Abstract
MicroRNAs (miRNAs) have been reported to contribute to the pathophysiology of the Parkinson’s disease (PD), an age related-neurodegenerative disorder. The aim of present study was to compare the expression profiles of a new set of candidate miRNAs related to aging and cellular senescence in peripheral blood mononuclear cells (PBMCs) obtained from the PD patients with healthy controls and then in the early and advanced stages of the PD patients with their controls to clarify whether their expression was correlated with the disease severity. We have also proposed a consensus-based strategy to interpret the miRNAs expression data to gain a better insight into the molecular regulatory alterations during the incidence of PD. We evaluated the miRNA expression levels in the PBMCs obtained from 36 patients with PD and 16 healthy controls by the reverse transcription-quantitative real-time PCR and their performance to discriminate the PD patients from the healthy subjects assessed using the receiver operating characteristic curve analysis. Also, we applied our consensus and integration approach to construct a deregulated miRNA-based network in PD with the respective targets and transcription factors, and the enriched gene ontology and pathways using the enrichment analysis approach were obtained. There was a significant overexpression of miR-885 and miR-17 and the downregulation of miR-361 in the PD patients compared to the controls. The blood expression of miR-885 and miR-17 tended to increase along with the disease severity. On the other hand, the lower levels of miR-361 in the early stages of the PD patients, as compared to controls, and its higher levels in the advanced stages of PD patients, as compared to the early stages of the PD patients, were observed. Combination of all three miRNAs showed an appropriate value of AUC (0.985) to discriminate the PD patients from the healthy subjects. Also, the deregulated miRNAs were linked to the known PD pathways and the candidate related target genes were presented. We revealed 3 candidate biomarkers related to aging and cellular senescence for the first time in the patients with PD. Our in-silico analysis identified candidate target genes and TFs, including those related to neurodegeneration and PD. Overall, our findings provided novel insights into the probable age-regulatory mechanisms underlying PD and a rationale to further clarify the role of the identified miRNAs in the PD pathogenesis.
Subject terms: Parkinson's disease, Predictive markers, Predictive markers, Predictive markers, Parkinson's disease
Introduction
Parkinson’s disease (PD) is the second most common age-related neurodegenerative disorder after Alzheimer’s disease, affecting the movement abilities. PD incidence is rare before the age of 50, but its occurrence rises from 5 to 10 folds in the age range of 60 to 901. Despite extensive studies, PD etiology and pathology have still remained largely unknown. Hence, a causal therapy for PD is currently not available, and its clinical diagnosis is mainly based on the presence of characteristic motor symptoms such as bradykinesia, muscle rigidity, resting tremor, and postural and gait impairment when neurodegeneration in substantia nigra (SN) pars compacta reaches the thresholds value of around 60%2,3.
Due to the heterogeneous nature of PD, a combination of aging, genetic and non-genetic risk factors are involved4; the current approach has been aimed to understand its underlying mechanisms at different levels of regulation. In this regard, microRNAs (miRNAs), a class of non-coding RNAs, have been reported to play an important role in PD5. Mature miRNAs are single-stranded RNAs of approximately 21–25 nucleotides in length, which are capable of post-transcriptionally regulating their target messenger RNAs (mRNAs) by mainly binding to the 3′ untranslated regions (3′UTR), leading to mRNA degradation or translational repression. A single 3′UTR may contain binding sites for various miRNAs or multiple sites for a single miRNA, representing the complex post-transcriptional regulation of the gene expression by miRNAs partaking in diverse cellular functions including cell proliferation, cell death, growth pathways, differentiation and immune system, which can be involved in the molecular mechanisms of various diseases. In human, there is at least one conserved miRNA-binding site in more than 60% of the protein-coding genes. Moreover, numerous non-conserved miRNA-binding sites are also present; therefore, most protein-coding genes may be under miRNA regulation6,7. MiRNAs also exist in a highly stable form in the peripheral blood and their expression profiles can be easily measurable, making them ideal biomarkers. In this regard, recently, a number of studies have investigated the expression profiles of several PD candidate miRNAs in the peripheral blood mononuclear cells (PBMCs) obtained from the PD patients, as compared to the healthy controls8–11.
Aging is recognized to be the greatest risk factor for developing idiopathic PD1. However, the specific age-related factors that trigger PD in the elderly are not still known. Therefore in this study, in order to better understand the possible contributing factors involved in these mechanisms, the expression of a set of candidate miRNAs including miR-17, miR-361 and miR-885 has been profiled in the PBMCs obtained from the PD patients and healthy controls; it has been previously demonstrated that as miRNAs are involved in aging and cellular senescence12–16. These miRNAs have also shown differential expression in the miRNA expression microarray dataset [GSE16658: consisted of 19 PD patients and 13 controls in the GEO database (https://www.ncbi.nlm.nih.gov/geo/)] in the PBMCs of the PD patients and controls11. Thus, in this study, we have not only confirmed the differential expression of these miRNAs in the PD patients compared to the healthy controls, but also presented them as a novel candidate set of aging and cellular senescence and evaluated associations of their PBMCs levels in relation to the disease severity via their expression profiling in both the early and advanced stages of the PD patients with their controls. Furthermore, miRNAs, as the regulatory molecules, are also under regulation, the most important of which is the miRNA-transcription factor (TF) regulatory network. In order to reach a more comprehensive view of the functional roles of these miRNAs in the PD pathology, we performed an enrichment analysis on the putative target genes and the TFs of these miRNAs.
Methods
Patients
This study was approved by the local ethics committee of Royan Institute (IR.ACECR.ROYAN.REC.1397.10). Meanwhile, all participants signed the written informed consent. All methods were performed in accordance with the relevant guidelines. Thirty-six patients with PD were recruited from Al-Zahra Hospital, Isfahan, Iran. The clinical diagnosis of PD was confirmed by a certified movement disorder neurologist, according to the UK Brain Bank criteria17. Subjects with other neurological diseases, atypical parkinsonism, drug-induced secondary parkinsonism, cognitive impairment, diabetes, renal dysfunction, infection, tumor, cardiovascular and cerebrovascular diseases were excluded. Exclusion criteria for the patients with cognitive impairment were those with Montreal Cognitive Assessment (MoCA) score less than 2618. The severity and stage of the PD symptoms were evaluated using the modified Hoehn and Yahr stage (HY)19. Thirteen patients with the unilateral motor impairment only (HY-1 stage) and fifteen patients with the bilateral or midline involvement in the absence of the impairment of balance (HY-2 stage) were classified as the early stage PD. In contrast, five patients with the postural reflexes impairment (HY-3 stage) and three patients with severe disability, who still were able to walk or stand unassisted (HY-4 stage), were classified as the advanced stage PD. In addition, we enrolled sixteen volunteer control subjects with no history of neurological or psychiatric diseases from spouses, unrelated caregivers and outpatient clinics.
PBMCs isolation
Five mL of the whole blood was collected in an EDTA-containing tube from all participants and processed within 6 hours of blood collection. PBMCs were isolated using centrifugation through a density gradient medium by Lymphodex (Inno-Train Diagnostik GmbH, Kronberg, Germany), according to the instructions of manufacturer. Cells were washed twice with phosphate buffered saline (PBS) (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), counted and resuspended at the desired concentration.
RNA extraction and quality control
Total RNAs, including small RNAs, were extracted from PBMCs using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol. We determined the RNA quality and quantity using a NanoDrop 2000/2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and the 260/280 nm absorbance ratio was evaluated. All samples showed an absorbance ratio between 1.8 and 2. In addition, to eliminate any potential genomic DNA (gDNA) contamination, the total RNA samples were treated with RNase-free DNase I (Fermentas, Thermo Fischer Scientific, Waltham, MA, USA), according to the manufacturer’s instructions.
Reverse transcription and the quantitative real-time PCR
Reverse transcription (RT) reaction was performed on 500 ng of the total RNA using the universal cDNA synthesis kit II (Exiqon, Vedbaek, Denmark) in a 10 μL reaction. The thermal cycling parameters were 60 minutes at 42 °C, 5 minutes at 95 °C for the heat-inactivation of the reverse transcriptase, and immediate cooling at 4 °C.
Subsequently, the real-time quantitative PCR (RT-qPCR) of hsa-miR-17-5p, hsa-miR-361-5p, hsa-miR-885-5p, and the candidate normalizer hsa-miR-191-5p was performed in triplicate with the specific primers on the Step One Plus Real-Time PCR thermal cycler (Applied Biosystems, Foster City, CA, USA) using SYBR Green Master Mix: SYBR Premix Ex TaqII (TaKaRa, Tokyo, Japan) and microRNA LNA™ primer sets (Exiqon, Vedbaek, Denmark); in accordance with the manufacturer’s instructions. The thermal cycling parameters were 10 minutes at 95 °C; this was followed by 40 cycles of 10 seconds at 95 °C and 1 minute at 60 °C.
Relative expression levels were calculated using the comparative CT method with hsa-miR-191-5p as the normalizing control.
Statistical analysis
Data are presented as the means ± standard error of the mean (SEM). The Shapiro-Wilk test was used to evaluate whether each variable followed the normal distribution; according to its results, normality was not proved. So, for the group-wise comparisons, the Mann-Whitney U test for 2 groups and the Kruskal-Wallis H test for n groups were used.
Receiver operating characteristic (ROC) curve analysis was used for each miRNA and using binomial logistic regression to combine miRNAs. The statistical analysis was performed using the IBM SPSS software, version 23.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad Software, USA). Significant differences were defined as p < 0.05.
microRNA target analysis
Five miRNA target databases were used, including TargetScan (release 7.2), microT-CDS (release 5.0) and miRDB (release 5.0); these consisted of computationally predicted miRNA targets20–22. On the other hand, miRTarBase (release 7.0) and TarBase (release 8.0) contained the experimentally validated miRNA targets23,24. Within them, we only retrieved the common predicted targets for miR-885-5p, miR-17-5p and miR-361-5p from TargetScan with a total context ++ score <−0.2, microT-CDS with an miTG score ≥0.8, and miRDB with a score >80. These strict thresholds were chosen to obtain a high-confidence list of the candidate miRNA-target interactions and reduce the number of false positives. We also extracted their validated targets with strong evidence (reporter assay, western blot, qPCR) from miRTarBase and TarBase.
Transcription factor-microRNA analysis
Transcription factors (TFs) regulating the transcription of miR-885, miR-17 and miR-361 were obtained from the TransmiR (release 2.0) database25. Within them, we only retrieved validated and more stringent evidence-level TF-miRNA interactions.
Regulatory network construction
We constructed a miRNA-based network which included miR-885-5p, miR-17-5p and miR-361-5p, their targets and TFs. The network was generated and visualized using the Cytoscape (version 3.4.0) software26. In addition, TFs and miRNAs involved in the feedback loops (FBLs), as well as the confidence level of their regulation based on TransmiR, were depicted in a subnetwork. Due to miRNA-target interactions, we also assumed that all miRNAs repressed their targets.
Functional enrichment and pathway analysis
In order to explore gene ontology (GO) biological processes, molecular function and cellular components predominantly shown in the miRNA-based network, we performed a functional annotation analysis using Enrichr (a comprehensive gene set enrichment analysis tool)27; the significantly enriched gene ontology terms (adjusted p-value < 0.05) were summarized using the REVIGO web server by removing the redundant GO terms and visualizing the remaining terms based on semantic similarity28. For the Go biological process, a 0.5 similarity was allowed (small list), while a 0.7 one was considered for the molecular function and cellular components (medium list). Moreover, significantly enriched PANTHER pathways29 (adjusted p-value < 0.001) were extracted from Enrichr and plotted using Microsoft Excel.
Ethics approval and consent to participate
Approval from the local ethics committee of Royan Institute (IR.ACECR.ROYAN.REC.1397.10) was obtained; all participants signed the written informed consent prior to the related studies.
Results
Characteristics of the subjects
The clinical and demographic characteristics of the PD patients and healthy subjects are summarized in Table 1. Statistical analyses showed no significant differences in terms of gender and age between the groups.
Table 1.
Demographic and clinical data of the PD patients and the control group.
| Controls | PD | Early stage PD | Advanced stage PD | p-Value | |||
|---|---|---|---|---|---|---|---|
| N | 16 | 36 | Hoehn & Yahr stage I | Hoehn & Yahr stage II | Hoehn & Yahr stage III | Hoehn & Yahr stage IV | — |
| 13 | 15 | 5 | 3 | ||||
|
Age (Year) |
62.5 ± 12.4 | 61.3 ± 11.4 | 60.9 ± 10.6 | 62.8 ± 14.6 |
0.741a 0.879b |
||
| F/M | 5/11 | 11/25 | 8/20 | 3/5 |
0.960c 0.889d |
||
| Disease Duration, (month) | — | 69.6 ± 67.2 | 58.8 ± 57.8 | 111 ± 88.5 | 0.232e | ||
Data are presented as mean ± SD. ap-value was calculated using a two-tailed Student’s t test. bp-value.
was calculated using one-way ANOVA. c,dp-values were calculated using chi-square test. ep-value.
was calculated using the Mann-Whitney test. As indicated, there was no significant difference between the controls and the PD patients based on the age and sexuality. PD stands for the Parkinson’s disease. F and M represent female and male, respectively.
microRNAs expression in the PD patients and controls
Quantitative analysis of the miRNA expression using RT-qPCR revealed that miR-885-5p and miR-17-5p expressions were significantly overexpressed, while the miR-361-5p expression was significantly reduced in the PBMCs of the PD patients, as compared to the controls (Fig. 1A). We also performed an exploratory expression analysis on the early and advanced stages in the PD patients (based on H & Y scale), as compared to the healthy controls. miR-885-5p demonstrated significant overexpression in both early and advanced stage patients, as compared to the control (Fig. 1B). On the other hand, miR-17-5p was significantly overexpressed in the advanced stage patients vs. healthy subjects and early-stage patients (Fig. 1D). Meanwhile, miR-361-5p revealed a significant reduction in the early-stage patients, as compared to the control group and significant overexpression in the advanced stage patients, in contrast to early-stage patients (Fig. 1C). In this regard, the mean difference (95% CI) for each miRNA within each group is represented (Table 2).
Figure 1.
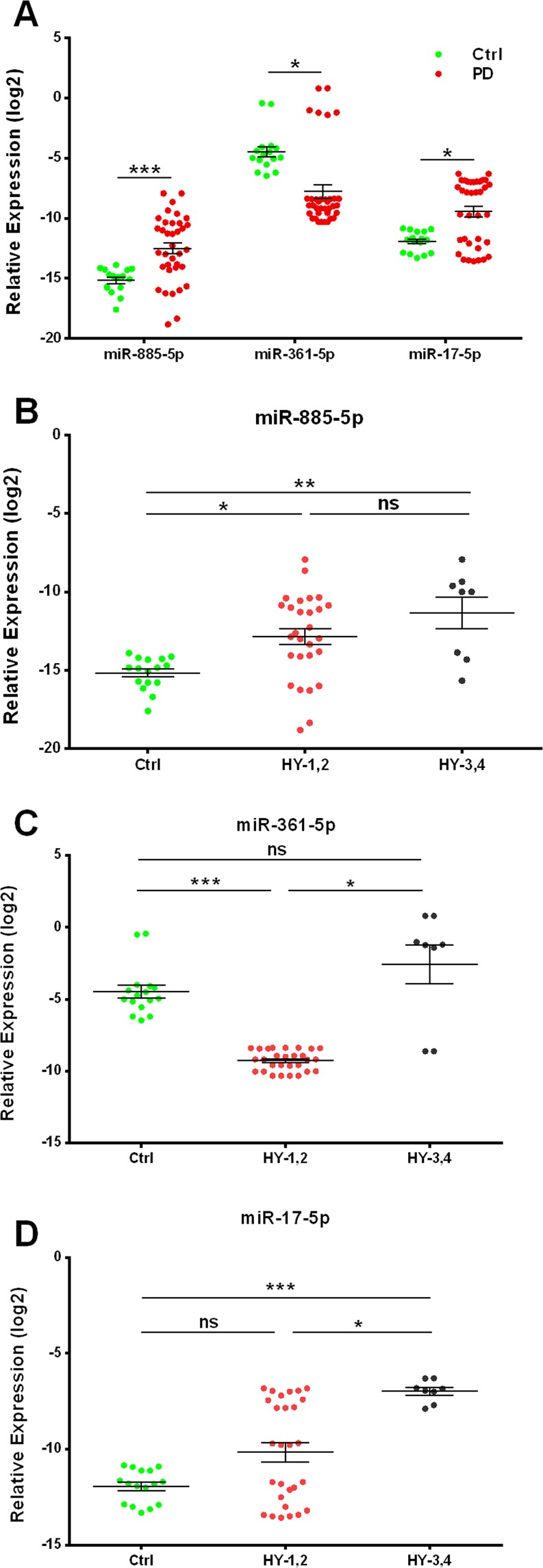
Assessment of the relative expression levels of miR-885-5p, miR-361-5p and miR-17-5p in the PBMCs of: (A) control subjects and PD patients. (B–D) Control subjects and early (HY-1,2) and advanced (HY-3,4) stage PD patients based on Hoehn & Yahr stages. Data are presented as the mean ± SEM. Differences were analyzed by the Mann-Whitney U test (A) and the Kruskal-Wallis H test (B–D) (***p ≤ 0.001, **p ≤ 0.01, *p ≤ 0.05, and ns p > 0.05). ns stands for the non-significance.
Table 2.
Results of the miRNA expression analysis.
| miRNA | Mean difference (95% CI) | p-value | |
|---|---|---|---|
| PD vs. controls | miR-885-5p | 2.58 (1.37, 3.79) | <0.001 |
| miR-361-5p | −3.31 (−4.93, −1.7) | 0.036 | |
| miR-17-5p | 2.48 (1.28, 3.68) | 0.034 | |
| Early stage PD patients vs. controls | miR-885-5p | 2.25 (0.15, 4.36) | 0.034 |
| miR-361-5p | −4.95 (−7.71, −2.19) | <0.001 | |
| miR-17-5p | 1.72 (−0.35, 3.79) | 0.116 | |
| Advanced stage PD patients vs. controls | miR-885-5p | 3.67 (0.79, 6.55) | 0.009 |
| miR-361-5p | 1.75 (−1.60, 5.10) | 0.332 | |
| miR-17-5p | 4.93 (2.25, 7.62) | <0.001 | |
| Advanced stage PD patients vs. Early stage PD patients | miR-885-5p | 1.41 (−1.27, 4.10) | 0.416 |
| miR-361-5p | 6.70 (3.06, 10.34) | 0.023 | |
| miR-17-5p | 3.22 (0.45, 5.98) | 0.021 |
Abbreviations: CI = confidence interval; miRNA = microRNA; PD = Parkinson’s disease. The results are referred to the expression data elaborated using a reference gene miR-191-5p.
microRNA as the predictive biomarkers
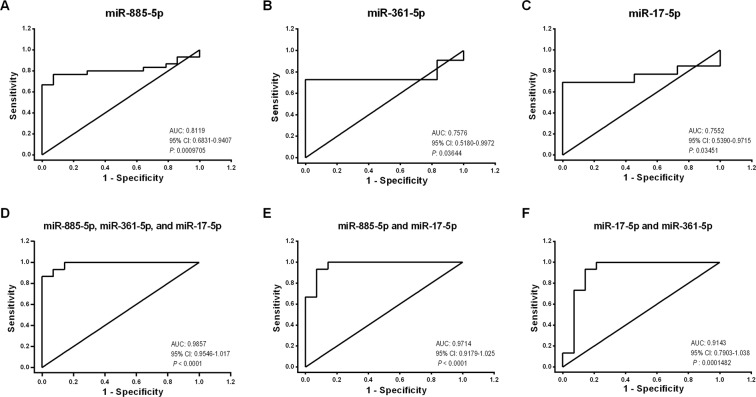
Receiver operating characteristic (ROC) analysis, which is defined as a sensitivity plotted against 1-specificity, was performed to assess the performance of significantly differentially expressed miR-885-5p, miR-361-5p and miR-17-5p to discriminate the PD patients from the healthy subjects (Fig. 2A–C). In this regard, miR-885-5p, miR-361-5p and miR-17-5p showed a fair separation between the PD and healthy controls samples with an area AUC of 0.811, 0.757 and 0.755, respectively.
Figure 2.
Receiver operating characteristic (ROC) curve analysis for: (A–C) miR-885-5p, miR-361-5p, and miR-17-5p in the PD patients versus the healthy controls. (D,E) combined microRNAs using binomial logistic regression in the PD patients versus the healthy controls.
We then applied the binomial logistic regression to identify the best discriminating combinations of miRNAs (Fig. 2D–F) in which the combination of all three miRNAs showed an appropriate value of AUC (0.985), as compared to the best individual miRNA (miR-885-5p, AUC: 0.811) and the best pair of miRNAs (miR-885-5p and miR-17-5p, AUC: 0.971) (Fig. 2D–F). Notably, the value of AUC for the combination of miR-885-5p and miR-361-5p was not satisfying (AUC: 0.638) and significant (p = 0.2057) (Supplemental Fig. 1).
Interaction analysis of miRNAs-target mRNAs
We then determined, predicted and validated the targets of three miRNAs. In this context, we found 126 predicted miRNA-target pairs via common-target screening with strict thresholds in TargetScan20, microT-CDS21 and miRDB22. Additionally, we extracted 133 validated miRNA-target pairs with strong validation methods from miRTarBase23 and TarBase24. There were some overlaps between the predicted and validated miRNA-target pairs. Hence, totally, we identified 247 miRNA-target interactions.
Transcription Factor-miRNA regulation
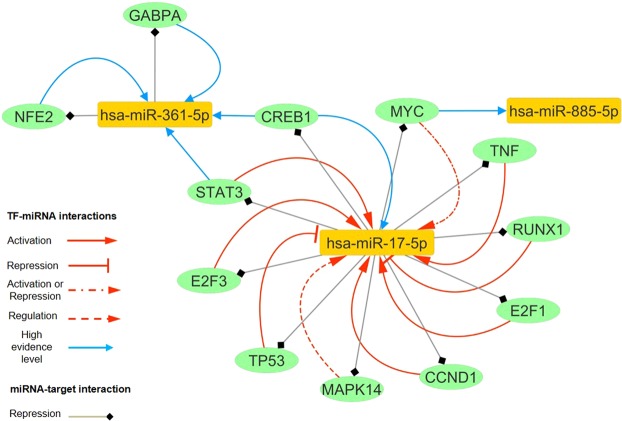
We explored transcription factors (TFs) regulating the 3 miRNAs from TransmiR25. By using TransmiR, an experimentally supported TF-miRNA regulatory relationship database, 94 TF-miRNA interactions were identified. Among them, 12 TFs were part of the feedback loops (FBLs) with miRNAs.
Construction of the miRNA-based regulatory network
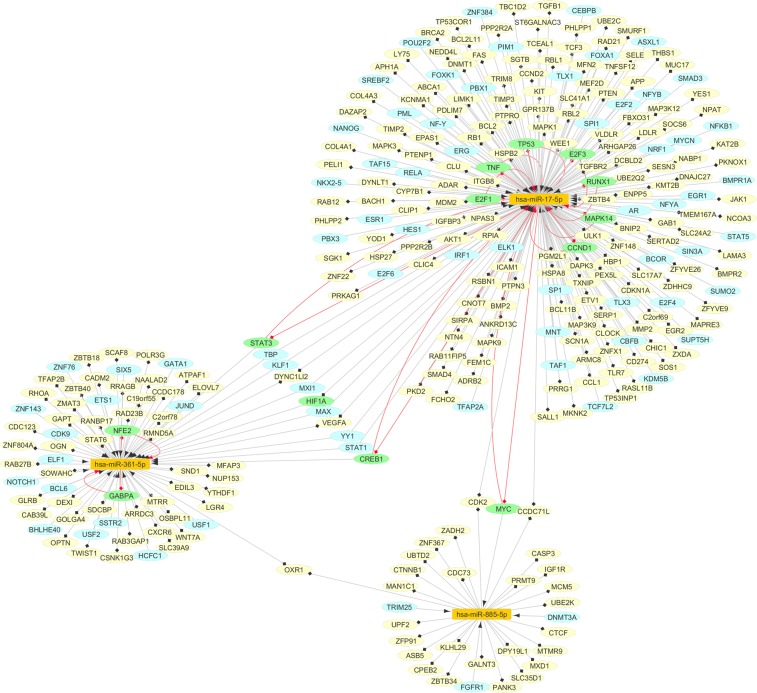
Further, we constructed a regulatory network based on 3 miRNAs, their TFs and target genes using the data obtained from the previous steps (Fig. 3). Our miRNA-based regulatory network consisted of 317 nodes (3 miRNAs, 84 TFs and 242 miRNA targets, while 12 TFs were also miRNA targets), and 341 directed edges (247 miRNA-target pairs and 94 TF-miRNA pairs). Overall, this network depicted 3 potential layers of the complex regulatory interactions based on 3 miRNAs, which could provide insight into the vast dysregulated network in PD. Furthermore, TFs were part of the FBLs with miRNAs, as illustrated in the subnetwork (Fig. 4).
Figure 3.
The miRNA-based network dysregulated in the PD. The orange nodes represent miRNAs, the yellow nodes represent the target genes, and the blue ones show the transcription factors (TFs); on the other hand, the green ones represent molecules that are TFs and also, the target genes. The red edges show the TF-miRNA feedback loops (FBLs).
Figure 4.
The nodes and edges involved in the feedback loops of the miRNA-based network. Orange nodes represent miRNAs, and the green ones show the molecules that are TFs and also, the target genes. Confidence level of TF-miRNA regulations is based on the TransmiR database. Regarding miRNA-target interactions, we also assumed that all miRNAs repressed their targets.
Functional enrichment and pathway analysis
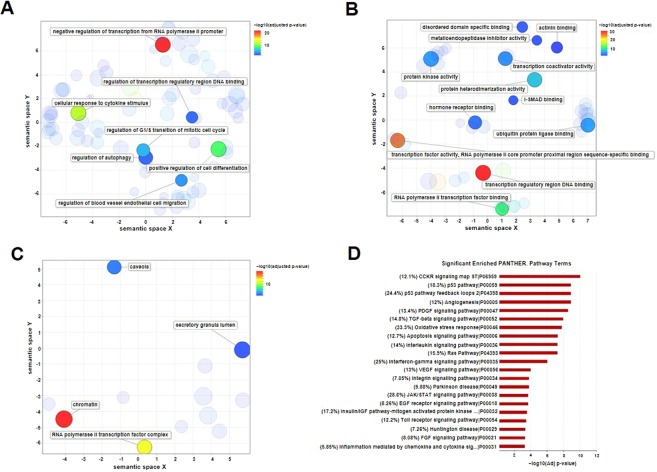
We performed a functional enrichment analysis for the Gene Ontology (GO) terms regarding the TFs and targets of the miRNA-based network using Enrichr27 to obtain a comprehensive view of the processes dysregulated during PD. In this context, significant enriched GO terms (adjusted p-value < 0.05) were summarized and visualized using REVIGO28 (Fig. 5A–C). Visualization of these results in the “biological processes” ontology pointed to the negative regulation of transcription from RNA polymerase II promoter, the cellular response to cytokine stimulus, the regulation of the transcription regulatory region DNA binding, the regulation of the blood vessel endothelial cell migration, the regulation of the G1/S transition of mitotic cell cycle, the regulation of autophagy, and the positive regulation of cell differentiation (Fig. 5A). Regarding “molecular functions” involving the transcription factor activity, transcription coactivator activity, protein kinase activity, metalloendopeptidase inhibitor activity, transcription regulatory region DNA binding, RNA polymerase II transcription factor binding, I-SMAD binding, disordered domain specific binding, actinin binding, protein heterodimerization activity, hormone receptor binding and ubiquitin protein ligase binding can be mentioned (Fig. 5B). These results indicated that three miRNAs were involved in the fundamental biological processes and molecular functions of PD pathophysiology. Furthermore, based on the analysis of terms in the “cellular component” ontology enriched for chromatin, caveola (including membrane raft) (Fig. 5C) were ones of the cellular components affected by PD.
Figure 5.
Gene Ontology (GO) and pathway enrichment analysis. (A–C) GO enrichment analysis was summarized and visualized as a scatter plot using REVIGO28. Scatter plot was generated in REVIGO by clustering the significant (adjusted p-value < 0.05) GO terms remaining after redundancy reduction for the biological processes (A), molecular functions (B) and cellular components (C) in a two-dimensional space (2D space), according to semantic similarities. The bubble color indicates -log10 (adjusted p-value) according to the legend. Bubble size refers to the frequency of the GO term in the underlying Homo sapiens database. Cluster representatives with the lowest dispensability value are labelled. (D) The bar plot shows −log10 (adjusted p-value) of the significantly enriched PANTHER pathways (adjusted p-value < 0.001) for all nodes of the microRNA-based network. Fold enrichment for each pathway is shown in parentheses as the percentage.
Finally, significantly enriched (adjusted p-value < 0.001) PANTHER29 pathways for the miRNA-based network nodes indicated the nodes in the network were part of the potential pathways associated with PD, including p53 signaling pathways, TGF-β signaling pathway, oxidative stress response, and immunological pathways such as the response to interleukin and interferon-gamma signaling pathway (Fig. 5D). It should be noted that dopaminergic pathway was not identified through this analysis.
Discussion
Aging is the greatest risk factor for PD30; however, the association between the molecular/cellular processes involved in physiologic aging and PD pathogenesis is still unclear. Therefore, focusing on aging regulators may provide more insight into the underlying PD pathophysiology that can be used for the future therapeutics or diagnostic studies.
In this study, we investigated differential expression (DE) of three miRNAs involved in aging and cellular senescence in the PBMCs obtained from the PD patients as the accessible tissue undergoing the inflammatory processes and cell death during the PD pathogenesis31. Then, logistic regression was used to develop a combined miRNA panel for a more specific and sensitive discrimination. Finally, a miRNAs-based network was created, and the potentially TF-miRNA feedback loops (FBLs) were determined. Based on this network, we were able to identify the gene ontology and pathways involved in PD.
In 2015, Prajapati et al. demonstrated the significant overexpression of miR-17 and eight other miRNAs in the TNF-α-treated SH-SY5Y cells (TNF: Tumor necrosis factor)32. In our miRNA-based network, miR-17 formed an FBL with TNF-α, which could transcriptionally activate it. It was visible on the network; miR-17 also downregulated E2F1 as a regulator of G1/S transition33 and WEE1 that is involved in G2/M transition34. Furthermore, a set of pro-proliferative and anti-proliferative target genes including CCND1, CDKN1A (p21), PTEN, BCL2L11 (BIM), RB1, RBL1 (p107) and RBL2 (p130) were shown to be suppressed by miR-17.
In 2010, Hackl et al. found the significant decrease of miR-17 in four different replicative cell aging and three organismal aging models, suggesting the use of miR-17 as a cellular aging biomarker. They also suggested the decreased miR-17 levels were associated with down regulation of E2F family members’ levels and the increased p53 activity in senescence12,35,36. As, E2F family members transcriptionally activate miR-17, while p53 represses it37,38. Notably, the same reduced expression pattern of miR-17 has been shown in PBMCs derived from the aging persons13. We know that lifelong exposure to exogenous and endogenous stressors can alter normal signaling mechanisms under pathological condition. For instance, deregulation of the E2F/Rb pathway, such as aberrant pRb expression and active E2F, has been detected in PD39,40. E2F/Rb pathway is critical for the cell-cycle regulation, in which pRb phosphorylation releases E2F family members to activate the expression of the genes involved in the regulation of DNA synthesis, cell cycle progression and apoptosis41. Interestingly, based on our miRNA-based network, miR-17 targets RBL1, RBL2 and RB1, which may result in the E2Fs activation. Also, miR-17 forms FBL with the activator E2Fs (E2F1 and E2F3), in which the activating effects of E2F1 and E2F3 probably overcome the inhibitory effect of miR-17 during the PD progression. We also found other transcription factors of miR-17, such as STAT3, CCND1, RELA, AR, ESR1, NANOG, SMAD3 and TFAP2A as its activators, and SPI1, MAX, MXI1, CEBPB, RUNX1 as its repressors. However, given the significant increased miR-17 levels in the advanced stage patients vs. the control and early stage patients in our study, it seems the miR-17 expression levels have been possibly regulated by the activator TFs during PD progression and further studies would be needed to confirm this.
Beside miR-17, the decreased miR-361 plasma levels were demonstrated during aging14. Chmielarz et al. (2017) also found the significant reduction of the miR-361 expression in the micro-dissected DA neurons while comparing old and young mice15. In our study, the expression levels of miR-361 were significantly reduced in the early stage patients in contrast to the control, and it was overexpressed in the advanced vs. early stage patients. Based on our miRNA-based network, miR-361 targets STAT6, a multifunctional cytokine that plays important roles through the modification of cell-specific differentiation, cell growth, gene regulation and induction of resistance to apoptosis42,43. Another target of miR-361 is GABPA (Nuclear Respiratory Factor 2), a transcription factor regulating the genes expression involved in the mitochondrial function44,45, immune response46, cell cycle progression47–49, cell survival50, hematopoietic stem and progenitor cells, and myeloid differentiation51,52. According to the network, GABPA also forms an FBL with miR-361, whose effects on each other during PD progression require further study. miR-361 also targets OPTN, a gene involved in protein trafficking53, the maintenance of the Golgi apparatus54, mediation of the autophagic flux, cell division, and NF-kB pathway. The increased midbrain OPTN expression with rotenone exposure in the pre-clinical and end-stage PD models has been reported55. Furthermore, Osawa et al. (2011) demonstrated the presence of optineurin-positive structures in various neurodegenerative diseases including PD56. However, the exact role of this OPTN alteration and how its levels are affected by miR-361 in the PD pathogenesis need to be clarified. Another target of miR-361, ZMAT3 (WIG1), has a stress-related function, which is a direct p53 target that is also a regulator of p53 mRNA57. Higashi et al. (2002) demonstrated the induction of the p53 expression by 6-OHDA treatment and the subsequent apoptosis-related morphological changes could be prevented with antisense Wig-1 cDNA in the catecholamine-containing cells (PC12 cells)58. Hence, the hypothetical downregulation of Wig-1 due to the overexpression of miR-361 could be protective.
Moreover, in our miRNA-based network, amongst the TFs of miR-361, BCL6 and HIF1A transcriptionally repressed miR-361, while GATA1 activated it. Interestingly, GATA1 is a potential key regulator of the PD-linked gene α-synuclein in the blood59.
Furthermore, in our study, miR-885, which is involved in the interference with cell cycle progression and cell survival and promotes cellular senescence and apoptosis in neuroblastoma cell lines16, demonstrated a significant overexpression in both early and advanced stage PD patients, as compared to their control counterparts. Based on our miRNA-based network, miR-885 targets IGF1R, which is involved in the cellular internalization of IGF-1 after binding to its receptor and activation of the downstream signaling pathways, such as PI3K/Akt/ GSK-3β signaling cascade60. miR-885 also targets CTNNB1, a key regulatory protein of canonical Wnt signaling, which regulates neurogenesis and neuronal survival. A significant reduction of the CTNNB1 protein expression in midbrain dopaminergic (DA) neurons of PD has been reported61. Its expression might be modified by the miR-885 downregulation. Another target of miR-885 is MAN1C1, which is involved in N-glycosylation, determining the proteins fate through mediating a role in the protein folding as well as in the misfolded protein degradation62. A significant down-regulation in MAN1C1 expression has been shown in the PD blood samples63. MAN1C1 deregulation affects N-glycosylation, which may lead to the ER stress. Hence, a hypothetical downregulation of miR-885 is likely to modify the MAN1C1 expression.
Moreover, OXR1 is a vital protein that controls cellular sensitivity to oxidative stress; as well, it is a potential neuroprotective factor in the neurodegenerative diseases64. OXR1 is a common target of miR-885-5p and miR-361-5p. Therefore, OXR1 overexpression due to the modification of these two miRNAs could result in the less susceptibility to the exogenous stress.
Overall, our results suggest that PBMCs miR-885-5p, miR-361-5p and miR-17-5p order their combination, can discriminate the PD samples from the controls. Furthermore, the significantly obtained biological process and the pathways of the miRNA-based network were reflective of the known processes and pathways in PD, such as the regulation of the G1/S transition of the mitotic cell cycle, the regulation of autophagy, cell differentiation, p53 pathway, the oxidative stress response, the interferon-gamma signaling pathway, and the JAK/STAT signaling pathway. Nevertheless, some limitations should be noted. First, we had limited access to subjects with the advanced stages of PD that referred to the outpatient clinics; this was probably since most of them were severely disabled to ambulate or their disease was complicated by other medical issues. Second, we were unable to include confounding factors such as PD-related medication due to the lack of access to de novo PD patients. Therefore, a larger sample size-based expression study would be needed to confirm the miRNA expression changes observed during the PD progression in this study and define the involvement of the PD-related medication in the observed differential expression of miRNAs.
Despite the preliminary character of our study, we have presented a novel candidate set of aging and cellular senescence-related miRNAs; also, a consensus-based strategy is proposed to interpret the miRNAs expression data in PD; this could contribute to the identification of the possible triggering age-related signatures in PD.
Conclusions
Overall, we revealed 3 candidate biomarkers related to aging and cellular senescence for the first time in the patients with PD; these included miR-885-5p, miR-361-5p and miR-17-5p. Also, we presented the molecular interactions of these miRNAs in two regulatory layers, thereby clarifying the potential biochemical mechanisms underlying PD. Our preliminary data provided a rationale to further clarify the role of these miRNAs in aging and PD development through their promising candidate target genes and TFs to validate their diagnostic potential.
Supplementary information
Acknowledgements
Part of this project was supported by the University of Isfahan and Royan Institute for Biotechnology to support the graduation of M.B. in the field of Cell and Molecular Biology (MSc. degree). The authors are grateful to the study participants for their participation and collaboration in this research project. The authors also acknowledge the help of Dr. Masoud Etemadifar (MD. Neurologist) in recruiting the study participants.
Author Contributions
M.B., M.P. and K.G. participated in designing research studies, conducting experiments, acquiring data, analyzing data, providing reagents, and writing the manuscript. M.S.H. and M.S. participated in designing the research studies, conducting experiments and acquiring the data. M.B., M.P., M.H.N.E. and K.G. contributed to acquiring and analyzing data, interpreting data and writing the manuscript. Finally, M.H.N.E. and K.G. helped in giving the final approval of the manuscript. All authors read and approved the final manuscript.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on a reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maryam Peymani, Email: m.peymani@iaushk.ac.ir.
Mohammad Hossein Nasr-Esfahani, Email: mh.nasr-esfahani@royaninstitute.org.
Kamran Ghaedi, Email: kamranghaedi@sci.ui.ac.ir.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50256-3.
References
- 1.Poewe W, et al. Parkinson disease. Nat Rev Dis Primers. 2017;3:17013. doi: 10.1038/nrdp.2017.13. [DOI] [PubMed] [Google Scholar]
- 2.Ayano G. Parkinson’s Disease: A Concise Overview of Etiology, Epidemiology, Diagnosis, Comorbidity and Management. Journal of Neurological Disorders (2016).
- 3.Braak H, et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24(2):197–211. doi: 10.1016/S0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 4.Horowitz M, Greenamyre J. Gene–environment interactions in Parkinson’s disease: the importance of animal modeling. Clinical Pharmacology & Therapeutics. 2010;88(4):467–74. doi: 10.1038/clpt.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma L, et al. Advances with microRNAs in Parkinson’s disease research. Drug design, development and therapy. 2013;7:1103. doi: 10.2147/DDDT.S48500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ha M, Kim VN. Regulation of microRNA biogenesis. Nature reviews Molecular cell biology. 2014;15(8):509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 7.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature Reviews Genetics. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Serafin A, et al. Overexpression of blood microRNAs 103a, 30b, and 29a in L-dopa-treated patients with PD. Neurology. 2015;84(7):645–53. doi: 10.1212/WNL.0000000000001258. [DOI] [PubMed] [Google Scholar]
- 9.Soreq, L et al. Small RNA sequencing-microarray analyses in Parkinson leukocytes reveal deep brain stimulation-induced splicing changes that classify brain region transcriptomes. Frontiers in molecular neuroscience, 6 (2013). [DOI] [PMC free article] [PubMed]
- 10.Margis R, Margis R, Rieder CR. Identification of blood microRNAs associated to Parkinsonis disease. J Biotechnol. 2011;152(3):96–101. doi: 10.1016/j.jbiotec.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Martins M, et al. Convergence of miRNA expression profiling, alpha-synuclein interacton and GWAS in Parkinson’s disease. PLoS One. 2011;6(10):e25443. doi: 10.1371/journal.pone.0025443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hackl M, et al. miR‐17, miR‐19b, miR‐20a, and miR‐106a are down‐regulated in human aging. Aging cell. 2010;9(2):291–6. doi: 10.1111/j.1474-9726.2010.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooten NN, et al. microRNA expression patterns reveal differential expression of target genes with age. PloS one. 2010;5(5):e10724. doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olivieri F, et al. Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mechanisms of ageing and development. 2012;133(11–12):675–85. doi: 10.1016/j.mad.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Chmielarz P, et al. Dicer and microRNAs protect adult dopamine neurons. Cell death & disease. 2017;8(5):e2813. doi: 10.1038/cddis.2017.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Afanasyeva EA, et al. MicroRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Differ. 2011;18(6):974–84. doi: 10.1038/cdd.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. Journal of Neurology, Neurosurgery & Psychiatry. 1992;55(3):181–4. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasreddine ZS, et al. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. Journal of the American Geriatrics Society. 2005;53(4):695–9. doi: 10.1111/j.1532-5415.2005.53221.x. [DOI] [PubMed] [Google Scholar]
- 19.Hoehn MM, Yahr MD. Parkinsonism: onset, progression and mortality. Neurology. 1967;17(5):427–42. doi: 10.1212/WNL.17.5.427. [DOI] [PubMed] [Google Scholar]
- 20.Agarwal, V., Bell, G. W., Nam, J. W., Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 4 (2015). [DOI] [PMC free article] [PubMed]
- 21.Paraskevopoulou MD, et al. DIANA-microT web server v5. 0: service integration into miRNA functional analysis workflows. Nucleic acids research. 2013;41(W1):W169–W73. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic acids research. 2014;43(D1):D146–D52.. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou CH, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karagkouni D, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46(D1):D239–D45. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Lu M, Qiu C, Cui Q. TransmiR: a transcription factor–microRNA regulation database. Nucleic acids research. 2009;38(suppl_1):D119–D22. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research. 2016;44(W1):W90–W7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS one. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic acids research. 2016;45(D1):D183–D9. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeve A, Simcox E, Turnbull D. Ageing and Parkinson’s disease: why is advancing age the biggest risk factor? Ageing Res Rev. 2014;14:19–30. doi: 10.1016/j.arr.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macchi B, et al. Inflammatory and cell death pathways in brain and peripheral blood in Parkinson’s disease. CNS & neurological disorders drug targets. 2015;14(3):313–24.. doi: 10.2174/1871527314666150225124928. [DOI] [PubMed] [Google Scholar]
- 32.Prajapati P, et al. TNF-α regulates miRNA targeting mitochondrial complex-I and induces cell death in dopaminergic cells. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2015;1852(3):451–61. doi: 10.1016/j.bbadis.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 33.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proceedings of the National Academy of Sciences. 1997;94(14):7245–50. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michael WM, Newport J. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science. 1998;282(5395):1886–9. doi: 10.1126/science.282.5395.1886. [DOI] [PubMed] [Google Scholar]
- 35.Dimri GP, Hara E, Campisi J. Regulation of two E2F-related genes in presenescent and senescent human fibroblasts. Journal of Biological Chemistry. 1994;269(23):16180–6. [PubMed] [Google Scholar]
- 36.Atadja P, Wong H, Garkavtsev I, Veillette C, Riabowol K. Increased activity of p53 in senescing fibroblasts. Proceedings of the National Academy of Sciences. 1995;92(18):8348–52. doi: 10.1073/pnas.92.18.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. Journal of Biological Chemistry. 2007;282(4):2130–4. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 38.Yan H, et al. Repression of the miR‐17‐92 cluster by p53 has an important function in hypoxia‐induced apoptosis. The EMBO journal. 2009;28(18):2719–32. doi: 10.1038/emboj.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jordan-Sciutto KL, Dorsey R, Chalovich EM, Hammond RR, Achim CL. Expression patterns of retinoblastoma protein in Parkinson disease. Journal of Neuropathology & Experimental Neurology. 2003;62(1):68–74. doi: 10.1093/jnen/62.1.68. [DOI] [PubMed] [Google Scholar]
- 40.Höglinger GU, et al. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson’s disease. Proceedings of the National Academy of Sciences. 2007;104(9):3585–90. doi: 10.1073/pnas.0611671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Black AR, Azizkhan-Clifford J. Regulation of E2F: a family of transcription factors involved in proliferation control. Gene. 1999;237(2):281–302. doi: 10.1016/S0378-1119(99)00305-4. [DOI] [PubMed] [Google Scholar]
- 42.Paul WE. Interleukin-4: a prototypic immunoregulatory lymphokine. Blood. 1991;77(9):1859–70. [PubMed] [Google Scholar]
- 43.Rothstein TL. Inducible resistance to Fas-mediated apoptosis in B cells. Cell research. 2000;10(4):245. doi: 10.1038/sj.cr.7290053. [DOI] [PubMed] [Google Scholar]
- 44.Ristevski S, et al. The ETS transcription factor GABPα is essential for early embryogenesis. Molecular and cellular biology. 2004;24(13):5844–9. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, Z.-F., Drumea, K., Mott, S., Wang, J., Rosmarin, A. G. GABP transcription factor (nuclear respiratory factor 2) is required for mitochondrial biogenesis. Molecular and cellular biology. MCB 00492–12 (2014). [DOI] [PMC free article] [PubMed]
- 46.Xue H-H, et al. The transcription factor GABP is a critical regulator of B lymphocyte development. Immunity. 2007;26(4):421–31. doi: 10.1016/j.immuni.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z-F, Mott S, Rosmarin AG. The Ets transcription factor GABP is required for cell-cycle progression. Nature cell biology. 2007;9(3):339. doi: 10.1038/ncb1548. [DOI] [PubMed] [Google Scholar]
- 48.Crook MF, et al. GA-binding protein regulates KIS gene expression, cell migration, and cell cycle progression. The FASEB Journal. 2008;22(1):225–35. doi: 10.1096/fj.07-8573com. [DOI] [PubMed] [Google Scholar]
- 49.Imaki H, et al. Cell cycle-dependent regulation of the Skp2 promoter by GA-binding protein. Cancer research. 2003;63(15):4607–13. [PubMed] [Google Scholar]
- 50.Wu H, et al. The Ets transcription factor GABP is a component of the hippo pathway essential for growth and antioxidant defense. Cell reports. 2013;3(5):1663–77. doi: 10.1016/j.celrep.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu S., Cui K., Jothi R., Zhao D.-M., Jing X., Zhao K., Xue H.-H. GABP controls a critical transcription regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells. Blood. 2010;117(7):2166–2178. doi: 10.1182/blood-2010-09-306563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ripperger T, et al. The heteromeric transcription factor GABP activates the ITGAM/CD11b promoter and induces myeloid differentiation. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2015;1849(9):1145–54. doi: 10.1016/j.bbagrm.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Park B, et al. Impairment of protein trafficking upon overexpression and mutation of optineurin. PLoS One. 2010;5(7):e11547. doi: 10.1371/journal.pone.0011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sahlender DA, et al. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. The Journal of cell biology. 2005;169(2):285–95.. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wise JP, Jr, Cannon J. From the cover: alterations in optineurin expression and localization in pre-clinical Parkinson’s disease models. Toxicological Sciences. 2016;153(2):372–81. doi: 10.1093/toxsci/kfw133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Osawa T, et al. Optineurin in neurodegenerative diseases. Neuropathology. 2011;31(6):569–74. doi: 10.1111/j.1440-1789.2011.01199.x. [DOI] [PubMed] [Google Scholar]
- 57.Vilborg A, et al. The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(37):15756–61. doi: 10.1073/pnas.0900862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higashi Y, et al. The p53-activated gene, PAG608, requires a zinc finger domain for nuclear localization and oxidative stress-induced apoptosis. J Biol Chem. 2002;277(44):42224–32. doi: 10.1074/jbc.M203594200. [DOI] [PubMed] [Google Scholar]
- 59.Scherzer CR, et al. GATA transcription factors directly regulate the Parkinson’s disease-linked gene alpha-synuclein. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(31):10907–12. doi: 10.1073/pnas.0802437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang L, Wang H, Liu L, Xie A. The Role of Insulin/IGF-1/PI3K/Akt/GSK3β Signaling in Parkinson’s Disease Dementia. Frontiers in neuroscience. 2018;12:73. doi: 10.3389/fnins.2018.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L, et al. Targeted methylation sequencing reveals dysregulated Wnt signaling in Parkinson disease. J Genet Genomics. 2016;43(10):587–92. doi: 10.1016/j.jgg.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 62.Hosokawa N, You Z, Tremblay LO, Nagata K, Herscovics A. Stimulation of ERAD of misfolded null Hong Kong alpha1-antitrypsin by Golgi alpha1,2-mannosidases. Biochem Biophys Res Commun. 2007;362(3):626–32. doi: 10.1016/j.bbrc.2007.08.057. [DOI] [PubMed] [Google Scholar]
- 63.Calligaris R, et al. Blood transcriptomics of drug-naive sporadic Parkinson’s disease patients. BMC Genomics. 2015;16:876. doi: 10.1186/s12864-015-2058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oliver PL, et al. Oxr1 is essential for protection against oxidative stress-induced neurodegeneration. PLoS genetics. 2011;7(10):e1002338. doi: 10.1371/journal.pgen.1002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on a reasonable request.