The goal was to investigate the mechanisms of colistin resistance and heteroresistance in Pseudomonas aeruginosa clinical isolates. Colistin resistance was determined by the broth microdilution method. Colistin heteroresistance was evaluated by population analysis profiling. Time-kill assays were also conducted. PCR sequencing was performed to detect the resistance genes among (hetero)resistant isolates, and quantitative real-time PCR assays were performed to determine their expression levels.
KEYWORDS: Pseudomonas aeruginosa, colistin, heteroresistance, lipid A modification, molecular mechanisms, resistance
ABSTRACT
The goal was to investigate the mechanisms of colistin resistance and heteroresistance in Pseudomonas aeruginosa clinical isolates. Colistin resistance was determined by the broth microdilution method. Colistin heteroresistance was evaluated by population analysis profiling. Time-kill assays were also conducted. PCR sequencing was performed to detect the resistance genes among (hetero)resistant isolates, and quantitative real-time PCR assays were performed to determine their expression levels. Pulsed-field gel electrophoresis and multilocus sequence typing were performed. Lipid A characteristics were determined via matrix-assisted laser desorption–ionization time of flight mass spectrometry (MALDI-TOF MS). Two resistant isolates and 9 heteroresistant isolates were selected in this study. Substitutions in PmrB were detected in 2 resistant isolates. Among heteroresistant isolates, 8 of 9 heteroresistant isolates had nonsynonymous PmrB substitutions, and 2 isolates, including 1 with a PmrB substitution, had PhoQ alterations. Correspondingly, the expression levels of pmrA or phoP were upregulated in PmrB- or PhoQ-substituted isolates. One isolate also found alterations in ParRS and CprRS. The transcript levels of the pmrH gene were observed to increase across all investigated isolates. MALDI-TOF MS showed additional 4-amino-4-deoxy-l-arabinose (l-Ara4N) moieties in lipid A profiles in (hetero)resistant isolates. In conclusion, both colistin resistance and heteroresistance in P. aeruginosa in this study mainly involved alterations of the PmrAB regulatory system. There were strong associations between mutations in specific genetic loci for lipid A synthesis and regulation of modifications to lipid A. The transition of colistin heteroresistance to resistance should be addressed in future clinical surveillance.
INTRODUCTION
Pseudomonas aeruginosa, a Gram-negative nonfermenting bacillus, is responsible for various nosocomial infections, such as pneumonia, urinary tract infections, surgical site infections, and bloodstream infections (1). The increased capacity to develop antibiotic resistance is due to improper and excessive use of antibiotics. Carbapenems were introduced to treat serious multidrug-resistant P. aeruginosa infections but eventually led to a rise of carbapenem-resistant isolates worldwide (2).
As a consequence, interest has been rekindled in “old” antibiotics such as the polymyxins (i.e., polymyxin B and colistin). Owing to its high activity against Gram-negative “superbugs,” including carbapenem-resistant P. aeruginosa, colistin is now being administered as last-resort therapy for patients with isolates against which none of the other available antibiotics is active (3). Although colistin maintains high antimicrobial activity against P. aeruginosa, colistin heteroresistance, a relatively poorly reported phenotype, requires more attention.
Heteroresistance, an intermediate situation, may have certain similarities and differences, compared with homogeneous resistance. It was first described for Haemophilus influenzae and refers to a phenotype characteristic involving the presence of resistant subpopulations among a susceptible population (4). This phenomenon was subsequently found for many antibiotics among both Gram-positive and Gram-negative bacteria (5). Colistin heteroresistance has been described (6). Multiple studies have indicated that the presence of this phenotype may account for unexplained treatment failures (7–9).
Colistin homogeneous resistance develops mainly due to mutations in the two-component regulatory systems (TCSs) (PhoPQ and PmrAB) (10, 11). Specific mutations trigger constitutive upregulation of the pmrHFIJKLM-ugd operon, which leads to the covalent attachment of 4-amino-4-deoxy-l-arabinose (l-Ara4N) to the lipid A component of the outer membrane lipopolysaccharide (LPS) (12–14). Recently, ParRS, CprRS, and ColRS TCSs have also been found to play a role in colistin homogeneous resistance in P. aeruginosa (15–17). Moreover, colistin heteroresistance mechanisms include activation of PmrAB and PhoPQ TCSs (Acinetobacter baumannii, Klebsiella pneumoniae, and Enterobacter cloacae) (9, 18, 19), soxRS-regulated overexpression of the acrAB-tolC efflux pump (Enterobacter asburiae and E. cloacae) (20), biofilm formation (Klebsiella pneumoniae) (21), and putrescine/YceI communication (Burkholderia cenocepacia) (22). However, only sporadic cases of colistin heteroresistance in P. aeruginosa have been reported (23), and their mechanisms of heteroresistance to colistin have not been investigated. Our aim was to determine and to compare the mechanisms that are responsible for resistance and heteroresistance to colistin in P. aeruginosa strains isolated from a Chinese teaching hospital.
RESULTS
Antibiotic susceptibility and homology characteristics of resistance and heteroresistance among P. aeruginosa isolates.
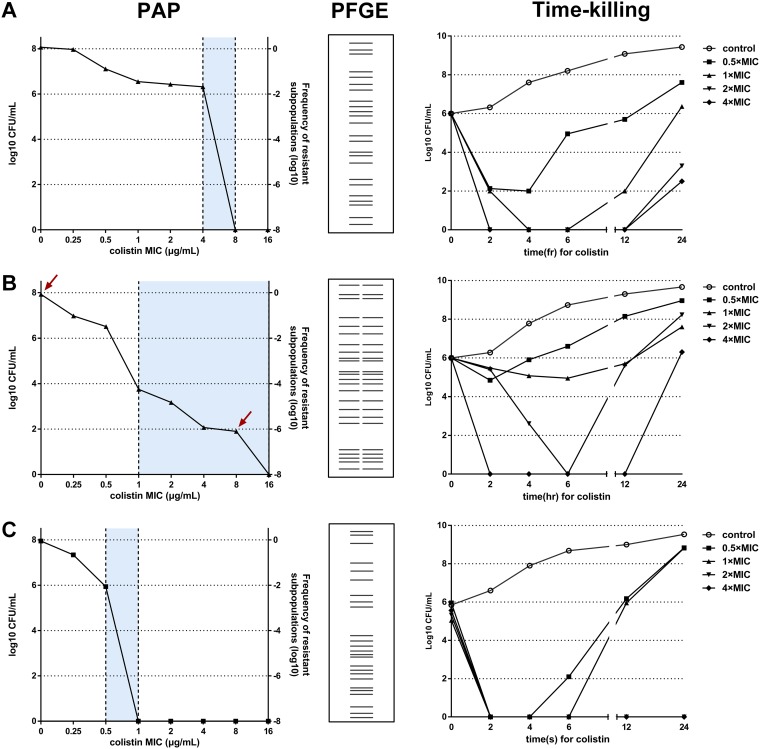
Of the 736 P. aeruginosa isolates, 2 isolates (TL1671 and TL2204) were resistant to colistin, with MICs of 8 mg/liter and 4 mg/liter, respectively. It is worth noting that some isolates displayed “skipped wells” in the first round of MIC testing, which indicated the possibility of heterogeneous bacterial behavior against colistin. Considering that not all heteroresistant strains exhibited skipped wells in routine susceptibility test, population analysis profiles (PAPs) were performed as the standard method for determining heteroresistance. Of 231 carbapenem-nonsusceptible P. aeruginosa isolates, 9 heteroresistant isolates (TL1722, TL1736, TL1744, TL2294, TL2314, TL2917, TL2967, TL3008, and TL3086) were detected. The results showed the growth of subpopulations with 4- to 32-fold higher colistin MICs than their parental populations (Fig. 1). The frequency of heteroresistant subpopulations ranged from 3.61 × 10−8 to 7.06 × 10−6. The colistin MICs against the resistant subpopulations remained the same after 1 week of subculturing in antibiotic-free medium. Five of 11 colistin-resistant or heteroresistant isolates exhibited multidrug resistance (Table 1).
FIG 1.
Verification of colistin heteroresistance, resistance, and susceptibility among P. aeruginosa isolates. (A) TL1671 (colistin-resistant clinical isolate). (B) TL1736 (colistin-susceptible and heteroresistant clinical isolates). (C) PAO1 (colistin-susceptible and control isolates). The arrows indicate the CFU of bacteria on antibiotic-free plates and plates with the highest concentration of colistin. The blue regions represent the differences in MICs between the native strain and the heterogeneous subpopulation.
TABLE 1.
Characteristics of the 11 Pseudomonas aeruginosa isolates studied
| Strain | Warda | ST | Broth MIC (mg/liter) | Highest concn for growth in PAPs (mg/liter) | Proportion of resistant subpopulation | MIC for resistant colonies after 1 wk of daily passages on colistin-free medium (mg/liter) | PAP | MIC (mg/liter)b
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | ATM | CAZ | CIP | GEN | IPM | LVX | TOB | FEP | MEM | ||||||||
| TL1671 | Endocrinology | 1020 | 8 | 8 | NA | NA | Resistant | 8 | 2 | 2 | 1 | 4 | 4 | 0.5 | 1 | 4 | 0.5 |
| TL2204 | EICU | 1129 | 4 | 8 | NA | NA | Resistant | 8 | 4 | 4 | 1 | 8 | 4 | 0.5 | 1 | 4 | 0.25 |
| TL1722 | NICU | 640 | 1 | 8 | 3.61 × 10−8 | 4 | Heteroresistant | 1 | 16 | 2 | 2 | 2 | 16 | 2 | 0.25 | 2 | 8 |
| TL1736 | NICU | 12 | 1 | 16 | 6.03 × 10−6 | 16 | Heteroresistant | 32 | 1 | 0.5 | 0.5 | 32 | 16 | 0.5 | 8 | 0.5 | 2 |
| TL1744 | ICU | 1971 | 0.5 | 8 | 1.04 × 10−6 | 4 | Heteroresistant | 4 | 32 | 32 | 8 | 64 | 16 | 8 | 64 | 8 | 16 |
| TL2294 | ICU | 1655 | 1 | 4 | 7.06 × 10−6 | 4 | Heteroresistant | 16 | 32 | 32 | 8 | 32 | 32 | 16 | 4 | 16 | 16 |
| TL2314 | ICU | 508 | 0.5 | 8 | 1.22 × 10−7 | 8 | Heteroresistant | 8 | 16 | 4 | 0.5 | 8 | 4 | 0.5 | 1 | 16 | 0.5 |
| TL2917 | Respiratory | 471 | 2 | 16 | 5.56 × 10−7 | 16 | Heteroresistant | 8 | 32 | 8 | 1 | 8 | 32 | 1 | 1 | 8 | 16 |
| TL2967 | EICU | 485 | 1 | 8 | 4.41 × 10−8 | 8 | Heteroresistant | 4 | 64 | 8 | 8 | 4 | 16 | 16 | 1 | 16 | >32 |
| TL3008 | NICU | 298 | 1 | 32 | 1.28 × 10−7 | 32 | Heteroresistant | 16 | 4 | 4 | 1 | 32 | 16 | 8 | 4 | 4 | 4 |
| TL3086 | NICU | 298 | 0.5 | 32 | 2.38 × 10−7 | 32 | Heteroresistant | 32 | 4 | 2 | 0.5 | 32 | 16 | 1 | 4 | 4 | 4 |
EICU, electronic intensive care unit; NICU, neonatal intensive care unit; ICU, intensive care unit; NA, not applicable.
For heteroresistant isolates, the objects for MIC detection were resistant subpopulations selected from PAPs.
Pulsed-field gel electrophoresis (PFGE) results confirmed the isogenic nature of resistant subpopulations and their respective native strains (Fig. 1). Multilocus sequence typing (MLST) analysis revealed that 11 (hetero)resistant isolates belonged to 10 different sequence types (STs), suggesting low homology among the investigated isolates (Table 1).
Time-kill kinetics of colistin-resistant and heteroresistant P. aeruginosa isolates.
Time-kill curves for colistin against P. aeruginosa isolates TL1671 (colistin resistant), TL1736 (colistin susceptible and heteroresistant), and PAO1 (colistin susceptible and not heteroresistant) are presented in Fig. 1. For PAO1, colistin showed rapid killing even at the lowest colistin concentration. Nevertheless, regrowth was observed at 4 to 6 h with 0.5×MIC to 1×MIC. In contrast, TL1671 and TL1736 both showed regrowth, after a reduction, at concentrations up to 4×MIC. However, there was a substantial difference in the killing of bacteria by colistin, with TL1736 regrowth being faster than TL1671 regrowth, particularly at 1× and 2× MIC.
Detection of colistin-(hetero)resistance-associated mutations.
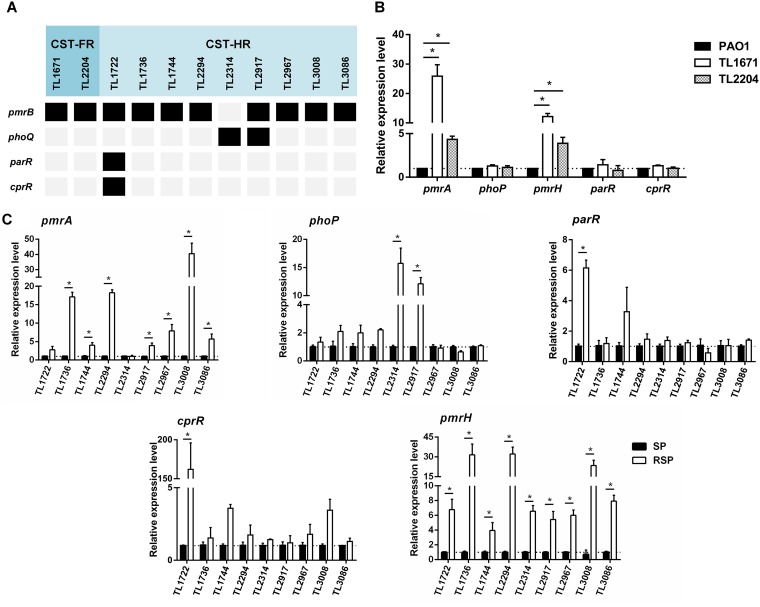
In this study, pmrA, pmrB, phoP, phoQ, oprH, parR, parS, cprR, cprS, colR, and colS were sequenced for 2 resistant isolates and 9 heteroresistant isolates, to investigate possible mechanisms. Major nonsynonymous substitutions in PmrB (i.e., V15I, S27R, D45E, G68S, G179D, V185A, A190G, V199I, P216S, S257N, Y345H, and I349V) were detected in all colistin-(hetero)resistant isolates except TL2917. The only V260G variation in phoQ was found in the heteroresistant isolates TL2314 and TL2917. It was noticeable that both pmrB and phoQ mutations were found in TL2917. In addition to PmrAB and PhoPQ TCSs, we observed ParR (R146H) and CprS (V181I and R209L) substitutions in TL1722 (Table 2). No mutation in colRS was detected among the isolates (data not shown).
TABLE 2.
Mutational analysis of the PmrAB, PhoPQ, ParRS, and CprRS regulatory pathways
| Strain | PmrB (477 aa)a
|
PhoQ (448 aa) | ParR (235 aa) | CprS (431 aa) | |||||
|---|---|---|---|---|---|---|---|---|---|
| TM1 (aa 15–37) | PD (aa 38–160) | TM2 (aa 161–183) | HAMP (aa 186–238) | HisKA (aa 239–304) | HATPase c (aa 344–459) | ||||
| Colistin-resistantb | |||||||||
| TL1671 | V15I | P216S | |||||||
| TL2204 | Y345H | ||||||||
| Colistin-heteroresistantc | |||||||||
| TL1722 | V199I | S257N | R146H | V181I R209L | |||||
| TL1736 | V185A | ||||||||
| TL1744 | V15I | G68S | |||||||
| TL2294 | G179D | I349V | |||||||
| TL2314 | V260G | ||||||||
| TL2917 | G179D | V260G | |||||||
| TL2967 | D45E | ||||||||
| TL3008 | A190G | ||||||||
| TL3086 | S27R | ||||||||
aa, amino acids.
For colistin-resistant isolates, the sequences were compared with the colistin-susceptible Pseudomonas aeruginosa strain PAO1.
For colistin-heteroresistant isolates, the comparisons were carried out between susceptible and resistant subpopulations.
Analysis of expression levels resulting from genetic mutations.
To connect the molecular genetic mutations described above with the transcriptional levels expected, the pmrA, phoP, parR, cprR, and pmrH expression levels of 11 colistin-(hetero)resistant isolates were analyzed (Fig. 2A). The results were presented in Fig. 2. All (hetero)resistant isolates (except TL2314) without mutation of pmrAB showed higher pmrA expression levels (2.8- to 40.5-fold higher levels) than the reference isolates, and only 1 isolate (TL1722) had no statistical significance (P < 0.05). The 2 isolates with phoPQ mutations (TL2314 and TL2917) showed increased expression of phoP (12.1- and 15.7-fold higher levels, respectively), which was found to be statistically significant (P < 0.05). TL1722, the colistin-heteroresistant isolate with substitutions in ParRS and CprRS, also showed significant increases in parR (6.15-fold) and cprR (161.75-fold) expression (P < 0.05). In addition, the pmrH gene, which encodes the enzyme responsible for biosynthesis of l-Ara4N and attachment to lipid A, showed significantly upregulated expression in all of studied isolates (P < 0.05) (Fig. 2B and C).
FIG 2.
Analysis of the mechanisms for colistin resistance and heteroresistance in P. aeruginosa clinical isolates. (A) Substitutions in PmrAB, PhoPQ, ParRS, and CprRS detected in this study. Black blocks represent amino acid substitutions, while gray blocks represent nonexistence. CST-FR, fully colistin-resistant isolates; CST-HR, colistin-heteroresistant isolates. (B) Expression levels of resistance genes in fully colistin-resistant isolates. (C) Expression levels of resistance genes in colistin-heteroresistant isolates. SP, susceptible population; RSP, resistant subpopulation. *, P < 0.05.
Identification of l-Ara4N addition to lipid A isolated from P. aeruginosa LPS.
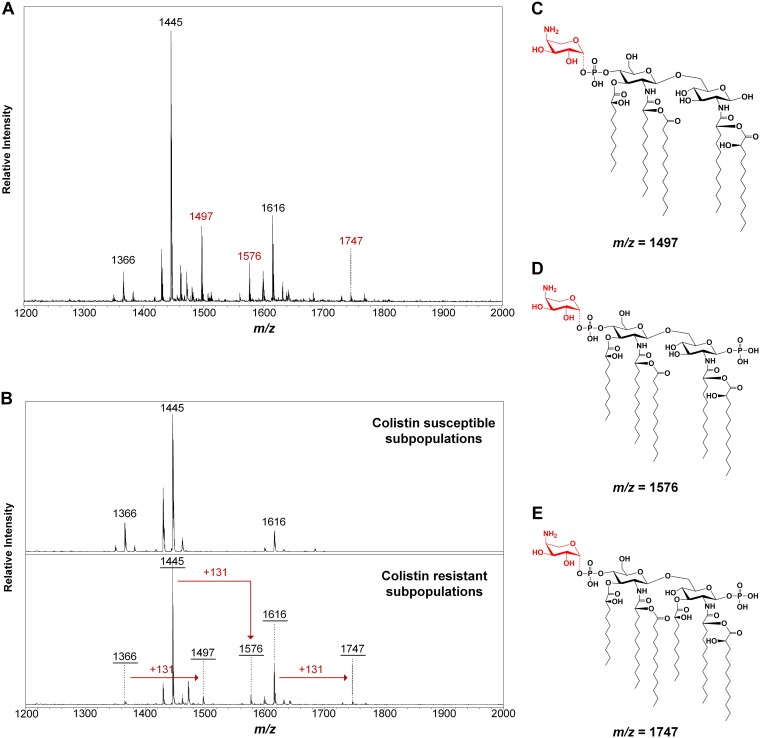
Lipid A extracted from (hetero)resistant strains was profiled using matrix-assisted laser desorption–ionization time of flight mass spectrometry (MALDI-TOF MS) to analyze modifications associated with observed genetic alterations. The structures of lipid A in P. aeruginosa in this study showed diversity (see Table S2 and Fig. S1B in the supplemental material). Among them, the pentaacylated form of lipid A (wild type; m/z 1,445) was predominant in all isolates (Fig. S1A). Lesser quantities of various modifications accompanied the wild-type lipid A, including dephosphorylation (−PO3), deacylation (−3-OH-C10), acylation (+3-OH-C10), palmitoylation (+C16), dehydroxylation (−OH), hydroxylation (+OH), and glycosylation (+l-Ara4N). The corresponding m/z values of these signature ions ware depicted. In the colistin-susceptible strain (PAO1) or subpopulations, the ions are present at m/z 1,195, 1,274, 1,366, 1,429, 1,445, 1,462, 1,600, 1,616, and 1,684. In comparison, mass spectra of colistin-resistant populations or subpopulations have additional ion peaks at m/z 1,497, 1,576, 1,731, and/or 1,747, indicating a mass shift of m/z +131, caused by modification of l-Ara4N to the pentaacylated or hexaacylated lipid A (m/z 1,366, 1,445, 1,600, and 1,616) (Fig. 3).
FIG 3.
MALDI-TOF MS of P. aeruginosa differential colistin susceptibility. The resistant isolate TL1671 and the heteroresistant pair TL1736 were grown overnight in LB culture at 37°C, and lipids were extracted and analyzed by MS. (A and B) In the heteroresistant pair, the colistin-susceptible subpopulation shows ions at m/z 1,366, 1,445, and 1,616, corresponding to base-pentaacylated or hexaacylated lipid A, while the resistant subpopulation shows ions at m/z 1,497, 1,576, and 1,747, indicating an l-Ara4N addition to the base structures. The colistin-resistant isolate TL1671 also shows ions at m/z 1,497, 1,576, and 1,747. (C to E) Molecular structures of the lipid A molecules found in mass spectra from heteroresistant and resistant isolates.
DISCUSSION
Colistin, regarded as a last-line antibiotic, has received increasing attention for treating multidrug-resistant Gram-negative pathogen infections reliably (24). Despite its favorable bacterial killing, resistance and heteroresistance to colistin have been described (3, 5). Heteroresistance is a phenomenon in which subpopulations of seemingly isogenic bacteria exhibit variable susceptibilities to a particular antibiotic (9). It has drawn extensive attention clinically, because the resistant proportion of bacterial isolates may survive and become predominant during therapy, leading to treatment failure and even lethal infections (25). In this study, 2 colistin-resistant isolates and 9 colistin-heteroresistant isolates were identified among P. aeruginosa clinical isolates by the broth microdilution method and PAPs, respectively.
Molecular mechanism analysis indicated that alteration of the PmrAB regulator system was mainly involved in colistin resistance and heteroresistance mechanisms, while other TCSs, such as PhoPQ, ParRS, and CprRS, also played a role in mediating colistin (hetero)resistance. Subsequently, we found that the lipid A from studied isolates displayed additional l-Ara4N modifications, corresponding to genetic findings.
It has been said that heteroresistance serves as an intermediate stage, which could transition from susceptibility to full resistance under certain conditions (26). We have attempted to uncover the underlying correlation between resistance and heteroresistance in P. aeruginosa through series studies.
Case reports showed that both resistant and heteroresistant isolates were collected from patients who had never been treated with colistin, thus indicating that the resistance and heteroresistance observed may not be related to prior exposure to colistin. Nevertheless, it was reported that colistin heteroresistance may act as a resistance reservoir, leading to the proliferation of resistant subpopulations upon exposure to colistin (27).
In the time-kill studies, colistin-heteroresistant isolates revealed stable growth at the MIC and showed evident regrowth after exposure to colistin at up to 4×MIC for 24 h. Pharmacokinetic studies demonstrated that plasma colistin concentrations in patients reached 1 to 4 mg/liter after intravenous administration of colistin methanesulfonate (24). Therefore, colistin-resistant isolates would require alternative treatment, while colistin-susceptible isolates harboring resistant subpopulations would proliferate rapidly and cause clinical treatment failure.
The molecular mechanisms of resistance to colistin in P. aeruginosa, such as substitutions in PmrAB, PhoPQ, ParRS, CprRS, and ColRS TCSs, have been characterized in detail (3). In this study, the substitutions (i.e., V15I, G68S, and S257N in PmrB and V260G in PhoQ) that had been reported previously for resistant isolates were detected in heteroresistant isolates (28–30). Therefore, there are some similarities between colistin resistance and heteroresistance. In addition, to our best of knowledge, this is the first report of the S27R, D45E, G179D, V185A, A190G, V199I, P216S, and I349V substitutions in PmrB, the R146H substitution in ParR, and the V181I and R209L substitutions in CprS that may mediate colistin heteroresistance. It was notable that TL1722 (PmrAB, ParRS, and CprRS) and TL2917 (PmrAB and PhoPQ) had more than one TCS involved in colistin heteroresistance. The interplay between several genes in heteroresistance should be further investigated.
The modification of lipid A, such as the addition of l-Ara4N, phosphoethanolamine, and galactosamine, was linked to homogeneous colistin resistance in various bacteria (12, 31). In P. aeruginosa, lipid A is modified with the addition of l-Ara4N through the pmrHFIJKLM operon and under the control of pmrAB and phoPQ, which leads to colistin resistance (32). However, there have been few studies of lipid A structure with respect to colistin heteroresistance. Research has demonstrated that E. cloacae lipid A is modified with l-Ara4N to induce colistin heteroresistance (14). Here we further analyzed the lipid A profiles to uncover the LPS-modified features in heteroresistant isolates and to determine the association between resistant and heteroresistant strains. Concordantly, the lipid A profiles for both colistin-resistant strains demonstrated the addition of l-Ara4N to the major hexa- and pentaacylated lipid A species (33). In addition, there were lipid A differences between colistin-heteroresistant pairs. Compared to those for susceptible subpopulations, the lipid A profiles for the resistant subpopulations displayed additional l-Ara4N modifications.
This study provides the first report of colistin-resistant and heteroresistant P. aeruginosa isolates. Comparative results did not show discrepancies for both colistin resistance and heteroresistance in P. aeruginosa being mainly caused by alterations in the PmrAB regulatory system, resulting in upregulation of the LPS modification system. The mechanisms involved in colistin heteroresistance are diverse and complicated, as has been described for several bacteria (9, 18–22) and shown for P. aeruginosa. The heteroresistance of bacteria is considered an indication of the mutator phenotype described in the literature (34, 35). The high mutation frequency may give rise to the emergence of resistance to antibiotics. Therefore, we screened for the mutator phenotype using rifampin plates (36), and we found that 5 of 9 heteroresistance isolates had the mutator phenotype, in contrast to PAO1 (see Table S3 in the supplemental material). Doßelmann et al. observed that mutations in mutS along with a mutator phenotype could facilitate resistance evolution (37). The possibility of amplification-driven heteroresistance mechanisms dependent on the genetic background of the isolate was noted (38). The evolutionary dynamics of heteroresistant P. aeruginosa isolates should be included in further study. Among other possibilities, the overexpression of efflux pump regulators in colistin heteroresistance cannot be ruled out. Two studies proved that efflux pumps could confer heteroresistance to colistin in Enterobacter spp. and A. baumannii (8, 20). In contrast, Chambers and Sauer showed that the MerR-like regulator BrlR could impair P. aeruginosa tolerance to colistin (39). The participation of efflux pumps in P. aeruginosa remains to be elucidated. In addition, biofilms constitute excellent niches for the emergence of heterogeneous variants, and Silva et al. showed that biofilm formation could trigger heteroresistance to colistin in K. pneumoniae (21). Interestingly, Pamp et al. indicated that colistin tolerance was related to heterogeneity within biofilms and depended on the pmr and mexAB-oprM genes (40). Hence, there seemed to be a complicated interplay between colistin heteroresistance and TCSs, efflux pumps, and biofilms. More studies are needed to better understand the extent of the colistin phenomenon.
MATERIALS AND METHODS
Bacterial strains.
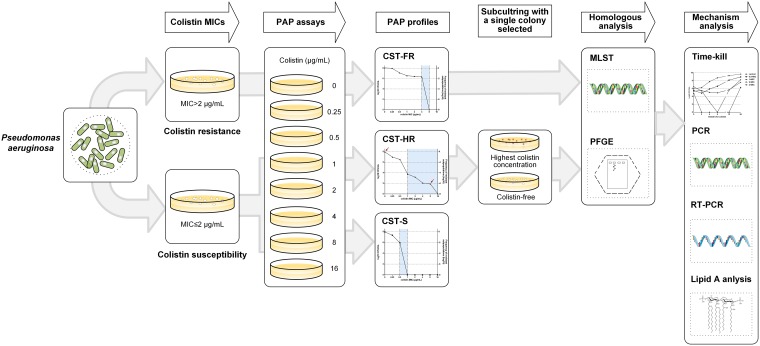
A total of 736 nonduplicated P. aeruginosa clinical isolates were recovered from the First Affiliated Hospital of Wenzhou Medical University, China, between 2015 and 2017. Each isolate represents a single sample from one patient. The isolates were identified as P. aeruginosa by the Vitek MS automated system (bioMérieux, Hazelwood, MO, USA). Colistin MICs were determined by broth microdilution in cation-adjusted Mueller-Hinton Broth (CAMHB) (Fig. 4). The CAMHB was prepared by adding appropriate amounts of Mg2+ and Ca2+ to Mueller-Hinton broth, to give final concentrations of 10 to 12.5 mg/liter and 20 to 25 mg/liter, respectively. The colistin MICs for P. aeruginosa were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines (susceptible, ≤2 mg/liter; resistant, >2 mg/liter) (45). ATCC 27853 was served as the quality control for susceptibility testing. The wild-type, colistin-susceptible, P. aeruginosa strain PAO1 was included as a control.
FIG 4.
Workflow for detecting colistin-resistant and heteroresistant P. aeruginosa clinical isolates and investigating mechanism differences in induction of colistin resistance and heteroresistance.
Population analysis profiles.
PAPs are used as the reference method to define antibiotic heteroresistance (9). The analysis was performed among 231 carbapenem-nonsusceptible isolates, based on previous analyses (23). Bacterial cultures were grown overnight to log phase, and then serial dilutions were plated on Luria-Bertani (LB) agar with or without various concentrations of colistin (0, 0.25, 0.5, 1, 2, 4, 8, or 16 mg/liter). Plates were then incubated at 37°C, and CFU were enumerated after 48 h. The limit of detection was 20 CFU/ml. Colistin heteroresistance was defined as a colistin-susceptible isolate (MIC of ≤2 mg/liter) with subpopulations growing in the presence of ≥2 mg/liter colistin (41). The rate of colistin resistance was calculated as the number of bacterial colonies that grew on colistin-containing plates divided by the number of bacteria that grew on LB broth without drug. For each isolate, a single colony was selected from the highest antibiotic concentration, and the colistin MIC was reassessed after serial passaging on antibiotic-free medium, to evaluate the stability of the heteroresistant phenotype. Cultures with resistant or susceptible subpopulations were isolated from the highest colistin concentration or drug-free medium separately for further studies. The details are shown in Fig. 4.
Antimicrobial susceptibility testing.
The MICs for clinical routine antimicrobial agents, including amikacin (AMK), aztreonam (ATM), ceftazidime (CAZ), ciprofloxacin (CIP), gentamicin (GEN), imipenem (IPM), levofloxacin (LVX), tobramycin (TOB), cefepime (FEP), and meropenem (MEM), were determined using the broth microdilution method, in accordance with CLSI guidelines.
Homology analysis.
PFGE of SpeI-digested genomic DNA of P. aeruginosa isolates was performed with a CHEF-DRIII system (Bio-Rad, Hercules, CA), and banding patterns were compared according to published criteria (42). MLST was carried out by sequencing seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) (43).
Time-kill kinetics.
The time-kill kinetics for colistin were examined according to a published protocol (6), with several modifications (Fig. 4). In brief, 1 resistant isolate (TL1671), 1 heteroresistant isolate (TL1736), and 1 control strain (PAO1) were selected as experimental strains. Tubes containing LB broth with colistin at 0×MIC, 0.5×MIC, 1×MIC, 2×MIC, or 4×MIC for the selected isolates were seeded with a log-phase bacterial inoculum of 107 CFU/ml. Viable cells were counted by plating 50-μl samples, after appropriate dilution with saline, on antibiotic-free Mueller-Hinton agar plates 0, 2, 4, 6, 12, and 24 h after antibiotic addition. The analysis was performed independently three times for these isolates, and the mean values of viable CFU were estimated and plotted on a semilogarithmic graph.
PCR and sequencing.
Whole-cell DNA of colistin-resistant and -heteroresistant isolates was extracted using the Bio-Spin bacterial genomic DNA extraction kit (BioFlux, Tokyo, Japan), according to the manufacturer’s instructions. The genes pmrA, pmrB, phoP, phoQ, oprH, parR, parS, cprR, cprS, colR, and colS in P. aeruginosa isolates were investigated by PCR using the primers and conditions described in Table S1 in the supplemental material. The amplicons of pmrA, pmrB, phoP, phoQ, oprH, parR, parS, cprR, cprS, colR, and colS were sequenced by Shanghai BGI Technology Co. and then analyzed with BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences of isolates with homogeneous resistance were compared with those of PAO1; for the isolates with heterogeneous resistance, comparisons were carried out between susceptible and resistant subpopulations (Fig. 4).
Quantitative real-time PCR.
Total RNAs of resistant isolates and heteroresistant isolates (including susceptible and resistant subpopulations) were extracted from the log-phase bacterial inocula using a bacterial RNA miniprep kit (Biomiga, Shanghai, China). An aliquot of RNA from each isolate was subjected to cDNA synthesis using the RevertAid first-strand cDNA synthesis kit (Thermo Fisher, Shanghai, China). Expression levels of pmrA, phoP, parR, cprR, and pmrH were performed by quantitative real-time PCR, as described previously (24). The 30S ribosomal rpsL gene served as the internal gene. The expression levels of five genes in heteroresistant isolates and resistant isolates were determined relative to their expression levels in P. aeruginosa PAO1. For colistin-heteroresistant isolates, the analysis of transcript levels was performed by comparing the susceptible and resistant subpopulations (Fig. 4). Experiments for each gene were conducted in triplicate. The primers used in this study are listed in Table S1.
Lipid A isolation from whole cells.
Lipid A was isolated by using an optimized large-scale protocol based on mild acid hydrolysis (44). Overnight cultures (200 ml at 37°C) in LB broth were harvested by centrifugation at 3,220 × g for 30 min. Bacterial pellets were washed with single-phase Bligh-Dyer mixture (chloroform/methanol/water, 1:2:0.8 [vol/vol]) and centrifuged at 3,220 × g for 15 min. The LPS pellets were suspended in sodium acetate buffer (50 mM [pH 4.5]) and incubated at 100°C for 30 to 45 min. Reactions were moved into a two-phase Bligh-Dyer mixture (chloroform/methanol/water, 1:1:0.9 [vol/vol]) and centrifuged at 3,220 × g for 15 min. The lower phases were removed to clean tubes and dried using rotary evaporation. The dried samples contained whole-cell extracts of lipid A.
Lipid A characterization by MALDI-TOF MS.
Dried lipid A samples were resuspended in 100 μl chloroform/methanol (1:1 [vol/vol]), and 3 μl 2,5-dihydroxybenzoic acid (DHB) matrix (20 mg/ml in TA30 solvent) was mixed with 3 μl lipid A. Aliquots of the mixture were spotted directly onto the well of the MALDI-TOF MS plate (ground steel). Mass spectra were recorded for optimal ion signals in negative-ion mode using a Bruker autoflex MALDI-TOF mass spectrometer (Bruker Daltonics Inc., Billerica, MA, USA). Data were acquired and processed by flexControl and flexAnalysis 3.4 (Bruker Daltonics Inc.).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Planned Science and Technology Project of Wenzhou (grant Y20170204) and the Health Department of Zhejiang Province of the People’s Republic of China (grant 2018KY123).
We declare no conflicts of interest.
T.Z. and J.C. conceived the project and all authors were involved in the design of the experiments. J.L. performed the experiments and wrote the manuscript, C.X., R.F., X.Z., and Y.Z. analyzed the results, and G.D. and Y.S. reviewed the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00556-19.
REFERENCES
- 1.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander HE. 1948. Mode of action of streptomycin on type B Hemophilus influenzae. Am J Dis Child 75:428–430. [PubMed] [Google Scholar]
- 5.El-Halfawy OM, Valvano MA. 2015. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pournaras S, Ikonomidis A, Markogiannakis A, Spanakis N, Maniatis AN, Tsakris A. 2007. Characterization of clinical isolates of Pseudomonas aeruginosa heterogeneously resistant to carbapenems. J Med Microbiol 56:66–70. doi: 10.1099/jmm.0.46816-0. [DOI] [PubMed] [Google Scholar]
- 8.Machado D, Antunes J, Simoes A, Perdigao J, Couto I, McCusker M, Martins M, Portugal I, Pacheco T, Batista J, Toscano C, Viveiros M. 2018. Contribution of efflux to colistin heteroresistance in a multidrug resistant Acinetobacter baumannii clinical isolate. J Med Microbiol 67:740–749. doi: 10.1099/jmm.0.000741. [DOI] [PubMed] [Google Scholar]
- 9.Charretier Y, Diene SM, Baud D, Chatellier S, Santiago-Allexant E, van Belkum A, Guigon G, Schrenzel J. 2018. Colistin heteroresistance and involvement of the PmrAB regulatory system in Acinetobacter baumannii. Antimicrob Agents Chemother 62:e00788-18. doi: 10.1128/AAC.00788-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol 186:575–579. doi: 10.1128/jb.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, Hoiby N, Moskowitz SM. 2011. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 55:5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung LM, Fondrie WE, Doi Y, Johnson JK, Strickland DK, Ernst RK, Goodlett DR. 2017. Identification of the ESKAPE pathogens by mass spectrometric analysis of microbial membrane glycolipids. Sci Rep 7:6403. doi: 10.1038/s41598-017-04793-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dortet L, Bonnin RA, Pennisi I, Gauthier L, Jousset AB, Dabos L, Furniss RCD, Mavridou DAI, Bogaerts P, Glupczynski Y, Potron A, Plesiat P, Beyrouthy R, Robin F, Bonnet R, Naas T, Filloux A, Larrouy-Maumus G. 2018. Rapid detection and discrimination of chromosome- and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: the MALDIxin test. J Antimicrob Chemother 73:3359–3367. doi: 10.1093/jac/dky330. [DOI] [PubMed] [Google Scholar]
- 14.Kang KN, Klein DR, Kazi MI, Guerin F, Cattoir V, Brodbelt JS, Boll JM. 2019. Colistin heteroresistance in Enterobacter cloacae is regulated by PhoPQ-dependent 4-amino-4-deoxy-l-arabinose addition to lipid A. Mol Microbiol 111:1604–1616. doi: 10.1111/mmi.14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez L, Jenssen H, Bains M, Wiegand I, Gooderham WJ, Hancock RE. 2012. The two-component system CprRS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of ParRS. Antimicrob Agents Chemother 56:6212–6222. doi: 10.1128/AAC.01530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowicki EM, O'Brien JP, Brodbelt JS, Trent MS. 2015. Extracellular zinc induces phosphoethanolamine addition to Pseudomonas aeruginosa lipid A via the ColRS two-component system. Mol Microbiol 97:166–178. doi: 10.1111/mmi.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Band VI, Satola SW, Burd EM, Farley MM, Jacob JT, Weiss DS. 2018. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio 9:e02448-17. doi: 10.1128/mBio.02448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong YK, Ko KS. 2019. PmrAB and PhoPQ variants in colistin-resistant Enterobacter spp. isolates in Korea. Curr Microbiol 76:644–649. doi: 10.1007/s00284-019-01672-1. [DOI] [PubMed] [Google Scholar]
- 20.Telke AA, Olaitan AO, Morand S, Rolain JM. 2017. soxRS induces colistin hetero-resistance in Enterobacter asburiae and Enterobacter cloacae by regulating the acrAB-tolC efflux pump. J Antimicrob Chemother 72:2715–2721. doi: 10.1093/jac/dkx215. [DOI] [PubMed] [Google Scholar]
- 21.Silva A, Sousa AM, Alves D, Lourenco A, Pereira MO. 2016. Heteroresistance to colistin in Klebsiella pneumoniae is triggered by small colony variants sub-populations within biofilms. Pathog Dis 74:ftw036. doi: 10.1093/femspd/ftw036. [DOI] [PubMed] [Google Scholar]
- 22.El-Halfawy OM, Valvano MA. 2013. Chemical communication of antibiotic resistance by a highly resistant subpopulation of bacterial cells. PLoS One 8:e68874. doi: 10.1371/journal.pone.0068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermes DM, Pormann Pitt C, Lutz L, Teixeira AB, Ribeiro VB, Netto B, Martins AF, Zavascki AP, Barth AL. 2013. Evaluation of heteroresistance to polymyxin B among carbapenem-susceptible and resistant Pseudomonas aeruginosa. J Med Microbiol 62:1184–1189. doi: 10.1099/jmm.0.059220-0. [DOI] [PubMed] [Google Scholar]
- 24.Zakuan ZD, Suresh K. 2018. Rational use of intravenous polymyxin B and colistin: a review. Med J Malaysia 73:351–359. [PubMed] [Google Scholar]
- 25.Hawley JS, Murray CK, Jorgensen JH. 2008. Colistin heteroresistance in Acinetobacter and its association with previous colistin therapy. Antimicrob Agents Chemother 52:351–352. doi: 10.1128/AAC.00766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Superti SV, Martins DdS, Caierão J, Soares FdS, Prochnow T, Zavascki AP. 2009. Indications of carbapenem resistance evolution through heteroresistance as an intermediate stage in Acinetobacter baumannii after carbapenem administration. Rev Inst Med Trop Sao Paulo 51:111–113. doi: 10.1590/s0036-46652009000200010. [DOI] [PubMed] [Google Scholar]
- 27.Yau W, Owen RJ, Poudyal A, Bell JM, Turnidge JD, Yu HH, Nation RL, Li J. 2009. Colistin hetero-resistance in multidrug-resistant Acinetobacter baumannii clinical isolates from the Western Pacific region in the SENTRY antimicrobial surveillance programme. J Infect 58:138–144. doi: 10.1016/j.jinf.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Lee JY, Ko KS. 2014. Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn Microbiol Infect Dis 78:271–276. doi: 10.1016/j.diagmicrobio.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 29.Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, Johansen HK, Hoiby N. 2012. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 56:1019–1030. doi: 10.1128/AAC.05829-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owusu-Anim D, Kwon DH. 2012. Differential role of two-component regulatory systems (phoPQ and pmrAB) in polymyxin B susceptibility of Pseudomonas aeruginosa. Adv Microbiol 2. doi: 10.4236/aim.2012.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powers MJ, Herrera CM, Tucker AT, Davies BW, Trent MS. 2019. Isolation of lipid cell envelope components from Acinetobacter baumannii. Methods Mol Biol 1946:233–252. doi: 10.1007/978-1-4939-9118-1_22. [DOI] [PubMed] [Google Scholar]
- 32.Han ML, Zhu Y, Creek DJ, Lin YW, Anderson D, Shen HH, Tsuji B, Gutu AD, Moskowitz SM, Velkov T, Li J. 2018. Alterations of metabolic and lipid profiles in polymyxin-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e02656-17. doi: 10.1128/AAC.02656-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ernst RK, Moskowitz SM, Emerson JC, Kraig GM, Adams KN, Harvey MD, Ramsey B, Speert DP, Burns JL, Miller SI. 2007. Unique lipid A modifications in Pseudomonas aeruginosa isolated from the airways of patients with cystic fibrosis. J Infect Dis 196:1088–1092. doi: 10.1086/521367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deiham B, Douraghi M, Adibhesami H, Yaseri M, Rahbar M. 2017. Screening of mutator phenotype in clinical strains of Acinetobacter baumannii. Microb Pathog 104:175–179. doi: 10.1016/j.micpath.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 35.Bulman ZP, Sutton MD, Ly NS, Bulitta JB, Holden PN, Nation RL, Li J, Tsuji BT. 2015. Emergence of polymyxin B resistance influences pathogenicity in Pseudomonas aeruginosa mutators. Antimicrob Agents Chemother 59:4343–4346. doi: 10.1128/AAC.04629-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 37.Doßelmann B, Willmann M, Steglich M, Bunk B, Nubel U, Peter S, Neher RA. 2017. Rapid and consistent evolution of colistin resistance in extensively drug-resistant Pseudomonas aeruginosa during morbidostat culture. Antimicrob Agents Chemother 61:e00043-17. doi: 10.1128/AAC.00043-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicoloff H, Hjort K, Levin BR, Andersson DI. 2019. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol 4:504–514. doi: 10.1038/s41564-018-0342-0. [DOI] [PubMed] [Google Scholar]
- 39.Chambers JR, Sauer K. 2013. The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ. J Bacteriol 195:4678–4688. doi: 10.1128/JB.00834-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- 41.Bergen PJ, Forrest A, Bulitta JB, Tsuji BT, Sidjabat HE, Paterson DL, Li J, Nation RL. 2011. Clinically relevant plasma concentrations of colistin in combination with imipenem enhance pharmacodynamic activity against multidrug-resistant Pseudomonas aeruginosa at multiple inocula. Antimicrob Agents Chemother 55:5134–5142. doi: 10.1128/AAC.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42:5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hankins JV, Madsen JA, Needham BD, Brodbelt JS, Trent MS. 2013. The outer membrane of Gram-negative bacteria: lipid A isolation and characterization. Methods Mol Biol 966:239–258. doi: 10.1007/978-1-62703-245-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing; 27th informational supplement. CLSI document M100-S27 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.