VT-1161 and VT-1598 are promising investigational tetrazole antifungals that have shown in vitro and in vivo activity against Candida and other fungi. Candida glabrata is a problematic opportunistic pathogen that is associated with high mortality in invasive infection, as well as both intrinsic and rapidly acquired antifungal resistance.
KEYWORDS: Candida glabrata, antifungal resistance, azole antifungals, tetrazole
ABSTRACT
VT-1161 and VT-1598 are promising investigational tetrazole antifungals that have shown in vitro and in vivo activity against Candida and other fungi. Candida glabrata is a problematic opportunistic pathogen that is associated with high mortality in invasive infection, as well as both intrinsic and rapidly acquired antifungal resistance. The MICs of VT-1161 and VT-1598 were determined by CLSI methodology to evaluate their in vitro activities against clinical C. glabrata isolates and strains containing individual deletions of the zinc cluster transcription factor genes PDR1 and UPC2A as well as the efflux transporter genes CDR1, PDH1, and SNQ2. Overall, both tetrazoles demonstrated relative activities comparable to those of the tested triazole antifungals against clinical C. glabrata isolates (MIC range, 0.25 to 2 mg/liter and 0.5 to 2 μg/ml for VT-1161 and VT-1598, respectively). Deletion of the PDR1 gene in fluconazole-resistant matched clinical isolate SM3 abolished the decreased susceptibility phenotype completely for both VT-1161 and VT-1598, similarly to the triazoles. UPC2A deletion also increased susceptibility to both triazoles and tetrazoles but to a lesser extent than PDR1 deletion. Of the three major transporter genes regulated by Pdr1, CDR1 deletion resulted in the largest MIC reductions for all agents tested, while PDH1 and SNQ2 deletion individually impacted MICs very little. Overall, both VT-1161 and VT-1598 have comparable activities to those of the available triazoles, and decreased susceptibility to these tetrazoles in C. glabrata is driven by many of the same known resistance mechanisms.
INTRODUCTION
Candida spp. are the fourth most commonly reported organism identified in health care-associated infections in the United States (1). Candida glabrata is the most common non-albicans Candida species in the United States and Northern Europe (2–5). Importantly, C. glabrata has recently seen the largest proportional increase in frequency in the United States, with estimates of mortality upwards of 50% in cases of nosocomial candidemia (6–9). While current guidelines now recommend the echinocandins as first-line agents in serious infections, such as candidemia, the azole antifungals remain essential frontline agents for treating many invasive Candida infections (10). However, C. glabrata, unlike Candida albicans, demonstrates inherently low susceptibility to fluconazole and can rapidly acquire resistance (6, 11, 12). Azole resistance in C. glabrata is primarily driven by Pdr1-dependent overexpression of the ATP-binding cassette (ABC) transporters Cdr1, Pdh1, and Snq2 (13–15). In addition, activating mutations in the zinc cluster transcription factor (ZCF) gene PDR1 leads to constitutive overexpression of all three transporters and is largely responsible for the azole resistance seen in C. glabrata (16–18).
The novel tetrazole antifungal agents VT-1161 and VT-1598 have been developed to possess high specificity for fungal CYP51 compared to human CYP enzymes (19–21). Specifically, by replacing the three-nitrogen-containing triazole ring with a tetrazole moiety, the affinity for the heme group iron in CYP enzymes, and therefore of nonselective CYP inhibition, is reduced, which may potentially translate into improved drug adverse effect profiles and lower potential for drug-drug interactions. In particular, VT-1161 had previously shown to potently bind fungal CYP51 without binding the human equivalent of the fungal target enzyme, whereas VT-1598 boasts a broader spectrum of antifungal activity (20, 22). Both agents have previously been shown to be effective against Candida strains, including azole-resistant strains, in vitro and in vivo (23–27).
SM1 and SM3 are a matched pair of clinical isolates taken from the same patient before and during treatment with fluconazole, respectively (28). While SM1 is considered fluconazole susceptible dose dependent (≤32 μg/ml), SM3 is a fluconazole-resistant isolate possessing an activating mutation in PDR1, leading to constitutive overexpression of the ABC transporters Cdr1, Pdh1, and Snq2. Previously, the individual contributions of these transporters to resistance against commonly available azole antifungals were reported (29). Upc2A is a transcription factor that regulates genes of the ergosterol biosynthesis pathway and has been shown to be essential for fluconazole resistance in C. glabrata. The relative importance, however, of the ABC transporter genes and the azole resistance-related genes PDR1 and UPC2A have not yet been determined for VT-1161 and VT-1598. In this study, we investigate the in vitro activities of these two tetrazoles against a collection of C. glabrata clinical isolates and compared them to those of fluconazole, itraconazole, posaconazole, and voriconazole. We also examine the activities of VT-1161 and VT-1598 against the matched clinical isolates SM1 and SM3 in addition to their derivative strains lacking the azole resistance genes PDR1, CDR1, PDH1, SNQ2, and UPC2A.
RESULTS
In vitro activity of VT-1161 and VT-1598 against clinical isolates of C. glabrata.
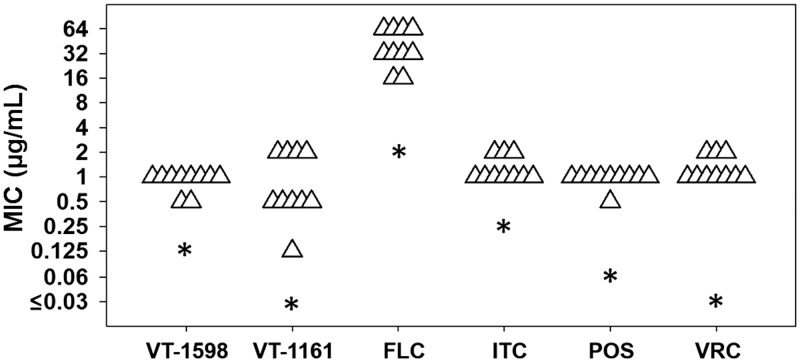
Susceptibility testing was performed against a mix of fluconazole-susceptible dose-dependent and fluconazole-resistant C. glabrata clinical isolates. Based on 24-h CLSI broth microdilution MICs (Table 1), 3 of 10 clinical isolates of C. glabrata were classified as susceptible dose dependent (≤32 μg/ml), and the remaining seven isolates were considered resistant (≥64 μg/ml) to fluconazole. Against voriconazole, all isolates were equal to or above the epidemiological cutoff value of 0.5 μg/ml (30). Overall, there was little variation in the MIC distribution of the clinical isolates for each drug tested. Only the fluconazole-susceptible dose-dependent isolate CG30 showed a decreased MIC to VT-1161 (0.25 μg/ml) and VT-1598 (0.5 μg/ml) compared to those of the other clinical isolates. All clinical isolates displayed elevated MICs to each agent tested compared to that of the susceptible dose-dependent isolate SM1 (Fig. 1).
TABLE 1.
CLSI MIC values for selected C. glabrata clinical isolates
| Isolate | MIC (μg/ml) for: |
|||||
|---|---|---|---|---|---|---|
| FLC | ITC | POS | VRC | VT-1161 | VT-1598 | |
| CG26 | 64 | 2 | 1 | 2 | 2 | 1 |
| CG27 | 64 | 2 | 1 | 2 | 2 | 1 |
| CG28 | 64 | 2 | 1 | 2 | 2 | 1 |
| CG29 | 64 | 2 | 1 | 4 | 2 | 2 |
| CG30 | 32 | 1 | 1 | 2 | 0.25 | 0.5 |
| CG31 | 32 | 1 | 1 | 1 | 0.5 | 1 |
| CG32 | 64 | 2 | 1 | 1 | 2 | 1 |
| CG33 | 32 | 2 | 1 | 1 | 2 | 1 |
| CG34 | 64 | 2 | 1 | 1 | 2 | 1 |
| CG35 | >64 | 4 | 2 | 2 | 2 | 2 |
FIG 1.
Susceptibility testing for novel tetrazoles, VT-1598 and VT-1161, compared to that for a panel of triazole antifungals. Each triangle represents an isolate from a collection of fluconazole-resistant clinical isolates. The asterisks represent the fluconazole-susceptible dose-dependent clinical isolate SM1. FLC, fluconazole; ITC, itraconazole; POS, posaconazole; VRC, voriconazole.
In vitro activity of VT-1161 and VT-1598 against the matched pair of clinical isolates SM1 and SM3 and derivative strains.
Table 2 shows the MICs of fluconazole, itraconazole, posaconazole, voriconazole, VT-1161, and VT-1598 against SM1 and SM3. Not surprisingly, the fluconazole MIC against the resistant isolate SM3 (64 μg/ml) was 32-fold higher than that of the matched susceptible dose-dependent isolate SM1 (2 μg/ml) (see Fig. S1 in the supplemental material). For itraconazole, posaconazole, and voriconazole, the MICs against isolate SM3 were 8-, 32-, and ≥64-fold higher than their respective MICs against SM1. By comparison, the VT-1161 MIC was 32-fold higher against SM3 than that against SM1, while the VT-1598 MIC against SM3 was only 8-fold higher than the MIC against SM1.
TABLE 2.
Twenty-four-hour CLSI MIC values for C. glabrata strains
| Strain | MIC (μg/ml) for: |
|||||
|---|---|---|---|---|---|---|
| FLC | ITC | POS | VRC | VT-1161 | VT-1598 | |
| SM1 | 2 | 0.25 | 0.06 | ≤0.03 | 0.03 | 0.125 |
| SM3 | 64 | 2 | 2 | 2 | 1 | 1 |
| SM1PDR1(SM3) | 64 | 2 | 1 | 2 | 1 | 1 |
| SM1Δpdr1 | 1 | 0.125 | ≤0.03 | ≤0.03 | ≤0.015 | 0.06 |
| SM3Δpdr1 | 1 | 0.125 | ≤0.03 | ≤0.03 | ≤0.015 | 0.06 |
| SM1Δupc2A | 1 | 0.25 | 0.125 | 0.06 | ≤0.015 | ≤0.015 |
| SM3Δupc2A | 4 | 0.5 | 0.125 | 0.06 | 0.125 | 0.125 |
| S1RPS3CDR1M2A | 8 | 0.5 | 0.125 | 0.06 | 0.06 | 0.25 |
| S1RPS3PDH1M2A | 64 | 2 | 1 | 2 | 1 | 1 |
| S1RPS3SNQ2M2A | 64 | 2 | 1 | 2 | 1 | 1 |
| S1RPS3CAPDH1M2A | 2 | 0.125 | ≤0.03 | ≤0.03 | 0.03 | 0.125 |
| S1RPS3CASNQ2M2A | 4 | 0.5 | 0.06 | 0.06 | 0.06 | 0.125 |
| S1RPS3SAPDH1M2A | 64 | 2 | 1 | 1 | 1 | 1 |
| S1RPS3CAPASNQ2M2A | 0.25 | 0.125 | ≤0.03 | ≤0.03 | ≤0.015 | ≤0.015 |
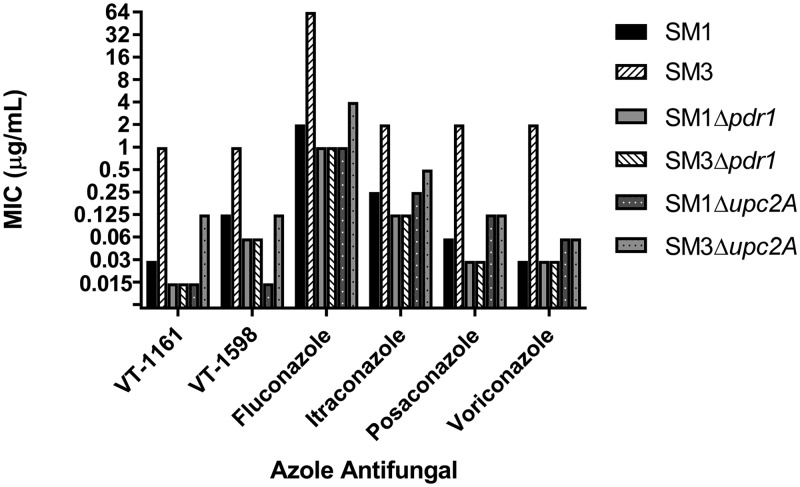
To determine the effects of the zinc cluster transcription factor Pdr1 on tetrazole susceptibility in these isolates, we obtained MICs for the previously reported strains SM1Δpdr1 and SM3Δpdr1, in which the PDR1 open reading frame (ORF) was deleted (31). As shown in Fig. 1, fluconazole resistance was completely abolished in SM3 when PDR1 was deleted. Moreover, deletion of PDR1 resulted in identical MICs against both SM1 and SM3 backgrounds for all agents tested. The MICs of VT-1161, posaconazole, and voriconazole against both SM1Δpdr1 and SM3Δpdr1 dropped to the lowest tested concentrations (≤0.015, ≤0.03, and ≤0.03 μg/ml, respectively). The MICs of VT-1598, fluconazole, and itraconazole against strains SM1Δpdr1 and SM3Δpdr1 decreased 2-fold compared to MICs against SM1.
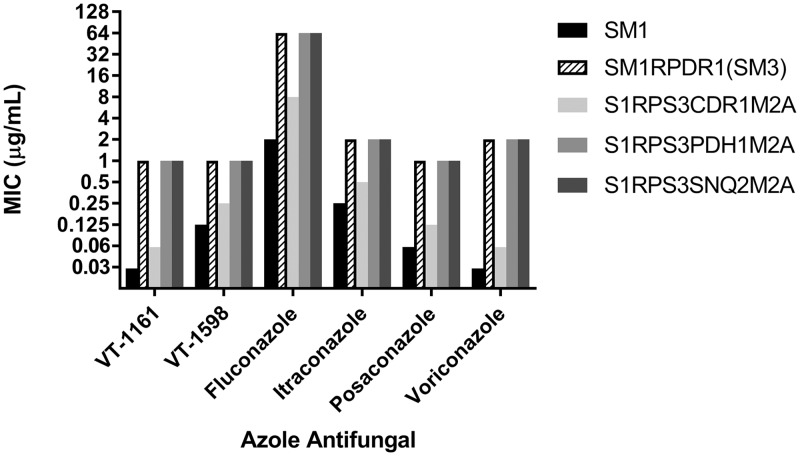
To elucidate the mechanisms by which PDR1 deletion increased susceptibility to VT-1161, VT-1598, and the triazoles, we tested these agents against individual deletions of the Pdr1-regulated ABC transporter genes CDR1, PDH1, and SNQ2 in an SM1 isolate derivative, SM1RPDR1(SM3), in which the native PDR1 open reading frame (ORF) was replaced with the PDR1 ORF from SM3 (Fig. 2). Deletion of CDR1 had the greatest effect on increasing triazole and tetrazole susceptibility in SM1RPDR1(SM3), in which the MIC decreased by 32-fold for voriconazole, 16-fold for VT-1161, 8-fold for fluconazole, posaconazole, and VT-1598, and 4-fold for itraconazole (Fig. 3). In contrast, individual deletion of PDH1 or SNQ2 showed no decrease in MICs against SM1RPDR1(SM3) for any antifungal agent.
FIG 2.
Twenty-four-hour CLSI MIC comparison of strains SM1Δpdr1, SM3Δpdr1, SM1Δupc2A, and SM3Δupc2A with the matched clinical isolates SM1 and SM3 for VT-1161, VT-1598, fluconazole, itraconazole, posaconazole, and voriconazole.
FIG 3.
Twenty-four-hour CLSI MIC comparison of the Δcdr1 deletion strain S1RPS3CDR1M2A, the Δpdh1 deletion strain S1RPS3PDH1M2A, and the Δsnq2 deletion strain S1RPS3SNQ2M2A in background SM1RPDR1(SM3) with SM1 and SM1RPDR1(SM3) for VT-1161, VT-1598, fluconazole, itraconazole, posaconazole, and voriconazole. Strain SM1RPDR1(SM3) is an SM1 derivative in which the PDR1 ORF of SM1 was replaced with the PDR1 ORF of SM3.
Tetrazole and triazole susceptibilities were also obtained against the different combinations of ABC transporter gene deletions, as well as a strain lacking all three transporter genes. In total, we tested four additional strains containing the double deletions of CDR1 and PDH1 (S1RPS3CBPDH1M2A), CDR1 and SNQ2 (S1RPS3CASNQ2M2A), PDH1 and SNQ2 (S1RPS3SAPDH1M2A), and the triple null mutant for CDR1, PDH1, and SNQ2 (S1RPS3CAPASNQ2M2A). Consistent with the susceptibilities of the individual transporter deletions, Cdr1 appears to be the major ABC transporter contributing to decreased azole susceptibility, as the deletion of PDH1 or SNQ2 in combination with the CDR1 deletion reduced the MIC between 4- and 64-fold for the various triazoles and between 8- and 32-fold for the tetrazoles (see Fig. S2 in the supplemental material). By comparison, strain S1RPS3SAPDH1M2A, which lacks only the PDH1 and SNQ2 genes, exhibited a 2-fold reduction in MIC compared to that of SM1RPDR1(SM3) only for voriconazole. The deletion of both PDH1 and SNQ2 did not alter the MIC for SM1RPDR1(SM3) against VT-1161, VT-1598, fluconazole, itraconazole, or posaconazole. Removing all three transporter genes resulted in MICs equal to or less than those for the PDR1 deletion of SM1 and SM3. For VT-1598 and fluconazole, the MIC decreased 4-fold between SM3Δpdr1 (0.06 and 1 μg/ml, respectively) and the triple null Δcdr1/Δpdh1/Δsnq2 mutant (≤0.015 and 0.25 μg/ml, respectively). No difference in MIC was observed between the triple mutant and SM3Δpdr1 for VT-1161, itraconazole, posaconazole, or voriconazole. However, the VT-1161, posaconazole, and voriconazole MICs for SM3Δpdr1 were already at the lowest concentration tested, so it is unknown if deletion of CDR1, PDH1, and SNQ2 would further increase susceptibility to these antifungals.
In addition to Pdr1 and the regulated ABC transporters, we also wanted to test the effects of the sterol-regulating transcription factor Upc2 on tetrazole susceptibility. Similar to the Δpdr1 deletion in SM1 and SM3, the deletion of UPC2A also resulted in reduced MICs for most of the triazoles and tetrazoles tested (Fig. 1). Compared to those of the fluconazole-resistant isolate SM3, the SM3Δupc2A MIC decreased 8-fold against VT-1598, VT-1161, and itraconazole; 16-fold against fluconazole and posaconazole; and 32-fold against voriconazole. Deleting UPC2A in SM1 did not show as great a reduction in MIC compared to that of the parent for all azoles tested. In SM1Δupc2A, the VT-1598 MIC decreased 4-fold and fluconazole and voriconazole MICs decreased 2-fold compared to that of the parent strain SM1. The itraconazole MICs did not change at all between SM1 and SM1Δupc2A, and the MIC actually showed a small 2-fold increase in the SM1Δupc2A strain compared to that of SM1.
DISCUSSION
In C. glabrata, the vast majority of reported fluconazole resistance originates from activating mutations in the zinc cluster transcription factor Pdr1, which leads to constitutive upregulated expression of the ABC transporters Cdr1, Pdh1, and Snq2, all of which have been shown to play a role in fluconazole resistance against the species (13–15, 17, 32–34). Based on CLSI breakpoints, SM1 is susceptible dose dependent to fluconazole (≤32 μg/ml) while SM3 is resistant (≥64 μg/ml). Similarly, SM1, but not SM3, is below the most recent epidemiological cutoff values for voriconazole (≥1 μg/ml). As a result of an L946S amino acid substitution in the zinc cluster transcription factor Pdr1 of SM3, the isolate overexpresses efflux pumps Cdr1, Pdh1, and Snq2 relative to SM1, which lacks any activating mutation in PDR1 (31).
As we might expect, SM3—and the PDR1SM3-containing strain SM1RPDR1(SM3)—was less susceptible than SM1 to every agent tested, including VT-1161 and VT-1598. Due to differences in agent-specific clinical breakpoints and epidemiological cutoff values, direct comparisons of MIC values are not useful between different antifungal agents. Instead, we compared the relative change in activity between the less susceptible SM3 and SM1 for each of the tested triazoles and tetrazoles by examining the fold increase in the MIC of SM3 over SM1. For example, for voriconazole, there was a sizeable difference (≥64-fold) between its MIC against SM1 (≤0.03 mg/liter) and that against SM3 (2 mg/liter). This relative difference was markedly less for both VT-1598 and itraconazole, both of which only showed an 8-fold increase in SM3 MICs compared to SM1 MICs. Fluconazole, posaconazole, and VT-1161 both exhibited 32-fold increases in SM3 MICs compared to those of SM1. In this regard, the tetrazole compounds were comparable to many of the clinically available azoles tested here against the SM3 resistance phenotype. Similarly, the relative activity of VT-1161 and VT-1598 against the ten C. glabrata clinical isolates, all of which overexpress CDR1 (data not shown), was comparable to that of the other tested triazoles. The increased MICs seen with all agents for SM3 and the clinical isolates suggest cross-resistance between the triazole and the newer tetrazole antifungals.
It had been previously shown that when comparing the relative contributions of Cdr1, Pdh1, and Snq2 to azole resistance in C. glabrata, deletion of CDR1 had the largest impact on Pdr1-mediated azole resistance (29). This also held true for the tetrazoles VT-1161 and VT-1598. While the single deletion of CDR1 in SM1RPDR1(SM3) lowered MICs by 4- to 32-fold compared to those of the wildtype for each antifungal tested, deletion of either PDH1 or SNQ2 resulted in little to no change (≤2-fold decrease) in MIC. Thus, for VT-1161 and VT-1598, as well as the other triazoles, Cdr1 is the key contributor of antifungal resistance in C. glabrata. This is supported by the fact that in SM1RPDR1(SM3) strains with combined deletions of two of the three ABC transporter genes, the Δcdr1/Δpdh1 and Δcdr1/Δsnq2 strains showed large decreases in MIC compared to that of the Δpdh1/Δsnq2 double deletion strain, which retained the CDR1 gene and the resistance phenotype.
The zinc cluster transcription factors PDR1 and UPC2A are also important in the potential development of resistance to VT-1161 and VT-1598 in C. glabrata. Earlier reports demonstrated that UPC2A is necessary for retaining a high-level triazole-resistant phenotype in C. glabrata and that deletion of UPC2A resulted in decreased cellular ergosterol content as well as reduced MICs to fluconazole, itraconazole, ketoconazole, and voriconazole (35). Deleting either the PDR1 gene or the UPC2A gene in SM3 and SM1 appeared to reduce the MICs against the tetrazoles compared to those of the wildtype isolates. Thus, PDR1 is important for reduced susceptibility to the tetrazoles VT-1161 and VT-1598, as has been established with the clinically available triazoles (17). Likewise, our results suggest that UPC2A contributes to susceptibility to the tetrazole antifungals as it does with the triazole antifungals. It should, however, be noted that increased ERG11 and activating mutations in UPC2A are not known phenomena driving azole resistance in clinical isolates of C. glabrata (18, 36).
In a randomized, double-blind phase 2 clinical trial evaluating oral VT-1161 in treating recurrent vulvovaginal candidiasis in 215 women, VT-1161 was found to be efficacious and safe compared to placebo with recurrence rates of between 0 and 7% through 48 weeks (24). VT-1598 has been effective in murine infection models with Candida, Coccidioides, and Cryptococcus species (23, 37, 38). However, reduced susceptibility has been observed to both VT-1161 and VT-1598 in clinical isolates of C. albicans, and this reduced in vitro susceptibility seems to be at least partly due to increased expression of the C. albicans ABC transporter Cdr1 (25, 39). This parallels our results in C. glabrata, where Cdr1 and its regulator Pdr1 appear to play an important role in reduced susceptibility to both VT-1161 and VT-1598, more so than either Pdh1 or Snq2.
Overall, the in vitro activities of VT-1161 and VT-1598 against C. glabrata are comparable to those of many of the commercially available triazole antifungals. This in combination with the potential for an improved adverse effect profile suggests that VT-1161 and VT-1598 could be useful in cases of C. glabrata infection where azole treatment is appropriate. However, our findings suggest that VT-1161 and VT-1598 would not be desirable alternatives in triazole-resistant C. glabrata infections, given that they seem to be substrates for the same efflux pumps involved in triazole resistance. Both tetrazoles appear to be affected by the same mechanisms driving resistance to the commonly used triazoles, though further study with VT-1161 and VT-1598 is warranted to determine cutoff values and susceptibility breakpoints relevant to clinical outcomes.
MATERIALS AND METHODS
Strains and growth medium.
All constructed strains used in this study are listed in Table S1 in the supplemental material. The clinical isolates and the PDR1 replacement strains have been described previously (28, 31). All strains were stored as frozen stocks at −80°C with 40% glycerol. Strains were routinely grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose) broth at 30°C in a shaking incubator except as indicated for specific experimental conditions. Clinical isolates were obtained from Daniel Diekema at the University of Iowa repository.
Construction of C. glabrata strain SM3Δpdr1.
For strain SM3Δpdr1, disruption of the PDR1 gene was performed via the previously described SAT1 flipper method (31, 40). Briefly, flanking sequences proximal and distal to the PDR1 ORF were PCR amplified and cloned into the pSFS2-derived plasmid vector pBSS2 containing the SAT1 nourseothricin selection marker and FLP recombinase. Plasmids were chemically transformed using a lithium acetate method previously described (29). Successful transformants were screened on YPD agar plates containing 200 mg/liter nourseothricin, and genomic integration was confirmed through Southern blot hybridization. Subsequent induction of the FLP recombinase was achieved through growth of the correct integrants in YPD medium for 24 h to recycle the SAT1 flipper cassette.
Susceptibility testing.
MICs of VT-1161, VT-1598, fluconazole, voriconazole, posaconazole, and itraconazole were measured using broth microdilution methods as described by the Clinical and Laboratory Standards Institute (41, 42). Briefly, 96-well microtiter plates containing RPMI 1640 medium (0.165 M morpholinepropanesulfonic acid [MOPS] with l-glutamine and without sodium bicarbonate, pH 7.0) with serially diluted concentrations of each azole were used to incubate strains. Drug concentrations ranged from 0.015 to 8 μg/ml for VT-1161 and VT-1598; 0.125 to 64 μg/ml for fluconazole; and 0.03 to 16 μg/ml for voriconazole, posaconazole, and itraconazole. C. glabrata strains and isolates were incubated at 35°C, and MIC values were determined after 24 h and interpreted as the minimum concentration required to reduce cell growth by approximately 50% or greater compared to drug-free control wells. MICs were performed in duplicate for clinical isolates. The large majority if duplicates (264/288 or 92%) were identical or within a single dilution of each other. If the duplicate readings were different, the higher of the MICs was used in this analysis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel Diekema of the University of Iowa for graciously providing the clinical isolates used in this study.
Additionally, we would like to recognize the generous support of this work by NIH grant R01 AI131620 to P.D.R.
N.P.W. has received research support to UT Health San Antonio from Astellas, bioMérieux, Cidara, F2G, Merck, and Viamet and has served on advisory boards for Merck, Astellas, Toyama, and Viamet. C.M.Y., R.J.S., and E.P.G. are employees of Viamet Pharmaceuticals, Inc. All other authors have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01304-19.
REFERENCES
- 1.Magill SS, O’Leary E, Janelle SJ, Thompson DL, Dumyati G, Nadle J, Wilson LE, Kainer MA, Lynfield R, Greissman S, Ray SM, Beldavs Z, Gross C, Bamberg W, Sievers M, Concannon C, Buhr N, Warnke L, Maloney M, Ocampo V, Brooks J, Oyewumi T, Sharmin S, Richards K, Rainbow J, Samper M, Hancock EB, Leaptrot D, Scalise E, Badrun F, Phelps R, Edwards JR, Emerging Infections Program Hospital Prevalence Survey Team. 2018. Changes in prevalence of health care-associated infections in U.S. hospitals. N Engl J Med 379:1732–1744. doi: 10.1056/NEJMoa1801550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sobel JD. 2010. Changing trends in the epidemiology of Candida blood stream infections: a matter for concern? Crit Care Med 38:990–992. doi: 10.1097/CCM.0b013e3181d16866. [DOI] [PubMed] [Google Scholar]
- 3.Azie N, Neofytos D, Pfaller M, Meier-Kriesche HU, Quan SP, Horn D. 2012. The PATH (Prospective Antifungal Therapy) Alliance registry and invasive fungal infections: update 2012. Diagn Microbiol Infect Dis 73:293–300. doi: 10.1016/j.diagmicrobio.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Pfaller MA, Jones RN, Castanheira M. 2014. Regional data analysis of Candida non-albicans strains collected in United States medical sites over a 6-year period, 2006–2011. Mycoses 57:602–611. doi: 10.1111/myc.12206. [DOI] [PubMed] [Google Scholar]
- 5.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, Rodloff A, Fu W, Ling TA, Global Antifungal Surveillance Group. 2010. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol 48:1366–1377. doi: 10.1128/JCM.02117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lockhart SR, Iqbal N, Cleveland AA, Farley MM, Harrison LH, Bolden CB, Baughman W, Stein B, Hollick R, Park BJ, Chiller T. 2012. Species identification and antifungal susceptibility testing of Candida bloodstream isolates from population-based surveillance studies in two U.S. cities from 2008 to 2011. J Clin Microbiol 50:3435–3442. doi: 10.1128/JCM.01283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamoth F, Lockhart SR, Berkow EL, Calandra T. 2018. Changes in the epidemiological landscape of invasive candidiasis. J Antimicrob Chemother 73:i4–i13. doi: 10.1093/jac/dkx444. [DOI] [PubMed] [Google Scholar]
- 8.Koehler P, Stecher M, Cornely OA, Koehler D, Vehreschild M, Bohlius J, Wisplinghoff H, Vehreschild JJ. 27 April 2019. Morbidity and mortality of candidaemia in Europe: an epidemiologic meta-analysis. Clin Microbiol Infect doi: 10.1016/j.cmi.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 9.Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, Messer S, Herwaldt L, Pfaller M, Diekema D. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis 37:1172–1177. doi: 10.1086/378745. [DOI] [PubMed] [Google Scholar]
- 10.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:e1–e50. doi: 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borst A, Raimer MT, Warnock DW, Morrison CJ, Arthington-Skaggs BA. 2005. Rapid acquisition of stable azole resistance by Candida glabrata isolates obtained before the clinical introduction of fluconazole. Antimicrob Agents Chemother 49:783–787. doi: 10.1128/AAC.49.2.783-787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glockner A, Cornely OA. 2015. Candida glabrata–unique features and challenges in the clinical management of invasive infections. Mycoses 58:445–450. doi: 10.1111/myc.12348. [DOI] [PubMed] [Google Scholar]
- 13.Sanglard D, Ischer F, Calabrese D, Majcherczyk PA, Bille J. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob Agents Chemother 43:2753–2765. doi: 10.1128/AAC.43.11.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyazaki H, Miyazaki Y, Geber A, Parkinson T, Hitchcock C, Falconer DJ, Ward DJ, Marsden K, Bennett JE. 1998. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob Agents Chemother 42:1695–1701. doi: 10.1128/AAC.42.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torelli R, Posteraro B, Ferrari S, La Sorda M, Fadda G, Sanglard D, Sanguinetti M. 2008. The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Mol Microbiol 68:186–201. doi: 10.1111/j.1365-2958.2008.06143.x. [DOI] [PubMed] [Google Scholar]
- 16.Vermitsky JP, Edlind TD. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob Agents Chemother 48:3773–3781. doi: 10.1128/AAC.48.10.3773-3781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vermitsky JP, Earhart KD, Smith WL, Homayouni R, Edlind TD, Rogers PD. 2006. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol Microbiol 61:704–722. doi: 10.1111/j.1365-2958.2006.05235.x. [DOI] [PubMed] [Google Scholar]
- 18.Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS, Rogers PD. 2016. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front Microbiol 7:2173. doi: 10.3389/fmicb.2016.02173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoekstra WJ, Garvey EP, Moore WR, Rafferty SW, Yates CM, Schotzinger RJ. 2014. Design and optimization of highly-selective fungal CYP51 inhibitors. Bioorg Med Chem Lett 24:3455–3458. doi: 10.1016/j.bmcl.2014.05.068. [DOI] [PubMed] [Google Scholar]
- 20.Warrilow AG, Hull CM, Parker JE, Garvey EP, Hoekstra WJ, Moore WR, Schotzinger RJ, Kelly DE, Kelly SL. 2014. The clinical candidate VT-1161 is a highly potent inhibitor of Candida albicans CYP51 but fails to bind the human enzyme. Antimicrob Agents Chemother 58:7121–7127. doi: 10.1128/AAC.03707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yates CM, Garvey EP, Shaver SR, Schotzinger RJ, Hoekstra WJ. 2017. Design and optimization of highly-selective, broad spectrum fungal CYP51 inhibitors. Bioorg Med Chem Lett 27:3243–3248. doi: 10.1016/j.bmcl.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 22.Hargrove TY, Garvey EP, Hoekstra WJ, Yates CM, Wawrzak Z, Rachakonda G, Villalta F, Lepesheva GI. 2017. Crystal structure of the new investigational drug candidate VT-1598 in complex with Aspergillus fumigatus sterol 14alpha-demethylase provides insights into its broad-spectrum antifungal activity. Antimicrob Agents Chemother 61:e00570-17. doi: 10.1128/AAC.00570-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Break TJ, Desai JV, Healey KR, Natarajan M, Ferre EMN, Henderson C, Zelazny A, Siebenlist U, Yates CM, Cohen OJ, Schotzinger RJ, Perlin DS, Garvey EP, Lionakis MS. 2018. VT-1598 inhibits the in vitro growth of mucosal Candida strains and protects against fluconazole-susceptible and -resistant oral candidiasis in IL-17 signalling-deficient mice. J Antimicrob Chemother 73:2089–2094. doi: 10.1093/jac/dky170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brand SR, Degenhardt TP, Person K, Sobel JD, Nyirjesy P, Schotzinger RJ, Tavakkol A. 2018. A phase 2, randomized, double-blind, placebo-controlled, dose-ranging study to evaluate the efficacy and safety of orally administered VT-1161 in the treatment of recurrent vulvovaginal candidiasis. Am J Obstet Gynecol 218:624.e1–624.e9. doi: 10.1016/j.ajog.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Nishimoto AT, Wiederhold NP, Flowers SA, Zhang Q, Kelly SL, Morschhäuser J, Yates CM, Hoekstra WJ, Schotzinger RJ, Garvey EP, Rogers PD. 2019. In vitro activities of the novel investigational tetrazoles VT-1161 and VT-1598 compared to the triazole antifungals against azole-resistant strains and clinical isolates of Candida albicans. Antimicrob Agents Chemother 63:e00341-19. doi: 10.1128/AAC.00341-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiederhold NP, Patterson HP, Tran BH, Yates CM, Schotzinger RJ, Garvey EP. 2018. Fungal-specific Cyp51 inhibitor VT-1598 demonstrates in vitro activity against Candida and Cryptococcus species, endemic fungi, including Coccidioides species, Aspergillus species and Rhizopus arrhizus. J Antimicrob Chemother 73:404–408. doi: 10.1093/jac/dkx410. [DOI] [PubMed] [Google Scholar]
- 27.Garvey EP, Hoekstra WJ, Schotzinger RJ, Sobel JD, Lilly EA, Fidel PL Jr. 2015. Efficacy of the clinical agent VT-1161 against fluconazole-sensitive and -resistant Candida albicans in a murine model of vaginal candidiasis. Antimicrob Agents Chemother 59:5567–5573. doi: 10.1128/AAC.00185-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magill SS, Shields C, Sears CL, Choti M, Merz WG. 2006. Triazole cross-resistance among Candida spp.: case report, occurrence among bloodstream isolates, and implications for antifungal therapy. J Clin Microbiol 44:529–535. doi: 10.1128/JCM.44.2.529-535.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whaley SG, Zhang Q, Caudle KE, Rogers PD. 2018. Relative contribution of the ABC transporters Cdr1, Pdh1, and Snq2 to azole resistance in Candida glabrata. Antimicrob Agents Chemother 62:e01070-18. doi: 10.1128/AAC.01070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaller MA, Andes D, Arendrup MC, Diekema DJ, Espinel-Ingroff A, Alexander BD, Brown SD, Chaturvedi V, Fowler CL, Ghannoum MA, Johnson EM, Knapp CC, Motyl MR, Ostrosky-Zeichner L, Walsh TJ. 2011. Clinical breakpoints for voriconazole and Candida spp. revisited: review of microbiologic, molecular, pharmacodynamic, and clinical data as they pertain to the development of species-specific interpretive criteria. Diagn Microbiol Infect Dis 70:330–343. doi: 10.1016/j.diagmicrobio.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Caudle KE, Barker KS, Wiederhold NP, Xu L, Homayouni R, Rogers PD. 2011. Genomewide expression profile analysis of the Candida glabrata Pdr1 regulon. Eukaryot Cell 10:373–383. doi: 10.1128/EC.00073-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanglard D, Ischer F, Bille J. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob Agents Chemother 45:1174–1183. doi: 10.1128/AAC.45.4.1174-1183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanguinetti M, Posteraro B, Fiori B, Ranno S, Torelli R, Fadda G. 2005. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob Agents Chemother 49:668–679. doi: 10.1128/AAC.49.2.668-679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai HF, Krol AA, Sarti KE, Bennett JE. 2006. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob Agents Chemother 50:1384–1392. doi: 10.1128/AAC.50.4.1384-1392.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whaley SG, Caudle KE, Vermitsky JP, Chadwick SG, Toner G, Barker KS, Gygax SE, Rogers PD. 2014. UPC2A is required for high-level azole antifungal resistance in Candida glabrata. Antimicrob Agents Chemother 58:4543–4554. doi: 10.1128/AAC.02217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whaley SG, Rogers PD. 2016. Azole resistance in Candida glabrata. Curr Infect Dis Rep 18:41. doi: 10.1007/s11908-016-0554-5. [DOI] [PubMed] [Google Scholar]
- 37.Garvey EP, Sharp AD, Warn PA, Yates CM, Schotzinger RJ. 2018. The novel fungal CYP51 inhibitor VT-1598 is efficacious alone and in combination with liposomal amphotericin B in a murine model of cryptococcal meningitis. J Antimicrob Chemother 73:2815–2822. doi: 10.1093/jac/dky242. [DOI] [PubMed] [Google Scholar]
- 38.Wiederhold NP, Shubitz LF, Najvar LK, Jaramillo R, Olivo M, Catano G, Trinh HT, Yates CM, Schotzinger RJ, Garvey EP, Patterson TF. 2018. The novel fungal Cyp51 inhibitor VT-1598 is efficacious in experimental models of central nervous system coccidioidomycosis caused by Coccidioides posadasii and Coccidioides immitis. Antimicrob Agents Chemother 62:e02258-17. doi: 10.1128/AAC.02258-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monk BC, Keniya MV, Sabherwal M, Wilson RK, Graham DO, Hassan HF, Chen D, Tyndall J. 2019. Azole resistance reduces susceptibility to the tetrazole antifungal VT-1161. Antimicrob Agents Chemother 63:e02114-18. doi: 10.1128/AAC.02114-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reuss O, Vik A, Kolter R, Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 41.Clinical and Laboratory Standards Institute. 2008. Reference method for broth microdilution antifungal susceptibility testing of yeasts; approved standard, 3rd ed. CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 42.Clinical and Laboratory Standards Institute. 2012. Reference method for broth microdilution antifungal susceptibility testing of yeasts; 4th informational supplement. CLSI document M27-S4. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.