Abstract
Malaria remains a major detrimental parasitic disease in the developing world, with more than 200 million cases annually. Widespread drug-resistant parasite strains push for the development of novel antimalarial drugs. Plant-derived natural products are key sources of antimalarial molecules. Euterpe oleracea Martius (“açaí”) originates from Brazil and has anti-inflammatory and antineoplasic properties. Here, we evaluated the antimalarial efficacy of three phenolic fractions of açaí; total phenolics (1), nonanthocyanin phenolics (2), and total anthocyanins (3). In vitro, fraction 2 moderately inhibited parasite growth in chloroquine-sensitive (HB3) and multiresistant (Dd2) Plasmodium falciparum strains, while none of the fractions was toxic to noncancer cells. Despite the limited activity in vitro, the oral treatment with 20 mg/kg of fraction 1 reduced parasitemia by 89.4% in Plasmodium chabaudi-infected mice and prolonged survival. Contrasting in vitro and in vivo activities of 1 suggest key antiplasmodial roles for polyphenol metabolites rather than the fraction itself. Finally, we performed haploinsufficiency chemical genomic profiling (HIP) utilizing heterozygous Saccharomyces cerevisiae deletion mutants to identify molecular mechanisms of açaí fractions. HIP results indicate proteostasis as the main cellular pathway affected by fraction 2. These results open avenues to develop açaí polyphenols as potential new antimalarial candidates.
Introduction
Caused by protozoa of the genus Plasmodium and transmitted by Anopheles mosquitoes, malaria is historically one of the major parasitic diseases to affect human populations. World Health Organization (WHO) estimates that more than 200 million cases of the disease and around 400 000 malaria-related deaths occurred in 2017.1 Efforts toward malaria elimination have been successful by using insecticide-treated nets in endemic regions, the development of rapid diagnostic tests, and the adoption of artemisinin-based combined therapies (ACTs). Despite the decreased number of malaria cases by 40% within the last decade, the incidence has significantly increased in the Americas between 2016 and 2017,1 keeping the long-term goal of eradication still unreachable.
One of the major drawbacks in malaria control is the development of parasite resistance to antimalarial drugs. Parasite resistance has been reported for all known classes of antimalarial drugs.2 Recently, Southeast Asia has recorded lower parasite susceptibility to ACTs, the front-line treatment indicated for severe cases of Plasmodium falciparum infection.3 Facing the imminent spread of ACT-resistant parasites, there is a renewed need for new compounds with antiplasmodial activity.
Natural extracts have played a major role in the discovery of antimalarial compounds, as antimalarial drugs such as quinine and artemisinin are derived from plants widely used in the traditional medicine.4,5 Biodiversity-rich regions like the Amazon are valuable sources of plants with potential antimalarial activity. Among Brazilian Amazon plants, açaí (Euterpe oleracea Mart.) is both one of the most consumed fruits and one of the major export products from the region,6 illustrating the cultural and economic importance of ethnobotanical-based approaches for natural compounds prospection. The use of açaí by local communities to treat malaria-related symptoms such as fever has been described.7 Açaí is known by its functional properties, mainly due to its polyphenolic composition, associated with antineoplasic activity and prevention of cardiovascular diseases.8,9
One of the main hurdles of working with natural products is the challenge of characterizing their components and respective modes of action. In this context, chemical genomic profiling of natural extracts aims to explore functional relationships between eukaryotic genes and bioactive compounds in response to pharmacological perturbation by quantification of yeast growth fitness in diploid Saccharomyces cerevisiae strains with a gene copy deleted.10
Preliminary studies have shown that açaí roots did not present in vitro antimalarial activity.11 However, the present study evaluated the antimalarial activity of polyphenolic açaí pulp fractions in vitro against CQ-sensitive and -resistant P. falciparum strains, besides in vivo antimalarial activity on P. chabaudi-infected mice. Finally, the mode of action of açaí polyphenols was investigated through yeast-based chemogenomics.
Results and Discussion
Chemical Characterization of Açaí Fractions
Three polyphenol-rich fractions were obtained from açaí pulp fractionation: total phenolics (1), nonanthocyanin phenolics (2), and total anthocyanins (3). Two major anthocyanins were identified in 1 and 3 in high relative abundance (Table 1). Cyanidin-3-rutinoside represented 62.7 and 69.3% of 1 and 3, respectively, while cyanidin-3-glucoside accounted for 14.9 and 15.0% of these two fractions. Compound 2 presented high relative abundance of phenolic acids with 36.9% of protocatechuic acid. Moreover, polyphenolic C-glycosides were also found in high proportions, as orientin and isoorientin represented 26.6 and 23.4% of its composition, respectively. The total phenolic concentration of each extract, quantified by Folin–Ciocalteu, was 26 932 ± 865 mg/L (1), 1343 ± 154 mg/L (2), and 17 820 ± 570 mg/L (3) gallic acid equivalent (GAE).
Table 1. Polyphenol Characterization of Açaí Fractions.
| relative
abundance (%) |
|||
|---|---|---|---|
| polyphenol | total phenolics (1) | nonanthocyanin phenolics (2) | total anthocyanins (3) |
| cyanidin-3-glucosidea | 14.9 | n.d. | 15.0 |
| cyanidin-3-rutinosidea | 62.7 | n.d. | 69.3 |
| protocatechuic acidb | 3.9 | 36.9 | 3.2 |
| orientinb | 5.8 | 26.6 | 3.0 |
| isoorientinb | 9.5 | 23.4 | 6.8 |
| isovitexinb | 1.7 | 6.5 | 1.7 |
| scoparinb | 1.6 | 6.6 | 1.1 |
Quantified as rutin equivalents.
Quantified as gallic acid equivalents; n.d. not detected.
The polyphenol composition of the açaí fractions used in this investigation is clearly diverse, while the anthocyanins cyanidin-3-rutinoside and cyanidin-3-glucoside were the most abundant polyphenols identified in both 1 and 3; these were not identified in 2. The major nonanthocyanin polyphenols found include protocatechuic acid, orientin, and isoorientin, considered flavonol-C-glucosides. These three compounds together account for about 85% of 2 composition, while in 1 and 3 fractions they represent less than 20%. These compounds are not highly bioavailable but are found as parental compounds in the blood stream,12 which could significantly contribute to their role in the antimalarial activity of fractions.
In Vitro Antimalarial Activity of Açaí Fractions
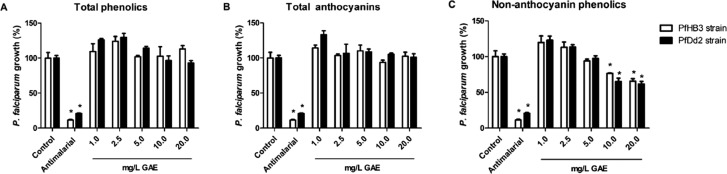
In vitro antimalarial activity was assessed using a PicoGreen-based fluorometric assay in P. falciparum cultures. The activity of 1, 2, and 3 in chloroquine-sensitive and -resistant P. falciparum strains is described as relative DNA content compared to untreated controls (Figure 1). Chloroquine was used at 30 nM as a positive control for HB3 (CQ-sensitive) strain and Artesunate also at 30 nM for Dd2 (CQ-resistant) strain. When tested from 1.0 to 20.0 mg/L GAE, none of the concentrations applied for 1 and 3 reduced DNA content, i.e., inhibited parasite growth, for either of the parasite strains assayed. Fraction 2, on the other hand, showed moderate antiplasmodial activity on both strains. Starting at 10.0 mg/L GAE of 2, the relative DNA content of HB3 strain was 76.5% of untreated control, while that of Dd2 strain was 65.1%, both showing statistically significant growth inhibition. At 20.0 mg/L GAE, relative DNA content of HB3 strain was 65.8% of untreated control, while that of Dd2 strain was 61.7%. Interestingly, contrary to our findings, it has been shown that antimalarial activity and total amount of anthocyanins are directly correlated.13 Hence, we hypothesize that within açaí fractions, another class of compounds might be responsible for antimalarial activity.
Figure 1.
Antimalarial activity in vitro of açaí fractions. Parasite growth (%) of chloroquine-sensitive (HB3) and multiresistant (Dd2) P. falciparum strains treated with açaí (A) total phenolics (1), (B) total anthocyanins (3), and (C) nonanthocyanin phenolics (2) for 48 h. Chloroquine 100 nM was used as antimalarial positive control for HB3 strain and Artesunate 70 nM for Dd2 strain. Each concentration tested was compared to its respective strain control (*p < 0.05, ANOVA-Dunnett).
Low proportion of 2 within 1 and 3 may explain the lack of antimalarial activity for the latter two fractions. Fraction 2 is composed of approximately 25% of isoorientin that has already been shown to present antimalarial activity in vitro in Ajuga laxmannii plant extracts.14 Also, protocatechuic acid, which accounts for approximately 37% of 2 composition, has been shown to have moderate antimalarial activity.15 Other plant-based extracts have been described as possessing in vitro antimalarial activity,16−18 including popular Amazonian plants such as guaraná (Paullinia cupana) and cajueiro (Anacardium occidentale).19
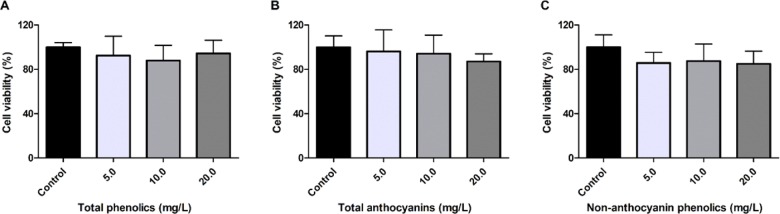
Cytotoxicity Assessment of Açaí Fraction
The cytotoxicities of 1, 2, and 3 were evaluated using the resazurin dye assay to quantify their effects on the proliferation of a murine macrophage cell line. Concentration range used for cytotoxicity assay was the same as that used for evaluation of antimalarial activity in vitro, up to 20.0 mg/L GAE. The 1, 2, and 3 fractions did not negatively influence viability of RAW264.7 cells (Figure 2). As our group has described the absence of cytotoxicity in nonmalignant colon myofibroblast cells by açaí compounds previously,20 there is validation on the safety of açaí polyphenols for distinct cell lines.
Figure 2.
Cell viability of RAW264.7 cells treated with açaí fractions. Murine macrophage cells were treated with (A) total phenolics, (B) total anthocyanins, and (C) nonanthocyanin phenolics for 24 h. No statistical differences were found between treatments and the control group (p > 0.05, ANOVA-Dunnett).
Evaluation of the in Vivo Antimalarial Activity of E. oleracea M
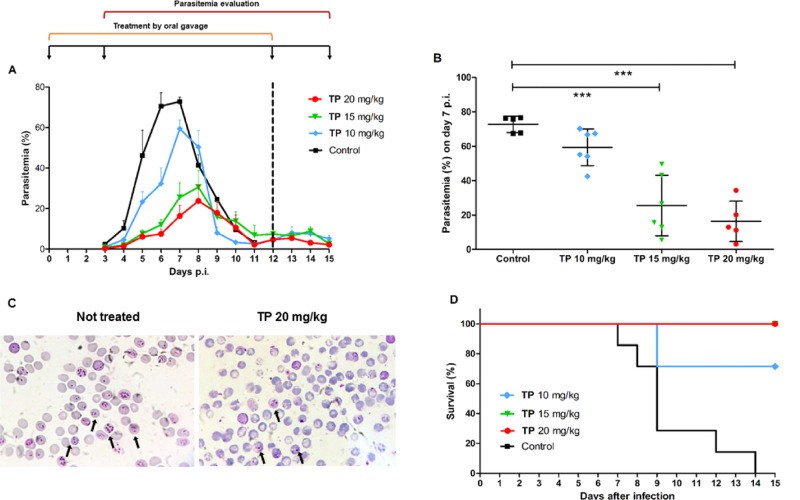
A murine model of infection was used to investigate whether açaí polyphenols could mitigate parasitemia in vivo. P. chabaudi-infected mice were treated orally with 1 (10, 15, and 20 mg/kg) on day 0 to day 12 post-infection. Results indicate that the treatment reduced parasitemia in a dose-dependent fashion (Figure 3A). During parasitemia peak (days 6 and 7), 20 mg/kg of 1 decreased parasite growth by 89.4 and 77.3% (Table 2), respectively, compared to the untreated control group (Figure 3B). Similarly, 15 mg/kg of 1 also diminished parasite burden and reduced parasitemia by 81 and 62.2% on days 6 and 7 p.i, respectively. Animals treated with 10 mg/kg of 1 also showed a significant parasitemia inhibition of 54.2% on day 6 p.i. Additionally, doses of 20 and 15 mg/kg of 1 allowed 100% of animal survival until day 15 p.i., in contrast to no survival observed for the untreated group. However, when treated with 10 mg/kg, 70% of the animals did not survive until day 9 p.i. due to infection (Figure 3D).
Figure 3.
In vivo effect of açaí total phenolics on murine Plasmodium parasites. Mice were infected intraperitoneally (i.p.) with 106P. chabaudi chabaudi AS-infected erythrocytes and treated with different doses of 1 administered by gavage for 13 consecutive days. (A) Parasitemia was determined daily until day 15 post-infection. (B) Dot plot of parasitemia on peak day (day 7) showing dose-dependent inhibition. (C) Microscopy of blood smears of mice untreated and treated orally with 20 mg/kg of 1. The arrows indicate infected erythrocytes. (D) Survival analysis throughout the study duration. The dotted line in the graph marks the end of açaí administration period. Results are expressed as mean ± standard deviation. (***p < 0.05 vs control, ANOVA-Dunnett). Data from one representative experiment.
Table 2. Parasitemiaa Inhibition (%) of P. chabaudi-Infected Mice Treated with Different Doses of the Total Phenolics (1) Fraction for 13 Days from Two Independent Experiments.
| mean parasitemia
inhibitiona (%) |
||||
|---|---|---|---|---|
| days post-infection | ||||
| doses (mg/(kg day)) | D5 | D6 | D7 | D8 |
| 10 | 49.42 ± 25.98c | 54.18 ± 27.45c | 18.46 ± 14.62 | NIb |
| 15 | 81.32 ± 8.53c | 81.03 ± 8.29c | 62.17 ± 25.99c | 28.66 ± 55.21 |
| 20 | 86.89 ± 10.13c | 89.40 ± 8.20c | 77.53 ± 16.12c | 42.55 ± 41.21 |
Values are expressed as the mean of parasitemia inhibition (%).
NI: no inhibition.
p < 0.05 vs control, ANOVA-Tukey.
In vivo antimalarial activity observed may be due to a few determinants: the presence of polyphenolic C-glycosides in 2, the activity of the microbiome in metabolizing açaí polyphenols, and the hepatic (postabsorption) metabolization. Most notably, better in vivo antimalarial results compared to in vitro ones for 1 suggests a role for compound metabolites rather than the compounds themselves.
We used 1 as an oral treatment to mice to represent a whole-fruit approach that is prevalent in malaria-endemic areas. In humans, clinical research has shown that the polyphenols in açaí are bioavailable29 and improve redox metabolism and oxidative status,30 besides retaining antioxidant and antigenotoxic activities even after digestion and gut microbiota fermentation.31
Yeast-Based Chemical Genomic Profiling
One of the main challenges of working with bioactive natural extracts is to identify their potential molecular targets. Therefore, seeking to characterize the antimalarial mechanism of açaí fractions, we assessed drug-induced hypersensitivity in a budding yeast collection of heterozygous deletion mutant strains, which has been shown to be a suitable approach for exploring the mode of action of crude extracts.21 To outline specific 1 and 2 signatures, both treatments were assayed due to their different patterns of Plasmodium inhibition in vitro and in vivo.
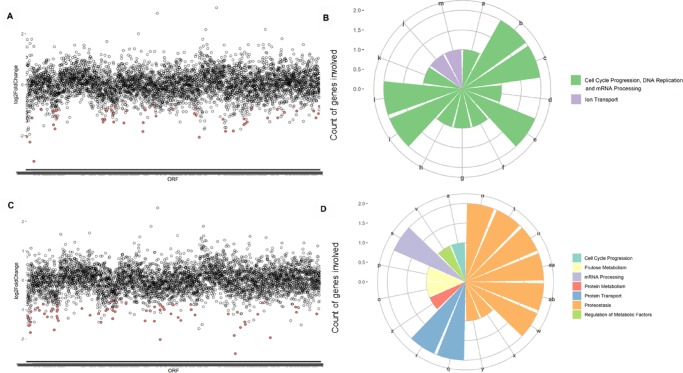
We identified 51 heterozygous yeast mutants depleted among the yeast collection treated with 1 (Figure 4A) and 67 mutants depleted in population treated with 2 (Figure 4C). The data set describing the sensitive mutants identified in our screens (p < 0.01) include information about their degree of differential growth (Supporting Information File 1). To the best of our knowledge, this is the first yeast-based chemical genomic profiling describing E. oleracea M.
Figure 4.
Haploinsufficiency profiling of heterozygous yeast strains treated with different açaí fractions. (A) Heterozygous yeast strains most depleted when treated with IC20 of 1. Differential growth (log 2 fold change) is plotted on the y-axis as a function of yeast strains organized by chromosome location of the respective deleted open reading frames (ORFs). The most negative log 2 fold change values represent the strains most susceptible to the treatment. The dots in red represent ORFs that fall within the cutoffs p-value <0.001 and log 2 fold change <0. (B) Biological processes identified as hits under treatment with 1 using the Reactome tool. Each bar represents a different biological process, whose name is represented by a letter (a to ab). The y-axis displays the count of genes involved in each process. Similar processes are grouped by color. (C) Yeast strains most depleted upon treatment with IC20 of 2. (D) Biological processes identified as hits under treatment with 2 using the Reactome tool. Biological processes: a, activation of kinases; b, activation of the mRNA upon binding of the cap-binding complex and eIFs; c, events during G2/M transition; d, DNA replication initiation; e, formation of the 43S complex; f, G2 phase; g, loss of binding ability of a transcriptional repression domain; h, phosphorylation of proteins involved in the G2/M transition; i, ribosomal scanning and start codon recognition; j, sodium-coupled sulfate, di- and tricarboxylate transporters; k, telomere C-strand synthesis initiation; l, translation initiation complex formation; m, zinc influx into cells; n, association of TriC/CCT with target proteins during biosynthesis; o, fructose catabolism; p, fructose metabolism; q, Cargo trafficking; r, Cargo trafficking to the membrane; s, mRNA splicing; t, β-folding of G-protein by TriC/CCT; u, cooperation of Prefoldin and TriC/CCT in actin and tubulin folding; v, dephosphorylation of key metabolic factors; w, prefoldin-mediated transfer of substrate to CCT/TriC, x, SUMO is conjugated to E1, y, SUMO is transferred from E1 to E2; z, hydrolysis of l-amino acids; aa, folding of actin by CCT/TriC; ab, formation of tubulin folding intermediates by CCT/TriC.
To identify enrichment in biological processes or functions within the strains negatively selected following treatment, we used the Gene Ontology (GO) tool “Go Term Finder”. However, no statistically significant functional enrichment was observed for neither 1 nor 2 based on this tool. In the absence of a tool that analyzes and connects gene ontology pathways in Plasmodium, data were analyzed using the Reactome tool, which describes the possible reactions occurring in a human cell when proteins used as input are present and active simultaneously. Hence, based on eukaryotic conservation of molecular functions, functional annotation of terms yielded by Reactome is taken from the roles of homologous proteins, i.e., human counterparts of yeast genes found to be depleted in the assay. For 1, control of cell cycle progression, DNA replication, and mRNA processing account for 10 of the 13 biological processes enriched (p < 0.05) among pathway hits (Figure 4B). The one strain heterozygous for an essential gene under 1 treatment was Δarb1/ARB1, involved in ribosome biogenesis and assembly.22 For 2, among the biological processes enriched based on the Reactome tool (p < 0.05), unfolded protein response and CCT/TRiC functions are responsible for 8 of the 16 processes identified (Figure 4D). Interestingly, mutants deleted for essential genes involved in proteostasis such as CCT2 and CCT523 and UBA2, related to protein sumoylation,24 were hypersensitive to 2 treatment. Considering the low proportion of 2 within the fraction 1, we confirmed distinct modes of action for the treatments.
As we believe that polyphenolic C-glycosides in 2 may be responsible for the antimalarial activity observed for 1 in vivo, we further investigated molecular mechanisms of 2. The cellular pathways most affected by fraction 2 relate to the CCT complex or TriC, which is an essential protein folding machinery involved in protein homeostasis that helps folding nascent polypeptides into biologically functional structures,25 regulating cellular response to proteotoxic stress.26 Hence, CCT plays a key role in proteostasis and cell survival and is highly conserved among eukaryotes.27 All CCT subunits have been identified in Plasmodium genome,28 and the complex is likely to play a critical role in parasite biology as the knockdown of one subunit leads to impaired growth in parasite asexual stages.29 CCT is also implicated in the export of PfEMP1, the most virulent factor of severe malaria.30 Moreover, transcriptomic analysis of P. falciparum isolates has linked increased levels of mRNAs for CCT/TriC to delayed parasite clearance upon artemisinin treatment.31 This finding suggests that 2 could present bioactive elements targeting protein homeostasis in a similar way to artemisinin, one of the most important antimalarial drugs in the clinic.
Two other elements listed among the pathways affected by treatment with 2 are Prefoldin and SUMO, both strongly correlated with proteostasis maintenance. Prefoldin is a cochaperonin that interacts with CCT, helping it to coordinate protein folding.32 SUMO, or small ubiquitin-like modifiers, is an essential family of proteins highly conserved among eukaryotes and known as competitive inhibitors of ubiquitin.33 Altogether, data indicate that 2 is likely to affect protein homeostasis in Plasmodium. Additionally, two strains mutant for genes from the glutaredoxin family (Grxs), GRX1 and GRX4, were hypersensitive to 2. Grx1 catalyzes reduction of disulfide bonds in response to oxidative stress, protecting cells against oxidative damage induced by reactive oxygen species via GSH metabolism.34 However, grx1 null mutants are more resistant to some oxidative stress agents, whereas grx4 mutants are hypersensitive to oxidative stress.35,36 Our findings reveal the action of 2 in disturbing the delicate redox balance of target cells, which require Grx proteins to be reestablished. Taken together, yeast genome-wide profiling of a potential molecular mechanism of 2 suggests that essential protein homeostasis and antioxidant pathways conserved in Plasmodium represent potential targets.
Collectively, the data presented here demonstrate the antimalarial potential of açaí pulp fractions in vitro and in vivo and indicate that 2 may account for this activity against the parasite by affecting proteostasis as major molecular target. Our findings indicate the potential to use açaí polyphenols as novel antimalarial compounds.
Experimental Section
Açaí Polyphenolic Extraction, Fractionation, and Chemical Analyses
Polyphenol-rich açaí fractions were obtained from clarified fruit pulp dried into a powder provided by Yakima Fruit Works (Moxee, WA) and Indústria de Produtos Alimenticios Café Campinho Ltda (Alfenas, MG, Brazil). Powdered açaí pulp was dissolved in distilled water and vacuum-filtered, followed by fractionation process, as previously described.37 Briefly, two activated C18 Sep-Pak 6cc cartridges (Waters Corporation, Milford, MA) were used to remove soluble solids such as sugars and acids and adsorb targeted polyphenolics. The first cartridge for total phenolics (1) was eluted with 100% methanol acidified with 0.1% formic acid. The second cartridge separated nonanthocyanin phenolics (2) by first eluting with 100% ethyl acetate followed by total anthocyanins (3) with 100% methanol acidified with 0.1% formic acid. Solvents were evaporated to dryness, and fractions redissolved in formic acid buffer (pH 3.5). All fractions were stored at −20 °C. Total phenolic quantification was performed using the Folin–Ciocalteu assay and expressed as gallic acid equivalents (GAEs).38 Fraction composition was determined as previously published37 by reverse-phase high-performance liquid chromatography coupled with mass spectrometry using a Thermo Finnigan LCQ Dexa XP Max MSn ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an ESI ion source.
Cultivation of P. falciparum-Infected Erythrocytes
Chloroquine-sensitive (HB3) and multidrug resistant (Dd2) strains of P. falciparum were cultured in erythrocytes from O+ healthy local donors. The culture media consisted of RPMI-1640 (Gibco, Waltham, MA) supplemented with 5% (w/v) AlbuMAX (Gibco, Waltham, MA), 25 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (Sigma-Aldrich, St. Louis, MO), 100 μM hypoxanthine (Sigma-Aldrich, St. Louis, MO), and 25 mM NaHCO3 (Sigma-Aldrich, St. Louis, MO). P. falciparum-infected erythrocytes were cultured at 2% hematocrit and incubated at modified atmosphere (5% CO2, 5% O2, 90% N2) and 37 °C, as previously described.39
In Vitro Antimalarial Activity by PicoGreen-Based Assay
A DNA-based fluorometric assay was used to assess the in vitro antimalarial activity of açaí pulp fractions in P. falciparum, as previously described.40,41 Parasites were synchronized with sorbitol and incubated with 1.0–20.0 mg/L GAE of fractions 1, 2, or 3 in 96-well plates at 2% hematocrit and 1% parasitemia. Negative (untreated) and positive controls (chloroquine or artesunate, IC50) were included. Incubation followed for 48 h in a candle jar at 37 °C. Inhibition of parasite growth was measured using a plate reader (BMG Labtech, Durhan, NC) at 485/520 nm (excitation/emission). Experiments were performed in triplicate.
Cytotoxicity Assay
The cytotoxicity of açaí fractions was assessed by a resazurin-based assay in murine macrophage cells (RAW264.7). Cells were obtained from American Type Culture Collection (ATCC, Manassas, VA) and cultured at 5% CO2 and 37 °C in Dulbecco’s modified Eagle’s medium (DMEM, ATCC, Manassas, VA) supplemented with 10% (v/v) fetal bovine serum (Gibco, Waltham, MA) and 1% (v/v) antibiotic mix (penicillin and streptomycin, Gibco, Waltham, MA). Cells (104/well) were seeded into 96-well plates and exposed to 5.0–20.0 mg/L GAE of fractions 1, 2, or 3 for 24 h. Untreated wells were also included. After incubation, resazurin (TOX8, Sigma-Aldrich, St. Louis, MO) was added. The fluorescence intensity was determined using a microplate reader (BMG Labtech Inc, Durhan, NC) at 560/590 nm (excitation/emission). Cell viability was calculated relative to the untreated group. Three independent experiments were performed.
In Vivo Antimalarial Activity of Açaí Polyphenols
In vivo antimalarial activity of açaí polyphenols was assessed in C57BL/6JUnib mice (4–6 weeks old) obtained from Centro de Bioterismo, UNICAMP, Brazil. All experiments and procedures were approved by the Ethical Committee for Animal Research at University of Campinas (protocol number 4367-1) before starting animal studies. Groups of five to seven female mice were intraperitoneally (i.p.) challenged with 106P. chabaudi chabaudi AS-infected erythrocytes. After infection, the animals were randomly divided into four groups and treated with vehicle (control) or fraction 1 (20, 15, and 10 mg GAE/(kg day)) by gavage twice a day for 13 consecutive days. Parasitemia was assessed daily from day 3 to day 15 post-infection (p.i.) by collecting mouse tail blood and counting 1000 erythrocytes in Giemsa-stained thin blood smears. Euthanasia was performed on day 15 p.i.
Yeast-Based Chemical Genomic Profiling
We used a collection of ∼6000 heterozygous S. cerevisiae diploid strains (Invitrogen cat. No. 95401.H4Pool), in which one copy of each of the predicted ORFs has been replaced by an antibiotic resistance marker (KANMX4), flanked both 5′ and 3′ by universal primer sequences flanking 20 bp DNA barcodes, unique to each of the deleted ORFs10,42
The IC20 determination was performed using yeast (BY4743) cultured in 384-well plates with initial OD600 of 0.1 in YPD (1% w/v yeast extract; 2% w/v peptone; 2% w/v glucose) in the presence of different concentrations of fractions 1 (starting at 552 mg/L GAE) and 2 (starting at 155 mg/L GAE). Growth was monitored by absorbance (OD600) for 24 h at 30 °C and 200 rpm double orbital shaking (BMG Clariostar, BMG Labtech Inc, Durhan, NC). Growth inhibition was calculated relatively to untreated control and plotted against log [compound] to determine IC20.43,44
For competition assays, log-phase cultures of the pool of barcoded heterozygous yeast strains42 were inoculated in quadruplicates in fresh YPD media with the IC20 concentration of 1 and 2 or drug-free (control) at final OD600 0.1 in 48-well plates. Plates were incubated for 12 h at 30 °C with shaking (200 rpm) allowing growth for about five generations. A fraction of this initial culture was diluted (1/20) and cultivated under the same conditions as above to obtain pools of cultures grown for ∼10, 15, and 20 generations. At each stage, samples were harvested and the pellets were stored at −80 °C in TE buffer. Genomic DNA was extracted with the Wizard Genomic DNA Purification Kit (Promega). DNA barcodes from the heterozygous yeast strain pools (grown for 5–20 generations) were sequenced using the platform Illumina HiSeq 2500 by the University of São Paulo Genomic Centre for Sequencing. Molecular barcodes of each of the 6000 heterozygous strains were PCR amplified using hybrid primers containing Illumina preadaptors and the sequences U1 or U2.
For sequencing analysis, a “virtual genome” was created based on the predicted barcode sequences. Reads of each sample were aligned to this virtual genome and quantified per barcode. Comparing the reads per barcode between 1 or 2 and controls, the heterozygous strains were ranked based on their compound sensitivity after 20 generations. Selection of strains most depleted in treated cell populations compared to untreated population (top hits, p-value <0.01) followed by Gene Ontology (GO) analysis45 and Reactome Tool46 suggested either direct targets or pathways associated with tolerance to fractions 1 or 2.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 6 (GraphPad, La Jolla, CA). Data were analyzed by ANOVA-Dunnett. Values were considered statistically significant when p < 0.05.
Acknowledgments
The authors thank Dr. Timothy J. C. Anderson (Texas Biomedical Research Institute, San Antonio, TX) for the technical assistance with P. falciparum cultivation in vitro and Dr. Gonçalo A. G. Pereira (Institute of Biology—UNICAMP) and his team for supporting this project with their expertise in bioinformatic analysis.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b02127.
Sensitive mutant yeast strains depleted under total phenolics (1) treatment (Table S1) and sensitive mutant yeast strains depleted under nonanthocyanin phenolics (2) treatment (Table S2) (PDF)
Author Contributions
L.T.F. and V.P.V. contributed to this work equally. F.T.M.C., S.U.M.-T., S.T.T., and L.A. designed and directed the study. V.P.V., L.C.C.A., and S.T.T. participated in extract purification and characterization. V.P.V., T.K., and L.C.C.A. performed in vitro assays. L.T.F. performed in vivo assays. L.T.F., T.A.T., and L.D.A. performed chemical genomic profiling assay. L.T.F., V.P.V., T.K., L.C.C.A., T.A.T., L.D.A., G.S.P., E.B., and S.T.T. analyzed data. L.T.F., V.P.V, T.A.T., E.B., P.S., S.T., S.U.M.-T., and F.T.M.C. participated in drafting the article and/or gave critical revision. All authors have given approval to the final version of the manuscript.
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP—Grants 2017/18611-7 and 2015/03553-6), FCT-FAPESP (Grant 2018/07007-4), Texas A&M University TAMU FAPESP Grant (2017/50400-6), Texas A&M AgriLife Vector Disease Program Funding, College Station, TX, Conselho Nacional de Desenvolvimento Científco e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES-TAMU 002/2014), and the Swedish Research Council (Grant 2016-05627). L.T.F. and T.A.T. were supported by a CAPES fellowship. L.D.A. was sponsored by a FAPESP fellowship (2017/01986-8). G.S.P. was sponsored by a FAPESP fellowship (2018/05328-8). F.T.M.C. is a CNPq researcher fellow level 1C.
The authors declare no competing financial interest.
Supplementary Material
References
- WHO. World Malaria Report 2018, 2018.
- Haldar K.; Bhattacharjee S.; Safeukui I. Drug Resistance in Plasmodium. Nat. Rev. Microbiol. 2018, 16, 156–170. 10.1038/nrmicro.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F.; Witkowski B.; Amaratunga C.; Beghain J.; Langlois A.-C.; Khim N.; Kim S.; Duru V.; Bouchier C.; Ma L.; et al. A Molecular Marker of Artemisinin-Resistant Plasmodium falciparum Malaria. Nature 2014, 505, 50–55. 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achan J.; Talisuna A. O.; Erhart A.; Yeka A.; Tibenderana J. K.; Baliraine F. N.; Rosenthal P. J.; D’Alessandro U. Quinine, an Old Anti-Malarial Drug in a Modern World: Role in the Treatment of Malaria. Malar. J. 2011, 10, 144. 10.1186/1475-2875-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y. The Discovery of Artemisinin (Qinghaosu) and Gifts from Chinese Medicine. Nat. Med. 2011, 17, 1217–1220. 10.1038/nm.2471. [DOI] [PubMed] [Google Scholar]
- Del Pozo-Insfran D.; Brenes C. H.; Talcott S. T. Phytochemical Composition and Pigment Stability of Açai (Euterpe oleracea Mart.). J. Agric. Food Chem. 2004, 52, 1539–1545. 10.1021/jf035189n. [DOI] [PubMed] [Google Scholar]
- Brandão M. G. L.; Grandi T. S. M.; Rocha E. M. M.; Sawyer D. R.; Krettli A. U. Survey of Medicinal Plants Used as Antimalarials in the Amazon. J. Ethnopharmacol. 1992, 36, 175–182. 10.1016/0378-8741(92)90018-M. [DOI] [PubMed] [Google Scholar]
- Noratto G. D.; Angel-morales G.; Talcott S. T.; Mertens-talcott S. U. Polyphenolics from Açaí (Euterpe oleracea Mart.) and Red Muscadine Grape (Vitis Rotundifolia) Protect Human Umbilical Vascular Endothelial Cells (HUVEC) from Glucose- and Lipopolysaccharide (LPS)-Induced Inflammation and Target MicroRNA-126. J. Agric. Food Chem. 2011, 59, 7999–8012. 10.1021/jf201056x. [DOI] [PubMed] [Google Scholar]
- Mertens-Talcott S. U.; Rios J.; Jilma-Stohlawetz P.; Pacheco-Palencia L. A.; Meibohm B.; Talcott S. T.; Derendorf H. Pharmacokinetics of Anthocyanins and Antioxidant Effects after the Consumption of Anthocyanin-Rich a??Ai Juice and Pulp (Euterpe oleracea Mart.) in Human Healthy Volunteers. J. Agric. Food Chem. 2008, 56, 7796–7802. 10.1021/jf8007037. [DOI] [PubMed] [Google Scholar]
- Giaever G.; Shoemaker D. D.; Jones T. W.; Liang H.; Winzeler E. A.; Astromoff A.; Davis R. W. Genomic Profiling of Drug Sensitivities via Induced Haploinsufficiency. Nat. Genet. 1999, 21, 278–283. 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- Oliveira D. R.; Krettli A. U.; Aguiar A. C. C.; Leitão G. G.; Vieira M. N.; Martins K. S.; Leitão S. G. Ethnopharmacological Evaluation of Medicinal Plants Used against Malaria by Quilombola Communities from Oriximiná, Brazil. J. Ethnopharmacol. 2015, 173, 424–434. 10.1016/j.jep.2015.07.035. [DOI] [PubMed] [Google Scholar]
- Hostetler G. L.; Ralston R. A.; Schwartz S. J. Flavones: Food Sources, Bioavailability, Metabolism and Bioactivity. Adv. Nutr. 2017, 8, 423–435. 10.3945/an.116.012948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Babili F.; Bouajila J.; Fouraste I.; Valentin A.; Mauret S.; Moulis C. Chemical Study, Antimalarial and Antioxidant Activities, and Cytotoxicity to Human Breast Cancer Cells (MCF7) of Argania Spinosa. Phytomedicine 2010, 17, 157–160. 10.1016/j.phymed.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Atay I.; Kirmizibekmez H.; Kaiser M.; Akaydin G.; Tasdemir D.; Atay I.; Kirmizibekmez H.; Kaiser M.; Akaydin G. Evaluation of in Vitro Antiprotozoal Activity of Ajuga Laxmannii and Its Secondary Metabolites. Pharm. Biol. 2016, 1–7. 10.3109/13880209.2015.1129542. [DOI] [PubMed] [Google Scholar]
- Garcia-Alvarez M. C.; Moussa I.; Njomnang Soh P.; Nongonierma R.; Abdoulaye A.; Nicolau-Travers M. L.; Fabre A.; Wdzieczak-Bakala J.; Ahond A.; Poupat C.; et al. Both Plants Sebastiania Chamaelea from Niger and Chrozophora Senegalensis from Senegal Used in African Traditional Medicine in Malaria Treatment Share a Same Active Principle. J. Ethnopharmacol. 2013, 149, 676–684. 10.1016/j.jep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Bankole A. E.; Adekunle A. A.; Sowemimo A. A.; et al. Phytochemical Screening and in Vivo Antimalarial Activity of Extracts from Three Medicinal Plants Used in Malaria Treatment in Nigeria. Parasitol. Res. 2016, 115, 299–305. 10.1007/s00436-015-4747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyo P.; Botha M. E.; Nondaba S.; Niemand J.; Maharaj V. J.; Eloff J. N.; Louw A. I.; Birkholtz L. In Vitro Inhibition of Plasmodium falciparum Early and Late Stage Gametocyte Viability by Extracts from Eight Traditionally Used South African Plant Species. J. Ethnopharmacol. 2016, 185, 235–242. 10.1016/j.jep.2016.03.036. [DOI] [PubMed] [Google Scholar]
- Muganza D. M.; Fruth B.; Nzunzu J. L.; Tuenter E.; Foubert K.; Cos P.; Maes L.; et al. In Vitro Antiprotozoal Activity and Cytotoxicity of Extracts and Isolated Constituents from Greenwayodendron Suaveolens. J. Ethnopharmacol. 2016, 193, 510–516. 10.1016/j.jep.2016.09.051. [DOI] [PubMed] [Google Scholar]
- Lima R. B.; Rocha Silva L. F.; Melo M. R. S.; Costa J. S.; Picanço N. S.; Lima E. S.; Vasconcellos M. C.; Boleti A. P. A.; Santos J. M. P.; Amorim R. C. N.; et al. In Vitro and in Vivo Anti-Malarial Activity of Plants from the Brazilian Amazon. Malar. J. 2015, 14, 508. 10.1186/s12936-015-0999-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos Dias M. M.; Martino H. S. D.; Noratto G.; Roque-Andrade A.; Stringheta P. C.; Talcott S.; Ramos A. M.; Mertens-Talcott S. U. Anti-Inflammatory Activity of Polyphenolics from Açai (Euterpe oleracea Martius) in Intestinal Myofibroblasts CCD-18Co Cells. Food Funct. 2015, 1–8. 10.1039/c5fo00278h. [DOI] [PubMed] [Google Scholar]
- Parsons A. B.; Lopez A.; Givoni I. E.; Williams D. E.; Gray C. A.; Porter J.; Chua G.; Sopko R.; Brost R. L.; Ho C. H.; et al. Exploring the Mode-of-Action of Bioactive Compounds by Chemical-Genetic Profiling in Yeast. Cell 2006, 126, 611–625. 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Dong J.; Lai R.; Jennings J. L.; Link A. J.; Hinnebusch A. G. The Novel ATP-Binding Cassette Protein ARB1 Is a Shuttling Factor That Stimulates 40S and 60S Ribosome Biogenesis. Mol. Cell. Biol. 2005, 25, 9859–9873. 10.1128/MCB.25.22.9859-9873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C.; Roe S. M.; McCormack E. A.; Beuron F.; Pearl L. H.; Willison K. R. The Crystal Structure of Yeast CCT Reveals Intrinsic Asymmetry of Eukaryotic Cytosolic Chaperonins. EMBO J. 2011, 30, 3078–3090. 10.1038/emboj.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heessen S.; Masucci M. G.; Dantuma N. P. The UBA2 Domain Functions as an Intrinsic Stabilization Signal That Protects Rad23 from Proteasomal Degradation. Mol. Cell 2005, 18, 225–235. 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Young J. C.; Agashe V. R.; Siegers K.; Hartl F. U. Pathways of Chaperone-Mediated Protein Folding in the Cytosol. Nat. Mol. Cell. Biol. 2004, 5, 781–791. 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Neef D. W.; Turski M. L.; Thiele D. J. Modulation of Heat Shock Transcription Factor 1 as a Therapeutic Target for Small Molecule Intervention in Neurodegenerative Disease. PLoS Biol. 2010, 8, e1000291 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U.; Bracher A.; Hayer-hartl M. Molecular Chaperones in Protein Folding and Proteostasis. Nature 2011, 475, 324–332. 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Mbengue A.; Vialla E.; Berry L.; Fall G.; Audiger N.; Demettre-Verceil E.; Braun-Breton C. New Export Pathway in Plasmodium falciparum-Infected Erythrocytes: Role of the Parasite Group II Chaperonin , PfTRiC. Traffic 2015, 16, 461–75. 10.1111/tra.12266. [DOI] [PubMed] [Google Scholar]
- Spillman N. J.; Beck J. R.; Ganesan S. M.; Niles J. C.; Daniel E.; Louis S. The Chaperonin TRiC Forms an Oligomeric Complex in the Malaria. Cell Microbiol. 2018, 19, 1–31. 10.1111/cmi.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batinovic S.; Mchugh E.; Chisholm S. A.; Matthews K.; Liu B.; Dumont L.; Charnaud S. C.; Schneider M. P.; Gilson P. R.; De Koning-ward T. F.; et al. An Exported Protein-Interacting Complex Involved in the Trafficking of Virulence Determinants in Plasmodium-Infected Erythrocytes. Nat. Commun. 2017, 8, 16044 10.1038/ncomms16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S.; Ashley E. A.; Ferreira P. E.; Zhu L.; Lin Z.; Chotivanich K.; Imwong M.; Pukrittayakamee S.; et al. Population Transcriptomics of Human Malaria Parasites Reveals the Mechanism of Artemisinin Resistance. Science 2015, 347, 431–435. 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert R.; Leroux M. R.; Scheufler C.; Hartl F. U.; Moarefi I. Structure of the Molecular Chaperone Prefoldin: Unique Interaction of Multiple Coiled Coil Tentacles with Unfolded Proteins. Cell 2000, 103, 621–632. 10.1016/S0092-8674(00)00165-3. [DOI] [PubMed] [Google Scholar]
- Johnson E. S. Protein Modification by SUMO. Annu. Rev. Biochem. 2004, 73, 355–382. 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Luikenhuis S.; Perrone G.; Dawes I. W.; Grant C. M. The Yeast Saccharomyces cerevisiae Contains Two Glutaredoxin Genes That Are Required for Protection against Reactive Oxygen Species. Mol. Biol. Cell 1998, 9, 1081–1091. 10.1091/mbc.9.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckers E.; Bien M.; Stroobant V.; Herrmann J. M.; Deponte M. Biochemical Characterization of Dithiol Glutaredoxin 8 from Saccharomyces cerevisiae: The Catalytic Redox Mechanism Redux. Biochemistry 2009, 48, 1410–1423. 10.1021/bi801859b. [DOI] [PubMed] [Google Scholar]
- Pujol-Carrion N.; De La Torre-Ruiz M. A. Glutaredoxins Grx4 and Grx3 of Saccharomyces cerevisiae Play a Role in Actin Dynamics through Their Trx Domains, Which Contributes to Oxidative Stress Resistance. Appl. Environ. Microbiol. 2010, 76, 7826–7835. 10.1128/AEM.01755-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco-Palencia L. A.; Mertens-Talcott S.; Talcott S. T. Chemical Composition, Antioxidant Properties, and Thermal Stability of a Phytochemical Enriched Oil from Açai (Euterpe oleracea Mart.). J. Agric. Food Chem. 2008, 56, 4631–4636. 10.1021/jf800161u. [DOI] [PubMed] [Google Scholar]
- Ainsworth E. A.; Gillespie K. M. Estimation of Total Phenolic Content and Other Oxidation Substrates in Plant Tissues Using Folin – Ciocalteu Reagent. Nat. Protoc. 2007, 2, 875–877. 10.1038/nprot.2007.102. [DOI] [PubMed] [Google Scholar]
- Trager W.; Jensen J. B. Human Malaria Parasites in Continuous Culture. Science 1976, 193, 673–675. 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Moneriz C.; Marín-García P.; Bautista J. M.; Diez A.; Puyet A. Haemoglobin Interference and Increased Sensitivity of Fluorimetric Assays for Quantification of Low-Parasitaemia Plasmodium Infected Erythrocytes. Malar. J. 2009, 8, 279. 10.1186/1475-2875-8-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quashie N. B.; de Koning H. P.; Ranford-Cartwright L. C. An Improved and Highly Sensitive Microfluorimetric Method for Assessing Susceptibility of Plasmodium falciparum to Antimalarial Drugs in Vitro. Malar. J. 2006, 5, 95. 10.1186/1475-2875-5-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce S. E.; Davis R. W.; Nislow C.; Giaever G. Genome-Wide Analysis of Barcoded Saccharomyces cerevisiae Gene-Deletion Mutants in Pooled Cultures. Nat. Protoc. 2007, 2, 2958–2974. 10.1038/nprot.2007.427. [DOI] [PubMed] [Google Scholar]
- Bilsland E.; Bean D. M.; Devaney E.; Oliver S. G. Yeast-Based High-Throughput Screens to Identify Novel Compounds Active against Brugia Malayi. PLoS Negl. Trop. Dis. 2016, 10, e0004401 10.1371/journal.pntd.0004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D.; Helliwell S. B.; Sadlish H.; Schuierer S.; Filipuzzi I.; Brachat S.; Bhullar B.; Plikat U.; Abraham Y.; Altorfer M.; et al. High-Resolution Chemical Dissection of a Model Eukaryote Reveals Targets, Pathways and Gene Functions. Microbiol. Res. 2014, 169, 107–120. 10.1016/j.micres.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Boyle E. I.; Weng S.; Gollub J.; Jin H.; Botstein D.; Michael J.; Sherlock G. GO::TermFinder—Open Source Software for Accessing Gene Ontology Information and Finding Significantly Enriched Gene Ontology Terms Associated with a List of Genes. Bioinformatics 2004, 20, 3710–3715. 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A.; Sidiropoulos K.; Viteri G.; Forner O.; Marin-garcia P.; Arnau V.; Eustachio P. D.; Stein L.; Hermjakob H. Reactome Pathway Analysis: A High- Performance in-Memory Approach. BMC Bioinf. 2017, 18, 1–9. 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.