Abstract
Synaptic scaffolding molecule (S-SCAM) is a synaptic membrane-associated guanylate kinase with inverted domain organization (MAGI) that interacts with NMDA receptor subunits and neuroligin. In epithelial cells, the non-neuronal isoform of S-SCAM (MAGI-1) is localized at tight or adherens junctions. Recent studies have revealed that the polarized targeting of MAGI-1 to the lateral membrane is mediated by its C-terminal region and that MAGI-1 interacts with β-catenin in epithelial cells. In this article, we report that S-SCAM interacts with β-catenin in neurons. β-Catenin is coimmunoprecipitated with S-SCAM from rat brain. Both S-SCAM and β-catenin are localized at synapses and are partially colocalized. The C-terminal region of S-SCAM binds to the C-terminal region of β-catenin. We have tested how the interaction between S-SCAM and β-catenin plays a role in the synaptic targeting of S-SCAM and β-catenin. S-SCAM is targeted to synapses via the C-terminal postsynaptic density-95/Dlg-A/ZO-1 (PDZ) domain. β-Catenin is targeted to synapses with armadillo repeats. The overexpressed C-terminal region of β-catenin blocks the synaptic targeting of S-SCAM. The overexpressed C-terminal region of S-SCAM is partially targeted to synapses and forms a small number of clusters. In the presence of overexpressed β-catenin, the C-terminal region of S-SCAM forms more clusters at synapses. These data suggest that the synaptic targeting of S-SCAM is mediated by the interaction with β-catenin.
Keywords: β-catenin, synaptic scaffolding molecule, membrane-associated guanylate kinase with inverted domain organization, NMDA receptor, armadillo repeats, PDZ domain
Synaptic scaffolding molecule (S-SCAM) has been identified as a protein interacting with a synaptic cytoskeleton adapter protein known as guanylate kinase (GK)-associated protein (GKAP) (also called synapse-associated protein 90/postsynaptic density-95-associated protein or discs-large tumor suppressor-associated protein) (Kim et al., 1996; Satoh et al., 1997;Takeuchi et al., 1997; Hirao et al., 1998). The same molecule is known as an atrophin-1-interacting protein or an activin receptor-interacting protein (Wood et al., 1998; Shoji et al., 2000). S-SCAM has six postsynaptic density-95/Dlg-A/ZO-1 (PDZ) domains (PDZ0 to PDZ5), one GK domain, and two WW domains. S-SCAM interacts with various molecules using these domains. GKAP binds to the GK domain. A neuronal cell adhesion molecule, neuroligin, and a GDP/GTP exchange factor for rap1 bind to the second PDZ domain (PDZ1) (Hirao et al., 1998; Ohtsuka et al., 1999). Protein tyrosine phosphatase and tensin homolog binds to the third PDZ domain (PDZ2) (Wu et al., 2000). NMDA receptor subunits, membrane-associated GK-interacting protein, neural plakophilin-related armadillo-repeat protein/δ-catenin, and activin receptor bind to the last PDZ domain (PDZ5) (Hirao et al., 1998; Ide et al., 1999a; Yao et al., 1999; Shoji et al., 2000). Atrophin-1 possibly interacts with the WW domains (Wood et al., 1998). S-SCAM also forms homodimers via the region containing PDZ4 and PDZ5 (Hirao et al., 2000). In epithelial cells, membrane-associated GK with inverted domain organization (MAGI)-1 (also called brain-specific angiogenesis inhibitor-associated protein-1), an isoform of S-SCAM, is localized at tight or adherens junctions (Dobrosotskaya et al., 1997; Shiratsuchi et al., 1998; Ide et al., 1999b; Dobrosotskaya and James, 2000; Nishimura et al., 2000). Recent studies have revealed that MAGI-1 interacts with β-catenin, megalin, and Mus musculus neuroepithelial cell transforming gene 1 (Dobrosotskaya and James, 2000; Dobrosotskaya, 2001; Patrie et al., 2001). The interaction with β-catenin is involved in the membrane association of MAGI-1 (Dobrosotskaya and James, 2000). These findings suggest that β-catenin determines the localization of MAGI-1 at epithelial junctions and subsequently recruits various molecules interacting with MAGI-1 to cell–cell contacts. In neurons, β-catenin is enriched at synapses and interacts with N-cadherin and other cadherin-like molecules (Uchida et al., 1996; Benson and Tanaka, 1998; Miskevich et al., 1998). Therefore, β-catenin may be involved in the synaptic targeting of S-SCAM. We actually obtained β-catenin from the yeast two-hybrid screening using S-SCAM as bait (Ide et al., 1999a). Recently, KIAA0707, which is homologous to S-SCAM, has been reported to bind to the glutathione S-transferase (GST)–β-catenin affinity column (Kawajiri et al., 2000). Here, we reveal that S-SCAM interacts with β-catenin in neurons and that the interaction with β-catenin is involved in the synaptic targeting of S-SCAM.
MATERIALS AND METHODS
Construction of expression vectors. Various expression vectors were constructed by conventional molecular biology techniques and the PCR method using pClneoMyc, pSind green fluorescent protein (GFP), and pSindMyc. The PCR product using pLGFPC (Clontech, Palo Alto, CA) as a template was ligated intoXbaI/SphI sites of pSindRep5 (Invitrogen, San Diego, CA) to generate pSindGFP. A linker was ligated toXbaI/SphI sites of pSindRep5 to generate pSindMyc. pClneoMyc S-SCAM-1, -4, -8, and -10 and pGex4T-1 S-SCAM-10 have been described previously (Hirao et al., 1998, 2000). pSindGFP S-SCAM-1, -4, -8, and -10 contain the same amino acid residues as pClneoMyc S-SCAM-1, -4, -8, and -10. pSindGFP S-SCAM-13 contains the amino acid residues 772–1277. pSindGFP S-SCAM-19 contains the amino acid residues 573–906 and 1134–1277. pSindMyc S-SCAM-1 contains the same amino acid residues as pClneoMyc S-SCAM-1. The constructs below contain the following amino acids of mouse β-catenin: pClneoMyc and pSindGFP β-catenin-1, 1–781; pClneoMyc β-catenin-3, 131–781; pClneoMyc β-catenin-4, 1–699; pClneoMyc β-catenin-5, 1–555; pClneoMyc β-catenin-6, 555–781; pClneoMyc β-catenin-7, 131–699; pSindGFP β-catenin-2, 131–781;pSindGFP β-catenin-6, 131–696; pSindGFP β-catenin-8, 696–781; pSindGFP β-catenin-9, 437–781; pSindGFP β-catenin-10, 1–367; and pSindGFP β-catenin-11, 368–696. pSindGFP NMDA receptor subunit 2A (NR2A) contains amino acid residues 839–1464 of rat NR2A.
Antibodies. The rabbit polyclonal anti-S-SCAM antibody against the WW domain has been described previously (Hirao et al., 1998; Ide et al., 1999b). Sheep polyclonal anti-Myc antibody was raised against the synthetic peptide Glu-Gln-Lys-Ile-Ser-Glu-Glu-Asp-Leu-Asn-Ser-Ala-Val-Asp. Other antibodies used in this study were as follows: mouse monoclonal anti-Myc-tag 9E10 (American Type Culture Collection, Manassas, VA); mouse monoclonal anti-synaptophysin (Roche Molecular Biochemicals, Basel, Switzerland); mouse monoclonal anti-β-catenin (BD Transduction Laboratories, San Diego, CA); and rhodamine-, fluorescein isothiocyanate-, and Cy5-conjugated second antibodies for dual labeling (Chemicon International Inc., Temecula, CA).
Immunoprecipitation from rat brain. S-SCAM was immunoprecipitated from the Triton X-100 extract of rat crude synaptosomes as described previously (Hirao et al., 2000). The immunoprecipitates were immunoblotted with either the anti-S-SCAM or the anti-β-catenin antibody.
Immunoprecipitation of various Myc-tagged constructs of S-SCAM from COScells. COS cells were cultured in DMEM supplemented with 10% (w/v) FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C under 5% CO2 and were transfected with DEAE dextran with various constructs of S-SCAM. COS cells from one 10 cm plate were homogenized in 200 μl of 20 mm HEPES/NaOH, pH 8.0, containing 200 mm NaCl, 1% (w/v) NP-40, and 2 mm EGTA, and were centrifuged at 100,000 × g for 15 min at 4°C. The supernatant was precleared with 7.5 μl of protein G Sepharose 4 fast-flow beads and then incubated with 2.5 μl of the anti-Myc ascites fixed on 5 μl of protein G Sepharose 4 fast-flow beads. After the beads were washed four times with 400 μl of 20 mm HEPES/NaOH, pH 8.0, containing 200 mm NaCl, 0.3% (w/v) NP-40, and 2 mm EGTA, the immunoprecipitates were immunoblotted with either the anti-Myc or the anti-β-catenin antibody.
Pull-down assay. COS cells were transfected with various constructs of β-catenin, and the extracts were prepared as described above. For each extract, 160 μl was incubated with either 200 pmol of GST–S-SCAM-10 containing PDZ4 and PDZ5 or 200 pmol of GST fixed on 14.5 μl of glutathione beads. After the beads were washed, the proteins on the beads were immunoblotted with the anti-Myc antibody.
Hippocampal neuron culture and hippocampal slice culture. Hippocampal neuron cultures were performed from embryonic day 18 embryos as described previously (Takeuchi et al., 1997; Goslin et al., 1998). Hippocampal slices were obtained from postnatal day 6 (P6) or P8 rats and cultured on Millicell CM culture plate inserts (Millipore, Bedford, MA) in Eagle's MEM containing 25% (v/v) HBSS, 6.5 gm/l glucose, 100 U/ml penicillin, 100 μg/ml streptomycin, and 25% (v/v) horse serum at 32°C under 5% CO2.
Sindbis virus production and infection. Baby hamster kidney cells were cultured in MEM supplemented with 5% (v/v) FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Capped in vitro transcripts and helper RNA were synthesized from various linearized pSindGFP constructs and DH(26S) template (Invitrogen) using a RiboMAX large-scale RNA production system (Promega, Madison, WI) and were transfected into baby hamster kidney cells by electroporation with a GenePulser (Bio-Rad, Hercules, CA). Two days later, the medium was collected and centrifuged at 400 × g for 5 min. The supernatant was centrifuged at 113,000 × g for 90 min. The pellet was collected, resuspended in 200 μl of the medium, and stocked at −80°C. Primary cultured hippocampal neurons were infected with 3–5 μl of the virus stock per 500 μl of the culture medium 10 d after plating. For the double infection, 3–5 μl of each virus stock was added to 500 μl of the culture medium at the same time. Hippocampal slices were infected with 1 μl of the virus stock per slice 7 d after plating.
Immunocytochemistry. Hippocampal neurons were fixed with 4% (w/v) paraformaldehyde for 15 min, blocked with 50 mmglycine in PBS for 30 min, and permeabilized with 0.25% (w/v) Triton X-100 in PBS for 5 min. After they had been blocked with PBS containing 10% (w/v) BSA, cells were incubated with the first antibody in PBS containing 3% (w/v) BSA overnight, washed with PBS, and incubated with the second antibody in PBS containing 3% (w/v) BSA for 1 hr. The images were obtained with confocal microscopes (Olympus FV300-BX;Olympus Optical, Tokyo, Japan) (Zeiss LSM510; Zeiss, Thornwood, NY).
RESULTS
Identification of β-catenin as a S-SCAM-interacting molecule
To identify S-SCAM-interacting molecules, we performed a yeast two-hybrid screening using S-SCAM as bait, as described previously (Ide et al., 1999a). We obtained 34 positive independent clones from a rat brain cDNA library. Among them, we found one clone encoding β-catenin. The interaction of β-catenin with MAGI-1 or KIAA0705 in epithelial cells has been reported previously (Dobrosotskaya et al., 2000; Kawajiri et al., 2000). To confirm the interaction of S-SCAM with β-catenin in neurons, we immunoprecipitated S-SCAM from rat brains. β-Catenin was coimmunoprecipitated with S-SCAM (Fig.1A). We subsequently determined whether S-SCAM was colocalized with β-catenin in neurons. In the primary cultured hippocampal neurons, S-SCAM formed clusters on dendrites apposed to those of synaptophysin, although some clusters of synaptophysin were free of S-SCAM (Fig. 1B). Similarly, S-SCAM was colocalized with β-catenin, but some clusters of β-catenin were free of S-SCAM (Fig. 1C). We determined how great a population of S-SCAM clusters was associated with β-catenin. We counted the clusters from three neurons. Of the S-SCAM clusters, 56.7 ± 7.1% were colocalized with β-catenin, whereas of the β-catenin clusters, 47.6 ± 9.8% were colocalized with S-SCAM.
Fig. 1.
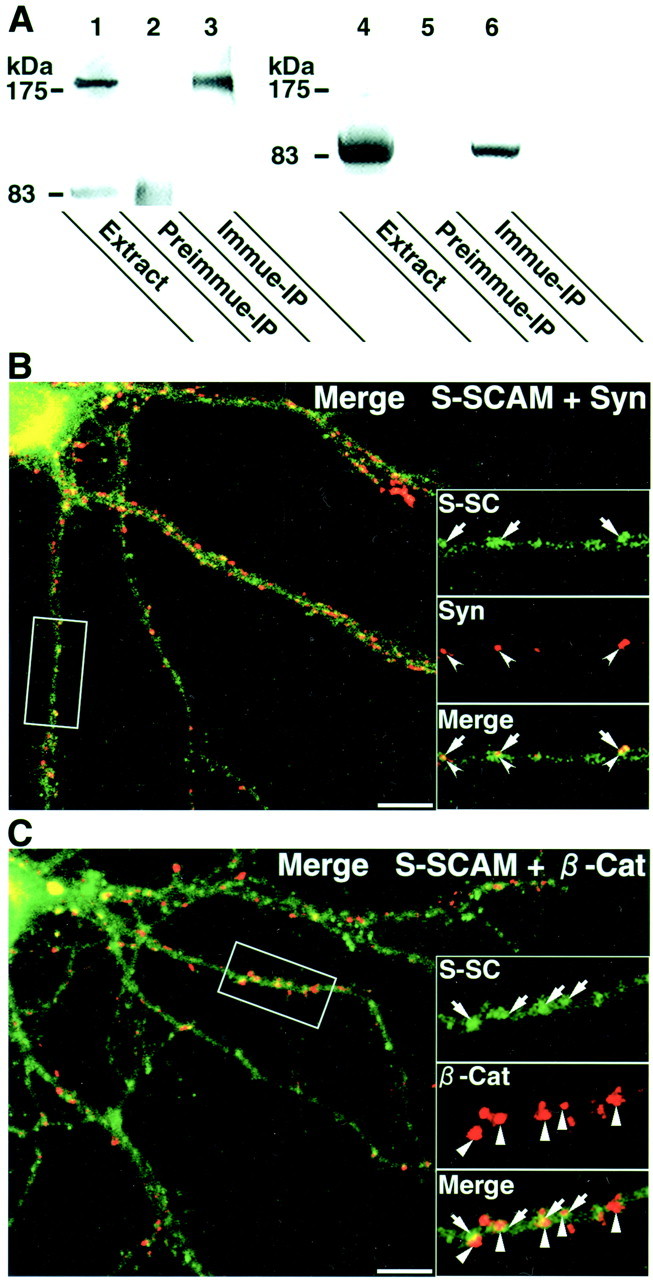
Interaction of S-SCAM and β-catenin in neurons.A, Coimmunoprecipitation of S-SCAM and β-catenin. S-SCAM was immunoprecipitated from rat brains, and the immunoprecipitate was immunoblotted with either the anti-S-SCAM antibody (lanes 1–3) or the anti-β-catenin antibody (lanes 4–6). Lanes 1, 4, The original rat brain extracts (Extract); lanes 2, 5, the immunoprecipitates with the preimmune serum (Preimmune-IP); lanes 3, 6, the immunoprecipitates with the anti-S-SCAM antibody (Immune-IP). B, Laser confocal image of a hippocampal neuron stained with the anti-S-SCAM and the anti-synaptophysin antibodies. Insets, Areas demarcated at higher magnification. S-SC, S-SCAM (arrows); Syn, synaptophysin (arrowheads); Merge, a superimposed image of S-SCAM and synaptophysin. Scale bar, 10 μm. C, Laser confocal image of a hippocampal neuron stained with the anti-S-SCAM and the anti-β-catenin antibodies. Insets, Areas demarcated at higher magnification. S-SC, S-SCAM (arrows); β-Cat, β-catenin (triangles); Merge, a superimposed image of S-SCAM and β-catenin. Scale bar, 10 μm.
β-Catenin-interacting region of S-SCAM and S-SCAM-interacting region of β-catenin
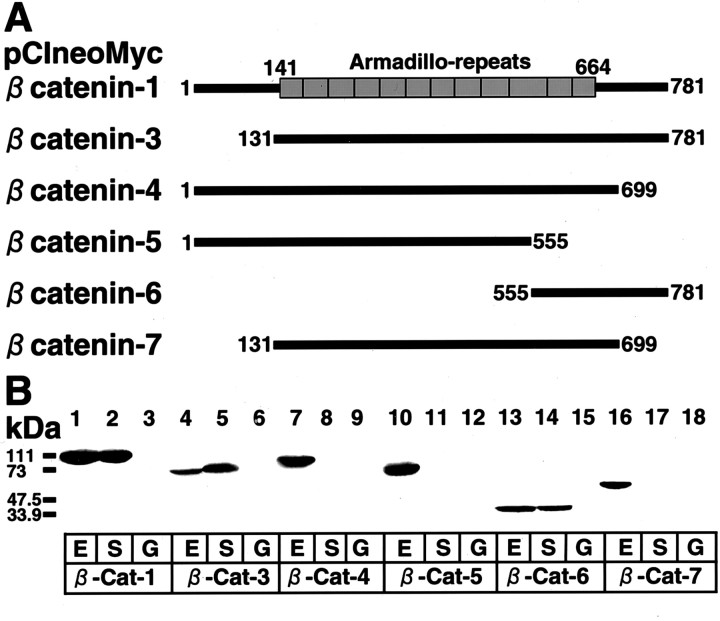
To determine the β-catenin-interacting region of S-SCAM, we prepared various Myc-tagged constructs of S-SCAM (Fig.2A). We expressed these proteins in COS cells, immunoprecipitated them with the anti-Myc antibody, and examined whether the endogenous β-catenin was coimmunoprecipitated. β-Catenin was detected in the immunoprecipitates from COS cells expressing S-SCAM-1, -10, and -12 but did not associate with S-SCAM-8 (Fig. 2B). To determine the S-SCAM-interacting region of β-catenin, we prepared various Myc-tagged constructs of β-catenin (Fig.3A). We expressed these proteins in COS cells and examined which constructs interacted with GST-fusion protein containing the C-terminal region of S-SCAM. β-Catenin-1, -3, and -6 interacted with S-SCAM, whereas β-catenin-4, -5, and -7 did not (Fig. 3B). These data suggest that the C-terminal region of S-SCAM interacts with the C-terminal region of β-catenin.
Fig. 2.
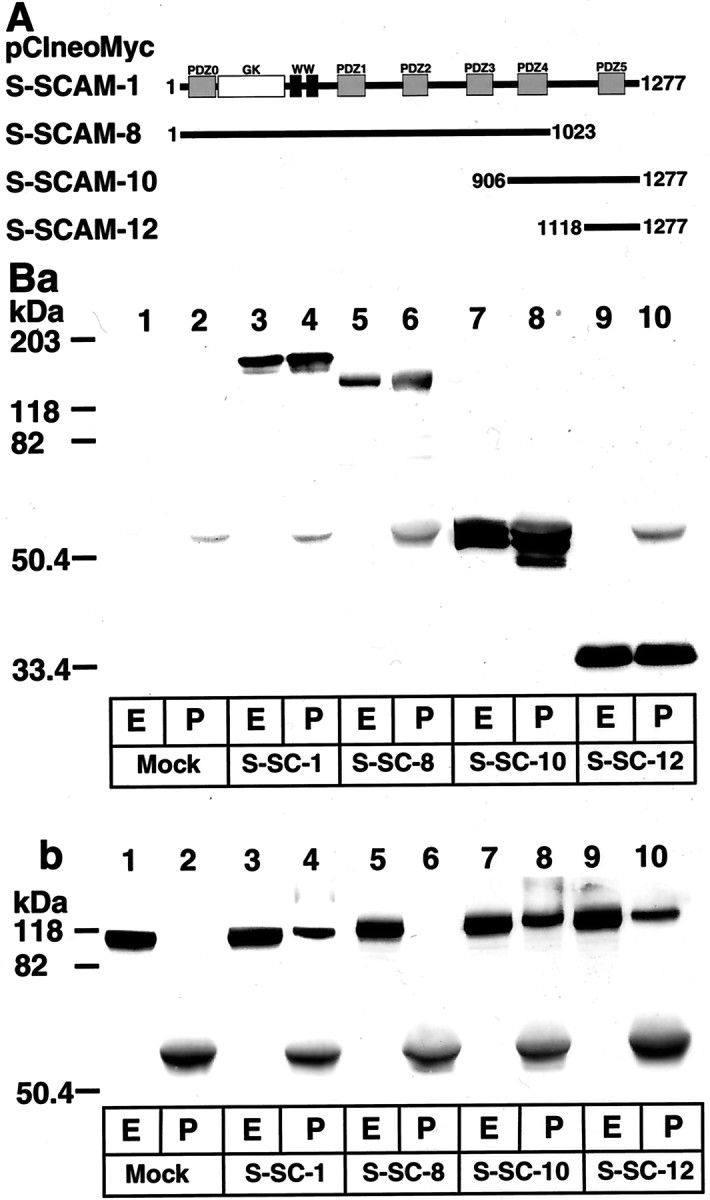
β-Catenin-interacting domain of S-SCAM.A, Schematic of various constructs of S-SCAM.Gray boxes, PDZ domains; white box, the GK domain; black boxes, WW domains. The numbers of the first and the last amino acid residues of each construct are shown. B, Interaction of the C-terminal region of S-SCAM with β-catenin. Various Myc-tagged S-SCAM proteins were expressed in COS cells and were immunoprecipitated with the anti-Myc antibody. The original extracts and the immunoprecipitates were immunoblotted with either the anti-Myc antibody (a) or the anti-β-catenin antibody (b). Lanes 1, 2, Transfected with mock; lanes 3, 4, pClneoMyc S-SCAM-1; lanes 5, 6, pClneoMyc S-SCAM-8; lanes 7, 8, pClneoMyc S-SCAM-10; lanes 9, 10, pClneoMyc S-SCAM-12.Lanes 1, 3, 5, 7, 9, The original extracts (E); lanes 2, 4, 6, 8, 10, the immunoprecipitates (P).
Fig. 3.
S-SCAM-interacting domain of β-catenin.A, Schematic of various Myc-tagged constructs of β-catenin. Boxes, Armadillo repeats. The numbers of the first and the last amino acid residues of each construct are shown.B, Interaction of the C-terminal region of β-catenin with the C-terminal region of S-SCAM. The extracts of COS cells expressing various Myc-tagged β-catenin proteins were incubated with either the GST–S-SCAM-10 containing PDZ4 and PDZ5 or the GST fixed on glutathione beads. The proteins on the beads were immunoblotted with the anti-Myc antibody. Lanes 1–3, pClneoMyc β-catenin-1; lanes 4–6, pClneoMyc β-catenin-3;lanes 7–9, pClneoMyc β-catenin-4; lanes 10–12, pClneoMyc β-catenin-5; lanes 13–15, pClneoMyc β-catenin-6; lanes 16–18, pClneoMyc β-catenin-7. Lanes 1, 4, 7, 10, 13, 16, The original extracts (E); lanes 2, 5, 8, 11, 14, 17, the precipitates with the GST–S-SCAM-10 (S); lanes 3, 6, 9, 12, 15, 18, the precipitates with the GST (G).
Localization of various GFP–S-SCAM constructs in rat hippocampal slice and primary cultured hippocampal neurons
To determine the region involved in the synaptic targeting of S-SCAM, we prepared GFP-tagged constructs containing various regions of S-SCAM (Fig. 4A). We expressed GFP–S-SCAM-1 in rat hippocampal slice neurons using Sindbis virus. The protein began to appear at 9 hr after the infection and was diffusely distributed in the soma and neurites (Fig.4B, a). At 12 hr after the infection, the clusters appeared on the dendrites, and the number of clusters increased (Fig. 4B, b and c). We also tested other GFP constructs. GFP–S-SCAM-8 lacking PDZ5 was diffusely distributed in the soma and neurites (Fig. 4C,a). GFP–S-SCAM-10 (PDZ4 and PDZ5) was diffusely distributed in most parts, although small clusters were observed in some parts (Fig. 4C, b). GFP–S-SCAM-4 (PDZ2, PDZ3, PDZ4, and PDZ5), -13 (PDZ3, PDZ4, and PDZ5), and -19 (PDZ2, PDZ3, and PDZ5) formed more clusters on dendrites (data not shown and Fig.4C, c and d). Therefore, PDZ5 is necessary for S-SCAM to form clusters on dendrites, although other regions may also be involved in the formation of clusters. We subsequently expressed GFP–S-SCAM-1, -10, -13, and -19 in rat primary cultured hippocampal neurons and examined whether these proteins were localized at synapses. The clusters of GFP–S-SCAM-1, -13, and -19 were apposed to those of synaptophysin, suggesting synaptic localization (Fig. 5A,C,D). GFP–S-SCAM-10 formed a few clusters in primary cultured hippocampal neurons, and these clusters were also apposed to those of synaptophysin (Fig. 5B).
Fig. 4.
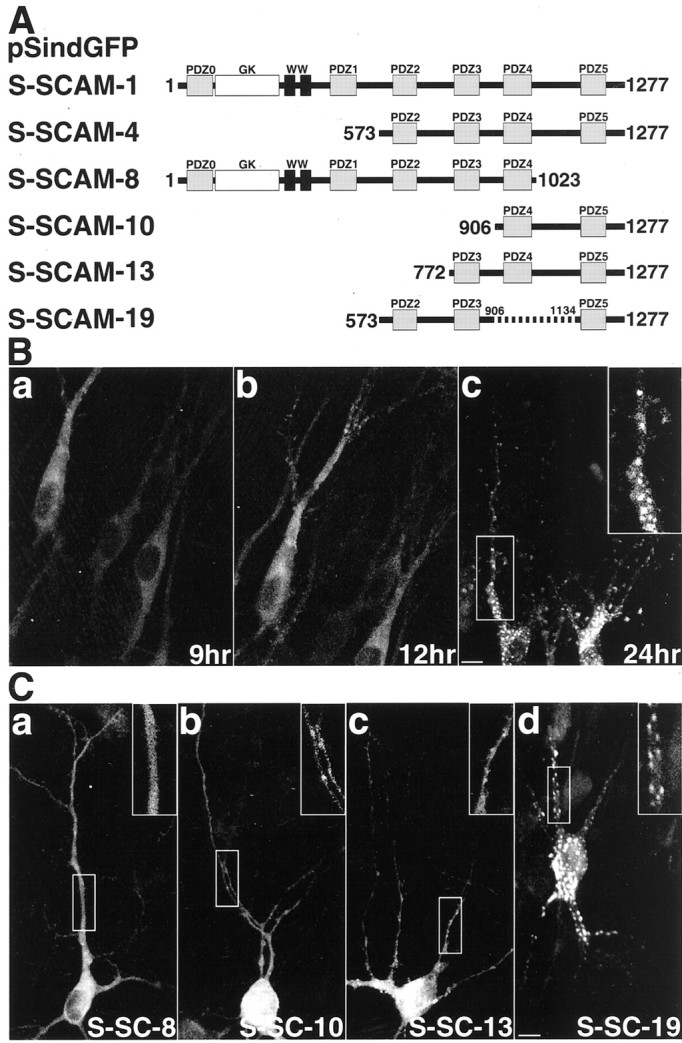
Various GFP–S-SCAM proteins expressed in hippocampal slice neurons. A, Schematic of various GFP constructs of S-SCAM. Gray boxes, PDZ domains;white box, GK domain; black boxes, WW domains. B, Temporal profile of the expression of GFP–S-SCAM-1 in hippocampal slice neurons. a, 9 hr after the infection; b, 12 hr after the infection;c, 24 hr after the infection. Insets, Demarcated areas at higher magnification to show the clusters of GFP–S-SCAM-1. Scale bar, 10 μm. C, Various GFP–S-SCAM proteins in hippocampal slice neurons.Insets, Demarcated areas at higher magnification.a, GFP–S-SCAM-8; b, GFP–S-SCAM-10;c, GFP–S-SCAM-13; d, GFP–S-SCAM-19. Scale bar, 10 μm.
Fig. 5.
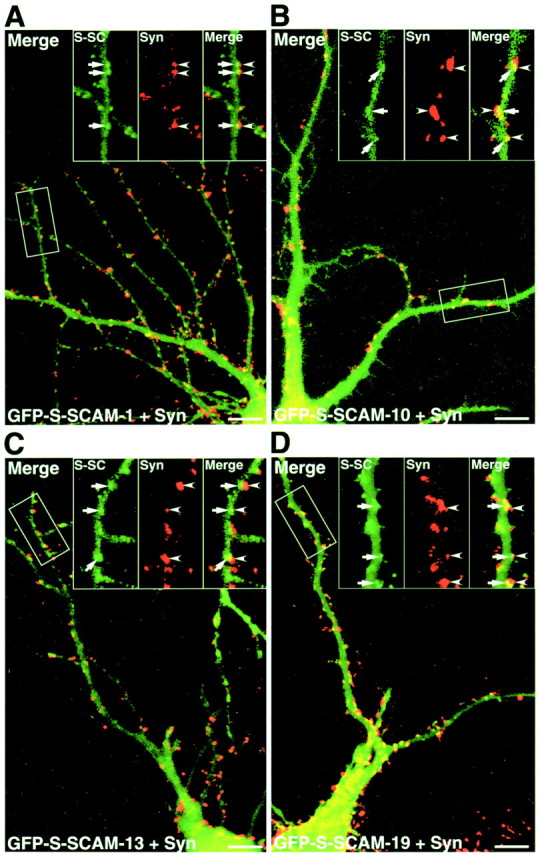
Various GFP–S-SCAM proteins expressed in primary cultured hippocampal neurons. A, GFP–S-SCAM-1. GFP–S-SCAM-1 formed clusters apposed to those of synaptophysin.B, GFP–S-SCAM-10. C, GFP–S-SCAM-13.D, GFP–S-SCAM-19. GFP–S-SCAM-10, -13, and -19 also formed clusters. Insets, Demarcated areas at higher magnification. S-SC, GFP–S-SCAM (arrows); Syn, synaptophysin (arrowheads); Merge, a superimposed image of S-SCAM and synaptophysin. Scale bars, 10 μm.
Localization of various GFP–β-catenin constructs in rat hippocampal slice and primary cultured hippocampal neurons
To determine the region involved in the synaptic targeting of β-catenin, we prepared GFP-tagged constructs containing various regions of β-catenin (Fig.6A). We expressed GFP–β-catenin-1 in rat hippocampal slice neurons using Sindbis virus. The protein began to express at 6 hr after the infection (Fig.6B). At that time, the protein was already clustered on dendrites. We also expressed other GFP constructs. GFP–β-catenin-2 lacking the N-terminal region was clustered on dendrites (Fig. 6C, a). GFP–β-catenin-6 containing all armadillo repeats also formed clusters on dendrites (Fig. 6C, b). In contrast, GFP–β-catenin-8 containing the C-terminal region and GFP–β-catenin-9 containing the last 4.5 armadillo repeats and the C-terminal region were diffusely distributed (Fig. 6C, c and d). GFP–β-catenin-10 containing the N-terminal region and the first five armadillo repeats was localized in the nucleus (Fig. 6C,e). GFP–β-catenin-11 containing the last six armadillo repeats was diffusely distributed (Fig. 6C,f). We compared the localization of GFP–β-catenin-1 and -6 with that of synaptophysin in rat primary cultured hippocampal neurons. Both GFP–β-catenin-1 and GFP–β-catenin-6 were colocalized with synaptophysin, suggesting that the C-terminal region involved in the interaction with S-SCAM is not necessary for the synaptic targeting of β-catenin (Fig.7A,B).
Fig. 6.
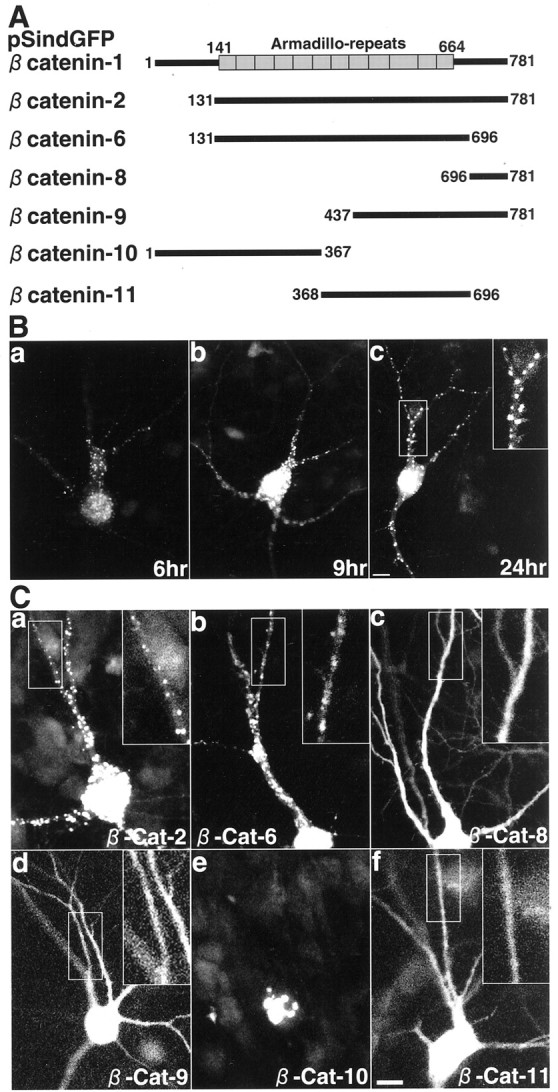
Various GFP–β-catenin proteins expressed in hippocampal slice neurons. A, Schematic of various GFP-tagged constructs of β-catenin. Boxes, Armadillo repeats. B, Temporal profile of the expression of GFP–β-catenin-1 in hippocampal slice neurons.a, 6 hr after the infection; b, 9 hr after the infection; c, 24 hr after the infection.Inset, Demarcated area at higher magnification. Scale bar, 10 μm. C, Various GFP–β-catenin proteins in hippocampal slice neurons. Insets, Demarcated areas at higher magnification. a, GFP–β-catenin-2;b, GFP–β-catenin-6; c, GFP–β-catenin-8; d, GFP–β-catenin-9;e, GFP–β-catenin-10; f, GFP–β-catenin-11. Scale bar, 10 μm.
Fig. 7.
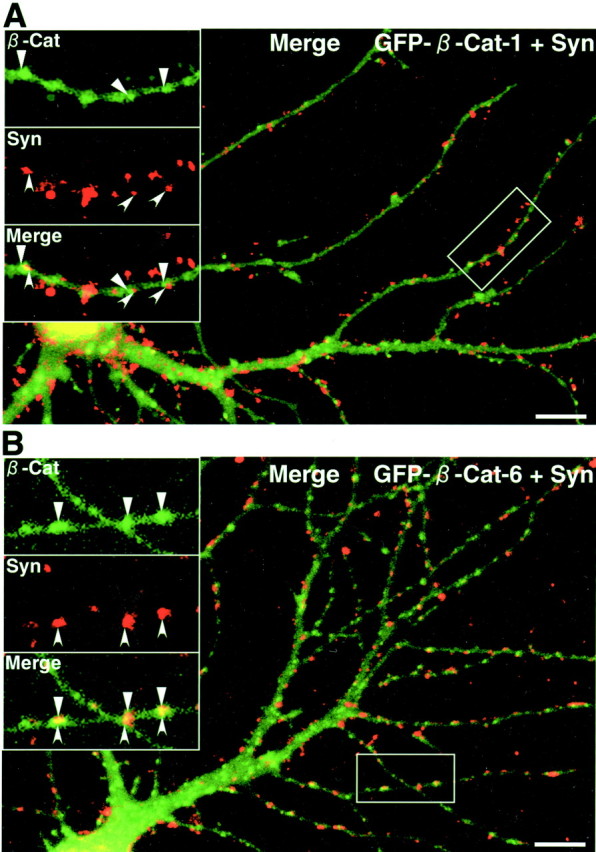
GFP–β-catenin proteins containing the armadillo repeats in primary cultured hippocampal neurons. A, GFP–β-catenin-1. B, GFP–β-catenin-6. Both GFP–β-catenin-1 and GFP–β-catenin-6 formed clusters colocalized with those of synaptophysin. Insets, Demarcated areas at higher magnification. β-Cat, GFP–β-catenin (triangles); Syn, synaptophysin (arrowheads); Merge, a superimposed image of GFP–β-catenin and synaptophysin. Scale bars, 10 μm.
The C-terminal region of β-catenin blocks the synaptic targeting of S-SCAM on dendrites, and the C-terminal region of S-SCAM was recruited to synapses with β-catenin
Next, we examined the effect of the overexpressed S-SCAM-interacting region of β-catenin on the localization of S-SCAM and that of the overexpressed β-catenin-interacting region of S-SCAM on the localization of β-catenin. For this purpose, we prepared the pSindMyc–S-SCAM-1 and -10. Myc–S-SCAM-1 containing full-length S-SCAM was concentrated at synapses such as GFP–S-SCAM-1 (Fig.8A). GFP–β-catenin-8 was diffusely distributed (Fig. 8B). When coexpressed with GFP–β-catenin-8, Myc–S-SCAM-1 became diffusely distributed (Fig. 8C). We also tested the effect of GFP–β-catenin-8 on the synaptic targeting of the endogenous S-SCAM. Forty-eight hours after the infection, we immunostained neurons with the anti-S-SCAM and the anti-synaptophysin antibodies and counted the clusters from three neurons. In neurons expressing GFP–β-catenin-8, the number of the clusters of S-SCAM decreased from 47.6 ± 5.5 to 30.3 ± 2.5 per 280 μm of the dendrite. Furthermore, 74.2 ± 5.1% of S-SCAM clusters were colocalized with synaptophysin in neurons without GFP–β-catenin-8, whereas only 32.3 ± 9.1% of the clusters were colocalized with synaptophysin in neurons expressing GFP–β-catenin-8 (Fig. 8D). In contrast, GFP–β-catenin-8 did not affect the colocalization of the clusters of NR2A with synaptophysin (Fig. 8E) or the number of the clusters of synaptophysin (65.0 ± 7.9 and 65.3 ± 8.1 per 280 μm of the dendrite in neurons with and without GFP–β-catenin-8, respectively). Myc–S-SCAM-10 containing PDZ4 and PDZ5 formed small clusters like GFP–S-SCAM-10 in some parts but did not form clusters in most parts (Fig.9A). When coexpressed with GFP–β-catenin-1, Myc–S-SCAM-10 did not block the synaptic targeting of GFP–β-catenin-1 (Fig. 9B, a). Conversely, Myc–S-SCAM-10 formed more and larger clusters apposed to those of synaptophysin (Fig. 9B, b). These data suggest that β-catenin is involved in the synaptic targeting of S-SCAM.
Fig. 8.
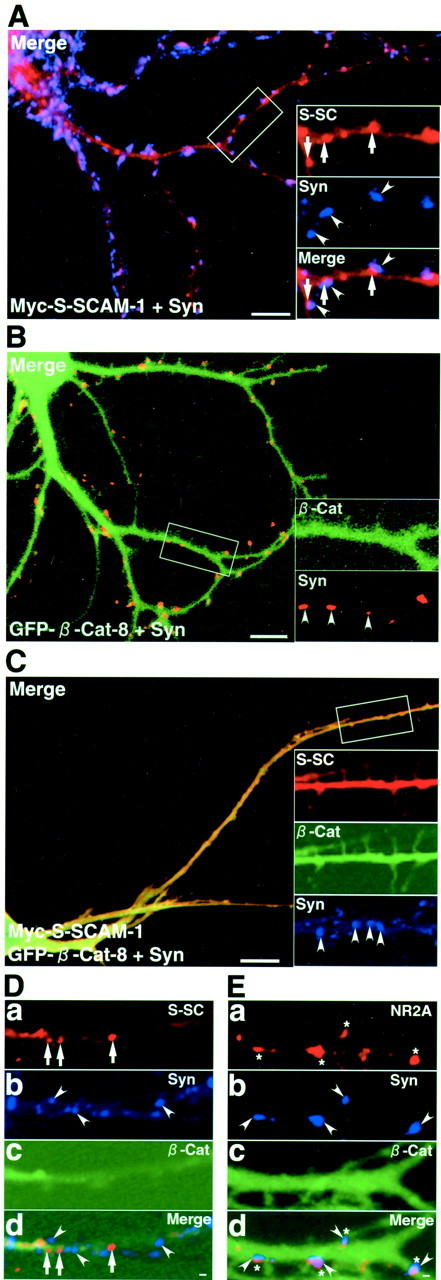
The C-terminal region of β-catenin inhibits the synaptic targeting of S-SCAM. A, Myc–S-SCAM-1 in a primary cultured hippocampal neuron. Hippocampal neurons expressing Myc–S-SCAM-1 were stained with the polyclonal sheep anti-Myc and the anti-synaptophysin antibodies. Myc–S-SCAM-1 formed clusters apposed to those of synaptophysin, such as GFP–S-SCAM-1. The large image shows the merged image of Myc–S-SCAM-1 and synaptophysin.Insets, Demarcated areas at higher magnification.S-SC, Myc–S-SCAM-1 (arrows);Syn, synaptophysin (arrowheads);Merge, a superimposed image of Myc–S-SCAM-1 and synaptophysin. Scale bar, 10 μm. B, GFP–β-catenin-8 in a primary cultured hippocampal neuron. GFP–β-catenin-8 was diffusely distributed as in hippocampal slice neurons. The large image shows the merged image of GFP–β-catenin-8 and synaptophysin. Insets, Demarcated areas at higher magnification. β-Cat, GFP–β-catenin-8;Syn, synaptophysin (arrowheads). Scale bar, 10 μm. C, Myc–S-SCAM-1 coexpressed with GFP–β-catenin-8 in a primary cultured hippocampal neuron. Myc–S-SCAM-1 became diffusely distributed when coexpressed with GFP–β-catenin-8. The large image shows the merged image of Myc–S-SCAM-1 and GFP–β-catenin-8. Insets, Demarcated areas at higher magnification. S-SC, Myc–S-SCAM-1; β-Cat, GFP–β-catenin-8;Syn, synaptophysin (arrowheads). Scale bar, 10 μm. D, The endogenous S-SCAM in neurons expressing GFP–β-catenin-8. The clusters of the endogenous S-SCAM were not apposed to those of synaptophysin in neurons expressing GFP–β-catenin-8. S-SC, S-SCAM; Syn, synaptophysin; β-Cat, GFP–β-catenin-8;Merge, a superimposed image of S-SCAM, synaptophysin, and GFP–β-catenin-8. Scale bar, 1 μm. E, NMDA receptors in neurons expressing GFP–β-catenin-8. The clusters of NMDA receptor subunit 2A were apposed to those of synaptophysin even in neurons expressing GFP–β-catenin-8. Syn, Synaptophysin; β-Cat, GFP–β-catenin-8;Merge, a superimposed image of NMDA receptor subunit 2A, synaptophysin, and GFP–β-catenin-8. Scale bar, 1 μm.
Fig. 9.
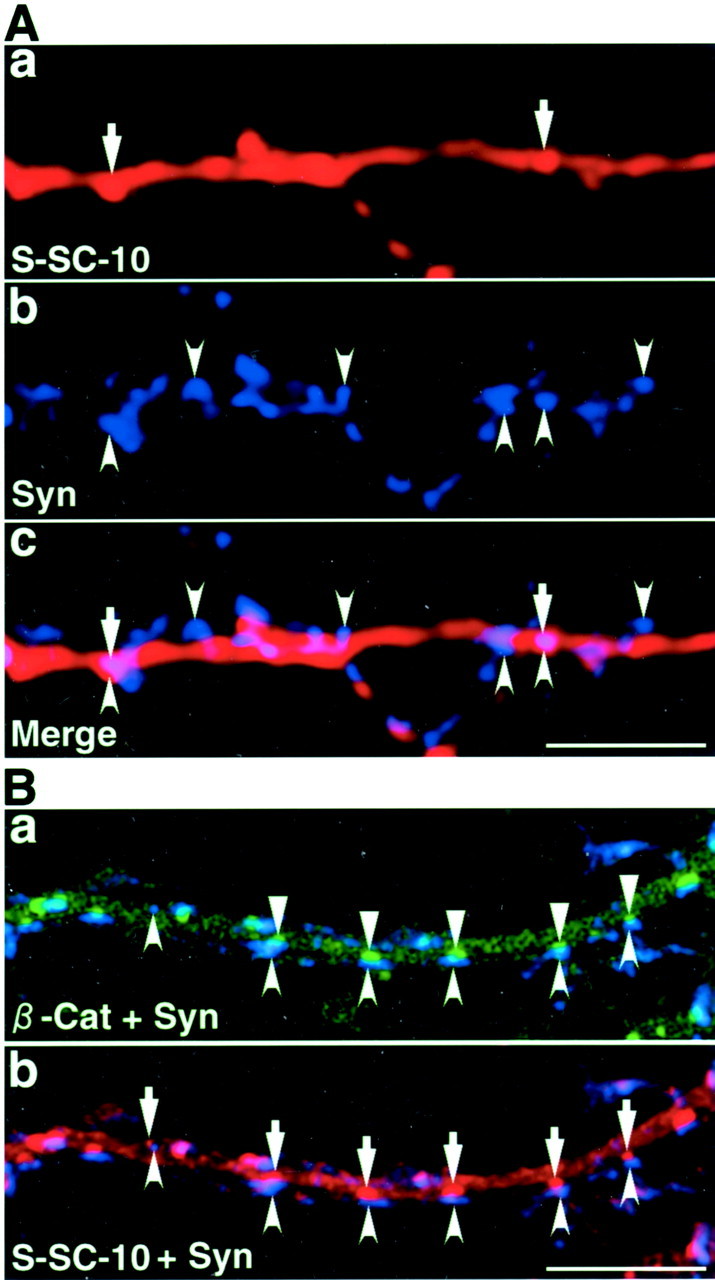
β-Catenin facilitates the accumulation of the C-terminal region of S-SCAM at synapses. A, Myc–S-SCAM-10 containing PDZ4 and PDZ5 in a primary cultured hippocampal neuron. Neurons expressing Myc–S-SCAM-10 were stained with the polyclonal sheep anti-Myc and the anti-synaptophysin antibodies. The distribution of Myc–S-SCAM-10 was similar to that of GFP–S-SCAM-10. The number of clusters formed by Myc–S-SCAM-10 was small. a, Myc–S-SCAM-10 (arrows);b, synaptophysin (arrowheads);c, a superimposed image of Myc–S-SCAM-10 and synaptophysin. Scale bar, 10 μm. B, Myc–S-SCAM-10 coexpressed with GFP–β-catenin-1. Primary cultured hippocampal neurons expressing Myc–S-SCAM-10 and GFP–β-catenin-1 were immunostained with the sheep anti-Myc and the mouse anti-synaptophysin antibodies. a, Merged image of GFP–β-catenin-1 (triangles) and synaptophysin (arrowheads); b, merged image of Myc–S-SCAM-10 (arrows) and synaptophysin (arrowheads). Scale bar, 1 μm.
The cytoplasmic region of NMDA receptor subunit 2A did not block the synaptic targeting of S-SCAM
Because PDZ5 of S-SCAM binds various molecules besides β-catenin, other molecules may also be involved in the synaptic targeting of S-SCAM. We tested whether the cytoplasmic region of NR2A blocked the synaptic targeting of S-SCAM. The distribution of the GFP-tagged cytoplasmic region of NR2A (GFP–NR2A) was similar to that of GFP–S-SCAM-10 (Fig.10A). It was diffusely distributed in most parts, although some clusters were observed. However, this protein did not block the synaptic targeting of Myc–S-SCAM-1, unlike GFP–β-catenin-8 (Fig. 10B). Thus, the cytoplasmic region of β-catenin is distinct from that of NR2A in the inhibition of the synaptic targeting of S-SCAM.
Fig. 10.
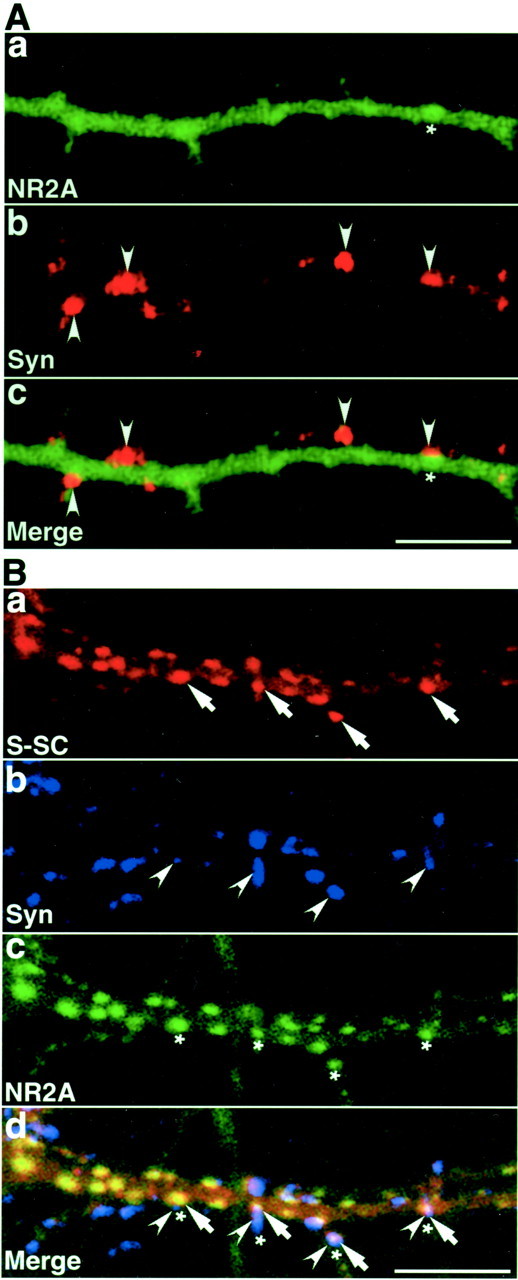
The cytoplasmic region of NMDA receptor subunit 2A does not inhibit the synaptic targeting of S-SCAM. A, GFP-tagged cytoplasmic region of NR2A (GFP–NR2A) in a primary cultured hippocampal neuron. The distribution of GFP–NR2A was similar to that of GFP–S-SCAM-10. The number of clusters formed by GFP–NR2A was small. a, GFP–NR2A (asterisk);b, synaptophysin (arrowheads);c, a superimposed image of GFP–NR2A and synaptophysin. Scale bar, 10 μm. B, Myc–S-SCAM-1 coexpressed with GFP–NR2A. Primary cultured hippocampal neurons expressing Myc–S-SCAM-1 and GFP–NP2A were immunostained with the sheep anti-Myc and the mouse anti-synaptophysin antibodies. GFP–NR2A did not inhibit the synaptic targeting of Myc–S-SCAM-1 and formed more clusters when coexpressed with Myc–S-SCAM-1. a, Myc–S-SCAM-1 (arrows); b, synaptophysin (arrowheads); c, GFP–NR2A (asterisks); d, a superimposed image of Myc–S-SCAM-1, synaptophysin, and GFP–NR2A. Scale bar, 10 μm.
DISCUSSION
In epithelial cells, β-catenin links E-cadherin to the cytoskeleton and regulates gene transcription. β-Catenin also binds lin-7 in Madin–Darby canine kidney cells, which fixes GABA transporter to the plasma membrane by blocking endocytosis (Perego et al., 1999,2000). In neurons, β-catenin is enriched at synapses and binds toN-cadherin and other cadherin-like molecules (Uchida et al., 1996; Benson and Tanaka, 1998; Miskevich et al., 1998). The role of β-catenin at synapses, however, is unclear. In this study, we reveal that S-SCAM interacts with β-catenin. β-Catenin is coimmunoprecipitated with S-SCAM from rat brain. β-Catenin is partially colocalized with S-SCAM in neurons. In vitrobinding assays indicate that the C-terminal region of S-SCAM binds to the C-terminal region of β-catenin. We have tried to clarify the meaning of this interaction.
We have determined which region of S-SCAM is involved in its synaptic targeting. The deletion construct of S-SCAM lacking the sixth PDZ domain (PDZ5) is not localized at synapses, whereas the constructs containing PDZ5 are localized at synapses. Because GFP-tagged PDZ5 did not express, we could not conclude that PDZ5 is sufficient for the synaptic targeting. Our findings indicate, however, that PDZ5 is necessary for the synaptic targeting. This result is consistent with the hypothesis that MAGI-1 is targeted to the lateral epithelial membrane via the C-terminal region (Nishimura et al., 2000). Because β-catenin binds to PDZ5, we hypothesized that the interaction with β-catenin is involved in the synaptic targeting of S-SCAM. The overexpressed C-terminal region of β-catenin is diffusely distributed in neurons and blocks the synaptic targeting of the coexpressed S-SCAM, supporting this model. We immunostained the endogenous β-catenin and S-SCAM with the antibodies at various stages of synaptogenesis to determine which molecule is targeted to synapses earlier, but we did not reach any conclusions because of the different sensitivities of the antibodies. However, GFP–β-catenin is concentrated at synapses earlier than GFP–S-SCAM when expressed in neurons, suggesting that β-catenin is targeted to synapses before S-SCAM. The C-terminal region of β-catenin did not inhibit the synaptic localization of NR2A or decrease or increase the number of the clusters of synaptophysin. These findings do not support the possibility that the C-terminal region of β-catenin affects synaptogenesis. It should be noted that we expressed GFP–β-catenin-8 in neurons 10 d after plating and could maintain them for only 2 d. If GFP–β-catenin-8 is expressed in neurons at an earlier stage and if neurons expressing GFP–β-catenin-8 are cultured for a longer time, the effect on the localization of other synaptic proteins may be detected.
The protein with PDZ4 and PDZ5 of S-SCAM (GFP–S-SCAM-10 or Myc–S-SCAM-10) can accumulate at synapses but is resistant to being targeted to synapses, compared with other constructs such as GFP–S-SCAM-1 (the full length), GFP–S-SCAM-13 (PDZ3, PDZ4, and PDZ5), and GFP–S-SCAM-19 (PDZ2, PDZ3, and PDZ5). Even so, we consider that the region covering PDZ4 and PDZ5 is sufficient for the synaptic targeting, because Myc–S-SCAM-10 is clustered effectively at synapses when coexpressed with β-catenin. The reason why the synaptic targeting of S-SCAM-10 depends on the overexpressed β-catenin while the targeting of other constructs, including GFP–S-SCAM-13 and -19, does not is currently unclear. PDZ3 may bind some molecule interacting with β-catenin, and the affinity of S-SCAM-10 for β-catenin may be lower than that of proteins with both PDZ3 and PDZ5. PDZ5 of S-SCAM binds various molecules. We have raised the question of whether β-catenin is specific in the inhibition of the synaptic targeting of S-SCAM. We have tested the C-terminal region of NR2A. Most of GFP–NR2A is diffusely distributed in neurons, but GFP–NR2A does not affect the synaptic localization of S-SCAM. This result supports the proposition that the C-terminal region of β-catenin has a specificity to block the synaptic localization of S-SCAM.
We have also determined that the region of the armadillo repeats of β-catenin is involved in the synaptic targeting. Among the constructs that we have tested, only those with all of the repeats are clustered at synapses. A recent study revealed that the cytoplasmic domain of E-cadherin interacts with all 12 armadillo repeats of β-catenin (Huber and Weis, 2001). Therefore, β-catenin is recruited to synapses by interaction with molecules such as N-cadherin, which requires all of the armadillo repeats to bind β-catenin, and the interaction with S-SCAM does not play an essential role in the synaptic localization of β-catenin. Our data establish a major role for β-catenin in recruiting S-SCAM to synapses. Although neuroligin and GKAP bind S-SCAM, the interactions with these molecules are not necessary for the synaptic targeting of S-SCAM.
Footnotes
This study was supported by grants-in-aid for Scientific Research and by special coordination funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science, and Technology; by a grant from the Novartis Foundation (Japan) for the Promotion of Science, 1999; and by a grant from the Yamanouchi Foundation for Research on Metabolic Disorders, 2000. We thank Prof. Shigetada Nakanishi (Kyoto University, Kyoto, Japan) for cDNA of rat NMDA receptor subunit 2A and Prof. Masatoshi Takeichi (Kyoto University) for cDNA of mouse β-catenin. pClneoMyc constructs of S-SCAM and β-catenin were prepared by Y.H. in the Takai Biotimer Project, Exploratory Research for Advanced Technology, headed by Prof. Yoshimi Takai (Osaka University, Osaka, Japan).
Correspondence should be addressed to Dr. Yutaka Hata, Department of Medical Biochemistry, Graduate School of Medicine, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo 113-8519, Japan. E-mail: yuhammch@med.tmd.ac.jp.
REFERENCES
- 1.Benson DL, Tanaka H. N-cadherin redistribution during synaptogenesis in hippocampal neurons. J Neurosci. 1998;18:6892–6904. doi: 10.1523/JNEUROSCI.18-17-06892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dobrosotskaya IY. Identification of mNET1 as a candidate ligand for the first PDZ domain of MAGI-1. Biochem Biophys Res Commun. 2001;283:969–975. doi: 10.1006/bbrc.2001.4880. [DOI] [PubMed] [Google Scholar]
- 3.Dobrosotskaya IY, James GL. MAGI-1 interacts with β-catenin and is associated with cell-cell adhesion structures. Biochem Biophys Res Commun. 2000;270:903–909. doi: 10.1006/bbrc.2000.2471. [DOI] [PubMed] [Google Scholar]
- 4.Dobrosotskaya IY, Guy RK, James GL. MAGI-1, a membrane-associated guanylate kinase with a unique arrangement of protein-protein interaction domains. J Biol Chem. 1997;272:31589–31597. doi: 10.1074/jbc.272.50.31589. [DOI] [PubMed] [Google Scholar]
- 5.Goslin K, Asmussen H, Banker G. Rat hippocampal neurons in low-density culture. In: Banker G, Goslin K, editors. Culturing nerve cells. MIT; Cambridge, MA: 1998. pp. 339–370. [Google Scholar]
- 6.Hirao K, Hata Y, Ide N, Takeuchi M, Irie M, Yao I, Deguchi M, Toyoda A, Sudhof TC, Takai Y. A novel multiple PDZ domain-containing molecule interacting with N-methyl-d-aspartate receptors and neuronal cell adhesion protein. J Biol Chem. 1998;273:21105–21110. doi: 10.1074/jbc.273.33.21105. [DOI] [PubMed] [Google Scholar]
- 7.Hirao K, Hata Y, Yao I, Deguchi M, Kawabe H, Mizoguchi A, Takai Y. Three isoforms of synaptic scaffolding molecule and their characterization. J Biol Chem. 2000;275:2966–2972. doi: 10.1074/jbc.275.4.2966. [DOI] [PubMed] [Google Scholar]
- 8.Huber AH, Weis WI. The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 9.Ide N, Hata Y, Deguchi M, Hirao K, Yao I, Takai Y. Interaction of S-SCAM with neural plakophilin-related armadillo-repeat protein/δ-catenin. Biochem Biophys Res Commun. 1999a;256:456–461. doi: 10.1006/bbrc.1999.0364. [DOI] [PubMed] [Google Scholar]
- 10.Ide N, Hata Y, Nishioka H, Hirao K, Yao I, Deguchi M, Mizoguchi A, Nishimori H, Tokino T, Nakamura Y, Takai Y. Localization of membrane-associated guanylate kinase (MAGI)-1/BAI-associated protein (BAP) 1 at tight junctions of epithelial cells. Oncogene. 1999b;16:7810–7815. doi: 10.1038/sj.onc.1203153. [DOI] [PubMed] [Google Scholar]
- 11.Kawajiri A, Itoh N, Fukata M, Nakagawa M, Yamada M, Iwamatsu A, Kaibuchi K. Identification of a novel β-catenin-interacting protein. Biochem Biophys Res Commun. 2000;273:712–717. doi: 10.1006/bbrc.2000.3002. [DOI] [PubMed] [Google Scholar]
- 12.Kim E, Naisbitt S, Hsueh YP, Rao A, Rothschild A, Craig AM, Sheng M. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1996;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miskevich F, Zhu Y, Ranscht B, Sanes JR. Expression of multiple cadherins and catenins in the chick optic tectum. Mol Cell Neurosci. 1998;12:240–255. doi: 10.1006/mcne.1998.0718. [DOI] [PubMed] [Google Scholar]
- 14.Nishimura W, Iizuka T, Hirabayashi S, Tanaka N, Hata Y. Localization of BAI-associated protein 1/membrane-associated guanylate kinase-1 at adherens junctions in normal rat kidney cells: polarized targeting mediated by the carboxyl-terminal PDZ domains. J Cell Physiol. 2000;185:358–365. doi: 10.1002/1097-4652(200012)185:3<358::AID-JCP6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 15.Ohtsuka T, Hata Y, Ide N, Yasuda T, Inoue E, Inoue T, Mizoguchi A, Takai Y. nRapGEP: a novel neural GDP/GTP exchange protein for Rap1 small G protein that interacts with synaptic scaffolding molecule (S-SCAM). Biochem Biophys Res Commun. 1999;265:38–44. doi: 10.1006/bbrc.1999.1619. [DOI] [PubMed] [Google Scholar]
- 16.Patrie KM, Drescher AJ, Goyal M, Wiggins RC, Margolis B. The membrane-associated guanylate kinase protein magi-1 binds megalin and is present in glomerular podocytes. J Am Soc Nephrol. 2001;12:667–677. doi: 10.1681/ASN.V124667. [DOI] [PubMed] [Google Scholar]
- 17.Perego C, Vanoni C, Villa A, Longhi R, Kaech SM, Frohli E, Hajnal A, Kim SK, Pietrini G. PDZ-mediated interactions retain the epithelial GABA transporter on the basolateral surface of polarized epithelial cells. EMBO J. 1999;18:2384–2393. doi: 10.1093/emboj/18.9.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perego C, Vanoni C, Massari S, Longhi R, Pietrini G. Mammalian Lin-7 PDZ proteins associate with beta-catenin at the cell-cell junctions of epithelia and neurons. EMBO J. 2000;19:3978–3989. doi: 10.1093/emboj/19.15.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satoh K, Yanai H, Send T, Kohu K, Nakamura T, Okumura N, Matsumine A, Kobayashi S, Toyoshima K, Akiyama T. DAP-1, a novel protein that interacts with the guanylate kinase-like domains of hDLG and PSD-95/SAP90. Genes Cells. 1997;2:415–424. doi: 10.1046/j.1365-2443.1997.1310329.x. [DOI] [PubMed] [Google Scholar]
- 20.Shiratsuchi T, Futamura M, Oda K, Nishimori H, Nakamura Y, Tokino T. Cloning and characterization of BAI-associated protein 1: a PDZ domain-containing protein that interacts with BAI1. Biochem Biophys Res Commun. 1998;247:597–604. doi: 10.1006/bbrc.1998.8603. [DOI] [PubMed] [Google Scholar]
- 21.Shoji H, Tsuchida K, Kishi H, Yamakawa N, Matsuzaki T, Liu Z, Nakamura T, Sugino H. Identification and characterization of a PDZ protein that interacts with activin type receptor. J Biol Chem. 2000;275:5485–5492. doi: 10.1074/jbc.275.8.5485. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi M, Hata Y, Hirao K, Toyoda A, Irie M, Takai Y. SAPAPs: a family of PSD-95/SAP90-associated proteins localized at postsynaptic density. J Biol Chem. 1997;272:11943–11951. doi: 10.1074/jbc.272.18.11943. [DOI] [PubMed] [Google Scholar]
- 23.Uchida N, Honjo Y, Johnson KR, Wheelock MJ, Takeichi M. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J Cell Biol. 1996;135:767–779. doi: 10.1083/jcb.135.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood JD, Yuan J, Margolis RL, Colomer V, Duan K, Kushi J, Kaminsky Z, Kleiderlein JJ, Jr, Sharp AH, Ross CA. Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol Cell Neurosci. 1998;11:149–160. doi: 10.1006/mcne.1998.0677. [DOI] [PubMed] [Google Scholar]
- 25.Wu X, Hepner K, Castelino-Prabhu S, Do D, Kaye MB, Yuan X-J, Wood J, Ross C, Sawyers CL, Whang YE. Evidence for regulation of the PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proc Natl Acad Sci USA. 2000;97:4233–4238. doi: 10.1073/pnas.97.8.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao I, Hata Y, Ide N, Hirao K, Deguchi M, Nishioka H, Mizoguchi A, Takai Y. MAGUIN: a novel neuronal membrane-associated guanylate kinase-interacting protein. J Biol Chem. 1999;274:11889–11896. doi: 10.1074/jbc.274.17.11889. [DOI] [PubMed] [Google Scholar]