p130Cas translocates into the nucleus upon fluid shear stress application and alleviates NF-κB activity in osteocytes.
Abstract
Mechanical loading plays an important role in bone homeostasis. However, molecular mechanisms behind the mechanical regulation of bone homeostasis are poorly understood. We previously reported p130Cas (Cas) as a key molecule in cellular mechanosensing at focal adhesions. Here, we demonstrate that Cas is distributed in the nucleus and supports mechanical loading–mediated bone homeostasis by alleviating NF-κB activity, which would otherwise prompt inflammatory processes. Mechanical unloading modulates Cas distribution and NF-κB activity in osteocytes, the mechanosensory cells in bones. Cas deficiency in osteocytes increases osteoclastic bone resorption associated with NF-κB–mediated RANKL expression, leading to osteopenia. Upon shear stress application on cultured osteocytes, Cas translocates into the nucleus and down-regulates NF-κB activity. Collectively, fluid shear stress–dependent Cas-mediated alleviation of NF-κB activity supports bone homeostasis. Given the ubiquitous expression of Cas and NF-κB together with systemic distribution of interstitial fluid, the Cas–NF-κB interplay may also underpin regulatory mechanisms in other tissues and organs.
INTRODUCTION
Mechanical forces are integral to normal cellular functions (1), disruption of which leads to loss of homeostasis. Although bones are known to respond to mechanical loading (2), molecular mechanisms underlying mechanical regulation of bone homeostasis are poorly understood.
The ubiquitously expressed adaptor molecule p130Cas (Crk-associated substrate, hereafter referred to as Cas), which is phosphorylated at focal adhesions upon extracellular matrix engagement, is involved in various cellular processes including migration, survival, transformation, and invasion (3). We previously demonstrated that Cas functions as an ion channel–independent cytoskeletal mechanosensor through force-dependent conformational changes (4, 5). However, it remains unclear whether Cas is involved in responses to mechanical forces in vivo.
Nuclear factor κB (NF-κB) is a transcription factor that plays a central role in regulating inflammation and aging (6). The classical NF-κB pathway involves activation of the IκB (inhibitor of κB) kinase (IKK) complex, which induces degradation of IκB, the inhibitor protein that masks nuclear localization signals of NF-κB molecules and sequesters them in the cytoplasm (7). In addition to being phosphorylated and translocated into the nucleus, RelA/p65 (a subunit of NF-κB, hereafter referred to as RelA) is subjected to acetylation. Whereas several lysine residues of RelA have been shown to be acetylated, acetylation at Lys310 is required for the complete transcriptional activity of NF-κB (8). Because NF-κB is also ubiquitously expressed and reportedly mechanoresponsive (9), we speculated that Cas might interact with NF-κB in the mechanical regulation of organismal homeostasis related to inflammation and aging.
Interstitial fluid is distributed throughout the entire body of all animals and comprises most extracellular fluid, occupying about four times as much volume as compared to circulating blood plasma. The skeleton, which is a metabolically active organ undergoing continuous remodeling, is not completely solid but a deformable porous body containing interstitial fluid. Several factors, including Earth’s gravity and physical exercise, apply mechanical load to the skeletal system and maintain bone mass (10). Osteocytes are the most common cells in mature bones (11). In lamellar bone, osteocytes and their highly interconnective dendritic processes reside inside spaces called lacunae and canaliculi, respectively. Bone deformation, which results from mechanical loading, generates an interstitial fluid flow in the lacuno-canalicular system inducing a shear stress on osteocytes (12). Ablation of osteocytes revealed their roles in bone metabolism and response to mechanical unloading (13). Their regulatory role is also highlighted by the fact that osteocytes secrete paracrine and systemic regulators that affect osteoblast-mediated bone formation and osteoclast-mediated bone resorption (14). Together, we hypothesized that the Cas–NF-κB interaction in osteocytes might be involved in mechanical regulation of bone homeostasis.
RESULTS
Cas and NF-κB are mechanically modulated in osteocytes in vivo and in vitro
To test our hypothesis on mechanical roles of Cas and NF-κB in bone metabolism, we used a mouse bone-unloading model, in which one of the hindlimbs of each individual mouse was subjected to sciatic and femoral nerve resection, while the other was exposed to a sham operation to serve as a control (15). Unloaded femurs exhibited a significant reduction of bone volume as compared to their contralateral controls (Fig. 1A). Unexpectedly, a substantial proportion of Cas was distributed in the nuclei of osteocytes in control bones, whereas most Cas was observed in the cytosol of osteocytes in unloaded bones (Fig. 1B). Furthermore, acetylation of RelA, which correlates with NF-κB activity (8), was increased in osteocytes of unloaded tibiae as compared to their contralateral controls (Fig. 1C, left and center). Notably, nuclear distribution of RelA in osteocytes was not significantly altered by unloading (Fig. 1C, right). Together, mechanical loading–dependent Cas nuclear distribution appeared inversely correlated with RelA acetylation in the nuclei of osteocytes. These in vivo findings suggest that Cas and NF-κB may be involved in the previously reported mechanosensory function of osteocytes (13) and contribute to mechanical loading–related bone homeostasis.
Fig. 1. Cas and NF-κB are mechanically modulated in osteocytes in vivo and in vitro.
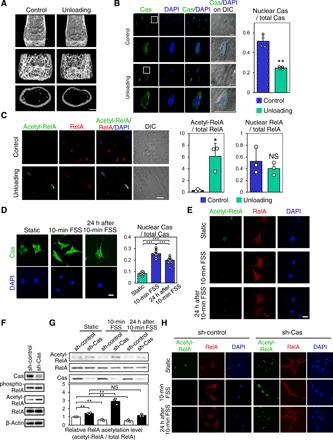
(A) Reduction in bone mass by hemilateral hindlimb unloading. Representative microcomputed tomography (μCT) images of mouse distal femurs are shown. Scale bar, 1 mm. (B) Loading-dependent change of Cas distribution in osteocytes in vivo. Confocal images of anti-Cas immunofluorescence (green) and nuclear [4′,6-diamidino-2-phenylindole (DAPI), blue] staining of osteocytes in midshaft tibiae are shown together with their DIC (differential interference contrast) images (left). The lower panels show higher-magnification images from regions highlighted by boxes in the upper panels. Scale bars, 10 μm. Quantitation of the nuclear distribution of Cas (right, n = 4 mice; each value was averaged from four to six osteocytes analyzed in each bone). (C) Increased RelA acetylation in osteocytes of unloaded bones. Confocal images of anti–acetylated RelA (acetyl-RelA, green), anti-RelA (RelA, red), and nuclear (DAPI) staining of osteocytes in midshaft tibiae are shown (left). Scale bar, 10 μm. RelA acetylation (center) and nuclear distribution (right) were quantified. RelA acetylation was scaled with the mean value of control bones (column 1) set at 1 (n = 3 mice per group, each value was averaged from four to six osteocytes analyzed in each bone). (D and E) FSS-induced increase of Cas nuclear distribution and decrease of RelA acetylation in MLO-Y4 osteocytes. Confocal images of anti-Cas (D, green), anti–acetylated RelA (E, green), and anti-RelA (E, red) immunofluorescence and nuclear (DAPI) staining of MLO-Y4 osteocytes in static culture, immediately and 24 hours after 10-min pulsatile FSS. Scale bar, 20 μm. Nuclear distribution of Cas was quantified as in (B) (n = 10 per group). (F) Cas-dependent decrease of RelA acetylation in MLO-Y4 osteocytes. RelA acetylation and phosphorylation in MLO-Y4 osteocytes infected with sh-Cas–expressing (sh-Cas) or its control (sh-control) retrovirus were analyzed by immunoblotting. Note that RelA phosphorylation was not affected by Cas knockdown. (G) Cas-dependent FSS-induced decrease of RelA acetylation in MLO-Y4 osteocytes. MLO-Y4 osteocytes with (sh-Cas) and without (sh-control) Cas knockdown, in static culture, and immediately or 24 hours after 10-min pulsatile FSS application, were analyzed by immunoblotting (top). The levels of RelA acetylation (acetyl-RelA/total RelA) were normalized against the static controls, which were set at 1 (bottom, n = 3). (H) Confocal images showing anti–acetylated RelA (acetyl-RelA) (green) and anti-RelA (red) immunofluorescence in MLO-Y4 osteocytes, with (sh-Cas) and without (sh-control) Cas knockdown, in static culture (top) or immediately (middle) and 24 hours (bottom) after 10-min pulsatile FSS. Nuclei were stained with DAPI (blue). Scale bar, 20 μm.
Simulative studies indicate that osteocytes are exposed to fluid shear stress (FSS) with a peak magnitude of 0.8 to 3 Pa, which arises from interstitial fluid flow in canaliculi generated upon mechanical loading on bones (12). Whereas nuclear distribution of Cas appeared minimal in MLO-Y4 osteocytes grown under static conditions, it significantly increased after applying pulsatile FSS (1 Pa on average, 1 Hz) for 10 min and remained increased even 24 hours after termination of FSS (Fig. 1D). In contrast, RelA acetylation decreased after FSS application and remained decreased for at least 24 hours (Fig. 1E). Furthermore, when Cas expression was retrovirally silenced in MLO-Y4 osteocytes (Fig. 1F), FSS did not decrease but rather increased RelA acetylation (Fig. 1G, compare lanes 2 and 4, and Fig. 1H). These results suggest that Cas mediates shear stress–induced attenuation of RelA acetylation via nuclear translocation in osteocytes in vitro.
Increased bone resorption and receptor activator of NF-κB ligand expression in osteocyte-specific Cas knockout mice
To determine whether Cas and NF-κB played physiological roles in regulating osteocytes’ function in vivo, we generated osteocyte-specific Cas conditional knockout mice (referred to herein as Cas cKO mice), which had markedly reduced Cas expression in osteocytes (Fig. 2A). Consistent with the inverse correlation between Cas nuclear distribution and RelA acetylation in the control and unloaded bones of wild-type (WT) mice (Fig. 1, B and C), we observed enhanced RelA acetylation, but not nuclear distribution, in osteocytes of Cas cKO mice (Fig. 2B). Although Cas cKO mice were born with no evident bone phenotype and developed normally with no apparent morphological abnormalities (fig. S1, A and B), 10-week-old Cas cKO mice exhibited significantly decreased bone volume and cortical thickness and increased cortical porosity (Fig. 2, C and D). Histomorphometric analysis revealed a significant increase in bone resorption (Fig. 2E) with unaltered bone formation (fig. S2) in Cas cKO mice. Furthermore, the expression level of sclerostin, a negative regulator of osteoblast proliferation and differentiation secreted by osteocytes (16), remained unchanged in bones of Cas cKO mice (Fig. 2F). Bone resorption is affected by the balance between receptor activator of NF-κB ligand (RANKL), a differentiation factor for osteoclasts (17), and osteoprotegerin (OPG), a decoy receptor that interferes with the binding of RANKL to its signal-transducing receptor RANK (18). Therefore, increased RANKL/OPG ratio, resulting from the increase in RANKL expression with the unaltered OPG expression in Cas cKO bones (Fig. 2F), accounts for their enhanced bone resorption (Fig. 2E). All in all, these results indicate that Cas primarily controls osteoclastic bone resorption through its regulation of RANKL expression in osteocytes.
Fig. 2. Increased bone resorption and RANKL expression in osteocyte-specific Cas knockout mice.
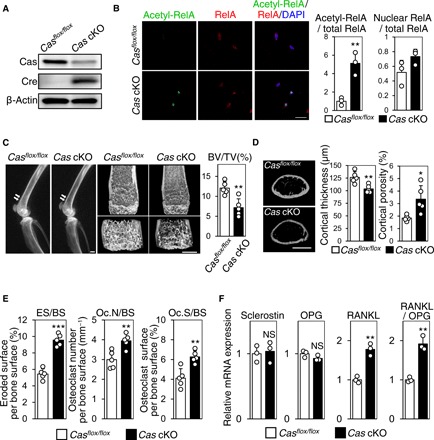
(A) Significant reduction of Cas expression in osteocyte fractions derived from Cas cKO mice. (B) Increased RelA acetylation in osteocytes of Cas cKO mice. Confocal images of anti–acetylated RelA (green) and anti-RelA (red) immunofluorescence images of midshaft tibiae of Casflox/flox mice and Cas cKO mice are shown with nuclear staining (left). RelA acetylation (center) and nuclear distribution (right) were quantified as in Fig. 1C. RelA acetylation was scaled with the mean value of control bones (column 1) set at 1 (n = 3 mice per group; each value was averaged from four to six osteocytes analyzed in each bone). Scale bar, 10 μm. (C) Reduced bone mass in Cas cKO mice. Representative radiographic images of the femurs (left; arrows point to apparent differences in bone density) and distal femur μCT (center) of Cas cKO mice and normal Casflox/flox littermates are shown. Scale bars, 1 mm. The ratio of bone volume over total volume (BV/TV) was calculated from μCT images (right, n = 5 mice per group). (D) Loss of cortical bone in Cas cKO mice. Representative μCT images of femoral cortices are shown. Scale bar, 1 mm (left). Cortical thickness (center) and cortical porosity (right) were calculated from μCT images (n = 5 mice per group). (E) Increased osteoclastic bone resorption in Cas cKO mice. Parameters for osteoclastic bone resorption in Cas cKO mice and their normal Casflox/flox littermates were determined by histomorphometric analysis (n = 5 mice per group). (F) Increased RANKL expression in osteocytes of Cas cKO mice. mRNA expression levels of sclerostin, OPG, and RANKL as well as RANKL/OPG ratio in the osteocyte fractions derived from Casflox/flox and Cas cKO mice were normalized against data from Casflox/flox mice, which were set at 1 (n = 3 mice per group).
Cas mediates mechanical loading–induced suppression of osteoclastic bone resorption and RANKL expression in osteocytes
To examine whether the bone loss exhibited in Cas cKO mice was relevant to mechanical regulation, we conducted mechanical unloading experiments. Unloaded bones of control mice exhibited significant reduction of bone volume (Fig. 3A) with significantly increased bone resorption parameters as compared to their contralateral controls (Fig. 3B). Although there was a significant decrease of bone volume in unloaded hindlimbs of Cas cKO mice (Fig. 3A, center), the extent of the unloading effect on bone volume was moderate and significantly less prominent in Cas cKO mice as compared to that in control mice (Fig. 3A, right). Notably, we did not observe significant differences in bone resorption parameters between control and unloaded bones of Cas cKO mice (Fig. 3B).
Fig. 3. Cas mediates mechanical loading–induced suppression of bone resorption and RANKL expression in osteocytes.
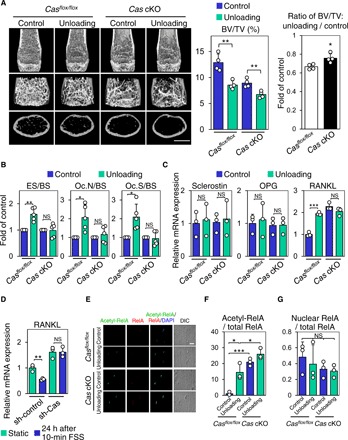
(A) Unloading effect on bone mass in control and Cas cKO mice. Representative μCT images of distal femurs of mice subjected to 2-week hemilateral hindlimb unloading are shown. Scale bars, 1 mm (left). BV/TV was determined as in Fig. 2C (center), and the ratio of unloading to control was calculated in each mouse (right) (n = 4 mice per group). (B) Unaltered bone resorption parameters in unloaded bones of Cas cKO mice. Data from tibiae of mice subjected to 2-week hemilateral hindlimb unloading are shown. Values from unloaded bones were normalized against data from their contralateral controls, which were set at 1 (n = 5 mice per group). (C) Unaltered RANKL expression in osteocytes in unloaded bones of Cas cKO mice. mRNA expression levels of sclerostin, OPG, and RANKL in the osteocyte fractions derived from mice subjected to 1-week hemilateral hindlimb unloading were analyzed as in Fig. 2F. Values were normalized against the control sham–operated hindlimbs (n = 3 mice per group). (D) Loss of FSS effect on RANKL expression in Cas-knockdown MLO-Y4 osteocytes. MLO-Y4 osteocytes with and without Cas knockdown were subjected to FSS experiments as in Fig. 1G. The levels of RANKL mRNA expression were normalized against the static controls, which were set at 1 (n = 3). (E) Loss of mechanical unloading effect on RelA acetylation in osteocytes of Cas cKO mice. Confocal images of anti–acetylated RelA (green), anti-RelA (red), and nuclear (DAPI) staining of osteocytes in midshaft tibiae are shown. Scale bar, 10 μm. (F and G) RelA acetylation (F) and nuclear distribution (G) were quantified as in Fig. 1C with the mean value from control (loaded) bones of control mice (column 1) set at 1 (n = 3 mice per group).
In control mice, RANKL mRNA expression in osteocyte fractions prepared from unloaded bones was increased as compared to those from their contralateral control bones (Fig. 3C, columns 1 and 2). However, RANKL expression in osteocyte fractions from Cas cKO mice was enhanced regardless of unloading (Fig. 3C, columns 3 and 4).
It is likely that bone formation reduced upon unloading (19) in both control and Cas cKO mice, contributing to their decrease in bone volume (Fig. 3A). Although we observed a similar tendency of reduced bone formation parameters in both control and Cas cKO mice, we were unable to detect significant differences between the control and unloaded bones in either of them due to a large variability within each parameter (fig. S3A).
In agreement with the in vivo results (Fig. 3C), RANKL expression was reduced in MLO-Y4 osteocytes 24 hours after FSS application (Fig. 3D, lanes 1 and 2), whereas it was significantly up-regulated in Cas-knockdown osteocytes irrespective of FSS (Figs. 1F and 3D, columns 3 and 4). These findings suggest that Cas in osteocytes plays a significant role in mechanical loading–induced decrease in RANKL-mediated bone resorption.
Cas is involved in mechanical loading–induced suppression of RelA acetylation in osteocytes
We next examined whether Cas was involved in the increase of RelA acetylation in osteocytes of unloaded bones. Consistent with our observations in WT mice (Fig. 1, B and C), Cas nuclear distribution was decreased and RelA acetylation was increased in the osteocytes of unloaded tibiae as compared to those of their contralaterals of control Casflox/flox mice (Fig. 3, E and F, and fig. S3B). However, RelA acetylation, but not nuclear distribution, was enhanced in those of Cas cKO mice regardless of unloading (Fig. 3, E to G). This suggests that Cas is involved in the mechanical loading–dependent decrease in RelA acetylation in osteocytes, supporting the physiological relevance of the Cas–NF-κB interplay.
Cas suppresses RANKL expression via down-regulation of RelA acetylation and intracellular reactive oxygen species level
Consistent with our in vivo findings from Cas cKO mice (Fig. 2F), Cas-knockdown MLO-Y4 osteocytes showed increased RANKL mRNA expression with no significant changes in sclerostin and OPG expressions (Fig. 4A). In addition, RelA acetylation was increased in both Cas cKO (Fig. 2B) and Cas-knockdown MLO-Y4 osteocytes (Fig. 1, F and G). Furthermore, RelA knockdown decreased RANKL expression, but not sclerostin or OPG, in MLO-Y4 osteocytes (Fig. 4B). These results indicate that suppression of RelA acetylation is responsible for Cas-dependent decrease in RANKL expression in MLO-Y4 osteocytes.
Fig. 4. Cas down-regulates NF-κB– and ROS-mediated RANKL expression in MLO-Y4 osteocytes.
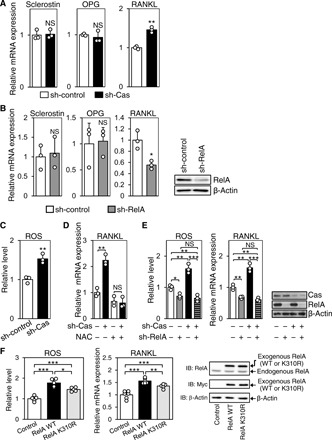
(A) Increased RANKL expression in Cas-knockdown MLO-Y4 osteocytes. mRNA expression levels of sclerostin, OPG, and RANKL in MLO-Y4 osteocytes with (sh-Cas) and without (sh-control) Cas knockdown were normalized against the mean of controls, which was set at 1 (n = 3). (B) Reduced RANKL expression in RelA-knockdown MLO-Y4 osteocytes. The mRNA expression levels of sclerostin, OPG, and RANKL in MLO-Y4 osteocytes infected with control (sh-control) or short hairpin RNA (shRNA) against RelA (sh-RelA)–expressing retroviruses were quantified as in (A) (left, n = 3). RelA expression level was evaluated by immunoblotting (right). (C) Increased intracellular ROS in Cas-knockdown MLO-Y4 osteocytes. Intracellular ROS level was measured as in fig. S4B (n = 3). (D) Loss of Cas-knockdown effect on RANKL expression in MLO-Y4 osteocytes by ROS inhibition. Control (sh-Cas −) and Cas-knockdown (sh-Cas +) MLO-Y4 osteocytes, either left untreated or treated with NAC (10 mM, 24 hours), were analyzed for RANKL expression as in (A) (n = 3). (E) Loss of Cas-knockdown effect on RANKL expression and intracellular ROS level in MLO-Y4 osteocytes by RelA knockdown. MLO-Y4 osteocytes infected with control or shRNA against Cas (sh-Cas)– and/or RelA (sh-RelA)–expressing retroviruses were analyzed by quantitative polymerase chain reaction (PCR) (left, n = 3), flow cytometry (center, n = 3), and immunoblotting (right). (F) Contribution of acetylation to RelA-mediated increases in intracellular ROS level and RANKL expression. MLO-Y4 cells infected with control, Myc-tagged RelA WT, or K310R-expressing retrovirus were subjected to measurement of intracellular ROS (left, n = 5) or reverse transcription PCR analysis to quantify RANKL mRNA expression (center, n = 6). Exogenous expressions of RelA proteins were evaluated by immunoblot (IB) analysis (right).
We next looked into the mechanism by which Cas–NF-κB interplay regulated RANKL expression in osteocytes. As we could not detect the binding of RelA to the promoter region of the RANKL gene by chromatin immunoprecipitation (ChIP) analysis (fig. S4A), we postulated that RANKL expression might not be directly induced by NF-κB and tested for intracellular reactive oxygen species (ROS). Increased ROS has been shown to stimulate RANKL expression in cells of osteoblastic lineage (20), and several molecules downstream of the classical NF-κB pathway, such as tumor necrosis factor–α (TNF-α), interleukin-1β (IL-1β), NADPH (reduced form of nicotinamide adenine dinucleotide phosphate) oxidase, and xanthine oxidase, enhance ROS production (21). ROS level was increased in Cas-knockdown MLO-Y4 osteocytes (Fig. 4C and fig. S4B), in which treatment with a ROS inhibitor, N-acetyl-l-cysteine (NAC), nullified the increase of RANKL expression (Fig. 4D). While silencing RelA alone moderately reduced RANKL expression (Fig. 4B), Cas knockdown did not increase RANKL expression and intracellular ROS level in RelA-knockdown MLO-Y4 osteocytes (Fig. 4E, compare columns 2 and 4 in each graph). Furthermore, exogenous expression of WT RelA in MLO-Y4 osteocytes increased intracellular ROS level and RANKL expression as compared to the expression of RelA K310R, the acetylation-defective mutant of RelA (Fig. 4F, compare columns 2 and 3 in each graph). This suggests the contribution of acetylation to RelA-mediated regulation of intracellular ROS level and RANKL expression in osteocytes. Collectively, these results show that Cas negatively regulates RelA acetylation, which, in turn, controls RANKL expression by increasing intracellular ROS level in osteocytes.
Cas down-regulates NF-κB activity by interfering with RelA-p300/CBP interaction in the nucleus
We next asked whether Cas affected the transcriptional activity of NF-κB and conducted various exogenous expression experiments. Exogenous expression of Cas suppressed RelA acetylation in human embryonic kidney (HEK) 293 cells (Fig. 5A, lanes 2 and 3) as well as in Cas−/− mouse embryonic fibroblasts (MEFs) (Fig. 5B), and therefore, Cas-dependent suppression for RelA acetylation did not appear to be osteocyte specific. Luciferase assay revealed that overexpression of Cas led to a reduction in the NF-κB transcriptional activity enhanced by overexpression of RelA (Fig. 5C). Furthermore, exogenous expression of Cas decreased expression levels of NF-κB target genes, IL-8 and TNF-α, both of which were increased by RelA overexpression (Fig. 5D). In line with this, Cas overexpression alleviated TNF-α–enhanced NF-κB activity (Fig. 5E). In contrast, Cas knockdown enhanced TNF-α–induced NF-κB activation (Fig. 5F) and expression of IL-8 and TNF-α (Fig. 5G). Coexpression of Cas hampered NF-κB activation by constitutively active form of IKKβ, IKKβSS/EE (Fig. 5H) (22). Notably, not only WT Cas (CasWT) but also Cas15YF, in which 15 phosphorylation-defective mutations are introduced to the substrate domain (CasSD) (4), tempered the IKKβ-induced NF-κB activation (Fig. 5H, top). These results indicate that Cas has an inhibitory effect downstream of the IKK complex in the NF-κB activation cascade, without requiring CasSD phosphorylation.
Fig. 5. Cas down-regulates NF-κB activity by interfering with RelA-p300/CBP interaction in the nucleus.
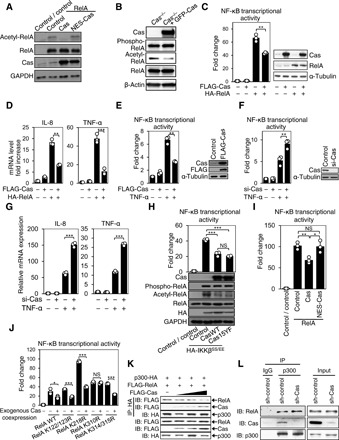
(A) Absence of suppressive effect of nuclear export signal (NES)–attached Cas on RelA acetylation. HEK293 cells transfected with Cas, NES-Cas, or their control vector were analyzed by immunoblotting. (B) GFP-Cas–dependent decrease in acetylation, but not phosphorylation, of RelA in MEFs. Cas-deficient MEFs infected with either control (Cas−/−) or GFP-tagged Cas (Cas−/− + GFP-Cas)–expressing retroviruses were analyzed by immunoblotting. (C) Alleviation of RelA-induced NF-κB activation by Cas. HEK293 cells cotransfected with the luciferase reporter vectors together with combinations of FLAG-Cas, HA (hemagglutinin)–RelA, and their respective controls were subjected to a luciferase assay. Values were normalized against the control data, which was set at 1 (left, n = 3). The expression levels of transfected genes were evaluated by immunoblotting (right). (D) Decreases in RelA-induced expression of NF-κB target genes by exogenous Cas expression. HEK293 cells, transfected with combinations of FLAG-Cas and HA-RelA as in (C) except for the luciferase reporter vectors, were analyzed for IL-8 and TNF-α mRNA expression. Values were normalized against the controls, which were transfected with parent vectors and set at 1 (n = 3). (E and F) Down-regulation of TNF-α–induced NF-κB activation by Cas. HEK293 cells cotransfected with pNF-κB Luc and phRL-TK together with FLAG-Cas (E) or Cas-targeted small interfering RNA (siRNA) (si-Cas) (F) or their respective controls, either left untreated or treated with TNF-α (10 ng/ml, 8 hours), were subjected to a luciferase assay as in (C) (n = 3). The expression level of Cas was evaluated by immunoblotting (E and F, right). (G) Down-regulation of TNF-α–induced NF-κB target gene expression by Cas. HEK293 cells, transfected with either control (si-Cas −) or Cas-targeted siRNA (si-Cas +) as in (F) except for the luciferase reporter vectors, were either left untreated or treated with TNF-α (10 ng/ml, 2 hours) and analyzed as in (D) (n = 3). (H) Cas-mediated down-regulation of NF-κB activation and acetylation by constitutively active IKKβ. HEK293 cells transfected with the luciferase reporter vectors together with combinations of HA-IKKβSS/EE, FLAG-CasWT, and FLAG-Cas15YF were subjected to a luciferase assay as in (C) (n = 3; top). HEK293 cells transfected likewise except for the luciferase reporter vectors were analyzed by immunoblotting (bottom). (I) Absence of suppressive effect of NES-Cas on RelA-enhanced NF-κB transcriptional activity. HEK293 cells, cotransfected with combinations of RelA, Cas, NES-Cas, and their respective controls as in (A), together with the luciferase reporter vectors, were subjected to a luciferase assay as in (C) (n = 3). (J) Absence of suppressive effect of exogenous Cas expression on the transcriptional activity of NF-κB in which Lys310 of RelA is mutated. HEK293 cells transfected with various RelA mutants and the luciferase reporter vectors, together with and without exogenous Cas, were subjected to a luciferase assay as in (C) (n = 3). (K) Competition of Cas with RelA for p300 binding. HEK293 cells, cotransfected with p300-HA and FLAG-RelA together with incremental FLAG-Cas, were analyzed by anti-HA immunoprecipitation (IP) and subsequent anti-FLAG and anti-HA immunoblotting. The levels of RelA and Cas recovered from coimmunoprecipitates with p300 were inversely correlated (compare the top and second rows). (L) Inverse correlation between endogenous RelA-p300 and Cas-p300 interactions in MLO-Y4 osteocytes. Note that the RelA-p300 association is enhanced in Cas-knockdown cells (compare lanes 2 and 3).
Whereas both phosphorylation and acetylation of RelA occur downstream of IKK complex activation (8), IKKβ-induced acetylation, but not phosphorylation, of RelA was significantly mitigated by CasWT and Cas15YF (Fig. 5H, bottom). Consistently, introducing GFP (green fluorescent protein)–Cas to Cas−/− MEFs suppressed acetylation, but not phosphorylation, of RelA (Fig. 5B). Together, Cas down-regulates NF-κB activity by inhibiting RelA acetylation.
We further tested whether the nuclear distribution of Cas was related to its attenuation of NF-κB activity. Nuclear accumulation of Cas was observed after treatment with an inhibitor of nuclear export, leptomycin B (fig. S5A). In our biochemical analysis, Cas was detectable in the nuclear fraction. Furthermore, the extent of nuclear distribution was comparable between CasWT and Cas15YF (fig. S5B), both of which ameliorated NF-κB activity and RelA acetylation (Fig. 5H). We next used nuclear export signal–attached Cas (NES-Cas) to determine whether translocation of Cas into the nucleus was necessary for its attenuation of NF-κB activity. NES-Cas was hardly detectable in the nuclear fraction (fig. S5C), and importantly, NES-Cas attenuated neither RelA acetylation (Fig. 5A, lanes 2 and 4) nor RelA-enhanced NF-κB activity (Fig. 5I, columns 2 and 4). These results suggest that nuclear distribution is required for Cas to hinder RelA acetylation and subsequently to alleviate NF-κB activity.
Although several lysine residues of RelA are known to be acetylated (23), only the mutation at Lys310 (K310R) rendered RelA resistant to Cas-mediated suppression of NF-κB activity (Fig. 5J and fig. S5D). This suggests that Lys310 of RelA (RelA K310) is the site of acetylation involved in the Cas-dependent modulation of NF-κB activity.
p300/CBP, which is distributed exclusively in the nucleus, has been reported to mediate RelA K310 acetylation (24). Coexpression experiments revealed that both RelA and Cas were coimmunoprecipitated with p300 (Fig. 5K and fig. S5E). Furthermore, increasing Cas expression inversely correlated with the coimmunoprecipitation of RelA with p300 (Fig. 5K), whereas coexpression with RelA hampered Cas-p300 coimmunoprecipitation (fig. S5E). In addition, endogenous RelA-p300 interaction was enhanced in Cas-knockdown MLO-Y4 osteocytes (Fig. 5L, compare lanes 2 and 3). Together with the inverse correlation between Cas nuclear distribution and RelA acetylation observed in osteocytes in vivo and in vitro (Fig. 1, B to E), these results suggest that nuclear Cas inhibits RelA acetylation by competing for interaction with p300/CBP in the nucleus.
Among various Cas truncation mutants (fig. S6A), those that contained amino acids 672 to 738 such as Cas672-874 attenuated NF-κB activity (fig. S6B) and coimmunoprecipitated with p300 (fig. S6C) similarly to full-length Cas. Furthermore, Cas672-874 appeared to interfere with the RelA-p300 interaction (fig. S6D, lanes 1 and 3). These results suggest that the amino acids 672 to 738 of Cas contain the region responsible for the inhibition of NF-κB activity.
Physiological role of the Cas–NF-κB interplay in bone homeostasis; consequences of Cas deletion in osteocytes are unaltered by heterozygous Rela deletion
Last, we analyzed Cas/Rela cKO mice to define the physiological relevance of the Cas–NF-κB interplay that we observed by these in vitro studies. To avoid enhanced apoptosis that has been shown to result from homozygous deletion of the Rela gene (25), we chose its heterozygous deletion. Whereas heterozygous Rela deletion alleviated bone loss caused by Cas knockout (Fig. 6A), the bone mass of heterozygous Rela cKO mice, which had intact Cas alleles, was not increased as compared to their control mice (fig. S7A). This suggests that the positive effect of heterozygous deletion of the Rela gene on bone mass is specific for Cas cKO. Furthermore, increased bone resorption parameters and RANKL expression in bones of Cas cKO mice were almost completely unaltered (Fig. 6, B and C) and bone formation parameters were not significantly altered (fig. S7B) by heterozygous deletion of the Rela gene. These in vivo results suggest that Cas–NF-κB interplay plays a physiological role in regulating bone resorption via RANKL expression in osteocytes.
Fig. 6. Increases in bone resorption and RANKL expression by Cas deletion in osteocytes are unaltered by heterozygous Rela deletion.
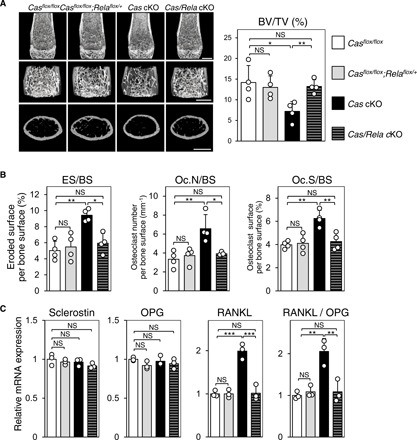
(A to C) Restoration of the low–bone mass phenotype (A), increased bone resorption (B), and RANKL expression in osteocytes (C) in Cas cKO mice by heterozygous Rela deletion. Representative μCT images of distal femurs. Scale bars, 1 mm (A, left). BV/TV was calculated from μCT images (n = 4 mice per group) (A, right). Histomorphometric parameters for bone resorption at tibiae [B; n = 4 mice per group, one-way analysis of variance (ANOVA) with post hoc Bonferroni test] and mRNA expression levels of sclerostin, OPG, and RANKL in the osteocyte fractions derived from femurs and tibiae (C; n = 3 mice per group, one-way ANOVA with post hoc Bonferroni test). Values, including RANKL/OPG ratio, were normalized against those from Casflox/flox mice, which were set at 1.
DISCUSSION
In this study, we demonstrate that Cas has a previously unidentified mechanoresponsive function whereby it translocates into the nucleus in response to shear stress and inhibits RelA acetylation. Deletion of full-length Cas withdraws not only this inhibitory effect on NF-κB activity but also other Cas functions, including force-dependent cytoskeletal association and phosphorylation (4, 5). Osteocyte-specific Cas KO mice may have mechanically relevant alterations arising independently of the NF-κB hyperactivation, particularly in relation to defective Cas phosphorylation. However, the bone phenotype of Cas cKO mice that we observed with our analysis (Fig. 2) was unaltered by heterozygous Rela deletion (Fig. 6); the phenotype can therefore be accounted for by a defect in the Cas–NF-κB interplay, which is distinct from Cas phosphorylation. Whereas the role of mechanoresponsive phosphorylation of Cas in bone homeostasis remains to be addressed, it may relate to other mechanosensory functions of osteocytes that do not involve suppression of NF-κB activity such as is mentioned below.
We demonstrate that RANKL expression in osteocytes decreases upon mechanical loading in vivo and FSS application in vitro (Fig. 3, C and D). In contrast, mechanical stretching of MLO-Y4 osteocytes with a hyperphysiological magnitude (10,000 με) has been reported to increase RANKL expression (26). Although the physiological relevance of this stretch-dependent increase of RANKL expression in osteocytes is yet to be determined, it may involve Cas phosphorylation and hamper homeostasis-disrupting events such as ectopic bone formation.
FSS imposed on the endothelium, which arises from blood flow, has been shown to regulate normal and pathological vascular functions, including morphogenesis, blood pressure, and atherosclerosis (27). The importance of FSS as a regulatory factor is not likely limited to the endothelium. Almost all cells in parenchymal tissues are bathed in interstitial fluid and can be exposed to FSS that arises from movement of interstitial fluid. Here, we demonstrate the significance of FSS-induced alleviation of NF-κB activity in bone metabolism. Given the ubiquitous expression of both Cas and NF-κB as well as the systemic distribution of interstitial fluid, shear stress–induced Cas-mediated down-regulation of NF-κB may underpin the regulation of organismal homeostasis in general, particularly that related to physical activity.
Although we find that FSS enhances nuclear distribution of Cas (Fig. 1D), the mechanism regulating Cas shuttling between the cytosol and the nucleus remains to be elucidated. Cas molecules associate with stretched cytoskeletal complexes (28) and appear to change their localization depending on cytoskeletal tension, which is modulated by fluid flow–induced shear stress (29). Together with the recently reported mechanical regulation of nuclear transport through nuclear pores (30), nuclear distribution of Cas may relate to cytoskeletal tension. Several mechanoresponsive proteins that bind to Cas without requiring CasSD phosphorylation, including FAK (focal adhesion kinase), zyxin, and DOCK180, are known to shuttle between the cytoplasm and the nucleus (3, 31). Interactions with these proteins may regulate the nuclear distribution of Cas, while further study is required to determine its detailed mechanism.
Amino acids 672 to 738 of Cas appear to include the region that is critical for binding to p300 and inhibiting NF-κB activity (fig. S6). Specific functions or interacting proteins of this region have yet to be reported. It has been shown that a C-terminal 31-kDa cleavage fragment of Cas is generated by caspase in apoptotic cells and translocated into the nucleus together with E2A protein (32). However, NES-Cas, in which NES was attached to the N terminus and thus the C terminus was not modulated, did not exhibit an inhibitory effect on NF-κB transcriptional activity and RelA acetylation (Fig. 5, A and I). Therefore, it is unlikely that the C-terminal cleavage fragment of Cas is responsible for the suppression of NF-κB activity.
Recent studies have revealed that mechanical forces are critical factors for vital cell processes (33) that underpin organismal functions. Nonetheless, the importance of mechanical loads exerted on, or generated in, local tissues and organs is poorly understood. Physical exercise is broadly recognized to be highly potent as a therapeutic or preventative procedure for numerous diseases and disorders, particularly those related to aging, as exemplified by the effectiveness of aerobic exercise in health promotion (34). Although bodily activities during exercise mostly generate mechanical loads on local tissues, molecular mechanisms behind the beneficial effects of physical exercise or activity remain largely unexplored from a mechanical perspective. Our present study clarifies the importance of cellular responses to mechanical stress in physical activity–mediated tissue homeostasis. We speculate that responses of tissues and organs to local mechanical loads, which are generated during bodily motion, may underlie the essence of physical exercise or activity as a positive measure toward well-being.
MATERIALS AND METHODS
Mice
Casflox/flox and Relaflox/+ mice of C57BL/6 background (35, 36) were mated with Dmp1-Cre transgenic mice, in which the Cre recombinase gene was specifically expressed in osteocytes (37). Dmp1-Cre+/−, Casflox/flox (Cas cKO) and Dmp1-Cre−/− Casflox/flox (normal littermates) mice were generated by mating Dmp1-Cre−/−, Casflox/flox male mice with Dmp1-Cre+/−, Casflox/+ female mice. Dmp1-Cre+/−;Casflox/flox;Relaflox/+(Cas/Rela cKO) and Dmp1-Cre−/−;Casflox/flox;Relaflox/+ (normal littermates) mice were generated by mating Dmp1-Cre+/−;Casflox/flox;Rela +/+ male mice with Dmp1-Cre−/−;Casflox/flox;Relaflox/+ female mice. Dmp1-Cre+/–Relaflox/+ (Rela cKO) and Dmp1-Cre−/−;Relaflox/+ (normal littermates) mice were generated by mating Dmp1-Cre+/−;Rela +/+ male mice with Dmp1-Cre−/−;Relaflox/flox female mice. All animals were housed under specific pathogen–free conditions and treated with humane care under approval from the Animal Care and Use Committee of Tokyo Metropolitan Institute of Gerontology, National Rehabilitation Center for Persons with Disabilities, and Nadogaya Hospital. For all mouse analyses except for those at 1 day after birth (fig. S1), male mice were used.
Cell lines
MLO-Y4 osteocytes (38) were maintained in α-MEM (Minimum Essential Medium Alpha Modification, Wako, Osaka, Japan) with 2.5% FBS (fetal bovine serum; Life Technologies), 2.5% iron-supplemented calf serum (HyClone Laboratories, Logan, UT, USA), and penicillin (100 IU/ml)/streptomycin (100 μg/ml) at 37°C. Cas-deficient MEFs and HEK293 cells (American Type Culture Collection, Manassas, VA, USA) were cultured in Dulbecco’s modified Eagle’s medium (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 10% FBS and penicillin (100 IU/ml)/streptomycin (100 μg/ml) at 37°C.
Analysis of bone phenotype
Radiography was performed using a high-resolution soft X-ray system (SOFTEX, Ebina, Japan). Microcomputed tomography (μCT) scanning was performed using a ScanXmate-L090 scanner (Comscan Techno, Yokohama, Japan). Three-dimensional (3D) microstructural image data were reconstructed, and structural indices were calculated, using TRI/3D-BON and TRI/3D-BON-C software (RATOC Systems, Osaka, Japan). Bone histomorphometric analyses involving double calcein labeling were performed as described previously (39). Symbols and abbreviations used for histomorphometric parameters were as follows (40): ES, eroded surface; BS, bone surface; Oc.N, osteoclast number; Oc.S, osteoclast surface; O.Th, osteoid thickness; Ob.S, osteoblast surface; BFR, bone formation rate; MS, mineralizing surface. BFR/BS was calculated as MAR (mineral apposition rate) × MS/BS.
Plasmids
Expression vectors for RelA (pcDNA3-FLAG-RelA and pcDNA3-HA-RelA), NF-κB–responsive reporter (pNF-κB Luc), and Renilla luciferase expression (phRL-TK) vectors were described previously (41). The HA (hemagglutinin)–IKKβ expression vector was provided by H. Ichijo (The University of Tokyo, Japan). The HA-IKKβSS/EE expression vector was constructed by site-directed mutagenesis. The expression vectors for FLAG-tagged Cas variants (Cas full-length, Cas15YF, Cas1-457, Cas1-638, Cas1-671, Cas1-738, Cas458-874, Cas639-874, Cas672-874, and Cas739-874) were constructed using pcDNA3-FLAG as a parent vector. The expression vector for p300 (CMVβ-p300-HA) (42) was a gift from R. H. Goodman (Oregon Health and Science University, OR, USA). The expression vector for NES-Cas was constructed by cloning an NES sequence (MNLVDLQKKLEELELDEQQ) (43) attached Cas into pcDNA3. NF-κB RelA K122/123R, K218R, K310R, and K314/315R were generated by site-directed mutagenesis and cloned into the pcDNA3-FLAG vector. The Myc-tagged RelA WT and K310R cloned in pcDNA3 (provided by J. Anrather) (44) were subcloned into pBabe-hygromycin to construct the retroviral expression vectors for them.
Antibodies
Polyclonal anti-Cas antibody (catalog no. 06-500, RRID:AB_310144) was purchased from Millipore (Burlington, MA, USA). Monoclonal anti-Cas antibody (catalog no. 610271, RRID:AB_397666) was purchased from BD Biosciences (San Diego, CA, USA). Monoclonal anti–α-tubulin (catalog no. ICN691251, RRID:AB_2335130) and anti-FLAG (catalog no. P2983, RRID:AB_439685) were purchased from Thermo Fisher Scientific (Waltham, MA, USA) and Sigma-Aldrich (St. Louis, MO, USA), respectively. Monoclonal anti-HA (catalog no. MMS-101P, RRID:AB_2314672) and anti-Cre (catalog no. MMS-106P-200, RRID:AB_291267) antibodies were from Covance (Denver, PA, USA). Monoclonal anti-RelA (catalog no. sc-71676, RRID:AB_1126642), anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase; catalog no. sc-32233, RRID:AB_627679), anti-p300 (catalog no. sc-56455, RRID:AB_2262185), and anti–c-Myc (catalog no. sc-40, RRID:AB_627268) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Monoclonal anti–phosphorylated RelA (Ser536) (catalog no. 3033, RRID:AB_331284), monoclonal anti–histone H3 (catalog no. 3638S, RRID: AB_1642229), and anti–β-actin (catalog no. 4967, RRID:AB_330288) antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). Polyclonal anti–acetylated RelA (Lys310) antibodies (catalog no. 3045S, RRID:AB_823580; catalog no. ab19870, RRID:AB_776753) were purchased from Cell Signaling Technology and Abcam (Cambridge, MA, USA).
Cellular fractionation
The cytosolic and nuclear fractions were prepared, following a protocol published previously (45) with some modifications. Cells were washed with ice-cold phosphate-buffered saline (PBS) twice and incubated with buffer A [10 mM Hepes·KOH (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.2% NP-40, 1 mM dithiothreitol (DTT), and protease inhibitor cocktail (Roche, Mannheim, Germany)] on ice for 10 min. Samples were then scraped, transferred to microtubes, and homogenized with vigorous shaking. The cytosolic fraction was yielded by collecting the supernatant after two consecutive centrifugations (1500g for 3 min followed by 20,000g for 10 min). The pellet after the first centrifugation was washed with wash buffer [10 mM Hepes·KOH (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, and protease inhibitor cocktail] twice, and the resultant pellet was resuspended in buffer B [20 mM Hepes·KOH (pH 7.9), 500 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and protease inhibitor cocktail], incubated on ice for 15 min, and homogenized with vigorous shaking. The nuclear fraction was yielded by collecting the supernatant after centrifugation at 20,000g for 10 min. To verify the nuclear/cytosolic fractioning and to obtain a loading control for each fraction, anti–histone H3 and anti–α-tubulin immunoblottings were conducted (fig. S5, B and C).
Immunofluorescence staining
For cultured cells, cells were washed with ice-cold PBS, fixed with 4% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 in PBS, stained using anti-Cas and anti-RelA as primary antibodies as well as Alexa Fluor 488 anti-rabbit immunoglobulin G (IgG) and Alexa Fluor 546 anti-mouse IgG (Invitrogen, Carlsbad, CA, USA) as secondary antibodies, and then viewed with a confocal microscope (Nikon A1R system). 4′,6-Diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA, USA) was used to stain the nucleus.
For osteocytes in mouse midshaft tibiae, mice were transcardially perfused with 4% paraformaldehyde in phosphate buffer (pH 7.4). Tibiae were immersed in the same fixative at 4°C overnight and decalcified in 10% EDTA (pH 7.4) at 4°C for 14 days. The samples were embedded in paraffin, sectioned with 5-μm thickness, and incubated with primary antibodies (anti-Cas, anti-RelA, and anti–acetylated RelA) at 4°C overnight. Alexa Fluor 488–conjugated goat anti-mouse IgG, Alexa Fluor 488 anti-rabbit IgG, and Alexa Fluor 594 anti-mouse IgG were used as secondary antibodies. Nuclei were counterstained using DAPI. Sections were mounted with Fluorescent Mounting Media (Dako, CA, USA). Quantitative 3D analysis of nuclear/total Cas was conducted using Imaris software (Bitplane, Zurich, Switzerland).
Coimmunoprecipitation to analyze association with p300
MLO-Y4 osteocytes or HEK293 cells were washed with ice-cold PBS, lysed with a lysis buffer [25 mM Hepes·Na (pH 7.2), 150 mM CH3COOK, 2 mM EDTA, 0.1% NP-40, 10 mM NaF, 1 mM DTT, and protease inhibitor cocktail] (46), scraped, and collected in a tube. After 10-min incubation on ice, samples were cleared of cell debris by centrifugation at 13,000g for 10 min and incubated with specific antibodies. Immunoprecipitation was conducted using PureProteome Magna Protein A Magnetic Beads (Millipore, Billerica, MA, USA) according to the manufacturer’s instruction, and proteins were recovered from resultant immunoprecipitates by 1× SDS sample buffer and subjected to SDS–polyacrylamide gel electrophoresis followed by immunoblotting.
RNA extraction and real-time reverse transcription polymerase chain reaction analysis
Total RNA was isolated using an RNA extraction kit (RNeasy, Qiagen or OMEGA Bio-Tech), and complementary DNA (cDNA), obtained using the First Strand cDNA Synthesis Kit (Qiagen or Fermentas), was amplified by quantitative polymerase chain reaction (PCR) using either QuantiTect SYBR Green PCR Master Mix (Qiagen) with a 7300 Real-Time PCR system (Applied Biosystems) or SsoFast EvaGreen Supermix with a CFX96 C1000 thermal cycler (Bio-Rad). All reactions were run in triplicate. The mRNA expression levels of individual genes were normalized to GAPDH or EF-1α (mouse) or β-actin (human) expression.
Primer sequences used for PCR
Primer sequences used were as follows: mouse GAPDH, 5′-GGATGCAGGGATGATGTTCT-3′ and 5′-AACTTTGCCATTGTGGAAGG-3′; mouse EF-1α, 5′-TGCTGCCATTGTTGATATGG-3′ and 5′-TCCACAGCTTTGATGACACC-3′; mouse sclerostin, 5′-TCCTGAGAAGAACCAGACCA-3′ and 5′-GCAGCTGTACTCGGACACATC-3′; mouse OPG, 5′-GTTTCCCGAGGACCACAAT-3′ and 5′-CCATTCAATGATGTCCAGGAG-3′; mouse RANKL, 5′-CAAGCTCCGAGCTGGTGAAG-3′ and 5′-CCTGAACTTTGAAAGCCCCA-3′; human IL-8, 5′-CTTGGCAGCCTTCCTGATTT-3′ and 5′-TTCTTTAGCACTCCTTGGCAAAA-3′; human TNF-α, 5′-TCTTCTCGAACCCCGAGTGA-3′ and 5′-CCTCTGATGGCACCACCAG-3′; human β-actin, 5′-GTACCACTGGCATCGTGATGGACT-3′ and 5′-CCGCTCATTGCCAATGGTGAT-3′.
Luciferase reporter assay
NF-κB–responsive reporter plasmid (pNF-κB Luc) and Renilla luciferase expression plasmid (phRL-TK) were cotransfected with the indicated expression plasmids into HEK293 cells. Twenty-four hours after transfection, cells were subjected to experiments. Luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega, Madison, WI, USA). Specific NF-κB Luc activity in individual samples was determined by normalizing firefly luciferase activity to Renilla luciferase activity. For all assays, experiments were performed in triplicate.
RNA interference
Short hairpin RNA introduction by retroviral gene transfer
The retroviral vectors pSuper-sh-Cas puro and pSuper-sh-RelA puro were cloned with the mouse Cas target sequence 5′-GCATGACATCTACCAAGTT-3′ and the mouse Rela target sequence 5′-GAAGAAGAGTCCTTTCAAT-3′, respectively, into a pSuper retro puro vector (Oligoengine, Seattle, WA). For double gene silencing of Cas and RelA, the retroviral vector pSuper-sh-RelA puro, in which the mouse Rela target sequence was cloned into a pSuper retro puro vector, was used together with pSuper-sh-Cas hygro. The plasmid harboring the retroviral construct was transiently transfected into the packaging cell line HEK293T or Plat-E (respectively provided by H. Ichijo and T. Kitamura, The University of Tokyo). Supernatants containing retroviral particles were collected 24 to 48 hours after transfection and used immediately to infect cell cultures. Subconfluent MLO-Y4 osteocytes were exposed to viral supernatants in the presence of polybrene (6 μg/ml) for 24 hours and then incubated in fresh culture medium for 2 days. Infected cells were selected by culturing them in the presence of puromycin (5 μg/ml) for at least 4 days. For double gene silencing, doubly infected cells were selected by culturing them in the presence of hygromycin (400 μg/ml) and puromycin.
Small interfering RNA transfection
To silence the endogenous expression of Cas in HEK293 cells, Cas-targeted small interfering RNA (siRNA) (Stealth RNAi, BCAR1-HSS114273, Thermo Fisher Scientific) was transfected using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s protocol. Twenty-four hours after transfection, cells were used for experiments. Similar results were obtained using another Cas-targeted siRNA (Stealth RNAi, BCAR1-HSS114272, Thermo Fisher Scientific).
Biochemical analysis of osteocytes in vivo and in vitro
Muscle, connective tissue, and periosteum were removed from femurs and tibiae to separate osteocytes from whole bone, and the bones were cut at the metaphyses. Hematopoietic cells and osteoblastic cells were removed from the diaphyses of femurs and tibiae by flushing with PBS and using a microinterdental brush (0.2 mm in diameter). The remaining bone was used as a source of osteocyte-enriched cells. The osteocyte-rich bones were frozen in liquid nitrogen and ground into a powder using a tissue pulverizer. The powdered bone was then subjected to extraction of RNA, DNA, and protein. MLO-Y4 osteocytes, MEFs, HEK293 cells, and osteocyte fractions were harvested, and cell lysates were biochemically analyzed.
Measurement of intracellular ROS level
The ROS level in MLO-Y4 osteocytes was measured using the DCFDA-Cellular Reactive Oxygen Species Detection Assay Kit (Abcam) according to the manufacturer’s instruction.
FSS experiments
MLO-Y4 osteocytes were exposed to FSS of 1 Pa at a frequency of 1 Hz, as described previously (47) with slight modifications. MLO-Y4 osteocytes were cultured in a 60-mm plastic dish (Sumitomo Bakelite, Akita, Japan) precoated with 0.1% bovine gelatin (Sigma-Aldrich). The experiments were conducted 1 or 2 days after the cells were seeded. To expose MLO-Y4 cells to FSS, a parallel plate flow chamber and roller pump (HV-07523-80, Masterflex) were used. The flow chamber, which was composed of a cell culture dish, a polycarbonate I/O unit, and a silicone gasket, generated a 35-mm-long, 26.7-mm-wide, 0.4-mm-high flow channel. The system was placed in a CO2 incubator to maintain pH and temperature of culture medium.
ChIP assay
The ChIP assay was performed using the EZ-ChIP Kit from Millipore according to the manufacturer’s instruction. The primers used for the ChIP assay were as follows:
mouse RANKL, 5′-GAGTTCTAGAATTTCCCCAAG-3′ and 5′-GTGCCTCTTTCACTCCCACAT-3′ (48); mouse TNF-α, 5′-TGTACCGCAGTCAAGATATG-3′ and 5′-TGCCTGTGTCTATTTCCTTT-3′.
Hemilateral hindlimb unloading experiments
Hemilateral hindlimb neurectomy was conducted by surgically resecting 5- to 6-mm segments of sciatic and femoral nerves, as described previously (15). As a control sham operation for neurectomy, the contralateral sciatic and femoral nerves were surgically exposed and left unresected. Neurectomy side (right or left) was chosen randomly. Operations were performed 1 week (for biochemical and immunohistochemical studies) or 2 weeks (for μCT and bone histomorphometric studies) before mice were sacrificed at the age of 10 weeks, and subjected to analyses.
Statistical analysis
Statistical comparisons were performed with two-tailed unpaired t test or analysis of variance (ANOVA) followed by post hoc Bonferroni test using SPSS (IBM) software, and each series of experiments was repeated at least three times. P values <0.05 were considered statistically significant. For all figures and supplementary figures, quantitative data are presented as means ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant by two-tailed unpaired t test unless specified otherwise. Adequate sample sizes of experiments were estimated on the basis of our pilot experiments and experience with similar measurements to ensure adequate power to detect a specified effect size. For animal analyses, at least three animals were assigned to each experimental group according to standard scientific conventions. We tried to reach a conclusion for each individual experiment, using as small a size of samples as possible. No data were excluded, whereas the investigators were not blinded to any group allocations, which were randomly performed for in vivo or in vitro experiments.
Supplementary Material
Acknowledgments
We thank R. M. Schmid for Relaflox/flox mice. Funding: This work was supported, in part, by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to T.M., H.K., T.O., and Y.S.); Precursory Research for Embryonic Science and Technology (PRESTO) (to M.S.); Project Grant from the Japanese Society for Musculoskeletal Medicine; MEXT-Supported Program for the Strategic Research Foundation at Private Universities, 2015–2019 from the Japanese Ministry of Education, Culture, Sports, Science and Technology (S1511017); and Seed Fund from the Mechanobiology Institute, Singapore (R-714-004-007-271) (to Y.S.). Author contributions: T.M. and Y.S. conceived the concept of this study, designed most experiments, and discussed the data. T.M., Z.Z., K.Sa., Y.I., M.S., and Y.S. wrote the manuscript. T.M. and I.H. generated conditional knockout mice and performed most of the in vivo analyses of the mice. Z.Z. performed most of the in vitro experiments to analyze the Cas–NF-κB interplay and prepared the manuscript. Y.I. carried out both in vivo and in vitro experiments to complete this work with help from I.H. D.Y. and Z.Z. conducted most of FSS experiments with support from K.Sa. and I.H. S.H. performed immunohistochemical analysis of mouse tibiae. S.M., H.Hi., K.K., K.A., K.Sh., S.T., T.I., H.K., M.N., and T.O. provided advice on project. L.F.B. provided Dmp1-Cre mice and MLO-Y4 cells. H.Ho. provided Casflox/flox mice. Y.S. directed and supervised the project. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All relevant data are available from the corresponding authors upon reasonable request. Dmp1-Cre and Relaflox/flox mice can be provided by L.F.B. and R. M. Schmid, respectively, pending scientific reviews and completed material transfer agreements. Requests for Casflox/flox mice should be submitted to RIKEN BioResource Research Center (Tsukuba, Japan: https://en.brc.riken.jp/).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaau7802/DC1
Fig. S1. Skeletal phenotype of Cas cKO mice is not distinct at birth.
Fig. S2. Bone formation parameters are not significantly altered in Cas cKO mice.
Fig. S3. Mechanical regulation of bone formation in control and Cas cKO mice and nuclear distribution of Cas in their osteocytes.
Fig. S4. ChIP analysis of RelA binding to the RANKL promoter region and measurement of intracellular ROS level.
Fig. S5. Cas suppresses RelA acetylation at Lys310 in the nucleus and alleviates NF-κB activity without requiring phosphorylation of its substrate domain.
Fig. S6. Cas672-738 contains the region responsible for the inhibition of NF-κB activity.
Fig. S7. Neither bone mass of mice with intact Cas alleles nor bone formation of Cas cKO mice is significantly altered by heterozygous deletion of Rela in osteocytes (Rela cKO).
REFERENCES AND NOTES
- 1.Fletcher D. A., Mullins R. D., Cell mechanics and the cytoskeleton. Nature 463, 485–492 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugiyama T., Meakin L. B., Browne W. J., Galea G. L., Price J. S., Lanyon L. E., Bones' adaptive response to mechanical loading is essentially linear between the low strains associated with disuse and the high strains associated with the lamellar/woven bone transition. J. Bone Miner. Res. 27, 1784–1793 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Defilippi P., Di Stefano P., Cabodi S., p130Cas: A versatile scaffold in signaling networks. Trends Cell Biol. 16, 257–263 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Sawada Y., Tamada M., Dubin-Thaler B. J., Cherniavskaya O., Sakai R., Tanaka S., Sheetz M. P., Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 127, 1015–1026 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamada M., Sheetz M. P., Sawada Y., Activation of a signaling cascade by cytoskeleton stretch. Dev. Cell 7, 709–718 (2004). [DOI] [PubMed] [Google Scholar]
- 6.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huxford T., Huang D.-B., Malek S., Ghosh G., The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell 95, 759–770 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Hayden M. S., Ghosh S., NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 26, 203–234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orr A. W., Sanders J. M., Bevard M., Coleman E., Sarembock I. J., Schwartz M. A., The subendothelial extracellular matrix modulates NF-κB activation by flow: A potential role in atherosclerosis. J. Cell Biol. 169, 191–202 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordström A., Karlsson C., Nyquist F., Olsson T., Nordström P., Karlsson M., Bone loss and fracture risk after reduced physical activity. J. Bone Miner. Res. 20, 202–207 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Knothe Tate M. L., Adamson J. R., Tami A. E., Bauer T. W., The osteocyte. Int. J. Biochem. Cell Biol. 36, 1–8 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Weinbaum S., Cowin S. C., Zeng Y., A model for the excitation of osteocytes by mechanical loading-induced bone fluid shear stresses. J. Biomech. 27, 339–360 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Tatsumi S., Ishii K., Amizuka N., Li M., Kobayashi T., Kohno K., Ito M., Takeshita S., Ikeda K., Targeted ablation of osteocytes induces osteoporosis with defective mechanotransduction. Cell Metab. 5, 464–475 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Komori T., Functions of the osteocyte network in the regulation of bone mass. Cell Tissue Res. 352, 191–198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aryal A. C., Miyai K., Hayata T., Notomi T., Nakamoto T., Pawson T., Ezura Y., Noda M., Nck1 deficiency accelerates unloading-induced bone loss. J. Cell. Physiol. 228, 1397–1403 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Winkler D. G., Sutherland M. K., Geoghegan J. C., Yu C., Hayes T., Skonier J. E., Shpektor D., Jonas M., Kovacevich B. R., Staehling-Hampton K., Appleby M., Brunkow M. E., Latham J. A., Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 22, 6267–6276 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson D. M., Maraskovsky E., Billingsley W. L., Dougall W. C., Tometsko M. E., Roux E. R., Teepe M. C., DuBose R. F., Cosman D., Galibert L., A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature 390, 175–179 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Lacey D. L., Timms E., Tan H.-L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y.-X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J., Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93, 165–176 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Kodama Y., Dimai H. P., Wergedal J., Sheng M., Malpe R., Kutilek S., Beamer W., Donahue L. R., Rosen C., Baylink D. J., Farley J., Cortical tibial bone volume in two strains of mice: Effects of sciatic neurectomy and genetic regulation of bone response to mechanical loading. Bone 25, 183–190 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Bai X. C., Lu D., Liu A. L., Zhang Z. M., Li X. M., Zou Z. P., Zeng W. S., Cheng B. L., Luo S. Q., Reactive oxygen species stimulates receptor activator of NF-κB ligand expression in osteoblast. J. Biol. Chem. 280, 17497–17506 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Zhang J., Wang X., Vikash V., Ye Q., Wu D., Liu Y., Dong W., ROS and ROS-mediated cellular signaling. Oxid. Med. Cell. Longev. 2016, 4350965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercurio F., Zhu H., Murray B. W., Shevchenko A., Bennett B. L., Li J., Young D. B., Barbosa M., Mann M., Manning A., Rao A., IKK-1 and IKK-2: Cytokine-activated IκB kinases essential for NF-κB activation. Science 278, 860–866 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Huang B., Yang X.-D., Lamb A., Chen L.-F., Posttranslational modifications of NF-κB: Another layer of regulation for NF-κB signaling pathway. Cell. Signal. 22, 1282–1290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L.-F., Greene W. C., Shaping the nuclear action of NF-κB. Nat. Rev. Mol. Cell Biol. 5, 392–401 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi H., Chang S. H., Mori D., Itoh S., Hirata M., Hosaka Y., Taniguchi Y., Okada K., Mori Y., Yano F., Chung U.-i., Akiyama H., Kawaguchi H., Tanaka S., Saito T., Biphasic regulation of chondrocytes by Rela through induction of anti-apoptotic and catabolic target genes. Nat. Commun. 7, 13336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakashima T., Hayashi M., Fukunaga T., Kurata K., Oh-Hora M., Feng J. Q., Bonewald L. F., Kodama T., Wutz A., Wagner E. F., Penninger J. M., Takayanagi H., Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat. Med. 17, 1231–1234 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Hahn C., Schwartz M. A., Mechanotransduction in vascular physiology and atherogenesis. Nat. Rev. Mol. Cell Biol. 10, 53–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawada Y., Sheetz M. P., Force transduction by Triton cytoskeletons. J. Cell Biol. 156, 609–615 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conway D. E., Breckenridge M. T., Hinde E., Gratton E., Chen C. S., Schwartz M. A., Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1. Curr. Biol. 23, 1024–1030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elosegui-Artola A., Andreu I., Beedle A. E. M., Lezamiz A., Uroz M., Kosmalska A. J., Oria R., Kechagia J. Z., Rico-Lastres P., Le Roux A. L., Shanahan C. M., Trepat X., Navajas D., Garcia-Manyes S., Roca-Cusachs P., Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 171, 1397–1410.e14 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Yi J., Kloeker S., Jensen C. C., Bockholt S., Honda H., Hirai H., Beckerle M. C., Members of the Zyxin family of LIM proteins interact with members of the p130Cas family of signal transducers. J. Biol. Chem. 277, 9580–9589 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Kim W., Kook S., Kim D. J., Teodorof C., Song W. K., The 31-kDa caspase-generated cleavage product of p130cas functions as a transcriptional repressor of E2A in apoptotic cells. J. Biol. Chem. 279, 8333–8342 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Wang N., Review of cellular mechanotransduction. J. Phys. D Appl. Phys. 50, 233002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warburton D. E., Nicol C. W., Bredin S. S., Health benefits of physical activity: The evidence. CMAJ 174, 801–809 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagai Y., Osawa K., Fukushima H., Tamura Y., Aoki K., Ohya K., Yasuda H., Hikiji H., Takahashi M., Seta Y., Seo S., Kurokawa M., Kato S., Honda H., Nakamura I., Maki K., Jimi E., p130Cas, Crk-associated substrate, plays important roles in osteoclastic bone resorption. J. Bone Miner. Res. 28, 2449–2462 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Algül H., Treiber M., Lesina M., Nakhai H., Saur D., Geisler F., Pfeifer A., Paxian S., Schmid R. M., Pancreas-specific RelA/p65 truncation increases susceptibility of acini to inflammation-associated cell death following cerulein pancreatitis. J. Clin. Invest. 117, 1490–1501 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Y., Xie Y., Zhang S., Dusevich V., Bonewald L. F., Feng J. Q., DMP1-targeted Cre expression in odontoblasts and osteocytes. J. Dent. Res. 86, 320–325 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Kato Y., Windle J. J., Koop B. A., Mundy G. R., Bonewald L. F., Establishment of an osteocyte-like cell line, MLO-Y4. J. Bone Miner. Res. 12, 2014–2023 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Miyazaki T., Iwasawa M., Nakashima T., Mori S., Shigemoto K., Nakamura H., Katagiri H., Takayanagi H., Tanaka S., Intracellular and extracellular ATP coordinately regulate the inverse correlation between osteoclast survival and bone resorption. J. Biol. Chem. 287, 37808–37823 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parfitt A. M., Drezner M. K., Glorieux F. H., Kanis J. A., Malluche H., Meunier P. J., Ott S. M., Recker R. R., Bone histomorphometry: Standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 2, 595–610 (1987). [DOI] [PubMed] [Google Scholar]
- 41.Kawauchi K., Tan W. W., Araki K., Abu Bakar F. B., Kim M., Fujita H., Hirata H., Sawada Y., p130Cas-dependent actin remodelling regulates myogenic differentiation. Biochem. J. 445, 323–332 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Ikeda A., Sun X., Li Y., Zhang Y.-k., Eckner R., Doi T. S., Takahashi T., Obata Y., Yoshioka K., Yamamoto K.-i., p300/CBP-dependent and -independent transcriptional interference between NF-κB RelA and p53. Biochem. Biophys. Res. Commun. 272, 375–379 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Araki K., Kawauchi K., Hirata H., Yamamoto M., Taya Y., Cytoplasmic translocation of the retinoblastoma protein disrupts sarcomeric organization. eLife 2, e01228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pejanovic N., Hochrainer K., Liu T., Aerne B. L., Soares M. P., Anrather J., Regulation of nuclear factor κB (NF-κB) transcriptional activity via p65 acetylation by the chaperonin containing TCP1 (CCT). PLOS ONE 7, e42020 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreiber E., Matthias P., Muller M. M., Schaffner W., Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17, 6419 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerritsen M. E., Williams A. J., Neish A. S., Moore S., Shi Y., Collins T., CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. U.S.A. 94, 2927–2932 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshino D., Sakamoto N., Takahashi K., Inoue E., Sato M., Development of novel flow chamber to study endothelial cell morphology: Effects of shear flow with uniform spatial gradient on distribution of focal adhesion. J. Biomech. Eng. 8, 233–243 (2013). [Google Scholar]
- 48.Byon C. H., Sun Y., Chen J., Yuan K., Mao X., Heath J. M., Anderson P. G., Tintut Y., Demer L. L., Wang D., Chen Y., Runx2-upregulated receptor activator of nuclear factor κB ligand in calcifying smooth muscle cells promotes migration and osteoclastic differentiation of macrophages. Arterioscler. Thromb. Vasc. Biol. 31, 1387–1396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaau7802/DC1
Fig. S1. Skeletal phenotype of Cas cKO mice is not distinct at birth.
Fig. S2. Bone formation parameters are not significantly altered in Cas cKO mice.
Fig. S3. Mechanical regulation of bone formation in control and Cas cKO mice and nuclear distribution of Cas in their osteocytes.
Fig. S4. ChIP analysis of RelA binding to the RANKL promoter region and measurement of intracellular ROS level.
Fig. S5. Cas suppresses RelA acetylation at Lys310 in the nucleus and alleviates NF-κB activity without requiring phosphorylation of its substrate domain.
Fig. S6. Cas672-738 contains the region responsible for the inhibition of NF-κB activity.
Fig. S7. Neither bone mass of mice with intact Cas alleles nor bone formation of Cas cKO mice is significantly altered by heterozygous deletion of Rela in osteocytes (Rela cKO).


