Abstract
Crx, an Otx-like homeobox gene, is expressed primarily in the photoreceptors of the retina and in the pinealocytes of the pineal gland. The CRX homeodomain protein is a transactivator of many photoreceptor/pineal-specific genes in vivo, such as rhodopsin and the cone opsins. Mutations inCrx are associated with the retinal diseases, cone–rod dystrophy-2, retinitis pigmentosa, and Leber's congenital amaurosis, which lead to loss of vision. We have generated transgenic mice, using 5′- and/or 3′-flanking sequences from the mouse Crxhomeobox gene fused to the β-galactosidase (lacZ) reporter gene, and we have investigated the promoter function of the cell-specific and developmentally regulated expression ofCrx. All of the independent transgenic lines commonly showed lacZ expression in the photoreceptor cells of the retina and in the pinealocytes of the pineal gland. We characterized the transgenic lines in detail for cell-specific lacZexpression patterns by 5-bromo-4-chloro-3-indolyl β-d-galactoside staining and lacZimmunostaining. The lacZ expression was observed in developing and developed photoreceptor cells. This observation was confirmed by coimmunostaining of dissociated retinal cells with thelacZ and opsin antibodies. The ontogeny analysis indicated that the lacZ expression completely agrees with a temporal expression pattern of Crx during retinal development. This study demonstrates that the mouse Crx5′-upstream genomic sequence is capable of directing a cell-specific and developmentally regulated expression of Crx in photoreceptor cells.
Keywords: photoreceptor, Crx, retina, transgenic mouse, transcriptional regulation, promoter
The retina is a very sensitive light detector, and the photoreceptor cells in the retina are essential for vision formation. The vertebrate retina contains two types of photoreceptors, rods and cones. Cones are responsible for daylight and color vision. Rods mediate dim light vision. Both rod and cone photoreceptors elaborate a specialized structure, the outer segment, to catch the light. The outer segments of rods and cones are filled with light-absorbing visual pigments, rhodopsin and cone opsins, respectively.
Considerable progress has been made toward elucidating the physiological and biochemical basis of the phototransduction pathway in photoreceptors. In contrast, the molecular basis of photoreceptor differentiation has been poorly understood. Most of the molecules involved in the phototransduction pathway are expressed specifically in the photoreceptors of the retina and in the pinealocytes of the pineal gland. Because defining the mechanisms of regulation of photoreceptor-specific genes is important to understanding the regulation of cell type specificity, mechanisms of transcriptional regulations of these photoreceptor-specific molecules have been studied extensively. However, the molecular basis, which confers photoreceptor specificity, is not well understood. CRX, an OTX-like homeoprotein, has been proposed to be a regulator of various photoreceptor-specific genes (Furukawa et al., 1997b). Regulation by CRX of many photoreceptor-specific genes has been proposed on the basis of CRX binding sites, their regulatory regions identified by protein–DNA binding assays and transient transfection assays (Chen et al., 1997; Furukawa et al., 1997b, 1999; Livesey et al., 2000). Interestingly, it recently has been reported that many photoreceptor-specific genes also are expressed in the pineal gland in which Crx is expressed (Blackshaw and Snyder, 1997). The pineal gland is related evolutionarily to the photoreceptors in the retina. In mammals the major function of the pineal gland is the secretion of melatonin. Melatonin is synthesized from serotonin by two key enzymes, N-acetyltransferase (NAT) and hydroxyindole-O-methyltransfererase (HIOMT). The regulatory regions of these genes contain multiple CRX binding sites (Li et al., 1998). Crx is a candidate factor to transactivate the expression of these two key enzymes in the melatonin synthesis pathway. Via the Crx knock-out mouse study we actually demonstrated that Crx is essential for the expression of NAT and other photoreceptor-specific genes in the pineal gland (Furukawa et al., 1999).
Mutations of various photoreceptor-specific genes have been shown to be responsible for the human genetic retinal disease retinitis pigmentosa (for review, see Dryja and Li, 1995). Mutations of human CRX, which also is expressed in photoreceptor cells specifically in the retina, have been demonstrated to be associated with three types of photoreceptor diseases: autosomal dominant cone–rod dystrophy-2 (adCRD2; Freund et al., 1997; Swain et al., 1997; Sohocki et al., 1998), retinitis pigmentosa (Sohocki et al., 1998), and Leber's congenital amaurosis (LCA; Freund et al., 1998; Sohocki et al., 1998). Patients of adCRD2 and retinitis pigmentosa undergo slow degeneration of photoreceptors, leading to blindness in later stages of their lives. LCA is a very severe type of photoreceptor disease that usually causes congenital blindness.
To understand the mechanisms of regulation of Crx expression in the development of rods and cones in the retina, we generated transgenic mice by using the 5′- and 3′-flanking mouse Crx(mCrx) sequence fused to the β-galactosidase (β-gal;lacZ) reporter gene. The present studies have indicated that the upstream mCrx genomic sequence directs, temporally and spatially, the lacZ expression in retinal photoreceptor cells corresponding to the mCrx expression pattern. In addition, we demonstrated that CRX transactivates itself to maintain its expression in vivo by positive feedback.
MATERIALS AND METHODS
Transgene vector and generation of transgenic mice.We obtained the Crx genomic clone from a 129SVJ mouse library (Stratagene, La Jolla, CA) by using a mouse Crx cDNA probe. We ligated and subcloned a 10 kb XhoI (partial digestion)–EcoRI fragment and a PCR-amplified 2 kbEcoRI–SmaI fragment containing exon 1 into a pβ-gal–Basic vector (Clontech, Palo Alto, CA) to make the Pcrx12k–lacZ construct (see Fig. 1A). Sequencing verified the 2 kb EcoRI–SmaI fragment. The Pcrx2k–lacZ vector contains the 2 kbEcoRI–exon 1 fragment and a 10 kbSmaI–EcoRI fragment that contains the first intron.
Fig. 1.
The Crxpromoter–lacZ transgene structure, the mouseCrx genomic sequence around the transcription initiation site, and Northern blot analysis of transgenic mice. A, Diagrammatic representation of the genomic structure of the mouseCrx gene and the Crx–lacZfusion constructs used for injection. Two transgenes that were used for the generation of transgenic mice are shown beneath the map of the mouse Crx genomic region. The mouse Crxgene is composed of four exons indicated by boxes on the genomic map. The homeodomain is indicated by the black box. Data are presented as the number of mice expressing the transgene in the photoreceptor-specific pattern through development per the number of transgenic mice that showed germline transmission.B, The nucleotide sequence around the transcription initiation site of the mouse Crx gene fused with β-galactosidase. An asterisk marks the transcription initiation site of the mouse Crx. The translation initiation site of β-galactosidase is underlined, and the connected site of the mouse Crx and the β-galactosidase is indicated by an arrow. The possible CRX binding sites are indicated by boxes.C, Northern blot analysis of total RNA (10 μg) isolated from cortex (lane 1), brainstem (lane 2), cerebellum (lane 3), and retina (lane 4) of Pcrx2k–lacZ (2kA) transgenic mice. The HindIII–ClaI 954 bp fragment of β-galactosidase was used as a labeled probe. 18S and28S are ribosomal RNAs used as marker RNA.
We extracted the Pcrx2k–lacZ and the Pcrx12k–lacZ from the recombinant plasmids by aNotI and SalI digestion. We fractionated theNotI–SalI fragments by electrophoresis on a 0.8% agarose gel and purified them by electroelution in dialysis tubes. We microinjected the DNA fragment into pronuclei of B6SJL/F2 C57BL/6 × SJL F2 hybrids. Then Southern blot hybridization of a HindIII–ClaI 954 bp fragment of the β-galactosidase or PCR verified the integration of thelacZ gene. In the PCR analysis we detected the transgene by using a sense primer (5′-TGCCGGTCTGGGAGGCATTGGTCTGGACACCAG-3′) and an anti-sense primer (5′-AGTTTGAGGGGACGACGACAGTATCGGCCTCAG-3′).
Antibodies. We acquired the following primary antibodies: mouse monoclonal antibodies against lacZ from Chemicon(Temecula, CA), against calbindin d-28k and against syntaxin (HPC-1) from Sigma (St. Louis, MO), and against vimentin from Zymed (San Francisco, CA); the Rho4D2 bovine monoclonal antibody against rhodopsin, a generous gift from Dr. R. S. Molday (University of British Columbia); rabbit polyclonal antibodies againstlacZ from Cortex Biochem (San Leandro, CA), chx10, a gift from Dr. R. McInnes (The Research Institute, Hospital for Sick Children, Toronto, Ontario), and against cone opsin blue and red/green, a generous gift from Dr. Y. Takada (Jikei Medical School of Tokyo, Tokyo, Japan). We used these antibodies at a 1:400–1:1000 dilution.
We purchased the following secondary antibodies: Cy3-conjugated donkey IgG against mouse IgG, Cy3-conjugated goat IgG against rabbit IgG, fluorescein isothiocyanate (FITC)-conjugated donkey IgG against rabbit IgG, and FITC-conjugated goat IgG against mouse IgG from Jackson ImmunoResearch Laboratories (West Grove, PA). We used these antibodies at a 1:2000 dilution.
Histochemistry and immunostaining. We used 3- to 4-week-old transgenic mice. While they were anesthetized with ketamine and xylazine (10:1), we enucleated the eyes and harvested the brains and the pineal glands. We performed 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) staining in a solution of 1 mg/ml X-Gal, 35 mmKFe3(CN)6, 35 mmK4Fe(CN)6–H2O, 2 mm MgCl2, 0.02% NP-40, and 0.01% Na-deoxycholate in PBS. We dissected the eyes at the limbus and fixed them with 0.5% glutaraldehyde in PBS for 12–24 hr and then removed the vitreous matter and fixed them for an additional 1 hr; meanwhile, the brains and pineal glands were immersed in the same fixative solution for 12–24 hr before X-Gal staining. We incubated the eyes, brains, and pineal glands in the X-Gal reaction solution for 48 hr at 37°C. We cryoprotected the samples after fixation with 30% sucrose in PBS, embedded them in optimal cutting temperature compound (Miles, Elkhart, IN), and froze them on dry ice. The samples were sectioned into 30-μm-thick sections with a cryostat at −27°C and were mounted on glass slides.
We dissected the eye at the limbus, isolated the retinas mechanically, and fixed them with 4% formaldehyde in PBS for immunofluorescence staining. The samples were cryoprotected, embedded, and frozen as above and sectioned into 18-μm-thick sections. We fixed the mounted sections with 4% formaldehyde in PBS and rinsed and preincubated them with blocking solution [2% normal goat serum, 2% normal donkey serum (Jackson ImmunoResearch), and 0.02% Triton X-100 (Sigma) in PBS] overnight. We incubated the sections with the primary antibodies at room temperature for 1 hr, rinsed them with blocking solution, and incubated them with the secondary antibodies for 30 min. We coverslipped the sections with Aqua Poly/Mount (Polysciences, Warrington, PA) after rinsing them with 0.02% Triton X-100 in PBS.
Cell dissociation and immunostaining. We dissected retinas from postnatal day 10 (P10) transgenic mice (2kB and 12kA) free from other tissues and incubated them in HBSS lacking Ca2+/Mg2+(Invitrogen, San Diego, CA), with trypsin (Worthington, Freehold, NJ) added to a final concentration of 1 mg/ml, at room temperature for 10 min. After trypsinization we added a soybean trypsin inhibitor (Sigma) to the final concentration of 2 mg/ml. We pelleted the cells by 2000 rpm centrifugation for 2 min, resuspended them, and triturated them into a single-cell suspension in HBSS with 100 μg/ml DNase I (Sigma). The cells were plated on poly-d-lysine-coated (Sigma) eight-well glass slides (Cel-Line Associates, Newfield, NJ) before fixation. We immunofluorescence-stained them as above.
β-gal assay. We dissected retinas from P10 transgenic mice (2kB) and dissociated the cells as described above. The cells were resuspended in 0.25 m Tris-HCl, pH 7.8, and alternately frozen in liquid nitrogen and thawed in a 37°C water bath, 3 min each, for a total of three freeze/thaw cycles. Both supernatant and cell extract were saved after a 14,000 rpm centrifugation for 15 min. We incubated the cell extracts at 37°C with 15 mmchlorophenol red-β-d-galactopyranoside (CPRG; Roche Bioscience, Palo Alto, CA) and measured the absorbance at 574 nm.
Northern blot analysis. We prepared total RNA from P21 retinas, brain cortices, cerebella, and brainstems from 2kA and 12kA transgenic mice. We used 10 μg of total RNA for electrophoresis and aHindIII–ClaI 954 bp fragment of β-galactosidase as a labeled probe. The electrophoresis, transfer, and hybridization were done as described previously (Furukawa et al., 1997a).
RESULTS
Generation of the mCrx–lacZ transgenic mice
To address whether the Crx upstream sequence is capable of directing its expression in a cell-specific and developmentally regulated manner, we fused the 5′-flanking region and/or the first intron of the mouse Crx gene to the lacZ reporter gene (Fig. 1A).
We first isolated phage clones encoding the mouse Crx locus as part of our effort to make a mouse knock-out of Crx(Furukawa et al., 1999). We have mapped the 5′-end of theCrx mRNA by using the RNA ligase-mediated rapid amplification of 5′-cDNA ends (RLM-RACE) method (Maruyama and Sugano, 1994; Volloch et al., 1994), and we determined the transcription initiation site (Fig. 1B). The mouse Crxupstream sequence lacks typical consensus TATA, GC, and CAT boxes. A sequence of the region from the murine Crxlocus is shown in Figure 1B. The transcription initiation site is indicated by an asterisk. We sequenced the entire 2 kb region and registered it in the GenBank database (accession numberAF301006). In this 2 kb region we found three CRX binding consensus sequences. Two of them are located proximal to the transcription initiation sites of the mouse Crx gene (Fig.1B).
For the Pcrx12k–lacZ construct the fusion gene consists of a 12 kb mCrx upstream genomic fragment, starting from 34 bp upstream of the mCrx translation initiation site; a 131 bp fragment containing the translation initiation site of theDrosophila melanogaster alcohol dehydrogenase gene; thelacZ gene; a small t antigen intron; and a polyadenylation site derived from the SV40 gene (Fig. 1A). For the Pcrx2k–lacZ construct, we used a 2 kb mCrxupstream genomic fragment and the 10 kb first intron fragment.
We have generated transgenic mice, using these two constructs, with the aim of identifying a region (or regions) that transactivatesCrx transcription specifically in photoreceptor cells. On blot hybridization analysis of tail DNAs, we identified six and four independent transgenic lines that passed their transgenes onto their offspring for the Pcrx12k–lacZ and the Pcrx2k–lacZ, respectively. Three of six Pcrx12k–lacZ lines (named 12kA, B, and F) and four of four Pcrx2k–lacZ lines (named 2kA, B, E, and G) exhibited an X-Gal-positive staining in the photoreceptor layers of the retina. Blot hybridization analysis of tail DNAs indicated that 12kA and 12kF each possess approximately five to six copies of the lacZ gene in the genomic DNA. 12kB has approximately seven to eight copies, 2kA and 2kG each have approximately one to two copies of the lacZgene, 2kB has ∼10–15 copies, and 2kE has ∼40–50 copies of thelacZ gene in the genomic DNA (data not shown).
We first analyzed tissue specificity of the lacZ expression by Northern blot hybridization of total RNA of the retina, cerebral cortex, cerebellum, and brainstem isolated from these transgenic animals (Fig. 1C). This analysis revealed that in these lines the lacZ mRNA was expressed specifically in the retina and pineal gland, but not in other parts of the brain (except in the pineal gland).
All Pcrx12K and Pcrx2K transgenic mice showing lacZexpression exhibited lacZ expression specifically in photoreceptor cells and in the pineal gland. Although we also examined tissue specificity of the lacZ expression by X-Gal staining of whole-mount embryos at embryonic day 12.5 (E12.5) and sections of various adult tissues, we did not observe any significant staining except in the retina and the pineal gland (data not shown). We therefore conclude that an overlapping 2 kb upstream region is responsible for the specific expression of mCrx in photoreceptors and pineal gland.
LacZ expression pattern in the retina
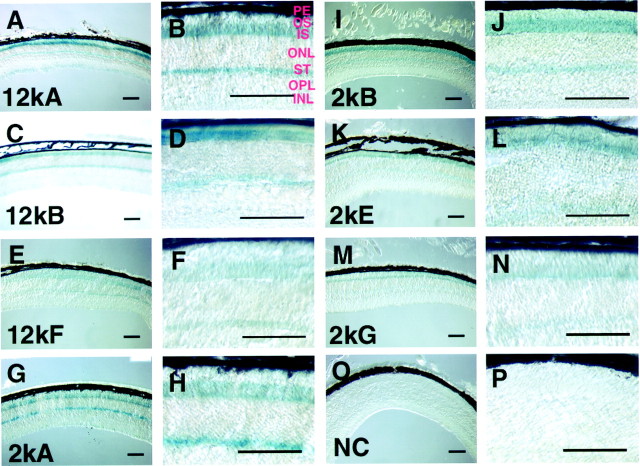
We examined an X-Gal staining pattern in the retinal section of the three Pcrx12k–lacZ and four Pcrx2k–lacZtransgenic lines at adult stage (3–4 weeks old). The densities of the X-Gal reaction product were not identical among these transgenic mice. The 12kA and 2kB retinal sections showed relatively strong and dense staining. All of these transgenic mice, however, exhibited the same spatial and developmental expression in the photoreceptor cell layer of the retina (Fig. 2). For additional analysis we focused our analysis on the 2kB line, which showed slightly stronger X-Gal staining. The retina consists of several different cell layers: the outer nuclear layer (ONL), composed of photoreceptor cell bodies; the inner nuclear layer (INL), consisting of bipolar, horizontal, and amacrine cells; and the ganglion cell layer (GCL), containing primarily ganglion cells. Photoreceptors, bipolar cells, and horizontal cells make synaptic connections in the outer plexiform layer (OPL). A differentiated mature photoreceptor is composed of the outer segment (OS), inner segment (IS), cell body, and synaptic terminus (ST). In all transgenic lines X-Gal staining was observed in the inner segment and synaptic terminus of photoreceptor cells (Fig. 2). Although a cytoplasmic β-galactosidase was used in this study, X-Gal-stained products were localized specifically in inner segments and synaptic termini. The mechanisms underlying this specific localization of the β-galactosidase are not yet clear.
Fig. 2.
Transverse retinal sections of the three Pcrx12k–lacZ (12kA, 12kB, and 2kF), the four Pcrx2k–lacZ (2kA, 2kB, 2kE, and 2kG) transgenic mice, and a nontransgenic mouse (NC) stained with X-Gal. Shown is the X-Gal staining pattern in lower magnification (A, C, E, G, I, K, M, O) and in higher magnification (B, D, F, H, J, L, N, P) from 12 kA (A, B), 12kB (C, D), 12kF (E, F), 2kA (G, H), 2kB (I, J), 2kE (K, L), 2kG (M, N), and nontransgenic (O, P) mice. The intensities of the X-Gal stain are different among the seven transgenic lines, but the staining is observed commonly in the inner segment and the axon terminals of the photoreceptor cells. X-Gal staining was observed all through the region of the retina. For negative control the retina of a nontransgenic littermate was stained with X-Gal. The X-Gal stain was observed slightly in the ganglion cell layer. PE, Pigment epithelium. Scale bars, 100 μm.
To investigate whether lacZ is expressed in rods under the control of the mCrx flanking sequences, we performed double-immunofluorescence staining with anti-rhodopsin antibody and anti-lacZ antibody on retinal transverse sections and examined the immunostaining pattern of lacZ expression via fluorescent microscopy. The immunofluorescent pattern of the lacZexpression was photoreceptor-specific, which was identical with the X-Gal staining patterns shown above (Fig.3). In addition, we also performed double-immunofluorescence staining with cell type-specific markers and anti-lacZ antibody to examine whether lacZ is expressed in nonphotoreceptors. Double-immunofluorescence staining of thelacZ and cell type-specific markers was performed by two different combinations of primary antibodies against rhodopsin (rod), chx10 (bipolar cell), calbindin (horizontal cell), vimentin (Müller glia), HPC-1 (amacrine cell), and lacZ. A polyclonal lacZ rabbit antibody and monoclonal cell type-specific mouse antibodies were used, except for chx10. For the staining of bipolar cells, a polyclonal chx10 antibody and a monoclonal lacZ mouse antibody were used. Rhodopsin-positive immunoreactivity andlacZ-positive immunoreactivity are overlapped. None of other markers overlap with lacZ-positive immunoreactivity. In summary, mCrx promoter-driven lacZ is expressed specifically in the photoreceptor layer of the retina.
Fig. 3.

Double-immunofluorescence staining oflacZ and cell type-specific markers in transverse retinal sections of the 2kB transgenic mice. Double-immunofluorescence staining of the lacZ and cell type-specific markers was performed by two different combinations of primary antibodies against rhodopsin (rod; A), chx10 (bipolar cell;D), calbindin (horizontal cell; G), vimentin (Müller glia; J), HPC-1 (amacrine cell; M), and lacZ (B, E, H, K, N). Overlaid images are displayed in C, F, I, L, O. A polyclonal lacZ rabbit antibody and monoclonal cell type-specific mouse antibodies were used, except for chx10. We used a polyclonal chx10 antibody and a monoclonal lacZ mouse antibody for the staining of bipolar cells. Rhodopsin-positive immunoreactivity andlacZ-positive immunoreactivity are overlapped. None of the other markers overlaps with lacZ-positive immunoreactivity. Scale bars, 50 μm.
Double-immunofluorescence staining in dissociated retinal cells
To characterize cell types expressing lacZ further, we performed double immunostaining of dissociated adult retinal cells (Fig. 4). By this method we can examine the lacZ expression at the single-cell level in greater detail than by immunofluorescence staining of sections. The retina of the 2kB mouse was dissociated by trypsin treatment, and dissociated cells were double immunostained with anti-lacZ and anti-rhodopsin antibodies. In a dissociated cell preparation there were manylacZ-positive/anti-rhodopsin-positive cells (Fig.4A–C). We then examined the expression oflacZ in other cell types for which the cell bodies are located in the inner nuclear layer, including horizontal cells (Fig.4D–F), Müller glia (Fig.4G–I), amacrine cells (Fig.4J–L), and bipolar cells (Fig.4M–O). We found that lacZ-expressing cells are negative for anti-calbindin, -vimentin, -HPC-1, and -chx10 antibodies (Fig. 4D–O). This result indicates thatlacZ is not expressed in these cell types.
Fig. 4.
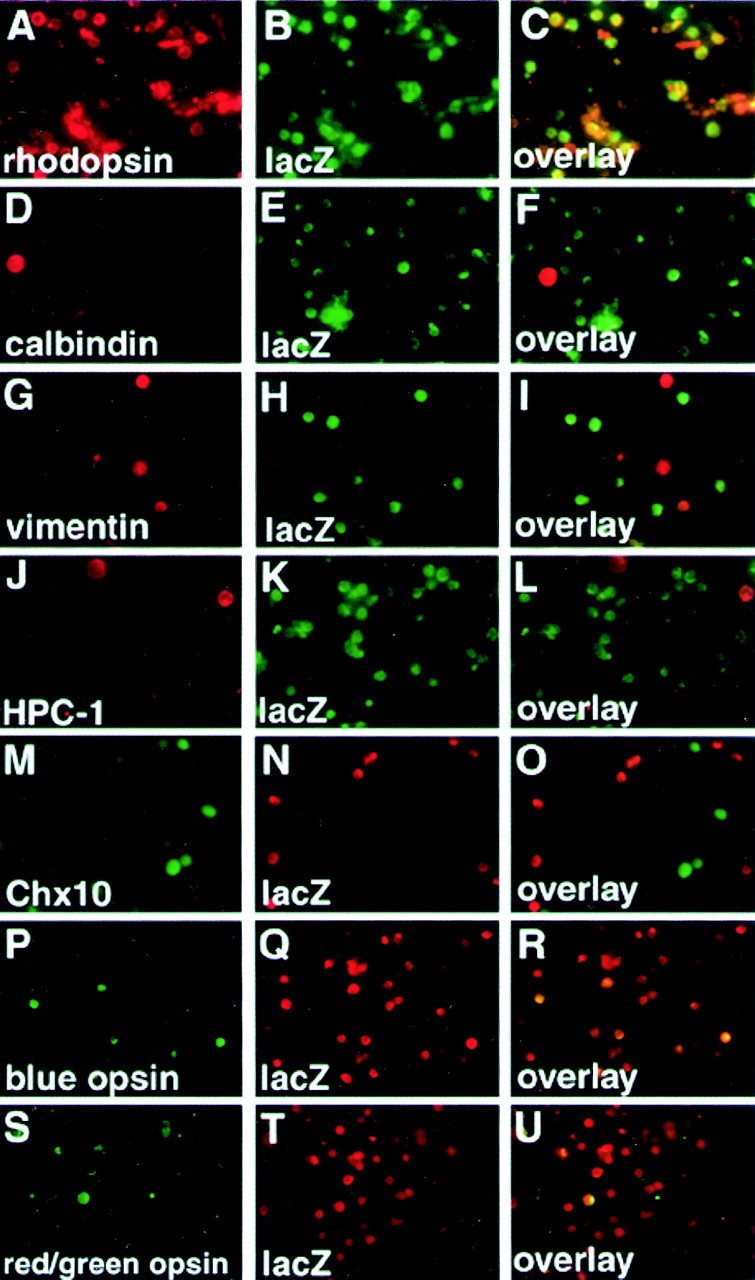
Double-immunofluorescence staining of dissociated retinal cells of the 2kB transgenic mice by anti-lacZ and cell type-specific markers. Double-immunofluorescence staining of thelacZ and cell type-specific markers was performed by two different combinations of primary antibodies against rhodopsin (rod;A), calbindin (horizontal cell; D), vimentin (Müller glia; G), HPC-1 (amacrine cell;J), chx10 (bipolar cell;M), and lacZ (B, E, H, K, N, Q, T). Overlaid images are displayed in C, F, I, L, O, R, U. Combinations of polyclonal anti-lacZ rabbit antibody (B, E, H, K) and monoclonal anti-cell-specific marker mouse antibodies (A, D, G, J), or of polyclonal anti-cell-specific marker rabbit antibodies (M, P, S) and monoclonal mouse anti-lacZ (N, Q, T) antibody were used for staining. There are many numbers of lacZ/rhodopsin-positive (C) and some lacZ/cone opsin-positive cells (R, U) seen in a dispersed preparation. However, lacZ-expressing cells are anti-calbindin, anti-vimentin, and anti-HPC-1, and chx10-negative (F, I, L, O), indicating that lacZ is not expressed in other cell types.
We also performed double immunostaining with the anti-lacZ and anti-cone opsin (blue or green/red) antibodies (Fig.4P–U). There were some anti-lacZ-positive/anti-cone opsin-positive cells. These results demonstrated that lacZ is expressed in both rods and cones. This completely agrees with the expression pattern of Crx, which is expressed both in rods and cones (Chen et al., 1997; Furukawa et al., 1997b).
LacZ expression during retinal development
The expression of mCrx initiates and develops during the embryonic and neonatal period in accordance with the development of photoreceptor cells in the retina (Furukawa et al., 1997b). We investigated the ontogeny of lacZ expression during retinal development in 12kA, 2kA, and 2kB transgenic mice by X-Gal staining. Because the lacZ expression patterns during development are very similar among these three lines (data not shown), we will present the lacZ expression pattern of 2kB (Figs.5, 6). The X-Gal reaction product was not detected at E11.5 (data not shown) and was detected first at E12.5 (Fig. 5A,B). This completely agrees with the observation that the mouse Crx mRNA first was detected by in situ hybridization in the developing mouse retina at E12.5 (Furukawa et al., 1997b). At E15.5 and E17.5 the X-Gal staining became denser and more uniform in the presumptive photoreceptor layer (Fig. 5E–H). In summary, the expression pattern oflacZ in the embryonic stages completely agrees with the expression pattern of Crx detected by in situhybridization.
Fig. 5.

Embryonic retinas stained with X-Gal. X-Gal staining was performed with transverse sections of embryonic retinas of the 2kB mice at E12.5 (A, B), E13.5 (C, D), E17.5 (E, F), and at E13.5 of nontransgenic mouse (NC, negative control; G, H). A red arrow indicates developing photoreceptor cells in the presumptive photoreceptor layer. Ared arrowhead indicates nonspecific staining at the ganglion cell layer of the retina from nontransgenic mouse. Scale bars, 100 μm.
Fig. 6.

Postnatal retinas stained with X-Gal. X-Gal staining was performed with transverse sections of postnatal retinas of the Pcrx2kB mice. A red arrow indicates developing photoreceptor cells in the photoreceptor layer. A red arrowhead indicates nonspecific staining at the ganglion cell layer. A slight nonspecific staining also is observed in the inner nuclear layer of the nontransgenic retina. PE, Pigment epithelium; IPL, inner plexiform layer. Scale bars, 100 μm.
After birth, X-Gal staining was detected in the developing photoreceptor layer and weakly in the inner nuclear layer and ganglion cell layer (Fig.6A–D). Because the X-Gal-stained retina from nontransgenic mice also showed weak signal in the inner nuclear layer and the ganglion cell layer (Fig.6G,H), all of or at least most of the X-Gal reaction products outside of the photoreceptor layer are considered to come from endogenous galactosidase activity in the mouse retina.
LacZ expression in pineal gland
Many photoreceptor-specific genes are known to be expressed in the pineal gland in which Crx also is expressed (Blackshaw and Snyder, 1997). The pineal gland is a small organ located on the midline in the dorsal cranium, which is related evolutionarily with photoreceptor cells in the retina. We also examined lacZexpression of the Crx promoter lacZ transgenic mice, 12kA and 2kB, in the pineal gland. The X-Gal reaction product was detected in the pinealocytes of the adult pineal gland (Fig.7). This result also agrees with the expression of Crx mRNA in the pineal gland (Chen et al., 1997; Furukawa et al., 1999).
Fig. 7.
Adult pineal gland stained with X-Gal. X-Gal staining was performed with transverse sections of the12kA (A) 2kB (B), and nontransgenic (C) mouse adult pineal gland. Intense X-Gal staining is seen in the pineal glands of the transgenic mice. Scale bars, 100 μm.
Crx itself is required for maintenance of the transgene expression
The existence of two CRX binding consensus motifs near the transcription initiation site suggests that CRX protein itself can bind and transactivate the Crx transcription (Fig.1B). To address the requirement for CRX function in the activity of the Crx regulatory sequences that have been characterized here, we examined the expression of aCrx–lacZ transgene in the Crx mutant background. The 2kB transgenic line was crossed with the Crxmutant mice (Furukawa et al., 1999). Animals hemizygous for theCrx mutant allele were crossed with Crxpromoter–lacZ heterozygous transgenic mice. The retinas of P10 pups were harvested and lysed; lacZ activity was measured by liquid assay. We chose to harvest P10 retinas because retinal degeneration of the Crx null mouse has not started at this stage. Compared with the mice in the wild-type background, the retinas of transgenic mice in the Crx null background exhibited only 31% of the lacZ activity (Fig.8A). Interestingly, retinas of transgenic mice in the Crx heterozygous mutant background also exhibited only 57% of the lacZ activity. This reduced lacZ activity in the Crxheterozygous mutants may reflect a Crx gene dosage effect for the phenotypes observed in the Crx heterozygous mutant mice (Furukawa et al., 1999). However, retinas of nontransgenic mouse in the Crx null background, the Crx heterozygous mutant background, and wild type exhibited 3.4, 3.4, and 3.7% of thelacZ activity, respectively. These activities are lower by far than those of transgenic mice. These data support a role of CRX protein in autoregulation.
Fig. 8.
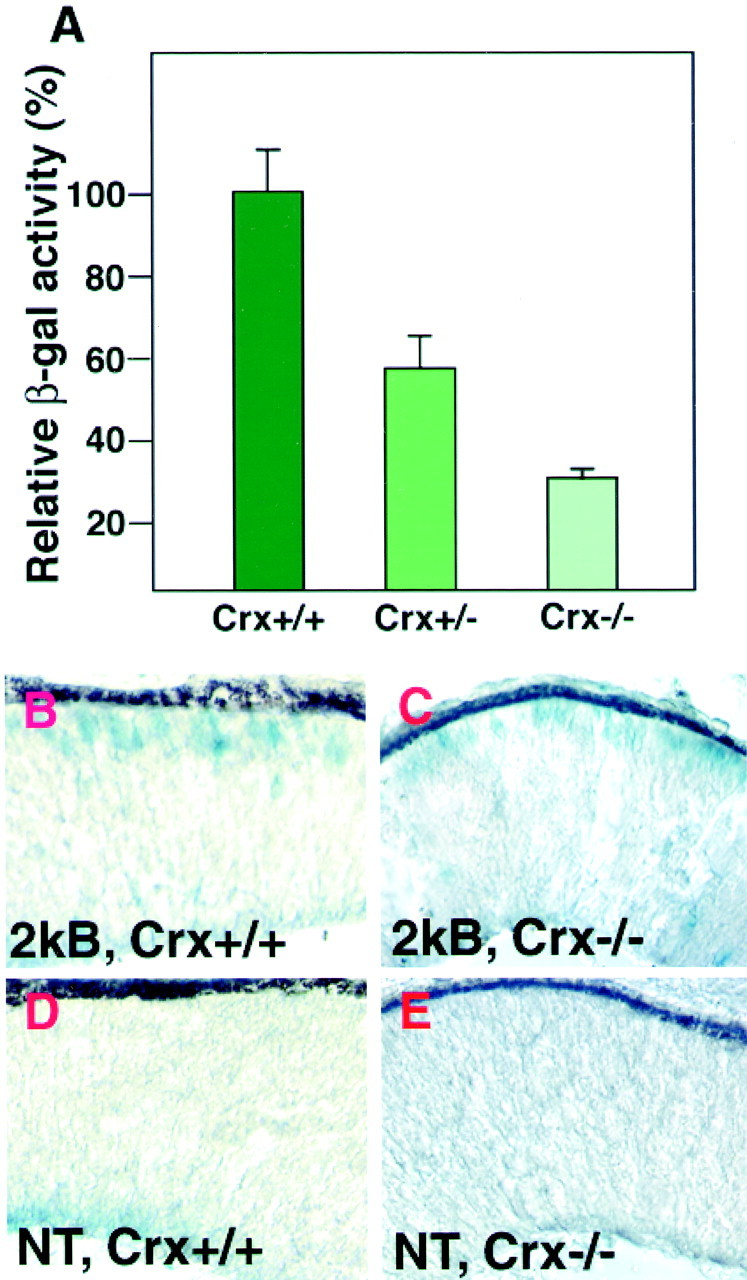
Requirement of the CRX function for the maintenance of Crx promoter–lacZtranscription. A, Relative β-galactosidase activity of retinas of 2kB transgenic mice in the wild-type,Crx+/−, orCrx−/− mutant background. Error bar indicates SEM. B, A cryosection of the retina from a 2kB transgenic mouse inCrx+/+ background at E12.5.C, A cryosection of the retina from a 2kB transgenic mouse in the Crx−/−background showing lacZ expression in the presumptive photoreceptor layer at E12.5. D, A cryosection of the retina from a nontransgenic wild-type mouse at E12.5. E, A cryosection of the retina from a nontransgenicCrx−/− mouse at E12.5.
We then examined to see whether CRX is required for the induction ofCrx transcription at E12.5, when Crxtranscription first is detected during development. We performed X-Gal staining on the retina of the 2kB line in the Crx null background (Fig. 8C). We observed X-Gal staining similar to the wild-type background shown in Figure 8B. We observed no X-Gal staining in the photoreceptor layer of the retina of nontransgenic mouse either in theCrx−/− background or in wild type (Fig. 8D,E). This result showed thatCrx itself is not essential for the initial induction of theCrx transcription.
DISCUSSION
Expression of Crx is restricted in large part in postmitotic differentiating photoreceptor cells, and its expression also is maintained in mature differentiated photoreceptor cells.Crx is the earliest known marker of photoreceptor identity in the developing retina. Crx mutant mice do not elaborate photoreceptor outer segments. There was a complete absence of rod and cone activity as assayed by electroretinogram. Expression of many photoreceptor and pineal-specific genes was found to be reduced inCrx mutants. Therefore, Crx is essential for proper differentiation of photoreceptor cells. Identification of the mechanisms of how Crx transcription is induced is a crucial step toward identifying the signaling pathways that control early cell fate determination of photoreceptor cells.
We have investigated the spatial and temporal expression patterns of the lacZ reporter gene under the regulation of flanking sequences of the mouse Crx gene. The results of this study have demonstrated that the 5′-flanking region of the mouseCrx encodes the sequence determinant that is necessary and sufficient for the photoreceptor-specific and developmentally regulated expression of the mouse Crx gene. First, all of the transgenic lines that were positive in the lacZ expression consistently showed the lacZ expression in the photoreceptor cells of the retina and in the pinealocytes of the pineal gland. Second, in a detailed characterization of the lacZexpression in retinal sections of the seven transgenic lines, thelacZ expression always was observed in photoreceptor cells. Furthermore, in a dissociated retinal cell preparation the opsin-immunoreactive cells were lacZ-positive. Third, the temporospatial pattern of the lacZ expression completely agreed with that of the mouse Crx expression during retinal development.
Although Crx expression is restricted primarily to the photoreceptor cells in the retina, it also has been suggested that bipolar cells may express Crx weakly in mammals. A recent report about the strong expression of a zebrafish Crx in the bipolar cells might support this (Liu et al., 2001). However, we did not detect any significant expression of the lacZ reporter in bipolar cells. Our transgenic mice may express lacZ in bipolar cells, but its expression level might be too low to be detected. Another possibility is that the Crx genomic regions used in this study may lack an enhancer element to driveCrx in bipolar cells.
Crx is an essential key transcription factor that governs development of the phototransduction pathway and outer segment formation. At P10 the expression of rhodopsin, cone opsins (blue and green/red), rod transducin α-subunit, cone arrestin, and recoverin clearly were reduced in Crx null mice. The levels of cone transducin, phosphodiesterase (PDE) β (rod and cone), rod cGMP-gated channel (RNG), rod arrestin, and peripherin RNAs were reduced weakly. In contrast, cone cGMP-gated channel (CNG) was upregulated inCrx null mutant (Furukawa et al., 1999). Crxregulates these photoreceptor-specific genes directly and/or indirectly. Therefore, the precise control of Crx expression is essential for proper photoreceptor differentiation. Many studies on gene regulation of photoreceptor-specific genes have been reported, including rhodopsin, cone opsins, interphotoreceptor retinoid-binding protein (IRBP), PDE, and rod arrestin (Wang et al., 1992; Kikuchi et al., 1993; Bobola et al., 1995; Chen and Zack, 1996;Di Polo et al., 1996; Kimura et al., 2000). We compared the sequences of the promoter regions of these genes with the 2 kb region of mouseCrx, but we did not find any significant homologous region. Therefore, this may suggest that cis-regulatory mechanisms of induction of Crx transcription are different from those of other photoreceptor-specific genes such as opsins.
We demonstrated that Crx itself plays an important role in maintaining its own expression in vivo. The β-galactosidase activity of the Pcrx2k transgenic mice is reduced to 31% in the Crx−/−background. However, Crx begins to be expressed at E12.5 in the Crx−/− background, showing that Crx is not essential for its initial induction. This leads us to hypothesize that there is an unknown factor (or factors) binding to the 2 kb region to induce Crxtranscription. Once Crx transcription is induced, the high-level expression of Crx is achieved by an autopositive feedback mechanism (Fig. 9). Then CRX upregulates various photoreceptor-specific genes, including opsins and transducins.
Fig. 9.
Transcriptional regulation of mouseCrx in photoreceptor cells. Crx is not expressed in retinal progenitors, but Crx begins to be expressed after they become postmitotic and start differentiation. An unknown factor (or factors) X initially induces Crxtranscription by interacting with the 2 kb region. ThenCrx itself contributes to upregulating and maintaining the expression of Crx by autopositive feedback.
The identification of the tissue-specific factor (or factors) interacting with the cis-acting element that we have described will contribute substantially toward understanding the mechanism of regulation of the Crx gene and, quite likely, the regulation of photoreceptor cell fate. Because Crx is the earliest known marker of photoreceptor identity in the developing retina, the mouse Crx promoter fragment that we have characterized in this study also will be a very useful tool, such as in the generation of developing and developed photoreceptor-specific transgenic mice, for various studies of development and function of photoreceptor cells.
Footnotes
This work was supported by the Howard Hughes Medical Institute and the National Institutes of Health. We thank Dr. R. S. Molday for Rho4D2 antibody, Dr. R. McInnes for chx10 antibody, and Dr. Y. Takada for antibodies against cone opsins.
Correspondence should be addressed to Takahisa Furukawa, Osaka Bioscience Institute, 6-2-4 Furuedai, Suita-city, Osaka 565-0874, Japan. E-mail:furukawa@obi.or.jp.
REFERENCES
- 1.Blackshaw S, Snyder SH. Developmental expression pattern of phototransduction components in mammalian pineal implies a light-sensing function. J Neurosci. 1997;17:8074–8082. doi: 10.1523/JNEUROSCI.17-21-08074.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bobola N, Hirsch E, Albini A, Altruda F, Noonan D, Ravazzolo R. A single cis-acting element in a short promoter segment of the gene encoding the interphotoreceptor retinoid-binding protein confers tissue-specific expression. J Biol Chem. 1995;270:1289–1294. doi: 10.1074/jbc.270.3.1289. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Zack DJ. Ret 4, a positive-acting rhodopsin regulatory element identified using a bovine retina in vitro transcription system. J Biol Chem. 1996;271:28549–28557. doi: 10.1074/jbc.271.45.28549. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. CRX, a novel Otx-like paired homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 5.Di Polo A, Rickman CB, Farber DB. Isolation and initial characterization of the 5′-flanking region of the human and murine cyclic guanosine monophosphate-phosphodiesterase β-subunit genes. Invest Ophthalmol Vis Sci. 1996;37:551–560. [PubMed] [Google Scholar]
- 6.Dryja TP, Li T. Molecular genetics of retinitis pigmentosa. Hum Mol Genet. 1995;4:1739–1743. doi: 10.1093/hmg/4.suppl_1.1739. [DOI] [PubMed] [Google Scholar]
- 7.Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis-Anagnostou A, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR. Cone–rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 8.Freund CL, Wang QL, Chen S, Muskat BL, Wiles CD, Sheffield VC, Jacobson SG, McInnes RR, Zack DJ, Stone EM. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis [letter]. Nat Genet. 1998;18:311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- 9.Furukawa T, Kozak CA, Cepko CL. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci USA. 1997a;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa T, Morrow EM, Cepko CL. Crx, a novel Otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997b;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 11.Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi T, Raju K, Breitman ML, Shinohara T. The proximal promoter of the mouse arrestin gene directs gene expression in photoreceptor cells and contains an evolutionarily conserved retinal factor-binding site. Mol Cell Biol. 1993;13:4400–4408. doi: 10.1128/mcb.13.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura A, Singh D, Wawrousek EF, Kikuchi M, Nakamura M, Shinohara T. Both PCE-1/RX and OTX/CRX interactions are necessary for photoreceptor-specific gene expression. J Biol Chem. 2000;275:1152–1160. doi: 10.1074/jbc.275.2.1152. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Chen S, Wang Q, Zack DJ, Snyder SH, Borjigin J. A pineal regulatory element (PIRE) mediates transactivation by the pineal/retina-specific transcription factor CRX. Proc Natl Acad Sci USA. 1998;95:1876–1881. doi: 10.1073/pnas.95.4.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Shen Y, Rest JS, Raymond PA, Zack DJ. Isolation and characterization of a zebrafish homologue of the cone–rod homeobox gene. Invest Ophthalmol Vis Sci. 2001;42:481–487. [PubMed] [Google Scholar]
- 16.Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- 17.Maruyama K, Sugano S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 18.Sohocki MM, Sullivan LS, Mintz-Hittner HA, Birch D, Heckenlively JR, Freund CL, McInnes RR, Daiger SP. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription factor gene. Am J Hum Genet. 1998;63:1307–1315. doi: 10.1086/302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, Sieving PA, Zack DJ. Mutations in the cone–rod homeobox gene are associated with the cone–rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- 20.Volloch V, Schweitzer B, Rits S. Ligation-mediated amplification of RNA from murine erythroid cells reveals a novel class of beta globin mRNA with an extended 5′-untranslated region. Nucleic Acids Res. 1994;22:2507–2511. doi: 10.1093/nar/22.13.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Macke JP, Merbs SL, Zack DJ, Klaunberg B, Bennett J, Gearhart J, Nathans J. A locus control region adjacent to the human red and green visual pigment genes. Neuron. 1992;9:429–440. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]