Abstract
The partitioning of the interphase nucleus into chromosome territories generally precludes DNA from making specific and reproducible inter-chromosomal contacts. However, with the development of powerful genomic and imaging tools for the analysis of the 3D genome, and with their application on an increasing number of cell types, it becomes apparent that regulated, specific, and functionally important inter-chromosomal contacts exist. Widespread and stereotypic inter-chromosomal interactions are at the center of chemosensation, where they regulate the singular and stochastic expression of olfactory receptor genes. In olfactory sensory neurons (OSNs) coalescence of multiple intergenic enhancers to a multi-chromosomal hub orchestrates the expression of a single OR allele, whereas convergence of the remaining OR genes from 18 chromosomes into a few heterochromatic compartments mediates their effective transcriptional silencing. In this review we describe the role of interchromosomal interactions in OR gene choice, and we describe other biological systems where such genomic interactions may contribute to regulatory robustness and transcriptional diversification.
Introduction
Despite the general positioning of chromatin in the interphase nucleus into distinct chromosome territories (Cremer 1982), imaging studies over a decade ago showed that interchromosomal interactions occur frequently (Branco Pombo 2006) albeit in a non stereotypic fashion. Over the past decade characterization of additional layers of chromatin organization into compartments, nuclear bodies and chromatin loops has been accelerated by advances in imaging as well as chromatin conformation capture-based technologies (3C, HiC, CaptureC). However, because inter-chromosomal interactions represent somewhat stochastic events that are not highly reproducible between cells, until recently, they have not been readily detected with unbiased methods like HiC (Rao, 2014) calling into question their regulatory significance.
The mouse olfactory system, however, where stochastic, singular and highly distributed expression of olfactory receptor (OR) genes is essential for odor perception, constitutes a striking exception. Olfactory sensory neurons (OSNs) stably express one out of ~1400 OR genes. The singular and stochastic choice of an OR gene is regulated by two types of inter-chromosomal interactions that are specific to OSNs: First, the aggregation of all OR loci from 18 chromosomes into a unique OR gene compartment maintains OR genes in a repressive environment, preventing transcription of multiple receptors even at low levels (Dalton 2014, Lyons, 2013). Second, a multi-chromosomal, multi-enhancer hub, formed by up to several dozen trans acting enhancers, drives the expression of the active OR allele. These two types of inter-chromosomal interactions occur in a developmentally regulated and coordinated sequence, and both are vital for regulating OR gene expression. This review will describe various contributions of inter-chromosomal interactions and their impact on nuclear organization, as they pertain to our current understanding of OSN nuclear architecture as well as other cell types.
De novo assembly of a nuclear compartment
OSNs are continuously regenerated from olfactory progenitor cells. While progenitor cells co-express multiple ORs at low levels (Tan L, 2015; Hanchate, 2015), OSNs converge on expressing one receptor at high levels. Singular OR expression occurs in the context of remodeling and reorganization of OR gene loci. More than 1000 OR genes residing in clusters on 18 chromosomes adopt H3K9me3 and H4K20me3 chromatin marks consistent with constitutive heterochromatin (Magklara 2011). This remodeling coincides with inversion of heterochromatin in the nucleus. Upon downregulation of lamin b receptor (LBR) heterochromatin detaches from the nuclear lamina and aggregates around the shell of pericentric heterochromatin that surrounds the nucleolus (Clowney EJ, 2012; Armelin-Correa 2014). This nuclear reorganization facilitates the aggregation of OR gene clusters in cis and in trans to form a unique OR gene compartment (Clowney EJ, 2012; **Monahan and Horta, 2019). However, the forces that drive OR compartmentalization are not known.
Several studies have shown that chromatin state is correlated with long range contacts (Yaffe, Tanay 2011;Rao, 2014). High resolution maps of human and mouse cell lines showed that the genome can be divided into six sub-compartments based on interactions, which are correlated with chromatin state, nuclear position and replication timing (Rao, 2014). Changes in chromatin state are accompanied by changes in compartment interactions. Recruitment of chromatin remodelers Ezh2 (which is responsible for H3K27me3 methylation at silenced genes) and Suv39h1 (which establishes H3K9me3 at constitutive heterochromatin) to an active genomic region resulted in the formation of new contacts with genomic regions that bear the respective chromatin marks (Wijchers, de Laat 2016). Interestingly, recruitment of a chromodomain mutant of Suv39h1 that cannot interact with HP1 did not induce new interactions with constitutive chromatin. This is consistent with findings that HP1 undergoes phase separation as it oligomerizes (Larson, 2017; Strom, 2017), providing a novel molecular mechanism by which HP1 may promote genomic compartmentalization. The role of HP1 in establishing heterochromatin compartments is especially pertinent to the olfactory system and raises the intriguing possibility that phase separation of heterochromatic OR clusters could be used to isolate and silence inactive OR genes. The finding that double knockout of G9a and GLP methyltransferases results in a loss of stochastic OR choice and singular OR expression (Lyons et al 2014), supports the important role of chromatin state and OR compartment formation in OR regulation.
Despite the fundamental role of compartments in nuclear organization, aside from OR clusters in OSNs and the nucleolus in every cell type, specific inter-chromosomal interactions are not readily captured with HiC. This is likely due to the fact that compartment contacts in individual cells are highly variable (Nagano, 2013). However, there may be additional technical reasons for which specific trans interactions are under-represented in HiC datasets including a study that suggests highly specific trans interactions may occur at longer distances than their cis counterparts (*Maass PG, 2018). A recently developed method (SPRITE) can detect such active an inactive compartment interactions by capturing clusters of multi-way genomic interactions. SPRITE detects inter-chromosomal interactions between chromatin of a similar type, organized around nuclear speckles (Quinodoz, 2018**). Such inter-chromosomal nuclear speckle association has been described for the globin genes in erythropoiesis (Brown JM 2006, Brown JM 2008) and may be an important feature for increasing transcription levels of active genes (Kim J, 2019). Another recently developed ligation-independent method, Genome Architecture Mapping (GAM) also detects multivalent long-range cis interactions occuring between genomic regions that contain super-enhancers and highly transcribed regions that span tens of megabases (Beagrie, 2017**). Similar hubs between super-enhancers in cis and in trans are only detected with HiC upon deletion cohesin subuits and thereby strengthening compartment interactions (Rao, 2017; Schwarzer, 2017), highlighting the advantage of ligation-free techniques for studying long-range cis and trans interations.
The multi-enhancer hub: a site of cooperative transcriptional activation
The convergence of OR clusters coincides in time with the formation of the second OSN-specific inter-chromosomal nuclear compartment--the OR multi-enhancer hub. OR enhancers exist in OR clusters and are defined by the co-binding of Ebf and Lhx2 to a stereotypically spaced Ebf and Homeodomain motif (Monahan, 2017). This ‘composite motif’ likely facilitates the cooperative binding of Ebf and Lhx2 and is highly enriched on OR enhancers relative to OR promoters and other EBF and Lhx2 co-bound sites genome wide. OR enhancers activate OR transcription as a multi-chromosomal enhancer hub that associates specifically with the active OR allele (Markenscoff-Papadimitriou et al 2014, **Monahan 2019). The requirement for coordinated action of OR enhancers in OR gene activation was first demonstrated by ectopic expression of Lbr in OSNs, which disrupted both OR compartments and OR enhancer hubs and resulted in a strong and widespread downregulation of OR gene expression (Clowney 2012). The significant regulatory role of these trans interaction in OR expression was further supported by deletion of Ldb1 (**Monahan and Horta 2019), a LIM domain protein that binds OR enhancers through its interaction with Lhx2, and likely Ebf, and facilitates long range interactions (Krivega, 2017). Loss of Ldb1 resulted in a specific disruption of the OR enhancer hub and an accompanying downregulation of OR expression. This necessity of OR enhancers to act as a complex to activate OR transcription seems contradictory with observation that deletion of individual OR enhancers only affects the expression of local, enhancer proximal OR genes (Fuss, 2007; Khan 2011; Nishizumi 2007). This paradox can be explained by the multifaceted role of OR enhancers. HiC analysis in mice with a triple OR enhancer deletion revealed that deletion of an enhancer diminishes both the ability of the corresponding OR gene cluster to recruit other enhancers in trans as well as reduces the overall contacts made by the OR cluster with the OR compartment (**Monahan and Horta 2019). Consequently, OR enhancers are essential in cis, since they allow the local OR genes to be recruited to OR compartments placing them in a nuclear position compatible with OR choice, and redundant in trans, since they can be replaced by any one of the 63 OR enhancers in a multi-enhancer hub.
The insufficiency of any single OR enhancer to determine OR expression raises questions about the composition of an OR enhancer hub that can overcome the repressive nature of the OR compartment to activate an OR allele. Genetic insertion of 5 OR enhancers in tandem into an OR cluster resulted in only an ~8 fold increase in the frequency of choice of proximal ORs, suggesting more than 6 OR enhancers are necessary to determine OR expression (Monahan 2017). Intriguingly, single cell HiC analysis, named Dip-C, recently showed that 8–12 OR enhancers converge into a single hub, providing the first approximation of the number of OR enhancers needed for OR activation (**Tan 2019). A steep thresholding requirement for number of OR enhancers to form an active hub is consistent with recent evidence that super-enhancers are sites of phase-separated compartments that concentrate transcription factors, co-factors and transcriptional machinery (Sabari BR, 2018; Boija A, 2018). OR enhancer hubs may act as multi-chromosomal super-enhancers that undergo a structural change only if a sufficient number of Lhx2/Ebf/Ldb1 bound enhancers coalesce with each other, generating a local high concentration of these transcription factors. A physical separation of the chosen OR gene and enhancer hub from the remaining heterochromatic OR genes in immiscible phase separated compartments could also explain the paradox that the active OR gene, one of the highest transcripts in OSNs, is in some cases only ~20 kb away from OR genes that are entirely transcriptionally silent despite common promoter elements. Such physical separation has been described for the nucleolus (Feric, 2016), but further studies are needed to determine the biophysical properties of OR compartments and the OR enhancer hub.
The molecular mechanisms that activate OR expression have many parallels in the cellular response to virus infection. The first stages of the response involve the monoallelic and stochastic transcriptional activation of IFN-β in only a fraction of infected cells (Zawatzky, 1985). Transcriptional activation is mediated by inter-chromosomal interaction between the IFN-β enhancer with three genomic loci known to bind NF-kB that serve to deliver limiting NF-kB to the IFN-β enhancer initiating enhanceosome assembly (Apostolou, 2008). The specificity to these inter-chromosomal interactions over the thousands of other NF-kB sites in the genome stems from the presence of a stereotypically positioned, GAGA-NFkB ‘composite’ motif, which marks sites of cooperative binding of NF-kB and ThPOK, which can oligomerize and is required for these long-range contacts (Nikopoulou, 2018). The parallels with OR choice suggest that inter-chromosomal hubs may be a robust mechanism for stochastic expression, activating transcription of a given allele at high levels in a low number of cells.
Inter-chromosomal Interactions at transcriptional hubs
A body of work has shown actively transcribed genes, including those from different chromosomes, coalesce around sites of highly concentrated transcription machinery (Osborne CS 2004, Schoenfelder, 2010). Inter-chromosomal interactions between active alleles have been observed with DNA-FISH and 3C and 4C-based methods (Spilianakis 2005, Apostolou E, 2013, Wei 2013). However, robust inter-chromosomal contacts are not detected using in situ HiC (Nagano 2015, Johanson, 2018). This discrepancy could be explained by the nature of contacts made by transcriptionally active chromatin--whether they form a stable hub or transiently co-localizing around sites of high transcription. Single molecule live imaging of Pol II showed that Pol II clusters are highly dynamic and short lived in a human osteosarcoma cell line (Cisse, 2013), whereas they can form larger more stable clusters in mouse ESC (*Cho WK, 2017) , that decrease in size upon differentiation to epiblast-like cells. This suggests that specialized, stable transcriptional hubs may only exist in certain cell types or under certain conditions. Recent work has shown that highly specific transcriptional hubs form in the yeast heat shock response: Hsf1 target genes from different chromosomes aggregate into foci in an Hsf1 dependent fashion. These interactions are highly specific, as other highly transcribed genes, as well as genes regulated by other heat shock induced transcription factors are not recruited into the foci (Chowdhary, 2017 and *Chowdhary, 2019). Similarly, research in pluripotent cells has shown inter-chromosomal contacts between Klf4 regulated genes (Wei, 2013, Stevens TJ, 2017). Identifying factors that promote stable and specific transcriptional hubs is an active area of research, but there is growing evidence that regulatory proteins with disordered low-complexity domains form aggregates in a concentration dependent fashion (Chong 2018, Boehning 2018).
That heat shock induced inter-chromosomal interactions were significantly reduced in the absence of Pol II (*Chowdhary, 2019) is intriguing in light of the emerging appreciation of the role RNA in nuclear architecture. LncRNAs have been shown to play an architectural role in nuclear organization and mediate inter-chromosomal interactions (Hacisuleyman, 2014; Maass, 2012). LncRNA Firre binds locally as well to several genomic loci in cis and in trans, that form frequent and stable interactions, as shown with CRISPR/Cas9 live imaging (CLING) (*Maass PG, 2018). Furthermore, forced transcription of the Neat1 ncRNA nucleates the formation of a nuclear body, the paraspeckle, through interactions between the nascent transcript and proteins that contain prion-like domains (Mao, 2011) that can phase-separate (Fox AH, 2018; Hennig S, 2015). This raises the question to which extent nascent transcripts regulate nuclear architecture. Nascent transcripts have been shown to enhance the binding of transcription factor YY1 to regulatory elements (Sigova, Young 2015) as well as enhance the acetyltransferase activity of chromatin modifying enzyme CBP (Bose 2017). New research also show that some chromatin loops are dependent on the RNA-binding activity of CTCF (Hansen AS, 2018; Saldana-Meyer R, 2019). Considering that the OR is one of the highest transcribed genes in an OSN, could its transcript be involved in stabilizing the enhancer hub, or bridging contacts between the active allele and OR enhancers? Additionally, since OR transcription occurs from multiple OR loci in progenitor cells, could it be involved in establishing the OR compartment? Only future experiments will tell if nuclear RNAs from OR or other non coding genomic regions contribute to the unique nuclear architecture of OSNs. However, it is intriguing that early deletion of Lhx2, which abolishes transcription of most OR genes, also results in complete disruption of OR compartments and OR enhancer hubs (Monahan and Horta 2018).
Diversity in Nuclear Architecture in OSN subtypes and beyond
The tight coupling between nuclear organization and transcriptional outcome is exemplified by OSN subtypes. A subset of ONSs express trace amine-associated receptors (TAARs) instead of ORs (Liberles S, 2006). The 15 genes encoding TAARs are all located in one gene cluster and like OR genes are expressed in a singular, monoallelic, stochastic fashion. They also share transcriptional regulators with ORs, as shown by the reduced expression of nearly all TAARs upon Lhx2 deletion (Zhang, 2015). However, despite the similarities in the mode of expression between TAARs and ORs, TAAR genes are not covered with H3K9me3 and H4K20me3 (Johnson MA, 2012) or recruited to the OR compartment upon heterochromatin inversion (Yoon KH, 2015). The different preference for both the chromatin state and nuclear position of their preferred genes between TAAR-OSNs and OR-OSNs sheds new light on the finding that OSNs expressing a non-functional TAAR allele will stochastically switch to express another TAAR instead of OR. This restriction in receptor choice suggests that the nuclear architecture in TAAR-OSNs could be incompatible with OR activation (Johnson MA, 2012). Furthermore, even within OR-expressing OSNs, there is a deterministic element to OR choice. Early experiments suggested that along the dorsoventral axis of the olfactory epithelium, each OR gene can be expressed in one out of 4 (and later increased to 5) zones of expression (Ressler 1993, Vassar 1993). The mechanism behind this deterministic restriction on the repertoire of receptors remained unknown for decades for two main reasons: First, it was difficult to make hypotheses about which regulators restrict OR choice without fully understanding the process, and second, a lack of zonal information for most OR genes impeded making comparisons about the genetic and epigenetic similarities and differences between OR genes expressed in the same or different zones, respectively. However, with significant progress in the understanding of OR gene regulation, and upon deciphering the zonal expression properties of most OR genes (Tan, 2018), we can make testable hypotheses on how zonality is established. For example, zonal differences in OR compartmentalization and OR heterochromatinization would be an elegant solution for providing restrictions in a stochastic process.
These data suggests a diversity of nuclear organizations in OSN subtypes. It is possible that other postmitotic or highly specialized cells have unique nuclear organization enabling their cellular functions and that cell types may differ in their frequency of specific trans interactions. This principle is exemplified in a recent study comparing the nuclear organization of two types of sensory neurons, olfactory and photoreceptor neurons from the mouse using an improved version of single cell HiC (Dip-C) (Tan, 2019**). Along similar lines, a study in plasma cells reported the coalescence of immunoglobulin genes from three different chromosomes to facilitate high levels of antibody production (Park SK, 2014). A recent HiC study in cardiomyocytes also detected formation of multi-chromosomal contacts which appear important for the proper splicing of cardiomyocyte-specific genes by Rbm20 (Bertero, 2019). Moreover, in situ HiC in FAC-sorted neocortical cells detects super long-range cis interactions (Bonev, 2017) and long-range cis and trans contacts have been reported between genomic regions that undergo large structural rearrangements linked to autism (Loviglio 2017). Finally, multi-chromosomal contacts were recently reported in Plasmodium, the parasite that causes malaria (Bunnik, 2018, 2019), centered around the var gene family of surface proteins, which like OR genes are expressed in a singular and stochastic fashion, and whose expression is tightly regulated towards generating antigenic variation for host immune evasion. Future work in different cell types and using a range of techniques in the genome organization toolkit will provide further clarity on how nuclear organization enables cellular function.
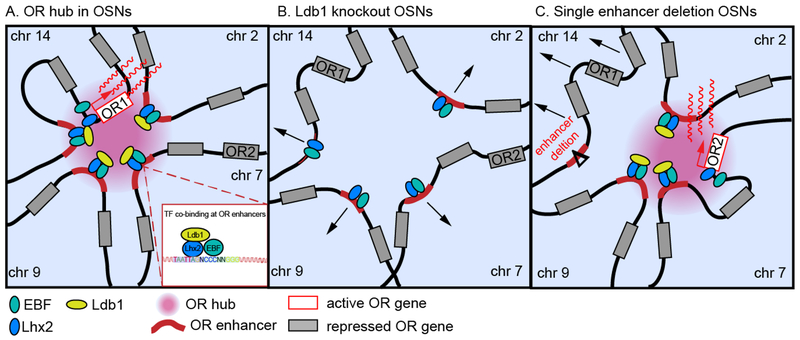
Figure 1:
Olfactory receptor enhancers drive expression of the active OR gene in cis and in trans
Intergenic olfactory receptor (OR) enhancers, residing in OR clusters aggregate in cis and in trans to drive the expression of the active OR allele (A) in olfactory sensory neurons (OSNs). Long-range enhancer interactions are mediated by Ldb1, and Ldb1 knockout OSNs don’t form an enhancer hub, precluding the expression of any OR gene (B). Deletion of a single enhancer abolishes its interaction with the enhancer hub and reduces the expression of local OR genes (C), illustrating the principle that OR enhancers are necessary in cis and redundant in trans.
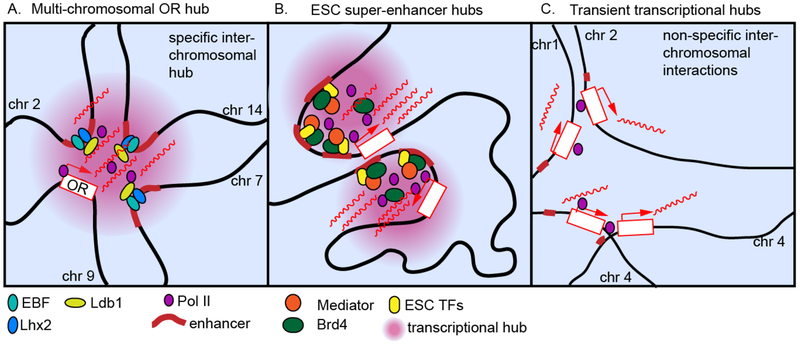
Figure 2:
Actively transcribed genes interact around sites of concentrated transcriptional machinery
Highly specific transcriptional hubs form in (A) olfactory sensory neurons (OSNs) when olfactory receptor (OR) enhancers co-bound by Lhx2, EBF and Ldb1 from multiple chromosomes coalesce in cis and in trans to drive expression of the active OR allele. Large and stable transcriptional hubs form at (B) embryonic stem cell (ESC) super-enhancers bound by ESC transcription factors and co-activators Mediator and Brd4, which can converge with other super-enhancers tens of megabases away. These hubs are distinct from the transient, non-specific interactions (C) between actively transcribed genes around highly dynamic pol II clusters.
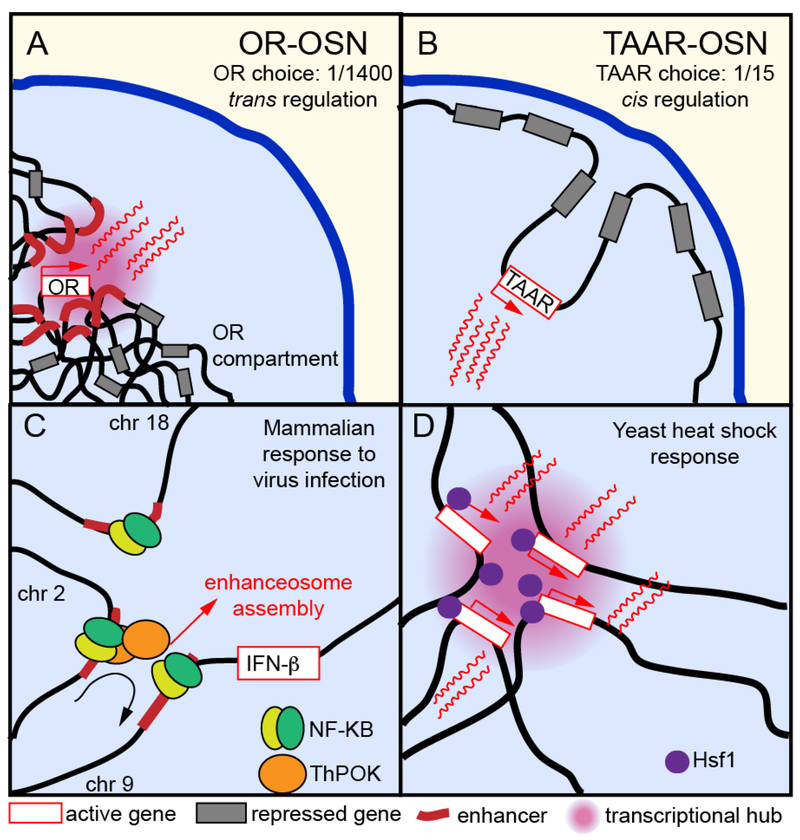
Figure 3:
Diverse nuclear architecture facilitate chemosensory receptor choice in olfactory sensory neurons (OSNs) and are involved in cellular responses to stimuli.
Singular expression of one olfactory receptor (OR) depends on the aggregation of silenced OR genes into a unique OR compartment and the interaction of the active OR allele with a multi-chromosomal, multi-enhancer hub (A), whereas singular expression of a trace amine-associated receptor (TAAR) is regulated locally by the repositioning of the active allele away from the nuclear lamina (B). Inter-chromosomal interactions facilitate the stochastic activation of IFN-β during cellular response to virus infection in mammalian cells (C) and play a crucial role in the yeast heat shock response (D), where HSF1 target genes aggregate for coordinated transcription.
Acknowledgments
We thank members of the Lomvardas lab for critical reading of the manuscript. This manuscript was supported by NIH grants R01DC015451 and U01DA040582 and the HHMI Faculty Scholar Award. The authors have no conflicting interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Citations:
- 1.Cremer T, Cremer C et al. Rabl’s model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments.Hum Genet 60(1),46–56 (1982). [DOI] [PubMed] [Google Scholar]
- 2.Branco MR, and Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-depen-dent associations. PLoS Biol 4(138) (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao SSP. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalton RP., et al. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 155, 321–332 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyons DB, et al. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 154(2), 325–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan L et al. Olfactory sensory neurons transiently express multiple olfactory receptors during development. Mol Syst Biol 11 (12):844 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanchate NK et al. Single cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science. 350(6265), 1251–1255 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magklara A, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell 145(4), 555–70 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clowney EJ et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 151 (4),724–737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armelin-Correa LM, Gutiyama LM, Brandt DY & Malnic B Nuclear compartmentalization of odorant receptor genes. Proc. Natl Acad. Sci. USA 111, 2782–2787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.**.Monahan K and Horta A et al. LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature 565(7740), 448–453 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; Monahan and Horta use in-situ HiC to investigate the nuclear architecture of olfactory lineage cells. They observe and the formation of the OR compartment and a multi-chromosomal, milti-enhancer hub that specifically interacts with the chosen OR allele in an Ldb1-dependent fashion.
- 12.Eitan Yaffe E, and Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nature Genetics 43(11), 1059–1065 (2011) [DOI] [PubMed] [Google Scholar]
- 13.Wijchers PJ. Cause and Consequence of Tethering a SubTAD to Different Nuclear Compartments. Mol Cell 61(3),461–473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larson AG et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strom AR et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyons DB, et al. Heterochromatin-Mediated Gene Silencing Facilitates the Diversification of Olfactory Neurons. Cell Reports Volume 9(3), 884–892 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagano T et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maass PG et al. Inter-chromosomal Contact Properties in Live-Cell Imaging and in Hi-C. Mol Cell. 69(6), 1039–1045(2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.**.Quinodoz SA, et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 174(3),744–757(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; Quinodoz et al develop a new method (SPRITE) for capturing long-range inter-chromosomal interactions. This method can detect multi-way interactions as well as interactions occurring at longer distances, around nuclear bodies.
- 20.Brown JM, et al. Coregulated human globin genes are frequently in spatial proximity when active.Journal of Cell Biology 172(2),177–87 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown JM, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. Journal of Cell Biology 182(6), 1083–97 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, et al. Transcription amplification by nuclear speckle association. Biorxiv doi: 10.1101/604298 (2019). [DOI] [Google Scholar]
- 23.**.Beagrie RA., et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543(7646), 519–524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; Beagrie et al develop a ligation-free method Genome Architecture Mapping (GAM) to probe nuclear architecture. In addition to detecting compartment interactions and TADs, they observe multi-way interactions occurring between super-enhancer containing TADs that span tens of megabases.
- 24.Rao SSP., et al. Cohesin Loss Eliminates All Loop Domains. Cell 171(2), 305–320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarzer W, et al. Two independent modes of chromatin organization revealed by cohesion removal. Nature 551(7678), 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monahan K et al. Cooperative interactions enable singular olfactory receptor expression in mouse olfactory neurons. Elife 6, 1–32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markenscoff-Papadimitriou E, et al. Enhancer interaction networks as a means for singular olfactory receptor expression. Cell 159(3), 543–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krivega I & Dean A LDB1-mediated enhancer looping can be established independent of mediator and cohesin. Nucleic Acids Res 45, 8255–8268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuss SH, Omura M & Mombaerts P Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell 130, 373–384 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Khan M, Vaes E & Mombaerts P Regulation of the probability of mouse odorant receptor gene choice. Cell 147, 907–921 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Nishizumi H, et al. Deletion of the core-H region in mice abolishes the expression of three proximal odorant receptor genes in cis. Proc. Natl Acad. Sci. USA 104, 20067–20072 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.**.Tan L., et al. Three-dimensional genome structures of single sensory neurons in mouse visual and olfactory systems. Nature Structural & Molecular Biology 26, 297–307(2019). [DOI] [PubMed] [Google Scholar]; Tan et al use diploid chromatin conformation capture (Dip-C) to profile the nuclear architecture in single cells from the retina and olfactory epithelium. In OSNs they observe multiple aggregates of OR genes and enhancers from different chromosomes within individual cells.
- 33.Sabari BR et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361(6400), (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boija A, et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175(7), 1842–1855 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apostolou E and Thanos D. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 134(1), 85–96 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Feric M, et al. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 165(7), 1686–1697 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zawatzky R, et al. Identification of individual interferon-producing cells by in situ hybridization. PNAS 82, 1136–1140 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikopoulou C, et al. The Transcription Factor ThPOK Orchestrates Stochastic Interchromosomal Interactions Required for IFNB1 Virus-Inducible Gene Expression. Molecular Cell 71(2), 352–361 (2018) [DOI] [PubMed] [Google Scholar]
- 39.Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nature Genetics 36, 1065–1071 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature Genetics 42(1),53–61 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spilianakis CG., et al. Interchromosomal associations between alternatively expressed loci. Nature 435(7042), 637–45 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Apostolou E, et al. Genome-wide Chromatin Interactions of the Nanog Locus in Pluripotency, Differentiation, and Reprogramming.Cell Stem Cell 12(6), 699–712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei Z, et al. Klf4 Organizes Long-Range Chromosomal Interactions with the Oct4 Locus in Reprogramming and Pluripotency. Cell Stem Cell 13(1), 36–47(2013) [DOI] [PubMed] [Google Scholar]
- 44.Stevens TJ, Iando D, Basu S, et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagano T et al. Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome Biol. 16, 175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johanson TM et al. Genome-wide analysis reveals no evidence of trans chromosomal regulation of mammalian immune development. PLoS Genet 14, e1007431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cisse II., et al. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 341:664–667 (2013) [DOI] [PubMed] [Google Scholar]
- 48.*.Cho WK et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361(6400) 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; Cho et al use live-cell superresolution and light-sheet imaging to study the dynamics of Mediator and RNA pol II. They observe stable RNA pol II and Mediator clusters that co-localize in a transcription dependent fashion. Stable Pol II clusters, observed in ESC, decrease in size and number upon differentiation to epiblast-like cells.
- 49.*.Chowdhary S, et al. Heat Shock Factor 1 Drives Intergenic Association of Its Target Gene Loci upon Heat Shock. Cell Reports 26, 18–28 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; Chowdhary et al use chromatin conformation capture and imaging to observe the coalescence of Hsp1 target genes into transcriptional foci upon induction of heat shock.
- 50.Chowdhary S, et al. Heat shock protein genes undergo dynamic alteration in their three-dimensional structure and genome organization in response to thermal stress. Mol Cell Biol 37(24), (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chong S et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361(6400), (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boehning M RNA polymerase II clustering through carboxy-terminal domain phase separation. Nature Structural & Molecular Biology 25, 833–840 (2018) [DOI] [PubMed] [Google Scholar]
- 53.Hacisuleyman E et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21(2),198–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maass PG, et al. A misplaced IncRNA causes brachydactyly in humans. J Clin Investigation 122(11), 3990–4002 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mao YS. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13(1), 95–101(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fox AH, et al. Paraspeckles: Where Long Noncoding RNA Meets Phase Separation. Trends in Biochemical Sciences 43(2), 124–135 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Hennig S, et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sigova AA, et al. Transcription factor trapping by RNA in gene regulatory elements. Science 350(6263), 978–81 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bose D, et al. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell 168(1-2), 135–149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen AS, et al. An RNA-binding region regulates CTCF clustering and chromatin looping. Biorxiv doi: 10.1101/495432 (2018). [DOI] [Google Scholar]
- 61.Saldana-Meyer R, et al. RNA interactions with CTCF are essential for its proper function. Biorxiv doi: 10.1101/530014 (2019). [DOI] [Google Scholar]
- 62.Liberies SD & Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature 442, 645–650 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Zhang G, et al. Lhx2 Determines Odorant Receptor Expression Frequency in Mature Olfactory Sensory Neurons. eNeuro 3(5),(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson MA et al. Neurons expressing trace amine-associated receptors project to discrete glomeruli and constitute an olfactory subsystem. PNAS 109 (33), 13410–13415 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoon KH et al. Olfactory receptor genes expressed in distinct lineages are sequestered in different nuclear compartments. Proc Natl Acad Sci 112(18), E2403–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ressler KJ, Sullivan SL & Buck LB A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73, 597–609 (1993). [DOI] [PubMed] [Google Scholar]
- 67.Vassar R, et al. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74(2), 309–318 (1993). [DOI] [PubMed] [Google Scholar]
- 68.Tan L and Xie X. A Near-Complete Spatial Map of Olfactory Receptors in the Mouse Main Olfactory Epithelium. Chem Senses. 43(6),427–432 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park SK, et al. Pronounced cohabitation of active immunoglobulin genes from three different chromosomes in transcription factories during maximal antibody synthesis. Genes Dev 28(11), 1159–64 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bertero A, et al. Dynamics of genome reorganization during human cardiogenesis reveal an RBM20-dependent splicing factory. Nature Communications 10(1), 1538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loviglio MN, et al. Chromosomal contacts connect loci associated with autism, BMI and head circumference phenotypes. Molecular Psychiatry. 22, 836–849 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bonev B, et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171(3), 557–572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bunnik EM., et al. Changes in genome organization of parasite-specific gene families during the Plasmodium transmission stages. Nature Communications 9(1), 1910 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bunnik EM., et al. Comparative 3D genome organization in apicomplexan parasites. PNAS 116 (8), 3183–3192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]