Abstract
The transcription factor NRF2 is a master regulator of cellular antioxidant and detoxification responses, but it also regulates other processes such as autophagy and pluripotency. In human embryonic stem cells (hESCs), NRF2 antagonizes neuroectoderm differentiation, which only occurs after NRF2 is repressed via a Primary Cilia-Autophagy-NRF2 (PAN) axis. However, the functional connections between NRF2 and primary cilia, microtubule-based plasma membrane protrusions that function as cellular antennae, remain poorly understood. For instance, nothing is known about whether NRF2 affects cilia, or whether cilia regulation of NRF2 extends beyond hESCs. Here, we show that NRF2 and primary cilia reciprocally regulate each other. First, we demonstrate that fibroblasts lacking primary cilia have higher NRF2 activity, which is rescued by autophagy-activating mTOR inhibitors, indicating that the PAN axis also operates in differentiated cells. Furthermore, NRF2 controls cilia formation and function. NRF2-null cells grow fewer and shorter cilia and display impaired Hedgehog signaling, a cilia-dependent pathway. These defects are not due to increased oxidative stress or ciliophagy, but rather to NRF2 promoting expression of multiple ciliogenic and Hedgehog pathway genes. Among these, we focused on GLI2 and GLI3, the transcription factors controlling Hh pathway output. Both their mRNA and protein levels are reduced in NRF2-null cells, consistent with their gene promoters containing consensus ARE sequences predicted to bind NRF2. Moreover, GLI2 and GLI3 fail to accumulate at the ciliary tip of NRF2-null cells upon Hh pathway activation. Given the importance of NRF2 and ciliary signaling in human disease, our data may have important biomedical implications.
Subject terms: Morphogen signalling, Transcriptional regulatory elements
Introduction
Primary cilia are microtubule-based plasma membrane protrusions that function as cell type-specific antennae by accumulating signal receptors and transducers. Structurally, the ciliary membrane is undergirded by the ciliary shaft, or axoneme, consisting of nine microtubule pairs emanating from the basal body, a membrane-anchored centriole1,2.
The presence of primary cilia is cell cycle and cell type-dependent3,4. In the cell cycle, cilia form during G1/G0 and disassemble before mitosis3. Moreover, specific transcription factors (e.g. RFX1-4, FOXJ1) promote ciliogenic gene expression in a cell type-specific manner4,5. Ciliogenesis and ciliary maintenance critically depend on a bidirectional trafficking system, known as intraflagellar transport (IFT), which moves cargoes up and down the axoneme1,2. To do this, cargoes bind adaptors such as IFT-B and IFT-A complexes, which in turn associate to microtubule motors moving toward (heterotrimeric kinesin-2) or from (cytoplasmic dynein-2) the ciliary tip.
In mammals, the complete absence of cilia is embryonic lethal, as seen in mice lacking essential ciliogenic genes such as Ift88 and Kif3a6,7. Less severe congenital ciliary defects lead to ciliopathies, a diverse group of human diseases causing blindness, cystic kidneys, obesity, sterility, polydactyly and brain malformations, among other symptoms1. Cilia defects acquired later in life are associated with ailments such as cancer and neurodegeneration8–11.
Disruption of Hedgehog (Hh) signaling, a cilia-dependent pathway controlling multiple aspects of embryogenesis and adult stem cell function7, explains many (but not all) ciliopathy symptoms and cilia-dependent cancers. Hh ligands, such as Sonic Hedgehog (SHH), act in paracrine fashion by binding to Patched (PTCH1), a ciliary transmembrane protein. Ligand binding prevents PTCH1 from opposing the ciliary accumulation of Smoothened (SMO), a seven transmembrane protein whose activation displaces another G protein-coupled receptor, GPR161, from cilia. This in turn lowers cAMP levels and PKA activity at the basal body, affecting phosphorylation of GLI2 and GLI3, the zinc finger transcription factors mediating Hh pathway output. PKA inactivation also causes GLI2 and GLI3 to accumulate at the ciliary tip, where they dissociate from SUFU, their repressor. From the ciliary tip, GLI2 and GLI3 travel to the ciliary base to meet different fates: GLI2 is processed into a transcriptional activator and translocates to the nucleus to activate target genes, whereas GLI3 is fully degraded in the proteasome, abolishing its basal activity as a transcriptional repressor. Thus, by promoting GLI2 activation and GLI3 non-repression, Hh ligands stimulate expression of target genes, including Ptch1 and Gli1, which provide negative (PTCH1) and positive (GLI1) feedback to the pathway7.
Primary cilia also modulate mTOR signaling and autophagy12–17. In the kidney, mechanosensory cilia in tubular epithelial cells respond to fluid flow by repressing mTOR, thereby promoting autophagy and reducing cell volume, a homeostatic mechanism perturbed in polycystic kidney disease13,15,18,19. Similar pathways occur in other cell types, such as fibroblasts and radial glia16,17. During human embryonic stem cell (hESC) differentiation, cell fate choice between mesendoderm and neuroectoderm is also controlled by cilium-dependent autophagy12. In the first stages of neuroectoderm progenitor specification, cell cycle lengthening allows cilia to emerge during G1. Cilia in turn activate autophagy, resulting in the inactivation of NRF2, which antagonizes neuroectoderm differentiation. Whether this so-called Primary Cilium-Autophagy-NRF2 (PAN) axis is conserved in other cell types, and whether mTOR is involved in it, has not been addressed12.
NRF2 (nuclear factor erythroid 2-related factor 2), encoded by the Nfe2l2 gene, is a basic region-leucine zipper (bZip) transcription factor best known as a master regulator of cellular antioxidant and detoxification responses20. Under normal conditions, NRF2 binding to KEAP1 targets the former for ubiquitin-dependent proteasome degradation. By modifying cysteine residues in KEAP1, oxidative stress, or electrophilic compounds like dimethyl fumarate (DMF), an FDA-approved drug for treatment of multiple sclerosis, disrupt KEAP1-NRF2 binding, leading to NRF2 accumulation20. NRF2 then translocates to the nucleus and activates expression of its multiple target genes (hundreds of them) by binding to antioxidant response elements (AREs) in their enhancer regions20. Many of these genes encode detoxification enzymes, such as heme oxygenase-1 (Hmox1) or the catalytic (Gclc) and modulatory (Gclm) subunits of glutamate-cysteine ligase, which catalyzes the first step in the biosynthesis of glutathione, a major cellular scavenger of reactive oxygen species (ROS)20. NRF2 also targets autophagy genes, which contribute to stress tolerance. Hence, NRF2 is both a regulator of autophagy and an autophagy-regulated protein12,21,22.
Likewise, primary cilia both control and are controlled by autophagy12,14,23,24. A cilia-specific form of autophagy, termed ciliophagy, plays a key role in lung damage caused by cigarette smoke. HDAC6, a histone deacetylase and ubiquitin-binding autophagy receptor, plays a key role in both ciliophagy and aggrephagy, the autophagic clearance of protein aggregates23,25,26. Moreover, autophagy promotes ciliogenesis and lengthens cilia by removing an inhibitory protein, OFD1, from the basal body vicinity24.
In view of these reciprocal cilia-autophagy and NRF2-autophagy connections, we wondered whether analogous reciprocal relationships exist between cilia and NRF2. Indeed, we find that fibroblasts lacking cilia have reduced autophagy and increased NRF2 activity, which we rescue with autophagy-activating mTOR inhibitors. Conversely, NRF2-null cells have defects in ciliogenesis and Hh signaling. These defects, rather than being caused by changes in redox stress or ciliophagy, are due to NRF2 promoting expression of multiple ciliogenic and Hh pathway genes.
Results
Primary cilia downregulate NRF2 activity via mTOR-dependent autophagy
Cells lacking KIF3A, a kinesin-2 subunit, or IFT88, an IFT-B complex component, are completely incapable of growing cilia and are commonly used to study the effects of cilia ablation1,6,7,27. We confirmed this using both Kif3a−/− and Ift88−/− mouse embryonic fibroblasts (MEFs). After 24 hours of starvation, most wild type MEFs displayed primary cilia, in which both axoneme (acetylated α-tubulin, AcTub) and ciliary membrane (ARL13B) markers are seen to emerge from the γ-tubulin-positive basal bodies. In contrast, Kif3a−/− and Ift88−/− MEFs failed to grow any cilia under the same conditions (Fig. 1a).
Figure 1.
Primary cilia downregulate NRF2 activity via mTOR-dependent autophagy. (a) Representative immunofluorescence images of ARL13B, acetylated α-tubulin (AcTub) and γ-tubulin (γTub) in control (Kif3a+/+, WT), Kif3a−/− and Ift88−/− MEFs starved 24 h to induce ciliogenesis. Nuclei were counterstained with DAPI. Scale bar, 5μm. (b) Expression of NRF2 target genes Hmox1 and Gclc was analyzed by RT-qPCR in Kif3a+/+ and Kif3a−/− MEFs starved for 24 h and then treated for 6 h in starvation medium with vehicle (DMSO) or 20 μM dimethyl fumarate (DMF), as indicated. Data are β-actin (Actb)-normalized relative mRNA levels (mean ± SEM, n = 4 independent experiments). (c) Same as in (b) using Ift88+/+ and Ift88−/− MEFs (mean ± SEM, n = 3 independent experiments). (d) Kif3a MEFs treated as in (b) were analyzed by Western blot with antibodies against NRF2 and α-tubulin, as loading control. Molecular weight markers in kilodaltons are on the right. Quantitation of NRF2/tubulin band intensity ratio is shown below. The compared bands were in the same gel and blot. See methods for details. Uncropped blots are shown in Supplementary Fig. S12. (e,f) Hmox1 (e) and Gclc (f) were analyzed as in (b) in Kif3a MEFs treated with DMF (20 μM), Torin-1 (250 nM) or Rapamycin (200 nM), as indicated. Data shown as in (b) (mean ± SEM, n = 6 independent experiments). (g) Kif3a MEFs were serum-starved for 24 h and then incubated for 4 h in Earle’s balanced salt solution (EBSS) with or without 10 μM Chloroquine and the indicated drugs, as in (e-f). Cell lysates were analyzed by Western blot with anti-LC3B antibodies. Each sample’s LC3B-II/LC3B-I ratio is shown below, together with relative autophagy flux (LC3B ratio increase by Chloroquine, normalized to control). (h) Kif3a MEFs treated as in (e-f) were analyzed by Western blot and quantitated as in (d). Statistical analysis in (b-c) and (e-f): two-way ANOVA followed by Tukey’s multiple comparisons tests. Asterisks indicate *p < 0.05, **p < 0.01 or ***p < 0.001.
To test whether cilia regulate NRF2 activity in MEFs, we measured mRNA levels of heme oxygenase-1 (Hmox1) and glutamate-cysteine ligase catalytic subunit (Gclc), two well-established transcriptional NRF2 targets20, in Kif3a−/−, Ift88−/− and their respective littermate control MEFs (Fig. 1b,c). We tested this in presence or absence of DMF, a known NRF2 activator20. Both with and without DMF, cilia-null MEFs displayed consistently higher Hmox1 and Gclc mRNA levels, an effect that was more obvious and significant in presence of DMF (Fig. 1b,c). As expected, DMF raised Hmox1 and Gclc, and it did so with similar fold increases in cilia-null MEFs (Fig. 1b,c). Since these results were observed for both Kif3a and Ift88 MEFs, this suggests that cilia downregulate NRF2 transcriptional activity. Accordingly, another NRF2 target gene, NADPH quinone oxidoreductase-1 (Nqo1), was also upregulated, together with Hmox1, in both Kif3a−/− and Ift88−/− MEFs, and this upregulation was seen more clearly in presence of another NRF2 activator, sulforaphane (Supplementary Fig. S1a,b)20. Furthermore, Hmox1 protein levels were also increased in Ift88−/− MEFs, and the effect was clearer in sulforaphane-treated cells (Supplementary Fig. S1c).
To see if this increase in NRF2 activity was due to increased NRF2 protein levels, we analyzed the latter by Western blot in Kif3a+/+ and Kif3a−/− MEFs treated or not with DMF. Relative to α-tubulin, NRF2 protein levels were strongly upregulated by DMF in both Kif3a+/+ and Kif3a−/− MEFs, without obvious differences between cell types (Fig. 1d). Hence, the effect of KIF3A on NRF2 activity is not due to changes in NRF2 protein levels.
Since primary cilia downregulation of NRF2 during neuroectoderm differentiation is mediated by autophagy, we next tested whether mTOR inhibitors, which are widely used to activate autophagy, could restore normal NRF2 activity in Kif3a−/− MEFs12,28. For this, we used both Torin-1 and rapamycin, an ATP-competitive and an allosteric mTOR inhibitor, respectively29. Indeed, gene expression of both Hmox1 and Gclc returned to wild type levels upon treatment of Kif3a−/− MEFs with either Torin-1 or rapamycin (Fig. 1e,f).
To assess whether changes in autophagic flux may explain the mTOR-dependent effects of cilia on NRF2 activity, we used Western blot to look at the relative levels of LC3B-I and LC3B-II in presence or absence of chloroquine, an autophagolysosome blocker. It is well-established that, during autophagy, LC3B-I is conjugated to phosphatidylethanolamine, thus generating LC3B-II and tethering it to autophagosomes, which evolve into autophagolysosomes where LC3B-II is degraded. Based on this, the chloroquine-induced increase in LC3B-II levels (normalized as LC3B-II/LC3B-I ratio) is a widely accepted measure of autophagic flux. In starved and DMF-treated Kif3a+/+ MEFs, we saw a clear chloroquine-induced increase in the LC3B-II/LC3B-I ratio, indicating active autophagy (Fig. 1g). Under the same conditions, autophagic flux in Kif3a−/− MEFs was reduced by more than 30% relative to the Kif3a+/+ control. This reduction, however, was rescued by both Torin-1 and rapamycin, in whose presence autophagic flux in Kif3a−/− MEFs was even higher than that in Kif3a+/+ cells (Fig. 1g). Since autophagy flux in these cells shows a strong inverse correlation with NRF2 target expression, these data are consistent with primary cilia downregulating NRF2 by promoting autophagy.
This correlation did not clearly extend to NRF2 protein levels, even though quantitation of NRF2/tubulin ratios showed a slight increase in Kif3a−/− relative to Kif3a+/+ MEFs, an increase that was no longer seen when Kif3a−/− MEFs were treated with mTOR inhibitors (Fig. 1h). However, this modest increase seems unlikely to explain the observed increase in NRF2 transcriptional activity.
Altogether, these data suggest that the PAN axis described in differentiating hESCs is also operative in more differentiated cell types, such as MEFs.
Ciliary repression of NRF2 does not involve Hh signaling
Since primary cilia are required for vertebrate Hh signaling, we also tested whether Hh pathway alterations might be involved in the higher NRF2 activity of Kif3a−/− MEFs. In these cells, lack of cilia perturbs proteolytic processing of GLI transcription factors7. Since GLI3 is processed into a transcriptional repressor (GLI3R) in a constitutive and cilia-dependent manner, Kif3a−/− MEFs fail to form GLI3R, leading to higher unstimulated expression of Hh target genes Gli1 and Ptch1 (Supplementary Fig. S2a).
KIF3A and cilia are also required for GLI2 processing, which forms a transcriptional activator (GLI2A) only upon Hh pathway stimulation. Hence, Kif3a−/− MEFs fail to form GLI2A and do not induce target gene expression in response to Hh pathway agonists. Nevertheless, Hh target gene expression can be stimulated in these cells by transfecting them with a constitutively active, oncogenic allele of GLI2 known as GLI2ΔN27. Indeed, transient expression of GLI2ΔN activated Hh target expression in Kif3a+/+ and, even more so, in Kif3a−/− MEFs, where GLI3R does not counteract its effect (Supplementary Fig. S2a).
However, despite strongly activating Hh signaling in Kif3a+/+ and Kif3a−/− MEFs, GLI2ΔN had no effect on Hmox1 and Gclc gene expression, indicating that NRF2 activity is not affected by GLI2ΔN-induced Hh pathway activation (Supplementary Fig. S2b).
Thus, GLI2 activation does not mediate the effects of cilia on NRF2. Nor does GLI2 inhibition, as GLI2 is transcriptionally inactive in unstimulated MEFs, as confirmed by our observation that GLI2 inhibitor GANT61 does not lower Hh target expression in MEFs not treated with Hh pathway agonists (data not shown)30.
Having ruled out GLI2, it was still possible that cilia repress NRF2 activity by promoting GLI3R synthesis. If this were the case, then GLI3R expression in Kif3a−/− MEFs should lower NRF2 activity, restoring it to normal. However, this is not the case (Supplementary Fig. S2c,d).
Overall, these data indicate that the negative impact of cilia on NRF2 activity is not related to Hh signaling.
Ciliogenesis is reduced in NRF2-null cells
We next wondered whether NRF2 affects cilia in addition to being affected by them. To test this, we first looked at ciliogenesis in Nfe2l2+/+ and Nfe2l2−/− MEFs. Although Nfe2l2−/− MEFs generated cilia that appeared normal by ARL13B, AcTub and γ-tubulin staining (Fig. 2a), the percentage of cells displaying primary cilia was reduced in half relative to Nfe2l2+/+ cells (Fig. 2b). Moreover, cilia in Nfe2l2−/− MEFs were on average 15–20% shorter than control cilia (Fig. 2c). Primary Nfe2l2−/− MEFs displayed similar ciliogenic defects, excluding MEF immortalization as their cause (Supplementary Fig. S3).
Figure 2.
Ciliogenesis is reduced in NRF2-null cells. (a) Representative immunofluorescence images for ARL13B, acetylated α-tubulin (AcTub) and γ-tubulin (γTub) in Nfe2l2+/+ and Nfe2l2−/− littermate MEFs starved 24 h to induce ciliogenesis. Nuclei were counterstained with DAPI. Scale bar, 10 μm. (b) Quantitation of percentage of ciliated cells from (a) (mean ± SEM of n = 3 independent experiments). (c) Quantitation of cilia length from (a) (mean ± SEM of n = 81–85 cilia measured per condition). (d) Representative immunofluorescence images for ARL13B, γ-tubulin and astrocyte marker GFAP in hippocampus of 3-month old Nfe2l2+/+ and Nfe2l2−/− mice. Nuclei were counterstained with DAPI. Boxed region in left is magnified on the right. Arrowheads point to cilia. Scale bars, 25 μm (left) and 10 μm (right). (e) Quantitation of percentage of ciliated cells from (d) (mean ± SEM, n = 3 mice). Asterisks in (b), (c) and (e): statistical significance in unpaired two-tailed t-tests with **p < 0.01, ***p < 0.001 or ****p < 0.0001.
We then examined whether NRF2 affects ciliogenesis in vivo. Since NRF2-null mice are viable and develop normally, we reasoned their embryonic cilia cannot be strongly perturbed31. In contrast, adult NRF2-null mice have phenotypes that could potentially be explained by ciliogenic defects. In particular, both NRF2 and cilia play important roles in the hippocampus, affecting cognitive functions11,32,33. Thus, we decided to look at hippocampal cilia in coronal sections of three month-old Nfe2l2+/+ and Nfe2l2−/− mouse brains. Moreover, since NRF2 plays a prominent role in astrocytes34–36, and ARL13B specifically labels astrocytic cilia in adult mouse brain37,38, we focused our study on this cell type, which can be readily identified with the GFAP marker38. In both Nfe2l2+/+ and Nfe2l2−/− hippocampus, ARL13B+ cilia were indeed only seen in close association with GFAP+ astrocytes, confirming the above reports (Fig. 2c)37,38. In control sections, about 50% of such GFAP+ cells contained a primary cilium, but this percentage was reduced to about 10% in equivalent Nfe2l2−/− sections (Fig. 2c,d).
To test whether the ciliogenic effects of NRF2 are cell type-specific, we also looked at the cilia of multiciliated ependymal cells in the brain ventricles of Nfe2l2+/+ and Nfe2l2−/− mice. For this, we stained coronal brain sections for AcTub and basal body marker CEP164. Plentiful cilia lined the ventricular surface in both control and NRF2-null mice, with no apparent differences between the two (Supplementary Fig. S4). This is consistent with the phenotype of NRF2-null mice, which show no signs of hydrocephalus, the usual outcome of ependymal cilia dysfunction4,31. Thus, our data suggest that NRF2 is required for optimal ciliogenesis of specific cell types, including astrocytes and fibroblasts.
Hh signaling is reduced in NRF2-null cells
Since Hh signaling is cilia-dependent, we next examined Hh pathway responsiveness in Nfe2l2−/− MEFs. To do this, we used Smoothened agonist (SAG), a well-established Hh pathway activator39. While SAG strongly induced Hh target genes Gli1 and Ptch1 in Nfe2l2+/+ cells, this response was much attenuated in Nfe2l2−/− MEFs, even if still significant (Fig. 3a). Very similar results were obtained in primary MEFs, confirming defective Hh responsiveness is not related to MEF immortalization (Supplementary Fig. S5). Moreover, differences in SAG responsiveness were also seen by Western blot of GLI1, whose protein levels were strongly upregulated by SAG in Nfe2l2+/+ MEFs but much less so in Nfe2l2−/− MEFs (Fig. 3b). Thus, NRF2-null cells display defects in both ciliogenesis and Hh signaling.
Figure 3.
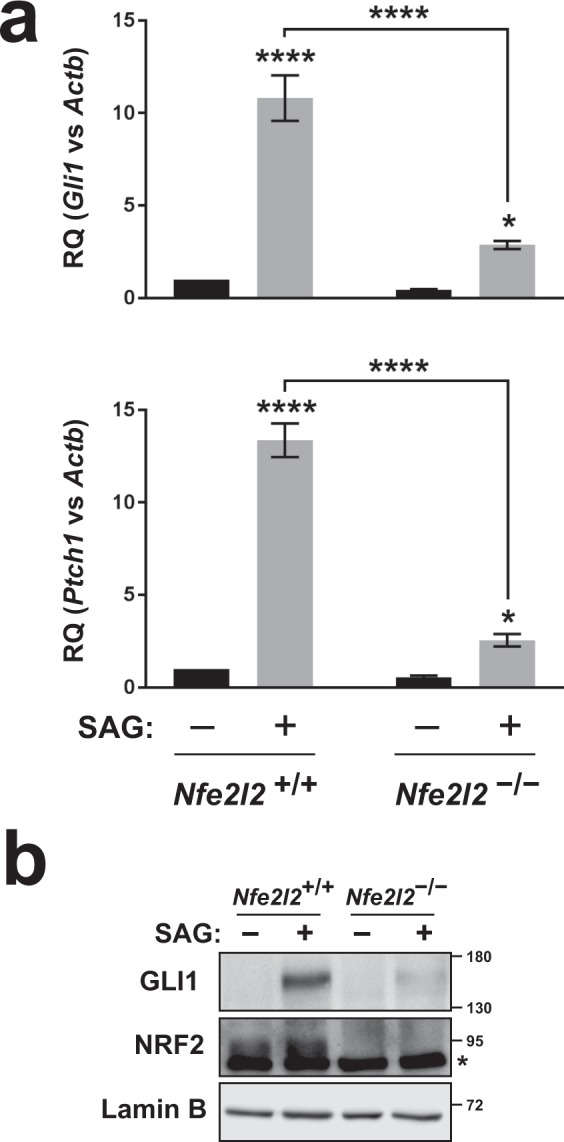
Hh signaling is reduced in NRF2-null cells. (a) Nfe2l2+/+ and Nfe2l2−/− MEFs were starved 24 h in presence of DMSO or SAG (200 nM) and expression of Hh target genes Gli1 (top) and Ptch1 (bottom) was analyzed by RT-qPCR. Data shown as β-actin (Actb)-normalized relative mRNA levels (mean ± SEM, n = 8–9 independent experiments). Statistical analysis: two-way ANOVA followed by Tukey’s multiple comparisons tests. Asterisks: *p < 0.05 or ****p < 0.0001. (b) Nfe2l2 MEFs treated as in (a) were analyzed by Western blot with antibodies for GLI1, NRF2 and Lamin-B as loading control. Molecular weight markers in kilodaltons are shown on the right. Asterisk denotes non-specific band (see methods on NRF2 antibody). Uncropped blots are shown in Supplementary Fig. S12.
ROS, HDAC6, mTOR and PKA inhibitors do not rescue Hh signaling in NRF2-null cells
We then asked whether increased oxidative stress in absence of NRF2 was the cause for the sharp decrease in Hh signaling. To address this, we used N-acetyl-cysteine (NAC), a glutathione precursor that promotes ROS scavenging, thereby reducing oxidative stress40. NAC treatment of Nfe2l2−/− MEFs did not affect their poor SAG responsiveness relative to wild type cells, indicating that the antioxidant functions of NRF2 are not the main reason why it promotes Hh signaling (Fig. 4).
Figure 4.

Hh signaling defects in NRF2-null cells are not rescued by HDAC6, PKA, mTOR or ROS inhibitors. Nfe2l2+/+ and Nfe2l2−/− MEFs were starved 24 h in presence of DMSO (vehicle), SAG (200 nM), Tubastatin A (10 μM), H89 (10 μM), Torin-1 (250 nM) or NAC (3 mM), as indicated, and expression of Hh target genes Gli1 and Ptch1 was analyzed by RT-qPCR. Data displayed as β-actin-normalized relative mRNA levels (mean ± SEM of n = 5 independent experiments). Statistical analysis: one-way ANOVA followed by Tukey’s multiple comparisons tests. Significance: *p < 0.05, ****p < 0.0001 or not significant (n.s.).
Another possible explanation for the effect of NRF2 on cilia and Hh signaling is that NRF2 affects ciliophagy, which critically depends on HDAC6, a protein that is upregulated in Nfe2l2−/− cells and functions as both a protein deacetylase and a ubiquitin-binding autophagy receptor23,25,26. If increased HDAC6-dependent ciliophagy was the cause of Nfe2l2−/− MEFs displaying ciliogenic and Hh signaling defects, then Tubastatin A, an HDAC6 inhibitor that blocks ciliophagy, should improve Hh responses in these cells23,41. However, this is not the case (Fig. 4).
Since PKA, by promoting generation of GLI repressors at the expense of GLI activators, is a negative regulator of Hh signaling, we also tested whether a PKA inhibitor, H89, restored high Hh responsiveness in Nfe2l2−/− MEFs7,42. The lack of such restoration indicates that NRF2 is required downstream of PKA in the Hh pathway (Fig. 4).
Lastly, given that NRF2 promotes autophagy21,22, we tested whether autophagy stimulation with mTOR inhibitor Torin-1 improved Hh signaling in Nfe2l2−/− MEFs, but it did not (Fig. 4).
Hence, oxidative stress, HDAC6-dependent ciliophagy, overactive PKA or reduced autophagy do not appear to explain the cilia and Hh phenotypes of Nfe2l2−/− MEFs.
Ciliogenic and Hh pathway gene expression are reduced in NRF2-null cells
NRF2 is a transcription factor with hundreds of known target genes, so we reasoned that its effects on cilia and Hh signaling might be mediated by changes in gene expression. To test this, we first looked at mRNA levels of ciliogenic genes in Nfe2l2+/+ and Nfe2l2−/− MEFs. These genes encode proteins such as ciliary microtubule motors (Dync2h1) and components of intraflagellar transport complexes IFT-B (Ift74, Ift88, Ift172) and IFT-A (Ift140). Interestingly, expression of all IFT and motor genes analyzed was significantly lower in Nfe2l2−/− relative to Nfe2l2+/+ MEFs, regardless of whether or not cells were treated with SAG (Fig. 5a). Similar results were obtained in primary MEFs (Supplementary Fig. S6a).
Figure 5.
Ciliogenic and Hh pathway gene expression is reduced in NRF2-null cells. (a) Nfe2l2+/+ and Nfe2l2–/– MEFs were starved 24 h in presence of DMSO or SAG (200 nM) and expression of the indicated ciliogenic genes was analyzed by RT-qPCR. (b) Expression of the indicated Hh pathway genes was analyzed as in (a). All data in (a-b) shown as β-actin-normalized relative mRNA levels (mean ± SEM of n = 3–5 independent experiments). Statistical analysis: two-way ANOVA. Significance between genotypes: *p < 0.05, **p < 0.01, ***p < 0.001 or ****p < 0.0001. SAG treatment did not cause significant differences. (c) Putative ARE sequences were identified for the human ciliogenic and Hh pathway genes indicated on the left. Table shows genomic location and sequences of these putative AREs, as well as maximum and relative scores. Only sequences with relative score above 0.8 were considered as hits. No such hits were found for DYNC2H1, SMO, SUFU or GLI1. Optimal consensus ARE sequence is (A/G)TGACTCAGCA, according to JASPAR database (see methods for details).
Nfe2l2−/− MEFs show a two-fold reduction in ciliogenesis (Fig. 2b), yet their Hh responsiveness is lower by at least 4-fold (Fig. 3a). This suggested that the remaining cilia in Nfe2l2−/− MEFs are not fully functional regarding Hh signal transduction. This, we hypothesized, might be due to defects in gene expression of Hh pathway components. To test this, we measured mRNA levels for several Hh pathway mediators, including positive regulators SMO, GLI2 and IFT27, and negative regulators GLI3, SUFU and GPR1617. All of them were significantly downregulated in Nfe2l2−/− MEFs independently of Hh pathway activation (Fig. 5b). Similar reductions in Hh pathway gene expression were observed in primary MEFs (Supplementary Fig. S6b).
Additionally, we measured gene expression of RFX transcription factors, some of which promote ciliogenesis by binding to X-box motifs in the regulatory regions of many ciliary genes1,4,5. Interestingly, two of them were significantly reduced (Rfx5 and Rfx7), one was strongly upregulated (Rfx8) and three were unaffected (Rfx1, Rfx2, Rfx3) in Nfe2l2−/− MEFs (Supplementary Fig. S7a). For Rfx4, absence of NRF2 made no difference in immortalized MEFs, yet it upregulated expression about 100-fold in primary MEFs (Supplementary Fig. S7b). However, as interesting as these changes may be, they seem to bear little or no relationship with our observed ciliogenic and Hh defects, as will be discussed later.
In contrast, the widespread reduction in ciliogenic and Hh pathway gene expression shown in Fig. 5a,b is likely to be a major cause of the ciliogenic and Hh signaling defects of Nfe2l2−/− MEFs.
DMF does not increase basal levels of ciliogenesis and Hh responsiveness
Since NRF2 loss in Nfe2l2−/− MEFs causes a reduction in ciliogenesis and Hh signaling, we wondered whether higher NRF2 levels would have the opposite effect, increasing ciliogenesis and Hh pathway output. To test this, we treated Nfe2l2+/+ MEFs (and Nfe2l2−/− MEFs as a negative control) with DMF, which raises NRF2 protein levels (Fig. 1d)20. DMF treatment did not significantly increase ciliogenesis in either Nfe2l2+/+ or Nfe2l2−/− MEFs, nor did it affect ciliary length (Supplementary Fig. S8a,b). Likewise, Hh responsiveness was not enhanced by DMF (Supplementary Fig. S8c). Thus, it appears that basal NRF2 levels are not limiting for ciliogenesis or Hh signaling in MEFs, at least under our study conditions.
DMF stimulates ciliogenic gene expression
Even though DMF did not enhance ciliogenesis or Hh signaling, its upregulation of NRF2 protein levels might still cause an increase in NRF2-dependent gene expression. We therefore tested if DMF affects expression of some of the ciliogenic and Hh pathway genes from Fig. 5a,b. Interestingly, DMF caused a significant increase in Dync2h1 and Ift88 mRNA levels in Nfe2l2+/+ but not Nfe2l2−/− MEFs, indicating that the effect depends on NRF2 (Supplementary Fig. S9a). As for Hh pathway genes, we checked mRNA levels of Smo, Gli2 and Gli3. Although we did not see any significant DMF-induced increase for any of these genes in control or NRF2-null MEFs, in the former there was a consistent upward trend, which might merit further study (Supplementary Fig. S9b). Altogether, these experiments suggest that NRF2 levels in control cells may be limiting for the expression of some genes but not others.
Identification of putative AREs in ciliogenic and Hh pathway genes
Given the effects of NRF2 upon ciliogenic and Hh pathway gene expression, we reasoned that some of these genes might be direct targets of NRF2. If so, they should contain ARE sequences in their regulatory regions. To assess this possibility, we analyzed the promoter regions of all ciliogenic and Hh pathway genes whose expression we found reduced in NRF2-null cells. Promoter sequences of the human genes were compared with consensus ARE sequences from the JASPAR database, as previously described22.
Remarkably, many of these genes contain putative ARE sequences with high scores relative to the consensus (Fig. 5c). In particular, two genes (GLI2 and GLI3) contain putative AREs with relative scores higher than 0.99, indicating a virtually perfect match with the consensus. Six other genes (IFT172, IFT140, IFT88, GPR161, PTCH1, and IFT27) contain putative AREs with scores above 0.9, whereas IFT74 has one above 0.8. In contrast, no putative AREs with scores above 0.8 were found in the promoter regions of DYNC2H1, GLI1, SUFU and SMO.
Altogether, our bioinformatic analysis supports the idea that some ciliogenic and/or Hh pathway genes are direct NRF2 transcriptional targets.
GLI2 and GLI3 ciliary localization and protein levels are reduced in NRF2-null cells
Of all the Hh pathway genes whose expression is reduced in NRF2-null cells, GLI2 and GLI3 are potentially the most significant, as they act downstream of all others, directly controlling Hh target gene expression7. We therefore looked at GLI2 and GLI3 proteins in Nfe2l2−/− MEFs. Both GLI2 and GLI3 are known to accumulate at the ciliary tip upon Hh pathway activation7,43. This accumulation is important for GLI2 to be processed into the transcriptionally active GLI2A, and for GLI3 to stop being processed into a repressor, GLI3R, and undergo full proteasomal degradation instead7,44–46. Thus, failure of GLI2 and/or GLI3 to accumulate at the ciliary tip might well explain the low Hh responsiveness of Nfe2l2−/− MEFs. To test this hypothesis, we performed immunofluorescence for GLI2 and GLI3 in Nfe2l2 MEFs treated with either vehicle or SAG. In Nfe2l2+/+ MEFs, SAG induced a strong accumulation of both GLI2 and GLI3 at the ciliary tip. By contrast, their accumulation in SAG-treated Nfe2l2−/− MEFs was strongly and significantly reduced (Fig. 6a–d).
Figure 6.
GLI2 and GLI3 ciliary tip accumulation and protein levels are reduced in NRF2-null cells. (a,b) Representative immunofluorescence images for ARL13B, γ-tubulin and GLI2 (a) or GLI3 (b) in Nfe2l2+/+ and Nfe2l2−/− littermate MEFs starved 24 h in presence of DMSO (vehicle) or SAG (200 nM). Nuclei were counterstained with DAPI. Top right of each image shows magnification of boxed area. Scale bars, 10 μm. (c,d) Quantification of GLI2 (c) or GLI3 (d) signal intensity at ciliary tip from (a,b). Data are mean ± SEM of at least 40 cilia per condition (A.U.: arbitrary units). Statistical analysis: two-way ANOVA followed by Tukey’s multiple comparisons tests. Asterisks: *p < 0.05, **p < 0.01, ***p < 0.001 or ****p < 0.0001. (e,f) Nfe2l2+/+ and Nfe2l2−/− MEFs were treated as in (a,b) and analyzed by Western blot with GLI2 (e) and GLI3 (f) antibodies. Most GLI2 and GLI3 are found in their processed forms (arrows), whose levels are reduced in mutant MEFs (and by SAG treatment for GLI3, as expected). Lamin B was used as loading control. Molecular weight markers in kDa are on the right.
Western blot analysis of GLI2 and GLI3 confirmed a strong reduction in the overall basal levels of these proteins, especially of their proteolytically processed lower molecular weight forms (Fig. 6e,f). Moreover, Nfe2l2−/− MEFs appeared to react differently to SAG than Nfe2l2+/+ MEFs. In control cells, as previously shown, SAG did not affect the pattern of GLI2 bands47, but caused a marked reduction in GLI3 levels48. On the other hand, Nfe2l2−/− MEFs responded to SAG by reducing the processed forms of GLI2 while having only a modest effect on GLI3 (Fig. 6e,f). Together with the immunofluorescence data, these results suggest that GLI2 and GLI3 ciliary accumulation, processing and protein levels are abnormal in NRF2-null cells.
Finally, we also carried out immunofluorescence and Western blot with IFT88 and SMO antibodies. Despite the observed lower mRNA levels in Nfe2l2−/− MEFs relative to control (Fig. 5a,b), we saw no such differences in either ciliary IFT88 intensity (Supplementary Fig. S10a,b), ciliary SMO intensity with or without SAG (Supplementary Fig. S10c,d), or total levels of these proteins as seen by Western blot in presence or absence of DMF (Supplementary Fig. S10e). This might reflect compensatory mechanisms that maintain protein homeostasis despite altered mRNA levels.
Discussion
In pluripotent hESCs, cell fate choice between mesendoderm and ectoderm lineages depends on the PAN axis, whereby early emergence of cilia in some cells stimulates autophagy, thereby downregulating NRF2 activity and committing those cells to neuroectoderm12. Here, we have shown data suggesting that the PAN axis is also at work in more differentiated cell types, including fibroblasts. In support of this, NRF2 transcriptional activity is higher in cells unable to grow cilia (Fig. 1a–c), whose autophagic flux is reduced (Fig. 1g). Moreover, mTOR inhibitors, which raise autophagic flux in cilia-null cells (Fig. 1g), fully rescue the abnormal NRF2 activity of these cells (Fig. 1e,f).
If the PAN axis is not restricted to hESCs, then what is its biological role beyond early embryogenesis? In stem cells, it may control cell fate choices in a similar way to what it does in hESCs, except that the relevant NRF2 target genes would differ in each case (in hESCs it promotes expression of pluripotency genes OCT4 and NANOG, which are epigenetically silenced in more differentiated cells12).
In terminally differentiated cells, the PAN axis might convey information about ciliary integrity or stress. Since genetic deletion of cilia reduces autophagy and raises NRF2 activity, something similar might occur by accidental or stress-induced cilia ablation or damage. If so, this would trigger NRF2-dependent cytoprotective responses, which might include cilia regeneration, consistent with our data that NRF2 promotes ciliogenic gene expression (Fig. 5a, Supplementary Figs S6a and S9a). Consistent with this, NRF2 plays a protective role in cigarette smoke-induced harm to airway cilia23. Also, flow sensation in renal mechanosensory cilia represses mTOR to promote autophagy and reduce cell size13,15. Thus, it would be interesting to test whether mechanical stress in renal cilia also modulates NRF2 activity. If so, this could have important implications, as disruption of this ciliary pathway causes polycystic kidney disease (PKD), the most common ciliopathy19. The fact that NRF2 dysregulation is involved in kidney cystogenesis in a mouse cancer model supports a potential connection between NRF2 and PKD49.
Mechanistically, many questions remain concerning how the PAN axis works. Our data indicate that GLI-dependent Hh signaling is not involved (Supplementary Fig. S2), but how cilia control autophagy and NRF2 remains poorly understood. For instance, our evidence shows cilia downregulate NRF2 activity but not its protein levels (Fig. 1d,h). Since NRF2 is often regulated through protein stability20, this raises the question of how cilia affect NRF2 in MEFs, and whether this mechanism also applies to hESCs, where NRF2 protein levels were not assessed12.
We have also found that NRF2-null cells grow fewer and shorter cilia than normal in some cell types (Fig. 2, Supplementary Figs S3 and S4), and display poor Hh signaling, a cilia-dependent pathway (Fig. 3). We investigated several possible causes for these effects. Hh responsiveness in Nfe2l2−/− MEFs was not improved by NAC, which acts in oxidatively stressed cells by replenishing their levels of glutathione, a major cellular antioxidant and ROS scavenger (Fig. 4)40. This largely rules out oxidative stress as a significant cause of Hh signaling defects in our model. And since Hh defects arise, at least partly, from ciliogenic defects, the lack of Hh signaling rescue by NAC also suggests, indirectly, that oxidative stress does not explain reduced ciliation in Nfe2l2−/− MEFs. Tubastatin A, an HDAC6 inhibitor, also failed to even partially rescue Hh signaling (Fig. 4). Thus, ciliophagy, the HDAC6-dependent autophagic resorption of cilia, does not account for the Hh defects either23. Likewise, lack of rescue by Torin-1 shows that mTOR is not involved in poor Hh responsiveness in Nfe2l2−/− MEFs, indicating that NRF2 affects cilia through mechanisms distinct from how cilia affect NRF2 (Fig. 4). We also failed to rescue Hh signaling with H89, a PKA inhibitor (Fig. 4). Since PKA acts as a Hh pathway repressor downstream of SMO but upstream of GLI2 and GLI3, these data pointed to Hh signaling disruption downstream of SMO and PKA7.
We then discovered that multiple ciliogenic and Hh pathway mRNAs are expressed at lower levels in Nfe2l2−/− MEFs (Fig. 5a,b). These moderate but highly significant reductions in gene expression, when combined, likely account for the ciliogenic and Hh defects of Nfe2l2−/− cells.
The evidence for this is particularly strong for the Hh pathway. GLI2 and GLI3 transcription factors are the main controllers of Hh target gene expression (as GLI1 is a Hh target gene whose expression is almost negligible before pathway stimulation)7. Hh signaling alters the transcriptional activities of GLI2 and GLI3, turning the former into an activator (GLI2A) and preventing the latter from turning into a repressor (GLI3R), its fate under basal conditions7. For these changes to occur, GLI2 and GLI3 need to accumulate at the ciliary tip, but this is strongly reduced in Nfe2l2−/− MEFs (Fig. 6a–d)7,44–46. In addition, GLI2 and GLI3 protein levels are strongly reduced in these mutants (Fig. 6e,f), as are their mRNAs (Fig. 5b). These changes in GLI2 and GLI3 can readily explain the observed Hh defects. And since GLI2 and GLI3 act downstream from SMO, GPR161, IFT27 and SUFU in the Hh pathway7, the lower mRNA expression observed for the latter genes is likely to be of little consequence in terms of Hh pathway output (Fig. 5b). In fact, despite the lower mRNA levels in Nfe2l2−/− MEFs, SMO protein levels and its SAG-induced cilia translocation appear unaffected in these cells (Supplementary Fig. S10c–e).
Regarding ciliogenesis, it is less clear which genes and their products are responsible for the observed defects. In principle, each of the genes studied in Fig. 5a might account for these defects, as they are all important in the process. Moreover, given that our analysis was far from exhaustive, many other ciliogenic genes might be affected in NRF2-null cells. Since IFT88 is absolutely required for ciliogenesis (Fig. 1a), we tested the role of IFT88 protein in the ciliogenic defects of Nfe2l2−/− MEFs. Intriguingly, neither total nor ciliary IFT88 protein levels were visibly affected in mutant relative to control MEFs (Supplementary Fig. S10a,b). This might indicate that compensatory mechanisms operate in Nfe2l2−/− MEFs to keep IFT88 and SMO protein levels constant, despite lowered mRNA levels.
Some RFX transcription factors promote ciliogenesis by stimulating expression of ciliogenic genes such as the ones in Fig. 5a, so a reduction in one or more pro-ciliogenic RFX factors could neatly explain why NRF2-null MEFs grow fewer and shorter cilia. RFX1-4 are well established pro-ciliogenic factors4, but none of them was reduced in Nfe2l2−/− MEFs. Instead, we found reductions in Rfx5 and Rfx7 (Supplementary Fig. S7a). RFX5 and RFX7 constitute a distinct RFX subfamily and both play important roles in immunity4,5,50, yet only RFX7 has recently been linked to ciliogenesis in the neural tube51. Whether RFX7 contributes to the ciliogenic phenotype of NRF2-null cells remains unaddressed.
Unlike Rfx5 and Rfx7, we found elevated levels of Rfx4 and Rfx8 in NRF2-null MEFs. This, however, cannot explain the ciliogenic defects, which were seen in both primary and immortalized Nfe2l2−/− MEFs (Fig. 2a–c and Supplementary Fig. S3), whereas Rfx4 upregulation only occurred in the former (Supplementary Fig. S7b). Additionally, higher RFX4 levels would be expected to upregulate ciliogenic genes, not reduce them. Virtually nothing is known about RFX8, except that it belongs to the same RFX subfamily as RFX44,5.
Since NRF2 stimulates its target genes by binding to ARE sequences in their promoters, we performed a bioinformatic search for putative ARE sequences in the human promoters of the ciliogenic and Hh pathway genes we found reduced in Nfe2l2−/− MEFs (Fig. 5c). Remarkably, the top two hits from this analysis were GLI2 and GLI3, whose promoters each contain a perfect match with the consensus ARE sequence (relative score = 0.997). Together with the above data, this suggests that GLI2 and GLI3 may be direct NRF2 targets, although this awaits experimental demonstration. Putative ARE sequences with relative scores in the 0.81–0.98 range were also found in IFT172, IFT140, IFT88, IFT74, GPR161, PTCH1 and IFT27. Consistent with these data, Ift74, Ift27 and Gpr161 promoter sequences were pulled down with NRF2 in a ChIP-seq study in sulforaphane-treated cells52. The presence of putative AREs in PTCH1 (but not GLI1) promoter raises the possibility that NRF2 directly upregulates Hh targets. If so, we would expect lower basal PTCH1 levels in unstimulated NRF2-null MEFs, which we did not detect. However, since basal PTCH1 levels are very low, detecting such a reduction, if it exists, could be very challenging. Using cells with higher basal PTCH1 levels could clarify this issue.
Since genetic deletion of NRF2 reduces ciliogenic and Hh pathway gene expression, we also tested whether NRF2 stabilization by DMF had the opposite effect. Interestingly, ciliogenic genes Dync2h1 and Ift88 were both significantly upregulated in DMF-treated Nfe2l2+/+ but not Nfe2l2−/− MEFs (Supplementary Fig. S9a). This confirms these genes are positively regulated by NRF2. For Gli2, Gli3 and Smo, although the data pointed in the same direction, DMF did not cause any statistically significant changes (Supplementary Fig. S9b). Likewise, DMF did not increase ciliogenesis or Hh responsiveness above basal levels, indicating that NRF2 levels in these cells are not limiting for these processes (Supplementary Fig. S8).
Beyond the mechanistic details, our work raises an important question: what is the biological significance of NRF2 affecting ciliogenesis and Hh signaling? Severe ciliogenesis and Hh signaling disruptions in mouse embryos lead to embryonic lethality and malformations7,53. In contrast, Nfe2l2−/− mice are viable, have no malformations, and do fairly well unless exposed to stress31,54. Hence, cilia and Hh signaling cannot be severely perturbed in Nfe2l2−/− mouse embryos. Together with our data, this suggests that NRF2 affects ciliogenesis and Hh signaling in a manner dependent on cell type, tissue and developmental stage. Comprehensive spatiotemporal studies would shed light on this issue.
Our data show NRF2 affects ciliogenesis in embryonic fibroblasts and hippocampal astrocytes, so one might expect NRF2-null mice to have cilia or Hh-dependent phenotypes related to these cell types. In this regard, we recently reported defects in hippocampal neurogenesis in NRF2-null mice, a process where cilia and Hh signaling also play important roles32,55–57. Thus, neurogenesis defects in NRF2-null hippocampus may be related to defective ciliation. If so, this might involve astrocyte cilia, but maybe also neuronal cilia, which are not labeled by ARL13B and thus were not visualized in our study37,38. On the other hand, we saw no ciliary defects in multiciliated ependymal cells of NRF2-null brain ventricles (Supplementary Fig. S4). This suggests that cilia defects in NRF2-null mice are cell type-specific, thereby explaining why these animals do not suffer from hydrocephalus, a pathology resulting from ependymal cilia malfunction4,31.
Why NRF2 affects ciliogenesis in some cell types more than others remains a mystery. Cell type-specific compensatory mechanisms may exist, which could act at many levels, such as gene expression, protein stability or enzyme activity, to name a few. Redundancy is another possibility: NFE2L1/NRF1, an NRF2-related transcription factor, acts redundantly with NRF2 during mouse development58. Thus, our data might be explained by NRF1 compensating for NRF2 loss in specific cell types. If so, one would expect Nfe2l1-Nfe2l2 double knockout mice to display cilia and Hh-related phenotypes. These embryos are delayed, show signs of massive oxidative stress and apoptosis, and die at midgestation, too early to assess for all but the most drastic defects in cilia and Hh signaling1,7,58. On the other hand, NRF1 is expressed ubiquitously, and Nfe2l1-Nfe2l2 double mutant MEFs show markedly higher apoptosis and stress sensitivity than Nfe2l2−/− MEFs, demonstrating that NRF1 is present and functional in the latter58,59. Therefore, lack of NRF1 function is unlikely to explain cilia and Hh defects in Nfe2l2−/− MEFs. Still, factors other than NRF1 might act redundantly with NRF2.
Another possibility is that Hh signaling participates in NRF2-dependent cytoprotective responses. Since NRF2 promotes Hh signaling, stress-induced NRF2 stabilization might enhance Hh signaling, which might protect cells from damage in some way. Although Hh signaling stimulation by oxidative stress and cytoprotective effects of Hh signaling are both reported in the literature60–65, our data showed no enhancement of Hh responsiveness by DMF, as mentioned above (Supplementary Fig. S8c). This, however, does not rule out that Hh responses play cytoprotective roles in vivo, particularly in cell types where basal NRF2 protein levels are low relative to what cells need for optimal Hh responsiveness.
Here, we have established a clear connection between NRF2, expression of ciliogenic and Hh pathway genes, and the processes controlled by these genes. The physiopathological contexts where changes in NRF2 activity affect cilia and Hh signaling remain to be elucidated.
Methods
Reagents, plasmids and antibodies
Reagents: Torin-1 (ApexBio, 250 nM), Rapamycin (Fisher Bioreagents, 200 nM), dimethyl fumarate (Sigma, 20 µM), Chloroquine (Acros Organics, 10 µM), SAG (Cayman, 200 nM), R,S-Sulforaphane (LKT Laboratories, 15 µM), N-acetyl-L-cysteine (Alfa Aesar, 3 mM), H89 dihydrochloride (BioVision, 10 µM) and Tubastatin A (Cayman, 10 µM). Plasmids: pcDNA3.1-mGli2ΔN and pcDNA3.1-hGLI3R were kind gifts of Drs. Victor L. Ruiz-Perez and Elisa Martí, respectively66. Antibodies: Acetylated α-tubulin (Sigma, T7451, 1:10,000), GLI1 (Cell Signaling, L42B10, 1:1000), GFAP (Sigma, G3893, 1:200), HMOX1 (Enzo Life Sciences, 1:2000), SMO (Proteintech, 66851-1-Ig, WB: 1:500), GAPDH (Proteintech, 60004-1-Ig, 1:1000), β-actin (Proteintech, 66009-1-Ig, 1:5000) and α-tubulin (Proteintech, 66031-1-Ig, 1:1000) were from mouse. GLI2 (R&D systems, AF3635, WB: 0.1 µg/ml, IF: 4 µg/ml), GLI3 (R&D systems, AF3690, WB: 1 µg/ml, IF: 4 µg/ml), Lamin B (Santa Cruz, sc-6217, 1:5000) and γ-tubulin (Santa Cruz, sc-7396, 1:200) were from goat. ARL13B (Proteintech, 17711-1-AP, 1:200), LC3B (Cell Signaling Technology, #2775, 1:500), SMO (Abcam, ab38686, IF: 1:1000), IFT88 (Proteintech, 13967-1-AP, WB: 1:1000, IF: 1:50), CEP164 (Proteintech, 22227-1-AP, 1:500) and NRF2 (described in21, 1:5000) were from rabbit. Due to their initial low specificity (see Fig. 3b and Supplementary Fig. S12), NRF2 antibodies were repeatedly incubated with membranes containing NRF2-null cell lysates, which led to greater specificity towards NRF2, as seen in Fig. 1d,h and Supplementary Fig. S12. Secondary antibodies were from donkey (Thermofisher, AlexaFluor 488, 555 or 647-conjugated) or from goat (Thermofisher, HRP-conjugated).
Animals
Colonies of Nfe2l2+/+ and Nfe2l2−/− C57BL/6 mice were established from founders kindly provided by Prof. Masayuki Yamamoto67. Animal procedures were performed according to protocols approved by the Ethical Committee for Research of the Spanish National Research Council (CSIC), in accordance with institutional, Spanish and European guidelines (Boletín Oficial del Estado, 18 March 1988; and European Council Directives 86/609/EEC and 2003/65/EC).
Cell culture
Kif3a+/+, Kif3a−/−, Ift88+/+ and Ift88−/− MEFs have been reported elsewhere68. Nfe2l2+/+ and Nfe2l2−/− littermate MEFs were derived from E11.5 mouse embryos and immortalized with SV40 large T antigen using previously described protocols43. All MEFs were grown in DMEM medium supplemented with 10% fetal bovine serum at 37 °C and 5% CO2 in a humidified atmosphere. All cell lines were mycoplasma-free, as ascertained by regular tests. To induce ciliogenesis and for drug treatments, MEFs were starved for 24 hours in OptiMEM medium (Thermofisher). For autophagy flux analysis, cells were starved for 4 hours in Earle’s Balanced Salts Solution (EBSS, Sigma).
RT-qPCR
Total cellular RNA was extracted using GeneJET RNA purification kit (Thermofisher) and reverse transcribed with MMLV Reverse Transcriptase (Promega) using oligo-dT18. Quantitative real time PCR was performed using PowerUp SYBR Green Master Mix (Thermofisher) and the primer pairs listed in Supplementary Fig. S11. Reactions were run on an Applied Biosystems StepOne Real Time PCR System and data analyzed using ΔΔCt method with β-actin as control gene.
Western blot and quantitation
Total cell lysates were prepared in buffer containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Igepal CA-630 and protease inhibitor cocktail (Thermofisher). Lysates were centrifuged at 20,000 × g for 10 min at 4 °C to obtain postnuclear supernatants, whose protein concentrations were equalized after measuring them with Pierce BCA Protein Assay kit. Samples were then processed for SDS-PAGE, run in 4–20% Novex Tris-Glycine gels (Thermofisher) and transferred to nitrocellulose membranes for immunostaining. Results were visualized using ECL chemiluminescence reagents and captured on either X-ray film or using an MF-ChemiBIS 3.2 system (DNR Bio-Imaging Systems). Images were then cropped and their brightness and contrast adjusted using Adobe Photoshop. Uncropped and unadjusted images are shown in Supplementary Fig. S12. For band quantitation, Fiji (Image J) software was used. Briefly, 8-bit grayscale images of blots were inverted and average pixel intensity measured for all bands of interest and their corresponding backgrounds. Total pixel intensity of each band was then obtained by multiplying average pixel intensity by pixel number. To this value, total pixel intensity of an equally-sized background area was subtracted in order to obtain the specific band signal, which was expressed as a ratio to the control band. For autophagy flux calculation, basal LC3B-II/LC3B-I ratio was subtracted from the same ratio in presence of chloroquine, and the resulting difference was normalized relative to control sample.
Immunofluorescence and quantitation
Cultured cells were grown on coverslips, fixed 5 min at RT in PBS with 4% paraformaldehyde, then 3 min at −20 °C in freezer-cold methanol. Cells were blocked and permeabilized in blocking solution (PBS + 0.1% Triton X100 + 2% donkey serum + 0.02% sodium azide) for 30–60 min at RT before proceeding with standard immunostaining procedures and imaging with a Nikon 90i fluorescence microscope. All cilia quantitations were performed with the aid of Fiji (Image J) software. For percent ciliation, one or more coverslips per condition were examined in each independent experiment. For each coverslip, seven representative cell fields were used to count cilia (identified by ARL13B and γ-tubulin costaining), whose number was divided by total cells (DAPI-labeled nuclei). For length measurements, cilia were identified as above and their length measured in the ARL13B channel. For ciliary intensity measurements, unsaturated 8-bit images were used. Total signal in the region of interest (ciliary tip region for GLI2-3 or whole cilium for IFT88 and SMO) was obtained for the relevant channel as the product of average pixel intensity and pixel number in the region. Background signal was likewise measured near each region of interest and subtracted from total signal to obtain the specific signal. For in vivo studies, 30 µm-thick coronal brain sections of 3-month-old mice were obtained and processed as previously described32, and imaged using a Leica TCS SP5 confocal microscope. Ependymal cilia were visualized in one mouse of each genotype. For quantitation of hippocampal samples, five sections were imaged for each of three mice. Fiji was then used to count cilia, identified as rod-shaped ARL13B+ structures, and GFAP+ astrocytes. Since ARL13B+ cilia were only seen in close association with astrocytes (consistent with reports indicating that ARL13B is an astrocytic ciliary marker in adult brain37,38), we plotted data as percentage of GFAP+ astrocytes carrying ARL13B+ cilia.
Bioinformatics
Putative ARE sequence identification was performed as previously described22. Briefly, we first identified putative AREs in the promoter regions of our genes of interest using the Encyclopedia of DNA Elements at UCSC (ENCODE) of the human genome (Feb. 2009), based on the ChIP data available for ARE-binding transcription factors BACH1 and MAFK. Subsequently, using a Python-based script, these putative AREs were compared with the position-specific scoring matrix (PSSM) derived from the frequency matrix of the consensus ARE sequence recognized by NRF2, obtained from JASPAR database. We considered only putative AREs with a relative score higher than 80%22.
Statistical analysis
GraphPad Prism 7 software was used to graph and statistically analyze data. The specific details of each experiment are provided in the corresponding figure legends.
Supplementary information
Acknowledgements
We thank members of the Garcia-Gonzalo and Cuadrado labs for helpful suggestions and Drs. Victor L. Ruiz-Perez, Elisa Martí, Masayuki Yamamoto and Bruno Sainz for sharing reagents, mice or equipment. This work was supported by European Regional Development Fund (ERDF)-cofunded grants from the Spanish Ministry of Economy and Competitiveness (MINECO) to FRGG (SAF2015-66568-R and RYC2013-14887) and to A.C. and I.L.B. (SAF2016-76520-R). AMH was supported by an ERDF-cofunded predoctoral contract from the Community of Madrid government. RMM, NRA and RB were supported by predoctoral grants from MINECO.
Author Contributions
I.L.B., A.C. and F.R.G.G. designed the study. A.M.H., R.M.M., N.R.A., R.B. and I.P.B. performed the experiments. All authors contributed to analyzing results. I.L.B., A.C. and F.R.G.G. supervised the work. F.R.G.G. wrote the manuscript with help from A.C., I.L.B., N.R.A. and A.M.H.
Data Availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50356-0.
References
- 1.Reiter JF, Leroux MR. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 2017;18:533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Gonzalo FR, Reiter JF. Scoring a backstage pass: mechanisms of ciliogenesis and ciliary access. J Cell Biol. 2012;197:697–709. doi: 10.1083/jcb.201111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Lei, Dynlacht Brian D. The regulation of cilium assembly and disassembly in development and disease. Development. 2018;145(18):dev151407. doi: 10.1242/dev.151407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choksi SP, Lauter G, Swoboda P, Roy S. Switching on cilia: transcriptional networks regulating ciliogenesis. Development. 2014;141:1427–1441. doi: 10.1242/dev.074666. [DOI] [PubMed] [Google Scholar]
- 5.Sugiaman-Trapman D, et al. Characterization of the human RFX transcription factor family by regulatory and target gene analysis. BMC Genomics. 2018;19:181. doi: 10.1186/s12864-018-4564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci USA. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bangs Fiona, Anderson Kathryn V. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harbor Perspectives in Biology. 2016;9(5):a028175. doi: 10.1101/cshperspect.a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eguether, T. & Hahne, M. Mixed signals from the cell’s antennae: primary cilia in cancer. EMBO Rep19, 10.15252/embr.201846589 (2018). [DOI] [PMC free article] [PubMed]
- 9.Hu L, Wang B, Zhang Y. Serotonin 5-HT6 receptors affect cognition in a mouse model of Alzheimer’s disease by regulating cilia function. Alzheimers Res Ther. 2017;9:76. doi: 10.1186/s13195-017-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaliszewski M, Knott AB, Bossy-Wetzel E. Primary cilia and autophagic dysfunction in Huntington’s disease. Cell Death Differ. 2015;22:1413–1424. doi: 10.1038/cdd.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armato U, Chakravarthy B, Pacchiana R, Whitfield JF. Alzheimer’s disease: an update of the roles of receptors, astrocytes and primary cilia (review) Int J Mol Med. 2013;31:3–10. doi: 10.3892/ijmm.2012.1162. [DOI] [PubMed] [Google Scholar]
- 12.Jang J, et al. Primary Cilium-Autophagy-Nrf2 (PAN) Axis Activation Commits Human Embryonic Stem Cells to a Neuroectoderm Fate. Cell. 2016;165:410–420. doi: 10.1016/j.cell.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Orhon I, et al. Primary-cilium-dependent autophagy controls epithelial cell volume in response to fluid flow. Nat Cell Biol. 2016;18:657–667. doi: 10.1038/ncb3360. [DOI] [PubMed] [Google Scholar]
- 14.Pampliega O, et al. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehlke C, et al. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat Cell Biol. 2010;12:1115–1122. doi: 10.1038/ncb2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foerster P, et al. mTORC1 signaling and primary cilia are required for brain ventricle morphogenesis. Development. 2017;144:201–210. doi: 10.1242/dev.138271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Struchtrup A, Wiegering A, Stork B, Ruther U, Gerhardt C. The ciliary protein RPGRIP1L governs autophagy independently of its proteasome-regulating function at the ciliary base in mouse embryonic fibroblasts. Autophagy. 2018;14:567–583. doi: 10.1080/15548627.2018.1429874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grantham JJ, Geiser JL, Evan AP. Cyst formation and growth in autosomal dominant polycystic kidney disease. Kidney Int. 1987;31:1145–1152. doi: 10.1038/ki.1987.121. [DOI] [PubMed] [Google Scholar]
- 19.Bergmann C, et al. Polycystic kidney disease. Nat Rev Dis Primers. 2018;4:50. doi: 10.1038/s41572-018-0047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuadrado A, et al. Transcription Factor NRF2 as a Therapeutic Target for Chronic Diseases: A Systems Medicine Approach. Pharmacol Rev. 2018;70:348–383. doi: 10.1124/pr.117.014753. [DOI] [PubMed] [Google Scholar]
- 21.Pajares M, et al. Transcription factor NFE2L2/NRF2 modulates chaperone-mediated autophagy through the regulation of LAMP2A. Autophagy. 2018;14:1310–1322. doi: 10.1080/15548627.2018.1474992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pajares M, et al. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12:1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam HC, et al. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J Clin Invest. 2013;123:5212–5230. doi: 10.1172/JCI69636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z, et al. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 2013;502:254–257. doi: 10.1038/nature12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 27.Wong SY, et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15:1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci USA. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci USA. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robledinos-Anton N, et al. Transcription factor NRF2 controls the fate of neural stem cells in the subgranular zone of the hippocampus. Redox Biol. 2017;13:393–401. doi: 10.1016/j.redox.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sterpka A, Chen X. Neuronal and astrocytic primary cilia in the mature brain. Pharmacol Res. 2018;137:114–121. doi: 10.1016/j.phrs.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen PC, et al. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson’s disease: Critical role for the astrocyte. Proc Natl Acad Sci USA. 2009;106:2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gan L, Vargas MR, Johnson DA, Johnson JA. Astrocyte-specific overexpression of Nrf2 delays motor pathology and synuclein aggregation throughout the CNS in the alpha-synuclein mutant (A53T) mouse model. J Neurosci. 2012;32:17775–17787. doi: 10.1523/JNEUROSCI.3049-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liddell Jeffrey. Are Astrocytes the Predominant Cell Type for Activation of Nrf2 in Aging and Neurodegeneration? Antioxidants. 2017;6(3):65. doi: 10.3390/antiox6030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kasahara K, Miyoshi K, Murakami S, Miyazaki I, Asanuma M. Visualization of astrocytic primary cilia in the mouse brain by immunofluorescent analysis using the cilia marker Arl13b. Acta Med Okayama. 2014;68:317–322. doi: 10.18926/AMO/53020. [DOI] [PubMed] [Google Scholar]
- 38.Sipos E, Komoly S, Acs P. Quantitative Comparison of Primary Cilia Marker Expression and Length in the Mouse Brain. J Mol Neurosci. 2018;64:397–409. doi: 10.1007/s12031-018-1036-z. [DOI] [PubMed] [Google Scholar]
- 39.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rushworth GF, Megson IL. Existing and potential therapeutic uses for N-acetylcysteine: the need for conversion to intracellular glutathione for antioxidant benefits. Pharmacol Ther. 2014;141:150–159. doi: 10.1016/j.pharmthera.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Butler KV, et al. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc. 2010;132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chijiwa T, et al. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J Biol Chem. 1990;265:5267–5272. [PubMed] [Google Scholar]
- 43.Garcia-Gonzalo FR, et al. Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling. Dev Cell. 2015;34:400–409. doi: 10.1016/j.devcel.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA. 2009;106:21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santos N, Reiter JF. A central region of Gli2 regulates its localization to the primary cilium and transcriptional activity. J Cell Sci. 2014;127:1500–1510. doi: 10.1242/jcs.139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Wang C, Pan Y, Bai Z, Wang B. Increased proteolytic processing of full-length Gli2 transcription factor reduces the hedgehog pathway activity in vivo. Dev Dyn. 2011;240:766–774. doi: 10.1002/dvdy.22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/S0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 49.Adam J, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castro W, et al. The transcription factor Rfx7 limits metabolism of NK cells and promotes their maintenance and immunity. Nat Immunol. 2018;19:809–820. doi: 10.1038/s41590-018-0144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manojlovic Z, Earwood R, Kato A, Stefanovic B, Kato Y. RFX7 is required for the formation of cilia in the neural tube. Mech Dev. 2014;132:28–37. doi: 10.1016/j.mod.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chorley BN, et al. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Gonzalo FR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci USA. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amador-Arjona A, et al. Primary cilia regulate proliferation of amplifying progenitors in adult hippocampus: implications for learning and memory. J Neurosci. 2011;31:9933–9944. doi: 10.1523/JNEUROSCI.1062-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berbari NF, et al. Hippocampal and cortical primary cilia are required for aversive memory in mice. PLoS One. 2014;9:e106576. doi: 10.1371/journal.pone.0106576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhee S, Kirschen GW, Gu Y, Ge S. Depletion of primary cilia from mature dentate granule cells impairs hippocampus-dependent contextual memory. Sci Rep. 2016;6:34370. doi: 10.1038/srep34370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- 59.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 60.Das S, et al. Loss of Merlin induces metabolomic adaptation that engages dependence on Hedgehog signaling. Sci Rep. 2017;7:40773. doi: 10.1038/srep40773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen KY, Chiu CH, Wang LC. Anti-apoptotic effects of Sonic hedgehog signalling through oxidative stress reduction in astrocytes co-cultured with excretory-secretory products of larval Angiostrongylus cantonensis. Sci Rep. 2017;7:41574. doi: 10.1038/srep41574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dai RL, et al. Sonic hedgehog protects cortical neurons against oxidative stress. Neurochem Res. 2011;36:67–75. doi: 10.1007/s11064-010-0264-6. [DOI] [PubMed] [Google Scholar]
- 63.He W, et al. Sonic hedgehog promotes neurite outgrowth of cortical neurons under oxidative stress: Involving of mitochondria and energy metabolism. Exp Cell Res. 2017;350:83–90. doi: 10.1016/j.yexcr.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 64.Kaushal JB, Popli P, Sankhwar P, Shukla V, Dwivedi A. Sonic hedgehog protects endometrial hyperplasial cells against oxidative stress via suppressing mitochondrial fission protein dynamin-like GTPase (Drp1) Free Radic Biol Med. 2018;129:582–599. doi: 10.1016/j.freeradbiomed.2018.10.427. [DOI] [PubMed] [Google Scholar]
- 65.Lin EH, et al. Hedgehog pathway maintains cell survival under stress conditions, and drives drug resistance in lung adenocarcinoma. Oncotarget. 2016;7:24179–24193. doi: 10.18632/oncotarget.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alvarez-Medina R, Cayuso J, Okubo T, Takada S, Marti E. Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development. 2008;135:237–247. doi: 10.1242/dev.012054. [DOI] [PubMed] [Google Scholar]
- 67.Itoh K, Mimura J, Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxidants & redox signaling. 2010;13:1665–1678. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 68.Kodani A, Salome Sirerol-Piquer M, Seol A, Garcia-Verdugo JM, Reiter JF. Kif3a interacts with Dynactin subunit p150 Glued to organize centriole subdistal appendages. EMBO J. 2013;32:597–607. doi: 10.1038/emboj.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.






