylation, polyA-mRNA, mass spectrometry
Rapid receptor-mediated control of local, postsynaptic protein synthesis is an intriguing mechanism for producing long-lasting changes in the efficiency of synaptic transmission. Moreover, there is now increasing evidence that activity-dependent control of protein synthesis, at the level of translation, is mediated at least partly by metabotropic glutamate receptor-triggered signaling pathways (Huber et al., 2000; Raymond et al., 2000; Job and Eberwine, 2001). These results support earlier findings describing polysome aggregation and loading of fragile X mental retardation protein (FMRP)-mRNA into small polysomes after metabotropic glutamate receptor (mGluR) stimulation in a synaptoneurosomal preparation (Weiler and Greenough, 1993; Weiler et al., 1997). Furthermore, an mGlu- induced increase in translation of a transfected GluR-2-mRNA was observed in isolated dendrites (Kacharmina et al., 2000). The mGluR-triggered signaling pathways responsible for these effects on translation are not known.
Localized changes in mRNA translation near activated synapses could be achieved by at least three different mechanisms: (1) receptor-mediated control of mRNA transport to dendrites, as has been described recently for Arc (Roberts et al., 1998; Steward et al., 1998), (2) receptor-triggered changes in the efficiency of translation of selected mRNAs that are already present in dendrites (Aakalu et al., 2001), or (3) receptor-induced changes in the accessibility of mRNAs by the translational machinery. Because all mRNAs are in complex with a number of proteins, it seems likely that some of those proteins associated with mRNAs are involved in the receptor-triggered mechanisms leading to altered translation. These trans-acting factors, known as messenger ribonucleoproteins (mRNPs), can control not only the secondary structure, stability, transport, and targeting of specific mRNAs (Krecic and Swanson, 1999; Sommerville, 1999; Mohr and Richter, 2001; Schwer, 2001), but they are also involved in translational repression or activation, as has been described for the cytoplasmic polyadenylation element binding protein (Wu et al., 1998). Moreover, those mRNAs that are to be transported out into dendrites might be masked to prevent premature translation in the cell body or inappropriate dendritic locations. A receptor-triggered mechanism that is able to unmask these mRNAs would cause a localized synthesis of the appropriate proteins. The subsequent alteration in the composition of synaptic proteins may then account for the observed long-lasting changes in synaptic function after an appropriate stimulus.
On the basis of the possibility that translational control at synapses leads to a selective rather than a general change in protein synthesis, we focused in this study on mRNPs that could serve as a detector for synapse activity and in response could locally affect the translation of particular mRNAs. Here, we demonstrate that a receptor for activated C kinase, RACK1, is associated with polyA-mRNAs and, furthermore, that in rat hippocampal slices mGluR stimulation controls the movement and binding of RACK1 together with activated protein kinase Cβ2 to mRNA/mRNP complexes.
MATERIALS AND METHODS
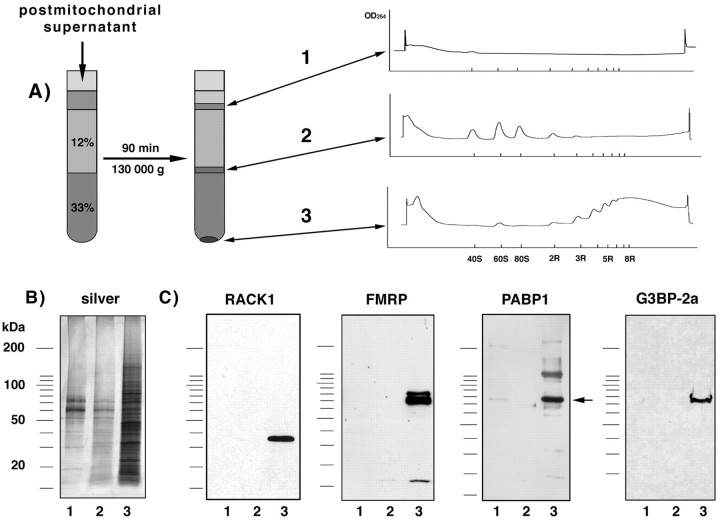
Preparation of mRNPs. Cortex or hippocampal slices of 15- to 19-d-old Long–Evans rats were homogenized in buffer A containing 125 mm NaCl, 100 mm sucrose, 50 mm HEPES, 2 mm potassium acetate, and 40 U/ml of an RNase-inhibitor, RNasin (Promega), and centrifuged for 2 min at 4000 × g (postnuclear supernatant) followed by 10 min at 14,000 × g (postmitochondrial supernatant). The supernatant was lysed (final concentration: 50 mmTris/HCl, pH 7.5, 1% NP40, 50 mm NaCl, 4 mm MgCl2, 45 μg/ml cycloheximide) and layered on a discontinuous sucrose gradient (4.5 ml of 12% sucrose and 4.5 ml of 33.5% sucrose). The tubes were centrifuged for 90 min at 41,000 rpm in a SW41 rotor (Beckman L8–70M centrifuge). To dissociate ribosomes/polysomes and release polyA-mRNA/mRNP complexes, the resulting pellet (monosomes and polysomes; see Fig. 1A) was resuspended in a solution (pellet buffer) containing 30 mm EDTA, 0.5% NP40, 20 mm Tris/HCl, pH 7.5, and kept on ice for 10 min. The interfaces were recovered and centrifuged again for 20 min at 400,000 ×g in a Beckman TL-100 ultracentrifuge. The resulting pellets, which contained mRNPs, free 40S, and 60S ribosomal subunits, and a small subfraction of monosomes (see Fig.1A) were resuspended in the pellet buffer and kept for 10 min at 4°C. The suspension was centrifuged for 2 min at 14,000 × g, and the supernatant was used for both the oligo(dT)-cellulose binding assay and for RACK1 immunoprecipitation (see below). The KCl concentration was adjusted to 200 mm and then 40 μl of prewashed oligo(dT)-cellulose (100 μg/ml; Sigma, St. Louis, MO) was added. After 90 min of constant rotation at 4°C, the cellulose was washed three times with 1 ml wash buffer (20 mmTris/HCl, pH 7.5, 100 mm KCl, 5 mm MgCl2). PolyA-mRNAs and attached mRNPs were eluted with 200 μl of 10 mmTris/HCl, pH 7.5, and proteins were then precipitated with 1 ml of 10% trichloroacetic acid in acetone. Precipitated proteins were washed with acetone and finally solubilized in 70 μl of SDS sample buffer (4% SDS, 250 mm Tris, 50 mmDTT, 3 mm EDTA, 20% glycerol, pH 8.0). The sample was boiled, and 25 μl of aliquot was loaded on a 5–20% polyacrylamide gel.
Fig. 1.
Preparation of mRNPs and detection of RACK1.A, Nontranslated mRNAs were separated from translated mRNAs by discontinuous sucrose density gradient centrifugation. Separation of the resulting interfaces and pellet by a 15–45% continuous sucrose gradient revealed that free mRNA/mRNP complexes and particles smaller than 40S were enriched in the first interface and free ribosomal subunits and monosomes were enriched in the second (12%–33% sucrose density) interface, whereas polysomes were found in the 33% sucrose pellet (right side). B, PolyA-mRNAs were purified from each fraction by binding to oligo(dT)-cellulose and, together with copurified proteins, eluted with 10 mm Tris/HCl, separated by SDS-PAGE, and silver-stained.C, Western blot analysis of the same samples using monoclonal antibodies recognizing RACK1, FMRP, PABP1 (location indicated by an arrow), and G3BP-2a. 1, Fraction 1; 2, fraction 2; 3, fraction 3.
Polysome profile. For each profile, one cortex of a 15- to 19-d-old rat was homogenized in 2 ml of buffer A. The homogenate was first centrifuged for 5 min at 4000 × g, and the supernatant was centrifuged again for 10 min at 14,000 ×g. The resulting postmitochondrial supernatant was then lysed as described above. The lysate was layered on 1.5 ml of 15% sucrose and centrifuged for 11 min at 400,000 × g. The resulting pellet was resuspended in 500 μl of a buffer containing 20 mm Tris/HCl, pH 7.5, 10 mmMgCl2, 100 mmNH4Cl, 2 mm KAc, 225 μg/ml cycloheximide. The suspension was then layered on 10.4 ml of a 15–45% continuous sucrose gradient in 20 mmTris/HCl, pH 9.0, 80 mm NaCl, 3 mm MgCl2, and 0.02% β-mercaptoethanol, and centrifuged for 90 min at 41,000 rpm. In some experiments, the resulting pellets from the discontinuous sucrose spin were used to check the ribosome composition in both fractions (see Fig.1A). The ribosomal subunits, monosomes, and polysomes within the sucrose gradient were detected at a wavelength of 254 nm using an ISCO spectrophotometer. For Western blot analysis, fractions (each 600 μl) were collected and treated with 800 μl of binding buffer (to adjust the final concentration of KCl to 200 mm, 30 mm EDTA, 0.05% NP40, 20 mm Tris/HCl, pH 7.5) and 40 μl of prewashed oligo(dT)-cellulose. PolyA-mRNA/mRNP complexes were purified as described above except that the proteins were eluted from oligo(dT)-cellulose with SDS sample buffer.
RACK1 immunoprecipitation. mRNA/mRNP complexes were released from polysomes on treatment with 30 mm EDTA as described above. After centrifugation the supernatant was adjusted to 20 mm TBS, pH 8.0, 0.5% NP40, and precleared by incubation with 50 μl of anti-IgM agarose (Sigma) or 40 μl of protein A agarose (Santa Cruz Biotechnology, Santa Cruz, CA). For immunoprecipitation, RACK1 monoclonal antibody (mAb) [3 μl, IgM (Transduction Laboratories, Lexington, KY) or IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA)] bound either to 30 μl of anti-IgM-agarose or to 30 μl of protein A-agarose was then added to the precleared supernatant and incubated, with constant agitation, for 2 hr at 4°C.
Samples were washed two times with 1 ml TBS/0.5% NP40 and two times with TBS, and the resulting pellet was boiled for 4 min with 70 μl of solubilizer. Anti-IgM-agarose or protein A-agarose was removed by centrifugation, and the resulting supernatant was used for Western blot analysis. For detection of RACK1-interacting proteins, RACK1-containing protein complexes were released from the anti-IgM-agarose beads after incubation with 60 μl of 0.1 m glycine, pH 3.0. Released proteins were either analyzed directly by mass spectrometry or solubilized and separated by SDS-PAGE and then analyzed by mass spectrometry. Coimmunoprecipitated proteins were cut out and identified by mass spectrometry.
Northwestern blot assay. RACK1 and coimmunoprecipitated proteins were separated by SDS-PAGE and blotted onto nitrocellulose. Detection of putative mRNA-binding proteins was performed as described by Holcik and Liebhaber (1997). Briefly, the nitrocellulose membrane was rinsed twice with PBS, and proteins were renatured by incubation in TNEDD buffer [10 mm Tris/HCl, pH 7.5, 50 mm NaCl, 1 mm EDTA, 1 mm DTT and 1×Denhardt's (0.2 mg/ml bovine serum albumin, 0.2 mg/ml Ficoll, 0.2 mg/ml polyvinylpyrrolidone) solution] three times for 45 min each, followed by a 5 min incubation in a hybridization solution (TNEDD buffer including 20 μg/ml tRNA and 5 μg/ml heparin). Total polyA-mRNA was prepared from rat cortex according to the manufacturer's protocol [Poly(A)Pure, Ambion, Austin, TX], labeled using α-32P ATP and poly(A) polymerase (USB Corp., Cleveland OH), precipitated with ammonium acetate/ethanol, washed, and finally resuspended in 10 mm Tris/HCl, pH 8.0. Labeled polyA-mRNA was added to the hybridization solution (final concentration ∼200,000 cpm/ml), and the nitrocellulose membrane was incubated in this solution for 120 min at room temperature. After a series of washes, the blot was dried, and bound radioactivity was detected by phosphoimaging (Fujix-Bas1000).
Yeast two-hybrid assay. A full-length ORF of RACK1 was amplified from cerebellar cDNA by the PCR, with EcoRI andBamHIII sites added at the end of the 3′ end primer and 5′ end primer, respectively. The construct was inserted into a TOPO-TA vector (Invitrogen) and subsequently subcloned into the pGBKT7 (bait) vector (Clonetech). To get the full-length ORF of PABP1, we amplified the 3′ end (1–707, an internal HindIII sequence) by PCR using a PABP1-pBSK+ vector (a generous gift from Dr. E. Mohr, University of Hamburg, Hamburg, Germany), with EcoRI andHindIII added to the primer. The PABP1-pBSK+ vector was digested with EcoRI and HindIII, and the resulting fragment PABP1–708-1911 was ligated with the PCR product (PABP1-1-707) and inserted into a pGAD424 (prey) vector (Clonetech). Proper orientation of the insert was tested by control digestion withHindIII–BamHI orKpnI–DraIII. Both vectors were transformed into yeast, and a possible direct protein interaction between RACK1 and PABP1 was measured using a β-galactosidase filter assay.
In vitro phosphorylation assay. mRNPs were prepared from the 33.5% sucrose pellet as described above. Washed oligo(dT)-cellulose containing bound proteins was resuspended in kinase buffer (20 mm Tris/HCl, pH 7.5, 10 mmMgCl2, 8% glycerol, 100 mm KCl). For detection of endogenous kinase activity, one aliquot of this sample was treated with 1 μCi 33P-γATP (final ATP concentration 50 μm). For PKCβ2 kinase assay, endogenous kinase activity was first blocked by heating the sample to 65°C for 5 min. Thereupon 40 ng of PKCβ2 (Calbiochem, La Jolla CA) was added, and the reaction started by addition of 1 μCi33P-γATP at 30°C. After 7 min, the reaction was stopped with 1 ml ice-cold wash buffer (20 mmTris/HCl, pH 7.5, 100 mm KCl), and the beads were washed again three times. Oligo(dT)-cellulose bound proteins were released and separated on a 5–20% polyacrylamide gel as described above, and phosphorylated proteins were detected by phosphoimaging (Fujix-Bas1000).
In situ phosphorylation of mRNP. Approximately 40 transverse 400-μm-thick hippocampal slices from 17-d-old rats were prepared and incubated in interface chambers in a medium containing (in mm): NaCl 134.0, KCl 6.24, MgSO4 1.3, CaCl2 2.0, NaHCO3 16, and glucose 10 (pH 7.4, 32°C). The medium was aerated with carbogen (95% O2, 5% CO2) throughout the experiment. After 60 min of preincubation, 200 μCi33Pi (final concentration in the medium 100 μCi/ml; Amersham) was added, and the slices were labeled for 90 min. During the last 30 min of slice incubation, 1 μm okadaic acid was present to inhibit phosphatase activities. Then slices were homogenized in 1.5 ml buffer A, and mRNP was prepared as described above except that 1 μm okadaic acid was added to all buffers.
Identification of mRNPs by mass spectrometry. In-gel digestion and extraction of the proteins was accomplished using a modification of the method of Wilm et al. (1996). Protein bands of interest were excised from the gel, diced into 1 mm2 squares, rinsed with 0.1N NH4HCO3 (AMBIC), dehydrated with acetonitrile (ACN), and dried in a vacuum centrifuge. The proteins were reduced (10 mm DTT in AMBIC, 56°C for 1 hr) and alkylated (55 mm iodoacetamide in AMBIC, room temperature for 45 min). The gel slices were then dehydrated (ACN) and dried (vacuum centrifuge). Gel pieces were then hydrated in a trypsin (sequencing grade, unlyophilized modified; Promega, Madison, WI) solution (12.5 ng/μl) for 45 min on ice. The trypsin solution was then replaced with 10 μl AMBIC. Digests were allowed to run overnight.
Extraction was accomplished by alternating dehydration–hydration steps, collecting, and pooling the solutions resulting from each step as follows. First the gel pieces were dehydrated (50% ACN, 5% formic acid) and dried in the vacuum centrifuge. The gel pieces were then rehydrated (AMBIC) and dehydrated (50% ACN, 5% formic acid) again. This was followed by two more steps of dehydration (100% ACN) and rehydration (AMBIC). The pooled supernatants were reduced to near dryness, reconstituted to 50 μl (1% acetic acid), reduced to near dryness again, and finally reconstituted to 40 μl with 1% acetic acid to give the final solution.
Aliquots (5 μl) of the gel digests were analyzed by nano-reverse phase HPLC-micro-electrospray ionization-mass spectrometry on an LCQ mass spectrometer (MS) (Finnigan, San Jose, CA). Data from ∼1 hr of run time (∼1000 MS/MS spectra per hour) were acquired on each sample. The data were acquired using data-dependent analysis in which one MS scan was followed by collisionally activated dissociation (CAD) of the top five most abundant ions present in the MS scan. Isotope exclusion was used to exclude analysis of isotope ions. Dynamic exclusion was used to reduce the redundancy of the data. Dynamic exclusion settings were repeat count 1, pre-exclude time 30 sec, exclude time 1 min. Filtering software was used to (1) reduce the number of low quality CADs, (2) determine the charge state of the peptides analyzed, and (3) recalibrate the parent mass. Data passing through the above filters were searched against the all nonredundant GenBank [National Center for Biotechnology Information (NCBI)] database using Sequest (Finnigan, San Jose, CA). All reported hits were verified manually.
Immunocytochemistry. Two female and two male 15-d-old Long–Evans rats were anesthetized deeply and perfused transcardially with 10 ml of heparin saline (1000 U/ml), followed first by 50 ml of fixative containing 3.75% acrolein and 2% paraformaldehyde in 0.1m phosphate buffer (PB), pH 7.4, and then with 200 ml of 2% paraformaldehyde in PB. The brains were removed, postfixed in the final fixative for 1 hr, and sectioned at 40 μm on a vibratome. The sections were collected in PB and then treated with 1% sodium borohydride for 30 min to improve antigenicity and reduce nonspecific immunolabeling. To further reduce the nonspecific labeling before incubation in primary antibody, the sections were treated for 30 min in a washing solution consisting of 0.8% BSA and 0.1% fish gelatin in 0.01 m PBS. Sections were incubated in primary anti-RACK1 antibody (1:2000) in washing solution for 24 hr at 4°C. Sections were rinsed in washing buffer and transferred for 30 min to a 1:50 dilution of goat anti-mouse gold-conjugated IgM (Amersham). The gold particles were enlarged by reaction with silver solution from an IntenS-EM kit (Amersham). For electron microscopy the sections were postfixed in 2% osmium tetroxide in 0.1m phosphate buffer, dehydrated, and flat-embedded between a slide and a coverslip that had been treated with liquid release agent (Electron Microscopy Sciences). Ultrathin sections from the CA1 subfield of the hippocampus, somatosensory cortex, and cerebellar cortex were collected on Formvar-coated slot grids, stained with uranyl acetate and lead citrate, and examined with a Philips CM-200 electron microscope.
RESULTS
RACK1 is a component of mRNP complexes associated with translated mRNAs
To characterize proteins that were part of the mRNP complex associated with translated (polysome-bound) or nontranslated (not ribosome-bound) mRNAs, we first separated polysomes from free ribosomal subunits/monosomes by discontinuous sucrose gradient centrifugation (Fig. 1A). The polyA-mRNAs/mRNP complexes were purified using oligo(dT)-cellulose and released by 10 mm Tris, pH 7.5, either for direct mass spectrometry analysis or for SDS-PAGE separation. Proteins, which were first separated by SDS-PAGE, were excised, eluted, and then identified by mass spectrometry or, alternatively, transferred onto nitrocellulose and detected by Western blot assay. With this approach we found, in addition to a number of known mRNA-binding proteins, such as heterogeneous nuclear (hn) RNP-A/B/C/D/E/G/H/K/l/Q3/R/U, HuA/B/C/D/R/, pRENT1, nucleolin, FMRP, staufen, and polyA binding protein 1 (unpublished observations), the RACK1 protein in fractions corresponding to translated mRNAs. The identity of RACK1 was determined from the detection of 13 different peptide sequences (Table1), all of which were specific for RACK1 protein as determined with BLAST2.0 using all nonredundant GenBank databases provided by NCBI. Furthermore, the presence of RACK1 in this fraction was confirmed by Western blot assay and is comparable to the localization of FMRP, which is already known to be associated with polyA-mRNAs in actively translating polysomes (Corbin et al., 1997) (Fig. 1C).
Table 1.
Detected amino acid sequences specific for RACK1 within the protein fraction associated with translated mRNAs
| Protein identity | Amino acid sequence | Residue number |
|---|---|---|
| gi 18543331 | KGHNGWVTQIATTPQFPDMILSASRD | 12–37 |
| gi 18543331 | DKTIIMWK | 37–44 |
| gi 18543331 | DETNYGIPQR | 48–57 |
| gi 18543331 | LWDLTTGTTTR | 89–99 |
| gi 18543331 | DVLSVAFSSDNR | 107–118 |
| gi 18543331 | LWNTLGVCK | 131–139 |
| gi 18543331 | FSPNSSNPIIVSCGWDK | 156–172 |
| gi 18543331 | VWNLANCK | 176–183 |
| gi 18543331 | DGQAMLWDLNEGK | 213–225 |
| gi 18543331 | YWLCAATGPSIK | 246–257 |
| gi 18543331 | IWDLEGK | 258–264 |
| gi 18543331 | KQEVISTSSKA | 271–281 |
| gi 18543331 | VWQVTIGTR | 309–317 |
Total protein coverage 48%.
Although the detection of PABP1 and other well known mRNPs in this fraction confirms the validity of the method used to enrich mRNPs, the simultaneous identification of RACK1 in this fraction was surprising, because RACK1 does not contain any known mRNA binding domains. However, by releasing the entire mRNP complex from polyA-mRNAs, we also might have obtained proteins that were linked to mRNAs via other proteins. One problem of using oligo(dT)-cellulose to purify proteins assumed to be bound to polyA-mRNAs is that a number of proteins bind in a polyA-mRNA-independent manner to oligo(dT)-cellulose, especially in fraction one (Fig. 2). Thus, treatment of the sample before oligo(dT)-cellulose binding with RNase A (10 μg/ml) did not prevent the binding of a number of proteins to the beads (data not shown). To reduce the presence of unspecific bound proteins, we focused therefore only on proteins that can be released from oligo(dT)-cellulose by 10 mm Tris/HCl, pH 7.5. Because the interaction between polyA-mRNA and oligo(dT)-cellulose is impaired in a salt-free buffer, mainly polyA-mRNAs together with associated proteins should be released under this condition. This procedure clearly reduced the presence of proteins that are not part of mRNP complexes, because after treatment with RNase-A, no proteins were present in the released fraction. However, it should be noted that there is not a complete release of polyA-mRNAs under this condition. Thus, mRNA-binding proteins such as PABP1 are still detectable among oligo(dT)-cellulose bound proteins after elution with 10 mm Tris/HCl (data not shown). To further determine whether RACK1 is in fact part of the mRNP complex and was not nonspecifically captured by the tracer cellulose, we added polyA and polyU oligonucleotides to compete with the binding of polyA-mRNAs to oligo(dT)-cellulose (Fig.3A). Furthermore, elution of polyA-mRNAs by 10 mm Tris/HCl from both oligo(dT)-cellulose and polyU-agarose coreleased the RACK1 protein, indicating that RACK1 does not simply bind nonspecifically to cellulose (Fig. 3B). That RACK1 binds in fact via polyA-mRNAs to oligo(dT)-cellulose is supported by the finding that RNase A (10 μg/ml) in the binding buffer completely abolished the binding of RACK1 to oligo(dT)-cellulose (Fig. 3B).
Fig. 2.
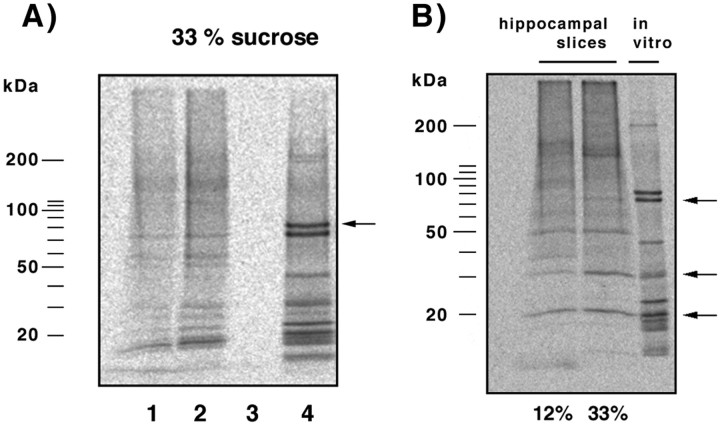
Comparison of oligo(dT)-cellulose bound proteins with proteins that can be eluted with polyA-mRNAs by 10 mm Tris/HCl. A postmitochondrial supernatant was stimulated with PS/DAG, and oligo(dT)-cellulose bound proteins were prepared from both interfaces and the 33% sucrose pellet and released either by SDS-sample buffer (4% SDS, 250 mm Tris, 50 mmDTT, 3 mm EDTA, 20% glycerol, pH 8.0) or by 10 mm Tris/HCl, pH 7.5. Although a number of proteins bind to oligo(dT)-cellulose, only a fraction of them can be eluted together with polyA-mRNAs by 10 mm Tris/HCl (silver stain). Thus, PKCβ2 (indicated by an arrow) binds probably nonspecifically to oligo(dT)-cellulose in fraction 1 because it cannot be released by 10 mm Tris/HCl; in contrast, the kinase can be coreleased with polyA-mRNAs in fraction 3. Comparable with the location of PKCβ2, RACK1 (location shown by anarrow) can be released from oligo(dT)-cellulose by 10 mm Tris/HCl from the fraction corresponding to translated polyA-mRNAs.
Fig. 3.
Binding of RACK1 to oligo(dT)-cellulose is mediated by polyA-mRNAs. A, PolyA and polyU oligonucleotides compete with the binding of RACK1 to oligo(dT)-cellulose. The locations of PABP1 and RACK1 in a silver-stained gel, as identified by mass spectrometry, are indicated by an arrow. Preincubation of oligo(dT)-cellulose with polyA (A) or incubation of the sample with polyU (U) during the binding resulted in decreased binding of a number of proteins, including PABP1 and RACK1, relative to control preparations (C). In contrast, the presence of polyG (G) did not affect the binding of PABP1 and RACK1 to oligo(dT)-cellulose. B, RNase A (10 μg/ml) treatment abolished the binding of RACK1 to oligo(dT)-cellulose. Under control conditions (top,first 3 lanes), RACK1 can be captured with oligo(dT)-cellulose from fraction 3 (Figs. 1, 2), which includes translated mRNAs. Treatment of these fractions with RNase A resulted in a complete absence of RACK1 among the proteins that are bound to oligo(dT)-cellulose (top, last 3 lanes).C, RACK1 can be released with polyA-mRNAs from both oligo(dT)-cellulose (first 3 lanes) and polyU-agarose (last 3 lanes). PolyA-mRNA/mRNP complexes were prepared from each fraction and incubated with oligo(dT)-cellulose or polyU-agarose and released with 10 mm Tris/HCl, pH 7.5, further indicating that RACK1 binds via polyA-mRNA and not nonspecifically to cellulose.
In an additional set of experiments we determined the localization of RACK1 within a 15–45% continuous sucrose gradient. From each fraction the polyA-mRNA/mRNP complexes were again purified using oligo(dT)-cellulose, and the presence of RACK1 and the polyA-binding protein 1 (PABP1) were detected by Western blot assay (Fig.4). In the presence of 3 mmMgCl2 within the sucrose gradient, which is crucial for the ribosome association, RACK1 is present in fractions containing 40S ribosomal subunits and larger components (Fig.4A). Omitting MgCl2 in the sucrose gradient led to a dissociation of ribosomes/polysomes, and therefore no polysomes were detectable. Under this condition, RACK1, comparable to PABP1, could be captured only in the first fraction containing the released polyA-mRNA/mRNP complexes as well as free mRNA/mRNP complexes (Fig. 4B). Therefore, we conclude that RACK1 is part of a protein complex that is associated with polyA-mRNAs.
Fig. 4.
RACK1 is preferentially associated with translated mRNAs. Seventeen fractions of a continuous sucrose gradient (15–45%) were collected, and those containing polyA-mRNAs/mRNP complexes were purified using oligo(dT)-cellulose. The presence of RACK1 and PABP1 (indicated by arrows) was determined by Western blot assay. For that the blot was cut (at ∼55 kDa); the top part was stained for PABP1 and the lower part for RACK1. A, Under the control condition, PABP1 was detectable in all fractions, whereas RACK1 appeared in the fraction that contained 40S and heavier particles (fractions 4-17).B, Absence of Mg2+ within the sucrose gradient caused a complete dissociation of polysomes, and therefore only one peak containing free ribosomal subunits was detectable. Under this condition RACK1 was not detectable in fractions higher than11. The distribution of PABP1 changed in a similar way, except that we could detect PABP1 in fraction 17, which might correspond to translationally arrested RNA granules (Krichevsky and Kosik, 2001).
RACK1 is in a complex with at least two mRNA-binding proteins
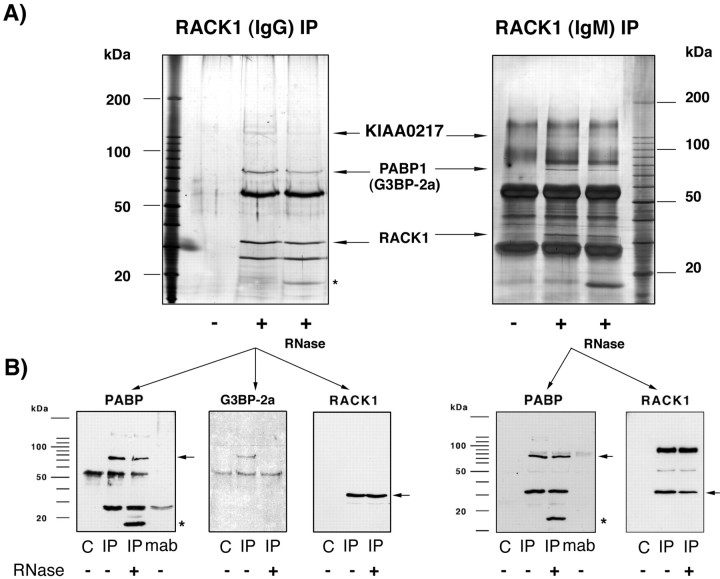
Although RACK1 does not contain any known mRNA binding domains, the protein binds to polyA-mRNAs as well as to RNA homopolymers (data not shown). However, this does not prove a direct interaction of RACK1 with mRNAs. It is also possible that RACK1 is linked by another protein to mRNAs. To search for possible candidates, we prepared the fraction used for the oligo(dT)-cellulose binding assay and performed an immunoprecipitation using a RACK1-specific antibody (clone 20, an IgM; Transduction Laboratories). To identify a possible polyA-mRNA binding protein within the RACK1-containing mRNP complex, we performed a Northwestern blot assay using 32P-labeled total polyA-mRNAs prepared from rat cortex. On the basis of this assay, RACK1 does not bind directly to polyA-mRNAs but is in a complex with at least two mRNA-binding proteins [RACK1-interacting mRNA-binding protein (Rimb)] with apparent molecular weights of 75 kDa (Rimb1) and 130 kDa (Rimb2), respectively (Fig. 5). Rimb1 and Rimb2, which coimmunoprecipitated with two different RACK1-specific monoclonal antibodies [clone 20 (Transduction Laboratories) and clone B-3, an IgG (Santa Cruz Biotechnology)] (Figs.5, 6), were excised from the gel, and the composition was analyzed by mass spectrometry. The Rimb1 band was identified as polyA-binding protein 1 (PABP1), HSP70, and the Ras-GTPase-activating-(GAP-120)-SH3-domain-binding protein 2a (G3BP-2a) (Table 2); the presence of the PABP1 and G3BP-2a within this 75 kDa band was confirmed by Western blot assay (Fig. 6). It remains unclear why G3BP-2a appears in our preparation at 75 kDa given the fact that the calculated molecular weight of G3BP-2a is 54 kDa (Kennedy et al., 1996).
Fig. 5.
RACK1 is in a complex with at least two polyA-mRNA binding proteins. A, To determine whether RACK1 binds directly or via another protein to polyA-mRNAs, a Northwestern blot analysis was performed. RACK1 and coimmunoprecipitated proteins were blotted on nitrocellulose, renatured, and incubated with32P-labeled polyA-mRNAs [−, immunoprecipitation was performed without the RACK1 IgM-mAb (negative control); +, immunoprecipitation with RACK1 IgM-mAb]. Two proteins, with an apparent molecular weight of 75 kDa (Rimb1) and 130 kDa (Rimb2), were found to bind polyA-mRNAs. RACK1, at ∼32 kDa, did not bind polyA-mRNAs in this assay. The presence of RNase A (last lane) did not prevent the interaction of these proteins with RACK1. B, To identify the mRNA-binding proteins, RACK1-IgM coimmunoprecipitated proteins were released from the agarose beads by 0.1 m glycine, pH 3.0, and silver-stained. The 75 kDa (Rimb1) and 130 kDa (Rimb2) bands were cut out and analyzed by mass spectrometry. The Rimb1 protein band contained (1) the polyA-binding protein (PABP1), (2) HSP70, and (3) the Ras-GTPase-activating protein (GAP120) SH3-domain-binding protein 2a (G3BP-2a) (Table 2). The Rimb2 protein band was found to contain the protein KIAA0217, a protein with an RNA-binding domain (Nagase et al., 1996), and a GPI-anchored protein (Table 3).
Fig. 6.
RACK1 coimmunoprecipitates with PABP1.A, RACK1 was immunoprecipitated from the protein fraction that was released from a polysomal pellet by 30 mmEDTA with two different monoclonal antibodies [left side, clone B-3, an IgG2a (Santa Cruz Biotechnology);right side, clone 20, an IgM (Transduction Laboratories)]. The immunoprecipitate was separated using SDS-PAGE and silver-stained. In confirmation of a previous experiment (Fig. 5), the additional protein at 75 kDa (left side,arrow) that was not detectable in the negative control (−, immunoprecipitation protocol using only the protein fraction but without RACK1 mAb) was identified by mass spectrometry as the PABP1 and G3BP-2a, and the 130 kDa protein was identified as KIAA0217 and a GPI-anchored protein. An asterisk indicates the localization of the added RNase A. B, An aliquot of the same sample used for silver staining was blotted onto nitrocellulose and immunostained [left 3 blots correspond to the RACK1-IgG immunoprecipitation (IP); right 2 blots correspond to RACK1-IgM IP]. By confirming the mass spectrometry results, we could detect both PABP1 and G3BP-2a as proteins that coimmunoprecipitate with RACK1 by Western blot assay.C, Negative control (immunoprecipitation without addition of a RACK1-mAb); mab, precipitation of the RACK1-mAb in the absence of the sample. The secondary antibody stained nonspecifically the RNase A (asterisk). The interaction between RACK1 and PABP1 was not abolished by treatment of the immunoprecipitate with 10 U/ml RNase A for 20 min at room temperature. In contrast, there was no coimmunoprecipitation of G3BP-2a when RNase A was added, indicating that both proteins are linked via mRNA to each other. RACK1 staining (arrow) checked the efficiency of the immunoprecipitation.
Table 2.
Identification of Rimb1 (75 kDa) (Fig. 5) that coimmunoprecipitates with RACK1 (clone B-3)
| Identified protein | Amino acid sequence | Residue number |
|---|---|---|
| Ras-GTPase-activating protein SH3-domain binding protein orthologs | ||
| gi 14727423 | AFSWASVTSK | 243–2522-a |
| gi 14727423 | NLPPSGTVSSSGIPPHVK | 253–2702-a |
| gi 6753930 | VLSLNFSECHTK | 65–762-b |
| gi 6753930 | FMQTFVLAPEGSVPNK | 108–1232-b |
| gi 6753930 | FYVHNDMFR | 124–1322-b |
| gi 6753930 | HLEELEEK | 217–2242-b |
| gi 6753930 | VDAKPEVQSQPPR | 245–2572-b |
| gi 6753930 | LPNFGFVVFDDSEPVQR | 338–3542-b |
| gi 6753930 | GIVGGGMMR | 406–4142-b |
| gi 7305075 | EAGEPGDVEPR | 319–3292-c |
| PolyA binding protein (rat): total coverage 28% | ||
| gi 19705459 | FSPAGPVLSIR | 31–41 |
| gi 19705459 | SLGYAYVNFQQPADAER | 51–67 |
| gi 19705459 | ALDTMNFDVIK | 68–78 |
| gi 19705459 | ALYDTFSAFGNILSCK | 114–129 |
| gi 19705459 | GYGFVHFETQEAAER | 139–153 |
| gi 19705459 | MNGMLLNDR | 158–166 |
| gi 19705459 | EFTNVYIK | 189–196 |
| gi 19705459 | NFGEDMDDER | 197–206 |
| gi 19705459 | GFGFVSFER | 232–240 |
| gi 19705459 | YQGVNLYVK | 291–299 |
| gi 19705459 | NLDDGIDDER | 300–309 |
| gi 19705459 | GFGFVCFSSPEEATK | 334–348 |
| gi 19705459 | LFPLIQAMHPSLAGK | 566–580 |
| gi 19705459 | VDEAVAVLQAHQAK | 607–620 |
| gi 19705459 | AVNSATGVPTV | 626–636 |
| Heat shock 70 kDa protein 8 (rat): total coverage 21% | ||
| gi 13242237 | VEIIANDQGNR | 26–36 |
| gi 13242237 | TTPSYVAFTDTER | 37–49 |
| gi 13242237 | NQVAMNPTNTVFDAK | 57–71 |
| gi 13242237 | VQVEYKGETK | 103–112 |
| gi 13242237 | MKEIAEAYLGK | 127–137 |
| gi 13242237 | DAGTIAGLNVLR | 160–171 |
| gi 13242237 | STAGDTHLGGEDFDNR | 221–236 |
| gi 13242237 | FEELNADLFR | 302–311 |
| gi 13242237 | LLQDFFNGK | 349–357 |
| gi 13242237 | LSKEDIER | 510–517 |
Summary of peptides identified by nano-HPLC-μESI-mass spectrometry.
Human similar to Ras-GTPase-activating protein SH3-domain binding protein 2.
Mouse Ras-GTPase-activating (GAPτ) SH3-domain binding protein 2.
Mouse Ras-GTPase-activating protein SH3-domain binding protein.
Rimb2 contains a number of protein sequences that correspond to a protein, KIAA0217, predicted from a partial cDNA D86971 (Nagase et al., 1996) as well as amino acid sequences that correspond to a glycosylphosphatidylinositol (GPI)-anchored membrane protein (Table3). On the basis of the fact that the known protein sequence of KIAA0217 possesses one eukaryotic RNA recognition motif (position aa171–255), we assume that this protein is Rimb2. In addition, characterization of all RACK1 coimmunoprecipitated (clone 20) proteins directly by mass spectrometry confirmed the presence of PABP1, G3BP-2a, and Rimb2 (KIAA0217) in addition to β-tubulin (detected amino acid sequences EIVHLQAGQCGNQIGAK, FPGQLNADLR, IREEYPDR, TAVCDIPPR, IREEYPDR) among proteins that interact with RACK1.
Table 3.
Identification of Rimb2 (130 kDa) (Fig. 5) that coimmunoprecipitates with RACK1 (clone B-3)
| Identified protein | Amino acid sequence | Residue number |
|---|---|---|
| No similarities to reported gene products: KIAA0217 (D86971) | ||
| gi 1504016 | SLPLVQVDEK | 151–1603-a |
| gi 1504016 | YLREEVK | 226–2323-a |
| gi 1504016 | SVQVNGAATELR | 570–5813-a |
| gi 18188796 | AEDLFENR | From RAT EST |
| gi 18188796 | LSSLIIGSSK | From RAT EST |
| gi 18188796 | SLSTDASTNTAPVVVPR | From RAT EST |
| GPI-anchored protein orthologs | ||
| gi 2137361 | ALKEIVER | 179–1863-b |
| gi 2137361 | QFMAETQFSSGEK | 245–2573-b |
| gi 2135305 | QILGVIDKK | 3–103-c |
| gi 2135305 | LDDYQER | 22–283-c |
| gi 2135305 | LNQDQLDAVSK | 35–453-c |
| gi 2135305 | YQEVTNNLEFAK | 46–573-c |
| gi 2135305 | SFMALSQDIQK | 62–723-c |
| gi 2135305 | EQLMREEAEQK | 81–913-c |
| gi 7949103 | DGYQQNFK | 615–6223-d |
Summary of selected peptides identified by nano-HPLC-μESI-mass spectrometry.
Human; no similarities to reported gene products.
Mouse GPI-anchored protein; fragment.
Human GPI-anchored protein p137 precursor.
Mouse GPI-anchored membrane protein 1.
To discern whether RACK1 is in the same protein complex as PABP1, G3BP-2a, and Rimb2 (KIAA0217), a parallel immunoprecipitation was performed in the absence or presence of RNase A (final concentration 10 μg/ml) using the same starting material. To avoid protein precipitation by RNase A treatment, we first performed the immunoprecipitation, washed the beads, and then added RNase A (10 μg/ml) for 20 min at room temperature. After this treatment, we were still able to coimmunoprecipitate Rimb1 and Rimb2 with RACK1, pointing to an mRNA-independent association of RACK1 to these proteins (Fig. 5). Western blot analysis using a specific monoclonal antibody for PABP1 (clone 10E10) or a polyclonal antiserum for G3BP-2a (rabbit poly-576) revealed that only PABP1 is associated with RACK1 in an mRNA-independent manner, whereas G3BP-2a is linked to RACK1 via mRNA (Fig. 6). However, it is not clear whether RACK1 and PABP1 interact physically with each other. Using a direct yeast two-hybrid screen with RACK1 fused to the DNA-binding domain (bait) and PABP1 fused to the activation domain (prey), we could not detect a direct interaction. Therefore, we conclude that RACK1 and PABP1 are together in one protein complex. In summary, we found that RACK1 is in a complex with two mRNA-binding proteins, PABP1 and Rimb2, and in addition is linked to another mRNA-binding protein, G3BP-2a, via mRNA, which further supports the conclusion that RACK1 is a component of mRNP particles associated with poly-mRNAs.
Activation of PKCβ2 leads to an increased association with mRNP complexes
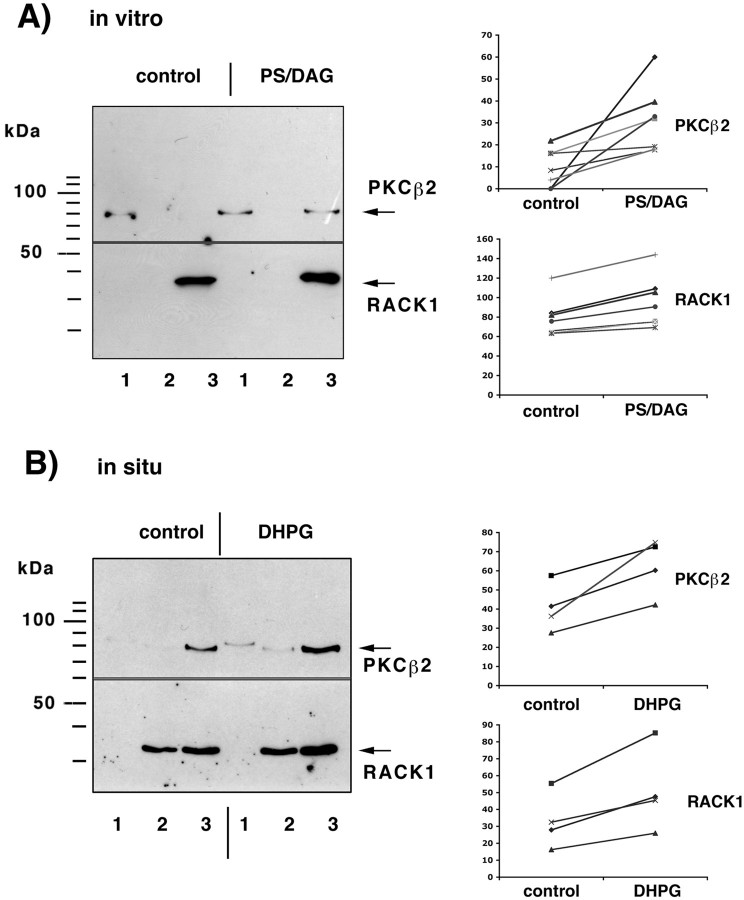
On the basis of these findings, we concluded that RACK1 binds to mRNAs in a complex that contains PABP1 and Rimb2 and might therefore be involved in targeting of two kinases, activated PKCβ2 and src-tyrosine kinase (Ron et al., 1994; Chang et al., 1998), to the mRNP complex. The presence of the src-tyrosine kinase within the mRNP complex could be not detected either by Western blot assay using two different antibodies [anti c-Src (clone H12; Santa Cruz Biotechnology) or anti-pp-60Src (clone GD11; Upstate Biotechnology)] or by mass spectrometry analysis (data not shown). Similarly, under control conditions PKCβ2 could scarcely be detected in fractions corresponding to translated mRNAs (Fig.7). However, after stimulation of PKC by addition of diacylglycerol (DAG) (10 μg/ml) and phosphatidylserine (PS) (50 μg/ml) to the postmitochondrial supernatant, the kinase colocalized with RACK1 and was detectable within the fractions corresponding to translated polyA-mRNAs (Fig. 7A). This binding is isoform specific: PKCβ1 could not be detected in the same fractions (data not shown). To confirm that this redistribution of PKCβ2 and RACK1 may in fact occur under in vivoconditions, we stimulated rat hippocampal slices with 0.1 mm (5)-3,5-dihydroxyphenylglycine (DHPG), an agonist for class 1 mGluR, for 10 min and determined the amount of RACK1 and PKCβ2 bound to polyA-mRNAs. Because of the small amount of starting material, we measured the total amount of RACK1 and PKCβ2 that can be purified with oligo(dT)-cellulose. In hippocampal slices incubated for at least 60 min in an interface chamber, RACK1 was found to be associated with mRNAs enriched from fractions 2 and 3, which correlates with the finding shown in Figures 2 and 4. However, in hippocampal slices incubated for >60 min, PKCβ2 was no longer detectable in fraction 1 but found consistently in fraction 3. This might reflect the previously described Ca2+-dependent redistribution of PKCβ from the cytosol to other compartments during slice incubation (Angenstein et al., 1997). Stimulation of these slices with 100 μm DHPG for 10 min resulted in an increased amount of RACK1 in these fractions. The amount of PKCβ2 increased to a comparable degree in the same mRNP fractions, indicating that activated PKCβ2 and RACK1 both redistribute to the mRNP complex associated with polyA-mRNAs (Fig. 7B).
Fig. 7.
Stimulation of PKCβ2 by phosphatidylserine/diacylglycerol (PS/DAG) causes an association with mRNPs associated with translated mRNAs.A, Activation of PKC by application of phosphatidylserine/diacyglycerol to a postmitochondrial supernatant resulted in an increased amount of RACK1 and PKCβ2 in fraction3 that could be released from oligo(dT)-cellulose by 10 mm Tris/HCl (polysome-bound mRNA/mRNP-complexes).Left, The amount of PKCβ2 and RACK1 associated with mRNAs was determined by Western blot assay; the blot was cut vertically, and the top part used for PKCβ2 staining and the bottom part for RACK1 staining (1, 2, and3 indicate the fraction of nontranslated mRNA/mRNP complexes, mRNA/mRNP complexes associated with 40S-80S ribosomal subunits, monosomes, and mRNA/mRNP particles bound to polysomes) (Fig.1). Right, Summary of the measured densities corresponding to the amount of PKCβ2 and RACK1 in fraction3 from seven independent experiments. B, Stimulation of hippocampal slices with 0.1 mm DHPG for 10 min increases in a comparable manner the amount of RACK1/PKCβ2 in the oligo(dT)-cellulose bound fraction prepared from polysome-bound mRNP complexes (fraction 3).
PKCβ2 can phosphorylate a set of polyA-mRNA bound proteinsin vitro and in situ
To search for putative substrates for PKCβ2, we performed anin vitro phosphorylation assay using the oligo(dT)-cellulose bound proteins prepared from fraction 3, the fraction that showed enhanced RACK1/PKCβ2 association after PS/DAG or DHPG stimulation. In this protein fraction an endogenous protein kinase activity could be detected that could not be substantially diminished by a PKC inhibitor peptide (PKC fragment 19–36) (Fig.8A). Therefore, additional kinases are present in this protein fraction. To inactivate these endogenous kinases, we first incubated the samples for 5 min at 65°C and then performed a PKCβ2 kinase assay. Exogenously added PKCβ2 (40 ng) phosphorylated a set of proteins with molecular weights of 200, 75, 46, 30, 23, 20, 19, and 17 kDa (Fig. 8A). The detected 75 kDa phosphoprotein colocalized exactly with the proteins identified previously by mass spectrometry as PABP1 and G3BP-2a. To determine which one of these proteins was phosphorylated under in vivo conditions, we labeled hippocampal slices with33Pi for 90 min and then prepared the oligo(dT)-cellulose bound fraction. We were able to detect endogenously phosphorylated proteins after electrophoretic separation by autoradiography. Four highly phosphorylated proteins (140, 50, 30, and 20 kDa) and three slightly phosphorylated proteins (75, 150, and 38 kDa) could be identified (Fig. 8B). On the basis of the fact that the 75, 30, and 20 kDa proteins are both excellent in vitro substrates for PKCβ2 and phosphoproteins in vivo, we propose that these proteins are putative endogenous substrates for PKCβ2.
Fig. 8.
PKCβ2 can phosphorylate a subset of polyA-mRNA-binding proteins. A, PolyA-mRNA/mRNP complexes were prepared from polysome-associated mRNAs and used for anin vitro phosphorylation assay. Addition of33P-γATP resulted in phosphorylation of a number of proteins, indicating the presence of endogenous kinases in this fraction (lane 2); lane 1, addition of a PKC inhibitor peptide (PKC fragment 19–36, 2 μg/ml) reduced endogenous kinase activity only slightly, pointing to additional kinases in this fraction; lane 3, heating the sample for 5 min at 65°C completely abolished the endogenous kinase activities;lane 4, addition of 40 ng PKCβ2 (localization of the phosphorylated kinase is indicated by an arrow) led to phosphorylation of a number of proteins, including a 75 kDa protein that comigrates exactly with Rimb1 (PABP1 and G3BP2a).B, Comparison of in situ phosphorylated mRNPs (first 2 lanes) with mRNPs phosphorylatedin vitro by PKCβ2 (lane three). Three endogenous phosphorylated proteins at 75, 30, and 20 kDa (arrows) are also in vitro substrates for PKCβ2 (last lane).
RACK1 is localized in dendrites and within dendritic spines
To further assess whether RACK1-dependent mechanisms might be involved in dendritic protein translation, we examined the cellular distribution of RACK1.
Light microscopic distribution of RACK1
In sections from the rat brain, RACK1 immunolabeling was found by light microscopy to be selectively associated with neuronal perikarya and dendrites in hippocampal CA1 (in both pyramidal neurons and interneurons in stratum radiatum). In somatosensory cortex, neurons in all layers had RACK1-immunopositive somata and prominent staining of apical dendrites (Fig. 9A), and the staining became more sparse and punctate farther away from the neuronal cell body. In cerebellar cortex, the Purkinje cell layer was especially densely labeled by RACK1 antibodies, although Golgi neurons, basket cells, and some granule cells were also positively stained for RACK1. Primary dendrites of Purkinje cells were immunopositive for RACK1, and staining became punctate higher in the molecular layer.
Fig. 9.

Subcellular localization of RACK1.A, Light micrograph illustrating immunogold-silver staining for RACK1 in a pyramidal neuron in somatosensory cortex. Strong immunolabeling is seen in the neuronal soma and the apical dendrite, and punctate staining in the neuropil suggests synaptic localization of RACK1. Scale bar, 50 μm. B,C, Immunogold-silver labeling for RACK1 (black arrows) in the neuronal cytoplasm (B) and the neuronal nucleus (C). In the cytoplasm the gold-silver particles are localized over ribosomal aggregates (B, white arrows). Scale bars:B, 0.5 μm; C, 1 μm. D, RACK1 immunolabeling in a postsynaptic spine (arrow) close to the postsynaptic density in the spine head. The labeled synapse is surrounded by nonlabeled ones (asterisks) in the neuropil of the somatosensory cortex. Scale bar, 0.5 μm.E, Localization of RACK1 in the cytoplasm of a dendrite. Labeling is associated with cisternae in dendritic cytoplasm. Scale bar, 0.5 μm.
Electron microscopic distribution of RACK1
RACK1 localization was studied in somatosensory cortex, in the CA1 subfield of the hippocampus, and in the molecular layer of the cerebellar cortex. In all areas, RACK1 immunolabeling was observed most often in dendrites and in somata, although occasional staining in neuronal nuclei was detected as well (Fig.9B,C). No staining was detected when the RACK1 antibody was omitted. RACK1 immunoreactivity was rarely observed in glial cells. In the soma, RACK1 was frequently colocalized with rough endoplasmic reticulum or polyribosomal clusters in Nissl bodies (Fig. 9B). In large and mid-size dendrites, RACK1 immunolabeling was scattered through the cytoplasm (Fig. 9E) and occasionally associated with cisterns of rough endoplasmic reticulum. Interestingly, dendritic spines were frequently labeled for RACK1, containing one or two immunogold-silver particles per spine (Fig. 9D). These particles were most often localized in the spine head. There was no distinct presynaptic or axonal RACK1 labeling.
DISCUSSION
Receptor-mediated activation of PKCβ2 is followed by a fast redistribution of the kinase from cytosol to other cell compartments such as the cell membrane, Golgi apparatus, or cytoskeleton. Translocation to these cellular compartments is controlled, at least partly, by PKC binding proteins such as RACK1 (Mochly-Rosen et al., 1991), PICK1 (Perez et al., 2001), or AKAP79 (Faux et al., 1999). Our finding that one of these PKC anchoring proteins, RACK1, is also associated with polyA-mRNAs points to mRNP complexes as a putative additional target of activated PKC after receptor stimulation. This, in combination with the finding that PKCβ2 can be detected, in fact, in an mRNP preparation after activation might indicate that this is part of a mechanism that is involved in activity-dependent translational control. This is supported by our observation that RACK1 is localized in dendritic spines where dendritic protein synthesis is known to occur.
Local changes in protein synthesis can be controlled by phosphorylation and subsequent activation/inhibition of (1) translation initiation factors (Clemens, 1996; Whalen et al., 1996), (2) elongation factors (Chang and Traugh, 1997; Marin et al., 1997; Scheetz et al., 1997), or (3) ribosomal proteins (Jefferies and Thomas, 1996). In addition, spatially restricted translation of particular mRNAs may be achieved by a selective alteration in their composition and the properties of proteins that are bound to it. Each mRNA is in a complex with a number of mRNA-binding proteins, which affect their stability/decay, localization/transport, and accessibility to translation initiation (Dreyfuss et al., 2002). The binding of these trans-elements depends on the presence of cis-elements within the mRNA, and therefore different mRNAs might be associated with different sets of mRNPs (Keene, 2001). This specificity might mediate the transport of only a small number of mRNAs into dendrites. Besides a controlled transport into dendrites, the targeting of a subset of mRNAs to activated synapses is an important mechanism for the control of localized protein synthesis. Candidates that are involved in targeting mRNAs into dendrites are Staufen, a somatodendritic localized double-stranded RNA-binding protein that has been shown recently to bind MAP2-mRNA (Kiebler et al., 1999; Monshausen et al., 2001), and zip-code binding protein 1, an actin-mRNA binding protein that is crucial for the movement of this mRNA into growth cones of developing neurons (Zhang et al., 2001). Both proteins are either tubulin-binding protein (Wickham et al., 1999) or appear coincident with microtubules (Zhang et al., 2001), an interaction that seems to be crucial for transporting and localizing mRNA/mRNP complexes. The cis-elements, except for the polyA tail, are stable, and consequently, if the binding of proteins depends solely on the cis-elements present, the composition of mRNA-binding proteins should remain constant. To enable a localized receptor-triggered translation of a specific mRNA, one or more of these mRNPs must be affected locally, for example, by receptor-induced second messenger systems. There are now indications that changes in the phosphorylation of mRNPs, such as hnRNP K, have functional consequences for translation (Habelhah et al., 2001). Our results demonstrate a number of phosphoproteins within the mRNP-complex.
The entire functional mRNP complex consists of primary mRNA-binding proteins and secondary proteins, which are bound to it. If we assume that different proteins can bind, possibly in a competitive way, to atrans-acting factor, this could allow changes in mRNP complex composition, corresponding to the relative concentration or the activation state of these proteins in different compartments within the cell. One of these proteins that could control the composition of the mRNP complex, and subsequently its function, is the RACK1 protein. RACK1, originally described as a receptor for activated C kinase (Ron et al., 1994), contains seven WD-repeat (Try-Asp) domains that are thought to be involved in protein–protein interactions (Neer et al., 1994; Smith et al., 1999). In addition, RACK1 has also been found to interact with the integrin β1 subunit, cAMP-specific phosphodiesterase PDE4D5, p120GAP, NMDA receptor 2B, fyn, and src kinase (Chang et al., 1998; Liliental and Chang, 1998; Yarwood et al., 1999; Koehler and Moran, 2001; Yaka et al., 2002). It is not known how many of these interactions may occur in parallel, but our results here indicate that RACK1 may be in a complex with activated PKCβ2, β-tubulin, PABP1, and a 130 kDa protein (KIAA0217, GPI-anchored protein). How RACK1 binds to PABP1 remains uncertain. The inability to detect a direct interaction of RACK1 with PABP1 using a yeast two-hybrid screen may indicate that the complex needs an additional shared partner such as Rimb2 or possibly that RACK1 needs activated PKCβ2 bound to it to interact with PABP1.
Our results imply that RACK1, like FMRP, is primarily associated with mRNAs engaged in translation. This could indicate that (1) RACK1 is also associated with ribosomes, (2) RACK1 binds to specific mRNA sequences that become accessible only during or after translation initiation, or (3) immediately after binding of RACK1 to the appropriate mRNAs, translation initiation is promoted.
Recently, mGluR-triggered mechanisms have been implicated in the control of protein synthesis (Weiler and Greenough, 1993; Weiler et al., 1997; Merlin et al., 1998; Raymond et al., 2000; Job and Eberwine, 2001), and it has also been shown that stimulation of hippocampal slices by 20 μm DHPG for 30 min causes a small increase in protein synthesis detected by35S-methionine incorporation (Raymond et al., 2000). Although indications for an involvement of group 1 mGluR in the regulation of local protein translation in dendrites, a mechanism probably involved in the maintenance of long-term potentiation (Raymond et al., 2000), long-term depression (Huber et al., 2000), and spine morphology (Vanderklish and Edelman, 2002), are accumulating, little is known about the mediating mechanisms. Group 1 mGluRs (mGluR1 and mGluR5) are coupled in a G-protein-dependent manner to the protein kinase A and protein kinase C phosphorylation systems and, in addition, in a G-protein-independent transduction pathway to the src-tyrosine kinase phosphorylation pathway (Heuss et al., 1999). Interestingly, two of the activated kinases, PKCβ2 and src, are able to bind RACK1. Although this paper focuses on the effect of PKCβ2 binding to mRNA-bound RACK1, we cannot exclude the possibility that the binding of src kinase to RACK1 may also be involved in mGluR-mediated, or more likely in growth factor-mediated, translational control. However, in our preparation we were not able to detect the src-kinase by either Western blot assay or mass spectrometry analysis.
One interesting property of RACK1 is the ability to bind PKCβ2 only if PKCβ2 is in an activated state (Ron et al., 1994), which can thereupon change the cellular distribution (Ron et al., 1999). According to our electron microscopy data, RACK1 is localized in dendrites and within dendritic spines, indicating that RACK1 is in a position to be used for mechanisms affecting dendritic translation. This, in combination with our finding that in hippocampal slices the amount of PKCβ2/RACK1 complexes bound to polyA-mRNAs is increased after mGluR1/5 stimulation, suggests that synaptic activity in fact can alter the mRNP composition in dendrites. Furthermore, at synapses where mGluR1/5 activation leads to an activation of PKCβ2, the activated kinase could not only translocate and bind to mRNP complex-associated polyA-mRNAs, but it could also modify the degree of phosphorylation of other proteins within the mRNP complex. Depending on the function of the affected mRNPs, this could cause (1) an interruption of mRNA transport, (2) a demasking of previously translationally silenced mRNAs, and (3) a spatially restricted change in the efficiency of translation (Fig. 10). Because PKCβ2 activation is triggered by receptor stimulation, the binding should occur only near activated synapses and consequently should only affect mRNAs that are close to these synapses. Therefore, we hypothesize that this mechanism leads to spatially restricted changes in the efficacy of translation by one of the mechanisms mentioned above. Although RACK1 can be in a complex with the PABP1 and therefore all polyA-mRNAs might be controlled by this mechanism, we cannot exclude the possibility that RACK1 binds only to a subset, such that the proposed mechanism affects only the translation of a subset of mRNAs. Indications of that come from the fact that mRNA–PABP1 complexes were also found in fractions that did not contain RACK1, such as fractions 1–3, which correspond to nontranslated mRNAs, and fraction 17 within the sucrose gradient without Mg2+ (Fig. 4), a fraction that very likely contains translationally arrested RNA granules (Krichevsky and Kosik, 2001).
Fig. 10.
Proposed model for an mGluR-mediated control of postsynaptic protein synthesis. 1, A polyA-mRNA/mRNP complex is transported along microtubules through the dendrite in both anterograde and retrograde directions. The mRNA might be masked by specific mRNPs to prevent premature translation. 2, Synaptic stimulation activates protein kinase Cβ2, via class 1 mGluR.3, Activated PKCβ2 and RACK1 form a complex that translocates and binds to a polyA-mRNA/mRNP complex localized near the stimulated synapse. The presence of activated PKCβ2 within the mRNP complex leads to increased phosphorylation of a subset of mRNPs.4, An alteration in the phosphorylation state of mRNPs may trigger (1) an interruption of mRNP transport that would increase the local amount of mRNAs, (2) a demasking of translational arrested mRNAs, leading to activity-dependent synthesis of specific proteins, or (3) a change in translation efficiency of a subset of postsynaptic localized mRNAs, which could shift the ratio of newly synthesized proteins.
The extent to which RACK1 might also be involved in sequestering of mRNAs to specific subcellular domains remains to be determined; an indication of such participation comes from the detection of β-tubulin within the RACK1 coimmunoprecipitated proteins. Interestingly, β1-integrin, a protein that also can interact with RACK1, recruits polyA-mRNAs and ribosomes to focal adhesion complexes by an as yet unknown mechanism (Chicurel et al., 1998). In addition, the reported ability of RACK1 to interact specifically with the NMDA receptor subunit 2B (Yaka et al., 2002) could be an attractive mechanism to localize RACK1/mRNP/mRNA complexes to synapses. The identification of additional RACK1 binding partners among cytoskeletal proteins might provide new mechanisms for the targeting of mRNAs.
Footnotes
This work was supported by the FRAXA-Research Foundation, Grants HD 37175 (W.T.G.), MH 35321 (W.T.G.), and GM 37537 (D.F.H.). We thank Dr. Susan Sesack (University of Pittsburgh, Pittsburgh, PA) for generous advice on immuno-EM methods, Dr. G. Dreyfuss (Howard Hughes Medical Institute and Department of Biochemistry and Biophysics, University of Pennsylvania School of Medicine, Philadelphia, PA) for the PABP1 (clone 10E10) antibody, Dr. D. Kennedy (Griffith University, Brisbane, Australia) for the G3BP-2a staining, Dr. E. Mohr (University of Hamburg, Hamburg, Germany) for the PABP1-pBSK+ vector, and Dr. J. Chen (Cell and Structural Biology, University of Illinois at Urbana/Champaign, Urbana, IL), Dr. C. Seidenbecher, and Dr. U. Thomas (Institute for Neurobiology, Magdeburg, Germany) for their generous help.
Correspondence should be addressed to Dr. Frank Angenstein, Institute for Neurobiology, Brenneckestrasse 6, 39120 Magdeburg, Germany. E-mail:angenstein@ifn-magdeburg.de.
REFERENCES
- 1.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 2.Angenstein F, Hirschfelder M, Staak S. Activation of metabotropic glutamate receptors increases endogenous protein kinase C substrate phosphorylation in adult hippocampal slices. Brain Res. 1997;745:46–54. doi: 10.1016/s0006-8993(96)01129-8. [DOI] [PubMed] [Google Scholar]
- 3.Chang BY, Conroy KB, Machleder EM, Cartwright CA. RACK1, a receptor for activated C kinase and a homolog of the b subunit of G proteins, inhibits activity of src tyrosine kinase and growth of NIH 3T3 cells. Mol Cell Biol. 1998;18:3245–3256. doi: 10.1128/mcb.18.6.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang Y-WE, Traugh JA. Phosphorylation of elongation factor 1 and ribosomal protein S6 by multipotential S6 kinase and insulin stimulation of translational elongation. J Biol Chem. 1997;272:28252–28257. doi: 10.1074/jbc.272.45.28252. [DOI] [PubMed] [Google Scholar]
- 5.Chicurel ME, Singer RH, Meyer CJ, Ingber DE. Integrin binding and mechanical tension induces movement of mRNA and ribosomes to focal adhesions. Nature. 1998;392:730–733. doi: 10.1038/33719. [DOI] [PubMed] [Google Scholar]
- 6.Clemens MJ. Protein kinases that phosphorylate eIF2 and eIF2B, and their role in eukaryotic cell translational control. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1996. pp. 139–172. [Google Scholar]
- 7.Corbin F, Bouillon M, Fortin A, Morin S, Rousseau F, Khandjian EW. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum Mol Genet. 1997;6:1465–1472. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- 8.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the message they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 9.Faux MC, Rollins EN, Edwards AS, Langeberg LK, Newton AC, Scott JD. Mechanism of A-kinase-anchoring protein 79 (AKAP79) and protein kinase C interaction. Biochem J. 1999;343:443–452. [PMC free article] [PubMed] [Google Scholar]
- 10.Habelhah H, Shah K, Huang L, Ostareck-Lederer A, Burlingame AL, Shokat KM, Hentze MW, Ronai Z. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat Cell Biol. 2001;3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 11.Heuss C, Scanziani M, Gahwieler BH, Gerber U. G-protein-independent signaling mediated by metabotropic glutamate receptors. Nat Neurosci. 1999;2:1070–1077. doi: 10.1038/15996. [DOI] [PubMed] [Google Scholar]
- 12.Holcik M, Liebhaber SA. Analysis of mRNP complexes assembled in vitro. In: Richter JD, editor. mRNA formation and function. Academic; San Diego: 1997. pp. 195–209. [Google Scholar]
- 13.Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- 14.Jefferies HBJ, Thomas G. Ribosomal protein S6 phosphorylation and signal transduction. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1996. pp. 389–409. [Google Scholar]
- 15.Job C, Eberwine J. Identification of sites for exponential translation in living dendrites. Proc Natl Acad Sci USA. 2001;98:13037–13042. doi: 10.1073/pnas.231485698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kacharmina JE, Job C, Crino P, Eberwine J. Stimulation of glutamate receptor protein synthesis and membrane insertion within isolated neuronal dendrites. Proc Natl Acad Sci USA. 2000;97:11545–11550. doi: 10.1073/pnas.97.21.11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keene JD. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc Natl Acad Sci USA. 2001;98:7018–7024. doi: 10.1073/pnas.111145598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy D, Wood SA, Ramsdale T, Tam PP, Steiner KA, Mattick JS. Identification of a mouse orthologue of the human ras-GAP-SH3-domain binding protein and structural confirmation that these proteins contain an RNA recognition motif. Biomed Pept Proteins Nucleic Acids. 1996;2:93–99. [PubMed] [Google Scholar]
- 19.Kiebler MA, Hemraj I, Verkade P, Kohrmann M, Fortes P, Marion RM, Ortin J, Dotti CG. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koehler JA, Moran MF. RACK1, a protein kinase C scaffolding protein, interacts with the PH domain of p120GAP. Biochem Biophys Res Commun. 2001;283:888–895. doi: 10.1006/bbrc.2001.4889. [DOI] [PubMed] [Google Scholar]
- 21.Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 22.Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 23.Liliental J, Chang DD. Rack1, a receptor for activated protein kinase C, interacts with integrin β subunit. J Biol Chem. 1998;273:2379–2383. doi: 10.1074/jbc.273.4.2379. [DOI] [PubMed] [Google Scholar]
- 24.Marin P, Nastiuk KL, Daniel N, Girault J-A, Czernik AJ, Glowinski J, Nairn AC, Premont J. Glutamate-dependent phosphorylation of elongation factor-2 and inhibition of protein synthesis in neurons. J Neurosci. 1997;17:3445–3454. doi: 10.1523/JNEUROSCI.17-10-03445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merlin LR, Bergold PJ, Wong RK. Requirement of protein synthesis for group I mGluR-mediated induction of epileptiform discharges. J Neurophysiol. 1998;80:989–993. doi: 10.1152/jn.1998.80.2.989. [DOI] [PubMed] [Google Scholar]
- 26.Mochly-Rosen D, Khaner H, Lopez J. Identification of intracellular receptor proteins for activated protein kinase C. Proc Natl Acad Sci USA. 1991;88:3997–4000. doi: 10.1073/pnas.88.9.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohr E, Richter D. Messenger RNA on the move: implications for cell polarity. Int J Biochem Cell Biol. 2001;33:669–679. doi: 10.1016/s1357-2725(01)00047-4. [DOI] [PubMed] [Google Scholar]
- 28.Monshausen M, Putz U, Rehbein M, Schweizer M, DesGroseillers L, Kuhl D, Richter D, Kindler S. Two rat brain staufen isoforms differentially bind mRNA. J Neurochem. 2001;76:155–165. doi: 10.1046/j.1471-4159.2001.00061.x. [DOI] [PubMed] [Google Scholar]
- 29. Nagase T, Seki N, Ishikawa K, Ohira M, Kawarabayasi Y, Ohara O, Tanaka A, Kotani H, Miyajima N, Nomura N. Prediction of the coding sequences of unidentified human genes. VI. The coding sequences of 80 new genes (KIAA0201-KIAA0280) deduced by analysis of cDNA clones from cell line KG-1 and brain. DNA Res 3 1996. 321 329, 341–354. [DOI] [PubMed] [Google Scholar]
- 30.Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 31.Perez JL, Khatri L, Chang C, Srivastava S, Osten P, Ziff EB. PICK1 targets activated protein kinase Cα to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raymond CR, Thompson VL, Tate WP, Abraham WC. Metabotropic glutamate receptors trigger homosynaptic protein synthesis to prolong long-term potentiation. J Neurosci. 2000;20:969–976. doi: 10.1523/JNEUROSCI.20-03-00969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts LA, Large CH, Higgins MJ, Stone TW, O'Shaughnessy CT, Morris BJ. Increased expression of dendritic mRNA following the induction of long-term potentiation. Mol Brain Res. 1998;56:38–44. doi: 10.1016/s0169-328x(98)00026-6. [DOI] [PubMed] [Google Scholar]
- 34.Ron D, Chen C-H, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. Cloning of an intracellular receptor for protein kinase C: a homolog of the β subunit of G proteins. Proc Natl Acad Sci USA. 1994;91:839–843. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ron D, Jiang Z, Yao L, Vagts A, Diamond I, Gordon A. Coordinated movement of RACK1 with activated βIIPKC. J Biol Chem. 1999;274:27039–27046. doi: 10.1074/jbc.274.38.27039. [DOI] [PubMed] [Google Scholar]
- 36.Scheetz AJ, Nairn AC, Constantine-Paton M. N-methyl-d-aspartate receptor activation and visual activity induce elongation factor-2 phosphorylation in amphibian tecta: a role for N-methyl-d-aspartate receptors in controlling protein synthesis. Proc Natl Acad Sci USA. 1997;94:14770–14775. doi: 10.1073/pnas.94.26.14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwer B. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat Struct Biol. 2001;8:113–116. doi: 10.1038/84091. [DOI] [PubMed] [Google Scholar]
- 38.Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biol Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 39.Sommerville J. Activity of cold-shock domain proteins in translational control. BioEssays. 1999;21:319–325. doi: 10.1002/(SICI)1521-1878(199904)21:4<319::AID-BIES8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 40.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 41.Vanderklish PW, Edelman GM. Dendritic spines elongate after stimulation of group 1 metabotropic glutamate receptors in cultured hippocampal neurons. Proc Natl Acad Sci USA. 2002;99:1639–1644. doi: 10.1073/pnas.032681099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiler IJ, Greenough WT. Metabotropic glutamate receptors trigger postsynaptic protein synthesis. Proc Natl Acad Sci USA. 1993;90:7168–7171. doi: 10.1073/pnas.90.15.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to transmitter activation. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whalen SG, Gingras A-C, Amankwa L, Mader S, Branton PE, Aebersold R, Sonenberg N. Phosphorylation of eIF4E on serine 209 by protein kinase C is inhibited by the translational repressors, 4E-binding proteins. J Biol Chem. 1996;271:11831–11837. doi: 10.1074/jbc.271.20.11831. [DOI] [PubMed] [Google Scholar]
- 45.Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilm M, Schevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 47.Wu L, Wells D, Tay J, Mendis D, Abbott M-A, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmatic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 48.Yaka R, Thornton C, Vagts AJ, Phamloung K, Boncy A, Ron D. NMDA receptor function is regulated by the inhibitory scaffolding protein, RACK1. Proc Natl Acad Sci USA. 2002;99:5710–5715. doi: 10.1073/pnas.062046299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yarwood SJ, Steele MR, Scotland G, Houslay MD, Bolger GB. The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J Biol Chem. 1999;274:14909–14917. doi: 10.1074/jbc.274.21.14909. [DOI] [PubMed] [Google Scholar]
- 50.Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of β-actin mRNP complex increases b-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]