Abstract
In vertebrates, a variety of light-stimulated genes are distributed in the retina, the pineal gland, and the suprachiasmatic nucleus, but acis-element(s) responsible for the light-dependent transcriptional regulation is left unexplored. Focusing on thepinopsin gene, a light-stimulated gene in the chick pineal gland, we performed a transcriptional analysis in the primary culture of the chick pineal cells that were transiently transfected with a luciferase reporter gene fused with various lengths of the 5′ upstream region of the pinopsin gene. Light-dependent enhancer activity was detectable in the construct with the upstream region between −1156 and +31. Introduction of mutations within the 18 bp sequence at positions −1103 to −1086 (TGGCACGTGGGGTTCCTC), including a CACGTG E-box sequence, elevated the transcriptional activity in the dark and thereby abrogated the light dependency, suggesting that the 18 bp sequence is essential for a reduction of the transcriptional activity in the dark. In an electrophoretic mobility-shift assay, we identified a pineal nuclear factor(s) capable of binding to the 18 bp element in a sequence-specific manner. When a 49 bp fragment (−1122 to −1074) including the 18 bp sequence was placed upstream of the simian virus 40 promoter, the transcriptional activity was dramatically suppressed regardless of light conditions in the chick pineal cells, and a more pronounced repression was observed in nonpineal/nonphotosensory LMH and NIH 3T3 cells. These results suggest that the 18 bp element in the pinopsin promoter constitutes the binding site of a ubiquitous factor that serves for the transcriptional repression that is required, although not sufficient, for the light-dependent expression of pinopsin gene in the chick pinealocytes.
Keywords: chicken, gene expression, light induction, pineal gland, pinopsin gene, transcriptional regulation
In animals, light plays an important role not only for vision but also for adaptation to changes in ambient conditions that greatly influence the expression of many genes. For example, light is a predominant time cue for entraining an endogenous circadian clock, which autonomously oscillates with a period of ∼24 hr (McWatters et al., 1999; King and Takahashi, 2000; Pando and Sassone-Corsi, 2001). In resetting the phase of the circadian clock in rodents, light stimulates the expression of a clock geneperiod (Per) in the suprachiasmatic nucleus (Albrecht et al., 1997; Shearman et al., 1997; Shigeyoshi et al., 1997), suggesting involvement of the induction of the Pergene in the phase-shift of the oscillator (Akiyama et al., 1999). The transcription of the Per gene is positively regulated by a heteromer of basic helix–loop–helix–Per-ARNT-Sim proteins, CLOCK and BMAL, through a CACGTG sequence (E box) in the Perpromoter (Gekakis et al., 1998). Recent studies (King and Takahashi, 2000) have revealed a basic mechanism underlying the circadian oscillation, but far less is known about the photic-entrainment pathway in animal clock systems.
Light also regulates the transcription of genes for retinal phototransduction proteins such as rhodopsin (Korenbrot and Fernald, 1989) and arrestin (McGinnis et al., 1994) to compensate for the protein degradation attributable to the circadian shedding of the photoreceptor cells. Several cis elements and transcription factors responsible for the photoreceptor cell-specific gene expression have been identified (Rehemtulla et al., 1996; Furukawa et al., 1997), whereas those important for light-dependent gene expression are unexplored. However, in plants, the regulatory processes of light-controlled genes triggering various photic events have been well characterized (Terzaghi and Cashmore, 1995; Heintzen et al., 2001).
Like those in plants, the light-responsive genes in animals would be regulated through a cis-DNA element, light-responsive element (LRE). To understand the mechanism underlying the light-dependent gene expression in the photosensory cells, we paid special attention to gene regulation of pinopsin that was identified in the chicken pineal gland as a pineal-specific photoreceptive molecule (Okano et al., 1994). The chicken pineal gland is a photosensitive neuroendocrine tissue; it has been widely used for biochemical and pharmacological studies on the circadian clock because it retains an intracellular phototransduction pathway regulating the phase of the intrinsic clock oscillator and because these functions are well maintained even in the dispersed cell culture (Deguchi, 1981; Zatz et al., 1988). We found previously that mRNA levels ofpinopsin are kept low in the dark and increase approximately sixfold with 6 hr of light exposure in chicks (Takanaka et al., 1998), in a time course similar to that of light-dependent Per2induction in the Xenopus retina (Steenhard and Besharse, 2000). Even in the isolated pineal organ culture, the expression ofpinopsin gene is upregulated by light, albeit to a less pronounced degree (∼1.5-fold) than in vivo. Here we show that an 18 bp element at positions −1103 to −1086 in the upstream region of pinopsin plays a key role in light responsiveness. This sequence represents the first example of elements that confer light-responsive gene expression in animal cells.
MATERIALS AND METHODS
Isolation of the pinopsin upstream region. Thepinopsin upstream DNA fragment was isolated using a long and accurate PCR in vitro cloning kit (TaKaRa, Otsu, Japan) with specific pinopsin primers R7 (at positions +320 to +336, relative to the transcription initiation site; 5′CGTGTTTGGCAGGAGGA3′) and R9 (at positions +27 to +46; 5′CCGATGTCCTCCAACAGCTC3′). The isolated fragments (2.5 kb) were subcloned into pCR 2.1 (Invitrogen, Carlsbad, CA) for sequence determination.
Animals and pineal cell culture. Animals were treated in accordance with the guidelines of the University of Tokyo. Newly hatched chicks were housed under a 12 hr light/dark cycle; the next day (day 1), the pineal glands were isolated from the chicks during the light period. The pineal cells were dispersed by passing them through a cell strainer (100 μm Nylon; Becton Dickinson, Franklin Lakes, NJ), and placed in 48 well plates (1 × 106 cell/well) with 1 ml of Medium 199 (Invitrogen) supplemented with 10 mm HEPES, pH 7.3, 100 U/ml penicillin G, 100 μg/ml streptomycin, 0.9 mg/ml NaHCO3, and 10% (v/v) fetal bovine serum (Invitrogen). The pineal cells were cultured at 37°C in 5% CO2 in a 12 hr light/dark cycle (lights on at 8:00 A.M. and off at 8:00 P.M.) with a light intensity of 250–300 lux at the level of culture plates in the CO2 incubator. The cultured cells were transfected with plasmids at 4:00 P.M. on day 3 and were harvested at 12:00 P.M. on day 4.
Plasmid construction and transfection. Promoter/reporter hybrid constructs were prepared by introducing each of several clones for the pinopsin upstream regions into theNheI/XhoI site of pGL3-Basic or by linking annealed oligonucleotide into the SmaI site of pGL3-promoter vector (Promega, Madison, WI). In vitro mutagenesis in thepinopsin promoter region was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The cells were transfected with these plasmids using LipofectAmine Plus (Invitrogen) according to the manufacturer's instructions. In every experiment, pRL-TK (Promega) was cotransfected as a control measure, and the luciferase activities were evaluated with the aid of the Dual-Luciferase reporter assay system (Promega).
Oligonucleotides. The synthetic DNA oligonucleotides used for the site-directed mutagenesis and for electrophoretic mobility-shift assay (EMSA) were pinopsin 49 bp (WT49) probe (at positions −1122 to −1074), 5′GATG-GAGCACATCCTGCTGTGG CACGTGGGGTTCCTCACTTTGTAC-GAA3′;δ-crystallin (DC5), 5′GATCTAAATATTCATTGTTGTTGCT CACCTACCATG3′ (Kamachi and Kondoh, 1993); DC5 mutation (mutDC5), 5′GATCTAAATATTCATTGTTGTTGCT CCAATACCATG3′ (Kamachi and Kondoh, 1993); arginine vasopressin (AVP), 5′GATCTCAGGCC CACGTGTGTCCCCAGGCC CACGTGTGTCCCCAGGCC CACGTGTGTCCCA3′ (Jin et al., 1999); AVP mutation (mutAVP), 5′GATCTCAGGCC GGACCTTGTCCCCAGGCC GGACCTTGTCCCCAGGCC GGACCTTGTCCCA3′ (Jin et al., 1999); chicken Per2 (cPer2), 5′GTGTCA CACGTGAGGCTTA3′ (Doi et al., 2001); and cPer 2 mutation (mutcPer2), 5′GTGTCA GGACCTAGGCTTA3′ (Okano et al., 2001).
Preparation of nuclear proteins. Newly hatched chicks were housed under 12 hr light/dark cycles for 2 d and the pineal glands were isolated during the light period (“light” sample), whereas the “dark” sample was prepared from chicks that were raised under constant dark conditions for 2 d and the pineal glands were isolated in the dark. The nuclear extracts were prepared according to the method of Gorski et al. (1986), with modifications. Briefly, 100 pineal glands (light or dark samples) or liver slices (∼1 gm; isolated from the chicks in the light) were homogenized with a glass–glass homogenizer on ice in 5 ml of ice-cold sucrose buffer I (10 mm HEPES-KOH, 15 mmKCl, 0.15 mm spermine, 0.5 mm spermidine, 1 mm EDTA, 2.2 m sucrose, 5% v/v glycerol, 0.5 mm DTT, 0.5 mm PMSF, and 14 μg/ml aprotinin, pH 7.6). These procedures for the dark sample were performed under dim red light (>640 nm) and subsequent manipulations were performed in the light. The homogenate was layered over a 6 ml cushion of ice-cold sucrose buffer II (10 mmHEPES-KOH, 15 mm KCl, 0.15 mm spermine, 0.5 mmspermidine, 1 mm EDTA, 2 msucrose, 10% v/v glycerol, 0.5 mm DTT, 0.5 mm PMSF, and 14 μg/ml aprotinin, pH 7.6) and centrifuged in an SW41Ti rotor (Beckman Coulter, Fullerton, CA) at 100,000 × g for 1 hr at 4°C. Then the pellet was resuspended in 240 μl of a nuclear lysis buffer (10 mm HEPES-KOH, 0.55 m KCl, 0.1 mm EDTA, 10% v/v glycerol, 3 mm MgCl2, 0.5 mm DTT, 0.1 mm PMSF, and 14 μg/ml aprotinin, pH 7.6). After gentle mixing for 30 min, the suspension was centrifuged in a TLA 100.3 rotor (Beckman Coulter) at 200,000 × g for 1 hr at 4°C. Solid (NH4)2SO4was added to the supernatant (0.3 gm/ml), followed by gentle mixing for 30 min. The proteins sedimented by centrifugation in a TLA 100.3 at 150,000 × g for 30 min at 4°C were dissolved in a dialysis buffer (25 mm HEPES-KOH, 50 mm KCl, 0.1 mm EDTA, 10% v/v glycerol, and 1 mm DTT, pH 7.6) for dialysis against the buffer at 4°C. This nuclear protein fraction was stored in liquid nitrogen until use.
Electrophoretic mobility-shift assay. The double-stranded oligonucleotide prepared as described by Okano et al. (2001) was radiolabeled at the 5′ end with [γ-32P]ATP using Megalabel (TaKaRa) and then incubated with 2.5 μg of nuclear proteins and 0.5 μg of poly(deoxyinosinic–deoxycytidylic) for 30 min at 37°C in a reaction buffer (25 mm HEPES-KOH, 50 mm KCl, 2.5 mmMgCl2, 0.1 mm EDTA, 10% v/v glycerol, and 1 mm DTT, pH 7.6) before electrophoresis. Competition EMSAs were performed by premixing nuclear proteins with a 50-fold molar excess of unlabeled double-stranded oligonucleotides (competitor). In the experiment shown in Figure5C, 1 μg of nuclear protein was incubated with radiolabeled WT49 probe and 2 μg of salmon sperm DNA in a reaction buffer (25 mm HEPES-KOH, 100 mm KCl, 7.5 mmMgCl2, 0.1 mm EDTA, 10% v/v glycerol, and 1 mm DTT, pH 7.6) in the presence or absence of competitors. These reaction mixtures were electrophoresed in a 6% w/v polyacrylamide gel with 0.5× Tris–borate–EDTA buffer containing (in mm): 44.5 Tris, 44.5 boric acid, and 1 EDTA; the radioactivity in the gel was detected by an image analyzer (FLA 2000; Fujifilm, Tokyo, Japan).
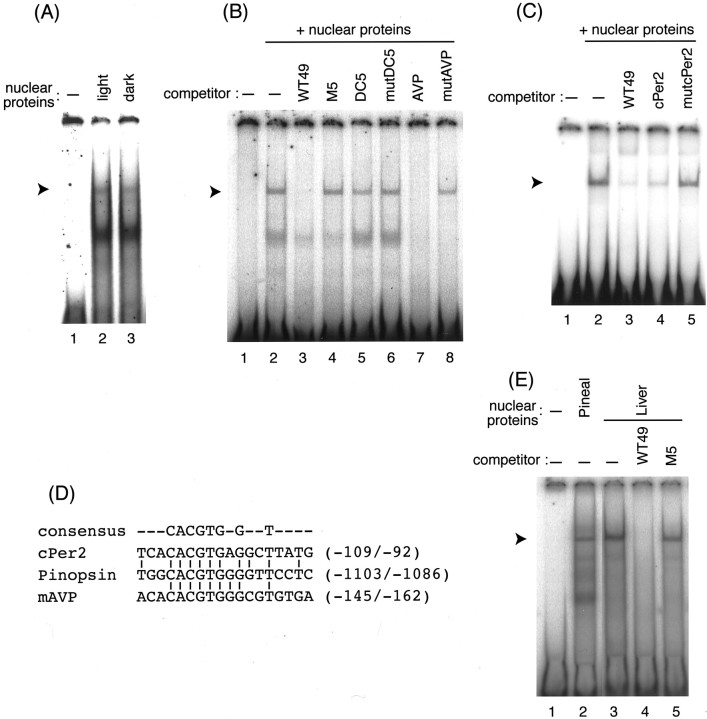
Fig. 5.
Characterization of the pinopsinLRE-binding protein by EMSA. A, EMSA using the WT49 probe in the presence of the pineal nuclear proteins prepared from light- or dark-adapted chicks. B, Competition EMSA with pineal nuclear proteins in the presence of competitors: WT49, M5, DC5, mutDC5, AVP, and mutAVP. C, Competition EMSA with pineal nuclear proteins in the presence of competitors: WT49, cPer2, and mutcPer2. D, Sequence comparison of the vicinity of the E-box region from chickenpinopsin, cPer2, and mouse AVP (mAVP).E, EMSA using the chick pineal and liver nuclear proteins. In each panel, the position of a band representing a specific interaction with the LRE is indicated by anarrowhead.
RESULTS
Functional characterization of the pinopsinpromoter region
Using oligonucleotide cassette-mediated PCR, we cloned a 2.5 kb chick genomic fragment including the pinopsin gene upstream region (Fig. 1A), in which a TATA box was found at positions −39 to −34 upstream from its transcription initiation site described in the GenBank database (accession number U87449). We also found at positions −94 to −90 an inverted pineal regulatory element (PIRE, TAATC/A) (Li et al., 1998), to which CRX (cone rod homeobox protein) binds for transactivation of photoreceptor cell-specific genes (Furukawa et al., 1997). Three copies of CACGTG E boxes were present at positions −1100 to −1095, −2009 to−2004, and −2023 to −2018 (Fig. 1A).
Fig. 1.
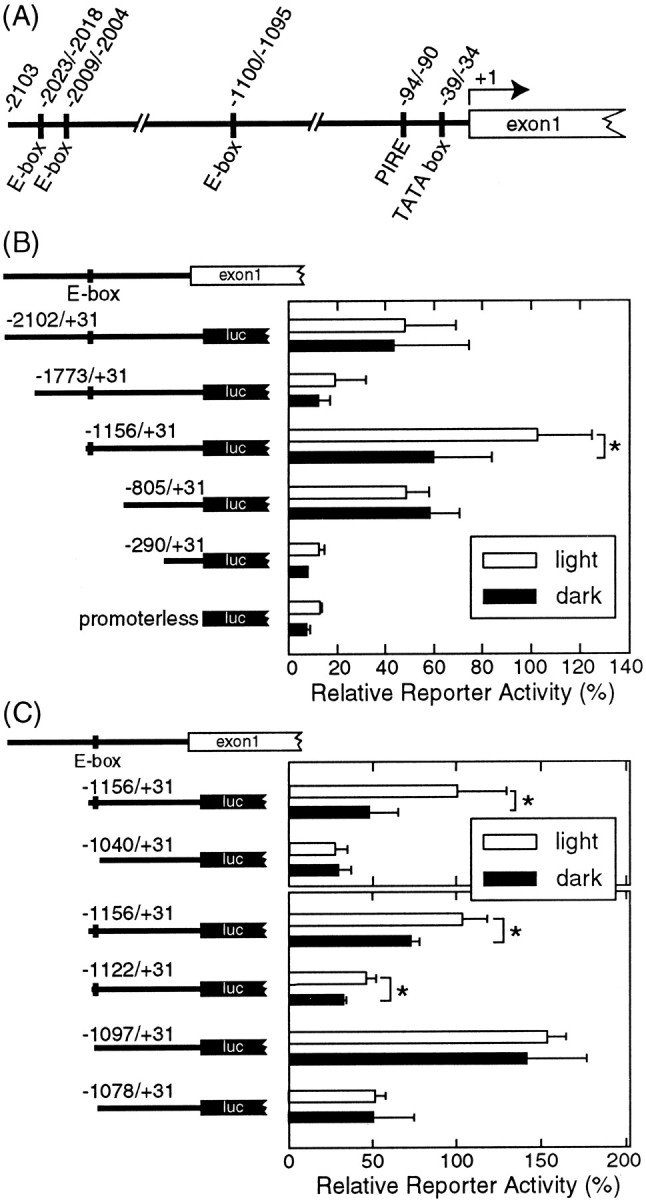
Transcriptional analysis of thepinopsin promoter region in cultured chick pineal cells under dark/light conditions. A, Schematic of thepinopsin promoter region. B, C, Luciferase (luc) reporter assays performed with constructs harboring various lengths of the pinopsin promoter region. The cultured chick pineal cells were transfected with each construct shown at the left and incubated in the light/dark cycles (light, open bars) or in constant darkness (dark,filled bars). Then the cells were harvested under the light or dark condition to measure the luciferase activities. A typical set of data out of three independent experiments is shown in this figure. Error bars indicate means ± SD of four replicated cell cultures. *p < 0.05; Student's ttest.
To search for the cis-DNA element(s) responsible for the light-dependent gene expression, we prepared five kinds of reporter constructs with various lengths of the 5′-flanking genomic fragments ofpinopsin gene each linked with the luciferase reporter gene (Fig. 1B). Chick pineal cells were cultured under 12 hr light/dark cycle conditions (lights on at 8:00 A.M. and off at 8:00 P.M.) for 2 d; they were transfected at 4:00 P.M. on the following day with each construct. One group of the cell plates was transferred to the constant dark at 8:00 P.M. (dark sample); the other was maintained under the light/dark cycle (light sample). After 16 hr (12:00 P.M. the next day) the cells were harvested under the dark or light conditions to measure the luciferase activities. We found that the construct harboring the region between position −1156 and +31 (−1156/+31) exhibited a light-stimulated reporter activity (Fig.1B), whereas the longer constructs (−2102/+31 and −1773/+31) showed no significant difference in reporter activity between the light and the dark conditions, suggesting regulation by an element(s) between position −2102 and −1156 that apparently masks the effect of light. Such a light-insensitive reporter activity was also observed for the shorter constructs −805/+31 and −290/+31; this suggests that at least one of the LREs indispensable for the light-dependent pinopsin gene expression is present in the region between position −1156 and −805. The shortest construct −290/+31 retains a PIRE-like sequence (Li et al., 1998) and a TATA box, but the reporter activity was very low and similar to that of a promoterless construct, indicating that these elements are insufficient not only for the light-dependent expression but also for the basal expression of the pinopsin gene.
To localize an LRE(s) between position −1156 and −805, we prepared another construct, −1040/+31. This construct no longer displayed the light-sensitive promoter activity (Fig. 1C), localizing the putative LRE(s) between −1156 and −1040. This region was also analyzed using three constructs (−1122/+31, −1097/+31, and −1078/+31), among which only the longest construct (−1122/+31) was capable of conferring the light responsiveness (Fig. 1C), suggesting the presence of the LRE in the vicinity of position −1122/−1097.
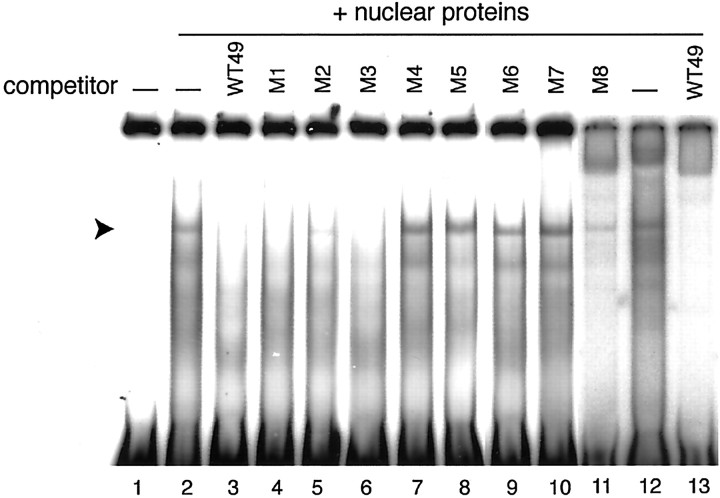
To determine the critical sequence for the light-dependent transactivation, mutations were introduced into the construct −1156/+31 at a region between position −1121 and −1082, producing eight constructs (Fig.2A, M1–M8), in the form of six consecutive base substitutions (M1–M7) or four base substitutions (M8). As shown in Figure 2B, M1, M2, M3, and M8 displayed light-sensitive reporter activities, whereas the activities of M4–M7 were insensitive to light (Fig.2B). Notably, M1–M3 showed reporter activities markedly higher than the wild-type −1156/+31 construct in the light and dark, suggesting that a transcriptional inhibitor(s) binds to this region (between −1121 and −1104) to suppress the basal transcriptional activity. More importantly, the light-insensitive reporter activities of the mutants M4–M7 were more comparable with that of the wild type detected in the light than that in the dark (Fig.2B); hence the 18 bp element at positions −1103 to −1086 (TGGCACGTGGGGTTCCTC) seemed to elicit transcriptional repression in the dark (see below).
Fig. 2.
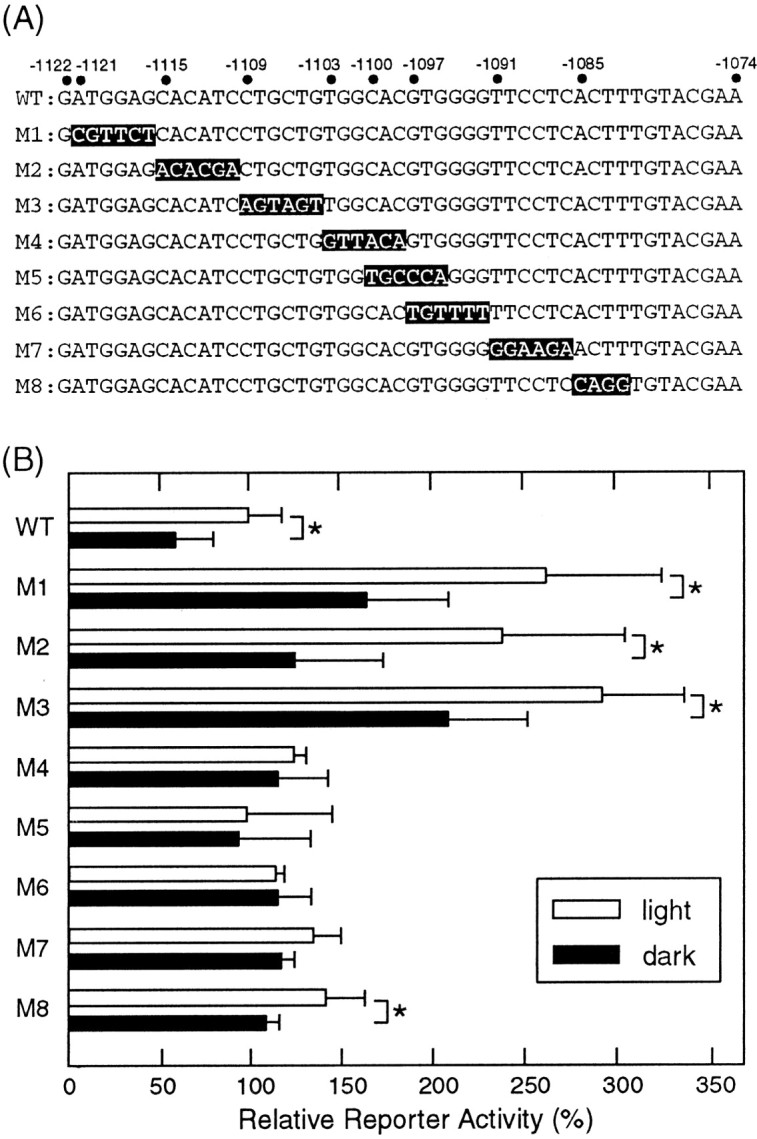
Scan of critical sequences for LRE gene expression. A, Positions of mutations introduced into the −1156/+31 construct. The numbers on thetop refer to base positions relative to the transcription initiation site. B, Luciferase assays performed with constructs harboring various mutations of thepinopsin promoter region. The cells were transfected with each construct and harvested in the light (open bars) or in the dark (filled bars). In eachpanel, a typical set of data out of three independent experiments is shown. Error bars indicate means ± SD of four replicated cell cultures. *p < 0.05; Student'st test.
Electrophoretic mobility-shift assay
Using a probe of the 49 bp DNA fragment [−1122/−1074, (WT49)] containing the 18 bp LRE, an EMSA was performed to explore a specific DNA binding protein(s) in the chick pineal nuclear extract. We detected a retarded band (Fig. 3, lanes 2 and 12, arrowhead), and the formation of this band was completely blocked by the addition of an excess amount of unlabeled WT49 (Fig. 3, lanes 3 and 13). The DNA sequence specificity of the interaction was examined by assessing the competitive effects of mutated WT49 fragments. The addition of an excess amount of M4–M7 had no competitive effect on the EMSA band, whereas M1, M2, M3, and M8 fragments retained significant binding capacities (Fig. 3, lanes 4–11). These binding patterns are in good agreement with the effects of mutations on the light responsiveness of the transcription evaluated by the luciferase assay (Fig. 2B). The recognition sequence of the WT49-binding factor lies in the 18 bp pinopsin LRE.
Fig. 3.
Competition EMSA of WT49 probe with the chick pineal nuclear proteins. Before mixing with end-labeled WT49 probe, pineal nuclear proteins prepared from light-adapted chicks were preincubated with the competitor shown on the top of each lane. The sequences of M1–M8 are shown in Figure2A; WT49 represents DNA fragment −1122 to −1074 of the pinopsin promoter region. An arrowheadindicates the position of a band representing an LRE-specific interaction.
Transcriptional repression through thepinopsin light-responsive element
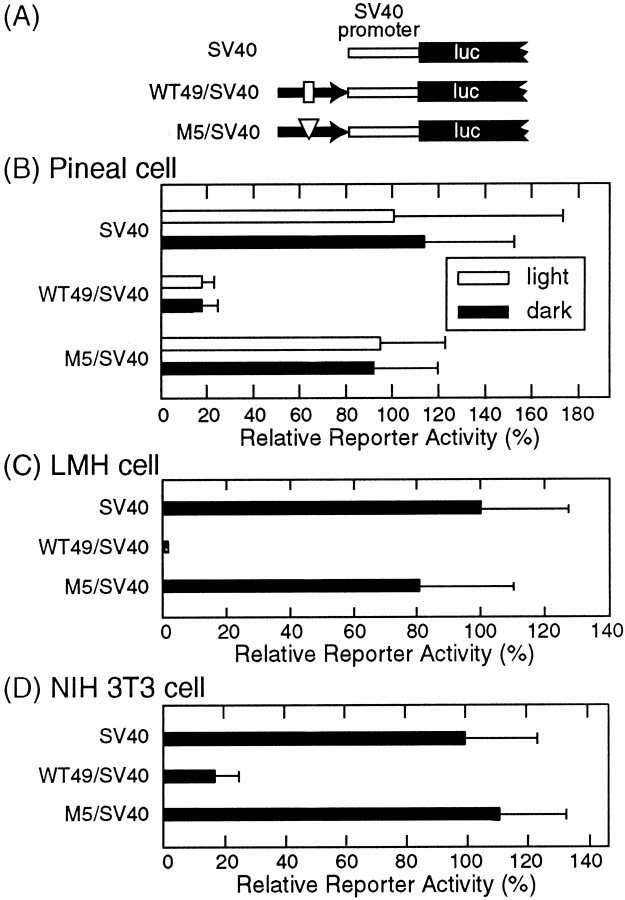
To investigate whether the short stretch of thepinopsin upstream region at positions −1122 to −1074 (WT49) can confer light-dependent gene expression, we generated a WT49/simian virus 40 (SV40) construct (Fig.4A) in which WT49 (−1122 to −1074) was linked to SV40 promoter (pGL3 promoter;Promega). The WT49/SV40 construct failed to show light-dependent promoter activity in the chick pineal cells (Fig.4B), but we found that the WT49 sequence strikingly suppressed the reporter activity governed by SV40 promoter regardless of light conditions (83–85% inhibition of the SV40 promoter activity) (Fig. 4B); this repression was completely restored by mutating the core CACGTG sequence (M5/SV40 construct in Fig.4B). These observations indicate that thepinopsin LRE by itself is insufficient for conferring light-dependent gene expression but is required for it by repressing the basal transcriptional activity. A similar or more pronounced inhibitory effect of WT49 on the reporter activity was seen not only in chicken hepatoma LMH cells (99% inhibition) (Fig.4C) but also in mammalian cells such as NIH 3T3 cells (83% inhibition) (Fig. 4D) and 293 Epstein–Barr nuclear antigen cells (79% inhibition) (data not shown), suggesting strongly that the pinopsin LRE is required for transcriptional repression mediated by a factor(s) that is widely distributed in nonphotosensory cells.
Fig. 4.
Transcriptional repression through thepinopsin LRE (−1122 to −1074). A, Schematic of the promoter/reporter constructs used in this experiment.luc, Luciferase. B–D, The reporter activities measured in the chick pineal cells (B), LMH cells (C), and NIH 3T3 cells (D). The cells transfected with each construct were harvested in the light (open bars inB) or in the dark (filled bars inB–D) to measure the luciferase activities. A typical set of data from three independent experiments is shown inB–D. Error bars indicate means ± SD of three replicated cell cultures.
Characterization of the nuclear protein bound to thepinopsin light-responsive element
We investigated whether the pinopsin LRE-binding protein(s) in the chick pineal nuclear extract varies quantitatively and/or qualitatively between the light and dark conditions. The EMSA revealed no significant difference in band pattern between the shifted bands derived from nuclear proteins in the light (Fig.5A, lane 2) and dark (Fig. 5A, lane 3).
One of the candidates for the pinopsin LRE-binding factor is δ-crystallin enhancer binding protein (δEF1)/zinc finger, E-box binding protein (ZEB), which was identified as a ubiquitous transcriptional repressor acting through the CACCT(G) E2 box (Funahashi et al., 1993; Genetta et al., 1994). This possibility was tested by the EMSA, in which the reaction mixture was supplied withδ-crystallin oligonucleotide harboring the δEF1/ZEB-binding site (DC5; Kamachi and Kondoh, 1993) or with its mutated oligonucleotide (mutDC5). However, these oligonucleotides had almost no effect on the formation of the retarded band (Fig.5B, lanes 5 and 6), indicating no functional relationship between the pinopsin LRE-binding factor(s) and δEF1/ZEB. In contrast, we observed competitive effects of E-box (CACGTG)-containing oligonucleotides corresponding to regions upstream of the mouse arginine vasopressin (mAvp) gene andcPer2 gene, each of which is transactivated through the E box by positive regulators, CLOCK and BMAL1 (Jin et al., 1999; Okano et al., 2001). These competitive effects were completely dependent on the CACGTG sequence (Fig. 5B, lanes 7 and8; C, lanes 4 and 5), indicating that the pinopsin LRE-binding factor can bind to these E boxes. The flanking sequences of these E boxes showed only weak similarities to each other (Fig. 5D); therefore, the core CACGTG sequence may be critical for the binding of the factor.
Using chick liver nuclear proteins, we detected a CACGTG-dependent formation of a retarded band with a mobility identical to that observed with the chick pineal nuclear proteins (Fig.5E). This is consistent with E-box-mediated transcriptional repression observed in chicken hepatoma-derived LMH cells (Fig.4C); it also supports the notion that the nuclear protein capable of binding to the pinopsin LRE functions as a ubiquitous repressor.
DISCUSSION
We found that an LRE for pinopsin gene regulation is present at positions −1103 to −1086 in the promoter region and that the light dependency of the promoter activity is completely lost by introducing mutations within these positions. Interestingly, the CACGTG sequence found in the pinopsin LRE completely matches the G box (CACGTGG), one of the LREs identified in plants (Donald and Cashmore, 1990), in which the element is not effective by itself and a combination with its specific minimal promoter is indispensable for the expression of light responsiveness. Such sequence conservation implies a mechanism for the light-dependent gene expression common to animal and plant cells. Among several transcription factors in plants, photomorphogenesis-promoting factor (HY5) (Chattopadhyay et al., 1998) and phytochrome interacting factor 3 (Martínez-García et al., 2000) inArabidopsis are well characterized as G-box-binding transcriptional activators in light-regulated gene expression. However, to date no example has been reported for a G-box-binding transcriptional repressor involved in light-dependent gene expression even in plants.
Although the pinopsin LRE in the construct −1156/+31 was required for the light dependency (Fig. 2), the 49 bp stretch encompassing the LRE was unable to confer light responsiveness even in the cultured pineal cells (Fig. 4B), indicating that the pinopsin LRE by itself is insufficient for conferring the light-dependent gene expression. One possible mechanism is that an additional LRE(s) within the region between −1156 and +31 would cooperatively function with the pinopsin LRE. In this case, another factor acting on this additional LRE would be required for light-dependent transactivation of the pinopsin promoter that is suppressed by the pinopsin LRE-binding protein. We have tried to identify this putative (second) LRE by transcriptional analyses of the other systematic series of deletion constructs in thepinopsin promoter region (−314 to +31), but this search was unsuccessful (data not shown). We now assume a contribution of multiple elements. Alternatively, the specific location of the LRE or its distance from the promoter region may absolutely be required for light-dependent transcriptional regulation.
In animals, a CACGTG E box has been shown to play a key role in the transcription/translation-based autoregulatory feedback loop of the circadian oscillator (Hao et al., 1997; Gekakis et al., 1998; Jin et al., 1999). Multiple copies of CACGTG E box were found in mousePer1 (mPer1) and human Per1 promoter, and each E box was shown to recruit the CLOCK–BMAL1 heteromer (Hida et al., 2000). At least a single copy of an E box with a similar function is present in the promoter region of cPer2 expressed in the pineal gland (Okano et al., 2001). Transcription of bothmPer1 and cPer2 genes is regulated by a circadian clock through the E box; it is also stimulated by light (Albrecht et al., 1997; Shearman et al., 1997; Akiyama et al., 1999; Okano et al., 2001), but very little information is available about thePer gene transcriptional regulation contributing to its light-dependent expression (Crosio et al., 2000). Our results clearly demonstrate that cPer2 and AVP oligonucleotides both harboring the E box have a competitive effect on the interaction between the pinopsin LRE and the pineal nuclear protein (Fig. 5B,C). This raised the possibility that a CACGTG E box(es) in the promoter region of cPer2 and AVPgenes may contribute to a light-regulatory mechanism similar to that underlying the light-dependent regulation of pinopsin gene expression through its LRE.
With respect to the transcriptional repressor in animals, δEF1/ZEB is known to bind directly to a subset of the CACCT(G) E box (Funahashi et al., 1993; Genetta et al., 1994), but δEF1/ZEB-like binding activity would not be involved in the pinopsin LRE-mediated negative regulation (Fig. 5B). Considering recent observations that the BMAL2 homodimer or the CLOCK–BMAL1 heterodimer potentially acts as a transcriptional inhibitor (Okano et al., 2001; Yu et al., 2002), a CLOCK–BMAL-containing complex might be involved in the light-dependent transcriptional regulation through the pinopsin LRE. Alternatively, as seen in Neurospora (Heintzen et al., 2001), a novel negative factor might modulate the transcription of light-regulated clock genes through the CACGTG E box. Like δEF1/ZEB and CLOCK–BMALs, the pinopsin LRE-binding factor seems to be expressed in a variety of cells not only for light-dependent gene expression but also for various aspects of gene regulation. Identification of this factor would be one of the ways to understand how the transcriptional apparatus in animal cells responds to light by using this general repressor through the pinopsin LRE.
Footnotes
This work was supported in part by grants-in-aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology. Y.T. is supported by Research Fellowships of the Japanese Society for the Promotion of Science for Young Scientists. We thank Prof. H. Kondoh and Dr. Y. Kamachi (Institute for Molecular and Cellular Biology, Osaka University, Osaka, Japan) for their kind gift of δ-crystallin oligonucleotides (DC5 and mutDC5) and for helpful advice and comments on this manuscript.
Correspondence should be addressed to Dr. Yoshitaka Fukada, Hongo 7-3-1, Bunkyo-ku, Tokyo 113, Japan. E-mail:sfukada@mail.ecc.u-tokyo.ac.jp.
REFERENCES
- 1.Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 3.Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- 5.Deguchi T. Rhodopsin-like photosensitivity of isolated chicken pineal gland. Nature. 1981;290:706–707. doi: 10.1038/290706a0. [DOI] [PubMed] [Google Scholar]
- 6.Doi M, Nakajima Y, Okano T, Fukada Y. Light-induced phase-delay of the chicken pineal circadian clock is associated with the induction of cE4bp4, a potential transcriptional repressor of cPer2 gene. Proc Natl Acad Sci USA. 2001;98:8089–8094. doi: 10.1073/pnas.141090998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donald RG, Cashmore AR. Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 1990;9:1717–1726. doi: 10.1002/j.1460-2075.1990.tb08295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. δ-crystallin enhancer binding protein δEF1 is a zinc finger-home-odomain protein implicated in postgastrulation embryogenesis. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 9.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 10.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 11.Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 13.Hao HL, Allen D, Hardin PE. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- 15.Hida A, Koike N, Hirose M, Hattori M, Sakaki Y, Tei H. The human and mouse Period1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics. 2000;65:224–233. doi: 10.1006/geno.2000.6166. [DOI] [PubMed] [Google Scholar]
- 16.Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 17.Kamachi Y, Kondoh H. Overlapping positive and negative regulatory elements determine lens-specific activity of the δ1-crystallin enhancer. Mol Cell Biol. 1993;13:5206–5215. doi: 10.1128/mcb.13.9.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci. 2000;23:713–742. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- 19.Korenbrot JI, Fernald RD. Circadian rhythm and light regulate opsin mRNA in rod photoreceptors. Nature. 1989;337:454–457. doi: 10.1038/337454a0. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Chen S, Wang Q, Zack DJ, Snyder SH, Borjigin J. A pineal regulatory element (PIRE) mediates transactivation by the pineal/retina-specific transcription factor CRX. Proc Natl Acad Sci USA. 1998;95:1876–1881. doi: 10.1073/pnas.95.4.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-García JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- 22.McGinnis JF, Austin BL, Stepanik PL, Lerious V. Light-dependent regulation of the transcriptional activity of the mammalian gene for arrestin. J Neurosci Res. 1994;38:479–482. doi: 10.1002/jnr.490380414. [DOI] [PubMed] [Google Scholar]
- 23.McWatters H, Dunlap JC, Millar AJ. Circadian biology: clocks for the real world. Curr Biol. 1999;9:R633–R635. doi: 10.1016/s0960-9822(99)80410-2. [DOI] [PubMed] [Google Scholar]
- 24.Okano T, Yoshizawa T, Fukada Y. Pinopsin is a chicken pineal photoreceptive molecule. Nature. 1994;372:94–97. doi: 10.1038/372094a0. [DOI] [PubMed] [Google Scholar]
- 25.Okano T, Yamamoto K, Okano K, Hirota T, Kasahara T, Sasaki M, Takanaka Y, Fukada Y. Chicken pineal clock genes: implication of BMAL2 as a bidirectional regulator in circadian clock oscillation. Genes Cells. 2001;6:825–836. doi: 10.1046/j.1365-2443.2001.00462.x. [DOI] [PubMed] [Google Scholar]
- 26.Pando MP, Sassone-Corsi P. Molecular clocks: a vivid loop of light. Nature. 2001;410:311–313. doi: 10.1038/35066655. [DOI] [PubMed] [Google Scholar]
- 27.Rehemtulla A, Warwar R, Kumar R, Ji X, Zack DJ, Swaroop A. The basic motif-leucine Zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci USA. 1996;93:191–195. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Jr, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 29.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 30.Steenhard BM, Besharse JC. Phase shifting the retinal circadian clock: xPer2 mRNA induction by light and dopamine. J Neurosci. 2000;20:8572–8577. doi: 10.1523/JNEUROSCI.20-23-08572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takanaka Y, Okano T, Iigo M, Fukada Y. Light-dependent expression of pinopsin gene in chicken pineal gland. J Neurochem. 1998;70:908–913. doi: 10.1046/j.1471-4159.1998.70030908.x. [DOI] [PubMed] [Google Scholar]
- 32.Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;5:445–475. [Google Scholar]
- 33.Yu W, Nomura M, Ikeda M. Interactivating feedback loops within the mammalian clock: BMAL1 is negatively autoregulated and upregulated by CRY1, CRY2, and PER2. Biochem Biophys Res Commun. 2002;290:933–941. doi: 10.1006/bbrc.2001.6300. [DOI] [PubMed] [Google Scholar]
- 34.Zatz M, Mullen DA, Moskal JR. Photoendocrine transduction in cultured chick pineal cells: effects of light, dark, and potassium on the melatonin rhythm. Brain Res. 1988;438:199–215. doi: 10.1016/0006-8993(88)91339-x. [DOI] [PubMed] [Google Scholar]