Abstract
Sepsis is characterized as life-threatening organ dysfunction caused by a dysregulated host immune response to infection. The purpose of this investigation was to determine the differential effect of sepsis on innate versus adaptive immunity, in humans, by examining RNA expression in specific immune cell subsets including monocytes/macrophages and CD4 and CD8 T cells. A second aim was to determine immunosuppressive mechanisms operative in sepsis that might be amenable to immunotherapy. Finally, we examined RNA expression in peripheral cells from critically-ill non-septic (CINS) patients and from cancer patients to compare the unique immune response in these disorders with that occurring in sepsis. Monocytes, CD4 T cells, and CD8 T cells from septic, CINS, patients with metastatic colon cancer, and healthy controls were analyzed by RNA-seq. Sepsis induced a marked phenotypic shift toward downregulation of multiple immune response pathways in monocytes suggesting that impaired innate immunity may be fundamental to the immunosuppression that characterizes the disorder. In the sepsis cohort, there was a much more pronounced effect on gene transcription in CD4 T cell than in CD8 T cells. Potential mediators of sepsis-induced immunosuppression included Arg-1, SOCS-1, and SOCS-3, which were highly upregulated in multiple cells types. Multiple negative co-stimulatory molecules including TIGIT, Lag-3, PD-1, and CTLA-4 were also highly upregulated in sepsis. Although cancer had much more profound effects on gene transcription in CD8 T cells, common immunosuppressive mechanisms were present in all disorders suggesting that immuno-adjuvant therapies that are effective in one disease may also be efficacious in the others.
Introduction
Sepsis is life-threatening organ dysfunction that results from the body’s response to invasive infection (1). Sepsis is the most common cause of death in intensive care units and is responsible for over a quarter of a million deaths annually in the United States alone (2–4). Although sepsis-induced death has historically been considered to be due to unbridled cytokine-mediated inflammation, there is a growing consensus that most of the deaths are due to impaired host immunity and failure to control invading pathogens (4–9). Many of the microbial organisms responsible for deaths in sepsis are weakly virulent and typically occur in patients with impaired immunity thereby underscoring the profound nature of immunosuppression in patients with protracted or recurrent sepsis (4–7). Additional evidence for immunosuppression in sepsis includes reactivation of latent viruses in patients with prolonged sepsis and autopsy studies documenting severe impairment of immune effector cell function (10). The fact that elderly patients who have age-related impairment in immunity, i.e., immunosenescence, have the highest morbidity and mortality in sepsis highlights the key role of immune competence as a critical factor in ability to survive sepsis. Many of these same factors associated with immunosuppression also play key roles in health and survival in patients with solid tumor cancers (11).
Numerous studies have examined gene expression in circulating immune cells in patients with sepsis and cancer to define the state of host immunity and to uncover mechanisms of immune dysregulation (12–14). A potential limitation of these previous investigations is that they did not differentiate the effects of sepsis on particular classes of immune cells, because the analyses were performed on whole blood rather than on specific cell subsets. Thus, results from these studies conducted in heterogeneous populations of immune cells from whole blood may confound and not differentiate the impact of sepsis on the various classes of immune cells comprising the innate and adaptive immune systems. This lack of cellular phenotypic discrimination is problematic, particularly in sepsis, given the current widely held paradigm that sepsis causes upregulation of effector functions (i.e. inflammatory cytokine production) in innate immune cells but downregulation of effector functions in adaptive immune cells (12, 15, 16). Also, findings from these studies may not reveal differences in immune response that exist in closely related cell types such as CD4 and CD8 T cells which play distinct roles in regulating host immunity.
Whole transcriptome shotgun sequencing, i.e., RNA-seq is a powerful method that enables detailed characterization of gene expression and provides a greater dynamic range at the lower and higher level range of expression when compared to hybridization-based (microarray genechip) approaches. To further increase the specificity and focus of the analysis, in this study we purified CD4 T cells, CD8 T cells, and monocytes from peripheral blood cells and performed RNA-seq on these individual cell populations.
Our goals were to determine the effect of sepsis on key immune cells and to discover immunosuppressive mechanisms and novel pathways operative in sepsis that might be amenable to the emerging class of immuno-adjuvant therapies that are transforming oncology. We also determined the differential effects of sepsis on CD4 versus CD8 T cells because of their unique roles in orchestrating host immunity and eliminating life-threatening pathogens. T cell IFN-γ production from septic patients was evaluated by ELISpot assay in order to relate the transcriptomic findings to the functional status of the cell. Finally, we compared the RNA expression profiles from immune cells from patients with cancer to those from patients with sepsis. Intriguingly, patients with sepsis and patients with cancer share many common immunosuppressive mechanisms including decreased MHC expression, impaired T cell IFN-γ production, increased myeloid derived suppressor and T regulatory cell signatures, and increased expression of inhibitory receptor ligands (11). Thus, insight into shared immunosuppressive mechanisms driving both disorders may be discerned by understanding the similarities and differences in the immune response in sepsis and metastatic cancer.
Materials and Methods
Study design, setting, and patient populations
This was a prospective trial performed at Barnes-Jewish Hospital, a 1,200-bed university-affiliated hospital in St. Louis, Missouri between 2015 and 2018. Data collection and analysis was approved by the Human Research Protection Office at Washington University. Informed consent for participation was provided by all patients or their legally authorized representatives.
Patients admitted to a medical or surgical intensive care unit (ICU) who were older than 18 years of age and who fulfilled a consensus panel definition of sepsis were included in the study (Table 1). Sepsis was defined as the presence of systemic inflammatory response syndrome (SIRS) and a known or suspected source of infection (17). Patients who had undergone bone marrow irradiation or who had received chemo- or radiation therapy within the last six months, patients with HIV infection, viral hepatitis, or who were receiving immunosuppressive medications (except corticosteroids at a dose of ≤300 mg hydrocortisone or equivalent per day) were excluded.
Table 1:
Patient Characteristics
| Septic | Critically-Ill | Cancer | Healthy Control | |
|---|---|---|---|---|
| Number | 29 | 22 | 5 | 20 |
| Age (Mean) (Q1,Q3) | 60 (49,66) | 59 (53,70) | 59(43,72) | 59 (49,67) |
| ICU Days (Mean) (Q1,Q3) | 12 (5,18) | 3 (1,3) | N/A | N/A |
| Patients on Ventilator (%) | 69 | 4.5 | N/A | N/A |
| Antibiotics (%) | 93 | 32 | N/A | N/A |
| Patients on Vasopressers (%) | 31 | 27 | N/A | N/A |
| Absolute Lymphocyte Count (Mean) (Q1,Q3) | 1.6 (0.8,2.2) | 1.3 (0.8,2.0) | 36.2 (20.5,46.8) | N/A |
| Absolute Monocyte Count (Mean) (Q1,Q3) | 1.0 (0.6,1.2) | 0.9 (0.6,1.0) | 11.9 (8.1,14.4) | N/A |
| Total White Cell Count (Mean) (Q1, Q3) | 15.0 (10.5,18.9) | 10.4 (8.6,11.6) | 6.6 (5.4,6.9) | 7.4 (6.2,7.9) |
| Primary Diagnosis | Septic Shock (4) | Trauma (7) | Colorectal Adenocarcinoma (Metastatic Colorectal tumors spread to liver tissue, 5) | |
| Trauma (4) | Post Spinal Surgery (5) | |||
| Necrotizing Fascitis (2) | Congestive Heart Failure | |||
| Neurologic Symptoms (4) | Gastro-Intestinal Symptoms | |||
| Cellulitis | Subdural hematoma | |||
| Peritonitis (5) | COPD Exacerbation | |||
| Infected Aortic Graft | Aortic Aneurism | |||
| Aortic dissection | Ventral Hernia | |||
| C. diff. Colitis | Subarachnoid Hemorrhage (2) | |||
| Adult Respiratory Distress Syndrome (2) | Ascites | |||
| Myocardial Infarction | Peripheral Vascular Disease | |||
The initial immune phenotype in sepsis is usually characterized by a cytokine storm mediated hyper-inflammatory innate immune response (2–5). If the patient remains septic, the immune response progresses to a more immunosuppressive phenotype. The focus of the present study was to define the more immunosuppressive phase of sepsis that occurs after the initial response had passed. Consequently, septic patient blood samples were obtained after at least 24-48 hours of sepsis onset and typically between two to 9 days after sepsis onset.
Critically-ill non-septic patients (CINS) who were admitted to the medical or surgical ICU and who were not suspected of having infection were included as a control population (Table 1). These patients consisted primarily of trauma patients and patients who underwent major surgical procedures requiring careful postoperative monitoring. Exclusion criteria were identical to that for patients with sepsis. Blood samples were typically obtained 24-48 hours after ICU admission.
Healthy age-matched control outpatients (controls) who were being evaluated for elective surgery in the preoperative clinic were included as another study population. Exclusion criteria were identical to that for patients with sepsis.
A cohort of patients with cancer was enrolled to compare and contrast the immunologic effects of cancer with those of sepsis. The cancer cohort consisted of patients who had colorectal tumors that had metastasized to the liver. Patients had to be >18 years of age and not received chemotherapy or radiation therapy within 8 weeks of blood sample collection.
Harvest of Peripheral Blood Mononuclear Cells (PBMC)
PBMCs were harvested by Ficoll-PaqueTM Plus (GE Healthcare, Uppsala, Sweden) gradient centrifugation, from blood collected in EDTA (18).
Isolation of Cell Types
Cells types were sequentially separated from patient PBMCs by positive selection first for CD8+ cells. The flow-through then underwent positive selection for CD14+ cells to obtain monocytes. Finally, the flow through of the CD14 selection underwent positive selection for CD4+ cells (EasySep by Cell Signalling Technology, Danvers, MA, USA). Following isolation, a small aliquot of cells were set aside for evaluation of purity and the bulk of the cell isolates were resuspended in QIAzolTM (QIAgen, Hilden, Germany). Isolated cell populations were stained with the following antibodies to test for purity of the population. CD4+ and CD8+ populations were stained with antiCD3-FITC (Clone: HIT3a, Cat. No. 300306), antiCD4-PerCP/Cy5.5 (Clone: RPA-T4, Cat. No. 300530), and antiCD8-APC/Cy7 (Clone:SK1, Cat. No. 344714). CD14+ populations were stained with antiCD14-PerCP/Cy5.5 (Clone:HCD14, Cat. No. 325622). Antibodies were sourced from BioLegend. Samples were acquired on a FACScan (BD Biosciences) which had been upgraded to 5 colors (CyTek, Freemont, CA. USA) Samples were analyzed using FlowJo v.10.4.1 (Bd Biosciences ).
ELISpot Assay
The ELISpot assay, which measures stimulated cytokine production of immune cells, was used in order to relate the findings from the RNA-seq to the functional status of the T cells. ELISpot assay was performed as previously reported (19) and per manufacturer’s instruction (R&D systems, Minneapolis, MN; catalog number SEL210).PBMCs were plated at a standardized density and incubated overnight with RPMI media containing anti-CD3 and anti-CD28 antibodies. IFN-γ was detected using a colorimetric reagent kit (Strep-AP and BCIP-NBT, R&D Systems, Minneapolic MO, USA, catalog number SEL002). ELISpot images were captured and analyzed on Cellular Technologies Ltd (Cleveland, OH, USA) ImmunoSpot 7.0 plate reader. In order to ensure that spot number was not just a function of varying numbers of lymphocytes in the assay patient to patient, the number of spots was divided by the number of lymphocytes input into the assay on a per-patient basis. Number of lymphocytes per assay was determined by multiplying the input number of PBMCs by the percentage of lymphocytes in the PBMC fraction.
Monocyte HLA-DR surface expression
Surface levels of HLA-DR was examined on monocytes as previously described (20). Briefly, whole blood was stained with QuantibriteTM Anti-HLADR-PE/Anti-Monocyte-PerCP/Cy5.5 (BD Biosciences, San Jose, CA, USA), red blood cells were lysed with RBC Lysis Buffer (BioLegend, San Diego, CA. USA), and samples were acquired on a FACScan (BD Biosciences) which had been upgraded to 5 colors (CyTek, Freemont, CA. USA) Samples were analyzed using FlowJo v.10.4.1 (Bd Biosciences ). Antibodies per Cell (ApC) were calculated using BD Quantibrite PE Beads as a reference in Excel (Microsoft Corp.).
Statistical Analysis of ELISpot and HLA-DR surface expression
Data from the ELISpot assay and monocyte HLA-DR surface expression was graphed in JMP 14.2.0 (SAS Institute Inc.). ELISpot and monocyte HLA-DR expression data were not normally distributed, as shown via residual plots and Box-Cox transformations. As both data sets followed a log-normal distribution, a log10 transformation was applied prior to data analysis. Both ELISpot and HLA-DR expression data were analyzed via one-way Analysis of Variance (ANOVA).
RNA Extraction, sequencing and analysis
RNA was extracted and barcoded libraries were prepared for analysis. Sequencing was performed on an Illumina HiSeq 3000 and on average, 57 Million paired read ends were uniquely mapped with an average coverage of 110 paired reads and quality checks were performed on the data prior to analysis. To assess potential sample cross-contaminations, we examined the expression of cell-type specific markers (CD3D, CD3E and CD4 for CD4+ T cells, CD8A and CD8B for CD8+ T cells, CD14 for CD14+ monocytes) on samples of individual cell types (See results). The expression of cell-type specific markers overall strongly agrees with the corresponding cell type. Eight samples present as outliers for cell-type specific marker expressions and were excluded from downstream analysis (Figure S1). An initial quality check of raw FASTQ data was performed using FastQC (21). Reads that were either less than 25bp, or had a maximum quality score below Q15, or average quality score below Q10 were discarded. The quality filtered reads were mapped to the rhesus macaque reference genome MacaM version 7.6.8 (22), with 16,048 genes and 18,753 transcripts, using STAR (23). The read counts were called using Subread (24). Differential gene expression analysis was performed for CD4+, CD8+ and CD14+ samples using DESeq2 in R Bioconductor (25).. Significant differentially expressed genes (DEGs) with fold change > 1.5 and false-discovery rate (FDR) adjusted P-value < 0.05 were identified comparing sepsis, CINS and cancer patients to age-matched healthy controls for each cell type. Pathway enrichment analysis for DEGs was performed using MetaCore (GeneGo) v5.0 (Thomson Reuters). RNAseq data are deposited at the National Centre for Biotechnology Information in the Gene Expression Omnibus databases (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133822). Additional methods are found in the supplementary section.
Results
Patient cohorts
A total of 99 subjects were enrolled between the septic, CINS, and control groups. Of these, 14 patients had no samples drawn as a result of withdrawal or transfer out of the intensive care unit prior to sample collection. Of the remaining 85 individuals, 13 patients were unevaluable due to cell counts which were too low to harvest sufficient amounts of RNA, or RNA quality was too low to continue: 5 Septic, 5 CINS, and 3 controls. The remaining 72 patients were grouped as follows: 29 septic, 23 CINS, and 20 controls (Table 1). Five patients with colon cancer which was metastatic to the liver were enrolled.
Peripheral blood samples from patients and healthy controls were sorted by magnetic bead separation into CD4+, CD8+ and CD14+ (monocyte) cell populations and the purity was confirmed by flow cytometry. Average purity as assessed by flow cytometry was 90.9 % CD4 T cells, 84.6 % CD8 T cells, and 77.8 % monocytes respectively (Supplemental Fig. 1). RNA was isolated from each cell type from each patient group and analyzed by RNA-seq. A further test of purity was performed using the RNA-seq results by examining gene expression of several cell type specific markers in each of the three cell types (Supplemental Fig. 1). Based on this analysis, eight samples (5 CD4+ samples, 2 CD8+ samples, and 1 CD14+ sample) were excluded.
Sepsis has more profound effects on CD4 T cells and monocytes compared to CD8 T cells
In order to compare the impact of sepsis on the 3 diverse cell types, we first examined the absolute number of genes that were altered in CD4 T cells, CD8 T cells, and monocytes in septic patients as compared to healthy age-matched controls. Surprisingly, sepsis induced approximately five times more differentially expressed genes in CD4 T cells and monocytes compared to changes in CD8 T cells. Specifically, CD4 T cells and monocytes from septic patients had 1,976 and 2,163 differentially expressed genes compared to healthy controls, while there were only 375 differentially expressed genes in CD8 T cells in septic versus controls (Fig. 1, Table 2).
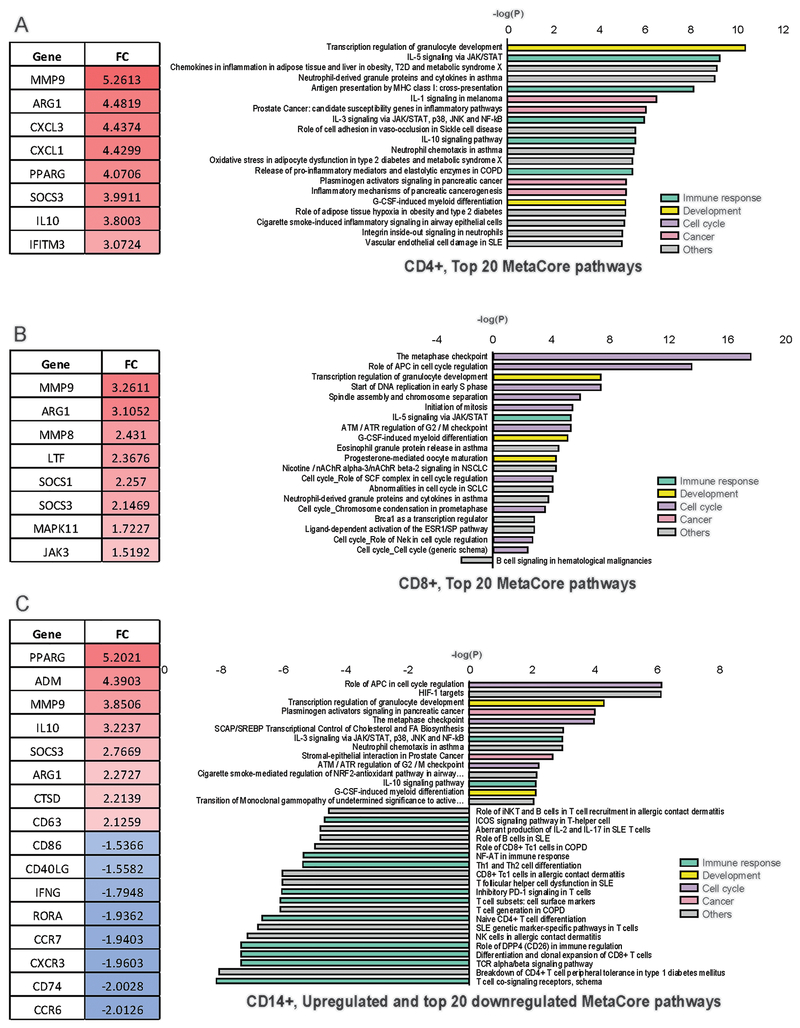
Fig1. Differentially expressed pathways and genes in septic PBMCs:
Several highly significant genes (left side) and the top MetaCore pathways (right side) found to be differentially expressed in CD4+ Tcells (A.), CD8+ Tcells (B.) and CD 14+ Monocytes (C.) of septic patients compared to healthy individuals are displayed. Genes are displayed with their fold change (FC) and pathways are displayed with their negative log (P). Color coding indicates functional categorization of the pathways (Right side). Monocytes had more considerably more downregulated pathways than upregulated pathways (C, right hand).
Table 2:
Numbers of differentially expressed genes (DEGs, A) and MetaCore pathways (B)
| DEGs (FC>1.5, FDR<0.05) | CD4 (Upregulated/Downregulated) | CD8 (Upregulated/Downregulated) | CD14 (Upregulated/Downregulated) |
|---|---|---|---|
| Sepsis vs Healthy * | 1976 (1586/390) | 375 (309/66) | 2163 (975/1188) |
| Crit-ill vs Healthy | 1477 (1255/222) | 65 (53/12) | 1264 (475/789) |
| Cancer vs Healthy | 3019 (1696/1323) | 4460 (2704/1756) | 2207 (812/1395) |
| Pathways (MetaCore, FDR<0.01) | |||
| Sepsis vs Healthy | 129/0 | 22/1 | 14/69 |
| Crit-ill vs Healthy | 175/0 | 2/0 | 46/66 |
| Cancer vs Healthy | 115/2 | 363/1 | 27/55 |
Pathway enrichment analysis was performed by MetaCore analysis and a similar differential effect was observed in the molecular and immunologic pathways of the cell types studied that were impacted in septic versus controls. There were 129 pathways activated in CD4 T cells and 83 pathways were activated in monocytes (Fig. 1A, Table 2). In contrast, only 23 pathways were activated in CD8 T cells (Fig. 1B,Table 2,). Importantly, the types of cellular molecular pathways activated in CD4 T cells were also distinctly different from those activated in CD8 T cells. The majority of the 129 pathways activated in CD4 T cells were related to cellular immune response. In contrast, almost all of the 23 pathways that were enriched in CD8 T cells were involved with cell cycle regulation and cell development, with few pathways related to modulation of immune response, (Fig. 1A&1B).
Sepsis causes marked downregulation of genes and immune response pathways in monocytes
Analysis of the differentially expressed genes in CD4 and CD8 T cells and monocytes demonstrated a marked difference in the response of T cells versus monocytes. There were 1,586 upregulated genes versus 390 downregulated genes in CD4 T cells. A similarly increased ratio of up to down regulated genes was seen in CD8 T cells in which there were 309 upregulated genes and 66 down regulated genes. In contrast to the predominant upregulation of genes in CD4 and CD8 T cells, approximately 55% of the differentially expressed genes in monocytes were downregulated (Table 2).
A second prominent finding in monocytes from septic patients compared to healthy control patients was the downregulation of pathways involved in mediating the cellular and immune response (Fig. 1C, Fig. 2 right hand panel). In CD4 and CD8 T cells the majority of immune response pathways were increased (Figs. 1A&B), whereas the majority in monocytes from septic patients were suppressed (Figs. 1C&2). Specifically, 10 immune response pathways were suppressed while only 2 immune response pathways were increased in monocytes from septic patients compared to controls. Furthermore, one of the two immune response pathways that was upregulated in monocytes in septic patients was the IL-10 pathway that is involved in immunosuppression. The dramatic difference in the impact of sepsis on immune response pathways in CD4 and CD8 T cells versus monocytes is further highlighted by noting the number of upregulating immune response pathways (Red color scale, left side of figure) in CD4 and CD8 T cells compared to the downregulated pathways (Blue color scale, right side of figure) for monocytes respectively (Fig. 2B). Those pathways that were upregulated in monocytes from septic versus control patients predominantly involved cell cycle and cell development (Fig. 1C). Please note that the lists of pathways in Figure 1 is not an exhaustive list of the pathways found to have altered expression, and instead lists only the top 20 up and down regulated pathways.
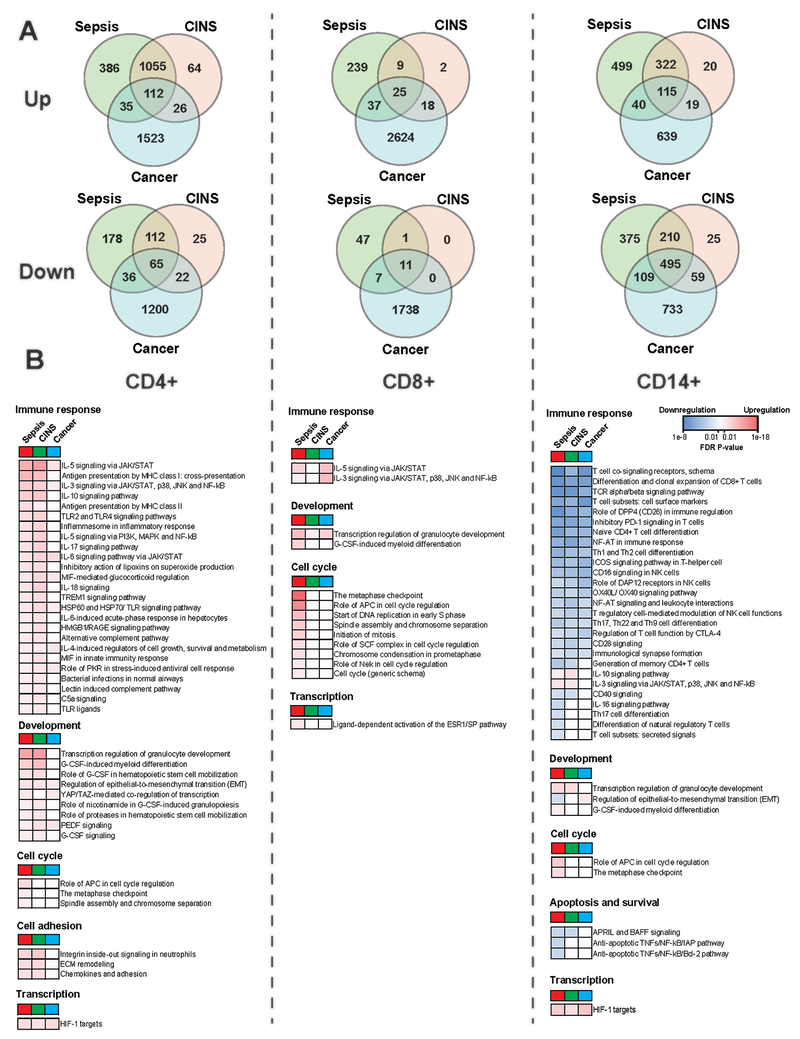
Fig2. Comparison of Differentially Expressed Genes DEGs and pathways across septic, Critically-Ill Non-Septic (CINS), and cancer groups:
A. Venn diagrams display numbers of DEGs from each cell type and group, with the overlap of found DEGs within each cell type across groupings. B. Many pathways found to be differentially expressed are displayed, and categorized into functional groupings. Comparisons of degree of significance across patient groups are shown for each of these pathways based upon False Discovery Rates (FDRs, Blue represents downregulation and red represents upregulation). FDRs for each group are as compared to the healthy group. (A) Note that there were significantly more changes in gene expression (both up and down regulated) in CD8 T cells in cancer patients as compared to septic or CINS patients. (B) Note the marked downregulation of immune response pathways in monocytes as compared to upregulation of pathways for CD4 and CD8 T cells, blue color shading for monocytes vs. red color for CD4 and CD8 T cells.
Sepsis suppresses monocyte co-stimulatory molecules, antigen presentation, and signaling
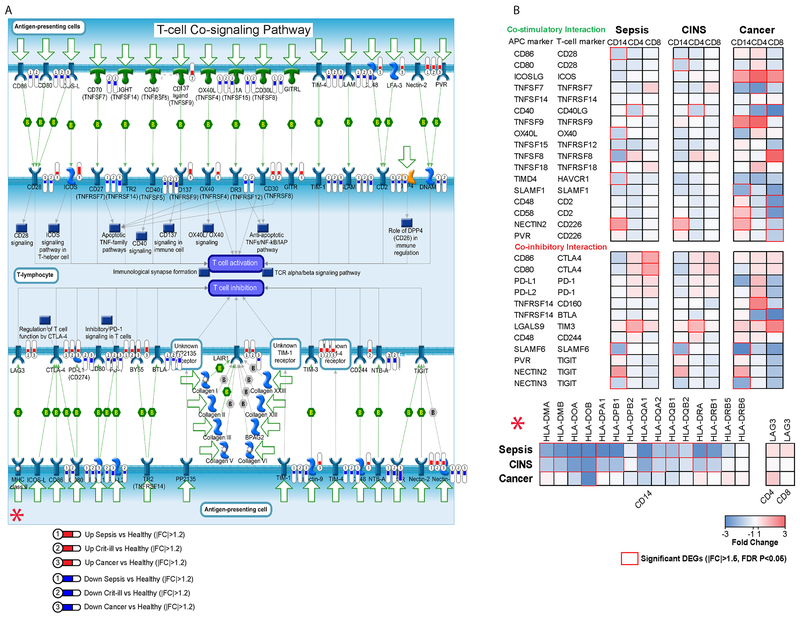
Intriguingly, we found numerous pathways and genes associated with co-stimulatory interactions of monocytes and T cells were downregulated in monocytes from sepsis patients compared with healthy controls (Figs. 1C, 2B, & 3). This finding is of particular interest given published studies of checkpoint inhibitors in sepsis (26–28). Representative downregulated genes included CD86, OX40L, and TIMD4. In addition to suppression of costimulatory pathways, several key signaling pathways (and representative genes) involving NF-AT (LCK, FASL, NFATC2, etc.) and ICOS (ICOS, PLCG1, ITK,) were also downregulated in monocytes from septic versus control patients. In addition, expression of antigen presentation genes were significantly decreased including 12 genes that function to regulate human leukocyte antigens (Fig. 3b, bottom panel). This finding of decreased expression of genes regulating antigen expression was consistent with results from flow cytometric analysis which demonstrated a significant decrease in monocyte HLA-DR expression in septic versus control patients (Supplemental Fig.2).
Fig3. Altered monocyte, CD4 Tcell interaction in sepsis, cancer, and critical illness:
Many of the molecules at the immunologic synapse showed altered expression, including several antigen presentation proteins. A. Bubbles represent that a gene was differentially expressed, numbers within them represent the patient population in which they were differentially expressed (1=Sepsis, 2=CINS, 3=Cancer). Blue colored bubbles represent downregulated genes while red colored bubbles represent upregulated genes. The amount of fill in the bubble indicates relative degree of altered expression, minimum 1.2 |FC|. B. Important immune synapse ligand/receptor pairs and antigen presentation molecules (HLAs) are shown with relative fold changes of specific genes. Red boxes represent significant DEGs (|FC|>1.5, FDR p<0.05). There was a more intense downregulation of antigen presentation molecules (HLAs) in septic and critically ill non-septic (CINS)patients vs cancer patients (lower right corner of figure). However, cancer patients had more intense downregulation of CD8 T cells vs septic or CINS patients (far upper right corner of figure).
Although most pathways were inhibited in monocytes from septic versus control patients, several pathways were upregulated. These upregulated pathways included cytokine mediated responses, e.g., IL-3 (PU.1, Cyclin A2, Bcl-6, JAK3, etc.) and IL-10 (IRS-2, HMOX1, FcgRI, etc.). Other upregulated pathways in septic versus control patients included those mediating myeloid differentiation (ITGAM, Myeloblastin, SOCS3, etc.) and cholesterol biosynthesis (SREBP2, SCD, FASN, etc.).
Sepsis upregulates key immunomodulatory and cell stress pathways
Findings of particular interest are presented because of the highly statistically significant nature of the results and because of the potential significant impact of these genes on mediating the immunosuppressive phenotype that is a hallmark of sepsis. Arginine is essential for T cell proliferation, zeta-chain peptide and T cell receptor (TCR) complex expression, and development of memory (26). Arginase, which breaks down arginine, is highly expressed on myeloid suppressor cells in sepsis and in cancer (29, 30). Arginase 1 (ARG1) was one of the most significantly upregulated genes in both CD4 (4.48 FC/6.68e-12 FDR) and CD8 T cells (3.11/2.03e-12), and, to a slightly lesser extent, in monocytes (2.27/1.28 e-5), from septic versus healthy control patients. Suppressors of cytokine signaling (SOCS) which are reported to be elevated in patients with sepsis are key inhibitors of the JAK-STAT signaling pathway that negatively regulate signaling by cytokine and toll-like receptors (31). SOCS1 and SOCS3 were significantly upregulated in both CD4 (1.82/1.28e-7, and 3.99/2.06e-25 respectively) and CD8 T cells (2.26/1.85e-9 and 2.15/6.39e-7 respectively) in septic patients. SOCS3 was also highly upregulated in monocytes (2.77/5.06e-17) from septic patients. Similarly, MMP9 was upregulated in CD4 Tcells, CD8 Tcells and monocytes from septic patients but not in CINS patients (Fig. 1).
The HIF-1α pathway has previously been shown to be upregulated in patients with sepsis and to play an important role in mediating functional re-programming of monocytes in sepsis from a pro-inflammatory to anti-inflammatory phenotype (32). While HIF-1α was not, itself, found to be differentially expressed in any of our samples, sepsis caused a highly significant upregulation in the hypoxia inducible factor 1 (HIF-1) targets pathway with an increase in expression of downstream targets of HIF-1 including TGM2, LDHA, CITED2, etc. in all 3 cell types (supplemental Fig. 3).
Histones play important roles in control of DNA expression by how tightly they bind and suppress DNA expression. Over twenty histone-related genes were upregulated in monocytes from septic versus control patients. Among these genes were histone deacetylases and methyltransferases.
Several pro- and anti-inflammatory cytokine signaling pathways (e.g. IL-5, IL-6, IL-8, IL-10, etc.) were significantly upregulated in CD4 T cells from septic versus healthy control patients. Other pathways and associated genes of interest that were enriched in CD4 T cells of septic patients involved immune function and response to cell stimulation , cell cycling (Aurora Kinase A and -B, PLK1, CDK1, etc.), and chemokines (4 C-C motif, and 5 C-X-C motif chemokines). A number of stress-related pathways also showed increased expression in CD4 T cells of septic versus control patients. These pathways included responses to both oxidative and hypoxic stress, for example, SOD2, HMOX1, APOE, GPX1, VEGFA, and MMP9, and to cell damage, for example, DDIAS, DRAM1, DDIT4, ICAM1, HGF, and Cyclin B2.
Critically-ill non-septic patients have extensive overlap in gene expression with septic patients
An additional goal of this study was to determine whether the transcriptional gene response in septic patients was unique to sepsis or also occurred in critically-ill non-septic (CINS) patients. Similar to findings in septic patients, RNA-seq analysis of samples from CINS patients showed that compared to healthy control patients there were marked changes in differentially expressed genes in CD4 T cells (1,477 genes: 1255 up/222 down) and monocytes (1,264 genes: 475 up/789 down) but not in CD8 T cells (65 genes: 53 up/12 down) (Table 2, Fig. 2A). Similarly, MetaCore analysis showed that CD8 T cells from CINS patients had far fewer activated pathways compared to CD4 T cells and monocytes. In this regard, 175 (175 up/0 down) 112 (46 up/66 down) pathways were enriched in CD4 T cells and monocytes respectively while only 2 (2 up/0 down) pathways were enriched in CD8 T cells of CINS patients versus healthy controls. Also consistent with the findings in patients with sepsis was the fact that most of the differentially expressed genes and pathways from monocytes of CINS patients were downregulated compared to healthy control patients.
There was a high degree of overlap in the genes that were upregulated in CD4 T cells in septic and CINS patients. Of the 1,583 genes that were upregulated in CD4 T cells in septic vs healthy controls, 1,167 or 73% were also upregulated in CINS patients (Fig 2A). Although not as extensive as the degree of upregulation, there was a 45% overlap in the downregulated genes in CD4 T cells in septic and CINS patients. Although sepsis caused far fewer genes to be upregulated in CD8 versus CD4 T cells, i.e., 310 versus 1,583 respectively, the increase in sepsis was much greater than in CINS in which only 54 genes were upregulated, (Fig. 2). There were numerous up and downregulated monocyte genes in both septic and CINS patients and, they also shared a high degree of overlap.
The extensive overlap in gene expression in CD4 T cells and monocytes in septic and CINS patients was reflected in the high degree of overlap in shared immune response pathways in these two conditions (Fig. 2b). All 25 of the top upregulated immune response pathways in CD4 T cells in septic patients were also upregulated in CINS patients (Fig. 2b, left hand panel). Similarly, all 25 pathways that were downregulated in monocytes from septic patients were also downregulated in CINS patients, (Fig. 2b, right hand panel). In contrast, sepsis had distinct effects on cell cycle pathways in both CD4 T cells and monocytes compared to CINS patients. In marked contrast to the high degree of overlap in pathways of CD4 T cells, there was almost no overlap in immune response pathways or cell cycle pathways for CD8 T cells of septic versus CINS patients (Fig. 2, middle panel).
One of the most striking findings was the extensive downregulation of HLA molecules occurring in both septic and CINS patients (Fig. 3). Eleven of the twelve HLA molecules that were downregulated in sepsis were also downregulated in CINS patients although usually to a much lower extent. Similarly, there was a very high degree of overlap in the impact of septic and non-septic critical illness on downregulation of costimulatory molecules in CD4 T cells and monocytes (Fig. 3). Upregulated pathways that were present in CD4 T cells of both septic and CINS included pro- and anti- inflammatory signaling pathways, e.g. IL-3 and IL-10 pathways, and targets of the hypoxic stress pathway associated with HIF-1α (Fig. 2b). HIF-1α downstream targets were upregulated in CD4 T cells and monocytes from both septic and CINS patients. SOCS3was upregulated in CD4 and CD8 T cells and monocytes in both septic and CINS patients.
Pathways that were upregulated in CD4 T cells of CINS but not septic patients were associated with glucocorticoid inhibition of signaling (IL-1RI, IL-1β, ICAM1, etc.) CCL signaling (CCL20, CXCL16, CCR1, MMP8, etc.) and oncostatin signaling (Oncostatin M, EGR1, c-FOS, etc.). The pathways selectively upregulated in monocytes of the CINS but not patients were involved with cell adhesion (ITGAM, Fibronectin, Thrombospondin-1, etc.) and IL-5 and IL-1 signaling.
Significant commonality in immune response in cancer, septic, and CINS patients
Analysis of the present results enables an overview of key similarities and differences in the phenotypes of circulating lymphocytes from colorectal cancer patients compared to those of sepsis and CINS conditions. The most remarkable finding is the intense downregulation of monocyte immune response pathways that occurred in cancer, septic, and CINS patients (Fig. 2b). Strikingly, over a third of the differentially expressed genes that were downregulated in monocytes from cancer patients were also downregulated in monocytes from septic and CINS patients (Fig. 2A, right hand panel).
Upregulated pathways common to all three conditions included numerous cytokine signaling pathways in CD8 T cells, HIF-1α targets pathways in CD4 T cells, and KRAS signaling in monocytes. Pathways that were downregulated in all three conditions included T cell signaling and co-signaling pathways in monocytes (Fig. 3). Relative levels of up-and down-regulation across the three patient groups can be seen in Fig. 3, across the three groups there are multiple instances of downregulation of costimulatory markers and upregulation of inhibitory markers, as well as large deficiencies in expression of HLA RNAs in monocytes (Fig. 3).
Pathways that were present in both sepsis and cancer samples but not CINS samples were few. In CD4 T cells, these included “IL-6 signaling in breast cancer cells”, “YAP/TZA-mediated co-regulation of transcription”, and “IGF family signaling in colorectal cancer”. There were other IL-6 related pathways that were found to be upregulated in CD4 T cells from CINS patients, likewise for IGF pathways. However, pathways including YAP/TZA were wholly absent from the CINS CD4 T cells. In CD8 T cells, there were 3 pathways that fit this criteria: Immune response_IL-3 signaling via JAK/STAT,p38/JNK and NF-kB, Neutrophil-derived granule proteins and cytokines in asthma, and Immune response_IL-5 signaling via JAK/STAT. There were no similar pathways relating to IL-3 nor IL-5 signaling, nor were there any pathways relating to neutrophil granule proteins in the CINS CD8 T cells. All pathways that were found to be common to sepsis and cancer in monocytes were found to also be common to CINS samples.
Unique effect of cancer on CD8 T cells compared to septic and CINS patients
The most apparent differences between cancer and sepsis on immune cells were evident in CD8 T cells. Sepsis had a major impact on gene expression in CD4 T cells and monocytes, but had comparatively minor effects on gene expression in CD8 T cells (Fig. 2). In contrast to findings in septic and CINS patients, CD8 T cells obtained from patients with metastatic cancer had marked changes in differentially expressed genes compared to healthy control patients (Fig. 2). The number of upregulated genes in CD8 T cells from cancer patients was approximately ten times greater than that in septic patients (Fig. 2). Similarly, the number of down regulated genes in CD8 T cells from cancer patients was approximately 25 times greater than that in septic patients. These abundant transcriptional changes are suggestive of immunoediting and a systemic anti-tumor T cell response.
Several altered pathways were unique to patients with sepsis
As previously noted, CD4 and CD8 T cells from septic individuals displayed upregulation of cell cycle pathways. These pathways were largely associated with mitosis checkpoints. In CD4 T cells: Metaphase Checkpoint, Spindle Assembly and Chromosome Separation, and the Anaphase-Promoting Complex Role in Cell Cycle Regulation were upregulated; these pathways were not altered in either the CINS nor cancer samples. The other sepsis-only pathway had to do with TGF-beta signaling via kinase cascades. The pathways found to be upregulated in CD8 T cells from septic patients versus controls were not differentially expressed in either of the other groups, although there were overlapping differentially expressed genes. As mentioned, the majority of these involved cell cycle related pathways. Other, non-cell cycle associated pathways unique to sepsis involved certain types of pro-inflammatory cytokine production, cell differentiation, DNA damage, granule protein release, and others. The only MetaCore pathway found downregulated in CD8 T cells in sepsis was B cell signaling in hematological malignancies.
In monocytes, pathways uniquely upregulated in sepsis involved myeloid differentiation, transcriptional control of cholesterol and fatty acid biosynthesis, and several cell cycle associated pathways. Those downregulated pathways that were sepsis specific included Bcl-2 and IAP anti-apoptotic pathways, H-RAS and TC21 regulatory pathways, several immune response pathways (including Treg differentiation, IL-16 signaling, secreted signals, and Th17 differentiation), rheumatoid arthritis, EMT, and Th2 cell migration pathways. In addition, there were greater than 20 genes upregulated in monocytes from sepsis, but not in monocytes from critically ill non-septic patients, associated with histones. This may indicate a role of chromatin alterations (epigenetic modifications) in sepsis contributing to the immunosuppressive phenotype of the monocytes.
Gene expression in functionally suppressed septic patients:
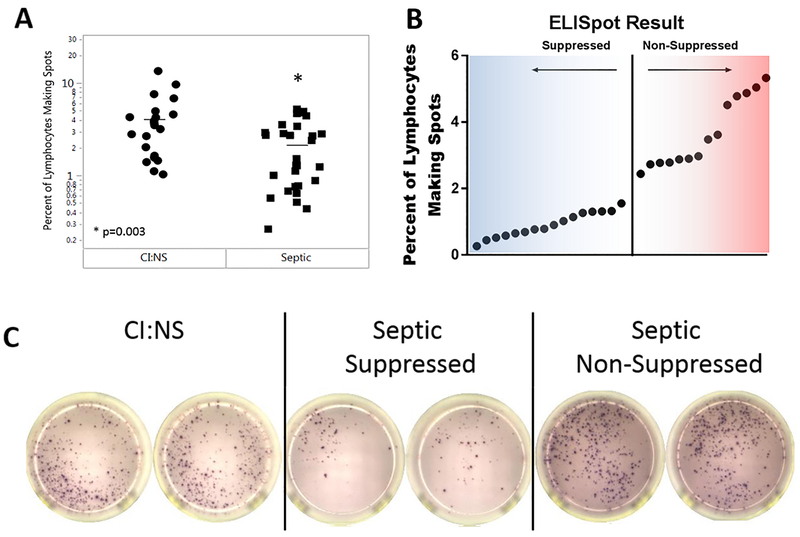
A hallmark of sepsis is impaired T cell function including reduced production of the key cytokine IFN-γ (5–7). Impaired T cell IFN-γ production is postulated to be an important pathophysiologic abnormality in sepsis and clinical trials of IFN-γ as an immune boosting therapy have been conducted (6, 9). To determine if patients with sepsis had impaired T cell function and to relate the putative decreased T cell function to gene expression, PBMCs from septic and CINS patients were stimulated with α-CD3/α-CD28 and IFN-γ production quantitated via ELISpot assay (Fig. 4). The percent of IFN-γ producing lymphocytes was approximately twice as great in CINS patients compared to septic patients (Fig. 4A). Examination of stimulated T cell IFN-γ in septic patients revealed a non-normal distribution (Shapiro-Wilk, p=0.0029) that appeared bimodal (Fig. 4A). These sepsis patients were subsequently stratified into two groups, i.e., those with a suppressed versus non-suppressed IFNγ response (Figure 4B).
Fig4.
Ability of Septic PBMCs to respond to stimulation as measured by IFNg ELISpot: Septic patient PBMCs were less able to respond to stimulation than similar CINS patients. A. When controlled for the number of lymphocytes in the assay, a larger percentage of the lymphocytes in PBMCs from CINS patients were able to mount a response to anti-CD3/anti-CD28 stimulation than those from septic patients as measured by ELISpot. B. Distribution of ELISpot results in the septic patient group was non-gaussian. The multi-modal distribution of response to stimulation allowed distinction between suppressed (i.e. little response to stimulation) and non-suppressed (i.e. greater response to stimulus). C. Representative ELISpot images from a CINS patient, a suppressed septic patient, and a non-suppressed septic patient.
A further analysis was carried out to determine what, if any, transcriptomic differences existed between patients who were unable to mount a response to stimulation and those who were able to mount a response. Between these groups, there were 486 and 206 DEGs in CD4 and CD8 T cells respectively. Classification of these differentially expressed genes demonstrated 12 pathways that were upregulated in CD4 T cells in non-responders versus responders (Table S1). These pathways included cell adhesion and inflammation GRO-1, GRO-2, IL8RA, IL8RB, TGF-α, etc. There were also pathways involving Th17 (MMP9, Leukocyte elastase [ELANE], etc.) and Th2 (MMP-12, CHI3L1, etc.) related functions. Interestingly, given the prior findings regarding the intense upregulation of ARG1, one of the differentially expressed genes that were upregulated in the non-responders included the immunosuppressive gene ARG1.
Discussion
A major goal of the present study was to determine the effect of sepsis on the host immune response using RNA sequencing of key immune cells. A more thorough understanding of the pathophysiologic effects of sepsis on the molecular mechanisms regulating immunity could provide insight into potential new therapeutic approaches. The present study using RNA-seq technology has a number of advantages compared to most previous studies that examined the sepsis transcriptome. As opposed to microarray analysis, the methodology that was employed in most previous studies, RNA-seq provides superior dynamic range and thus is able to more precisely define the magnitude of the effect of sepsis to increase or decrease gene expression. Additionally, RNA-seq does not depend on probe capture, and thus will identify all transcripts, including those that may have polymorphisms or be unexpected or unknown. An additional strength of the present study is the inclusion of patients with three different clinical conditions thereby enabling the ability to identify and compare immuno-regulatory mechanisms occurring in all three diverse conditions. Another advantage of the present study is the identification of the effects of sepsis on individual cellular components, i.e., CD4 T cells, CD8 T cells, and monocytes. Most previous studies that have examined the impact of sepsis on gene expression have examined whole blood obtained by Pax gene tubes. Whole blood gene analysis may not enable an accurate assessment of the differential effect of sepsis on the two key arms of immunity, that is, innate and adaptive immunity.
Although there are a number of novel findings in the present study, perhaps the most surprising finding was the degree of down regulation of monocyte molecular pathways involved in the immune response in patients with sepsis compared to healthy controls (Fig. 1 & Fig. 2 right panel). This finding of sepsis-induced suppression of monocyte immune pathways contrasts with the widely held concept that sepsis induces upregulation of the innate immune response (5–7). Comparing changes in gene expression in monocytes from septic patients to healthy controls showed 2,165 differentially expressed genes with 976 genes upregulated and 1,189 genes down regulated. Although the number of up versus down regulated genes was comparable, the impact of gene changes on immune response pathways was markedly shifted towards immune suppression (Figs 1&2B). Furthermore, one of the few upregulated immune response pathways in monocytes shown in Fig. 2B, is the IL-10 signaling pathway which has an immunosuppressive effect. Immune pathways that were upregulated in monocytes of septic patients did include several pro-inflammatory pathways including, for example, chemotaxis and IL-3 signaling. Undoubtedly, both pro- and anti-inflammatory pathways are activated in sepsis but the present results show a marked predominance of immunosuppressive mechanisms. Numerous studies of circulating and tissue monocytes from septic patients consistently show impaired secretion of pro-inflammatory cytokines, e.g., TNF-α, and decreased antigen expression (6–10). Thus, these functional studies of monocytes support the present transcriptomic results indicating that sepsis results in a predominant immunosuppressive phenotype in monocytes.
A puzzling question is why does the present study show extensive downregulation of genes involved in control of innate immunity given the current widely held belief that sepsis induces intense upregulation of innate immunity (5–7, 12–16). One probable explanation for this discrepancy likely relates to the timing of our patient sampling. The purpose of the present study was to examine mechanisms responsible for the immunosuppressive phase of sepsis, i.e., after the initial hyper-inflammatory phase of sepsis had passed. The timing of sampling of patients was at least 24-48 hours after sepsis onset and typically 2-7 days after admission to the ICU for sepsis management. Many previous studies that have examined gene expression in trauma and sepsis investigated the early initial phase of the disorder which would correspond to the hyper-inflammatory phase. The present transcriptomic findings showing suppressed innate immunity are consistent with numerous studies that have documented impaired function of monocytes from patients with sepsis as indicated by global impairment in stimulated monocyte cytokine secretion (33). These functional studies showing impaired monocyte activity provide support for the present transcriptomic findings which are indicative of marked and widespread gene suppression. Undoubtedly, there are inflammatory mechanisms that are still operative in monocyte/macrophages in patients with prolonged sepsis. However, the present results are consistent with an overall profound degree of suppression of innate immunity as well as adaptive immunity.
The shift of monocytes to a more immunosuppressive phenotype was also present in critically ill non-septic patients and cancer patients (Fig. 2B). Interestingly, the overlap in the number of differentially expressed genes in septic, critically ill non-septic, and cancer patients is much more profound for monocytes than for CD4 or CD8 T cells (Fig. 2A, upper right panel). This overlap in the monocyte transcriptome for these 3 states suggests that monocytes respond to diverse insults by activating common molecular pathways. In this regard, it should be noted that monocytes from patients with sepsis, trauma, and cancer share many immune phenotypic features including, for example, decreased HLA expression, increased production of IL-10, and increased myeloid derived suppressor cells (11).
A major goal of the present study was to discover potential mechanisms responsible for the immune suppression that is a hallmark of patients with sepsis. Several findings were particularly noteworthy because of both the magnitude of their increase and their potential relevance to the field. The profound upregulation of the transcriptome for ARG1 105 −1012 fold in monocytes, CD4 T cells, and CD8 T cells of septic patients is striking. Further, when patient samples were tested for their ability to respond to stimulus, ARG1 was found to have higher expression in those samples displaying functional suppression than those who were not suppressed. Previously, arginase has been implicated in mediating immune suppression in both sepsis and cancer and is postulated to be a major mechanism by which myeloid-derived suppressor cells impair host immunity (29, 30). Arginase depletes cells of arginine which is essential for T cell proliferation, zeta-chain peptide and T cell receptor (TCR) complex expression, and development of memory (30). Recently, arginase 1 has been associated with increased morbidity and mortality in sepsis (29). The potential significance of the present results is underscored by the fact that arginase inhibitors are currently undergoing testing in multiple clinical trials in patients with a variety of solid tumors. Given the extensive preclinical studies implicating the role of arginine deficiency in the pathophysiology of sepsis, if arginase inhibitors are shown to be safe and efficacious in oncology clinical trials, they might be considered in clinical trials in sepsis as well.
Additional potential mediators of sepsis-induced immunosuppression that were markedly upregulated in sepsis and have potent abilities to modulate immunity were members of the suppressors of cytokine signaling (SOCS) family. SOCS are pleiotropic inhibitors of the immuno-inflammatory response which block the JAK-STAT signaling pathway and prevent activation of pathogen recognition and cytokine receptors (31, 34). SOCS1 was one of the most highly upregulated genes in patients with sepsis; i.e., 107 and 1025 fold upregulation in CD4 and CD8 T cells respectively. The present findings parallel recent work by Alvarez et al. who documented increased SOCS1 mRNA expression in blood leukocytes from pediatric patients with sepsis (31). Inhibition of SOCS1 has been reported to be either beneficial or detrimental in various animal models of sepsis and likely depends upon the severity of the inflammatory response and virulence of the pathogen (34). SOCS3 was also highly upregulated (107 - 1025 fold upregulation) in all three cell types in patients with sepsis. Intriguingly, SOCS3 was also highly upregulated in all three cell types in cancer and non-septic critical illness, consistent with a potential important role for SOCS mediated cytokine suppression across all three diverse conditions.
Hypoxia inducible factor 1α (HIF-1α) has previously been characterized in sepsis (13). Shalova et al. found HIF-1α to be upregulated in monocytes from septic patients and, further, that those monocytes both acquired an immunosuppressive phenotype but could still function in wound-healing and anti-microbial capacities (32). In the current study, HIF-1α was not found to be upregulated in the monocytes from septic patients, nor the CD4 or CD8 T cells, however down-stream targets of the HIF-1 pathway were found to be upregulated in monocytes and CD4 T cells from all groups (Fig S3). It’s known that immune responses can tend to lead to increased activity in the HIF-1 pathway, and though there are studies suggesting that HIF-1 leads to increased levels of inflammation, the overall picture gained in the current study agrees with the data of Shalova that indicate a more suppressive phenotype in the monocytes (32). Shalova et al. also found an upregulation of IRAKM which can function to negatively regulate toll-like receptor (TLR) signaling. Like HIF-1α, IRAKM was not found in the current study to be differentially expressed in sepsis monocytes; however, several TLRs were found to be downregulated in septic monocytes. Thus, while some of the details may vary between particular genes in the two studies, both show overlapping pathways that would tend to contribute toward the same phenotype.
Other potential mechanisms for immune suppression in sepsis were a decreased expression of a broad array of antigen presentation molecules, downregulation of numerous positive co-stimulatory molecules, and upregulation of inhibitory receptor/ligands (Fig 3). Specifically, transcripts for the inhibitory receptors LAG-3, CTLA-4, PD-1, and TIGIT were increased in CD4 T cells, CD8 T cells, and/or monocytes from patients with sepsis (Fig 3). Previous flow cytometric studies of circulating immune cells from septic patients have demonstrated increased expression of all four of these inhibitory receptors thereby highlighting the potential clinical relevance of the present studies (35–37). Furthermore, antibodies which block the inhibitory receptors Lag-3, CTLA-4, PD-1, and TIGIT have restored immune effector cell function and improved survival in animal models of sepsis, chronic viral infection, and/or cancer (35–38). Anti-PD-1 and anti-CTLA-4 antibodies are now widely used clinically to treat cancer patients and anti-LAG3 antibodies are undergoing clinical trials.
An additional finding in the present study that provides insight into the pathophysiology of sepsis is the observation that CD8 T cells have a much reduced transcriptomic response to sepsis compared to CD4 T cells. This diminished CD8 T cell response in sepsis compared to the robust CD4 T cell response is likely because the vast majority of pathogens that are responsible for inducing sepsis in ICU patients are extracellular and not intracellular microbes. Although CD8 T cells are major producers of IFN-γ, which is beneficial for activation of macrophages in sepsis, CD8 T cells play a reduced role compared to CD4 T cells in controlling extracellular organisms. In contrast to the relative paucity of transcriptomic changes in sepsis, CD8 T cells from patients with cancer had a marked increase in their transcriptome, which exceeded that occurring in CD4 T cells in cancer. In addition, the transcriptomic response of CD8 T cells in cancer dwarfed that occurring in sepsis or non-septic critical illness (Fig. 2A). Several cytokine signaling pathways were upregulated in CD8 T cells including the IL-3 and IL-5 signaling pathways (Fig. 2A). It is interesting to postulate that this increase in gene expression of CD8 T cells from cancer patients is related to their important cytotoxic role in killing tumor cells.
Limitations:
There are a number of limitations to the present study. The phenotypic immune response in sepsis is shaped by many factors including the virulence and number of invading pathogens, host genetics, underlying patient co-morbidities, site of infection, and duration of sepsis. In particular, the host response in sepsis changes over time if sepsis persists. The initial host response in sepsis is characterized by an early cytokine storm mediated robust innate immune response which is often manifested by fever and shock. If the patient remains septic, the innate immune response progresses to a more immunosuppressive phenotype. Thus, depending upon when the septic immune response is examined, different findings may result. The focus of the present study was to define the immune phenotype of septic patients after the initial response had passed and during the more sustained phase. Therefore, the present results may not reflect the early initial immune response in sepsis that occurs during the first 24-48 hours of sepsis. Furthermore, because of this extended time period of sampling of blood from septic patients, the study population may include patients with considerable differences in their immune status ranging from late hyperinflammation/early immunosuppression (day 2) to sustained immunosuppression without hyperinflammatory component (day 9 and beyond). This may confound interpretation of the results. A second limitation is that the observed changes in RNA transcription may not necessarily be accompanied by comparable changes in the corresponding proteins because of translational regulation. This post transcriptional regulation is particularly true for cytokines. However, it is gratifying that the transcriptional changes of HLA molecules and the co-stimulatory and co-inhibitory receptor/ligands that were detected by RNA-seq in the present study are associated with corresponding changes in the accompanying cellular proteins as reported in the literature for patients with sepsis (32–33). Finally, a third limitation is the fact that only five patients with cancer were included in the study population and this number is much lower than the number of septic and critically ill non-septic patients. Unlike sepsis, in which there is great variability in the pathogens, effected organs, and severity of illness, the patients included in the cancer arm of the present study were more homogeneous. All patients had cancer due to a single disease, i.e., colorectal cancer that had metastasized to the liver, and were otherwise free of major organ dysfunction. The number of patients in the cancer arm of the study is comparable to other studies which reported on data from similar numbers of patients (39, 40). The fact that there were no outliers in the patients in the cancer cohort and that the transcriptomic results of the cancer patients were highly statistically different from the septic and critically ill non-septic patients supports the findings from these patients.
Conclusions:
There are a number of conclusions and implications from the present study. The phenotypic shift toward downregulation of multiple immune response pathways in monocytes suggests that the immunosuppression that is a hallmark of sepsis is due not only to defective T cell function but also to defective monocyte-mediated innate immunity. Thus, restoration of host immunity in patients with sepsis is likely to benefit not only from immuno-adjuvant therapies targeting T cells but also from monocyte directed immune-based therapies. Such monocyte targeted therapies are now undergoing clinical trials in oncology patients. Potential mediators of the sepsis-induced immunosuppression that were broadly expressed across multiple cells at high levels included Arg-1 and SOCS-1 and SOCS-3. Arginase inhibitors, which have shown efficacy in animal models of sepsis and which are currently in oncologic clinical trials, might have beneficial effects to restore sepsis-induced defects in both T cells and monocytes. Other potential therapeutic targets in sepsis include the several negative co-stimulatory molecules TIGIT, Lag-3, CTLA-4, and PD-1/L1, which were highly upregulated in septic patients. There is extensive overlap in the changes in gene expression in patients with sepsis and cancer in CD4 T cells and monocytes but not in CD8 T cells. This overlap in gene expression includes numerous immunosuppressive pathways indicating that immuno-adjuvant therapies that are effective in one disorder may be efficacious in both. Thus, the advances that are taking place in cancer immunotherapy may have important implications for the therapeutic approach to sepsis.
Supplementary Material
Key Points.
Sepsis induces transcription of suppressive genes in innate and adaptive immune cells
Sepsis, trauma, and cancer share many common immunosuppressive mechanisms
Monocytic transcriptomic changes during sepsis are markedly suppressive
Acknowledgments:
The authors gratefully acknowledge the nurses and staff of the 4400 Surgical Intensive Care Unit at Barnes Jewish Hospital.
This work was supported by grants from the National Institutes of Health GM 126928 (to R.S.H.) and from the National Cancer Institute P50CA196510 and from research support by GlaxoSmithKline
REFENCES
- 1.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, and Angus DC. 2016. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 315:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotchkiss RS, and Karl IE. 2003. The pathophysiology and treatment of sepsis. N. Engl. J. Med 348:138–150. [DOI] [PubMed] [Google Scholar]
- 3.Angus DC, and van der Poll T. 2013. Severe sepsis and septic shock. N. Engl. J. Med 369:840–851. [DOI] [PubMed] [Google Scholar]
- 4.Hotchkiss RS, and Opal S. 2010. Immunotherapy for sepsis--a new approach against an ancient foe. N. Engl. J. Med 363:87–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hotchkiss RS, Monneret G, and Payen D. 2013. Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect Dis. 13:260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hotchkiss RS, Monneret G, and Payen D. 2013. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. 13:862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hotchkiss RS, Moldawer LL, Opal SM, Reinhart K, Turnbull IR, and Vincent JL. 2016. Sepsis and septic shock. Nat Rev Dis Primers. 2:16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fattahi F, and Ward PA. 2017. Understanding Immunosuppression after Sepsis. Immunity. 47:3–5. [DOI] [PubMed] [Google Scholar]
- 9.Venet F, Rimmele T, and Monneret G. 2018. Management of Sepsis-Induced Immunosuppression. Crit. Care Clin 34:97–106. [DOI] [PubMed] [Google Scholar]
- 10.Walton AH, Muenzer JT, Rasche D, Boomer JS, Sato B, Brownstein BH, Pachot A, Brooks TL, Deych E, Shannon WD, Green JM, Storch GA, and Hotchkiss RS. 2014. Reactivation of multiple viruses in patients with sepsis. PLoS One. 9:e98819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hotchkiss RS, and Moldawer LL. 2014. Parallels between cancer and infectious disease. N. Engl. J. Med 371:380–383. [DOI] [PubMed] [Google Scholar]
- 12.Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H, Hayden DL, Hennessy L, Moore EE, Minei JP, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Brownstein BH, Mason PH, Baker HV, Finnerty CC, Jeschke MG, Lopez MC, Klein MB, Gamelli RL, Gibran NS, Arnoldo B, Xu W, Zhang Y, Calvano SE, McDonald-Smith GP, Schoenfeld DA, Storey JD, Cobb JP, Warren HS, Moldawer LL, Herndon DN, Lowry SF, Maier RV, Davis RW, and Tompkins RG. 2011. A genomic storm in critically injured humans. J. Exp. Med 208:2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davenport EE, Burnham KL, Radhakrishnan J, Humburg P, Hutton P, Mills TC, Rautanen A, Gordon AC, Garrard C, Hill AV, Hinds CJ, and Knight JC. 2016. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med. 4:259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer M, Giamarellos-Bourboulis EJ, Kortgen A, Moller E, Felsmann K, Cavaillon JM, Guntinas-Lichius O, Rutschmann O, Ruryk A, Kohl M, Wlotzka B, Russwurm S, Marshall JC, and Reinhart K. 2016. A Transcriptomic Biomarker to Quantify Systemic Inflammation in Sepsis - A Prospective Multicenter Phase II Diagnostic Study. EBioMedicine. 6:114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Efron PA, Mohr AM, Bihorac A, Horiguchi H, Hollen MK, Segal MS, Baker HV, Leeuwenburgh C, Moldawer LL, Moore FA, and Brakenridge SC. 2018. Persistent inflammation, immunosuppression, and catabolism and the development of chronic critical illness after surgery. Surgery. 164:178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loftus TJ, Mira JC, Stortz JA, Ozrazgat-Baslanti T, Ghita GL, Wang Z, Brumback BA, Ungaro RF, Bihorac A, Leeuwenburgh C, Moore FA, Moldawer LL, Brakenridge SC, Efron PA, and Mohr AM. 2019. Persistent inflammation and anemia among critically ill septic patients. J Trauma Acute Care Surg. 86:260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, and Sibbald WJ. 1992. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest 35. Physicians/Society of Critical Care Medicine. Chest. 101:1644–1655. [DOI] [PubMed] [Google Scholar]
- 18.Grossman JG, Nywening TM, Belt BA, Panni RZ, Krasnick BA, DeNardo DG, Hawkins WG, Goedegebuure SP, Linehan DC, and Fields RC. 2018. Recruitment of CCR2(+) tumor associated macrophage to sites of liver metastasis confers a poor prognosis in human colorectal cancer. Oncoimmunology. 7:e1470729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thampy LK, Remy KE, Walton AH, Hong Z, Liu K, Liu R, Yi V, Burnham CD, and Hotchkiss RS. 2018. Restoration of T Cell function in multi-drug resistant bacterial sepsis after interleukin-7, anti-PD-L1, and OX-40 administration. PLoS One. 13:e0199497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francois B, Jeannet R, Daix T, Walton AH, Shotwell MS, Unsinger J, Monneret G, Rimmele T, Blood T, Morre M, Gregoire A, Mayo GA, Blood J, Durum SK, Sherwood ER, and Hotchkiss RS. 2018. Interleukin-7 restores lymphocytes in septic shock: the IRIS-7 randomized clinical trial. JCI Insight. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews S 2010. FastQC: A quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- 22.Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, Meehan DT, Wipfler K, Bosinger SE, Johnson ZP, Tharp GK, Marcais G, Roberts M, Ferguson B, Fox HS, Treangen T, Salzberg SL, Yorke JA, and Norgren RB Jr. 2014. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct. 9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Y, Smyth GK, and Shi W. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guignant C, Lepape A, Huang X, Kherouf H, Denis L, Poitevin F, Malcus C, Cheron A, Allaouchiche B, Gueyffier F, Ayala A, Monneret G, and Venet F. 2011. Programmed death-1 levels correlate with increased mortality, nosocomial infection and immune dysfunctions in septic shock patients. Crit Care. 15:R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patera AC, Drewry AM, Chang K, Beiter ER, Osborne D, and Hotchkiss RS. 2016. Frontline Science: Defects in immune function in patients with sepsis are associated with PD-1 or PD-L1 expression and can be restored by antibodies targeting PD-1 or PD-L1. J. Leukoc. Biol 100:1239–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hotchkiss RS, Colston E, Yende S, Angus DC, Moldawer LL, Crouser ED, Martin GS, Coopersmith CM, Brakenridge S, Mayr FB, Park PK, Ye J, Catlett IM, Girgis IG, and Grasela DM. 2019. Immune Checkpoint Inhibition in Sepsis: A Phase 1b Randomized, Placebo-Controlled, Single Ascending Dose Study of Antiprogrammed Cell Death-Ligand 1 (BMS-936559). Crit. Care Med [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badurdeen S, Mulongo M, and Berkley JA. 2015. Arginine depletion increases susceptibility to serious infections in preterm newborns. Pediatr. Res 77:290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zea AH, Rodriguez PC, Atkins MB, Hernandez C, Signoretti S, Zabaleta J, McDermott D, Quiceno D, Youmans A, O’Neill A, Mier J, and Ochoa AC. 2005. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 65:3044–3048. [DOI] [PubMed] [Google Scholar]
- 31.Pineros Alvarez AR, Glosson-Byers N, Brandt S, Wang S, Wong H, Sturgeon S, McCarthy BP, Territo PR, Alves-Filho JC, and Serezani CH. 2017. SOCS1 is a negative regulator of metabolic reprogramming during sepsis. JCI Insight. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nastase A, Teo JY, Heng HL, Ng CC, Myint SS, Rajasegaran V, Loh JL, Lee SY, Ooi LL, Chung AY, Chow PK, Cheow PC, Wan WK, Azhar R, Khoo A, Xiu SX, Alkaff SM, Cutcutache I, Lim JQ, Ong CK, Herlea V, Dima S, Duda DG, The BT, Popescu I, and Lim TK 2017. Genomic and proteomic characterization of ARID1A chromatic remodeler in amullary tumors. Am. J. Cancer Res 7: 484–502. [PMC free article] [PubMed] [Google Scholar]
- 33.Munoz C, Carlet J, Fitting C, Misset B, Bleriot JP, and Cavaillon JM. 1991. Dysregulation of in vitro cytokine production by monocytes during sepsis. J. Clin. Invest 88:1747–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chikuma S, Kanamori M, Mise-Omata S, and Yoshimura A. 2017. Suppressors of cytokine signaling: Potential immune checkpoint molecules for cancer immunotherapy. Cancer Sci. 108:574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boomer JS, To K, Chang KC, Takasu O, Osborne DF, Walton AH, Bricker TL, Jarman SD 2nd, Kreisel D, Krupnick AS, Srivastava A, Swanson PE, Green JM, and Hotchkiss RS. 2011. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA. 306:2594–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boomer JS, Shuherk-Shaffer J, Hotchkiss RS, and Green JM. 2012. A prospective analysis of lymphocyte phenotype and function over the course of acute sepsis. Crit Care. 16:R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng Y, Zhao Z, Zhu W, Yang T, Deng X, and Bao R. 2017. CD155 blockade improves survival in experimental sepsis by reversing dendritic cell dysfunction. Biochem. Biophys. Res. Commun 490:283–289. [DOI] [PubMed] [Google Scholar]
- 38.Anderson AC, Joller N, and Kuchroo VK. 2016. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity. 44:989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uniken Venema WT, Voskuil MD, Vila AV, van der Vries G, Jansen BH, Jabri B, Faber KN, Dijkstra G, Xarvier RJ, inmenga C, Graham DB, Weersma RK, Festen EA. 2019. Single-Cell RNA Sequencing of Blood and Ileal T Cells from Patients with Crohn’s Disease Reveals Tissue-Specific Characteristics with Drug Targets. Gastroenterology 156:812–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen T, Aronow BJ, Rochman Y, Rochman M, Kc K, Dexheimer PJ, Putnam P, Mukkada V, Foote H, Rehn K, Darko S, Doued D, Rothenberg ME. 2019. Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. Journal of Clinical Investigation 129:2014–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.