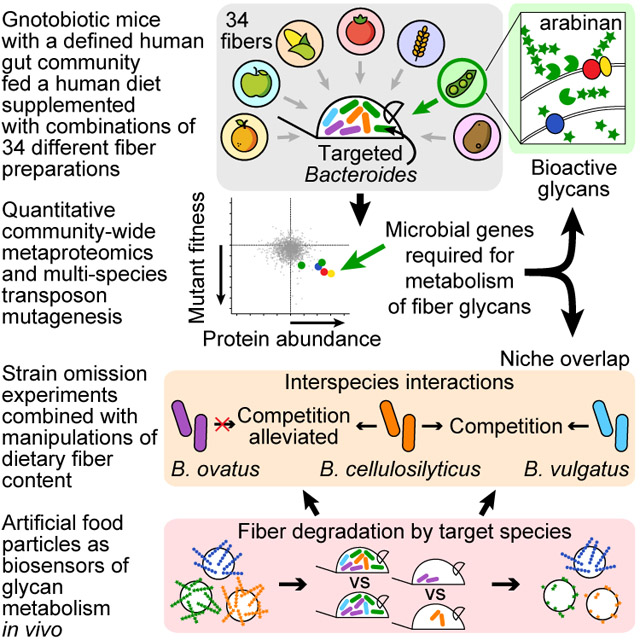
Summary
Development of microbiota-directed foods (MDFs) that selectively increase the abundance of beneficial human gut microbes, and their expressed functions, requires knowledge of both the bioactive components of MDFs and the mechanisms underlying microbe-microbe interactions. Here, gnotobiotic mice were colonized with a defined consortium of human gut-derived bacterial strains and fed different combinations of 34 food-grade fibers added to a representative low-fiber diet consumed in the USA. Bioactive carbohydrates in fiber preparations targeting particular Bacteroides species were identified using community-wide quantitative proteomic analyses of bacterial gene expression coupled with forward genetic screens. Deliberate manipulation of community membership combined with administration of retrievable artificial food particles, consisting of paramagnetic microscopic beads coated with dietary polysaccharides, disclosed the contributions of targeted species to fiber degradation. Our approach, including the use of bead-based biosensors, defines nutrient harvesting strategies that underlie, as well as alleviate, competition between Bacteroides and control the selectivity of MDF components.
Keywords: human gut microbiota-directed foods, polysaccharide utilization, bioactive components of dietary fibers, interspecies competition and acclimation, artificial food particles as biosensors
Graphical Abstract
Introduction
Increasing evidence that the gut microbiota impacts multiple features of human biology has stimulated efforts to develop microbiota-directed interventions that improve health status. Microbiota-directed foods (MDFs) are an obvious choice, as diet has pronounced and rapid effects on microbial community configuration. Dietary carbohydrates provide an important source of energy for gut bacteria, with the products of their metabolism benefiting primary microbial consumers, their syntrophic partners, and the host (Porter and Martens, 2017). Consumption of plant polysaccharides in the form of dietary fiber has been linked to a number of health benefits (Desai et al., 2016; Menni et al., 2017; Zhao et al., 2018). In addition, the diminished diversity of complex polysaccharides in the diets of those living in industrialized countries has been associated with loss of bacterial diversity in their microbiota (Fragiadakis et al., 2018; Sonnenburg et al., 2016).
Achieving a better understanding of the mechanisms by which human gut bacterial species interact with dietary polysaccharides, and with one another, should facilitate development of fiber-based interventions that establish, restore, and/or sustain health-promoting microbiota functions. The vast majority of studies of the biological effects of fibers are performed with preparations whose biochemical features are largely uncharacterized, in part because identifying the bioactive components of fibers is a formidable challenge. Painstaking linkage analysis is required to reveal the structures of polysaccharide constituents (Atmodjo et al., 2013), and the difficulty in separating these structures biochemically hinders efforts to ascribe in vivo activity to a particular class of glycan. Even after identifying a fiber’s component glycan structures, knowledge that a targeted organism possesses a suitable arsenal of genes for acquiring and processing these glycans is generally not sufficient to predict the organism’s response to the fiber without considering ‘community context’. Microbial community members can have overlapping capacities to metabolize a given class of glycans, which may lead to competition. Competitive environments can generate unstable states that result in exclusion (and extirpation) of species over time (Hibbing et al., 2010). Therefore, understanding the mechanisms used by organisms to compete with one another for fiber components as well as to avoid competition could yield prebiotic and synbiotic formulations that more precisely and effectively manipulate the properties of different human gut communities.
In the present study, we describe an in vivo approach for identifying fibers and their bioactive components that selectively increase the fitness of a group of human gut Bacteroides, and the different mechanisms these organisms deploy when encountering these nutrient resources and one another. The bacterial targets for fiber-based manipulation originated from our previous study of twins stably discordant for obesity (Ridaura et al., 2013). Fecal microbiota from these twin pairs transmitted discordant adiposity and metabolic phenotypes to recipient germ-free mice. Cohousing mice shortly after they received microbial communities from lean (Ln) or obese (Ob) cotwins prevented recipients of the Ob donor microbiota from developing obesity and associated metabolic abnormalities. Analysis of their gut communities revealed that invasion of Bacteroides species from Ln into Ob microbiota, notably B. thetaiotaomicron, B. vulgatus, B. caccae, and B. cellulosilyticus, correlated with protection. Invasion was diet-dependent, occurring when animals consumed a human diet designed to represent the lower tertile of consumption of saturated fats and upper tertile of consumption of fruits and vegetables (high in fiber) in the USA, but not when they consumed a diet representing the upper tertile of saturated fat and lower tertile of fruit and vegetable consumption (HiSF-LoFV).
Here we identify dietary fibers and constituent bioactive components that increase the fitness of targeted Bacteroides (B. thetaiotaomicron, B. vulgatus, B. caccae, and/or B. cellulosilyticus) in vivo in the HiSF-LoFV diet context. To do so, we first colonized germ-free mice with a defined collection of sequenced bacterial strains cultured from a Ln donor, and fed them diets generated by supplementing the HiSF-LoFV formulation with 34 different food-grade fibers. Armed with a consortium that contained targeted Bacteroides species, each in the form of a library of tens of thousands of transposon (Tn) mutant strains, we subsequently characterized the effects of selected fibers on the fitness of Tn mutants and on the community’s expressed proteome. By identifying polysaccharide-processing genes whose expression was increased and that functioned as key fitness determinants, we inferred which components of the fibers were bioactive. Time series proteomic analyses of the complete community and derivatives lacking one or more Bacteroides, revealed nutrient harvesting strategies resulting in, as well as alleviating, interspecies competition for fiber components. Finally, administering artificial food particles coated with dietary polysaccharides to gnotobiotic mice with deliberately varied community membership further established the contributions of individual Bacteroides species to glycan processing in vivo.
Results
In vivo screen for fiber preparations that target specific human gut microbes
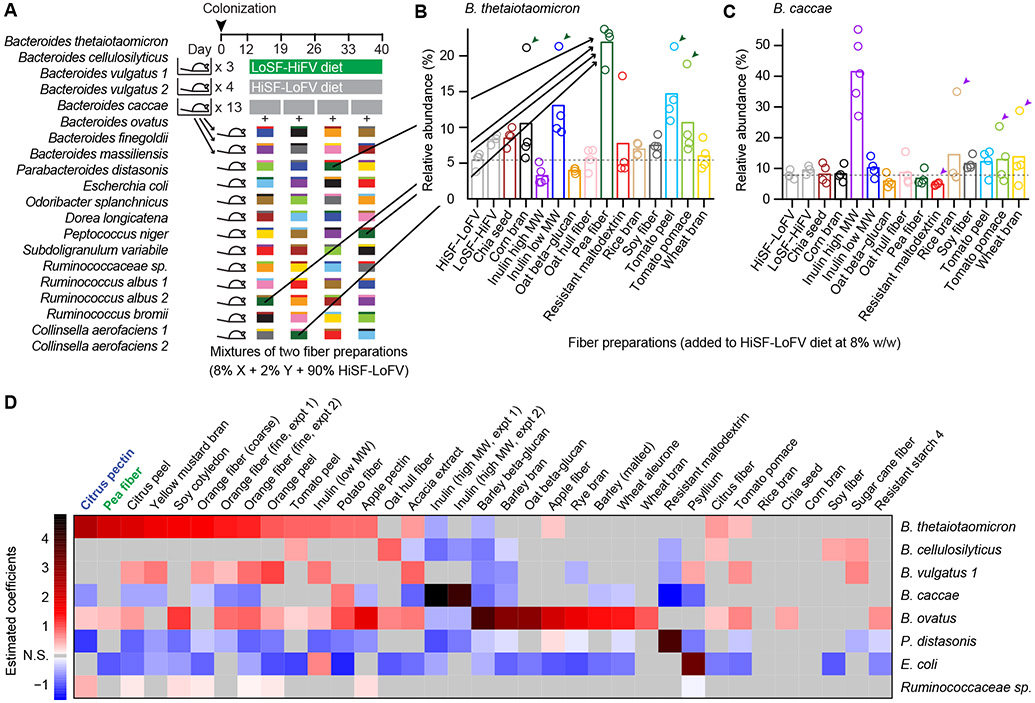
Three screening experiments (Figure 1A; Table S2D) were performed to complete an analysis of the effects of 34 fiber preparations on community structure. These fibers were obtained from diverse plant sources including fruits, vegetables, legumes, oilseeds, and cereals. Each mouse was gavaged with a 20-member consortium of sequenced bacterial strains cultured from a single Ln co-twin donor. Each animal received a different fiber-supplemented diet each week for a total of four weeks. Each of the 144 unique diets tested contained one fiber type present at a concentration of 8% (w/w) and another fiber type at 2%. These two concentrations were systematically paired to maximize the number of fibers tested and mitigate potential hysteresis effects. Control groups were monotonously fed the unsupplemented HiSF-LoFV or LoSF-HiFV diet.
Figure 1. In vivo screen of the effects of food-grade fiber preparations on members of a defined human gut microbiota.
(A) Schematic design of screen (one of three similar screens). Gnotobiotic mice harboring a consortium of bacterial strains obtained from a single human donor. Animals received a series of supplemented HiSF-LoFV diets, each containing one fiber type at 8% (w/w) and another at 2% (w/w) (colored boxes). Control animals received the unsupplemented HiSF-LoFV or LoSF-HiFV diet monotonously for four weeks.
(B,C) Average relative abundance values for B. thetaiotaomicron and B. caccae during administration of the indicated fiber. Bars show mean values. Circles denote individual mice. Arrowheads mark mice that received pea fiber (B) or high molecular weight (MW) inulin (C) as the minor fiber type (2% w/w).
(D) Estimated coefficients from linear models (with at least one estimated coefficient > 0.4) for bacterial strains across the three screening experiments. Statistically significant coefficients (P < 0.01; ANOVA) are shaded according to the color bar.
We analyzed the relative abundance of each member of the defined community by collecting fecal samples and performing 16S rRNA gene sequencing. Fifteen of the strains consistently colonized animals at >0.1% relative abundance. Binning the data according to the fiber type present at 8% concentration revealed potent and specific effects on distinct taxa (Figures 1B and 1C; Table S2). To analyze the independent effects of the two fibers administered during each diet treatment, we generated a linear mixed-effects model for each bacterial taxon using the data from the last two days of consumption of each diet. The coefficient estimates in these models describe the slope of the predicted dose response curve for each fiber’s effect on each community member (Table S2E). Twenty-one fiber preparations had significant estimated coefficients of >1 (Figure 1D). Large coefficients were observed in the B. thetaiotaomicron models for citrus pectin (b=2.6) and pea fiber (2.1). The B. ovatus models revealed pronounced effects of barley beta-glucan (3.9) and barley bran (3.1), consistent with previous in vitro studies (Martens et al., 2011). Estimated coefficients for high molecular weight inulin (4.5, B. caccae model; in accord with a previous in vivo analysis; Sonnenburg et. al., 2010), resistant maltodextrin (3.8, P. distasonis model), and psyllium (3.4, E. coli model) were notable with 8% fiber administration driving the relative abundances of these community members from 10-20% to nearly 50%. High molecular weight inulin and fine orange fiber were tested across two separate experiments; the results established that the effects on the relative abundances of community members were reproducible (coefficients were highly correlated between these independent experiments, R2=0.96; Table S2E). The even distributions of residuals around the fitted values in the models indicated that there were no pronounced threshold or saturation effects of these fibers at the concentrations tested. For bacterial species that exhibited notable responses to fiber (at least one coefficient >1), the average R2 value of the models was 0.82. We also repeated our analyses using DNA yield from each fecal sample to estimate the absolute abundance of each organism as a function of fiber type. The estimated coefficients obtained from these two measures were highly correlated (R2 = 0.88) (Table S2F). Together, results obtained from this screen illustrate the specificity of the effects of different types of dietary fiber on community configuration.
Proteomics and forward genetics identify bioactive polysaccharides in fiber preparations
Several possible mechanisms could account for the increase of a target Bacteroides in response to fiber administration, including indirect effects involving other species. Therefore, we sought to determine which polysaccharides in the fiber preparations caused the target species to expand and whether they acted directly on those species by serving as nutrient sources for their growth. To do so, we simultaneously quantified community-wide protein expression and assessed the contributions of proteins to bacterial fitness using a forward genetic screen. The screen was based on genome-wide transposon (Tn) mutagenesis and a method known as multi-taxon INsertion Sequencing (INSeq), which allows simultaneous analysis of Tn mutant libraries generated from different Bacteroides species in the same recipient gnotobiotic mouse. We employed five INSeq libraries constructed using type strains corresponding to four Bacteroides species present in the Ln co-twin donor culture collection. The quality and performance of these libraries had been characterized previously in vitro and in vivo (Hibberd et al., 2017; Wu et al., 2015). Additionally, we simplified the community used in these experiments by omitting six strains from the original 20-member consortium that were not robust colonizers in the HiSF-LoFV diet context (Faith et al., 2014; Ridaura et al., 2013). All mice were colonized with the resulting 15-member community (Table S3) and fed the base HiSF-LoFV diet or, two days after gavage, switched to the HiSF-LoFV diet containing one of the fibers identified in the screen. We tested pea fiber, citrus pectin, orange peel, and tomato peel, each at a concentration of 10% (w/w), based on their ability to increase the representation of one or more of the targeted Bacteroides (Figure 1D). All diets were administered ad libitum and given monotonously for the duration of the experiment (Tables S3A-S3C). DNA isolated from fecal samples was subjected to short read shotgun DNA sequencing (COmmunity PROfiling by Sequencing, COPRO-Seq; (Hibberd et al., 2017; McNulty et al., 2013) to quantify each community member as a function of fiber treatment, including the combined abundance of all INSeq mutants for a given species. Our previous studies had established that in aggregate, a population of INSeq mutants behaves similarly to the corresponding wild-type parental strain (Hibberd et al., 2017; Wu et al., 2015).
Consistent with results obtained from seven days of fiber administration in the screening experiments, we observed a statistically significant expansion of B. thetaiotaomicron in mice consuming pea fiber (ANOVA, P <0.05; Figure 2B; Tables S3A-S3C). In addition, the relative abundance of B. ovatus was significantly greater in the pea fiber-treated group (Figure 2C), while cellulosilyticus and B. vulgatus did not exhibit significant changes during this time period (Figures 2D and 2E). Citrus pectin induced significant expansion of a set of species (B. cellulosilyticus, Bacteroides finegoldii, and a member of the Ruminococcaceae) that was distinct from the set affected by pea fiber (Tables S3A, S3B, and S3D). Although the fiber screen predicted an increase in the abundance of B. thetaiotaomicron in response to citrus pectin, this was not observed during monotonous feeding until later in the time course, indicating a difference between the strains employed or the effect of different community context. Orange peel significantly increased the representation of B. vulgatus, but otherwise had a minimal effect on community structure (Table S3A). Tomato peel did not significantly increase any members of this community, which may indicate the strain-dependency of a given species’ response to a certain fiber when the effect size of a given fiber is low (Table S2E, S2F, S3A). We selected pea fiber and citrus pectin for more detailed functional studies of their utilization by community members since both had pronounced effects on distinct sets of taxa.
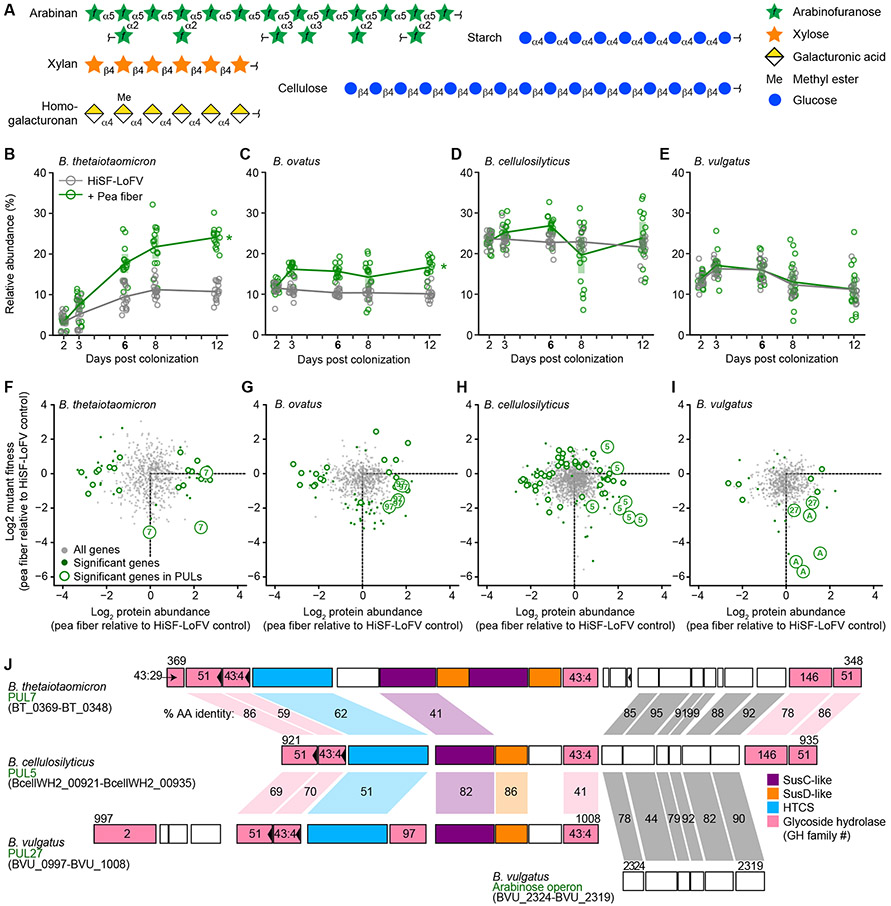
Figure 2. Proteomics and forward genetics identify arabinan in pea fiber as a nutrient source for multiple species.
(A) Schematic representation of polysaccharide structures detected in pea fiber based on monosaccharide and linkage analyses (with stereochemistry of anomeric carbon inferred).
(B-E) Gnotobiotic mice harboring a 15-member community consisting of INSeq libraries representing four Bacteroides species together with 10 additional bacterial strains used in screening experiments (Figure 1). Relative abundance is shown for each bacterial strain at each of the indicated time points in mice monotonously fed the HiSF-LoFV diet with (green) or without (grey) addition of 10% (w/w) pea fiber. Key: circles, individual mice; lines, mean values; shading, 95%CI (n=15 individually-caged mice per group; Tables S3A-S3C). *, P < 0.05; (pea fiber diet versus control HiSF-LoFV diet; ANOVA).
(F-I) Proteomic and INSeq analyses of fecal samples collected on experimental day 6. On the x-axis, the position of each dot denotes the mean value for the abundance of a single bacterial protein in samples obtained from mice fed a HiSF-LoFV + pea fiber diet (relative to control diet). The y-axis indicates the mean value of the differential enrichment of mutant strains with Tn disruptions in the gene encoding each protein in the pea fiber versus HiSF-LoFV diet groups. All genes represented in both the protein dataset and INSeq mutant pool are plotted as grey dots. Green dots highlight genes that are significantly affected by pea fiber (P < 0.05, ∣fold change∣ > log2(1.2); limma or limma-voom) as judged by levels of their protein products or their contribution to fitness; open circles mark the subset of these genes that are encoded by PULs. Genes that are present in the three homologous arabinan-processing PULs (see below) are labeled with their PUL number (Table S7). Genes in an arabinose-processing operon in B. vulgatus are labeled with an ‘A’. Genes in the B. ovatus RGI-processing PUL97 are also labeled.
(J) Alignment of B. thetaiotaomicron PUL7, B. cellulosilyticus PUL5, B. vulgatus PUL27, and the B. vulgatus arabinose operon. The direction of transcription is left to right (unless marked by a leftward pointing arrowhead). The first and last genes are labeled above with their locus tag number. Genes are color-coded according to their functional annotation (see key). GH families for enzymes in the CAZy database are shown as numbers inside the gene boxes. Shaded regions connecting genes denote significant BLAST homology (E-value <10−9); the percent amino acid identity of their protein products is shown.
Structural analyses of lead fibers –
We used permethylation and gas-chromatography-mass spectrometry to analyze the monosaccharide composition and glycosidic linkages of polysaccharides present in pea fiber and citrus pectin. After accounting for starch and cellulose, the most abundant polysaccharide in pea fiber was arabinan, consisting of a linear 1,5-linked arabinose backbone with arabinose residues as side chains at position 2 or 3 (Table S1B; Figure 2A). Linear xylan (4-linked xylose), homogalacturonan (4-linked galacturonic acid) and rhamnogalacturonan I (2- and 2,4-linked rhamnose) were also detected in pea fiber. Homogalacturonan with a high degree of methyl esterification was the major polysaccharide in citrus pectin (88.6% galacturonic acid), with arabinan, 1,4-linked galactan and RGI present as minor components (Table S1C).
Proteomic analysis of community gene expression –
The results of these biochemical analyses raised the possibility that metabolism of arabinan in pea fiber and methylated homogalacturonan in citrus pectin were involved in the responses of target Bacteroides. To test this hypothesis, we turned to high-resolution shotgun proteomic analysis, focusing on fecal samples obtained on day 6 of the monotonous feeding experiment. We calculated a z-score for each expressed protein from each bacterial species using the abundances of all proteins assigned to that individual species in a given sample. This allowed us to determine changes in the abundance of each protein irrespective of changes in the abundance of that species in the community. Linear models were constructed using limma (Smyth, 2004; Ting et al., 2009) and significant effects were identified between bacterial protein abundances and supplementation of the control diet with pea fiber and citrus pectin (245 and 450 proteins, respectively; ∣fold-change∣ > log2(1.2), P < 0.05, FDR corrected, Tables S4A and S4B). Bacteroides contain multiple polysaccharide utilization loci (PULs) in their genomes. PULs provide a fitness advantage by endowing a species with the ability to sense, import, and process complex glycans using their encoded carbohydrate-responsive transcription factors, SusC/SusD-like transporters, and carbohydrate active enzymes (CAZymes) (Glenwright et al., 2017; Kotarski and Salyers, 1984; Martens et al., 2011; McNulty et al., 2013; Shepherd et al., 2018). Eighty-five of the proteins whose levels were significantly altered by pea fiber and 134 that were significantly affected by citrus pectin were encoded by PULs (Terrapon et al., 2018).
Ranking proteins by the pea-fiber induced increase in their abundance disclosed that in B. thetaiotaomicron, 6 of the top 10 were encoded by PULs 7, 73, and 75. PUL7 is known to be involved in arabinan metabolism (Lynch and Sonnenburg, 2012; Schwalm et al., 2016), and encodes characterized and predicted arabinofuranosidases in glycoside hydrolase (GH) family 43, GH51, and GH146 (Table S4A). PUL75 carries out the degradation of rhamnogalacturonan I (RGI) (Luis et al., 2018), but its expression is also triggered by exposure to purified arabinan in vitro (Martens et al., 2011). PUL73 processes homogalacturonan (Luis et al., 2018) and encodes CAZymes that cleave linked galacturonic acid residues and remove methyl and acetyl esters from galacturonic acid [polysaccharide lyase (PL)1, GH105, GH28, CE8, CE12 family members]. B. ovatus proteins encoded by predicted RGI-processing PULs (PUL97) (Luis et al., 2018) were among the most increased by pea fiber administration in this species (Tables S4A and S4B). Citrus pectin resulted in increased abundance of proteins encoded by a B. cellulosilyticus PUL that is induced by homogalacturonan in vitro (PUL83; Tables S4A and S4B). In addition, citrus pectin induced expression of proteins in several B. finegoldii PULs (PUL34, 35, 42, and 43) that encode galacturonan-processing enzymes (GH28, GH105, GH106, PL11 subfamily 1, CE8 and CE12) (Tables S4A and S4B). This latter finding correlates with the organism’s citrus pectin-driven expansion (Tables S3A and S3B).
Combining proteomic and INSeq analyses –
As noted above, we colonized mice with INSeq libraries and then fed them the base HiSF-LoFV diet for two days before switching the experimental groups to fiber-supplemented diets. We measured the abundances of Tn mutant strains, and calculated log ratios between fecal samples collected on experimental day 6 (posttreatment) and day 2 (pre-treatment); results were expressed relative to the reference HiSF-LoFV treatment arm to focus on fitness effects significantly impacted by fibers (P < 0.05, FDR corrected; see STAR Methods; 223 genes, 24% in PUls; Table S5A). Genes exhibiting a significant positive fold-change in protein abundance and negative effect on fitness when mutated appear in the bottom right quadrant of the orthogonal protein-fitness plots shown in Figures 2F-2I.
Genes in PULs were ranked by the magnitude of pea-fiber-dependent decreases in strain fitness when they were disrupted by a Tn insertion. The results revealed genes in three PULs (PUL7 in B. thetaiotaomicron, PUL5 in B. cellulosilyticus, and PUL27 in B. vulgatus; Figures 2F, 2H, and 2I) that were affected by pea fiber. These three PULs are homologous as judged by a BLASTp comparison of their encoded proteins against the genomes of other community members (Figure 2J). Genes in a conserved arabinose utilization operon, present within the B. thetaiotaomicron and B. cellulosilyticus PULs, but at a site distant from PUL27 in B. vulgatus, had the greatest effect on B. vulgatus fitness of any genes represented in the mutant library (Figure 2I; Tables S4A, S4B, and S5A). We subsequently compared the genomes of five strains of B. thetaiotaomicron, and found that PUL7 was highly conserved with the exception of a single gene of unknown function (BT _0352) that was present in two of the strains (Figure S1). PUL27 in B. vulgatus was also well conserved across 6 strains with the exception of variability in the gene lengths of the hybrid two-component system and SusC-like transporter.
The decreased fitness produced by mutations in B. ovatus RGI-processing PUL97, but not the B. thetaiotaomicron RGI-processing PUL75, indicated that these species prioritize different carbohydrates in pea fiber (RGI and arabinan, respectively; Figure 2G; Tables S4A, S4E, and S5A). In contrast, the overlapping reliance on arabinan degradation pathways in B. thetaiotaomicron, B. vulgatus, and B. cellulosilyticus raised the possibility that these species were engaged in competition with one another for arabinan in pea fiber.
A parallel analysis of mice monotonously fed citrus pectin revealed that five genes encoded by galacturonan-processing PUL83 in B. cellulosilyticus were among the most abundantly expressed and most important for fitness compared to the base diet condition (Figure S2H). B. vulgatus did not expand during citrus pectin feeding (Figure S2E); nevertheless, it contained galacturonan-processing PULs (PUL5/6, PUL31, and PUL42/43) with genes involved in hexuronate metabolism whose protein products increased in abundance and, when mutated, conveyed decreased fitness when exposed to this fiber preparation (Figure S2I). Consistent with increased reliance on citrus pectin, the abundance of B. vulgatus proteins involved in starch utilization (PUL38) was decreased in the presence of this fiber (Table S4).
Together, our proteomic and INSeq datasets revealed the bacterial genes required during fiber-driven expansion, highlighted the polysaccharides that contributed to the fitness effects of these fibers and provided evidence for functional overlap in the nutrient harvesting strategies of B. cellulosilyticus and B. vulgatus, in two distinct fiber conditions. The dominance of B. cellulosilyticus in diverse diet contexts led us to ask whether this species directly competes with other community members for polysaccharides.
Interspecies competition underlies the selective effects of dietary fibers
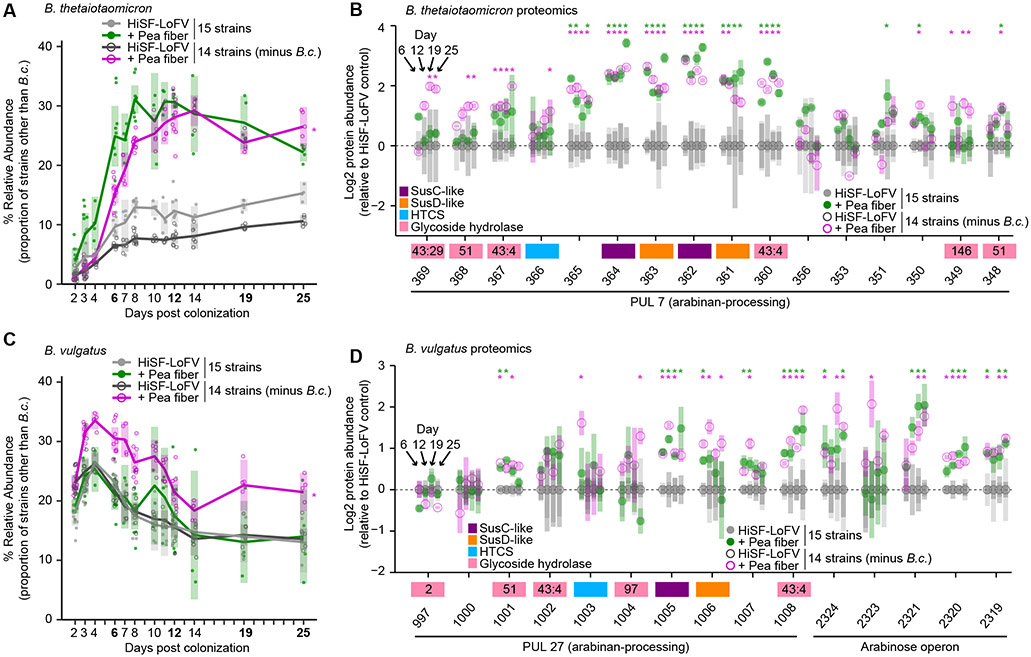
We performed a direct test for interactions between B. cellulosilyticus and other species by comparing the defined 15-member community to the derivative 14-member community lacking B. cellulosilyticus. Using an experimental design that mimicked the monotonous feeding study described above, groups of germ-free mice were colonized with these two communities and fed the HiSF-LoFV diet with or without 10% (w/w) pea fiber or citrus pectin (Tables S3B and S3C). COPRO-Seq was used to determine the abundance of each strain as a proportion of all strains other than B. cellulosilyticus, to control for the compositional effect of removing this species. Defined this way, the abundance of B. thetaiotaomicron did not increase upon omission of B. cellulosilyticus in the presence of pea fiber, suggesting minimal competition between these two species for arabinan (Figure 3A; Tables S3B and S3C). Proteomic analysis of fecal samples collected on experimental days 6, 12, 19, and 25 demonstrated that proteins in B. thetaiotaomicron PUL7 whose abundances were increased by pea fiber in the complete community context, were not further increased in the absence of B. cellulosilyticus (Tables S4B-S4E; Figure 3B). B. vulgatus was the only species that expanded with pea fiber administration in the absence of B. cellulosilyticus(p < 0.05, ANOVA, FDR corrected; Figure 3C; Tables S3B and S3C). Proteomic analysis of serially collected fecal samples disclosed that the abundances of proteins encoded by B. vulgatus PUL27, as well as its arabinose operon, were persistently increased during exposure to pea fiber, regardless of whether B. cellulosilyticus was included in the community (Tables S4B-S4E; Figure 3D). Citrus pectin provided a second example of fiber driven expansion of B. vulgatus in the absence of B. cellulosilyticus (Figure S3B; Tables S3B and S3D). Expression of proteins encoded by B. vulgatus ’ galacturonan-processing PULs 5, 6, 31, 42, and 43, were also induced by citrus pectin, irrespective of B. cellulosilyticus (Table S4B-S4E; Figure S3D). Odoribacter splanchnicus expanded in the absence of B. cellulosilyticus; this effect was repressed by both pea fiber and citrus pectin administration.
Figure 3. Deliberate manipulation of community composition demonstrates interspecies competition for pea fiber arabinan.
(A,B) Relative abundance of each bacterial strain in fecal samples from gnotobiotic mice harboring the 15-member community. Mice were fed the control HiSF-LoFV diet in the presence (light grey, closed circles), or absence (dark grey, open circles) of B. cellulosilyticus (B.c.), or fed the HiSF-LoFV diet + 10% (w/w) pea fiber in the presence (green, closed circles) or absence (magenta, open circles) of B.c. Key: circles, individual mice; lines, mean values; shading, 95%CI (n=4-10 mice per group). *, P < 0.05; (diet-by-community interaction; ANOVA). (C,D) Proteomics analysis of fecal communities sampled on experimental days 6, 12, 19, and 25. Genes in PULs of interest are shown along the x-axis (as locus tag number only; BT_XXXX or BVU_XXXX). Mean values ± SD (vertical shading) are indicated (n=5 animals/treatment group). Genes are color-coded according to their functional annotation (see key). GH families for enzymes in the CAZy database are shown as numbers inside the gene boxes. Key for circles is identical to that used in panels A and B. *, P < 0.05, ∣fold change∣ > log2(1.2) (HiSF-LoFV + pea fiber versus HiSF-LoFV diet; limma).
These results demonstrate negative interactions between B. vulgatus and B. cellulosilyticus and suggest that the suppression of B. vulgatus when B. cellulosilyticus is present occurs due to persistent competition between these organisms for arabinan in pea fiber and homogalacturonan in citrus pectin.
Artificial food particles as biosensors of community glycan degradative activities
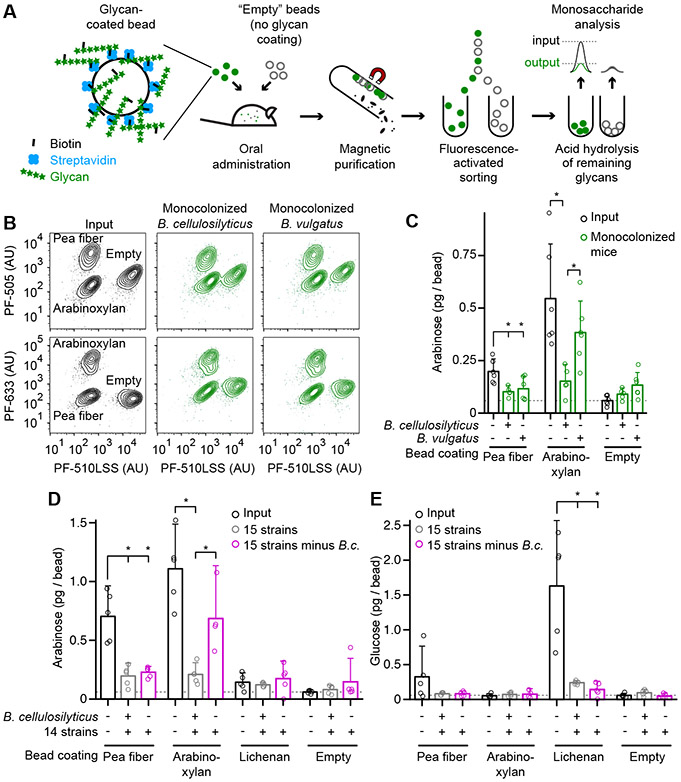
To directly test the capacity of competing Bacteroides to process the same nutrient substrate in vivo, we developed a bead-based glycan degradation assay (Figure 4A). We selected two polysaccharides of interest: (i) a soluble, starch-depleted fraction of pea fiber polysaccharides composed predominantly of arabinose (83% of monosaccharides) with little xylose (4%), and (ii) wheat arabinoxylan (38% arabinose/62% xylose). The latter was used as a control given its established ability to support growth (in vitro) of B. cellulosilyticus (McNulty et al., 2013) but not B. vulgatus (Tauzin et al., 2016). We biotinylated these polysaccharides and attached each product to a distinct population of microscopic (20μm diameter) streptavidin-coated paramagnetic glass beads, generating carbohydrate-coated artificial ‘food particles’ that could be recovered from mouse intestinal contents using a magnetic field. We also labeled each population of beads with a distinct biotinylated fluorophore so that several types of polysaccharide-beads could be pooled, administered at the same time to the same mouse, recovered from the gut lumen or feces and then sorted into their original groups using a flow cytometer (Figure 4B). ‘Empty’ beads that had not been incubated with polysaccharides, but were labeled with a unique biotinylated fluorophore, served as negative controls. The sorted beads were subjected to acid hydrolysis and the hydrolysis products were assayed by gas chromatography-mass spectrometry (GC-MS) to quantify the levels of bead-bound carbohydrate present before and after transit through the mouse gut.
Figure 4. Characterizing glycan processing as a function of community membership with artificial food particles.
(A) Schematic depiction of the bead-based in vivo glycan degradation assay.
(B) Fluorescence intensities of pooled bead types before and after transit though the guts of mice representing two colonization conditions. Axes are labeled with the fluorophore detected in each channel.
(C) Gnotobiotic mice, mono-colonized with either B. cellulosilyticus (B.c.) or B. vulgatus, were gavaged with a pool of glycan-coated and ‘empty’ uncoated beads. Beads were purified from cecal and colonic contents four hours after gavage. The mass of arabinose associated with each bead type is plotted before (black) and after (green) passage through the intestine. Circles denote individual animals. Bars show mean values and 95%CI.
(D,E) Glycan degradation in mice colonized with derivatives of the 15-member community (with or without B.c.) fed the HiSF-LoFV diet (Table S3B). The mass of bead-associated arabinose (panel D) or glucose (panel E) is plotted before (black) and after collection from cecal and colonic contents on experimental day 12 (grey, 15-member community group; magenta, minus B.c. group). Circles denote individual mice. Mean values + 95%CI are shown (n=3-6 animals/group). *, P < 0.05 (Mann-Whitney U test).
Germ-free mice were colonized with either B. cellulosilyticus or B. vulgatus alone and fed the HiSF-LoFV diet plus 10% (w/w) pea fiber. Seven days after colonization, all mice were gavaged with an equal mixture of the three bead types (5×106 of each type/animal, n=5-6 animals). Mice were euthanized 4 h later, beads were recovered from their cecum and colon, and the mass of monosaccharides on the different purified bead types was quantified. The fluorescent signal present on all bead types persisted after intestinal transit, confirming that the biotin-streptavidin interactions were stable under these conditions (Figure 4B). Pea fiber-beads recovered from both groups of mice had significantly reduced arabinose [26.1±3.4% (mean±SD) and 29.1±0.7% of levels in input beads, respectively]. In contrast, levels of arabinose were only significantly decreased on arabinoxylan-coated beads recovered from mice colonized with B. cellulosilyticus (Figure 4C; Table S6B).
We performed a follow-up experiment of identical design except that animals fed HiSF-LoFV supplemented with pea fiber were gavaged 12 days rather than seven days after colonization with a collection of four rather than three types of beads. These beads were either empty (no glycan bound) or coated with (i) the soluble, starch-depleted fraction of pea fiber, or wheat arabinoxylan, or lichenan from Icelandic moss, a control glycan low in arabinose (81% glucose / 8% mannose / 6% galactose / 2% arabinose). Beads were recovered, purified by flow cytometry and analyzed using GC-MS. The degradation of bead-bound pea fiber and arabinoxylan was similar to that observed on day 7.
To control for microbe-independent polysaccharide degradation, germ-free mice were given a gavage of arabinoxylan-coated, pea-fiber coated, lichenan-coated, and empty beads (n=13 animals). We collected all fecal samples produced during an 8 h period (from 4 to 12 hours after gavage). Assays of arabinoxylan-, pea fiber-, and lichenan-coated beads purified from fecal samples obtained from each germ-free animal revealed no significant degradation of these polysaccharides after passage through their intestines (Figures S3C and S3D; Tables S6A and S6B). Together, these results provide a direct, in vivo demonstration of the overlapping capacities of competing Bacteroides species to degrade arabinan present in pea fiber.
Given our observation that several species can metabolize pea fiber arabinan in vivo, we assessed whether the absence of B. cellulosilyticus would compromise the efficiency with which the community carried out this function. Mice consuming the HiSF-LoFV diet were given pea fiber-coated, arabinoxylan-coated, lichenan-coated, and empty beads 12 days after colonization with (i) the 15-member consortium or (ii) the derivative 14-member community lacking B. cellulosilyticus. Analysis of beads recovered from the cecal and colonic contents of these mice disclosed that the level of pea fiber degradation was not affected by the absence of B. cellulosilyticus (Figure 4D). In a separate group of mice fed the HiSF-LoFV plus pea fiber diet, degradation of bead-bound pea-fiber was also similar whether or not B. cellulosilyticus was present (Figures S3E and S3F). Thus, consistent with our detection of multiple species exploiting pea fiber arabinan as a nutrient source (Figures 2 and 3), this community can compensate for the loss of B. cellulosilyticus-mediated arabinan degradation.
Acclimation to the presence of a potential competitor alleviates resource conflict
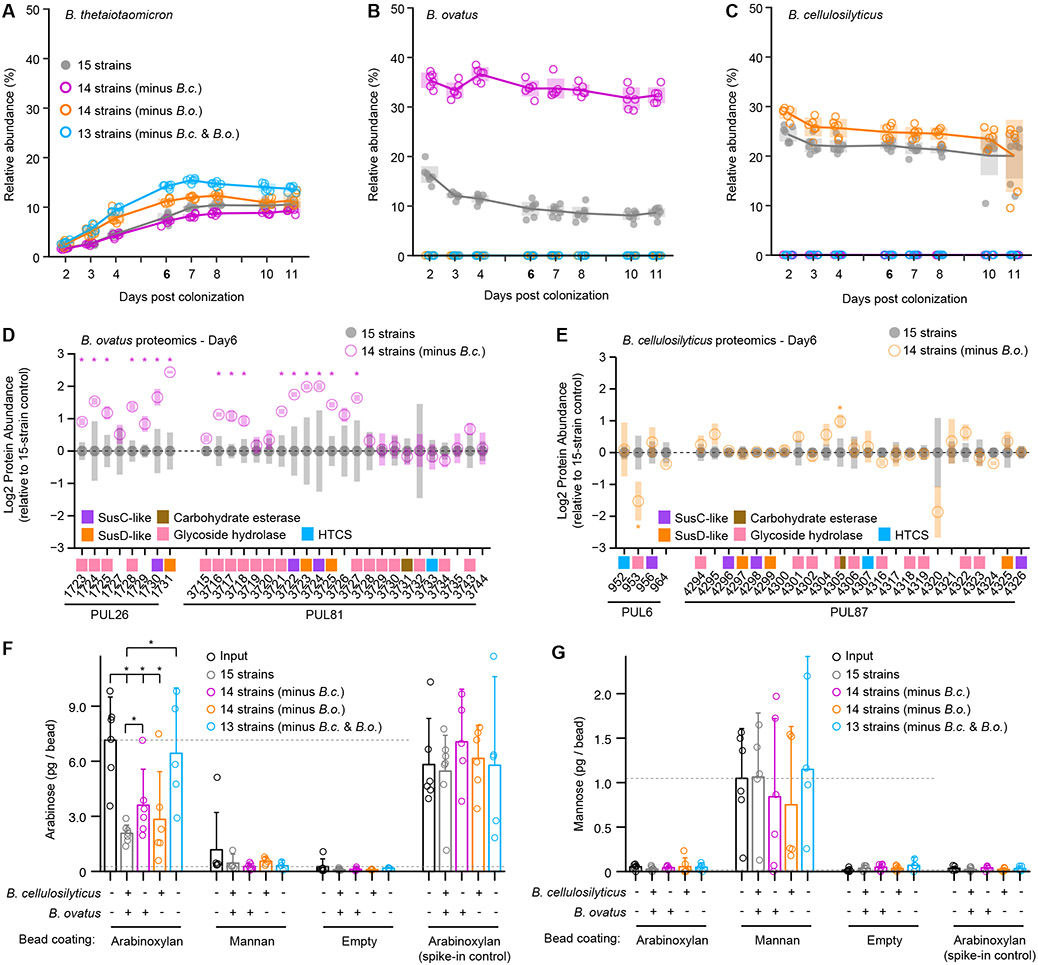
The in vivo degradation assay revealed that in contrast to arabinan, the capacity of the community to process arabinoxylan was not fully rescued by other species in the absence of B. cellulosilyticus (Figure 4D; Table S6). This was unexpected, given that B. cellulosilyticus omission resulted in a significant increase in the relative abundance of B. ovatus (Figure 5A; Table S3), which encodes PULs capable of arabinoxylan breakdown (Martens et al., 2011; Rogowski et al., 2015). We examined whether these results could arise from a type of interspecies relationship between B. cellulosilyticus and B. ovatus distinct from that observed between B. cellulosilyticus and B. vulgatus.
Figure 5. Detecting acclimation to the presence of a potential competitor using proteomics and forward genetics.
(A,B) Relative abundance of each bacterial strain in fecal samples from gnotobiotic mice harboring derivatives of the 15-member community. Mice received the control HiSF-LoFV diet in the presence (grey closed circles) or absence of B. cellulosilyticus (B.c.) or B. vulgatus (open circles; magenta and brown respectively). Key: circles, individual mice; lines, mean values; shading, 95%CI.
(C,D) Protein abundance and INSeq data for genes in arabinoxylan PULs shown along the x-axis (as locus tag number only; Bovatus_0XXXX). Mean values ± SD (vertical shading) are indicated (n=5 animals/treatment group). Genes are color-coded according to functional annotation. Key for circles: grey, 15-member community; magenta or brown, mice harboring communities without B.c. or B. vulgatus, respectively. *, P < 0.05, ∣fold change∣ > log2(1.2) [plus B.c. versus minus B.c.); limma or limma-voom].
(E) Mean protein abundance in fecal samples from day 6 in the minus B.c. group relative to the plus B.c. group; proteins whose levels are significantly different between the groups and encoded by genes in PULs are highlighted with open circles, while those encoded by genes in arabinoxylan processing PULs are labeled with their PUL number.
(F) INSeq analysis showing the mean change in abundance of mutant strains from experimental day 2 to day 6 in the minus B. c. group relative to the plus B. c. group. Genes in PULs that have a significant effect on fitness are highlighted with open circles; those located in arabinoxylan processing PULs are labeled with their PUL number.
As discussed above, the abundances of B. vulgatus proteins involved in pea fiber or citrus pectin degradation were unchanged upon removal of its competitor B. cellulosilyticus. In contrast, B. ovatus exhibited metabolic flexibility, with proteins encoded by two arabinoxylan-processing PULs (PUL26 and PUL81) predominating among those whose abundances were increased when B. cellulosilyticus was absent versus present (Figures 5C and 5E; Table S4B-S4G). This effect was apparent regardless of whether mice were fed the pea fiber, citrus pectin, or only the control HiSF-LoFV diet, consistent with the presence of arabinoxylan in the HiSF-LoFV diet (Table S4B-S4E). When we analyzed the contributions of genes to the fitness of B. ovatus (by calculating the changes in the abundance of Tn mutant strains from day 2 to day 6), we found that those in these two arabinoxylan PULs were the most affected by omission of B. cellulosilyticus (Figures 5D and 5F; Table S5B). This result indicates that B. ovatus exhibits a marked decrease in its reliance on arabinoxylan in the full 15-member community context. Omission of B. vulgatus did not induce changes in the relative abundance of B. ovatus, the levels of proteins encoded by its PULs, or in the fitness cost associated with mutations in its PULs (Figures 5A, 5C, and 5D; Tables S3D, S4F, and S5B).
Monosaccharide and linkage analysis verified that arabinoxylan was present in the HiSF-LoFV diet; this conclusion was based on finding abundant 4-linked xylose with branching 4,3-linked xylose, and terminal arabinose (Table S1). We also detected small amounts of 3-linked glucose (indicative of hemicellulose beta-glucans), galacturonic acid and rhamnose, consistent with the increase in abundance of proteins in B. ovatus PULs shown or predicted to process beta-glucan, rhamnogalacturonan, and host glycan when B. cellulosilyticus is present (Figure S5; Table S4).
Based on these results, we reasoned that metabolic flexibility allows B. ovatus to acclimate to the presence of B. cellulosilyticus by shifting its nutrient harvesting strategies, de-emphasizing arabinoxylan degradation, and mitigating competition between the two species. To test this notion further, we performed an experiment omitting B. cellulosilyticus, B. ovatus, or both species from the 15-member consortium introduced into micefed the base HiSF-LoFV(Table S3E). Confirming our earlier results, COPRO-Seq revealed that the abundance of B. ovatus was increased in the absence of B. cellulosilyticus (Figure 6B; Tables S3B-S3E). Proteomics analysis of fecal samples obtained on experimental day 6 also revealed an increase in the abundance of 16 proteins encoded by arabinoxylan-processing PULs 26 and 81 in B. ovatus when B. cellulosilyticus was removed (Figure 6D; Tables S4B-S4G). In contrast, the abundance of B. cellulosilyticus as a proportion of the remaining strains did not increase (Figure 6C; Table S3E), and one protein specified by the arabinoxylan-processing PUL in B. cellulosilyticus (PUL 87) increased in abundance when B. ovatus was absent (Figure 6E; Table 5G). These results, combined with the observation that arabinoxylan-processing genes are important for fitness of B. ovatus only when B. cellulosilyticus is absent (Figure 5E), indicate that the metabolic flexibility of B. ovatus mitigates competition between two species that have the capacity to process the same dietary fiber resource.
Figure 6. Alleviation of competition between arabinoxylan-consuming Bacteroides.
(A-C) Relative abundance of each bacterial strain in fecal samples from gnotobiotic mice harboring derivatives of the 15-member community. Animals fed the control HiSF-LoFV diet in the presence (closed circles) or absence of B. cellulosilyticus (B.c.) or B. ovatus (B.o.) or both species (open circles; magenta, orange, or cyan respectively). Key: circles, individual mice; lines, mean values; shading, 95%CI.
(D,E) Mean protein abundances in fecal samples obtained on experimental day 6. Genes in B.o. and B. c. arabinoxylan-processing PULs are shown along the x-axis (as locus tag number only; Bovatus_0XXXX or BcellWH2_0XXXX) relative to full 15-member community condition. Mean values ± SD (vertical shading) are indicated (n=5-7 animals/treatment group). Genes are color-coded according to functional annotation (see key). Key for circles: grey, 15-member community; magenta, orange, or cyan, mice harboring communities without B.c., B.o., or both species, respectively. *, P < 0.05 [15-member community versus 14-member (minus B. cellulosilyticus); limma].
(F,G) Bead-based assay of polysaccharide degradation in mice fed the HiSF-LoFV diet and colonized with the complete 15-member community, with or without B.c., B.o., or both species. (Table S3E). The mass of bead-associated arabinose (panel F) or mannose (panel G) is plotted before (black) and after exposure to the indicated communities (grey, complete 15-member community; magenta, minus B.c. group; orange, minus B.o. group; cyan, minus B.c. and B.o. group). Circles denote individual mice. Mean values + 95%CI are shown (n=5-7 animals/group). *, P < 0.05 (Mann-Whitney U test).
We sought to directly measure the functional outcome of metabolic flexibility in B. ovatus and establish that this species degraded arabinoxylan in the community lacking B. cellulosilyticus. Arabinoxylan-beads, as well as empty and yeast alpha-mannan-coated control beads, were administered to the four groups of mice described above. In the absence of B. cellulosilyticus, significant degradation of arabinoxylan was detected (Figure 6F), consistent with our previous observations (Table S6). No change in degradation occurred when B. ovatus was omitted (Figure 6F), as expected based on the expression of arabinoxylan PULs by B. cellulosilyticus. However, arabinoxylan-coated beads recovered from mice lacking B. ovatus and B. cellulosilyticus were indistinguishable from input beads (Figure 6F). In addition, omission of both B. cellulosilyticus and B. ovatus did not produce significant increases in the proportions of the remaining strains relative to one another (Table S3E), suggesting that these other species were unable to take advantage of the available arabinoxylan resources in the diet. None of the community contexts examined produced significant decreases in bead-bound mannan, controlling for non-specific polysaccharide degradation (Figure 6G). As an additional ‘spike-in’ control, we added arabinoxylan beads to cecal and fecal samples obtained from all groups of mice immediately after they were euthanized and processed them in parallel with the orally administered beads. The preservation of carbohydrate on spike-in beads established that B. cellulosilyticus/B. ovatus-dependent degradation occurred during intestinal transit and not sample processing (Figure 6F).
Together, these experiments show that, in contrast to the persistent competition for arabinan and homogalacturonan exhibited by B. vulgatus, B. ovatus avoids competition for arabinoxylan via acclimation to the presence of its potential competitor, B. cellulosilyticus. This conclusion is based on several observations; (i) the HiSF-LoFV diet contains arabinoxylan polysaccharides (ii) omission of B. ovatus did not cause detectable expansion of B. cellulosilyticus, (iii) proteins encoded by B. ovatus arabinoxylan PULs were significantly increased when B. cellulosilyticus was absent, (iv) genes in B. ovatus arabinoxylan PULs were more important for fitness when B. cellulosilyticus was absent, and (v) B. ovatus was responsible for the residual arabinoxylan degradation that took place in the absence of B. cellulosilyticus.
Discussion
In this era of precision medicine, as knowledge of the effects of the gut microbial community on human biology expands, and appreciation of how microbiomes exhibit substantial interpersonal variation emerges, understanding how to precisely and effectively manipulate community properties becomes an important goal. Combining high resolution proteomics, forward genetic screens, and glycan-coated artificial food particles in gnotobiotic mice fed ‘representative’ high-fat, low-fiber USA diet has allowed us to characterize how human gut Bacteroides with distinct nutrient harvesting capacities respond to food-grade fibers. Our approach identified bioactive components in compositionally complex fibers that impact specific members of the microbiota. Obtaining this type of information could inform efforts to enrich for these active components through judicious selection of cultivars of a given food staple, food processing methods, or existing waste streams from food manufacturing.
Deliberately manipulating the membership of a consortium of cultured, sequenced human-donor derived microbes introduced into gnotobiotic mice provides an opportunity to determine whether and how organisms compete and what mechanisms they use to avoid competition. Simultaneous harvest of a particular dietary resource by two species is theoretically possible whenever they both contain a genetic apparatus sufficient for metabolism of that resource. We provide evidence that competition for particular glycans in fibers is realized in such a model community, since glycan-degrading genes were expressed and required for fitness in both species, and negative interactions were observed in strain omission experiments. These omission experiments disclosed distinct relationships between B. vulgatus, B. ovatus and B. cellulosilyticus; namely, the ability of B. ovatus to acclimate to the presence of a competitor (B. cellulosilyticus) as opposed to the persistent competition between B. vulgatus and B. cellulosilyticus for the same resource. A healthy human gut microbiota has great strain-level diversity. Determining which strains representing a given species to select as a lead candidate probiotic agent, or for incorporation into synbiotic (prebiotic plus probiotic) formulations, is a central challenge for those seeking to develop next generation microbiota-directed therapeutics. Identifying organisms with metabolic flexibility, as opposed to those that are more prone to competing with other community members, could contribute to understanding how certain strains are capable of coexisting with the residents of diverse human gut communities.
Particles present in foods prior to consumption, or generated by physical and biochemical/enzymatic processing of foods during their transit through the gut, provide community members with opportunities to attach to their surfaces, and harvest surface-exposed nutrient resources. The ability of organisms to adhere to such particles and the physical partitioning their component nutrients can be envisioned as affecting competition, conflict avoidance, and cooperation. The ability of a given gut microbial community to degrade different fiber components was quantified in our studies using artificial food particles composed of fluorescently labeled, paramagnetic microscopic beads coated with different polysaccharides. This approach provides an additional dimension for characterizing the functional properties of a microbial community, and has a number of advantages. First, the measurement of polysaccharides coupled to magnetic beads is not confounded by the presence in the gut of structurally similar (or even identical) dietary, host, or microbial polysaccharides. Second, this technology, when applied to gnotobiotic mice, permits simultaneous testing of multiple glycans in the same animal, allowing a direct comparison of the degradative capabilities of different assemblages of human gut microbes in vivo. For example, we were able to demonstrate non-redundant arabinoxylan degradation carried out by B. cellulosilyticus in this community, despite the presence of another arabinoxylan consumer, B. ovatus. Third, applied directly to humans, these diagnostic ‘biosensors’ could be used to quantify functional differences between their gut microbiota, and physical associations between carbohydrates and strains of interest, as a function of host health status, nutritional status/interventions, or other perturbations. As such, results obtained with these biosensors could facilitate ongoing efforts to use machine learning algorithms that integrate a variety of parameters, including biomarkers of host physiologic state and features of the microbiota, to develop more personalized nutritional recommendations (Zeevi et al., 2015). Lastly, this technology could be used to advance food science. The bead coating strategy we employed was successful with over 30 commercially available polysaccharides and we have extended the assay to measure the degradation of other biomolecules, including proteins. Particles carrying components of food that have been subjected to different processing methods, or particles bearing combinations of nutrients designed to attract different sets of primary (and secondary) microbial consumers could also be employed in preclinical models to develop and test food prototypes optimized for processing by microbiota representative of different targeted human populations.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeffrey I. Gordon (jgordon@wustl.edu).
Experimental Model and Subject Details
Gnotobiotic mice –
All experiments involving mice were carried out in accordance with protocols approved by the Animal Studies Committee of Washington University in St. Louis. For screening different fiber preparations, germ-free male C57BL/6J mice (10-16 weeks-old) were singly housed in cages located within flexible plastic isolators. Cages contained paper houses for environmental enrichment. Animals were maintained on a strict light cycle (lights on at 0600 h, off at 1900 h). Mice were fed a LoSF-HiFV diet for five days prior to colonization. After colonization, the community was allowed to stabilize on the LoSF-HiFV diet for an additional five days. One group of control mice remained on this diet for the rest of the experiment and a second control group was switched to the HiSF-LoFV diet for the rest of the experiment.
Mice in the experimental group first received an introductory diet containing equal parts of all fiber preparations employed in a given screen (totaling 10% of the diet by weight), and then received a series of diets containing different fiber preparations as described in Figure 1A. A 10 g aliquot of a given diet/fiber mixture was hydrated with 5 mL sterile water in a gnotobiotic isolator; the resulting paste was pressed into a feeding dish and placed on the cage floor. Food levels were monitored nightly, and a freshly hydrated aliquot of that diet was supplied every two days (preventing levels from dropping below roughly one third of the original volume). Bedding (Aspen Woodchips; Northeastern Products) was replaced after each 7-day diet period to prevent any spilled food from being consumed during the next diet exposure. Fresh fecal samples were collected from each animal within seconds of being produced on days 1, 3, 6, and 7 of every diet period, and placed in liquid nitrogen within 45 min. Pre-colonization fecal samples were collected to verify the germ-free status of mice.
For monotonous feeding experiments, germ-free male C57BL/6J mice (10-16 weeks-old) were fed the control HiSF-LoFV diet in its pelleted form for two weeks prior to colonization. Two days after colonization, mice were switched to paste diets containing 10% of the powdered fiber preparation mixed into the base diet (or the base diet in paste form without added fiber) for the remainder of the experiment. As noted above, these diets were delivered in freshly hydrated aliquots every two days. Fecal samples, including those obtained prior to colonization, were collected on the days indicated in the supplemental tables.
Defined microbial communities –
The screening experiments used cultured, sequenced bacterial strains obtained from a fecal sample that had been collected from a lean co-twin in an obesity-discordant twin-pair [Twin Pair 1 in (Ridaura et al., 2013); also known as F60T2 in (Faith et al., 2013)]; Bacteroides caccae TSDC17.2-1.2, Bacteroides finegoldii TSDC17.2-1.1, Bacteroides intestinalis TSDC17.2-1.1, Bacteroides massiliensis TSDC17.2-1.1, Bacteroides ovatus TSDC17.2-1.1, Bacteroides thetaiotaomicron TSDC17.2-2.2, Bacteroides vulgatus TSDC17.2-1.1, Bacteroides vulgatus TSDC17.2-2.1, Collinsella aerofaciens TSDC17.2-1.1, Collinsella aerofaciens TSDC17.2-2.1, Dorea longicatena TSDC17.2-1.1, Peptococcus niger TSDC17.2-1.1, Escherichia coli TSDC17.2-1.2, Odoribacter splanchnicus TSdC17.2-1.2, Parabacteroides distasonis TSDC17.2-1.1, Ruminococcaceae TSDC17.2-1.2, Ruminococcus albus TSDC17.2-1.1, Ruminococcus albus TSDC17.2-1.4, Ruminococcus bromii TSDC17.2-1.1, Subdoligranulum variabile TSDC17.2-1.1. Isolates were grown to stationary phase at 37° C in TYGS medium (Goodman et al., 2009) in an anaerobic chamber (atmosphere; 75% N2, 20% CO2, 5% H2). Equivalent numbers of organisms were pooled (based on OD600 measurements). The pool was divided into aliquots that were frozen in TYGS/15% glycerol, and maintained at −80° C until use. On experimental day 0, aliquots were thawed, the outer surface of their tubes were sterilized with Clidox (Pharmacal) and the tubes were introduced into gnotobiotic isolators. The bacterial consortium was administered through a plastic tipped oral gavage needle (total volume, 400μL per mouse). Based on inconsistent colonization observed in screening experiment 1 (Table S2A), one isolate (Enterococcus fecalis; average relative abundance, 2.1%) was not included in screening experiments 2 and 3.
Model communities containing INSeq libraries –
Ten strains selected from the human donor-derived community described above were colony purified, and each frozen in 15% glycerol and TYGS medium. Recoverable CFUs/mL were quantified by plating on brain-heart-infusion (BHI) blood agar. The identity of strains was verified by sequencing full-length 16S rRNA amplicons. On the day of gavage, stocks of these strains were thawed in an anaerobic chamber and mixed together along with each of five multi-taxon INSeq libraries (B. thetaiotaomicron VPI-5482, B. thetaiotaomicron 7330, B. cellulosilyticus WH2, B. vulgatus ATCC-8482, B. ovatus ATCC-8483) whose generation and characterization have been described in earlier publications (Hibberd et al., 2017; Wu et al., 2015). An aliquot of this mixture was administered by oral gavage to germ-free mice housed in gnotobiotic isolators (2×106 CFUs of each donor organism plus an OD600 0.5 of each INSeq library per mouse recipient; total gavage volume, 400 μL). For B. cellulosilyticus, B. vulgatus, B. ovatus, or B. cellulosilyticus and B. ovatus omission experiments, gavage mixtures were prepared in parallel without these organisms. The absence of one or both of these strains was verified by COPRO-Seq analysis of both the gavage mixture and fecal samples collected throughout the experiment from recipient mice.
Method Details
Fiber-rich food ingredient mixtures –
HiSF-LoFV and LoSF-HiFV diets were produced using human foods, selected based on consumption patterns from the National Health and Nutrition Examination Survey (NHANES) database (Ridaura et al., 2013). Diets were milled to powder (D90 particle size, 980 mm), and mixed with pairs of powdered fiber preparations [one preparation at 8% (w/w) and the other preparation at 2% (w/w)]. Fiber content was defined for each preparation [Association of Official Agricultural Chemists (AOAC) 2009.01], as was protein, fat, total carbohydrate, ash, and water content [protein AOAC 920.123; fat AOAC 933.05; ash AOAC 935.42; moisture AOAC 926.08; total carbohydrate (100 - (Protein + Fat + Ash + Moisture)]. The powdered mixtures were sealed in containers and sterilized by gamma irradiation (20-50 kilogreys, Steris, Mentor, OH). Sterility was confirmed by culturing the diet under aerobic and anaerobic conditions (atmosphere, 75% N2, 20% CO2, 5% H2) at 37°C in TYG medium, and by feeding the diets to germ-free mice followed by COPRO-Seq analysis of their fecal DNA.
Monosaccharide and linkage analysis of polysaccharides –
For fiber preparations, uronic acid (as GalA) was measured using the m-hydroxybiphenyl method (Thibault, 1979). Sodium tetraborate was used to distinguish GlcA and GalA (Filisetti-Cozzi and Carpita, 1991). The degree of methylation of galacturonic acid (pectins) in the sample was estimated as previously described (Levigne et al., 2002). Samples were hydrolyzed with 1M H2SO4 for 2h at 100°C and individual neutral sugars were analyzed as their alditol acetate derivatives (Englyst and Cummings, 1988) by gas chromatography. To fully release glucose from cellulose, a prehydrolysis step was carried out by incubation in 72% H2SO4 for 30 minutes at 25°C prior to the hydrolysis step. Linkage analysis was performed after carboxyl reduction of uronic acid with NaBD4/NaBH4 according to a previously published procedure (Pettolino et al., 2012) with minor modifications (this procedure allows galactose, galacturonic acid and methylesterified galacturonic acid to be distinguished). Methylation of carboxyl-reduced samples was performed as described in (Buffetto et al., 2015).
Polysaccharides from the HiSF-LoFV diet were isolated by sequential alkaline extractions (Pattathil et. al., 2012). Briefly, lipids were removed from a sample of powdered HiSF-LoFV by sequential incubation in 80% ethanol, 100% ethanol, and acetone. The dried precipitate was suspended in 1M KOH containing 0.5%(w/w) NaBH4 and stirred overnight. The solution was neutralized and the supernatant was collected by centrifugation (this material is referred to as fraction 1 (F1)). The insoluble material was suspended in 1M KOH/ 0.5%(w/w) NaBH4 overnight, and the supernatant was collected (referred to as F2). The insoluble material was suspended in 4M KOH/0.5%(w/w) NaBH4 overnight and the supernatant was collected (referred to as F3). Each fraction was dialyzed (SnakeSkin 3.5K MWCO, Thermo Scientific) in water, lyophilized, and then treated for 4 hours at 37° C with amyloglucosidase (36 units/mg) and alpha-amylase (100 units/mg; both enzymes from Megazyme). Enzymes were inactivated by boiling and samples were dialyzed and lyophilized. Measurement of the dry mass of each fraction before and digestion revealed that the total starch content of the base HiSF-LoFV diet was 22% (w/w) (note a comparable analysis the pea fiber yielded a value of 3.6%, meaning that HiSF-LoFV diet supplemented with 10% pea fiber contains a total starch content of 20% by weight).
HiSF-LoFV diet polysaccharides were analyzed by the Center for Complex Carbohydrate Research at the University of Georgia in Athens. Glycosyl composition analysis was performed by combined GC-MS of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methyl glycosides produced from the sample by acidic methanolysis (Santander et al., 2013). Briefly, samples (300-500 μg) were heated with methanolic HCl in a sealed screw-top glass test tube for 17 h at 80 °C. After cooling and removal of the solvent under a stream of nitrogen, samples were derivatized with Tri-Sil® (Pierce) at 80 °C for 30 min. GC-MS analysis of the TMS methyl glycosides was performed on an Agilent 7890A GC interfaced to a 5975C mass selective detector (MSD), using a Supelco Equity-1 fused silica capillary column (30 m × 0.25 mm ID).
Glycosyl linkage analysis of HiSF-LoFV diet polysaccharides was performed as previously described with slight modification (Heiss et. al., 2009). Samples were permethylated, depolymerized, reduced and acetylated, and the resulting partially methylated alditol acetates (PMAAs) were analyzed by GC-MS. About 1 mg of the sample was used for linkage analysis. The sample was suspended in 200 μL of dimethyl sulfoxide and left to stir for 1 day. Permethylation of the sample was affected by two rounds of treatment with sodium hydroxide (15 minutes) and methyl iodide (45 minutes). The permethylated material was hydrolyzed using 2 M TFA (2 hours in sealed tube at 121 °C), reduced with NaBD4, and acetylated using acetic anhydride/TFA. The resulting PMAAs were analyzed on an Agilent 7890A GC interfaced to a 5975C MSD (electron impact ionization mode); separation was performed on a 30 m Supelco SP-2331 bonded phase fused silica capillary column.
V4-16S rRNA gene sequencing –
DNA was isolated from fecal samples by first bead-beating the sample with 0.15mm-diameter zirconium oxide beads and a 5mm-diameter steel ball in 2X buffer A (200 mM NaCl, 200 mM Tris, 20 mM EDTA), followed by extraction in phenol:chloroform:isoamyl alcohol, and further purification (QiaQuick 96 purification kit; Qiagen, Valencia, CA). PCR amplification of the V4 region of bacterial 16S rRNA genes was performed as described (Bokulich et al., 2013). Amplicons with sample-specific barcodes were pooled for multiplex sequencing using an Illumina MiSeq instrument. Reads were demultiplexed and rarefied to 5000 reads per sample. Reads sharing ≥99% nucleotide sequence identity [99% ID operational taxonomic units (OTUs)], that mapped to a reference OTU in the GreenGenes 16S rRNA gene database (McDonald et al., 2012) were assigned to that OTU. The 16S rRNA gene could not be amplified in multiple fecal DNA samples from mice fed 8% cocoa fiber. A small subset of reads (<5%) representing additional V4-16S rDNA amplicon sequences produced from colony-purified stocks of Bacteroides ovatus, Parabacteroides distasonis, Dorea longicatena, and Collinsella aerofaciens were omitted from our analyses of fecal DNA samples. Streptococcus thermophilus, an organism heavily used in cheese processing, was also omitted based on its detection in DNA isolated from samples of the sterile HiSF-LoFV diet.
COPRO-Seq analyses of bacterial species abundances –
Libraries were prepared from fecal DNA using sonication and addition of paired-end barcoded adaptors (McNulty et al., 2013) or by tagmentation using the Nextera DNA Library Prep Kit (Illumina) and combinations of custom barcoded primers (Adey et al., 2010). Libraries were sequenced using an Illumina NextSeq instrument [1,011,017± 314,473 reads/sample (mean ± SD) across experiments]. Reads were mapped to bacterial genomes with previously published custom Perl scripts (see below and in Key Resources Table) adapted to use Bowtie II for genome alignments (Hibberd et al., 2017); samples represented by less than 150,000 uniquely mapped reads were omitted from the analysis.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Bacteroides caccae TSDC17.2-1.2, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Bacteroides finegoldii TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Bacteroides intestinalis TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Bacteroides massiliensis TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Bacteroides ovatus TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Bacteroides thetaiotaomicron TSDC17.2-2.2, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Bacteroides vulgatus TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Bacteroides vulgatus TSDC17.2-2.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Collinsella aerofaciens TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Collinsella aerofaciens TSDC17.2-2.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Dorea longicatena TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Peptococcus niger TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Escherichia coli TSDC17.2-1.2, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Odoribacter splanchnicus TSDC17.2-1.2, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Parabacteroides distasonis TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Ruminococcaceae TSDC17.2-1.2, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Ruminococcus albus TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Ruminococcus albus TSDC17.2-1.4, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Ruminococcus bromii TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| Subdoligranulum variabile TSDC17.2-1.1, from bacterial culture collection, donor fecal sample F60T2 | Faith, et al. 2013; Ridaura et al., 2013; Ahern, Faith, et al. 2014 | N/A |
| INSeq library (B. thetaiotaomicron VPI-5482) | Wu, et al. 2015; | N/A |
| INSeq library (B. cellulosilyticus WH2) | Wu, et al. 2015; | N/A |
| INSeq library (B. thetaiotaomicron 7330) | Wu, et al. 2015; | N/A |
| INSeq library (B. ovatus ATCC-8483) | Wu, et al. 2015; | N/A |
| INSeq library (B. vulgatus ATCC-8482) | Hibberd, et al. 2017 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Wheat arabinoxylan | Megazyme | Cat# P-WAXYL |
| Lichenan | Megazyme | Cat# P-LICHN |
| Yeast alpha-mannan | Sigma | Cat# M7504 |
| TFPA-PEG3-Biotin | ThermoFisher | Cat# 21303 |
| Myo-Inositol-D6 | CDN Isotopes | Cat# D3019 |
| Biotin PF-415 | Promofluor | Cat# PK-PF415-6-01 |
| Biotin PF-505 | Promofluor | Cat# PK-PF505-6-01 |
| Biotin PF-510-LSS | Promofluor | Cat# PK-PF510LSS-6-01 |
| Biotin PF-633P | Promofluor | Cat# PK-PF633P-6-01 |
| MmeI | NEB | Cat# R0637L |
| T4 DNA Ligase | NEB | Cat# NEM0202M |
| Pfx Polymerase | ThermoFisher | Cat# INV11708013 |
| T4 Polynucleotide kinase | ThermoFisher | Cat# PRM4101 |
| Phusion HF master mix | ThermoFisher | Cat# F531LPM |
| Critical Commercial Assays | ||
| Nextera DNA library prep kit | Illumina | Cat# 20018705 |
| Microplate BCA assay kit | ThermoFisher (Pierce) | Cat# 23235 |
| MinElute Gel Extraction Kit | Qiagen | Cat# 28604 |
| ZEBA spin desalting columns, 7K | ThermoFisher | Cat# 89882 |
| CountBright absolute counting beads | ThermoFisher | Cat# C36950 |
| Percoll PLUS | GE Healthcare | Cat# 17-5445-02 |
| Quant-iT dsDNA assay kit, high sensitivity | ThermoFisher | Cat# Q33120 |
| Deposited Data | ||
| V4-16S rRNA sequences | This study | PRJEB26564 |
| COPRO-Seq shotgun sequences | This study | PRJEB26564 |
| INSeq insertion site sequences | This study | PRJEB26564 |
| Whole-community LC-MS/MS proteomics data | This study | MSV000082287; PXD009535 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J mice (re-derived germ-free) | The Jackson Laboratory | Cat# 000664 |
| Oligonucleotides | ||
| MmeI oligo M12 (double stranded) CTG TCC GTT CCG ACT ACC CTC CCG AC | Wu, et al. 2015 | N/A |
| INSeq PCR (forward) CAA GCA GAA GAC GGC ATA CG | Wu, et al. 2015 | N/A |
| INSeq PCR (reverse) AAT GAT ACG GCG ACC ACC GAA CAC TCT TTC CCT ACA CGA | Wu, et al. 2015 | N/A |
| COPRO-Seq PCR (forward) AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC T | Hibberd, et al. 2017 | N/A |
| COPRO-Seq PCR (reverse) CAA GCA GAA GAC GGC ATA CGA GAT CGG TCT CGG CAT TCC TGC TGA ACC GCT CTT CCG ATC T | Hibberd, et al. 2017 | N/A |
| Software and Algorithms | ||
| QIIME v1.9.0 | Caporaso, et al. 2010 | http://qiime.org |
| COPRO-Seq pipeline | Hibberd, et al. 2017 | https://github.com/nmcnulty/COPRO-Seq |
| INSeq pipeline | Wu, et al. 2015 | https://github.com/mengwu1002/Multi-taxon_analysis_pipeline |
| MyriMatch v2.2 | Tabb, et al. 2007 | N/A |
| IDPicker v3.0 | Clarkson, et al. 2017 | N/A |
| R | The R foundation | https://www.r-project.org/ |
| FlowJo v10.5.3 | FlowJo LLC | www.flowjo.com |
| Prism v7.0 | Graphpad | www.graphpad.com |
| Other | ||
| HiSF-LoFV and LoSF-HiFV mouse diets | Ridaura, et al. 2013 | N/A |
| Citrus pectin | Creamsa | Cat# Creampectin RS 47000 |
| Pea fiber | Rettenmaier | Cat# Pea Fiber EF 100 |
| Citrus peel | Herbafoods Ingredients Gmbh | Cat# Citrus Peel |
| Yellow mustard | Sakai Spice Corporation | Cat# Yellow Mustard Bran |
| Soy cotyledon | Solae Europe S.A. | Cat# Fibrim 1270 IP Soy Fiber |
| Orange fiber (Coarse) | Fiber Star | Cat# CitriFi 100 |
| Orange fiber (Fine) | Fiber Star | Cat# CitriFi 100 M40 |
| Orange peel | Herbafoods Ingredients Gmbh | Cat# Herbacel Classic Orange |
| Tomato peel | Mondelēz | Cat# Experimental Tomato Peel |
| Inulin, low molecular weight | Beneo/ Orafti | Cat# P-95 Oligo- Fructose |
| Potato Fiber | Avebe Food | Cat# Paselli FP |
| Apple pectin | Herbafoods Ingredients Gmbh | Cat# Herbapek SF 50 LV |
| Oat hull fiber | Canadian Harvest | Cat# Oat Fiber 770 |
| Acacia extract | Nexira | Cat# Fibergum B |
| Inulin, high molecular weight | Beneo/ Orafti | Cat# Inulin, High Mw |
| Barley beta-glucan | DKSH | Cat# Glucagel |
| Barley bran | PolyCell Technologies | Cat# Concentrated Barley Beta Glucan |
| Oat beta-glucan | DSM | Cat# Oat β-Glucan |
| Apple fiber | Herbafoods Ingredients Gmbh | Cat# Herbacel AQ Plus Apel A |
| Rye bran | Dr. Suwelack | Cat# Micronized Rye Bran |
| Barley malted | Mondelēz | Cat# Experimental Milled Spent Grains |
| Wheat aleurone | Kampffmeyer | Cat# Goldlader Weizen Aleuron |
| Wheat bran | Mondelēz | Cat# Experimental Wheat Bran |
| Resistant maltodextrin | Tate & Lyle | Cat# Resistant Maltodextrin |
| Psyllium | Rettenmaier | Cat# Psyllium (P-95) |
| Cocoa | Barry Callebaut | Cat# Fat Reduced Cocoa Powder |
| Citrus fiber | Creamsa | Cat# Creamfibre 7000 F |
| Tomato pomace | Mondelēz | Cat# Experimental Tomato Pomace |
| Rice bran | RiBran Technologies | Cat# RiBran 100 |
| Chia seed | Glanbia Nutritionals | Cat# White Chia |
| Corn bran | Bunge | Cat# Corn Bran |
| Soy fiber | Canadian Harvest | Cat# Soy Fiber S-200 |
| Sugar cane fiber | Rettenmaier | Cat# Sugarcane SF 601 |
| Resistant starch 4 | Penford | Cat# Penfibre RS |
| PureProteome Streptavidin beads | MilliporeSigma | Cat# LSKMAGT |
Community-wide quantitative proteomics –
Lysates were prepared from fecal samples by bead beating in SDS buffer (4% SDS, 100 mM Tris-HCl, 10 mM dithiothreitol, pH 8.0) using 0.15 mm diameter zirconium oxide beads, followed by centrifugation at 21,000 x g for 10 minutes. Precleared protein lysates were further denatured by incubation at 85°C for 10 minutes, and adjusted to 30 mM iodoacetamide to alkylate reduced cysteines. After incubation in the dark for 20 minutes at room temperature, protein was isolated by chloroform-methanol extraction. Protein pellets were then washed with methanol, air dried, and re-solubilized in 4% sodium deoxycholate (SDC) in 100 mM ammonium bicarbonate (ABC) buffer, pH 8.0. Protein concentrations were measured using the BCA (bicinchoninic acid) assay (Pierce). Protein samples (250 μg) were then transferred to a 10 kDa MWCO spin filter (Vivaspin 500, Sartorius), concentrated, rinsed with ABC buffer, and digested in situ with sequencing-grade trypsin (Clarkson et al., 2017). The tryptic peptide solution was then passed through the spin-filter membrane, adjusted to 1% formic acid to precipitate the remaining SDC, and the precipitate removed from the peptide solution with water-saturated ethyl acetate. Peptide samples were concentrated using a SpeedVac, measured by BCA assay and analyzed by automated 2D LC-MS/MS using a Vanquish UHPLC with autosampler plumbed directly in-line with a Q Exactive Plus mass spectrometer (Thermo Scientific) outfitted with a 100 μm ID triphasic back column [RP-SCX-RP; reversed-phase (5 μm Kinetex C18) and strong-cation exchange (5 μm Luna SCX) chromatographic resins; Phenomenex] coupled to an in-house pulled, 75 μm ID nanospray emitter packed with 30 cm Kinetex C18 resin. For each sample, 12 μg of peptides were autoloaded, desalted, separated and analyzed across four successive salt cuts of ammonium acetate (35, 50, 100 and 500 mM), each followed by a 105-minute organic gradient. Eluting peptides were measured and sequenced by data-dependent acquisition on the Q Exactive Plus (Clarkson et al., 2017).
MS/MS spectra were searched with MyriMatch v.2.2 (Tabb et al., 2007) against a proteome database derived from the genomes of the strains in the defined model community concatenated with major dietary protein sequences, common protein contaminants, and reversed entries to estimate false-discovery rates (FDR). Since the relative abundance of B. thetaiotaomicron 7330 was low on day 6 [0.05% ± 0.041% (mean ± SD) for all groups], we chose to analyze all peptides that mapped to the B. thetaiotaomicron VPI-5482 proteome, regardless of whether they also mapped to B. thetaiotaomicron 7330. Peptide spectrum matches (PSM) were required to be fully tryptic with any number of missed cleavages, and contain a static modification of 57.0214 Da on cysteine and a dynamic modification of 15.9949 Da on methionine. PSMs were filtered using IDPicker v.3.0 (Ma et al., 2009) with an experiment-wide FDR <1% at the peptide-level. Peptide intensities were assessed by chromatographic area-under-the-curve (label-free quantification option in IDPicker). To remove cases of extreme sequence redundancy, the community meta-proteome was clustered at 100% sequence identity post-database search [UCLUST; (Edgar, 2010)] and peptide intensities were summed to their respective protein groups/seeds to estimate overall protein abundance. Proteins were included in the analysis only if they were detected in more than 3 biological replicates in at least one experimental group. After considering only peptides that uniquely mapped to a single seed protein, the summed abundances proteins advanced to quantitative analysis yielded 59% from community members, 36% from mouse, and 2% from diet. Missing values were imputed to simulate the limit of detection of the mass spectrometer, using mean minus 2.2 x standard deviation with a width of 0.3 x standard deviation. Four additional imputed distributions produced results that were in general agreement with this approach in terms of fold-abundance change induced by fiber treatment and statistical significance.
Multi-taxon INSeq –
Multi-taxon INSeq allows simultaneous analysis of multiple mutant libraries in the same recipient gnotobiotic mouse owing to the fact that the mariner Tn vector contains MmeI sites at each end plus taxon-specific barcodes. MmeI digestion cleaves genomic DNA at a site 20-21 bp distal to the restriction enzyme’s recognition site so that the site of Tn insertion and the relative abundance of each Tn mutant can be defined in given diet/community contexts by sequencing the flanking genomic sequence and taxon-specific barcode (Wu et al., 2015). Purified fecal DNA was processed as described previously (Wu et al., 2015). DNA was digested with MmeI and the products were ligated to sample-specific barcoded adaptors. Sequencing was performed on an Illumina HiSeq 2500 instrument, with a custom indexing primer providing the strain-specific barcode for the insertion. Analysis of mutant strain frequencies was carried out using custom software (see below and in Key Resources Table). Log ratios of the abundances of Tn mutant strains on experimental days 6 and 2 (corresponding to the period of fiber treatment compared to just prior to fiber exposure) were calculated for each mouse.
PUL nomenclature and homology –
All PUL assignments used in the current study are summarized in Table S7; assignments were made based on “new assembly” genomes present in the CAZy PUL database (www.cazy.org/PULDB) (Terrapon et al., 2018). All boundaries of PULs were algorithmically defined (listed as ‘predicted PUL’ in PULDB). The algorithmically defined boundaries of B. thetaiotaomicron PUL7 were extended to include the adjacent arabinose operon based on previously published experimental datasets (Schwalm et al., 2016). A cluster of three or more adjacent CAZymes was defined as a ‘polysaccharide utilization complement’. Homology between genes in PULs was determined using a reciprocal BLASTp approach with an E-value threshold of 1 × 10−9, querying each protein product contained within a CAZy-annotated PUL against reference genomes from other species in the community.
Generation of glycan-coated magnetic beads –
Wheat Arabinoxylan and Icelandic Moss Lichenan were purchased from Megazyme (P-WAXYL, P-LICHN) and yeast alpha-mannan was purchased from Sigma-Aldrich (M7504). Polysaccharides were solubilized in water (at a concentration of 5mg/mL for pea fiber and 20 mg/mL for arabinoxylan and lichenan), sonicated and heated to 100°C for 1 minute, then centrifuged at 24,000 x g for 10 minutes to remove debris. TFPA-PEG3-biotin (Thermo Scientific), dissolved in DMSO (10 mg/mL) was added to the polysaccharide solution at a ratio of 1:5 (v/v). The sample was subjected to UV irradiation for 10 minutes (UV-B 306 nm, 7844 mJ total), and then diluted 1:4 to facilitate desalting on 7 kD Zeba spin columns (Thermo Scientific).
Biotinylated polysaccharide was mixed with one of several biotinylated fluorophores (PF- 505, PF-510LSS, PF-633, PF-415; all at a concentration of 50 ng/mL; all obtained from Promokine). A 500 μL aliquot of this preparation was incubated with 107 paramagnetic streptavidin-coated silica beads (LSKMAGT, Millipore Sigma) for 24 hours at room temperature. Beads were washed by centrifugation three times with 1 mL HNTB buffer (10mM HEPES, 150mM NaCl, 0.05% Tween-20, 0.1% BSA) followed by addition of 5 μg/mL streptavidin (Jackson Immunoresearch) in HNTB (30 min incubation at room temperature). Beads were washed as before and then incubated with 250 μL of the biotinylated polysaccharide preparation. The washing, streptavidin, and polysaccharide incubation steps were repeated three times. Bead preparations were assessed using an Aria III cell sorter (BD Biosciences) to confirm adequate labeling, and then analyzed by GC-MS (see below) to quantify the amount of carbohydrate bound.
Administration and recovery of beads –
Beads were incubated with 70% ethanol for 1 minute in a biosafety cabinet, then washed three times with 1 mL sterile HNTB using a magnetic stand. The different bead types were combined, diluted, and aliquoted to 107 beads per 650 μL HNTB in sterile Eppendorf microcentrifuge tubes. The number of beads in each aliquot was counted using an Aria III cell sorter and CountBright fluorescent microspheres (BD Bioscience). Tubes containing beads were introduced into gnotobiotic isolators and the beads were administered by oral gavage (600 μL per mouse). Separate aliquots of control beads, used to establish input carbohydrate content were stored in the dark at 37°C until collection of experimental beads from mouse fecal or cecal samples had been completed.
For germ-free mouse experiments, animals were fed the HiSF-LoFV diet for two weeks and then gavaged with beads; all fecal pellets were collected during the 4- to 12-hour interval that followed gavage. During this time period, bedding was removed and mice were placed on grated cage bottoms (with access to food and water); cage bottoms were placed just above a 0.5 cm deep layer of sterile water on the floor of the cage, to prevent pellets from drying. For colonized animals, cecal and colonic contents were collected four hours after administration of beads at the time of euthanasia. Recovered samples were immediately placed in sterile water on ice.
Fecal, cecal, and input samples were vortexed and filtered through nylon mesh (100 μm pore-diameter). The resulting suspension of luminal contents was layered over sterile Percoll Plus (GE Health Care) and centrifuged for 5 minutes at 500 x g. Beads were collected from underneath the Percoll layer and washed four times using a magnetic stand, each time with 1 mL fresh HNTB. Recovered beads were counted by flow cytometry as before, filtered through nylon mesh (40 μm pore diameter, BD Biosciences) and stored at 4°C overnight. Beads were sorted back into their polysaccharide types based on fluorescence using an Aria III sorter (average sort purity, 96%). Sorted samples were centrifuged (500 x g for 5 minutes) to pellet beads and the beads were transferred to a 96-well plate. All bead samples were incubated with 1% SDS / 6M Urea / HNTB for 10 minutes at room temperature to remove exogenous components, washed three times with 200 μL HNTB using a magnetic plate rack, and then stored overnight at 4°C prior to monosaccharide analysis.
Analysis of bead-bound glycan by GC-MS –
The number and purity of beads in each sorted sample was determined by taking an aliquot for analysis on the Aria III cell sorter. Equal numbers of beads from each sample were transferred to a new 96-well plate and the supernatant was removed with a magnetic plate rack. For acid hydrolysis, 200 μL of 2M trifluoroacetic acid and 250 ng/mL myo-inositol-D6 (CDN Isotopes; spike-in control) were added to each well, and the entire volume was transferred to 300 μL glass vials (ThermoFisher; catalog number C4008-632C). Another aliquot was taken to verify the final number of beads in each sample. Monosaccharide standards were included in separate wells and subjected to the hydrolysis protocol in parallel with the other samples. Vials were crimped with Teflon-lined silicone caps (ThermoFisher) and incubated at 100°C with rocking for 2 h. Vials were then cooled, spun to pellet beads, and their caps were removed. A 180 μL aliquot of the supernatant was collected and transferred to new 300 μL glass vials. Samples were dried in a SpeedVac for 4 hours, methoximated in 20 μL O-methoxyamine (15mg/mL pyridine) for 15 h at 37°C, followed by trimethylsilylation in 20 μL MSTFA/TMCS [N-Methyl-N-trimethylsilyltrifluoroacetamide/2,2,2-trifluoro-N-methyl-N-(trimethylsilyl)-acetamide, chlorotrimethylsilane] (ThermoFisher) for 1 h at 70° C. One half volume of heptane (20 μL) was added before loading the samples for injection onto a 7890B gas chromatography system coupled to a 5977B MS detector (Agilent). The mass of each monosaccharide detected in each sample of sorted beads was determined using monosaccharide standard curves. This mass was then divided by the final count of beads in each sample to produce a measurement of mass of recoverable monosaccharide per bead.
Quantification and Statistical Analysis
Using data from days 6 and 7 of each diet treatment, a mixed effects model was generated in the R programming environment for each species in each of three fiber screening experiments. The relative abundance of that species in feces (or the relative abundance scaled by fecal DNA yield) was used as the dependent variable, and the concentration of administered fiber (10 to 13 fibers tested per experiment), as well as experimental day were used as independent variables. Mixed effects models incorporated terms to describe repeated measures of individual mice. In rare cases where B. cellulosilyticus failed to colonize (7 of 49 mice), the animals were not considered biological replicates since they harbored a distinct microbiota; they were omitted from the models (data from these animals is available in Table S2). ANOVA (with Satterthwaite approximation for degrees of freedom) was performed to evaluate the significance of individual terms in models (FDR corrected P value cutoff of 0.01). Models were evaluated based on conditional R2 values (incorporating random factors) and plots of the residuals and Cook’s distance (no samples were excluded based on these assessments).
For COPRO-Seq analyses, differences between groups were assessed using mixed-effect models with time as a categorical variable, including day 2 as a pre-treatment time point. For omission experiments, the abundance of each strain as a proportion of all other strains except the omitted strain or strains was used for statistical tests. Significant terms in models were identified using ANOVA (FDR corrected P value cutoff of 0.05). Mann-Whitney Utest was used for analyses of individual time-points of interest.
For quantitative proteomics, significant differences in protein abundance were determined using limma (Ting et al., 2009). For multi-taxon INSeq analyses, mutant strain abundances were analyzed using limma-voom (Law et al., 2014) after quantile normalization. The general linear model framework in limma-voom allowed us to perform moderated t-tests to determine the statistical significance (P <0.05, FDR corrected) of differences in fitness in the context of the control versus fiber-supplemented diets. A Mann-Whitney U test was used to calculate significant differences in monosaccharide abundance between bead samples. All tests were two-tailed. The exact value of n (corresponding to the number of individual animals) for each experiment can be found in the supplementary tables describing that experiment, and the definitions of center and variance in figures is listed in the figure legends.
Data and Code Availability
Datasets of V4-16S rRNA sequences in raw format prior to post-processing and data analysis, plus COPRO-Seq and INSeq datasets have been deposited at the European Nucleotide Archive under study accession PRJEB26564. All LC-MS/MS proteomic data have been deposited into the MassIVE data repository under accession numbers MSV000082287 (MassIVE) and PXD009535 (ProteomeXchange). INSeq software: https://github.com/mengwu1002/Multi-taxon_analysis_pipeline. COPRO-Seq software: https://github.com/nmcnulty/COPRO-Seq.
Supplementary Material
Figure S1. Strain-level variability across arabinan-utilization loci in Bacteroides species (related to Figure 2).
(A,B,C) Alignment of B. thetaiotaomicron PUL7 (panel A), B. cellulosilyticus PUL5 (panel B), and B. vulgatus PUL27 (panel C) across multiple strains of each species. The direction of transcription is indicated by the arrowhead. The genes are labeled with their locus tag number and color-coded according to their functional annotation (see key). Shaded regions connecting genes denote (i) significant BLAST homology (E-value <10−5) and the percent amino acid identity (>50%) of their protein products (see key).
Figure S2. Proteomics and forward genetics identify homogalacturonan in citrus pectin as a nutrient source for multiple species (related to Figure 2).
(A) Schematic representation of polysaccharide structures detected in citrus pectin based on monosaccharide and linkage analyses (with stereochemistry of anomeric carbons inferred).(B-E) Adult C57BL/6J germ-free mice were colonized with a 15-member community consisting of INSeq libraries representing four Bacteroides species together with 10 additional bacterial strains used in screening experiments depicted in Figure 1. Relative abundance is shown for each bacterial strain at each of the indicated time points in mice fed the control HiSF-LoFV diet (grey), or the HiSF-LoFV diet supplemented with 10% (w/w) citrus pectin (blue). Circles denote individual mice, lines the mean value and shading the 95% CI (n=15 individually caged mice per group; results pooled from three independent experiments). *, P < 0.05 (Mann-Whitney Utest). (F-I) Proteomic and INSeq analyses of fecal samples collected on experimental day 6. On the x-axis, each dot denotes the mean value for the abundance of a single bacterial protein in samples from animals monotonously fed the citrus pectin-supplemented HiSF-LoFV diet (relative to controls fed the unsupplemented diet). The y-axis indicates the mean value for the differential enrichment of mutants with Tn disruptions in the gene encoding each protein in the citrus pectin versus HiSF-LoFV diet groups. Blue dots represent genes that are significantly affected by citrus pectin (P < 0.05, ∣fold change∣ > log2(1.2); limma or limma-voom) as judged by levels of their protein products or their contribution to fitness while open circles mark the subset of these genes that are encoded by PULs. Genes present in predicted homogalacturonan-processing PULs in B. thetaiotaomicron, B. cellulosilyticus and B. vulgatus are labeled with their PUL number as it appears in PULDB (Terrapon et al., 2018).
Figure S3. Deliberate manipulation of community composition demonstrates interspecies competition for homogalacturonan in citrus pectin (related to Figure 3).
(A,B) Adult C57BL/6J germ-free mice were colonized with the same defined community used for the experiments in Figure 2, with or without B. cellulosilyticus (B.c.). Relative abundance B. thetaiotaomicron and B. vulgatus in fecal samples obtained at the indicated time points from mice fed the control HiSF-LoFV diet or the diet supplemented with 10% (w/w) citrus pectin. These mice were colonized with a community that contained B. cellulosilyticus or one where it had been omitted (see key). Circles, individual mice; lines, mean values; shading, 95%CI (n=4-10 mice per group). *, P < 0.05 (diet-by-community interaction; ANOVA).
(C,D) Proteomics analysis of fecal communities obtained on experimental days 6, 12, 19, and 25. Genes in predicted homogalacturonan-processing PULs are shown along the x-axis (as locus tag number only; BT_XXXX, BVU_XXXX) according to the order in which they appear in the genome. Mean values ± SD (vertical lines) are indicated (n=5 animals/treatment group). Genes are color-coded according to functional annotation (see key). GH families for enzymes in the CAZy database are shown as numbers inside the gene boxes. Key for circles is identical to that used in panels A and B. *, P < 0.05, ∣fold change∣ > log2(1.2) (citrus pectin supplemented versus unsupplemented-HiSF-LoFV diet; limma)
Figure S4. In vivo bead-based assay of glycan degradation as a function of community and diet context (related to Figure 4).
(A,B) Gnotobiotic mice, mono-colonized with either B. cellulosilyticus or B. vulgatus, were gavaged with three types of polysaccharide-coated beads together with empty uncoated beads. Beads were purified from cecal and colonic contents 4 hours after gavage. The mass of arabinose (A) or glucose (B) associated with beads is plotted before (black) and after (green) their transit through the gut. Circles denote individual animals. Bars show mean values with 95%CI.(C,D) Adult C57BL/6J germ-free mice were gavaged with four beads types (labeled on the x-axis). Beads were isolated from fecal samples collected from 4 to 12 hours after gavage. The mass of arabinose (panel C) and glucose (panel D) associated with beads is plotted before (black) and after (blue) their transit through the gut. Circles denote individual mice. Bars show the mean values + 95%CI (n=13 animals).
(E,F) Polysaccharide degradation in mice colonized with the 15-member community (with B. cellulosilyticus), or the 14-member community lacking B. cellulosilyticus fed the HiSF-LoFV diet +10% pea fiber (see Table S3C for experimental design). Beads were recovered from cecal and colonic contents. The mass of bead-associated arabinose (E) or glucose (F) is plotted before (black) and after transit through the gut (green, 15-member community group; magenta, minus B. cellulosilyticus group). In panels A, B, E and F of this Figure, and in Figure 4D and 4E, input beads are shared for all plots, since all six groups of mice were analyzed in the same experiment. The presence or absence of B. cellulosilyticus in each group of mice is noted along the x-axis. Circles denote individual mice. Mean values + 95%CI are shown (n=3-6 animals/group). *, P < 0.05 (Mann-Whitney U test).
Figure S5. Evidence for B. cellulosilyticus-dependent glycan use by B. ovatus in the HiSF-LoFV diet context (related to Figure 6).
(A,B,C) Proteomics analysis of fecal communities sampled on experimental days 6, 12, 19, and 25. Genes, color-coded according to their functional annotation including GH family assignments, in the indicated PULs are shown along the x-axis together with their locus tag numbers (Bovatus_0XXXX). The abundance of their expressed protein products (mean values ± SD) is plotted along the y-axis (n=5 animals/treatment group). Key for circles: grey, 15-member community; magenta, mice harboring communities without B. cellulosilyticus. *, P < 0.05, ∣fold change∣ > log2(1.2), [15-member community versus 14-member (minus B. cellulosilyticus); limma].
Table S1 (related to Figure 1). Analyses of dietary fiber preparations and the base HiSF-LoFV diet. (A) Compositional analysis of food-grade dietary fiber preparations used in screening experiments 1, 2, and 3. (B) Linkage analysis of pea fiber preparation. (C) Linkage analysis of citrus pectin preparation. (D) Monosaccharide and glycosyl linkage analyses of polysaccharides in the HiSF-LoFV diet.
Table S2 (related to Figure 1). V4-16S rRNA-based analysis of the effects of different diets on the relative abundances of members of the defined community employed for the in vivo screen of 144 different diets. (A) Screening experiment 1. (B) Screening experiment 2. (C) Screening experiment 3. (D) Mixtures of fiber preparations administered during fiber screens. (E) Estimated Coefficients from linear mixed effect models generated using relative abundance. (F) Estimated Coefficients from linear mixed models generated using DNA-scaled abundance.
Table S3 (related to Figures 2, 3, 5, and 6) COPRO-Seq results for monotonous feeding experiments involving mice harboring a 15-member model microbial community. (A) Monotonous feeding of the unsupplemented HiSF-LoFV diet or the diet supplemented with one of four different fiber preparations. (B) Monotonous feeding of the unsupplemented HiSF-LoFV diet or the HiSF-LoFV diet supplemented with pea fiber or citrus pectin to mice colonized with the community with or without B. cellulosilyticus. (C) Monotonous feeding of the HiSF-LoFV with or without pea fiber to mice colonized with the community with or without B. cellulosilyticus. (D) Monotonous feeding of HiSF-LoFV with or without citrus pectin to mice colonized with a community with or without B. cellulosilyticus or B. vulgatus. (E) Monotonous feeding of the unsupplemented HiSF-LoFV diet to mice harboring communities with or without B. cellulosilyticus and/or B. ovatus.
Table S4 (related to Figures 2, 3, 5, and 6). Community meta-proteomic data from the monotonous feeding experiments outlined in Table S3. Data are shown for proteins whose abundances were significantly altered by supplementing the HiSF-LoFV diet with pea fiber (Ai), citrus pectin (Aii), orange peel (Aiii), or tomato peel (Aiv) in the experiment shown in Table S3A. For day 6 of the experiment outlined in Table S3B, proteins are shown whose abundances were significantly altered in response to pea fiber or citrus pectin (Bi, Bii, respectively), altered in response to omission of B. cellulosilyticus in mice fed the unsupplemented, pea fiber-supplemented, and citrus pectin-supplemented HiSF-LoFV diet (Biii, Biv, Bv, respectively), or altered in response to pea fiber or citrus pectin in mice colonized with the community lacking B. cellulosilyticus (Bvi, Bvii). Proteins whose abundances were significantly altered by these seven conditions on day 12 of this experiment are listed in Ci-Cvii, on day 19 in Di-Dvii, and on day 25 in Ei-Evii, respectively. For experiments shown in Table S3D, proteins whose abundances were significantly altered by omission of B. cellulosilyticus or B. vulgatus during administration of the unsupplemented HiSF-LoFV diet are listed in Fi and Fii, and by citrus pectin administration to mice harboring the complete community, a community lacking B. cellulosilyticus or B. vulgatus in Fiii-Fv, respectively. Proteins whose abundances were significantly altered by omission of B. cellulosilyticus, B. ovatus, or both in mice fed the unsupplemented HiSF-LoFV diet described in Table S3E are shown in Gi-Giii respectively.
Table S5 (related to Figures 2 and 5). Fitness determinants identified by INSeq analysis in the monotonous feeding experiments. Data are shown for genes with significant contributions to fitness during administration of pea fiber (Ai), citrus pectin (Aii), orange peel (Aiii), or tomato peel (Aiv) in the monotonous feeding experiment outlined in Table S3A. Genes with a significantly increased or decreased contribution to fitness during omission of B. cellulosilyticus (Bi) or B. vulgatus (Bii) in the context of a HiSF-LoFV diet in the experiment outlined in Table S3D. Genes with a significant contribution to fitness during citrus pectin administration to mice harboring the complete community (Biii) as outlined in Table S3D.
Table S6 (related to Figures 4 and 6). Monosaccharide analysis of beads administered to gnotobiotic mice (A) Mice mono-colonized with individual Bacteroides species. (B) Mice colonized with the 15-member community, or a derivative lacking B. cellulosilyticus, or with individual Bacteroides species. (C) Germ-free mice. (D) Mice colonized with the 15-member community or derivatives lacking B. cellulosilyticus, B. ovatus, or both.
Table S7 (related to Figure 2). PUL numbering showing correspondence between each ‘predicted PUL’, ‘literature PUL’ and its gene locus tags.
In vivo screen for fibers targeting specific human gut taxa in a defined community
Proteomics and forward genetics identify bioactive nutrients and their utilization
Interspecies competition controls the outcome of fiber-based microbiota manipulation
Artificial food particles as biosensors of community-wide glycan degradation
Acknowledgements:
We thank David O’Donnell, Maria Karlsson, Marty Meier, Justin Serugo, Sabrina Wagoner, and J. Hoisington-López, for superb technical assistance, Janaki Lelwala-Guruge for help with anaerobic microbiology, Brigitte Laillet and Marie-Jeanne Crépeau for technical assistance in fiber characterization, and Michael Barratt, Darryl Wesener, and Jennifer Bando for helpful discussions. Pierre Aymard, Dominic Vellucci and Luca Dimartino sourced fibers, processed freeze-dried diets, and assisted with proximate analysis of fibers. This work was supported by grants from the NIH (DK070977, DK078669, F32DK107158), Mondelez International, and the U.S. Department of Energy grant (DE-SC0015662).
Footnotes
Declaration of interests:
J.I.G. is a co-founder of Matatu, Inc., a company characterizing the role of diet-by-microbiota interactions in animal health. Elements of this report are the subject of patent applications that are currently being submitted.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Adey A, Morrison HG, Asan Xun, X., Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, et al. (2010). Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol 11, R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Hao Z, and Mohnen D (2013). Evolving views of pectin biosynthesis. Annu Rev Plant Biol 64, 747–779. [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, and Caporaso JG (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature Methods 10, 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffetto F, Cornuault V, Rydahl MG, Ropartz D, Alvarado C, Echasserieau V, Le Gall S, Bouchet B, Tranquet O, Verhertbruggen Y, et al. (2015). The Deconstruction of Pectic Rhamnogalacturonan I Unmasks the Occurrence of a Novel Arabinogalactan Oligosaccharide Epitope. Plant Cell Physiol 56, 2181–2196. [DOI] [PubMed] [Google Scholar]
- Clarkson SM, Giannone RJ, Kridelbaugh DM, Elkins JG, Guss AM, and Michener JK (2017). Construction and Optimization of a Heterologous Pathway for Protocatechuate Catabolism in Escherichia coli Enables Bioconversion of Model Aromatic Compounds. Applied and Environmental Microbiol 83, e01313–01317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. (2016). A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 167, 1339–1353 e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Englyst HN, and Cummings JH (1988). Improved Method for Measurement of Dietary Fiber as Non-Starch Polysaccharides in Plant Foods. J Assoc Official Analytical Chemists 71, 808–814. [PubMed] [Google Scholar]
- Faith JJ, Ahern PP, Ridaura VK, Cheng J, and Gordon JI (2014). Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med 6, 220ra211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. (2013). The long-term stability of the human gut microbiota. Science 341, 1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filisetti-Cozzi TM, and Carpita NC (1991). Measurement of uronic acids without interference from neutral sugars. Anal Biochem 197, 157–162. [DOI] [PubMed] [Google Scholar]
- Fragiadakis GK, Smits SA, Sonnenburg ED, Van Treuren W, Reid G, Knight R, Manjurano A, Changalucha J, Dominguez-Bello MG, Leach J, et al. (2018). Links between environment, diet, and the hunter-gatherer microbiome. Gut Microbes, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenwright AJ, Pothula KR, Bhamidimarri SP, Chorev DS, Basle A, Firbank SJ, Zheng H, Robinson CV, Winterhalter M, Kleinekathofer U, Bolam DN, et al. (2017). Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 541, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, and Gordon JI (2009). Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host & Microbe 6, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd MC, Wu M, Rodionov DA, Li X, Cheng J, Griffin NW, Barratt MJ, Giannone RJ, Hettich RL, Osterman AL, et al. (2017). The effects of micronutrient deficiencies on bacterial species from the human gut microbiota. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, and Peterson SB (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nature Reviews Microbiology 8, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss C, Klutts JS, Wang Z, Doering TL, and Azadi P (2009). The structure of Cryptococcus neoformans galactoxylomannan contains beta-D-glucuronic acid. Carbohydr Res 344, 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotarski SF, and Salyers AA (1984). Isolation and characterization of outer membranes of Bacteroides thetaiotaomicron grown on different carbohydrates. J Bacteriol 158, 102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levigne S, Thomas M, Ralet MC, Quemener BC, and Thibault JF (2002). Determination of the degrees of methylation and acetylation of pectins using a C18 column and internal standards. Food Hydrocolloids 16, 547–550. [Google Scholar]
- Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Basle A, Cartmell A, Terrapon N, Stott K, et al. (2018). Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nature Microbiol 3, 210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JB, and Sonnenburg JL (2012). Prioritization of a plant polysaccharide over a mucus carbohydrate is enforced by a Bacteroides hybrid two-component system. Mol Microbiol 85, 478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZQ, Dasari S, Chambers MC, Litton MD, Sobecki SM, Zimmerman LJ, Halvey PJ, Schilling B, Drake PM, Gibson BW, et al. (2009). IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J Proteome Res 8, 3872–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, et al. (2011). Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biology 9, e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, and Hugenholtz P (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6, 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, et al. (2013). Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biology 11, e1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menni C, Jackson MA, Pallister T, Steves CJ, Spector TD, and Valdes AM (2017). Gut microbiome diversity and high-fibre intake are related to lower long-term weight gain. Int J Obes 41, 1099–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Miller JS, and Hahn MG (2012). Immunological approaches to plant cell wall and biomass characterization: Glycome Profiling. Methods Mol Biol 908, 61–72. [DOI] [PubMed] [Google Scholar]
- Pettolino FA, Walsh C, Fincher GB, and Bacic A (2012). Determining the polysaccharide composition of plant cell walls. Nature Protocols 7, 1590–1607. [DOI] [PubMed] [Google Scholar]
- Porter NT, and Martens EC (2017). The critical roles of polysaccharides in gut microbial ecology and physiology. Annu Rev Microbiol 71, 349–369. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Basle A, Morland C, Day AM, Zheng H, et al. (2015). Glycan complexity dictates microbial resource allocation in the large intestine. Nature Commun 6, 7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santander J, Martin T, Loh A, Pohlenz C, Gatlin DM 3rd, and Curtiss R 3rd (2013). Mechanisms of intrinsic resistance to antimicrobial peptides of Edwardsiella ictaluri and its influence on fish gut inflammation and virulence. Microbiology 159, 1471–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalm ND 3rd, Townsend GE 2nd, and Groisman EA (2016). Multiple signals govern utilization of a polysaccharide in the gut bacterium Bacteroides thetaiotaomicron. MBio 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, and Sonnenburg JL (2018). An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, Article3. [DOI] [PubMed] [Google Scholar]
- Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, and Sonnenburg JL (2016). Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, and Sonnenburg JL (2010). Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, Fernando CG, and Chambers MC (2007). MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res 6, 654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauzin AS, Laville E, Xiao Y, Nouaille S, Le Bourgeois P, Heux S, Portais JC, Monsan P, Martens EC, Potocki-Veronese G, et al. (2016). Functional characterization of a gene locus from an uncultured gut Bacteroides conferring xylo-oligosaccharides utilization to Escherichia coli. Mol Microbiol 102, 579–592. [DOI] [PubMed] [Google Scholar]
- Terrapon N, Lombard V, Drula E, Lapebie P, Al-Masaudi S, Gilbert HJ, and Henrissat B (2018). PULDB: the expanded database of Polysaccharide Utilization Loci. Nucleic Acids Res 46, D677–D683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault JF (1979). Automatisation du dosage des substances pectiques par la méthode au métahydroxydiphényle. Lebensml Wiss Technol 12, 247–251. [Google Scholar]
- Ting L, Cowley MJ, Hoon SL, Guilhaus M, Raftery MJ, and Cavicchioli R (2009). Normalization and statistical analysis of quantitative proteomics data generated by metabolic labeling. Mol Cell Proteomics 8, 2227–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, McNulty NP, Rodionov DA, Khoroshkin MS, Griffin NW, Cheng J, Latreille P, Kerstetter RA, Terrapon N, Henrissat B, et al. (2015). Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science 350, aac5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, et al. (2015). Personalized Nutrition by Prediction of Glycemic Responses. Cell 163, 1079–1094. [DOI] [PubMed] [Google Scholar]
- Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, et al. (2018). Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Strain-level variability across arabinan-utilization loci in Bacteroides species (related to Figure 2).
(A,B,C) Alignment of B. thetaiotaomicron PUL7 (panel A), B. cellulosilyticus PUL5 (panel B), and B. vulgatus PUL27 (panel C) across multiple strains of each species. The direction of transcription is indicated by the arrowhead. The genes are labeled with their locus tag number and color-coded according to their functional annotation (see key). Shaded regions connecting genes denote (i) significant BLAST homology (E-value <10−5) and the percent amino acid identity (>50%) of their protein products (see key).
Figure S2. Proteomics and forward genetics identify homogalacturonan in citrus pectin as a nutrient source for multiple species (related to Figure 2).
(A) Schematic representation of polysaccharide structures detected in citrus pectin based on monosaccharide and linkage analyses (with stereochemistry of anomeric carbons inferred).(B-E) Adult C57BL/6J germ-free mice were colonized with a 15-member community consisting of INSeq libraries representing four Bacteroides species together with 10 additional bacterial strains used in screening experiments depicted in Figure 1. Relative abundance is shown for each bacterial strain at each of the indicated time points in mice fed the control HiSF-LoFV diet (grey), or the HiSF-LoFV diet supplemented with 10% (w/w) citrus pectin (blue). Circles denote individual mice, lines the mean value and shading the 95% CI (n=15 individually caged mice per group; results pooled from three independent experiments). *, P < 0.05 (Mann-Whitney Utest). (F-I) Proteomic and INSeq analyses of fecal samples collected on experimental day 6. On the x-axis, each dot denotes the mean value for the abundance of a single bacterial protein in samples from animals monotonously fed the citrus pectin-supplemented HiSF-LoFV diet (relative to controls fed the unsupplemented diet). The y-axis indicates the mean value for the differential enrichment of mutants with Tn disruptions in the gene encoding each protein in the citrus pectin versus HiSF-LoFV diet groups. Blue dots represent genes that are significantly affected by citrus pectin (P < 0.05, ∣fold change∣ > log2(1.2); limma or limma-voom) as judged by levels of their protein products or their contribution to fitness while open circles mark the subset of these genes that are encoded by PULs. Genes present in predicted homogalacturonan-processing PULs in B. thetaiotaomicron, B. cellulosilyticus and B. vulgatus are labeled with their PUL number as it appears in PULDB (Terrapon et al., 2018).
Figure S3. Deliberate manipulation of community composition demonstrates interspecies competition for homogalacturonan in citrus pectin (related to Figure 3).
(A,B) Adult C57BL/6J germ-free mice were colonized with the same defined community used for the experiments in Figure 2, with or without B. cellulosilyticus (B.c.). Relative abundance B. thetaiotaomicron and B. vulgatus in fecal samples obtained at the indicated time points from mice fed the control HiSF-LoFV diet or the diet supplemented with 10% (w/w) citrus pectin. These mice were colonized with a community that contained B. cellulosilyticus or one where it had been omitted (see key). Circles, individual mice; lines, mean values; shading, 95%CI (n=4-10 mice per group). *, P < 0.05 (diet-by-community interaction; ANOVA).
(C,D) Proteomics analysis of fecal communities obtained on experimental days 6, 12, 19, and 25. Genes in predicted homogalacturonan-processing PULs are shown along the x-axis (as locus tag number only; BT_XXXX, BVU_XXXX) according to the order in which they appear in the genome. Mean values ± SD (vertical lines) are indicated (n=5 animals/treatment group). Genes are color-coded according to functional annotation (see key). GH families for enzymes in the CAZy database are shown as numbers inside the gene boxes. Key for circles is identical to that used in panels A and B. *, P < 0.05, ∣fold change∣ > log2(1.2) (citrus pectin supplemented versus unsupplemented-HiSF-LoFV diet; limma)
Figure S4. In vivo bead-based assay of glycan degradation as a function of community and diet context (related to Figure 4).
(A,B) Gnotobiotic mice, mono-colonized with either B. cellulosilyticus or B. vulgatus, were gavaged with three types of polysaccharide-coated beads together with empty uncoated beads. Beads were purified from cecal and colonic contents 4 hours after gavage. The mass of arabinose (A) or glucose (B) associated with beads is plotted before (black) and after (green) their transit through the gut. Circles denote individual animals. Bars show mean values with 95%CI.(C,D) Adult C57BL/6J germ-free mice were gavaged with four beads types (labeled on the x-axis). Beads were isolated from fecal samples collected from 4 to 12 hours after gavage. The mass of arabinose (panel C) and glucose (panel D) associated with beads is plotted before (black) and after (blue) their transit through the gut. Circles denote individual mice. Bars show the mean values + 95%CI (n=13 animals).
(E,F) Polysaccharide degradation in mice colonized with the 15-member community (with B. cellulosilyticus), or the 14-member community lacking B. cellulosilyticus fed the HiSF-LoFV diet +10% pea fiber (see Table S3C for experimental design). Beads were recovered from cecal and colonic contents. The mass of bead-associated arabinose (E) or glucose (F) is plotted before (black) and after transit through the gut (green, 15-member community group; magenta, minus B. cellulosilyticus group). In panels A, B, E and F of this Figure, and in Figure 4D and 4E, input beads are shared for all plots, since all six groups of mice were analyzed in the same experiment. The presence or absence of B. cellulosilyticus in each group of mice is noted along the x-axis. Circles denote individual mice. Mean values + 95%CI are shown (n=3-6 animals/group). *, P < 0.05 (Mann-Whitney U test).
Figure S5. Evidence for B. cellulosilyticus-dependent glycan use by B. ovatus in the HiSF-LoFV diet context (related to Figure 6).
(A,B,C) Proteomics analysis of fecal communities sampled on experimental days 6, 12, 19, and 25. Genes, color-coded according to their functional annotation including GH family assignments, in the indicated PULs are shown along the x-axis together with their locus tag numbers (Bovatus_0XXXX). The abundance of their expressed protein products (mean values ± SD) is plotted along the y-axis (n=5 animals/treatment group). Key for circles: grey, 15-member community; magenta, mice harboring communities without B. cellulosilyticus. *, P < 0.05, ∣fold change∣ > log2(1.2), [15-member community versus 14-member (minus B. cellulosilyticus); limma].
Table S1 (related to Figure 1). Analyses of dietary fiber preparations and the base HiSF-LoFV diet. (A) Compositional analysis of food-grade dietary fiber preparations used in screening experiments 1, 2, and 3. (B) Linkage analysis of pea fiber preparation. (C) Linkage analysis of citrus pectin preparation. (D) Monosaccharide and glycosyl linkage analyses of polysaccharides in the HiSF-LoFV diet.
Table S2 (related to Figure 1). V4-16S rRNA-based analysis of the effects of different diets on the relative abundances of members of the defined community employed for the in vivo screen of 144 different diets. (A) Screening experiment 1. (B) Screening experiment 2. (C) Screening experiment 3. (D) Mixtures of fiber preparations administered during fiber screens. (E) Estimated Coefficients from linear mixed effect models generated using relative abundance. (F) Estimated Coefficients from linear mixed models generated using DNA-scaled abundance.
Table S3 (related to Figures 2, 3, 5, and 6) COPRO-Seq results for monotonous feeding experiments involving mice harboring a 15-member model microbial community. (A) Monotonous feeding of the unsupplemented HiSF-LoFV diet or the diet supplemented with one of four different fiber preparations. (B) Monotonous feeding of the unsupplemented HiSF-LoFV diet or the HiSF-LoFV diet supplemented with pea fiber or citrus pectin to mice colonized with the community with or without B. cellulosilyticus. (C) Monotonous feeding of the HiSF-LoFV with or without pea fiber to mice colonized with the community with or without B. cellulosilyticus. (D) Monotonous feeding of HiSF-LoFV with or without citrus pectin to mice colonized with a community with or without B. cellulosilyticus or B. vulgatus. (E) Monotonous feeding of the unsupplemented HiSF-LoFV diet to mice harboring communities with or without B. cellulosilyticus and/or B. ovatus.
Table S4 (related to Figures 2, 3, 5, and 6). Community meta-proteomic data from the monotonous feeding experiments outlined in Table S3. Data are shown for proteins whose abundances were significantly altered by supplementing the HiSF-LoFV diet with pea fiber (Ai), citrus pectin (Aii), orange peel (Aiii), or tomato peel (Aiv) in the experiment shown in Table S3A. For day 6 of the experiment outlined in Table S3B, proteins are shown whose abundances were significantly altered in response to pea fiber or citrus pectin (Bi, Bii, respectively), altered in response to omission of B. cellulosilyticus in mice fed the unsupplemented, pea fiber-supplemented, and citrus pectin-supplemented HiSF-LoFV diet (Biii, Biv, Bv, respectively), or altered in response to pea fiber or citrus pectin in mice colonized with the community lacking B. cellulosilyticus (Bvi, Bvii). Proteins whose abundances were significantly altered by these seven conditions on day 12 of this experiment are listed in Ci-Cvii, on day 19 in Di-Dvii, and on day 25 in Ei-Evii, respectively. For experiments shown in Table S3D, proteins whose abundances were significantly altered by omission of B. cellulosilyticus or B. vulgatus during administration of the unsupplemented HiSF-LoFV diet are listed in Fi and Fii, and by citrus pectin administration to mice harboring the complete community, a community lacking B. cellulosilyticus or B. vulgatus in Fiii-Fv, respectively. Proteins whose abundances were significantly altered by omission of B. cellulosilyticus, B. ovatus, or both in mice fed the unsupplemented HiSF-LoFV diet described in Table S3E are shown in Gi-Giii respectively.
Table S5 (related to Figures 2 and 5). Fitness determinants identified by INSeq analysis in the monotonous feeding experiments. Data are shown for genes with significant contributions to fitness during administration of pea fiber (Ai), citrus pectin (Aii), orange peel (Aiii), or tomato peel (Aiv) in the monotonous feeding experiment outlined in Table S3A. Genes with a significantly increased or decreased contribution to fitness during omission of B. cellulosilyticus (Bi) or B. vulgatus (Bii) in the context of a HiSF-LoFV diet in the experiment outlined in Table S3D. Genes with a significant contribution to fitness during citrus pectin administration to mice harboring the complete community (Biii) as outlined in Table S3D.
Table S6 (related to Figures 4 and 6). Monosaccharide analysis of beads administered to gnotobiotic mice (A) Mice mono-colonized with individual Bacteroides species. (B) Mice colonized with the 15-member community, or a derivative lacking B. cellulosilyticus, or with individual Bacteroides species. (C) Germ-free mice. (D) Mice colonized with the 15-member community or derivatives lacking B. cellulosilyticus, B. ovatus, or both.
Table S7 (related to Figure 2). PUL numbering showing correspondence between each ‘predicted PUL’, ‘literature PUL’ and its gene locus tags.
Data Availability Statement
Datasets of V4-16S rRNA sequences in raw format prior to post-processing and data analysis, plus COPRO-Seq and INSeq datasets have been deposited at the European Nucleotide Archive under study accession PRJEB26564. All LC-MS/MS proteomic data have been deposited into the MassIVE data repository under accession numbers MSV000082287 (MassIVE) and PXD009535 (ProteomeXchange). INSeq software: https://github.com/mengwu1002/Multi-taxon_analysis_pipeline. COPRO-Seq software: https://github.com/nmcnulty/COPRO-Seq.