Abstract
In three alternative splice variants of Homer 1 transcripts, Homer 1a mRNA has been shown to be upregulated selectively and rapidly by neural stimulation and represents a member of the immediate early gene (IEG) family. We investigated the mechanism underlying Homer 1a mRNA induction in cerebellar granule cell culture. All Homer 1 variants were expressed in cultured granule cells as analyzed by RNA blotting and immunochemical characterization. Glutamate stimulation of granule cells selectively upregulated Homer 1a mRNA via NMDA receptor-mediated influx of extracellular calcium. The induction of Homer 1a mRNA was much slower (peaked at 4 hr) and sustained longer than that of the typical IEG c-fos mRNA. Actinomycin D and cycloheximide experiments have revealed that, despite the presence of the mRNA-destabilizing AU-rich motif, transcriptional activation is a main determinant for selective Homer 1a mRNA induction. Inhibitor analysis as well as immunochemical characterization has indicated that the MEK (MAPK/ERK kinase)–ERK (extracellular signal-regulated kinase) cascade plays an indispensable role in glutamate-stimulated induction of Homer 1a mRNA. Consistent with this observation, brain-derived neurotrophic factor, which is known to activate the ERK cascade, similarly upregulated Homer 1a mRNA. These results demonstrate that MAPK (mitogen-activated protein kinase) is a key mediator that links distinct extracellular stimuli to the transcriptional activation of Homer 1a mRNA.
Keywords: granule cell culture, Homer 1, NMDA receptor, brain-derived neurotrophic factor, calcium signaling, MAP kinase, transcriptional activation
Homer 1 (also termedvesl) is an immediate early gene (IEG) that originally was isolated as a neural activity-regulated gene product from seizure-stimulated rat hippocampus (Brakeman et al., 1997;Kato et al., 1997). The Homer family consists of three distinct genes, namely Homer 1, 2, and 3. They share a conserved N-terminal Ena/VASP homology 1 (EVH1) domain that binds to group I metabotropic glutamate receptors (mGluRs), inositol triphosphate receptors, and Shank family proteins (Tu et al., 1998, 1999; Xiao et al., 1998). Homer 1 and Homer 2 comprise three and two splice variants, named Homer 1a, 1b, and 1c, and Homer 2a and 2b, respectively (Xiao et al., 1998). All but Homer 1a are expressed constitutively and possess a C-terminal coiled coil (CC) domain that serves to form a multimeric Homer complex and facilitates the assembly of signaling complexes. In contrast, Homer 1a lacks the C-terminal CC domain and is upregulated dynamically by various synaptic activities associated with neural plasticity, seizure, visual stimuli, and cocaine administration (Brakeman et al., 1997; Kato et al., 1997; Park et al., 1997). Induction of Homer 1a by neural stimulation therefore is believed to compete with the constitutive CC–Homers and thus to disassemble the association of multiple CC–Homer complexes.
Recent studies have shown a role of Homer complexes in receptor surface expression (Ciruela et al., 1999, 2000; Roche et al., 1999), receptor clustering (Tadokoro et al., 1999), and mGluR coupling to ion channels (Tu et al., 1998; Kammermeier et al., 2000). However, little is known about how neural activity selectively upregulates Homer 1a expression. The 3′-untranslated region (3′-UTR) of Homer 1a mRNA, but not Homer 1b/c mRNA, possesses a characteristic repeat of the AU-rich motif (Xiao et al., 1998), which is implicated in the destabilization of short-lived mRNAs (for review, see Chen and Shyu, 1995). Because Homer 1a and Homer 1b/c represent products of the same gene, the dynamic expression of Homer 1a has been suggested to be regulated by turnover of the Homer 1 transcripts, splicing, or transcript termination (Xiao et al., 1998). However, the mechanism underlying activity-dependent Homer 1a mRNA induction remained to be determined.
This study has focused on the regulatory mechanism of Homer 1a mRNA induction. To gain insights into the mechanism of Homer 1a mRNA expression, we used in vitro culture of cerebellar granule cells with a high uniformity and abundance of specific neuronal cells. This study has indicated that the activation of NMDA receptors and brain-derived neurotrophic factor (BDNF) selectively upregulated Homer 1a mRNA in cultured granule cells. We also report that the downstream signals of these extracellular stimuli converge into activation of the mitogen-activated protein kinase (MAPK) and lead to transcriptional induction of Homer 1a mRNA.
MATERIALS AND METHODS
Culture of cerebellar granule cells. Primary cultures of cerebellar granule cells were prepared from 8-d-old (P8) ICR mice (Japan SLC, Hamamatsu, Japan) according to the procedures described byFischer (1982) with minor modifications. Briefly, cerebella from P8 mice were treated with 0.1% trypsin and 0.05% DNase I in Ca2+/Mg2+-free HBSS at 37°C for 15 min and tritiated by pipetting in DNase I containing Ca2+-free HBSS. Dissociated cells were suspended in growth medium composed of basal medium Eagle (Sigma, St. Louis, MO) supplemented with 5% horse serum, 100 U/ml penicillin, and 0.1 mg/ml streptomycin (all from Life Technologies, Rockville, MD), 1 mg/ml bovine serum albumin (BSA), 10 μg/ml insulin, 0.1 nml-thyroxine, 0.1 mg/ml transferrin, 1 μg/ml aprotinin, 30 nm selenium, 0.25% glucose, and 2 mg/ml sodium bicarbonate (all from Sigma). Cells were plated at a density of 2 × 105cells/cm2 onto poly-d-lysine-coated 100 mm culture dishes (Becton Dickinson, Franklin Lakes, NJ). On the following day the cultures were switched to serum-free growth medium. Immunocytological characterization with an antibody against β-tubulin type III showed that the cultures contained ∼90% neuronal cells. In some experiments the cells were cultured in the medium containing 25 mm KCl with or without 5% fetal calf serum and 5% horse serum and were subjected to glutamate stimulation as described below.
Cell stimulation. At 5 d after plating the cultured cells were washed and preincubated at 37°C for 20 min in Mg2+-free Locke's solution consisting of (in mm) 154 NaCl, 5.6 KCl, 3.6 NaHCO3, 1.3 CaCl2, 5.6 glucose, and 5 HEPES, pH 7.3, plus 1 μm tetrodotoxin and 10 μm glycine. Cultures were maintained at 37°C in the humidified atmosphere of 5% CO2. Cells were stimulated by the addition of glutamate (final concentration, 10 μm) or BDNF (final concentration, 100 ng/ml; Life Technologies) to the preincubation solution; BDNF was dissolved in the preincubation medium containing 0.1% BSA. At the end of glutamate or BDNF stimulation the cells were washed with PBS and subjected to RNA extraction by the guanidine isothiocyanate method (TRIzol Reagent; Life Technologies). Cycloheximide, actinomycin D, and kinase/phosphatase inhibitors were added for 30 min before glutamate stimulation, and other antagonists were added for 20 min. Drugs were purchased from the following sources:d-2-amino-5-phosphonovalerate (AP5), 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), MK-801, and AMPA from Tocris (Bristol, UK); tetrodotoxin from Sankyo (Tokyo, Japan); nifedipine from Nacalai Tesque (Kyoto, Japan); cycloheximide from Wako (Osaka, Japan); NMDA and actinomycin D from Sigma; KN-62 from Seikagaku (Tokyo, Japan); PD98059, U0126, SB203580, FK506, and bisindolylmaleimide I from Calbiochem (San Diego, CA).
RNA blotting. Total RNA (10 μg) was electrophoresed on a formalin-containing 0.8% agarose gel and blotted onto a nylon membrane. [32P]-labeled DNA probes were synthesized by using a random priming method. Hybridization was performed in QuikHyb hybridization solution (Stratagene, La Jolla, CA) at 68°C for at least 2 hr. The membrane was washed twice with 0.1× SSC (15 mm NaCl plus 1.5 mm sodium citrate) and 0.1% SDS at 60°C for 30 min and then exposed to a BAS 5000 Imaging Plate (Fuji, Tokyo, Japan). Radioactivities were quantified with a BAS 5000 Bioimaging Analyzer (Fuji). Amounts of loaded RNA were determined by ethidium bromide staining of 18S ribosomal RNA. Linearity of hybridization signals was verified with a standard RNA sample. Probes used for RNA blotting were as follows: Homer 1a, nucleotide residues 1342–2139 (GenBank accession number AF093257); Homer 1b/c, residues 785–1396 (GenBank accession number AF093258); pan-Homer 1, residues 592–1055 (GenBank accession number U92079); c-fos, residues 1609–2693 (GenBank accession number V00727) from which an intron sequence of residues 1850–2254 was deleted. All of these cDNA fragments were synthesized by RT-PCR and subcloned into pBluescript II (Stratagene). Human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) probe was purchased from Clontech (Palo Alto, CA).
Reverse transcription-PCR. RT-PCR of granule cell RNA was performed as described previously (Minoshima and Nakanishi, 1999). The 5′ and 3′ primers used were as follows: for Homer 1a, 5′-primer, 5′-CAATTCAGCAATCATGATTA-3′ (residues 1342–1361); 3′-primer, 5′-ATCCTATGCCCCTATTAATG-3′ (residues 2120–2139); for Homer 1b and 1c, 5′-primer, 5′-AAGTCGCAGGAGAAGATGGA-3′ (residues 498–517); 3′-primer, 5′-CTCAGTGACCCGCTTGTGCA-3′ (residues 826–845). Amplification was conducted with 35 cycles of 95, 59, and 72°C for 1 min each, followed by further incubation at 72°C for 8 min. The amplified products were electrophoresed on a 2.0% agarose gel containing 0.5 μg/ml ethidium bromide. A single DNA fragment with an approximate size of 800 bp (Homer 1a) and doublet DNA fragments with approximate sizes of 350 bp (Homer 1b) and 380 bp (Homer 1c) were identified, extracted, and sequenced with ABI PRISM 310 Genetic Analyzer (PerkinElmer, Foster City, CA).
Immunocytochemistry. Cells were fixed with 10% formalin in PBS for 15 min and incubated with primary antibodies for 1 hr at room temperature, followed by incubation with appropriate secondary antibodies. For counterstaining of cell nuclei, DAPI (10 μg/ml; Sigma) was included in the secondary antibody solution. All antibodies were diluted with 2% normal goat serum, 0.1% Triton X-100, and 0.02% sodium azide in PBS. The primary antibodies that were used were obtained from the following sources and diluted as indicated in parentheses: rabbit anti-pan-Homer 1 antibody (1:2000), a gift from Dr. P. F. Worley (Johns Hopkins University, Baltimore, MD); rabbit and mouse anti-β-tubulin type III antibodies from Babco (1:600 and 1:300, respectively; Richmond, CA); mouse anti-glial fibrillary acidic protein (GFAP) antibody from Sigma (1:300); and mouse anti-phospho-ERK 1/2 antibody from New England Biolabs (1:400; Beverly, MA). The secondary antibodies that were used were fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG antibody (1:1000; Cappel, Durham, NC) and Texas Red X-conjugated anti-mouse IgG antibody (1:1000; Molecular Probes, Eugene, OR). Fluorescent images were acquired via a Carl ZeissAxiophot epifluorescence microscope equipped with a 63× Plan-APOCHROMAT objective (numerical aperture 1.40) and a cooled CCD camera (Roper, Trenton, NJ). Images for presentation were prepared with IPLab software (Scanalytics, Fairfax, VA).
Western blotting. Total cell lysates were prepared from cultures by boiling in SDS-PAGE sample buffer. An aliquot of the lysate was separated by SDS-PAGE and blotted onto a polyvinylidene difluoride membrane. The Homer 1 proteins were immunoblotted with rabbit anti-pan-Homer 1 antibody (1:2000). ERK 1/2 and phospho-ERK 1/2 were immunoblotted with rabbit anti-ERK 1/2 antibody and rabbit anti-phospho-ERK 1/2 antibody (1:1000; New England Biolabs), respectively. The immunoblots were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody and developed with ECL reagents (Amersham, Buckinghamshire, UK). The band intensity was quantified with GS-710 Calibrated Imaging Densitometer (Bio-Rad, Hercules, CA).
Cell survival assay. Cell viability was measured by using LIVE/DEAD Viability/Cytotoxicity Kit (Molecular Probes). Briefly, cells were stained with 2 μm calcein-AM and 4 μmethidium homodimer-1 (EthD-1) in PBS for 30 min at room temperature. Calcein-AM-stained cells were judged as live cells and cells stained with EthD-1 as dead cells. Cell viability was calculated at randomly chosen fields, and ∼1000 cells were counted in each culture.
Statistical analysis. Data were analyzed statistically by Student's t test.
RESULTS
Homer 1 expression in cultured cerebellar granule cells
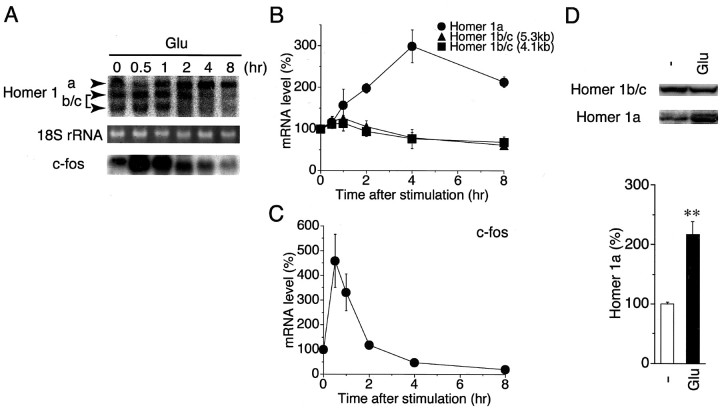
Expression of Homer 1 mRNA in cultured cerebellar granule cells was examined first to test for their feasibility in studying the regulation of Homer 1a expression. Granule cells were prepared from 8-d-old mice and maintained in culture with a serum-free growth medium. Total RNA was extracted from the culture at 5 d in vitro, and expression of Homer 1 mRNA was examined by RNA blotting. Hybridization with a Homer 1a-specific probe gave rise to a major band with an estimated size of ∼6.8 kb (Fig.1A). This size corresponded to that reported previously for Homer 1a mRNA (Xiao et al., 1998). Levels of Homer 1a mRNA in cultured granule cells were comparable with those in postnatal and adult cerebella (Fig.1A). A Homer 1b/c-specific probe yielded two bands with estimated sizes of ∼5.3 and 4.1 kb from both cultured cells and brain tissues (Fig. 1B). A probe covering a common sequence of the Homer 1 splice variants provided all three bands with sizes of 6.8, 5.3, and 4.1 kb (see Fig. 2A). The results indicate that all splice variants of Homer 1 mRNA are expressed in cultured cerebellar granule cells. We used the pan-Homer 1 probe for the subsequent experiments.
Fig. 1.

Expression of Homer 1 in cultured cerebellar granule cells. A, RNA blotting with the Homer 1a probe. Total RNAs (10 μg per lane) were prepared from cultured granule cells (GC) and 8-d-old (P8) and adult (Ad) cerebella and hybridized with a Homer 1a probe. The sizes of an RNA ladder are indicated by kilobases (kb) on the right. An arrowhead indicates Homer 1a mRNA. B, RNA blotting with the Homer 1b/c probe. The arrowheads show Homer 1b/c mRNAs.C, Total cell lysate from cultured granule cells (GC) was immunoblotted with anti-pan-Homer 1 antibody or without the addition of the primary antibody. A doublet of Homer 1a (H1a) and a band of Homer 1b/c (H1b/c) are seen in GC lysates at the appropriate positions as compared with the molecular sizes (kDa) of the marker proteins indicated on the right. D, Homer 1 immunoreactivity (green) was detected as puncta on soma and neurites of granule cells. Granule cells were labeled with anti-β-tubulin type III antibody (red). Cell nuclei were counterstained with DAPI (blue). Scale bar, 10 μm.
Fig. 2.
Temporal induction patterns of mRNAs and the increase of Homer 1a protein in cultured granule cells after glutamate stimulation. A–C, Cells were treated with 10 μm glutamate (Glu) for the indicated times, and levels of variant Homer 1 mRNAs and c-fosmRNA were quantified by RNA blotting of total extracted RNAs (10 μg per lane). A, A representative blot of variant Homer 1 mRNAs (top) and c-fos mRNA (bottom). The middle panel shows a reference of RNA loading as analyzed by ethidium bromide staining of 18S rRNA. B, C, The induction time courses of variant Homer 1 mRNAs and c-fos mRNA are indicated. InB, glutamate upregulated Homer 1a mRNA, but not Homer 1b/c mRNAs, of either 5.3 or 4.1 kb. In C,c-fos mRNA was induced more rapidly and transiently after glutamate stimulation. Data represent mean ± SEM (n = 3). D, Cultured cells were incubated with or without 10 μm glutamate for 8 hr, and levels of Homer 1a and 1b/c were quantified by immunoblotting of total cell lysates with anti-pan-Homer 1 antibody. Representative blots of Homer 1a and 1b/c are indicated at the top; data at the bottom show mean ± SEM (n= 3; **p < 0.01).
To verify the expression of Homer 1 protein in granule cells, we subjected cell lysates to Western blotting with an anti-pan-Homer 1 antibody. The antibody gave rise to a 28/29 kDa doublet of Homer 1a and a 45 kDa band of Homer 1b/c (Fig.1C; Brakeman et al., 1997; Xiao et al., 1998). Immunostaining of cultured cerebellar cells with anti-pan-Homer 1 antibody showed that Homer 1 immunoreactivity is localized in granule cells that are immunostained with the antibody against neuronal marker β-tubulin type III (Fig. 1D), but not in astrocytes labeled by the glial marker anti-GFAP antibody (data not shown). A punctate pattern of pan-Homer 1 immunoreactivity was seen on soma and along neurites of granule cells (Fig. 1D). The results indicate that both inducible and noninducible Homer 1 mRNAs are expressed and translated into the corresponding isoforms in cultured granule cells.
During preparation of this manuscript Ango et al. (2000) have reported that none of the Homer 1 variant proteins is expressed in cultured mouse cerebellar granule cells under the nonstimulatory condition. We further examined the expression of Homer 1 mRNA in cultured granule cells. We sought to synthesize cDNA fragments of individual Homer 1 mRNAs from nonstimulatory granule cells, using RT-PCR techniques with appropriate primers (see Materials and Methods). The sequence determination of resultant cDNA products revealed appreciable amounts of expression of all three variant Homer 1 mRNAs under the nonstimulatory condition. In our experiments the granule cells were cultured in the serum-free medium with a low concentration of KCl (5.4 mm), thus avoiding an unnecessary depolarization of granule cells (Bessho et al., 1994). In contrast, Ango et al. (2000) used the serum-containing medium with a high concentration of KCl (25 mm). Therefore, we cultured granule cells with high KCl and found that all three Homer 1 variant mRNAs were expressed in the medium containing 25 mm KCl also, regardless of the presence and absence of 5% horse serum and 5% fetal calf serum (data not shown). The different observations between the two investigations remain to be clarified but may result from the different sensitivity in detecting Homer 1 immunoreactivity or the difference in mouse strains that were used, or otherwise different culture conditions not yet identified.
Homer 1a mRNA is upregulated preferentially by glutamate
To address whether Homer 1a mRNA expression is regulated in an activity-dependent manner, we stimulated cells with glutamate (10 μm) and determined mRNA levels of Homer 1 splice variants by RNA blotting (Fig.2A,B). Homer 1a mRNA was induced as early as 1 hr after glutamate stimulation. This induction peaked at 4 hr and was sustained up to 8 hr after stimulation. In contrast, changes of Homer 1b/c mRNA levels were minimal; Homer 1b/c mRNAs exhibited a fairly small increase at 1 hr after glutamate stimulation, returned to the basal level at 2 hr, and slightly decreased below the basal level at 8 hr. The observed temporal pattern of Homer 1a mRNA induction seemed to be relatively slow as compared with those of other IEGs (Szekely et al., 1989). As an example, c-fos mRNA rapidly and transiently increased after glutamate stimulation, peaked within 1 hr, and fell back to basal levels at 2 hr (Fig. 2A,C). In control, levels of the housekeeping GAPDH mRNA were found not to be altered after glutamate stimulation (data not shown). The results demonstrate that Homer 1a mRNA is upregulated preferentially with a temporal profile distinct from a typical IEG mRNA in cultured granule cells after glutamate stimulation.
The culture of granule cells at high KCl (25 mm) is known to promote their survival (Bessho et al., 1994). Under our experimental condition with low KCl, cell viability was unchanged during glutamate stimulation; percentages of viable cells/total cells were calculated to be 87 ± 1.6, 88 ± 1.0, and 84 ± 2.9% at 0, 4, and 8 hr after the addition of glutamate, respectively (mean ± SEM;n = 4). Furthermore, similar extents of glutamate-stimulated Homer 1a mRNA induction were observed under culture conditions with high KCl (25 mm).
Western blot analysis showed that treatment of granule cells with glutamate for 8 hr increased Homer 1a, but not Homer 1b/c (Fig.2D). Similarly, Ango et al. (2000) have reported recently that the addition of NMDA and kainate increased the levels of Homer 1a protein in granule cells cultured in the medium containing high KCl together with horse and fetal calf sera. These results indicate that glutamate stimulation selectively upregulates Homer 1a mRNA and its translation product in cultured granule cells. In this and subsequent experiments, Homer 1b/c mRNAs of 5.3 and 4.1 kb showed a common profile not only in expression patterns but also in responsiveness to various activators and inhibitors that were tested. The data of Homer 1b/c mRNA of 5.3 kb are presented in the subsequent analysis.
Role of NMDA receptors and calcium influx in Homer 1a mRNA induction
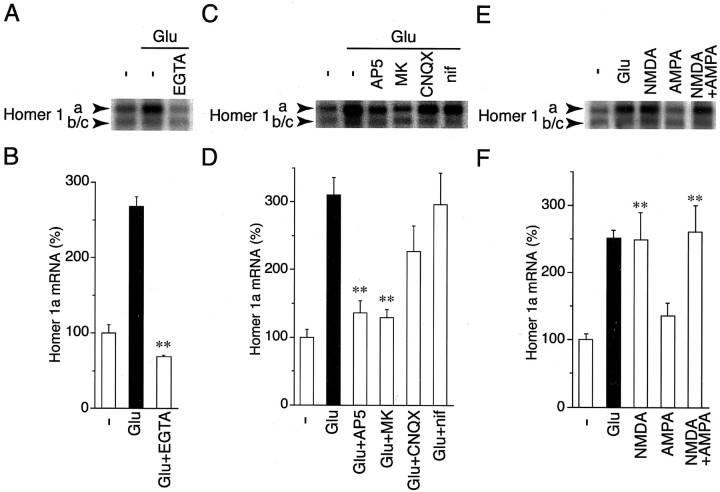
A number of studies have revealed that calcium influx plays a crucial role in activity-regulated gene expression in neuronal cells (for review, see Ghosh and Greenberg, 1995; Bito et al., 1997). To address whether Homer 1a mRNA upregulation is controlled by calcium influx, we stimulated cultured cells by glutamate in an EGTA-containing medium for 4 hr. Chelation of extracellular calcium by EGTA abrogated Homer 1a mRNA induction by glutamate (Fig.3A,B), indicating that an influx of extracellular calcium into neuronal cells is necessary for Homer 1a mRNA induction.
Fig. 3.
Homer 1a mRNA induction by NMDA receptor-mediated calcium influx. A,C, and E show representative blots of Homer 1a mRNA and Homer 1b/c mRNA of 5.3 kb. B,D, and F indicate quantitative data of RNA blotting analysis. A, B, Cells were preincubated for 20 min in medium containing 2 mm EGTA (EGTA) or without EGTA (−) and further incubated with or without a treatment of 10 μm glutamate for 4 hr. Chelation of extracellular calcium with EGTA abolished the glutamate-induced Homer 1a mRNA increase (n = 3; **p < 0.01;Glu vs Glu+EGTA). C, D, Cultured cells were preincubated for 20 min with 100 μmAP5, 10 μm MK-801 (MK), 30 μm CNQX, or 10 μm nifedipine (nif) and further incubated with 10 μm glutamate for 4 hr (n = 3–4). AP5 and MK-801 suppressed Homer 1a mRNA induction up to unstimulated levels (**p < 0.01; Glu vsGlu+AP5 or Glu+MK). CNQX slightly reduced Homer 1a mRNA induction, but this reduction was not statistically significant (p = 0.12).E, F, Cultured cells were incubated with glutamate (10 μm), NMDA (30 μm), AMPA (30 μm), or NMDA (30 μm) plus AMPA (30 μm) for 4 hr. Homer 1a mRNA was induced by NMDA, but not by AMPA, and this induction was not enhanced by the addition of AMPA (n = 4–7; **p < 0.01; unstimulated vs NMDA or NMDA+AMPA).
To define subtypes of glutamate receptors and a route of calcium entry responsible for Homer 1a mRNA induction, we treated the cells with several antagonists selective to calcium-permeating receptors and ion channels. Homer 1a mRNA induction was blocked completely by two different types of NMDA receptor antagonists, AP5 and MK-801 (Fig.3C,D). In contrast, a non-NMDA receptor antagonist, CNQX, and an L-type calcium channel blocker, nifedipine, showed only a partial or no inhibitory effect on Homer 1a mRNA induction, respectively (Fig. 3C,D). Consistent with this observation, the addition of NMDA (30 μm) upregulated Homer 1a mRNA (Fig. 3E,F). In contrast, AMPA (30 μm) neither induced Homer 1a mRNA significantly nor enhanced NMDA-stimulated Homer 1a mRNA upregulation (Fig.3E,F). These results demonstrate that NMDA receptor-mediated calcium influx specifically upregulates Homer 1a mRNA in cultured granule cells.
Transcriptional regulation of Homer 1a mRNA induction
Gene expression generally is controlled by either transcriptional activation or stabilization of mRNA, or both. As shown in Figure4, A and B, the addition of the RNA polymerase inhibitor actinomycin D (10 μg/ml) prevented glutamate-stimulated Homer 1a mRNA induction, suggesting that transcriptional activation is required for glutamate-evoked Homer 1a mRNA upregulation. The 3′-UTR of Homer 1a mRNA, but not Homer 1b/c mRNA, contains a repeat of the AU-rich motif (Xiao et al., 1998). This motif has been implicated in the destabilization of short-lived mRNAs (for review, see Chen and Shyu, 1995). To address whether Homer 1a mRNA is either short-lived or stable, we examined the turnover of Homer 1a mRNA by actinomycin D chase experiments. Granule cells were treated with actinomycin D (10 μg/ml) in the presence and absence of glutamate, and temporal changes in levels of splice variant Homer 1 mRNAs as well as c-fos mRNA were determined by RNA blotting analysis (Fig. 4C–F). Homer 1a mRNA was relatively stable, becoming ∼80% of control levels at 2 hr after actinomycin D treatment, regardless of the presence and absence of glutamate (Fig.4C–E). This stability of Homer 1a mRNA was comparable with that of Homer 1b/c mRNAs (Fig. 4C–F) and in marked contrast to a rapid degradation of c-fos mRNA (Fig.4C,D).
Fig. 4.
The effect of actinomycin D on the induction and turnover of Homer 1 mRNAs. A, B, Cultured cells were preincubated with or without 10 μg/ml actinomycin D (Act D) for 30 min and further incubated for 4 hr in the presence and absence of 10 μm glutamate. Actinomycin D treatment abrogated Homer 1a mRNA induction by glutamate (n = 3; **p < 0.01;Glu vs Glu+Act D). C–F, Cultured cells were incubated either with 10 μm glutamate for 4 hr or without glutamate for 20 min. Then mRNA synthesis was inhibited by the addition of 10 μg/ml actinomycin D. Incubation was continued further, and total RNAs were prepared at the indicated times after the addition of actinomycin D. C,Top and middle panels show representative blots of variant Homer 1 mRNAs and c-fos mRNA, respectively, which were prepared from glutamate-untreated cells.C, Bottom, A representative blot of variant Homer 1 mRNAs prepared from cells treated with glutamate for 4 hr and then with actinomycin D for the indicated times.D, Shown is the turnover of Homer 1a, Homer 1b/c, andc-fos mRNAs in glutamate-untreated cells; the turnover is expressed as percentages of mRNA levels in cells immediately before (0 min) the addition of actinomycin D (n = 3–4). E, F, The turnover of Homer 1a and Homer 1b/c mRNAs, respectively, in cells with or without the addition of glutamate. The data are plotted as described in D (n = 3–4).
In many cases mRNAs that possess an AU-rich repeating motif and exhibit rapid turnover are superinduced by an inhibitor that blocks ongoing translation that is required for rapid mRNA degradation (for review, see Chen and Shyu, 1995). To address whether ongoing protein synthesis also is involved in the regulation of Homer 1 mRNA expression, we examined the effects of the translation inhibitor cycloheximide (30 μg/ml) on Homer 1a mRNA levels in granule cells. At this concentration, cycloheximide was found to inhibit >95% of protein synthesis as measured by [3H]-leucine incorporation into protein (data not shown). Cycloheximide alone slightly increased Homer 1a mRNA levels (Fig.5A,B). However, this effect was not specific to Homer 1a mRNA, and a comparable increase of Homer 1b/c mRNAs was observed by cycloheximide treatment (Fig.5A,B). In contrast, a notable increase of c-fosmRNA levels was observed by the addition of cycloheximide (Fig.5A,B).
Fig. 5.
Effects of cycloheximide on Homer 1a mRNA expression. A, B, Cultured cells were treated with or without 30 μg/ml cycloheximide (CHX) for 4 hr. mRNA levels were quantified by RNA blotting analysis (n = 3–4). Cycloheximide slightly increased both Homer 1a and Homer 1b/c mRNAs, and no selective accumulation of Homer 1a mRNA was observed. In contrast, the c-fos mRNA level was elevated markedly by the same treatment. C, D, Cultured cells were preincubated with or without 30 μg/ml cycloheximide for 30 min and incubated further for 4 hr in the presence or absence of 10 μm glutamate. Induction of Homer 1a mRNA by glutamate was not blocked by cycloheximide treatment (n = 3–4).
Homer 1a mRNA was induced more slowly and sustained longer than that ofc-fos mRNA (see Fig. 2B,C). This finding raised the possibility that Homer 1a mRNA may require the previous induction of certain IEGs. However, cycloheximide treatment during glutamate stimulation had no inhibitory effect on the upregulation of Homer 1a mRNA (Fig. 5C,D). Rather, cycloheximide increased Homer 1a mRNA levels (Fig. 5C,D), probably reflecting a common effect of cycloheximide on the expression of Homer 1 splice variants (Fig. 5A,B). The result shows that the induction of Homer 1a mRNA by glutamate is regulated by preexisting transcriptional machinery. Collectively, the results of both actinomycin D and cycloheximide indicate that glutamate preferentially activates the transcription of Homer 1a mRNA in cultured granule cells.
Extracellular signal-regulated kinase (ERK) signaling in Homer 1a mRNA induction
An elevation of intracellular calcium is known to activate protein kinase/phosphatase signaling cascades and lead to the activation of gene transcription in neuronal cells. These calcium-dependent signaling cascades include the activation of calmodulin-dependent protein kinases (CaMK), ERK (a member of the MAPK superfamily), protein kinase C (PKC), and the calcineurin phosphatase (Bading et al., 1993; Bito et al., 1996; Xia et al., 1996; Graef et al., 1999). We addressed whether any of these calcium-dependent signaling cascades are involved in the glutamate-stimulated Homer 1a mRNA induction. Cultured granule cells were treated with various kinase/phosphatase inhibitors together with glutamate for 4 hr, and the effects of these inhibitors on Homer 1a mRNA induction were examined. Inhibition of the MEK (MAPK/ERK kinase)–ERK cascade by the MEK inhibitors PD98059 (30 μm) and U0126 (5 μm) both completely abrogated Homer 1a mRNA induction by glutamate (Fig.6), indicating that the MEK–ERK pathway is involved in Homer 1a mRNA induction. The inhibitory effect on Homer 1a mRNA induction seemed to be ERK-specific, because such potent inhibition was not observed by the addition of SB203580 (5 μm), which is a specific inhibitor for another member of the MAPK superfamily, p38. Also, Homer 1a mRNA induction was not inhibited significantly by treatments with either the CaMK-selective inhibitor KN-62 (10 μm), the calcineurin inhibitor FK506 (1 μm), or the PKC inhibitor bisindolylmaleimide I (1 μm).
Fig. 6.
Blockade of Homer 1a mRNA induction by inhibitors of the ERK cascade. A–D, Cultured cells were preincubated for 30 min with the following inhibitors: 30 μm PD98059 (PD), 10 μm KN-62 (KN), 5 μm U0126 (U), 5 μm SB203580 (SB), 1 μm FK506 (FK), and 1 μm bisindolylmaleimide I (Bis I). These cells were incubated further for 4 hr with the addition of 10 μm glutamate in the presence of the respective inhibitors. B, D, Levels of Homer 1a mRNA were quantified by RNA blotting. The ERK cascade inhibitors PD98059 and U0126 blocked Homer 1a mRNA induction by glutamate (n = 3–5; **p < 0.01;Glu vs Glu+PD orGlu+U).
To examine further the importance of the ERK cascade for NMDA receptor-mediated Homer 1a mRNA induction, we monitored the enhancement of ERK phosphorylation by immunostaining with the anti-phospho-ERK 1/2 antibody that specifically recognizes the activated form of ERK 1/2. Glutamate stimulation rapidly increased an immunoreactivity of phospho-ERK 1/2 in almost all β-tubulin type III-immunopositive granule cells (Fig. 7A). This enhancement of ERK 1/2 phosphorylation was blocked by the MEK inhibitor PD98059 (Fig. 7A). The time course of ERK 1/2 phosphorylation, as analyzed by immunoblotting, showed that the glutamate-stimulated ERK 1/2 phosphorylation was rapid, peaking by 5 min after the addition of glutamate and sustained at levels higher than the basal level up to at least 30 min (Fig. 7B,C). In control, no increase in the level of ERK 1/2 protein was observed during this time course (Fig. 7B). In addition, immunoblotting showed that ERK 1/2 phosphorylation was enhanced by treatment of the granule cells with NMDA (30 μm) for 5 min (Fig. 7D). Furthermore, glutamate-stimulated ERK 1/2 phosphorylation was blocked by either depletion of extracellular calcium with EGTA (2 mm) or addition of the NMDA receptor antagonist MK-801 (10 μm) (Fig. 7D). The results support the view that ERK activation is a key event linking calcium influx via NMDA receptors to transcriptional activation of Homer 1a mRNA.
Fig. 7.

ERK phosphorylation by glutamate stimulation.A, Cultured cells were preincubated with either 30 μm PD98059 (PD) or vehicle for 30 min and treated with or without 10 μm glutamate for 5 min. These cells were fixed and immunostained with anti-phospho-ERK 1/2 (red) and anti-β-tubulin type III antibodies (green). Cell nuclei were counterstained with DAPI (blue). B, Cells were treated with 10 μm glutamate for the indicated times, and cell lysates were immunoblotted with anti-phospho-ERK 1/2 and anti-ERK 1/2 antibodies. C, Levels of phospho-ERK 2 were quantified by immunoblotting. Values shown represent mean ± SEM (n = 3–4) and are expressed as percentages of the control values obtained from glutamate-unstimulated conditions.D, Cells were incubated with 10 μmglutamate, 30 μm NMDA, or vehicle for 5 min (left 3 lanes). Cells were preincubated for 20 min with 10 μm MK-801 (MK) or in the medium containing 2 mm EGTA (EGTA) and incubated further with or without 10 μm glutamate for 5 min (right 4 lanes). Cell lysates were immunoblotted with anti-phospho-ERK 1/2 and anti-ERK 1/2 antibodies.
BDNF-stimulated Homer 1a mRNA induction via a convergent ERK signaling
It has been shown that BDNF stimulates TrkB receptors and activates the ERK cascade in cerebellar granule cells (Zirrgiebel et al., 1995; Bonni et al., 1999). Both immunostaining and immunoblotting showed that ERK 1/2 phosphorylation was enhanced by the treatment of cultured granule cells with 100 ng/ml BDNF for 5 min (Fig.8A). We addressed whether BDNF has a similar ability to upregulate Homer 1a mRNA in cultured granule cells. Granule cells were treated with 100 ng/ml BDNF for 4 hr, and the effects of BDNF on Homer 1a mRNA induction were examined. BDNF treatment significantly and selectively upregulated Homer 1a mRNA (Fig. 8B,C). Homer 1a mRNA induction by BDNF was insensitive to the NMDA receptor antagonist, indicating that BDNF acts independently from the activation of NMDA receptors. A combined addition of glutamate and BDNF further enhanced Homer 1a mRNA levels. Because a high concentration of glutamate (100 μm) killed cultured granule cells, the maximum capability of glutamate to induce Homer 1a mRNA remains to be determined. Importantly, the MEK inhibitor blocked Homer 1a mRNA induction by both BDNF treatment alone and a combined addition of BDNF and glutamate (Fig. 8B,C). The results demonstrate that the ERK activation is indispensable for the upregulation of Homer 1a mRNA induced by the two distinct extracellular stimuli.
Fig. 8.

Homer 1a mRNA induction by the convergent ERK cascade after NMDA receptor and BDNF stimulation. A, Cells were treated with or without 100 ng/ml BDNF for 5 min, and cell lysates were immunoblotted with anti-phospho-ERK 1/2 and anti-ERK 1/2 antibodies. B, C, Cultured cells were preincubated for 30 min with 30 μm PD98059 (PD), 5 μm U0126 (U), 100 μmAP5, or vehicle and further incubated for 4 hr by the addition of 100 ng/ml BDNF, 10 μm glutamate, or both BDNF and glutamate as indicated. Levels of Homer 1a mRNA were quantified by RNA blotting. BDNF significantly upregulated Homer 1a mRNA, and this induction was blocked by PD98059, but not by AP5. Costimulation with BDNF (100 ng/ml) and glutamate (10 μm) enhanced Homer 1a mRNA induction, and this induction was blocked by U0126 (n = 3–5; **p < 0.01; unstimulated vs BDNF, BDNF vsBDNF+PD, or Glu+BDNF vsGlu+BDNF+U).
DISCUSSION
This study concerns the mechanism that underlies Homer 1a mRNA induction in granule cell culture in vitro. The results indicate that glutamate triggers a rapid and specific upregulation of Homer 1a mRNA in the members of Homer 1 splice variants. This selective induction is mediated by NMDA receptors and is dependent on extracellular calcium influx via the activation of NMDA receptor channels. The selective and rapid induction of Homer 1a mRNA in cultured granule cells is consistent with a dynamic upregulation of this mRNA in seizure-stimulated hippocampus (Brakeman et al., 1997;Kato et al., 1997; Xiao et al., 1998). However, induction levels of Homer 1a mRNA in cultured cells are much lower than those of seizure-induced Homer 1a mRNA in vivo (Xiao et al., 1998). Seizure may trigger complex neuronal responses and could transduce multiple signaling to activate the Homer 1 transcription maximally in vivo. Inhibitor analysis of calcium-relevant kinases/phosphatases has indicated that the MEK–ERK cascade plays a pivotal role in NMDA receptor-mediated induction of Homer 1a mRNA. Consistent with this observation, BDNF similarly induces Homer 1a mRNA via the MEK–ERK cascade. This investigation demonstrates that MAPK is a key mediator that links the extracellular stimuli to the transcriptional activation of Homer 1a mRNA.
The results of actinomycin D and cycloheximide experiments have indicated that, despite the presence of the AU-rich motif, transcriptional activation is a main determinant for selective Homer 1a mRNA induction. Furthermore, the induction of Homer 1a mRNA is regulated by a preexisting transcriptional machinery that responds to the activation of NMDA receptors and does not require protein synthesis. Therefore, although NMDA receptor stimulation has been shown to activate the BDNF gene (Bessho et al., 1993; Favaron et al., 1993), the induction of Homer 1a mRNA by glutamate results from the activation of the downstream ERK cascade rather than the secondary effect derived from the NMDA receptor-induced BDNF. Stimulation of NMDA receptors also upregulates a number of IEGs encoding transcription factors (for review, see Sheng and Greenberg, 1990). Importantly, the induction of Homer 1a mRNA by glutamate is slower and sustained longer than that of the typical IEG c-fos mRNA. Furthermore, unlike many other IEGs encoding transcription factors, Homer 1a mRNA is not superinduced by cycloheximide. More specifically, both induction kinetics and the effect of cycloheximide in Homer 1a mRNA upregulation resemble those for BDNF mRNA induction (Shieh et al., 1998; Tao et al., 1998). These similarities suggest that the mechanisms of Homer 1a mRNA regulation may be shared with those of BDNF mRNA, although they may differ, in part, from those of typical IEGs such as c-fosmRNA.
The mechanisms underlying ERK-mediated IEG induction have been well characterized in cultured neuronal cells as well as in vivo(Xia et al., 1996; Sgambato et al., 1998; Davis et al., 2000). In neuronal cells the ERK signaling is transmitted to the nucleus once it has been activated via serial phosphorylation events and can regulate transcriptional activity of many IEGs. Two DNA regulatory elements are, at least, crucial for this transcriptional activation: the cAMP response element (CRE) and the serum response element (SRE). The CRE site is controlled by the CRE-binding protein (CREB), whereas the SRE site is targeted by the ternary complex factor Elk-1. It therefore would be of interest to address whether Homer 1a mRNA is regulated coordinately via the interaction of these transcription factors with their targeting DNA elements.
Both NMDA receptors and BDNF as well as their downstream ERK signals have been shown to play a critical role in synaptic plasticity and long-term memory formation (for review, see Bliss and Collingridge, 1993; Impey et al., 1999; Schinder and Poo, 2000). These long-lasting changes in synaptic efficacy are thought to result from alterations not only in neuronal cell responsiveness but also in synaptic architectures (for review, see Lüscher et al., 2000). Recently, Homer proteins have been shown to regulate a supracomplex formation of synaptic proteins. They are thought to play a crucial role in signal transduction, synaptogenesis, and receptor trafficking at synapses (for review, see Xiao et al., 2000). Importantly, Homer 1a lacks the C-terminal CC domain that is involved in the assembly of protein complexes. It thus possesses the potential ability to compete with the actions of constitutive CC–Homer proteins. We therefore believe that the selective and sustained upregulation of Homer 1a expression by the extracellular stimuli can provide an effective, activity-dependent mechanism that allows for alterations in synaptic architectures of neuronal cells.
Footnotes
This work was supported in part by research grants from the Ministry of Education, Science, and Culture of Japan. M.S. is a fellow of the Japan Society for the Promotion of Science. We are grateful to Paul Worley for providing anti-pan-Homer 1 antibody. We also thank Haruhiko Bito and Hiroshi Kawasaki for invaluable advice and Kumlesh K. Dev for a careful reading of this manuscript.
Correspondence should be addressed to Shigetada Nakanishi, Department of Biological Sciences, Kyoto University Faculty of Medicine, Yoshida, Sakyo-ku, Kyoto 606-8501, Japan. E-mail:snakanis@phy.med.kyoto-u.ac.jp.
REFERENCES
- 1.Ango F, Pin JP, Tu JC, Xiao B, Worley PF, Bockaert J, Fagni L. Dendritic and axonal targeting of type 5 metabotropic glutamate receptor is regulated by Homer 1 proteins and neuronal excitation. J Neurosci. 2000;20:8710–8716. doi: 10.1523/JNEUROSCI.20-23-08710.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- 3.Bessho Y, Nakanishi S, Nawa H. Glutamate receptor agonists enhance the expression of BDNF mRNA in cultured cerebellar granule cells. Mol Brain Res. 1993;18:201–208. doi: 10.1016/0169-328x(93)90190-z. [DOI] [PubMed] [Google Scholar]
- 4.Bessho Y, Nawa H, Nakanishi S. Selective up-regulation of an NMDA receptor subunit mRNA in cultured cerebellar granule cells by K+-induced depolarization and NMDA treatment. Neuron. 1994;12:87–95. doi: 10.1016/0896-6273(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 5.Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca2+- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- 6.Bito H, Deisseroth K, Tsien RW. Ca2+-dependent regulation in neuronal gene expression. Curr Opin Neurobiol. 1997;7:419–429. doi: 10.1016/s0959-4388(97)80072-4. [DOI] [PubMed] [Google Scholar]
- 7.Bliss TVP, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 8.Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras–MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 9.Brakeman PR, Lanahan AA, O'Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- 10.Chen C-YA, Shyu A-B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 11.Ciruela F, Soloviev MM, McIlhinney RAJ. Co-expression of metabotropic glutamate receptor type 1α with Homer-1a/Vesl-1S increases the cell surface expression of the receptor. Biochem J. 1999;341:795–803. [PMC free article] [PubMed] [Google Scholar]
- 12.Ciruela F, Soloviev MM, Chan W-Y, McIlhinney RAJ. Homer-1c/Vesl-1L modulates the cell surface targeting of metabotropic glutamate receptor type 1α: evidence for an anchoring function. Mol Cell Neurosci. 2000;15:36–50. doi: 10.1006/mcne.1999.0808. [DOI] [PubMed] [Google Scholar]
- 13.Davis S, Vanhoutte P, Pagès C, Caboche J, Laroche S. The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. J Neurosci. 2000;20:4563–4572. doi: 10.1523/JNEUROSCI.20-12-04563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Favaron M, Manev RM, Rimland JM, Candeo P, Beccaro M, Manev H. NMDA-stimulated expression of BDNF mRNA in cultured cerebellar granule neurones. NeuroReport. 1993;4:1171–1174. [PubMed] [Google Scholar]
- 15.Fischer G. Cultivation of mouse cerebellar cells in serum-free, hormonally defined media: survival of neurons. Neurosci Lett. 1982;28:325–329. doi: 10.1016/0304-3940(82)90079-9. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh A, Greenberg ME. Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 17.Graef IA, Mermelstein PG, Stankunas K, Neilson JR, Deisseroth K, Tsien RW, Crabtree GR. L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature. 1999;401:703–708. doi: 10.1038/44378. [DOI] [PubMed] [Google Scholar]
- 18.Impey S, Obrietan K, Storm DR. Making new connections: role of ERK/MAP kinase signaling in neuronal plasticity. Neuron. 1999;23:11–14. doi: 10.1016/s0896-6273(00)80747-3. [DOI] [PubMed] [Google Scholar]
- 19.Kammermeier PJ, Xiao B, Tu JC, Worley PF, Ikeda SR. Homer proteins regulate coupling of group I metabotropic glutamate receptors to N-type calcium and M-type potassium channels. J Neurosci. 2000;20:7238–7245. doi: 10.1523/JNEUROSCI.20-19-07238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato A, Ozawa F, Saitoh Y, Hirai K, Inokuchi K. vesl, a gene encoding VASP/Ena family-related protein, is upregulated during seizure, long-term potentiation, and synaptogenesis. FEBS Lett. 1997;412:183–189. doi: 10.1016/s0014-5793(97)00775-8. [DOI] [PubMed] [Google Scholar]
- 21.Lüscher C, Nicoll RA, Malenka RC, Muller D. Synaptic plasticity and dynamic modulation of the postsynaptic membrane. Nat Neurosci. 2000;3:545–550. doi: 10.1038/75714. [DOI] [PubMed] [Google Scholar]
- 22.Minoshima T, Nakanishi S. Structural organization of the mouse metabotropic glutamate receptor subtype 3 gene and its regulation by growth factors in cultured cortical astrocytes. J Biochem. 1999;126:889–896. doi: 10.1093/oxfordjournals.jbchem.a022531. [DOI] [PubMed] [Google Scholar]
- 23.Park HT, Kang EK, Bae KW. Light regulates Homer mRNA expression in the rat suprachiasmatic nucleus. Mol Brain Res. 1997;52:318–322. doi: 10.1016/s0169-328x(97)00292-1. [DOI] [PubMed] [Google Scholar]
- 24.Roche KW, Tu JC, Petralia RS, Xiao B, Wenthold RJ, Worley PF. Homer 1b regulates the trafficking of group I metabotropic glutamate receptors. J Biol Chem. 1999;274:25953–25957. doi: 10.1074/jbc.274.36.25953. [DOI] [PubMed] [Google Scholar]
- 25.Schinder AF, Poo M-m. The neurotrophin hypothesis for synaptic plasticity. Trends Neurosci. 2000;23:639–645. doi: 10.1016/s0166-2236(00)01672-6. [DOI] [PubMed] [Google Scholar]
- 26.Sgambato V, Pagès C, Rogard M, Besson M-J, Caboche J. Extracellular signal-regulated kinase (ERK) controls immediate early gene induction on corticostriatal stimulation. J Neurosci. 1998;18:8814–8825. doi: 10.1523/JNEUROSCI.18-21-08814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 28.Shieh PB, Hu S-C, Bobb K, Timmusk T, Ghosh A. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron. 1998;20:727–740. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 29.Szekely AM, Barbaccia ML, Alho H, Costa E. In primary cultures of cerebellar granule cells the activation of N-methyl-d-aspartate-sensitive glutamate receptors induces c-fos mRNA expression. Mol Pharmacol. 1989;35:401–408. [PubMed] [Google Scholar]
- 30.Tadokoro S, Tachibana T, Imanaka T, Nishida W, Sobue K. Involvement of unique leucine-zipper motif of PSD-Zip45 (Homer 1c/vesl-1L) in group 1 metabotropic glutamate receptor clustering. Proc Natl Acad Sci USA. 1999;96:13801–13806. doi: 10.1073/pnas.96.24.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 32.Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 33.Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- 34.Xia Z, Dudek H, Miranti CK, Greenberg ME. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao B, Tu JC, Petralia RS, Yuan JP, Doan A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ, Worley PF. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of Homer-related synaptic proteins. Neuron. 1998;21:707–716. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- 36.Xiao B, Tu JC, Worley PF. Homer: a link between neural activity and glutamate receptor function. Curr Opin Neurobiol. 2000;10:370–374. doi: 10.1016/s0959-4388(00)00087-8. [DOI] [PubMed] [Google Scholar]
- 37.Zirrgiebel U, Ohga Y, Carter B, Berninger B, Inagaki N, Thoenen H, Lindholm D. Characterization of TrkB receptor-mediated signaling pathways in rat cerebellar granule neurons: involvement of protein kinase C in neuronal survival. J Neurochem. 1995;65:2241–2250. doi: 10.1046/j.1471-4159.1995.65052241.x. [DOI] [PubMed] [Google Scholar]