Abstract
Despite the previous development of single-gene knock-out mice that exhibit alterations in aggressive behavior, very little progress has been made toward identifying the natural gene variants (alleles) that contribute to individual or strain differences in aggression. Whereas most inbred mouse strains show an intermediate level of inter-male aggression in the resident-intruder or dangler behavioral tests, NZB/B1NJ mice are extremely aggressive and A/J mice are extremely unaggressive. We took advantage of the large phenotypic difference between these strains and used an outcross–backcross breeding protocol and a genome-wide scan to identify aggression quantitative trait loci (QTLs) on distal chromosome 10 (Aggr1;p = 6 × 10−7) and proximal chromosome X (Aggr2; p = 2.14 × 10−5). Candidate genes forAggr1 and Aggr2, respectively, include the diacylglycerol kinase α subunit gene (Dagk1) and the glutamate receptor subunit AMPA3 gene (Gria3). This is the first report of significant aggression QTLs established through a genome-wide scan in any mammal. The mapping of these QTLs is a step toward the definitive identification of mouse alleles that affect aggression and may lead, ultimately, to the discovery of homologous alleles that affect individual differences in aggression within other mammalian species.
Keywords: aggression, quantitative trait locus, QTL, individual differences, genetics, complex trait analysis, inbred mouse strain, NZB/B1NJ, A/J, behavior
Aggressive behaviors are symptoms associated with certain human neuropsychiatric disorders (Swanson et al., 1990; Torrey, 1994; Tardiff et al., 1997; Steadman et al., 1998;Taylor et al., 1998) and are also frequent occurrences among members of the general population (Torrey, 1994; Steadman et al., 1998). The many biological and psychosocial factors that may contribute to multifarious aggressive behaviors are only beginning to be elucidated (Cadoret et al., 1995; Steadman et al., 1998; Riggins-Caspers and Cadoret, 1999). Consequently, current strategies for preventing and treating extreme aggressive behaviors are empirical and often ineffective.
Despite the complexity of these behaviors, there is accumulating evidence from twin studies that genetic factors play a role, together with environment, in predisposing humans toward aggression (Rushton et al., 1986; Tellegen et al., 1988; Coccaro et al., 1993; Coccaro et al., 1997; Eley et al., 1999; Vernon et al., 1999). Some initial strides have been made toward identifying the many genes and gene–environment interactions that are likely to be involved (Brunner et al., 1993;Manuck et al., 1999), but progress has been slow. The dissection of the genetic component of aggressive behaviors is confounded by genetic heterogeneity, polygenic inheritance, incomplete penetrance, and strong environmental influences on expression.
Experimental animals are useful for dissecting complex traits because of the greater degree of control they afford over genetic and environmental variables. Tests of inter-male aggression in the mouse include the “resident-intruder” and “dangler” tests (Kessler et al., 1977; Maxson, 1992). Using these tests, and variants of them, investigators have clearly demonstrated that genes influence aggressive behavior in mice. There are robust differences among inbred mouse strains in levels of aggressive behavior; for example, NZB/B1NJ (NZB) mice are extremely aggressive, A/J (A) mice are markedly unaggressive, and other strains show intermediate levels of aggression (Roubertoux et al., 1999). Moreover, knock-out of any of a number of genes, including genes encoding nitric oxide synthase 1, monoamine oxidase A, the 5-hydroxytryptamine (serotonin) 1B receptor, and regulator of G-protein signaling 2 protein (RGS2), has marked affects on levels of aggressive behavior in mice (Maxson, 2000;Oliveira-Dos-Santos et al., 2000).
Despite the success in creating single-gene mutations that affect mouse aggression, little progress has been made toward identifying the naturally occurring alleles that account for the differences in aggression among inbred mouse strains. Several studies, using reciprocal F1 hybrids and congenics, have implicated regions of the Y chromosome in inter-male aggression (Maxson, 1992; Roubertoux et al., 1994; Guillot et al., 1995; Maxson, 2000), but no whole genome scan has been reported that has revealed statistically significant quantitative trait loci (QTLs) (Lander and Kruglyak, 1995).
We report here a QTL analysis of aggressive behavior in NZB and A mice. We used a two-generation outcross–backcross breeding protocol to generate several hundred second-generation animals that were tested for aggression. Genotyping of aggressive N2 animals was performed at ∼20 centimorgan (cM) intervals. This analysis led to the identification of two significant QTLs that affect aggression.
MATERIALS AND METHODS
Animal housing and breeding protocol. NZB/B1NJ, A/J, and 129T2/SvEmsJ mice were obtained from The Jackson Laboratory (Bar Harbor, ME) at 7–8 weeks age. Mice obtained from The Jackson Laboratory that were to be used for behavioral testing were individually housed in a “home room” for 4 weeks before behavioral testing. 129T2/SvEmsJ males were housed four per cage in the home room for 1 week before being used as opponents in behavioral tests. Additional NZB and A mice, as well as (NZB × A)F1, (A × NZB)F1, and [(NZB × A) × A]N2 mice, were bred at Princeton University in a separate breeding colony room. For breeding, one male and one female mouse were housed together. The male was removed from the cage before the birth of the pups. Litters were culled to no more than five pups during the first 3 postnatal days. Pups were weaned at 27–30 d of age. Male pups were individually housed immediately after weaning and remained individually housed through the end of behavioral testing. They were moved in their cages from the breeding colony room to the home room several weeks before behavioral testing, and behavioral testing was performed in the home room. With the exception of a weekly cage change, the mice were not handled until after the completion of all behavioral tests. Animals were housed in temperature-controlled rooms with a 14/10 hr light/dark cycle (lights on at 5:00 A.M.). They were given Purina 5015 Lab Chow (Ralston Purina Company, St. Louis, MO) and water ad libitum. All animal procedures were in strict accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Princeton University Animal Care and Use Committee.
Measurement of aggressive behavior. Only male mice (i.e., resident mice) were tested for aggressive behavior. Behavioral testing of mice bred at Princeton University was begun when the mice were 63–70 d of age. Behavioral testing of mice bred at The Jackson Laboratory and shipped to Princeton University was begun when the mice were 11–12 weeks of age. Opponents used in the aggression test were all males of the inbred strain 129T2/SvEmsJ that were received from The Jackson Laboratory at 49–56 d of age and were 56–63 d of age on the first day that they were used in aggression testing. Each resident mouse was tested once per day on 3 consecutive days, and testing was always conducted between 7:00 A.M. and 1:00 P.M. The order in which mice were tested was varied randomly from day to day over the 3 days. Testing was conducted in a small area of the home room that was enclosed by a curtain and that was dimly lit (2 lux). Aggression was measured, one mouse at a time, using a modification of the dangler test (Scott and Fredericson, 1951; Kessler et al., 1977). Each resident mouse was taken in its home cage from the housing rack in the home room to a table behind the curtain. An investigator (E.S.B.) sat in the area behind the curtain to observe the behavior of each mouse. The cage top was removed from the home cage and replaced with a Plexiglas top, and then the resident mouse was allowed to adjust to the dim lighting and the Plexiglas cage top for 2 min. Then, an opponent mouse was lowered through a hole in the Plexiglas cage top. The opponent's tail was taped to the Plexiglas cage top, so that the opponent's front two paws were on the cage bottom (in the middle of the long side of the cage), and its back two paws were lifted off of the cage bottom and were resting against the side of the cage. The test began immediately after the opponent's tail had been taped in place. The resident mouse was observed to see whether it would display aggressive behavior toward the opponent within 300 sec. All aggressive displays (tail rattles) and behaviors (lunges and bites) were observed and recorded. Several other behaviors were recorded, including time to first contact with the opponent and numbers of nonaggressive contacts (e.g., sniffing). Only unequivocal bites and/or lunges of at least moderate intensity were considered to be “attacks,” whereas tail rattles, small nips that occurred during grooming, or lower-intensity pushes were not classified as attacks. Residents that attacked the opponent in at least two of three tests were classified as being aggressive; those residents that either attacked in only one of three tests or never attacked were classified as being unaggressive. Aggression was thus analyzed as a threshold trait (Lynch and Walsh, 1998), i.e., a binary variable, that was defined by the presence or absence of reproducible attack behavior; we chose not to measure aggression as a score based on a composite of various behaviors, because other behaviors, such as tail rattling, are only weakly correlated with attack behavior and may have a different underlying genetics (Roubertoux et al., 1999). The test was stopped in 300 sec or within 3 sec of the start of an attack, whichever came sooner. No opponent was used in more than one test per day. Each resident mouse was tested with three different opponent males over the course of 3 consecutive days of testing. Opponents were used in only one 3 d series of behavioral testing. The body weight of each resident and each 129T2/SvEmsJ mouse was measured on the day of the third opponent test, after the completion of the testing.
Phenotypic analysis. The percentages of mice classified as aggressive were compared between Princeton-bred and The Jackson Laboratory-bred A mice, between Princeton-bred and The Jackson Laboratory-bred NZB mice, between inbred strains, and between reciprocal F1 hybrids using two-tailed Fisher's exact tests. The body weight of aggressive versus nonaggressive mice was compared using at test.
Genotypic analysis. Genomic DNA from each mouse was prepared from tail tissue by phenol–chloroform extraction and was amplified with microsatellite marker primers developed at the Whitehead Institute/Massachusetts Institute of Technology Center for Genome Research (Cambridge, MA) and obtained from Research Genetics(Huntsville, AL). Microsatellite markers spaced at distances of ∼20 cM across the genome were used for genotypic analysis. Chromosomal map positions of microsatellite markers were obtained from The Jackson Laboratory website (www.informatics.jax.org/searches/marker_form.shtml). PCR was performed using protocols suggested by the manufacturer (Research Genetics). PCR products were run in 2.5% Metaphor agarose gels (BioWhittaker Molecular Applications, Rockland, ME). The bands were visualized using ethidium bromide staining and ultraviolet transillumination.
Linkage analysis. Selective genotyping of the aggressive backcross animals was performed (Silver, 1995). The number of aggressive animals with A/N versus A/A genotypes at each marker were compared and tested for a significant departure from a 1:1 ratio with the χ2 test. A χ2 value of 15 (p< 1 × 10−4; df = 1) was set as the threshold for significance (Lander and Kruglyak, 1995). This analysis allowed the identification of markers linked to QTLs. Classical interval mapping is a form of regression (Liu, 1998) that cannot be performed with threshold (binary) traits; we therefore performed a simple analysis to determine whether it was likely that the relevant gene might map in the intervals between adjacent, closely spaced markers in the vicinity of QTLs. Recombinants were eliminated between adjacent markers within the QTLs, and the χ2 value was recalculated for each interval between adjacent markers. If the χ2 value was higher once recombinants were eliminated, this was considered suggestive evidence that a gene affecting the phenotype might lie within this interval, because it would be unlikely to get a higher χ2value when the total n was reduced. To rule out the possibility that there was segregation distortion at the identified QTLs, 98 backcross animals chosen randomly (including both aggressive and nonaggressive animals at random) were genotyped at one marker within each QTL (D10Mit180 and DXMit140), and the genotypes (A/N vs. A/A) were tested for a significant departure from a 1:1 ratio with the χ2 test. The threshold for significance for these tests of segregation distortion was set at p < 0.05. To determine whether there was epsistasis between identified QTLs, a χ2 test was performed to detect significant deviation from the expected ratios of genotypes under the null hypothesis of an additive interaction between the loci. The threshold for significance was set at p < 0.05.
RESULTS
Aggression in inbred strains, F1 hybrids, and backcross mice
We took several steps to control the effect of environmental variables on aggressive behavior in this study. First, to control the effects of early social experiences on aggression (Maxson, 1992), fathers were removed from breeding cages before the birth of pups, and all male mice were individually housed for at least 4 weeks before behavioral testing. Second, to reduce the variability among opponent mice used for aggression testing, we used males of a relatively nonaggressive inbred strain, the 129T2/SvEmsJ strain, as standard opponents (Roubertoux and Carlier, 1987; Maxson, 1992) in aggression tests, and we measured aggressive behavior using a modification of the dangler paradigm (Kessler et al., 1977), in which the opponent mouse is restricted to one part of the cage and thus restricted in its ability to provoke an attack. Aggression was analyzed as a threshold trait (Lynch and Walsh, 1998), i.e., a binary variable that was defined by the presence or absence of reproducible attack behavior; we chose not to measure aggression as a score based on a composite of various behaviors, because other behaviors, such as tail rattling, are only weakly correlated with attack behavior and may have a different underlying genetics (Roubertoux et al., 1999). Only unequivocal bites and/or lunges of at least moderate intensity were considered to be attacks, whereas small nips that occurred during grooming or lower-intensity pushes were not considered to be attacks. Mice that attacked the opponent in at least two of three tests were classified as being aggressive, whereas mice that attacked only once, or not at all, during the course of three tests were classified as being unaggressive.
NZB mice were highly aggressive. Of all NZB mice tested, including The Jackson Laboratory- and Princeton-bred animals, 19 of 22 mice (86%) were classified as aggressive. Eight of 10 (80%) The Jackson Laboratory-bred NZB mice were aggressive, and 11 of 12 (92%) Princeton-bred NZB mice were aggressive; this difference in aggression between Princeton-bred and The Jackson Laboratory-bred NZB mice was not statistically significant. In sharp contrast, A mice were strikingly unaggressive. Of all A mice tested, including The Jackson Laboratory- and Princeton-bred animals, 0 of 29 (0%) were classified as aggressive (Table 1). We tested F1 hybrid animals for aggression and sought to determine whether reciprocal F1 hybrid animals differed in aggression. Seventeen of 22 (77%) (NZB × A)F1 animals were aggressive, whereas 11 of 20 (55%) (A × NZB)F1 animals were aggressive; this difference was not statistically significant. Sixty-four of 470 (14%) backcross animals [(NZB × A) × A] were aggressive (Table 1).
Table 1.
Aggression frequency in parental strains and crosses
| Genotype or cross | Breeding location | # aggr | n | % aggr |
|---|---|---|---|---|
| NZB | JAX | 8 | 10 | 80 |
| NZB | Princeton | 11 | 12 | 92 |
| NZB | JAX or Princeton | 19 | 22 | 86 |
| A | JAX | 0 | 10 | 0 |
| A | Princeton | 0 | 19 | 0 |
| A | JAX or Princeton | 0 | 29 | 0 |
| (A × NZB)F1 | Princeton | 11 | 20 | 55 |
| (NZB × A)F1 | Princeton | 17 | 22 | 77 |
| (NZB × A) × A | Princeton | 64 | 470 | 14 |
JAX, The Jackson Laboratory;
aggr, number of mice that were classified as aggressive (attacked in at least 2 tests); % aggr, percentage of mice that were classified as aggressive (attacked in at least 2 tests).
NZB and A mice differ in body weight at 10 weeks of age (mean ± SEM body weights for NZB and A mice, respectively, were 28.1 ± 0.7 and 20.9 ± 0.3 gm). To determine whether the larger body size of NZB mice accounted for their greater aggressiveness, we measured the body weight of all 470 backcross animals at ∼10 weeks of age (immediately after the last of three aggression tests) and compared the body weight of aggressive backcross mice with the body weight of nonaggressive backcross mice by t test. There was no significant difference in body weight between the aggressive and nonaggressive mice.
Aggression QTL on chromosome 10
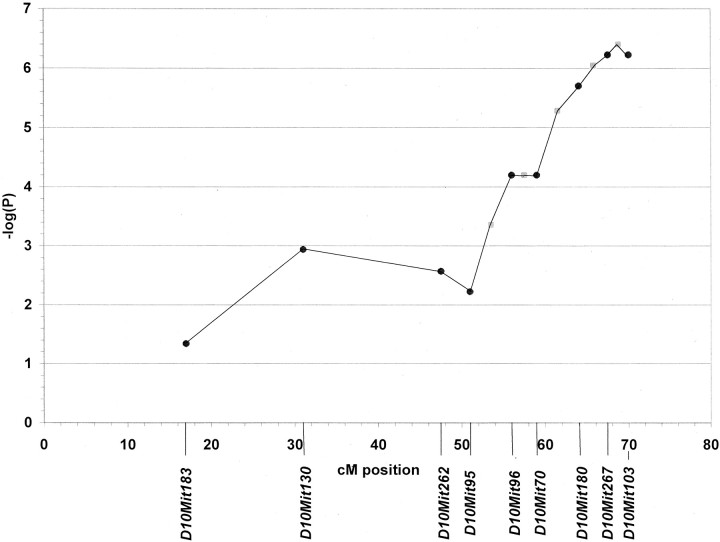
We tested 470 backcross animals for aggressive behavior, and 64 were classified as aggressive. Selective genotyping of the aggressive backcross animals was performed (Silver, 1995). Because the aggression phenotype was <100% penetrant in NZB mice (Table 1), the backcross mice that were nonaggressive could not be said to have a phenotype more similar to either the A strain or the NZB strain, and therefore genotype information from the nonaggressive backcross animals would not add significantly to the power to detect QTLs. The aggressive backcross animals were genotyped at microsatellite markers spaced at ∼20 cM intervals throughout the genome. The χ2test was used to identify marker loci that showed a significant departure from the 1:1 ratio expected for A/N and A/A genotypes in the absence of linkage. A χ2 value of 15 (p < 1 × 10−4; df = 1) was set as the threshold for significance for the presence of a QTL (Lander and Kruglyak, 1995). This analysis revealed a QTL on the distal end of chromosome 10, which we name Aggression-1 (Aggr1). The strongest linkage was found at marker loci D10Mit267 at 67.5 cM (χ2 = 25; p = 6 × 10−7) and D10Mit103 at 70 cM (χ2 = 25; p = 6 × 10−7). When recombinants between these two markers were eliminated, a higher χ2value and lower p value were found, which provides support for the mapping of the relevant gene between these two markers (Fig.1).
Fig. 1.
Localization of the Aggr1 locus on chromosome 10. p values associated with individual microsatellite marker loci (D10Mit183,D10Mit130, D10Mit262,D10Mit95, D10Mit96,D10Mit70, D10Mit180,D10Mit267, and D10Mit103) were determined by the χ2 test, using the genotypes of the 64 aggressive backcross mice. p values associated with chromosomal intervals between marker loci were determined by eliminating recombinants between adjacent marker loci and then recalculating the χ2 value for each interval. The results across 53 cM of chromosome 10 are presented in −(log10) converted form. The −log(P) for marker loci are graphed as blackcircles, and the −log(P) for intervals are graphed as gray squares in the middle of each interval. The centimorgan scale along the x-axis starts at the centromere. Chromosomal map positions of microsatellite markers were obtained from The Jackson Laboratory website (www.informatics.jax.org/searches/marker_form.shtml).
To rule out the possibility that this was a false positive result attributable to segregation distortion at or near this locus on chromosome 10, 98 backcross animals were chosen randomly (including both aggressive and nonaggressive animals at random) and were genotyped at a marker within the locus (D10Mit180). The genotyping results were tested for a significant departure from a 1:1 ratio (A/N vs A/A) with the χ2 test, and the threshold for significance was set at p < 0.05. There was no significant departure from a 1:1 ratio and thus no evidence of segregation distortion.
Aggression QTL on chromosome X
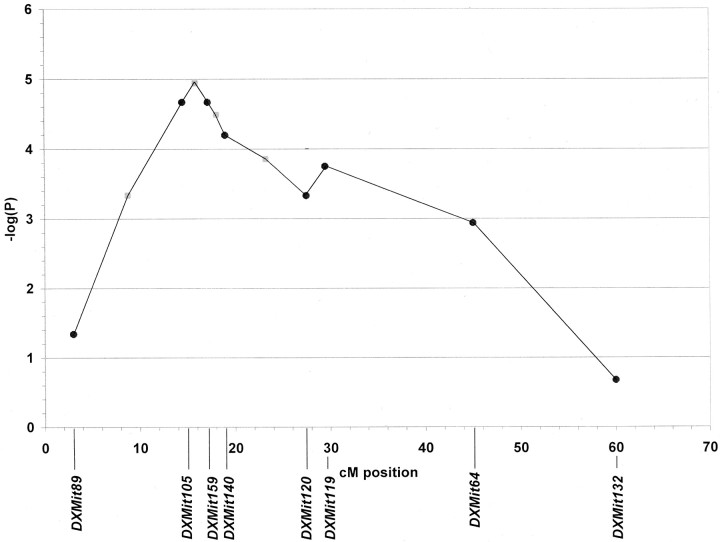
The genome scan also revealed a QTL on chromosome X, which we name Aggression-2 (Aggr2). The strongest linkage was found at marker loci DXMit105 at 14.5 cM (χ2 = 18.1; p = 2.14 × 10−5) andDXMit159 at 17.2 cM (χ2 = 18.1; p = 2.14 × 10−5). Elimination of recombinants between these markers and recalculation of the χ2 value was performed as described above. This analysis provides support for the mapping of the relevant gene between these two markers (Fig. 2). Segregation distortion was ruled out at a marker within the locus (DXMit140) using the method described above. By the χ2 test, there was no evidence of an epistatic interaction between Aggr1 and Aggr2; rather, there was a simple additive interaction between the two loci in their effect on the aggression phenotype.
Fig. 2.
Localization of the Aggr2 locus on chromosome X. p value calculations and data display format are the same as described for Figure 1.
DISCUSSION
We identified two loci, Aggr1 and Aggr2, that affect inter-male aggression in NZB and A mice. Aggr1maps to chromosome 10 in a region that shows conserved synteny with the human chromosomal region 12q13-q15. Possible candidate genes forAggr1 include diacylglycerol kinase α subunit (Dagk1) at 71 cM, kinesin family member 5A (Kif5a) at 70 cM, and olfactory receptor 9 (Olfr9) at 69.5 cM. Diacylglycerol kinases phosphorylate the second-messenger diacylglycerol to phosphatidic acid; thus, these kinases are involved in the phosphatidylinositol signal transduction system that is important in neurotransmission in the brain. Diacylglycerol kinases are expressed at high levels in the hippocampus, cerebellum, olfactory bulb, and retina (Topham and Prescott, 1999).Kif5a is expressed throughout the mouse brain and is a molecular motor protein involved in the transport of membranous organelles (including synaptic vesicle precursors) along the axons of nerve cells, toward the synapse (anterograde transport) (Hirokawa, 1997). Olfactory receptors, expressed on olfactory neurons, may have a minor role in the detection of pheromones that are important in aggression and other social behavior; however, a separate family of receptors, expressed on vomeronasal sensory neurons, appear to be primarily involved in pheromone detection in the mouse (Bargmann, 1997). Interestingly, the arginine vasopressin receptor 1A gene (Avpr1a) maps to 12q14 in the human genome, in the region that shows conserved synteny with the chromosome 10 QTL reported here, although Avpr1a has not yet been mapped in the mouse, to our knowledge. Avpr1a has been implicated in aggressive behavior in rodents (Ferris et al., 1997) and is expressed in rodent and primate brain regions thought to play a role in mediating aggression, including the amygdala, septum, and hypothalamus (Young et al., 1997, 1999).
The Aggr2 locus maps to chromosome X in a region that shows conserved synteny with the human chromosomal region Xq24-q27. Possible candidate genes for Aggr2 include the glutamate receptor subunit AMPA3 gene (Gria3) at 13.3 cM and the hypoxanthine guanine phosphoribosyl transferase gene (Hprt) at 17 cM. Mouse Gria3 encodes a subunit of the ionotropic glutamate receptor AMPA3, which is involved in excitatory neurotransmission throughout the brain (Ozawa et al., 1998). Mutations of the humanHprt gene, which is involved in purine metabolism, have been associated with the Lesch-Nyhan syndrome, characterized by aggressive and self-injurious behavior, hyperuricemia, mental retardation, movement abnormalities, and deficiencies of brain dopamine levels (Ernst et al., 1996). Hprt-deficient mutant mice also show brain dopamine depletion, the magnitude of which varies depending on the genetic background of the mouse strain used (Jinnah et al., 1999). However, mutations of Hprt on a 129 or C57BL/6 mouse strain background do not seem to cause self-injurious or aggressive behaviors, except when an inhibitor of adenine phosphoribosyltransferase was administered (Wu and Melton, 1993).
Several genes, whose products have been implicated in aggression, map further from the interval between DXMit105 andDXMit159 on mouse chromosome X and therefore are less likely candidate genes for Aggr2. These include the following: the monoamine oxidase A gene (5.2 cM from the centromere), which has been implicated in both human (Brunner et al., 1993) and mouse aggression (Cases et al., 1995); the gene encoding the α 3 subunit of the GABA-A receptor (28.5 cM from the centromere), which is involved in inhibitory neurotransmission in the brain; and the androgen receptor gene (36 cM from the centromere). To our knowledge, the only gene that is known to affect aggression levels when mutated and that maps anywhere near either Aggr1 or Aggr2 is monoamine oxidase A (5.2 cM from the centromere of chromosome X). The markedly low levels of aggression and impulsivity in A mice are not easily explained by low circulating levels of androgens, because the A strain has been shown to have levels of testosterone that are comparable with that of other inbred strains (Hampl et al., 1971).
Using evidence from reciprocal hybrids, congenic strains, and other crosses, previous investigators have suggested that a gene that affects aggression may lie on the Y chromosome, and both the sex-determining region on chromosome Y (Sry) (Maxson, 2000) and the steroid sulfatase gene (Sts) in the pseudoautosomal region (at 75 cM on chromosome X) (Roubertoux et al., 1994) have been suggested as candidates. In our study, however, we did not find evidence for the effect of either of these genes on aggression. If there were a gene in the non-pseudoautosomal region of the Y chromosome that accounts for increased aggression in NZB mice, one would have expected higher levels of aggression in (A × NZB)F1 hybrids versus (NZB × A)F1 hybrids, but in fact we found the reverse, although this difference between reciprocal F1 hybrids was not statistically significant. Moreover, the QTL we identified on chromosome X does not cover the pseudoautosomal region of chromosomes X and Y, which containsSts. Our results may differ from those of previous investigators because we crossed NZB mice to a different mouse strain from those used previously (to A rather than CBA/H) (Roubertoux et al., 1994), we socially isolated the mice for longer than many other groups, and we used a modified version of the dangler test of aggression rather than the resident-intruder test used by some other groups (Maxson, 1992; Roubertoux et al., 1999). Also, it is worth noting that the use of inbred strains simplifies QTL mapping, because the number of genes that account for a phenotypic difference between two strains is likely to be smaller than the number of genes that cause phenotypic variation among outbred mice. On the other hand, QTL mapping using two inbred strains will miss some of the loci that affect aggression in outbred mice or in crosses of other inbred strains.
The mapping of Aggr1 and Aggr2 is an important step toward the identification of naturally occurring alleles that affect aggression in the mouse. The effort to identify such alleles, in conjunction with the study of gene mutations and knock-outs that affect mouse aggression, may make important contributions to our understanding of the biology of mammalian aggression. This, in turn, may ultimately enhance our understanding of the complex interactions among biological, psychological, and social factors that affect human aggression.
Footnotes
This study was supported by National Institutes of Health, National Research Service Award 1 F32 MH12203-01 from the National Institute of Mental Health and National Institute of Child Health and Human Development Grant R37 HD20275-17. E.S.B. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences. We thank Jeremy L. Peirce for his comments on this manuscript.
Correspondence should be addressed to Lee M. Silver at the above address. E-mail: lsilver@princeton.edu.
REFERENCES
- 1.Bargmann CI. Olfactory receptors, vomeronasal receptors, and the organization of olfactory information. Cell. 1997;90:585–587. doi: 10.1016/s0092-8674(00)80518-8. [DOI] [PubMed] [Google Scholar]
- 2.Brunner HG, Nelen M, Breakefield XO, Ropers HH, van Oost BA. Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science. 1993;262:578–580. doi: 10.1126/science.8211186. [DOI] [PubMed] [Google Scholar]
- 3.Cadoret RJ, Yates WR, Troughton E, Woodworth G, Stewart MA. Genetic-environmental interaction in the genesis of aggressivity and conduct disorders. Arch Gen Psychiatry. 1995;52:916–924. doi: 10.1001/archpsyc.1995.03950230030006. [DOI] [PubMed] [Google Scholar]
- 4.Cases O, Seif I, Grimsby J, Gaspar P, Chen K, Pournin S, Muller U, Aguet M, Babinet C, Shih JC, de Maeyer E. Aggressive behavior and altered amounts of brain serotonin and norepinephrine in mice lacking MAOA. Science. 1995;268:1763–1766. doi: 10.1126/science.7792602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coccaro EF, Bergeman CS, McClearn GE. Heritability of irritable impulsiveness: a study of twins reared together and apart. Psychiatry Res. 1993;48:229–242. doi: 10.1016/0165-1781(93)90074-q. [DOI] [PubMed] [Google Scholar]
- 6.Coccaro EF, Bergeman CS, Kavoussi RJ, Seroczynski AD. Heritability of aggression and irritability: a twin study of the Buss-Durkee aggression scales in adult males subjects. Biol Psychiatry. 1997;41:273–284. doi: 10.1016/s0006-3223(96)00257-0. [DOI] [PubMed] [Google Scholar]
- 7.Eley TC, Lichtenstein P, Stevenson J. Sex differences in the etiology of aggressive and nonaggressive antisocial behavior: results from two twin studies. Child Dev. 1999;70:155–168. doi: 10.1111/1467-8624.00012. [DOI] [PubMed] [Google Scholar]
- 8.Ernst M, Zametkin AJ, Matochik JA, Pascualvaca D, Jons PH, Hardy K, Hankerson JG, Doudet DJ, Cohen RM. Presynaptic dopaminergic deficits in Lesch-Nyhan disease. N Engl J Med. 1996;334:1568–1572. doi: 10.1056/NEJM199606133342403. [DOI] [PubMed] [Google Scholar]
- 9.Ferris CF, Melloni RH, Koppel G, Perry KW, Fuller RW, Delville Y. Vasopressin/serotonin interactions in the anterior hypothalamus control aggressive behavior in golden hamsters. J Neurosci. 1997;17:4331–4340. doi: 10.1523/JNEUROSCI.17-11-04331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillot P-V, Carlier M, Maxson SC, Roubertoux PL. Intermale aggression tested in two procedures, using four inbred strains of mice and their reciprocal congenics: Y chromosomal implications. Behav Genet. 1995;25:357–360. doi: 10.1007/BF02197285. [DOI] [PubMed] [Google Scholar]
- 11.Hampl R, Ivanyi P, Starka L. Testosterone and testosterone binding in murine plasma. Steroidologia. 1971;2:113–120. [PubMed] [Google Scholar]
- 12.Hirokawa N. The mechanisms of fast and slow transport in neurons: identification and characterization of the new kinesin superfamily motors. Curr Opin Neurobiol. 1997;7:605–614. doi: 10.1016/s0959-4388(97)80079-7. [DOI] [PubMed] [Google Scholar]
- 13.Jinnah HA, Jones MD, Wojcik BE, Rothstein JD, Hess EJ, Friedmann T, Breese GR. Influence of age and strain on striatal dopamine loss in a genetic mouse model of Lesch-Nyhan disease. J Neurochem. 1999;72:225–229. doi: 10.1046/j.1471-4159.1999.0720225.x. [DOI] [PubMed] [Google Scholar]
- 14.Kessler S, Elliott GR, Orenberg EK, Barchas JD. A genetic analysis of aggressive behavior in two strains of mice. Behav Genet. 1977;7:313–321. doi: 10.1007/BF01066801. [DOI] [PubMed] [Google Scholar]
- 15.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 16.Liu B-H. Statistical genomics: linkage, mapping, and QTL analysis, pp 417–458. CRC; Boca Raton, FL: 1998. [Google Scholar]
- 17.Lynch M, Walsh B. Genetics and analysis of quantitative traits, pp 727–744. Sinauer; Sunderland, MA: 1998. [Google Scholar]
- 18.Manuck SB, Flory JD, Ferrell RE, Dent KM, Mann JJ, Muldoon MF. Aggression and anger-related traits associated with a polymorphism of the tryptophan hydroxylase gene. Biol Psychiatry. 1999;45:603–614. doi: 10.1016/s0006-3223(98)00375-8. [DOI] [PubMed] [Google Scholar]
- 19.Maxson SC. Genetic influences on aggressive behavior. In: Pfaff DW, Berrettini WH, Joh TH, Maxson SC, editors. Genetic influences on neural and behavioral functions. CRC; Boca Raton, FL: 2000. pp. 405–416. [Google Scholar]
- 20.Maxson SC. Methodological issues in genetic analyses of agonistic behavior (offense) in male mice. In: Goldowitz D, Wahlsten D, Wimer RE, editors. Techniques for the genetic analysis of brain and behavior. Elsevier Science; New York: 1992. pp. 349–373. [Google Scholar]
- 21.Oliveira-Dos-Santos AJ, Matsumoto G, Snow BE, Bai D, Houston FP, Whishaw IQ, Mariathasan S, Sasaki T, Wakeham A, Ohashi PS, Roder JC, Barnes CA, Siderovski DP, Penninger JM. Regulation of T cell activation, anxiety, and male aggression by RGS2. Proc of the Natl Acad Sci USA. 2000;97:12272–12277. doi: 10.1073/pnas.220414397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozawa S, Kamiya H, Tsuzuki K. Glutamate receptors in the mammalian central nervous system. Prog Neurobiol. 1998;54:581–618. doi: 10.1016/s0301-0082(97)00085-3. [DOI] [PubMed] [Google Scholar]
- 23.Riggins-Caspers K, Cadoret RJ. Detecting and measuring gene-environment interaction in human temperament (aggressivity) and personality deviation (conduct disorder, antisocial personality). In: Jones BC, Mormede P, editors. Neurobehavioral genetics: methods and applications. CRC; New York: 1999. pp. 163–185. [Google Scholar]
- 24.Roubertoux PL, Carlier M. Difference between CBA/H and NZB mice on intermale aggression. Maternal effects. Behav Genet. 1987;2:175–184. doi: 10.1007/BF01067839. [DOI] [PubMed] [Google Scholar]
- 25.Roubertoux PL, Carlier M, Degrelle H, Haas-Dupertuis MC, Phillips J, Moutier R. Co-segregation of the pseudoautosomal region of the Y chromosome with aggression in mice. Genetics. 1994;135:254–263. doi: 10.1093/genetics/136.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roubertoux PL, Le Roy I, Mortaud S, Perez-Diaz F, Tordjman S. Measuring aggression in the mouse. In: Crusio WE, Gerlai RT, editors. Handbook of molecular-genetic techniques for brain and behavior research. Elsevier; Amsterdam: 1999. pp. 696–709. [Google Scholar]
- 27.Rushton JP, Fulker DW, Neale MC, Nias DKB, Eysenck HJ. Altruism and aggression: the heritability of individual differences. J Pers Soc Psychol. 1986;50:1192–1198. doi: 10.1037//0022-3514.50.6.1192. [DOI] [PubMed] [Google Scholar]
- 28.Scott JP, Fredericson E. The causes of fighting in mice and rats. Physiol Zool. 1951;24:273–309. [Google Scholar]
- 29.Silver LM. Mouse genetics: concepts and applications. Oxford UP; New York: 1995. [Google Scholar]
- 30.Steadman HJ, Mulvey EP, Monahan J, Robbins PC, Appelbaum PS, Grisso T, Roth LH, Silver E. Violence by people discharged from acute psychiatric inpatient facilities and by others in the same neighborhoods. Arch Gen Psychiatry. 1998;55:393–401. doi: 10.1001/archpsyc.55.5.393. [DOI] [PubMed] [Google Scholar]
- 31.Swanson JW, Holzer CE, Ganju VK, Jono RT. Violence and psychiatric disorder in the community: evidence from the Epidemiologic Catchment Area surveys. Hosp Community Psychiatry. 1990;41:761–770. doi: 10.1176/ps.41.7.761. [DOI] [PubMed] [Google Scholar]
- 32.Tardiff K, Marzuk PM, Leon AC, Portera L. A prospective study of violence by psychiatric patients after hospital discharge. Psychiatr Serv. 1997;48:678–681. doi: 10.1176/ps.48.5.678. [DOI] [PubMed] [Google Scholar]
- 33.Taylor PJ, Leese M, Williams D, Butwell M, Daly R, Larkin E. Mental disorder and violence: a special (high security) hospital study. Br J Psychiatry. 1998;172:218–226. doi: 10.1192/bjp.172.3.218. [DOI] [PubMed] [Google Scholar]
- 34.Tellegen A, Lykken DT, Bouchard TJ, Wilcox KJ, Segal NL, Rich S. Personality similarity in twins reared apart and together. J Pers Soc Psychol. 1988;54:1031–1039. doi: 10.1037//0022-3514.54.6.1031. [DOI] [PubMed] [Google Scholar]
- 35.Topham MK, Prescott SM. Mammalian diacylglycerol kinases, a family of lipid kinases with signaling functions. J Biol Chem. 1999;274:11447–11450. doi: 10.1074/jbc.274.17.11447. [DOI] [PubMed] [Google Scholar]
- 36.Torrey EF. Violent behavior by individuals with serious mental illness. Hosp Community Psychiatry. 1994;45:653–662. doi: 10.1176/ps.45.7.653. [DOI] [PubMed] [Google Scholar]
- 37.Vernon PA, McCarthy JM, Johnson AM, Jang KL, Harris JA. Individual differences in multiple dimensions of aggression: a univariate and multivariate genetic analysis. Twin Res. 1999;2:16–21. doi: 10.1375/136905299320566068. [DOI] [PubMed] [Google Scholar]
- 38.Wu CL, Melton DW. Production of a model for Lesch-Nyhan syndrome in hypoxanthine phosphoribosyltransferase-deficient mice. Nat Genet. 1993;3:235–240. doi: 10.1038/ng0393-235. [DOI] [PubMed] [Google Scholar]
- 39.Young LJ, Toloczko D, Insel TR. Localization of vasopressin (V1a) receptor binding and mRNA in the rhesus monkey brain. J Neuroendocrinol. 1999;11:291–297. doi: 10.1046/j.1365-2826.1999.00332.x. [DOI] [PubMed] [Google Scholar]
- 40.Young LJ, Winslow JT, Nilsen R, Insel TR. Species differences in V1a receptor gene expression in monogamous and nonmonogamous voles: behavioral consequences. Behav Neurosci. 1997;111:599–605. doi: 10.1037//0735-7044.111.3.599. [DOI] [PubMed] [Google Scholar]