Abstract
Experimental mouse chimeras have proven useful in analyzing the cell lineages of various tissues. Here we use experimental mouse chimeras to study cell lineage of the hippocampus. We examined clonal architecture and lineage relationships of the hippocampal pyramidal cells, dentate granule cells, and GABAergic interneurons. We quantitatively analyzed like-genotype cohorts of these neuronal populations in the hippocampus of the most highly skewed chimeras to provide estimates of the size of the progenitor pool that gives rise to these neuronal groups. We also compared the percentage chimerism across various brain structures to gain insights into the origins of the hippocampus relative to other neighboring regions of the brain. Our qualitative analyses demonstrate that like-genotype cohorts of pyramidal cells are aligned in radial arrays across the pyramidal cell layer, whereas like-genotype cohorts in the C-shaped dentate gyrus colonize either the outer shell or inner core of the granule cell layer in a symmetrical manner. Clonally related populations of GABAergic interneurons are dispersed throughout the hippocampus and originate from progenitors that are separate from either pyramidal or granule cells. Granule and pyramidal cells, however, are closely linked in their lineages. Our quantitative analyses yielded estimates of the size of the progenitor pools that establish the pyramidal, granule, and GABAergic interneuronal populations as consisting of 7000, 400, and 40 progenitors, respectively, for each side of the hippocampus. Last, we found that the hippocampal pyramidal and granule cells share a lineage with cortical and diencephalic cells, pointing toward a common lineage that crosses the di-telencephalic boundaries.
Keywords: dentate gyrus, cell lineage, granule cell, pyramidal cell, GABAergic interneuron, chimera
At the junction where the dorsal telencephalon meets the diencephalon, the hippocampal rudiments form, eventually developing into the dentate gyrus and hippocampus proper (collectively termed the hippocampus in this paper). The hippocampus has been at the forefront of research in the neurosciences, with numerous studies examining its development and how its simple structure subserves the complex functions of learning and memory. However, to our knowledge, the early origins of the hippocampus have never been analyzed thoroughly. Cell lineage analysis is the principal means to explore the cellular dynamics involved in the formation of a tissue. To date, lineage-based dissection of hippocampal development has been limited to one retroviral study showing that clonally related neurons can cross cytoarchitectonic regions within the hippocampus proper (Grove et al., 1992).
In recent years, molecular approaches to understanding mammalian forebrain development have begun to demonstrate distinct organizational patterns of gene expression and function throughout embryogenesis (for review, see Rubenstein et al., 1998). For the hippocampus, this has led to a growing number of genes linked to hippocampal development [Gli3, BETA2/NeuroD, Tlx, and members of the gene families: BMP, LIM, EMX, OTX, WNT, and LEF1/TCF (Boncinelli et al., 1993; Monaghan et al., 1995, 1997;Morita et al., 1995; Pellegrini et al., 1996; Acampora et al., 1997; Furuta et al., 1997; Porter et al., 1997; Suda et al., 1997; Yoshida et al., 1997; Grove et al., 1998; Galceran et al., 1999,2000; Grove and Tole, 1999; Theil et al., 1999; Zhao et al., 1999; Lee et al., 2000; Liu et al., 2000; Tole et al., 2000a,b)]. This evolving molecular picture therefore suggests a unique set of genes defining hippocampal development.
To elucidate the patterned development and lineal origins of the hippocampus, and to complement the emerging molecular organization, we used aggregation and blastocyst-injection chimeras to retrospectively analyze like-genotype cohorts of the three major cell types in the adult mammalian hippocampus. We examined the underlying clonal architecture and lineage relationships of the pyramidal cells, granule cells, and GABAergic interneurons of the hippocampus. We found that the hippocampus proper follows a radial clonal architecture, whereas the dentate gyrus follows an outside-in neurogenetic gradient in its clonal organization. Furthermore, the two sides of the hippocampus arise from distinct progenitor populations. We also demonstrate that the GABAergic interneuronal population arises separately from the principal hippocampal neurons, and their progenitors contribute to the dentate gyrus and hippocampus proper alike. Quantitative analyses of like-genotype cells in the hippocampus provide upper estimates for the size of the progenitor pools of each of the three major hippocampal cell types. Finally, observations of like-genotype cells across brain regions in several chimeras indicate that the hippocampus shares lineage with telencephalic and diencephalic structures. These new insights into the clonal architecture of the hippocampus form a developmental foundation for the emerging molecular events that are critical for understanding hippocampal development and function.
MATERIALS AND METHODS
Aggregation chimeras. Aggregation chimeras were made by a standard method of fusing two four- to eight-cell embryos (Goldowitz et al., 1992). The original stocks of mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and maintained in our colony at the University of Tennessee Animal Care Facility. We used the B6;129S-Gtrosa26 (ROSA26) transgenic mouse line to mark cells in chimeras, because this line has a constitutive expression of β-galactosidase (β-gal) in CNS neurons that permits the analysis of cell lineage (Friedrich and Soriano, 1991; Goldowitz et al., 2000). The other component of chimeras came from either hybrid 129/B6 or ICR mice. The use of two genetically distinct embryos in combination with the ROSA26 embryo to produce chimeras provided added validity to our findings derived from the chimeric combinations. Embryos were harvested from the oviducts of the donor mothers 2 d after the appearance of a vaginal plug. The embryos were then subjected to a light pronase treatment to remove the zonae pellucidae and aggregated and cultured overnight (37°C, 5% CO2) in drops of medium (Mullen and Whitten, 1971) covered with paraffin oil. The following afternoon, successfully fused embryos were transplanted into the uterine horns of pseudopregnant ICR females. Avertin was administered intraperitoneally as the general anesthetic for the ICR host females before transplantation. All surgical procedures and animal care were in accordance with National Institutes of Health guidelines for animal welfare and The Society for Neuroscience policy on the use of animals in research.
Embryonic stem cell, blastocyst-injection chimeras. Blastocyst-injection chimeras were made from an embryonic stem (ES) cell line derived from the H253 transgenic mouse line to mark cells in chimeras. This line has an X-linked lacZ marker on both X chromosomes (Tan et al., 1993; Sturm et al., 1997). Individual host blastocysts were each injected with one ES cell (passages 21–23). Both the ES cells and the host blastocysts were derived from the same F1 strain (C57BL/6 × DBA/2). ES cell-containing blastocysts were then implanted into the uterine horns of pseudopregnant host mothers as described above.
Processing of aggregation chimeras. Six- to 8-week-old chimeras were anesthetized with avertin and transcardially perfused with physiologic saline followed by 4% paraformaldehyde in 0.1m PBS, pH 7.2, for 20 min. The brains were removed and cryoprotected in 30% sucrose in 0.1m PBS and then sectioned in the sagittal plane at 12 μm in a cryostat and mounted on glass slides.
Processing of blastocyst-injection chimeras. Eight- to 16-week-old chimeras were fixed by transcardial perfusion with 4% paraformaldehyde and 0.5% glutaraldehyde in 0.1m Sorensen's phosphate buffer, pH 7.4, with 2 mm MgCl2 and 5 mm EGTA. The brains were then removed and cryoprotected in 30% sucrose after post-fixation for at least 1 hr. Brains were sectioned in the coronal plane at 35 μm thickness using a freezing microtome. Free-floating sections were then processed for histochemistry.
Histochemistry. The ROSA26 strain of mice and H253 ES cells are transgenic for a bacterial lacZ marker gene. All neurons from these transgenic mice and ES cells therefore express β-galactosidase and can be visualized using a histochemical reaction with 5-bromo-4-chloro-3-indolyl B-d-galactopyranoside (X-gal) as the substrate. The β-gal-positive neurons have a blue precipitate in their cytoplasm (Tan et al., 1998; Goldowitz et al., 2000). Nuclear Red was used as a counterstain. For the analysis of GABAergic interneurons, low percentage aggregation and ES cell chimeras were selected for double staining with X-gal and anti-GABA antibodies. Two different antibodies against GABA, ∝GABA KLH [provided by J. G. Hildebrand (University of Arizona) and T. G. Kingan (University of California, Riverside) (Kingan and Hildebrand, 1985)] and ∝GAD67 (Chemicon polyclonal antiserum K2) (Dupuy and Houser, 1996), were used. After incubation in primary antisera, tissues were exposed to an anti-rabbit biotinylated secondary antibody and subsequently visualized using the ABC reaction (Vectastain kit, Vector Laboratories). Double-labeled cells were identified by the brown reaction product of diaminobenzidine along with the blue-colored stain from β-galactosidase. Sections were then mounted and dehydrated through ascending alcohol concentrations before being cleared with xylenes and coverslipped with Permount.
Analysis of cohort size and total cell number. The seven lowest-percentage chimeras were subjected to detailed analysis to detect clonal patterns of development. Serial coronal sections from each chimera were traced using the Neurolucida imaging system (MicroBrightField) for subsequent determination of the area and volume of cell layers and reconstruction of like-genotype cohorts.
Direct counts of all β-gal-positive neurons were made for the pyramidal, granule, and GABAergic neurons in serial sections. To determine the percentage of labeled cells for the pyramidal and granule cell populations, we calculated the total pyramidal and granule cell number for each section by multiplying the area of each individual layer by its packing density (∼4 cells/1000 μm2 for the pyramidal cell layer and 16 cells/1000 μm2 for the granule cell layer). Numerical estimates of hippocampal interneurons were obtained by direct counts of all GABAergic cells across a sampling of the hippocampus. The total number of hippocampal interneurons was estimated by multiplying the average number of GABAergic perikarya per section by the total number of sections through each hippocampus (∼200). This total number was then corrected for perikarya that may have been counted twice using the Abercrombie correction factor (Abercrombie, 1946).
Analysis of lineage relationships. To identify lineage relationships that may exist between the cell populations of the hippocampus and those of other brain regions, the percentages of X-gal-labeled cells for the pyramidal, granule, and GABAergic cells were compared with each other and to rough visual estimates of the percentage chimerism of the following brain regions: motor cortex; somatosensory cortex; piriform cortex; auditory cortex; visual cortex; entorhinal, ectorhinal, and perirhinal cortices; basal ganglia; globus pallidus; substantia nigra; subthalamic nucleus; amygdala; reticular nucleus; zona incerta; nucleus reunions; medial dorsal thalamus; lateral dorsal thalamus; ventral thalamus; habenula; lateral geniculate nucleus; superior colliculus; hypothalamus; mammillary bodies, and subiculum. These estimates were subjected to analysis using the Pearson product moment correlation coefficient of the natural log10. Because some of the data consisted of zero values with which the natural log function cannot be used, a value of 1 was added to each of the estimates before determining the natural log. Relationships were considered significant at p < 0.01.
On the basis of our comparisons of percentage chimerism across various brain regions and the Pearson values obtained above, we made note of the frequency of specific patterns of labeling that we observed in 24 low-percentage chimeras. Specifically, we looked for the frequency of occurrence of four patterns of label: a match between the principal cells of the hippocampus, overlying neocortex, and underlying mediodorsal diencephalon, an exclusive match between the principal cells of the hippocampus and overlying neocortex, an exclusive match between the principal cells of the hippocampus and underlying mediodorsal diencephalon, and a mismatch of the principal hippocampal cells and the above two brain regions.
RESULTS
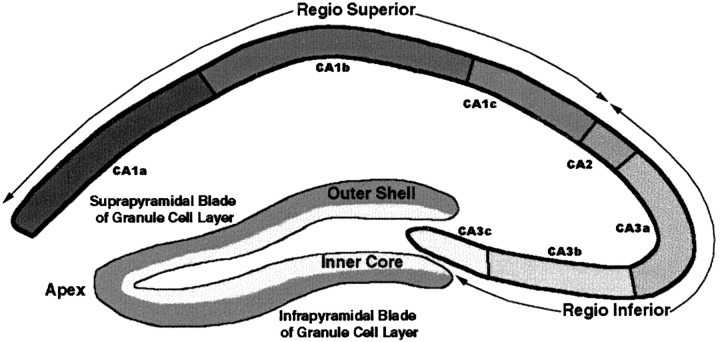
In our lineage analysis of the hippocampal pyramidal cells, dentate granule cells, and GABAergic interneurons, we adhere to the divisions of the hippocampus as depicted in Figure1. The hippocampus proper [cornu ammonis (CA)] is subdivided into CA regions 1 and 2 comprising regio superior and CA region 3 comprising regio inferior (Lorente de No, 1934). The granule cell layer of the dentate gyrus is subdivided into an outer shell and inner core based on the time of origin of the granule cells (Altman and Bayer, 1990b).
Fig. 1.
Illustration of the hippocampus outlining the divisions of the hippocampus proper and dentate gyrus based on the original description by Lorente de No (1934).
Brain sections from adult, chimeric mice were analyzed after X-gal histochemistry. A total number of 21 aggregation and 22 blastocyst-injection mice were found to have mixtures of β-gal-positive and -negative neurons in the hippocampus. Six of the 21 aggregation chimeras and 16 of the 22 blastocyst-injection chimeras were determined to have a low percentage of β-gal-positive cells in the hippocampus (<20%; see Materials and Methods). Most of our analyses focused on these low-percentage chimeras. Of the 22 low-percentage chimeras, 9 were studied in detail (Table1).
Table 1.
Estimated percentages of β-gal-positive cells in the hippocampus of nine low-percentage chimeras
| Chimera number1-a | Pyramidal cells | Granule cells | Interneurons1-b |
|---|---|---|---|
| 1R | 0.5 | 0 | 2.6 |
| 1L | 0.5 | 0 | 2.4 |
| 2R | 0.5 | 0 | 4 |
| 2L | 1.5 | 0.5 | 2.5 |
| 3R | 1.5 | 0.5 | 0 |
| 3L | 0.5 | 0 | 0 |
| 4R | 7.5 | 3 | 0 |
| 4L | 0.5 | 0 | 4.1 |
| 5R | 0 | 0 | 0 |
| 5L | 0.5 | 0 | * |
| 6R | 18 | 12 | * |
| 6L | 3.5 | 2 | * |
| 7R | 2 | 7 | * |
| 7L | 2 | 8 | * |
| 8R | 3.5 | 5 | * |
| 8L | 1 | 0.5 | * |
| 9R | 13 | 33 | * |
| 9L | 50 | 70 | * |
Each number represents a separate chimera. R, Right hemisphere; L, left hemisphere.
Interneuron percentages are based on direct counts versus estimates for pyramidal and granule cell populations. Only chimeras with the lowest-percentage chimerism of interneuron populations were counted.
indicates not counted.
Qualitative examination of chimeras
Observations of the hippocampal pyramidal cells
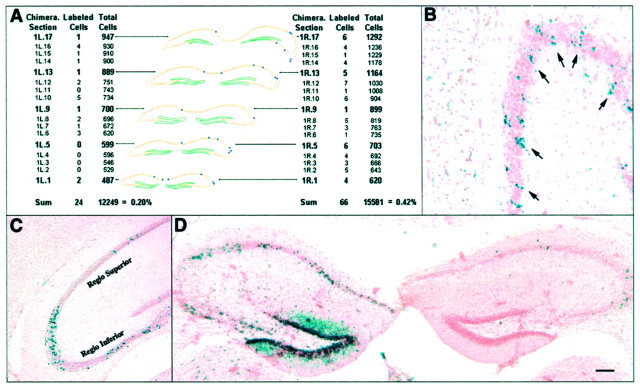
Like-genotype cohorts of hippocampal pyramidal cells were examined to determine clonal relationships within this cell population, as well as lineage relationships that may exist with other hippocampal cell populations. Through this analysis, the following three observations were made. The first observation was that small cohorts of pyramidal cells were always found in spatially restricted clusters that were aligned in the radial dimension (Fig.2B). These cohorts varied in their septal–temporal spread, with the largest clusters spanning the entire hippocampal axis. The spatial extent of like-genotype clusters in the mediolateral domain, as determined from serial reconstruction of clusters, was also dependent on the size of the cohort. As might be expected, the spatial extent of the larger cohorts was increased in chimeras with higher percentages of “blue” cells. A three-dimensional reconstruction of our three smallest clusters in two different chimeras containing an average of 25 cells per cluster revealed that the septotemporal spread of labeled pyramidal cells covered ∼500 μm, but was far more restricted in the mediolateral dimension (Fig. 2A). This septotemporal spread of labeled pyramidal cells represents ∼0.5 mm of the total 7 mm of the septotemporal axis (as determined in measurements of whole dissected hippocampi).
Fig. 2.
Observations of the patterns of X-gal-labeled hippocampal pyramidal cells. A, Three-dimensional reconstruction of the hippocampus of a low-percentage chimera (chimera 1) illustrating the spatially restricted clustering of like-genotype cohorts of pyramidal cells across a series of coronal sections (blue circles represent individual labeled neurons). The most anterior section is at the front/bottom of the reconstruction. Thetables to the left andright show the number of X-gal-labeled cells and the estimated total number of pyramidal cells in each analyzed section. The overall percentage of X-gal-labeled cells of the total pyramidal cell population is shown at the bottom of eachtable. B, Example of radial arrays of X-gal-labeled cells across the hippocampal pyramidal cell layer of chimera 13 (arrows). C, Example of cell-mixing across cytoarchitectonic boundaries of regio inferior and regio superior in the pyramidal cell layer of chimera 14.D, Example of the asymmetrical colonization of cells of the hippocampus shown in this section of chimera 11. Scale bars: B, 70 μm; C,D, 135 μm.
The second observation was that most cohorts were found in either regio superior or regio inferior and seemed to comply for the most part with the major cytoarchitectonic boundaries of the hippocampus. However, we found some examples of small and isolated clusters of blue pyramidal cells that crossed cytoarchitectonic boundaries. This is illustrated in Figure 2C, in which a cluster of like-genotype cells is superimposed on Lorente de No's divisions of the hippocampus (Lorente de No, 1934). In this example, a single cluster of β-gal-positive cells from a low-percentage chimera was found to spread over the boundary between regio superior and regio inferior. Another example is illustrated in the three-dimensional reconstruction of the left hippocampus of chimera 1, in which labeled cells could be found in both regio superior and regio inferior (Fig. 2A). This particular example is significant because the labeled cells in this hippocampus may be derived from a single clone (see quantitative observations, below).
The third observation was that the colonization of pyramidal cells in the two sides of the hippocampus was asymmetrical. This is illustrated by several of our chimeras that demonstrate a large mismatch in hippocampal labeling patterns in opposing sides. Although slight variation in labeling patterns across hemispheres can be attributed to random allocation of cells from a common precursor pool, large and frequent mismatches in the colonization of each side of the hippocampus are indicative of separate lineages. In fact, six of the nine chimeras analyzed in detail had threefold or greater differences in the percentages of β-gal-positive cells that colonized each side of the hippocampus (Figs. 2D, 5A,B, Table 1).
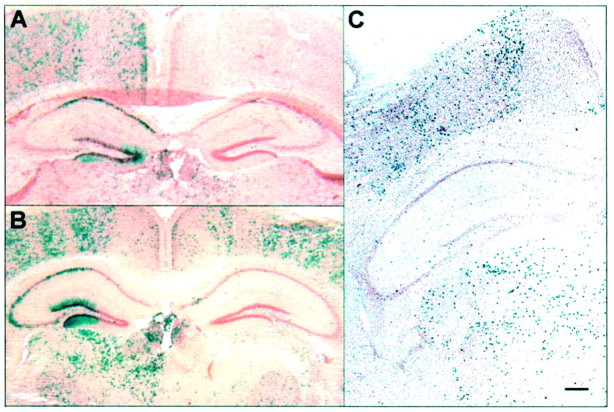
Fig. 5.
Representative chimeras illustrating labeling patterns between the hippocampus and other brain structures. A, Example of one chimera with a similar percentage of marked cells between the hippocampus and overlying neocortex. B, Example of a chimera with a similar percentage of marked cells between the hippocampus and underlying mediodorsal diencephalon. C, Example of a chimeric half-brain with a very small percentage of marked cells in the hippocampus, but a moderate percentage of marked cells in both the overlying neocortex and underlying mediodorsal diencephalon. Scale bar:A, B, 235 μm; C, 120 μm.
Observations of the dentate granule cells
The clonal architecture of the dentate granule cells was examined in a manner similar to the hippocampal pyramidal cell population. Four observations about lineage relationships in the dentate gyrus were made. The first observation was that in all 22 of our low-percentage chimeras, cohorts of blue neurons clearly divided the dentate granule cell layer into an outer shell and inner core. In fact, in some of our lowest-percentage chimeras, the outer shell contained cells from the β-gal-positive lineage, whereas the inner core contained no labeled cells or vice versa (Fig.3A,B). Interestingly, in all low-percentage blastocyst-injection chimeras, labeled granule cells consistently and preferentially colonized the inner core, whereas in low-percentage aggregation chimeras, cells from the β-gal-positive lineage were found in either the inner core or outer shell of the dentate gyrus. This preferential colonization may be attributable to a late incorporation of the embryonic stem cells into the inner cell mass of the chimera or to intrinsic genetic differences between the cell types used in this chimeric combination. It is known that the outer shell of the granule cell layer forms before the inner core (Altman and Das, 1965; Angevine, 1965; Bayer, 1980; Altman and Bayer, 1990b). We have found similar differences in aggregation chimeras made between Mus musculus and Mus caroli, presumably reflecting a difference in developmental timing (Goldowitz, 1989). This unique pattern of colonization in the dentate gyrus between blastocyst-injection and aggregation chimeras was the only notable difference in the clonal architecture of the hippocampus between these chimera types.
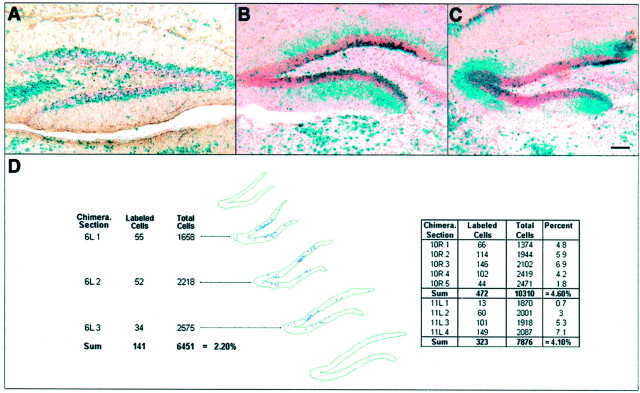
Fig. 3.
Observations of the patterns of X-gal-labeled dentate granule cells. A, Example of a chimera (chimera 12) with many X-gal-labeled cells in the outer shell of the dentate granule cell layer and very few X-gal-labeled cells in the inner core.B, Example of a chimera with many X-gal-labeled cells in the inner core of the dentate granule cell layer and no X-gal-labeled cells in the outer shell (chimera 6). The diffuse bluelabel in the molecular layer is associated with the dendrites of labeled granule cells. This is a characteristic of the LacZ reaction product in the H253 transgenic line used for blastocyst-injection chimeras. C, Example of a mirrored-image pattern of like-genotype granule cells in the inner core of each blade of the dentate gyrus. In this chimera (chimera 9), X-gal-labeled granule cells in the apex and blade tips of the granule cell layer form mirrored-images of each other. D, Three-dimensional reconstruction of the left dentate gyrus of a low-percentage chimera (chimera 6) demonstrating the spatial restrictions of like-genotype cohorts of granule cells. Thetable on the left shows the number of X-gal-labeled cells and the estimated total number of granule cells in each analyzed section. The overall percentage of X-gal-labeled cells of the total granule cell population is shown at thebottom. The percentage of X-gal labeling of the total granule cell population for similar segments of the dentate gyrus of two other low-percentage chimeras (chimeras 10 and 11) is shown in thetable on the right. Scale bar (A–C): 110 μm.
The second observation of the granule cells was that β-gal-positive cells of the inner core of one blade formed a mirrored image of the opposite blade (Fig. 3C). In the chimera illustrated in Figure 3C, several β-gal-labeled cells are observed in the tips of each blade as well as in the apex of the dentate. In other chimeras, the specific locus of blue cells differed (such as labeled cells in the middle of each blade, and few or no labeled cells in the apex or blade tips; like-genotype cells in the blade tips only; labeled cells only in the apex), but each blade consistently mirrored the pattern of like-genotype cohorts of the other.
The third observation was that like the hippocampal pyramidal cell populations, the granule cells of each side of the dentate gyrus appeared to originate from distinct progenitor pools. This is made evident by the dramatic mismatches observed between each side of the dentate gyrus in several chimeras (Figs. 2D,5A,B, Table 1).
Finally, the last observation relative to the granule cells was the septotemporal and mediolateral spread of like-genotype neurons. Within the septotemporal domain, three-dimensional reconstruction of our smallest cohort cluster, containing an average of 35 granule cells per section, showed the spread of like-genotype cohorts to be ∼800 μm (Fig. 3D). Within the mediolateral domain, these clusters are restricted to the apex, mid-blade region, or blade tips of the dentate gyrus.
Interneurons of the hippocampus
The GABAergic interneurons were analyzed to determine colonization patterns of like-genotype cohorts as well as their lineage relationships to the hippocampal pyramidal cells and dentate granule cells. β-gal-positive cells that were also labeled immunocytochemically for GABA were found to have a widespread and apparently random dispersion pattern throughout the entire hippocampus from septal to temporal poles, in contrast to the clear clustering of the pyramidal and granule cells. Like-genotype cohorts of GABAergic interneurons were also observed across the entire mediolateral extent of the hippocampus. These findings were consistent in all low-percentage chimeras. A three-dimensional reconstruction of one low-percentage chimera illustrates this finding in Figure4A.
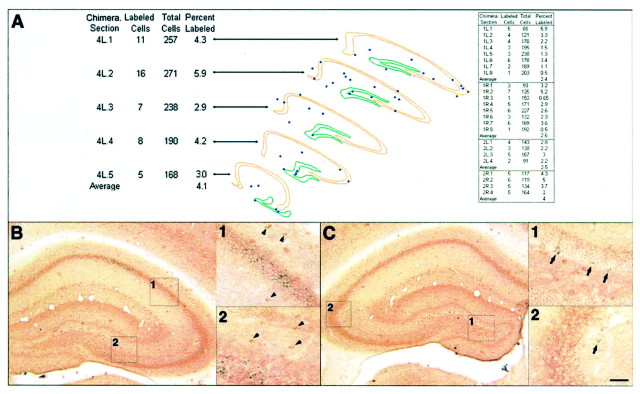
Fig. 4.
Observations of the patterns of X-gal-labeled GABAergic interneurons of the hippocampus. A, Three-dimensional reconstruction of the left hippocampus of a low-percentage chimera (chimera 4) demonstrating the widespread and apparent random dispersion of like-genotype cohorts of the GABAergic interneurons. Table on left shows the percentage of X-gal-labeled cells of the total GABAergic interneuron population. Table on right shows the percentage of X-gal-labeled cells of the total GABAergic interneuron population of four other low-percentage hippocampi (from chimeras 1 and 2). B, Example of a low-percentage chimera with no X-gal-labeled GABAergic interneurons (see arrowheads ininsets 1 and 2) but X-gal-labeled pyramidal cells. C, Example of a low-percentage chimera with no X-gal-labeled pyramidal or granule cells, but X-gal-labeled GABAergic interneurons (see arrows in insets 1 and 2). Scale bar: B,C, 175 μm; insets in B,C, 85 μm.
The analysis also suggests that interneurons of the hippocampus proper and dentate gyrus share a common lineage. This was evident from our examination of five chimeras. In three chimeras, there were no β-gal-positive interneurons in either the hippocampus proper or dentate gyrus on one or both sides of the hippocampus. On the other side of the hippocampus in two of the above three chimeras, and on both sides of two more chimeras, β-gal-positive interneurons colonized both the hippocampus proper and the dentate gyrus. Thus labeled interneurons were never found in only the hippocampus proper or dentate gyrus.
Although interneurons across various hippocampal regions in each side of the hippocampus appear to share a common lineage, it is clear that they do not share this lineage with the pyramidal or granule cells. This was apparent from three low-percentage hippocampi with β-gal-positive pyramidal and/or granule cells, but no β-gal-positive interneurons, and from four other low-percentage hippocampi with β-gal-positive interneurons, but no β-gal-positive pyramidal or granule cells (Fig.4B,C, Table 1). These findings indicate that the interneurons comprise a distinct lineage from the principal neurons of the hippocampus, as has been found in the cerebral cortex (Anderson et al., 1997; Tan et al., 1998).
Quantitative analysis of low-percentage chimeras
In determining the number of progenitors that are allocated to establish a specific cell lineage, chimeras have proven to be an invaluable tool (Rossant 1987). To derive an estimated number of progenitors that give rise to each of the neuronal subtypes of the hippocampus, we quantitatively analyzed the smallest like-genotype cohorts of β-gal-positive pyramidal, granule, and interneurons in a group of our lowest-percentage chimeras. The minimum contribution of a single founder cell to a specific cell population is assumed to be the lowest number of like-genotype cells contributing to that population over a series of chimeras. This lowest number of like-genotype cells, or minimum descendant clone, is 1/n, where n is the total number of progenitor cells (Rossant, 1984). Thus, clonal cohorts larger than the minimum descendant clone should be multiples of “1/n” cells (i.e., 2/n, 3/n,…n/n). In addition, with estimates of cell cycle length and the time course of neurogenesis, the time when the progenitor pool is allocated can be estimated. We applied these quantitative methods to the analysis of the hippocampal pyramidal, dentate granule, and GABAergic interneuron populations.
Numerical analysis of the hippocampal pyramidal cells
Counts of labeled and unlabeled pyramidal cells were made from chimeric brains that had sections with labeled cells bracketed by sections containing no labeled cells. Thus, we were confident that these counts represented the full expansion of like-genotype cohorts. Figure 2A shows the percentage of the total number of pyramidal cells that were β-gal positive in our most highly skewed chimera (chimera 1). From this chimera, we determined that the smallest like-genotype cohort (in the left hippocampus) spanned 0.5 mm of the septotemporal extent of the hippocampus and consisted of ∼0.2% of the total number of pyramidal cells in that region of the hippocampus. The percentage of β-gal-positive pyramidal cells on the other side (in the right hippocampus) of this chimera was 0.42% of the total pyramidal cell population within the region subtended by the labeled cells (Fig. 2A). In a third low-percentage hippocampus from chimera 2, 1% of the total pyramidal cell population within the region subtended by the labeled cells was β-gal positive. These latter percentages of β-gal-positive pyramidal cells are intriguing multiples of the percentage of labeled cells in the left hippocampus in chimera 1. Also, one of our low-percentage hippocampi (the right side of chimera 5) contained no β-gal-positive pyramidal cells, indicating that we sampled a range of chimeras that included the lowest extent of chimerism for the pyramidal cell lineage (Table 1). Taken together, these findings suggest that the hippocampal progenitor pool is made up of at least 500 progenitors (1/0.002) for a segment of the hippocampal formation that comprises 0.5 mm of the unilateral septotemporal axis, making the total progenitor pool of the pyramidal cells for each side of the hippocampus to be ∼7000 cells.
Given that the total hippocampal pyramidal cell population for one side of the hippocampus in the adult mouse is somewhere between 200,000 and 500,000 cells (Wimer et al., 1980; Abusaad et al., 1999) (L. Lu and R. W. Williams, personal communication), each progenitor cell generates anywhere from 32 to 72 progeny between initial cell allocation and the end of neurogenesis on E18 (Angevine, 1965; Stanfield and Cowan, 1979). The cell cycle length found for mouse neocortex varies from 8 to 20 hr from the beginning to the end of corticogenesis (Caviness et al., 1995). Using these times as a model for pyramidal cell cycle length, and matching the longest time with the slowest period of proliferation and the shortest time with the fastest proliferative period, we estimate the pyramidal cell population to have an average cell cycle length of 12 hr. Working backward from the end of neurogenesis using the total hippocampal pyramidal cell number (200,000–500,000) (Wimer et al., 1980; Abusaad et al., 1999) (Lu and Williams, personal communication) along with the average cell cycle length, cell allocation was determined to occur within 1 or 2 d of the major onset of pyramidal cell production at around embryonic day (E) 12 (Angevine, 1965; Stanfield and Cowan, 1979). Interestingly, previous reports have demonstrated that molecular markers of the hippocampal primordium first appear at this same time (Grove and Tole, 1999; Galceran et al., 2000).
Numerical analysis of the dentate granule cells
For the dentate granule cell population, we again analyzed the lowest-percentage chimeras to determine the smallest like-genotype cohort that contributes to the granule cell lineage. Figure3D shows the percentage of the total number of granule cells that were β-gal positive in three highly skewed chimeras (chimeras 6, 10, and 11). Chimera 6 had the smallest like-genotype cohort of β-gal-positive granule cells, spanning 0.8 mm of the septotemporal extent of the hippocampus and consisting of ∼2.2% of the total granule cell population in this region. The percentages of chimerism in the other two lowest-percentage chimeras were 4.1 and 4.6, apparent twofold increases over the lowest percentage of labeled cells in chimera 6. As with the pyramidal cells, it appears that we have sampled a range of chimeras that has reached the lowest extent of chimerism for dentate granule cell lineage, with seven dentate gyri from five different chimeras containing no β-gal-positive granule cells (Table1). Our findings indicate that the pool of cells that establishes 0.8 mm of the dentate granule cell population is composed of at least 45 progenitors (1/0.022). Therefore, ∼400 cells comprise the progenitor pool that establishes the granule cell population for each side of the dentate gyrus.
Considering that the total granule cell population for one side of the dentate gyrus in the adult mouse is ∼300,000–450,000 cells (Wimer and Wimer, 1989; Abusaad et al., 1999; Lu et al., 2001), each initial progenitor cell produces ∼750 dentate granule cells. This number is much higher than that for the pyramidal cells because of the extended length of granule cell neurogenesis, which begins around E12 and continues through the first 20 d postnatally (Angevine, 1965;Stanfield and Cowan, 1979). We again used the cell cycle length of the neocortex along with the proliferative rate of the dentate granule cells to estimate the cell cycle length of granule cells to be 16 hr. Through calculations similar to those for the pyramidal cells, the allocation of the granule cell progenitor pool was estimated to occur ∼2 d before the beginning of neurogenesis.
Numerical analysis of the GABAergic interneuron population
The contribution of the smallest like-genotype cohort to the GABAergic interneuron population of the hippocampus was determined through counts of double-labeled (β-gal positive and anti-GABA/anti-GAD positive) and single-labeled (antibody positive only) cells in five hippocampi from three low-percentage chimeras (Fig.4A). In these chimeras, we estimated the total number of double-labeled and single-labeled cells throughout the full septotemporal extent of the hippocampus (see Materials and Methods). The total number of GABAergic interneurons (double-labeled + single-labeled cells) for each side of the hippocampus was determined to be ∼25,000 (±3000) cells. Because like-genotype cohorts of GABAergic interneurons were found to disperse throughout the hippocampus from septal to temporal poles (see interneuron observations), a sampling of the percentage chimerism in an evenly spaced series of sections through the hippocampus provides a reasonable estimate of the overall percentage chimerism in the GABAergic population.
We found that both the left and right hippocampi from chimera 1 and the left hippocampus of chimera 2 contained the smallest like-genotype cohorts of double-labeled interneurons, comprising 2.4, 2.6, and 2.5% of the total GABAergic population, respectively. Like-genotype cohorts from two other hippocampi shown in Figure 4A contain 4.0 and 4.1% of the total number of hippocampal GABAergic interneurons. These percentages may constitute a doubling of the smallest like-genotype cohort. Also, four other low-percentage chimeras were analyzed that had no X-gal-labeled GABAergic interneurons in the hippocampus, indicating that we have again sampled the low ends of chimerism, this time for the hippocampal interneurons (Table 1). From the average of the lowest percentages of chimerism, the number of progenitors that establish the GABAergic population for each side of the entire hippocampus was determined to be at least 40 cells (1/0.025). With our own estimate of 25,000 interneurons for each side of the hippocampus, every founder cell gives rise to ∼625 progeny. Given the period of neurogenesis for the hippocampal interneurons in the mouse from E11 to E15 [inferred from Angevine (1965)], and using the same cell cycle length as for the pyramidal cells, we calculate that cell allocation for the GABAergic interneuron progenitor pool of the hippocampus occurs approximately six to seven cell cycle lengths before the beginning of neurogenesis.
Lineage relationships of hippocampal cell populations
The examination of a series of chimeric brains should consistently yield a similar percentage contribution of marked cells between related cell populations in different brain regions. Our detailed analysis of nine low-percentage chimeras using the Pearson correlation coefficient indicated a relationship between the two principal cell types of the hippocampus, granule cells and pyramidal cells, and a relationship between these principal neurons of the hippocampus and cells of cortex and mediodorsal diencephalon (see Materials and Methods;p < 0.01). In our subsequent visual evaluation of 22 low-percentage chimeras, 7 of the 44 half-brains had similar percentages of marked cells that existed exclusively between the hippocampal pyramidal and dentate granule cell populations and the cells of the overlying neocortex (Fig.5A), whereas in 6 other half-brains, the same was true for the hippocampal pyramidal and dentate granule cell populations and the cells of the mediodorsal diencephalon (Fig. 5B). Four other chimeric half-brains had similar percentages of marked cells between all three of the above brain regions that stood in contrast to the percentage chimerism of surrounding brain regions. We also found 6 of the 44 chimeric half-brains with few or no marked cells in the hippocampus but a substantial amount of marked cells in cortex and diencephalon (Fig.5C). The results from this analysis point to four patterns of neuronal relationships between the principal neurons of the hippocampus and other structures: (1) the hippocampus has origins similar to mediodorsal diencephalon and cortex, (2) the hippocampus has origins similar just to the cortex, (3) the hippocampus has origins similar just to dorsal diencephalon, and (4) the cortex and diencephalon can have patterns of labeled cells that are not shared with the hippocampus.
Through comparisons of the GABAergic interneurons of the hippocampus with other brain structures made in 14 chimeras, we observed that β-gal-positive interneurons of the hippocampus always coexisted with β-gal-positive interneurons of the neocortex, even in our most highly skewed chimeras. Also, in those chimeras with no β-gal-positive interneurons in the hippocampus, no β-gal-positive interneurons were found in the neocortex. Together, these findings indicate a lineage relationship between these interneuron populations.
DISCUSSION
Overview
We examined like-genotype cohorts of hippocampal pyramidal, granule, and interneuronal cells and revealed that organization of clonal cohorts is different in each of these populations. Pyramidal cohorts, for example, are tightly clustered in the radial dimension and somewhat restricted septotemporally, whereas granule cell cohorts are expansively spread along the neurogenetic gradient mediolaterally but similarly restricted septotemporally. In contrast to these patterns of clonal development, interneuronal cohorts are completely dispersed throughout the hippocampal formation, consistent with reports for interneurons in the neocortex (Tan et al., 1998). Although most of the pyramidal cohorts are confined within major cytoarchitectonic boundaries, some cell mixing occurs between CA regions, providing confirmation of a previous lineage analysis that suggested cell mixing across these boundaries (Grove et al., 1992).
The colonization of pyramidal and granule cells in each hippocampal side can be independent of the opposite side, unlike the symmetrical lineage of other brain structures (i.e., retina) (Williams and Goldowitz, 1992; Goldowitz et al., 1996). Although slight differences between hemispheres can be attributed to random progenitor cell distribution, marked differences over several chimeras can best be accounted for by independent lineages.
Although it is reasonable to question lineage results derived from a single set of chimeras (Goldowitz, 1989; Kuan et al., 1997), a unique feature of the present experiments is that we examined two types of chimeras, aggregation and blastocyst-injection, that used two different marker constructs. With one interesting exception (concerning dentate gyrus colonization; see Results), consonant results were obtained. This strengthens the validity of our conclusions in that they extend beyond possible vagaries associated with a single chimera methodology.
Dentate granule cell lineage
The clonal structure of the dentate gyrus has revealed two intriguing results. First, we have demonstrated that the colonization of like-genotype cohorts divides the granule cell layer into an outer shell and an inner core, with separate progenitors for each subdivision. This pattern recapitulates the neurogenetic development of the granule cell layer, where the outer shell forms first from the earliest-born granule cells followed by the development of the inner core by the later-born granule cells (Altman and Das, 1965; Angevine, 1965; Bayer, 1980). Previous studies have shown that the progenitors of these clonally distinct granule cells originate in the germinal zone of the primary dentate neuroepithelium, where they initially migrate to the secondary dentate matrix just beneath the ventricular zone (Altman and Bayer, 1990a). Interestingly, differences in the time of neurogenesis, migratory path, and germinal zone location of the outer shell and inner core granule cells give early indications of their clonal distinction. The granule cells of the outer shell originate from the secondary dentate matrix and migrate in a subpial route to the granule cell layer. The granule cells of the inner core, however, originate at a later time point, from the tertiary dentate matrix in the hilar region of the dentate gyrus. This tertiary dentate matrix is established by inner core progenitors from the secondary dentate matrix that migrate through a stream located above the initial subpial stream between the developing infrapyramidal blade of the dentate and the pyramidal layer of the hippocampus proper (Fig.6A) (Altman and Bayer, 1990b).
Fig. 6.
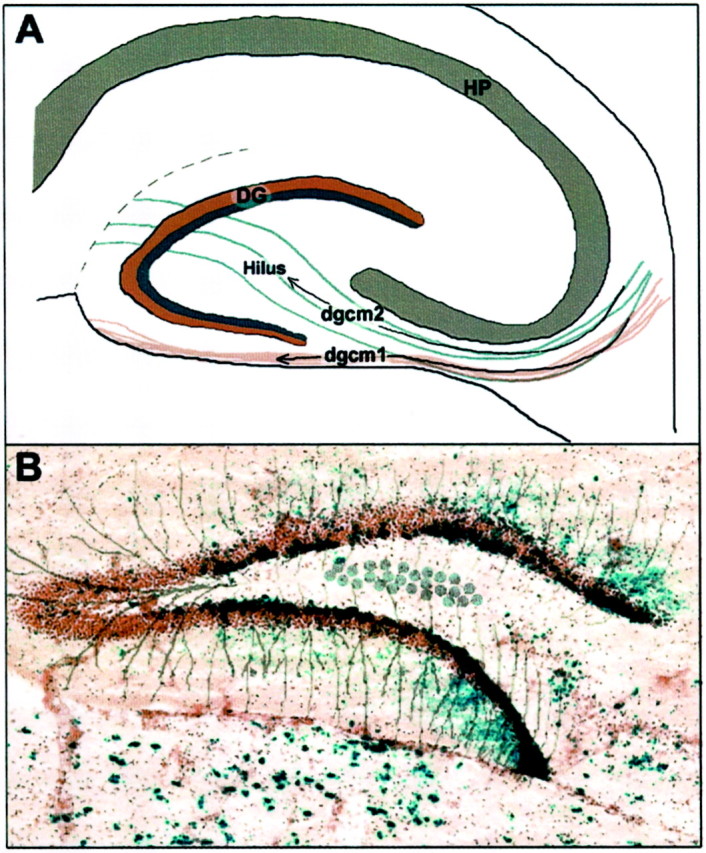
Illustrations depicting the distribution of radial glial processes in the dentate gyrus and the migratory streams of the dentate granule cells that may follow these processes.A, Schematic of a postnatal hippocampus with an overlay of the early developmental patterns of radial glia and the two separate dentate granule cell migrations [adapted from Rickmann et al. (1987);Altman and Bayer (1990b)]. DG, Dentate gyrus;HP, hippocampus proper. The granule cells of the outer shell (red) arrive through the first granule cell migration (dgcm1), probably by route of the fimbrial radial glia bundle (light red). The granule cells of the inner core (blue) are established from the progenitors of the dentate hilus, which arrive through the second granule cell migration (dgcm2), most likely by following the arrangement of radial glia (light blue) that traverse through the dentate hilus. B, Example of the early developmental radial glia patterns of the dentate gyrus superimposed over a chimera with a mirrored inner core granule cell layer demonstrating how this unique arrangement of radial glia would allow for the symmetric development of each dentate blade from a common progenitor pool in the hilus.
Second, the inner core of one blade mirrors the labeling pattern of the opposite blade. This indicates that corresponding parts of each blade arise from the same progenitors and that cells derived from these progenitors are either spatially programmed for a specific position along the dentate blades or follow radial glial paths to opposite blades from their origin in the hilus. On the basis of previous reports illustrating a unique arrangement of radial glia in the hilus of the dentate anlage with processes extending out through the granule cell layer in all directions (Fig. 6B), the latter appears to be a more plausible hypothesis (Eckenhoff and Rakic, 1984; Rickmann et al., 1987).
The radial glia patterns in the rat dentate gyrus demonstrated byRickmann et al. (1987, their Fig. 19) appear to overlap with the migratory patterns of dentate granule cells shown by Altman and Bayer (1990b). In fact, the “fimbrial bundle” of radial glial processes coincides with the initial subpial granule cell migration, and a more loosely arranged grouping of processes extending from the fimbria through the dentate hilus coincides with the second granule cell migration (Fig. 6A) (Rickmann et al., 1987;Altman and Bayer, 1990b). The unique organization of radial glia in the developing dentate and the clonal patterning of the granule cell layer give us insight into the intricate relationship between these two cell types because it appears that radial glia are involved in all levels of dentate development. Furthermore, the independent lineage of the outer shell and inner core granule cells along with their migratory routes and the location of radial glial processes can help explain the unique outside-in development of the dentate gyrus. That is, it appears that the earlier developing outer shell is formed by progenitors from a region near the dentate neuroepithelium that migrate along radial glial processes to the future granule cell layer, and the later developing inner core is formed by granule cell progenitors located within the dentate hilus which generate progeny that migrate along a unique set of radial glia based in the hilus with extensions through the granule cell layer.
GABAergic cell lineage
The interneurons of the hippocampus proper and dentate gyrus share a common lineage with each other but are independent from pyramidal and granule cells. This finding parallels the neocortex, where separate progenitors have been demonstrated for glutamatergic and GABAergic cells (Parnavelas et al., 1991; Luskin et al., 1993; Mione et al., 1994, 1997; Tan et al., 1998). The hippocampal interneurons, however, do appear to share a common lineage with the interneuron population of the neocortex. It has been shown previously that most neocortical interneurons are derived from the ganglionic eminences and migrate into the neocortex (Anderson et al., 1997; Tamamaki et al., 1997; Zhu et al., 1999). Recent experimental studies have also shown that these two interneuronal populations stem from the same source (Pleasure et al., 2000). Thus, our findings, in conjunction with those of Pleasure et al. (2000), demonstrate that the hippocampal interneurons are lineally related to the interneurons of the neocortex.
Progenitor pools and time of cell allocation
For pyramidal, granule, and interneuronal cells, there is quite a variation in the size of each progenitor pool and the time of pool allocation. Two factors that are important when considering progenitor pool size at the time of allocation are the amount of time between allocation and the beginning of neurogenesis, and the length of neurogenesis. For pyramidal cells, allocation occurs 1–2 d before neurogenesis, allowing little time for doubling of the progenitor pool before neurogenesis begins. Also, neurogenesis is limited to ∼7–8 d (Angevine, 1965). Therefore, a large progenitor pool (∼7000) is required to generate the pyramidal population. In comparison, progenitor allocation to the granule population also occurs shortly before neurogenesis begins; however, neurogenesis lasts almost 4 weeks (Angevine, 1965). Thus, the granule progenitor pool (∼400) is much smaller than the pyramidal progenitor pool. The hippocampal interneurons have a short period of neurogenesis (Angevine, 1965), but because their progenitor pool is established 6–7 d before neurogenesis, their progenitor number (∼40) is relatively small at allocation, allowing the pool to double several times before neurogenesis.
The dynamics of pool size and ontogeny for hippocampal interneurons resembles dynamics previously established for the cerebellar Purkinje cells. It has been found that 65–80 progenitors give rise to ∼80,000 Purkinje cells on one side of the cerebellum (Baader et al., 1996;Mathis et al., 1997; Hawkes et al., 1998), and neurogenesis takes place from E11 to E13 with an average cell cycle length of 9 hr (Miale and Sidman, 1961; Korr, 1980). Allocation of progenitors therefore occurs at about E8.5, or seven cell divisions before neurogenesis (Baader et al., 1996). The early allocation of two GABAergic neuronal populations, the Purkinje cells and hippocampal interneurons, stands in contrast to the late allocation of two glutamatergic populations, the hippocampal pyramidal and granule cells. Differences in cell allocation might pertain to a role for early allocated cells as a substrate for development and later-allocated cells as a reservoir for developmental and evolutionary change, possibly suggesting a generalized role for GABAergic and glutamatergic cells in neuronal development.
Hippocampal pyramidal and granule cell lineage
A synthesis of the four types of chimeric patterns that are seen [cortex, dorsal diencephalon, and hippocampus are of similar clonal composition; cortex or dorsal diencephalon share an exclusive lineage pattern with the hippocampus (Fig.5A,B); and the hippocampus can be excluded from the establishment of both cortex and dorsal diencephalon (Fig. 5C)] suggests at least two hypothetical means by which the cells that give rise to the hippocampus are related to the development of cortex and dorsal diencephalon. One possible lineage mode is that there is an initial common pool of cells that diverge, early on, into cortical, hippocampal, and dorsal diencephalic lineages. The common lineage that is observed between these structures is testimony to this early common origin. However, the various outcomes that are seen indicate that this initial pool is relatively small, giving rise to rather striking clonal patterns based on the random assortment of cells.
A second scenario also posits a common progenitor pool that is distinguished by the expression of molecules in a gradient manner centered on the hippocampal formation. In this case, two relatively independent precursor populations at the opposite ends of the gradient would give rise to neocortex and mediodorsal diencephalon. Toward the center of the gradient, precursor cells would have a greater likelihood of being shared between the hippocampus and the brain region on the respective end of the gradient. Precursors residing nearest the center of the gradient would therefore be shared between all three brain regions. The common origins that we observe for hippocampus, neocortex and mediodorsal diencephalon find support from gene expression and knock-out phenotypes that specifically involve these brain regions [Gli3 (Theil et al., 1999; Tole et al., 2000b), Emx (Pellegrini et al., 1996; Yoshida et al., 1997;Tole et al., 2000a), Otx (Acampora et al., 1997; Suda et al., 1997), Wnt3a (Grove et al., 1998; Lee et al., 2000), and Lhx2 (Porter et al., 1997)]. The analysis of a larger set of chimeras and the application of other lineage tracing protocols will help distinguish between these possibilities and other scenarios.
Footnotes
This research was supported by a University of Tennessee Health Science Center, Center for Neuroscience Predoctoral Fellowship (L.A.M.) and by a grant from Cure Autism Now. We thank Dr. Dave Airey for statistical advice, Richard Cushing for technical support, and the anonymous reviewers for key comments and suggestions.
Correspondence should be addressed to Dan Goldowitz, Department of Anatomy and Neurobiology, University of Tennessee Health Science Center, 855 Monroe Avenue, Memphis, TN 38163. E-mail:dgold@nb.utmem.edu.
REFERENCES
- 1.Abbie AA. The relations of the fascia dentata, hippocampus and neocortex, and the nature of the subiculum. J Comp Neurol. 1937;66:307–333. [Google Scholar]
- 2.Abercrombie M. Estimation of nuclear population from microtome sections. Anat Rec. 1946;94:239–247. doi: 10.1002/ar.1090940210. [DOI] [PubMed] [Google Scholar]
- 3.Abusaad I, MacKay D, Zhao J, Stanford P, Collier DA, Everall IP. Stereological estimation of the total number of neurons in the murine hippocampus using the optical dissector. J Comp Neurol. 1999;408:560–566. doi: 10.1002/(sici)1096-9861(19990614)408:4<560::aid-cne9>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 4.Acampora D, Avantaggiato V, Tuorto F, Simeone A. Genetic control of brain morphogenesis through Otx gene dosage requirement. Development. 1997;124:3639–3650. doi: 10.1242/dev.124.18.3639. [DOI] [PubMed] [Google Scholar]
- 5.Altman J, Bayer SA. Mosaic organization of the hippocampal neuroepithelium and the multiple germinal sources of dentate granule cells. J Comp Neurol. 1990a;301:325–342. doi: 10.1002/cne.903010302. [DOI] [PubMed] [Google Scholar]
- 6.Altman J, Bayer SA. Migration and distribution of two populations of hippocampal granule cell precursors during the perinatal and postnatal periods. J Comp Neurol. 1990b;301:365–381. doi: 10.1002/cne.903010304. [DOI] [PubMed] [Google Scholar]
- 7.Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965;124:319–336. doi: 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- 8.Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 9.Angevine JB., Jr Time of neuron origin in the hippocampal region. An autoradiographic study in the mouse. Exp Neurol Suppl. 1965;2:1–70. [PubMed] [Google Scholar]
- 10.Baader SL, Schilling ML, Rosengarten B, Pretsch W, Teutsch HF, Oberdick J, Schilling K. Purkinje cell lineage and the topographic organization of the cerebellar cortex: a view from X inactivation mosaics. Dev Biol. 1996;174:393–406. doi: 10.1006/dbio.1996.0083. [DOI] [PubMed] [Google Scholar]
- 11.Bayer SA. Development of the hippocampal region in the rat. I. Neurogenesis examined with [3H] thymidine autoradiography. J Comp Neurol. 1980;190:87–114. doi: 10.1002/cne.901900107. [DOI] [PubMed] [Google Scholar]
- 12.Boncinelli E, Gulisano M, Broccoli V. Emx and Otx homeobox genes in the developing mouse brain. J Neurobiol. 1993;24:1356–1366. doi: 10.1002/neu.480241008. [DOI] [PubMed] [Google Scholar]
- 13.Caviness VS, Jr, Takahashi T, Nowakowski RS. Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- 14.Dupuy ST, Houser CR. Prominent expression of two forms of glutamate decarboxylase in the embryonic and early postnatal rat hippocampal formation. J Neurosci. 1996;16:6919–6932. doi: 10.1523/JNEUROSCI.16-21-06919.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckenhoff MF, Rakic P. Radial organization of the hippocampal dentate gyrus: a Golgi, ultrastructural, and immunocytochemical analysis in the developing rhesus monkey. J Comp Neurol. 1984;223:1–21. doi: 10.1002/cne.902230102. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 17.Furuta Y, Piston DW, Hogan BL. Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development. 1997;124:2203–2212. doi: 10.1242/dev.124.11.2203. [DOI] [PubMed] [Google Scholar]
- 18.Galceran J, Farinas I, Depew MJ, Clevers H, Grosschedl R. Wnt3a−/−-like phenotype and limb deficiency in Lef1(−/−)Tcf1(−/−) mice. Genes Dev. 1999;13:709–717. doi: 10.1101/gad.13.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JL, Grosschedl R. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development. 2000;127:469–482. doi: 10.1242/dev.127.3.469. [DOI] [PubMed] [Google Scholar]
- 20.Goldowitz D. Cell allocation in mammalian CNS formation: evidence from murine interspecies aggregation chimeras. Neuron. 1989;3:705–713. doi: 10.1016/0896-6273(89)90239-0. [DOI] [PubMed] [Google Scholar]
- 21.Goldowitz D, Moran H, Wetts R. Mouse chimeras in the study of genetic and structural determinants of behavior. In: Goldowitz D, Wahlsten D, Wimer RE, editors. Techniques for the genetic analysis of brain and behavior: focus on the mouse. Elsevier; Amsterdam: 1992. pp. 271–290. [Google Scholar]
- 22.Goldowitz D, Rice DS, Williams RW. Clonal architecture of the mouse retina. Prog Brain Res. 1996;108:3–15. doi: 10.1016/s0079-6123(08)62528-5. [DOI] [PubMed] [Google Scholar]
- 23.Goldowitz D, Hamre KM, Przyborski SA, Ackerman SL. Granule cells and cerebellar boundaries: analysis of Unc5h3 mutant chimeras. J Neurosci. 2000;20:4129–4137. doi: 10.1523/JNEUROSCI.20-11-04129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grove EA, Tole S. Patterning events and specification signals in the developing hippocampus. Cereb Cortex. 1999;9:551–561. doi: 10.1093/cercor/9.6.551. [DOI] [PubMed] [Google Scholar]
- 25.Grove EA, Kirkwood TB, Price J. Neuronal precursor cells in the rat hippocampal formation contribute to more than one cytoarchitectonic area. Neuron. 1992;8:217–229. doi: 10.1016/0896-6273(92)90289-p. [DOI] [PubMed] [Google Scholar]
- 26.Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- 27.Hawkes R, Faulkner-Jones B, Tam P, Tan SS. Pattern formation in the cerebellum of murine embryonic stem cell chimeras. Eur J Neurosci. 1998;10:790–793. doi: 10.1046/j.1460-9568.1998.00085.x. [DOI] [PubMed] [Google Scholar]
- 28.Kingan TG, Hildebrand JG. Gamma-aminobutyric acid in the central nervous system of metamorphosing and mature Manduca sexta. Insect Biochem. 1985;15:667–675. [Google Scholar]
- 29.Korr H. Proliferation of different cell types in the brain. Adv Anat Embryol Cell Biol. 1980;61:1–72. doi: 10.1007/978-3-642-67577-5. [DOI] [PubMed] [Google Scholar]
- 30.Kuan CY, Elliott EA, Flavell RA, Rakic P. Restrictive clonal allocation in the chimeric mouse brain. Proc Natl Acad Sci USA. 1997;94:3374–3379. doi: 10.1073/pnas.94.7.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SM, Tole S, Grove E, McMahon AP. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127:457–467. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- 32.Lorente de No R. Studies on the structure of the cerebral cortex. II. Continuation of the study of the ammonic system. J Psychol Neurol. 1934;46:113–177. [Google Scholar]
- 33. Liu M, Pleasure SJ, Collins AE, Noebels JL, Naya FJ, Tsai MJ, Lowenstein DH. Loss of BETA2/NeuroD leads to malformation of the dentate gyrus and epilepsy. Proc Natl Acad Sci USA 97 2000. 865 870 [Erratum (2000) 97:5679] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu L, Airey DC, Williams RW. Complex trait analysis of the hippocampus: mapping and biometric analysis of two novel gene loci with specific effects on hippocampal structure in mice. J Neurosci. 2001;21:3503–3514. doi: 10.1523/JNEUROSCI.21-10-03503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luskin MB, Parnavelas JG, Barfield JA. Neurons, astrocytes, and oligodendrocytes of the rat cerebral cortex originate from separate progenitor cells: an ultrastructural analysis of clonally related cells. J Neurosci. 1993;13:1730–1750. doi: 10.1523/JNEUROSCI.13-04-01730.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathis L, Bonnerot C, Puelles L, Nicolas JF. Retrospective clonal analysis of the cerebellum using genetic laacZ/lacZ mouse mosaics. Development. 1997;124:4089–4104. doi: 10.1242/dev.124.20.4089. [DOI] [PubMed] [Google Scholar]
- 37.Miale I, Sidman RL. An autoradiographic analysis of histogenesis in the mouse cerebellum. Exp Neurol. 1961;4:277–296. doi: 10.1016/0014-4886(61)90055-3. [DOI] [PubMed] [Google Scholar]
- 38.Mione MC, Danevic C, Boardman P, Harris B, Parnavelas JG. Lineage analysis reveals neurotransmitter (GABA or glutamate) but not calcium-binding protein homogeneity in clonally related cortical neurons. J Neurosci. 1994;14:107–123. doi: 10.1523/JNEUROSCI.14-01-00107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mione MC, Cavanagh JFR, Harris B, Parnavelas JG. Cell fate specification and symmetrical/asymmetrical divisions in the developing cerebral cortex. J Neurosci. 1997;17:2018–2029. doi: 10.1523/JNEUROSCI.17-06-02018.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monaghan AP, Grau E, Bock D, Schutz G. The mouse homolog of the orphan nuclear receptor tailless is expressed in the developing forebrain. Development. 1995;121:839–853. doi: 10.1242/dev.121.3.839. [DOI] [PubMed] [Google Scholar]
- 41.Monaghan AP, Bock D, Gass P, Schwager A, Wolfer DP, Lipp HP, Schutz G. Defective limbic system in mice lacking the tailless gene. Nature. 1997;390:515–517. doi: 10.1038/37364. [DOI] [PubMed] [Google Scholar]
- 42.Morita T, Nitta H, Kiyama Y, Mori H, Mishina M. Differential expression of two zebrafish emx homeoprotein mRNAs in the developing brain. Neurosci Lett. 1995;198:131–134. doi: 10.1016/0304-3940(95)11988-9. [DOI] [PubMed] [Google Scholar]
- 43.Mullen RJ, Whitten WK. Relationship of genotype and degree of chimerism in coat color to sex ratios and gametogenesis in chimeric mice. J Exp Zool. 1971;178:165–176. doi: 10.1002/jez.1401780203. [DOI] [PubMed] [Google Scholar]
- 44.Parnavelas JG, Barfield JA, Franke E, Luskin MB. Separate progenitor cells give rise to pyramidal and nonpyramidal neurons in the rat telencephalon. Cereb Cortex. 1991;1:463–468. doi: 10.1093/cercor/1.6.463. [DOI] [PubMed] [Google Scholar]
- 45.Pellegrini M, Mansouri A, Simeone A, Boncinelli E, Gruss P. Dentate gyrus formation requires Emx2. Development. 1996;122:3893–3898. doi: 10.1242/dev.122.12.3893. [DOI] [PubMed] [Google Scholar]
- 46.Pleasure JP, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JLR. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- 47.Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, Alt F, Westphal H. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- 48.Rickmann M, Amaral DG, Cowan WM. Organization of radial glial cells during the development of the rat dentate gyrus. J Comp Neurol. 1987;264:449–479. doi: 10.1002/cne.902640403. [DOI] [PubMed] [Google Scholar]
- 49.Rossant J. Somatic cell lineages in mammalian chimeras. In: Le Douarin N, McLaren A, editors. Chimeras in developmental biology. Academic; London: 1984. pp. 89–109. [Google Scholar]
- 50.Rossant J. Cell lineage analysis in mammalian embryogenesis. Curr Top Dev Biol. 1987;23:115–146. doi: 10.1016/s0070-2153(08)60622-5. [DOI] [PubMed] [Google Scholar]
- 51.Rubenstein JL, Shimamura K, Martinez S, Puelles L. Regionalization of the prosencephalic neural plate. Annu Rev Neurosci. 1998;21:445–477. doi: 10.1146/annurev.neuro.21.1.445. [DOI] [PubMed] [Google Scholar]
- 52.Stanfield BB, Cowan WM. The development of the hippocampus and dentate gyrus in normal and reeler mice. J Comp Neurol. 1979;185:423–459. doi: 10.1002/cne.901850303. [DOI] [PubMed] [Google Scholar]
- 53.Sturm KS, Berger CN, Zhou SX, DunWoodie SL, Tan SS, Tam PPL. Unrestricted lineage differentiation of parthenogenetic ES-cells. Dev Genes Evol. 1997;206:377–388. doi: 10.1007/s004270050067. [DOI] [PubMed] [Google Scholar]
- 54.Suda Y, Matsuo I, Aizawa S. Cooperation between Otx1 and Otx2 genes in developmental patterning of rostral brain. Mech Dev. 1997;69:125–141. doi: 10.1016/s0925-4773(97)00161-5. [DOI] [PubMed] [Google Scholar]
- 55.Tamamaki N, Fujimori KE, Takauji R. Origin and route of tangentially migrating neurons in the developing neocortical intermediate zone. J Neurosci. 1997;17:8313–8323. doi: 10.1523/JNEUROSCI.17-21-08313.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tan SS, Williams EA, Tam PP. X-chromosome inactivation occurs at different times in different tissues of the post-implantation mouse embryo. Nat Genet 3 1993. 170 174 [Erratum (1993) 4:320] [DOI] [PubMed] [Google Scholar]
- 57.Tan SS, Kalloniatis M, Sturm K, Tam PP, Reese BE, Faulkner-Jones B. Separate progenitors for radial and tangential cell dispersion during development of the cerebral neocortex. Neuron. 1998;21:295–304. doi: 10.1016/s0896-6273(00)80539-5. [DOI] [PubMed] [Google Scholar]
- 58.Theil T, Alvarez-Bolado G, Walter A, Ruther U. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development. 1999;126:3561–3571. doi: 10.1242/dev.126.16.3561. [DOI] [PubMed] [Google Scholar]
- 59.Tole S, Goudreau G, Assimacopoulos S, Grove EA. Emx2 is required for growth of the hippocampus but not for hippocampal field specification. J Neurosci. 2000a;20:2618–2625. doi: 10.1523/JNEUROSCI.20-07-02618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tole S, Ragsdale CW, Grove EA. Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J). Dev Biol. 2000b;217:254–265. doi: 10.1006/dbio.1999.9509. [DOI] [PubMed] [Google Scholar]
- 61.Williams RW, Goldowitz D. Structure of clonal and polyclonal cell arrays in chimeric mouse retina. Proc Natl Acad Sci USA. 1992;89:1184–1188. doi: 10.1073/pnas.89.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wimer CC, Wimer RE. On the sources of strain and sex differences in granule cell number in the dentate area of house mice. Brain Res Dev Brain Res. 1989;48:167–176. doi: 10.1016/0165-3806(89)90073-4. [DOI] [PubMed] [Google Scholar]
- 63.Wimer RE, Wimer CC, Chernow CR, Balvanz BA. The genetic organization of neuron number in the pyramidal cell layer of hippocampal regio superior in house mice. Brain Res. 1980;196:59–77. doi: 10.1016/0006-8993(80)90716-7. [DOI] [PubMed] [Google Scholar]
- 64.Yoshida M, Suda Y, Matsuo I, Miyamoto N, Takeda N, Kuratani S, Aizawa S. Emx1 and Emx2 functions in development of dorsal telencephalon. Development. 1997;124:101–111. doi: 10.1242/dev.124.1.101. [DOI] [PubMed] [Google Scholar]
- 65.Zhao Y, Sheng HZ, Amini R, Grinberg A, Lee E, Huang S, Taira M, Westphal H. Control of hippocampal morphogenesis and neuronal differentiation by the LIM homeobox gene Lhx5. Science. 1999;284:1155–1158. doi: 10.1126/science.284.5417.1155. [DOI] [PubMed] [Google Scholar]
- 66.Zhu Y, Li H, Zhou L, Wu JY, Rao Y. Cellular and molecular guidance of GABAergic neuronal migration from an extracortical origin to the neocortex. Neuron. 1999;23:473–485. doi: 10.1016/s0896-6273(00)80801-6. [DOI] [PubMed] [Google Scholar]