Abstract
The ability of neurons to modify synaptic connections based on activity is essential for information processing and storage in the brain. The induction of long-lasting changes in synaptic strength requires new protein synthesis and is often mediated by NMDA-type glutamate receptors (NMDARs). We used a dark-rearing paradigm to examine mRNA translational regulation in the visual cortex after visual experience-induced synaptic plasticity. In this model system, we demonstrate that visual experience induces the translation of mRNA encoding the α-subunit of calcium/calmodulin-dependent kinase II in the visual cortex. Furthermore, this increase in translation is NMDAR dependent. One potential source for newly synthesized proteins is the translational activation of dormant cytoplasmic mRNAs. To examine this possibility, we developed a culture-based assay system to study translational regulation in neurons. Cultured hippocampal neurons were transfected with constructs encoding green fluorescent protein (GFP). At 6 hr after transfection, ∼35% of the transfected neurons (as determined by in situ hybridization) expressed detectable GFP protein. Glutamate stimulation of the cultures at this time induced an increase in the number of neurons expressing GFP protein that was NMDAR dependent. Importantly, the glutamate-induced increase was only detected when the 3′-untranslated region of the GFP constructs contained intact cytoplasmic polyadenylation elements (CPEs). Together, these findings define a molecular mechanism for activity-dependent synaptic plasticity that is mediated by the NMDA receptor and requires the CPE-dependent translation of an identified mRNA.
Keywords: protein synthesis, synaptic plasticity, CPEB, NMDA receptor, dendrites, visual cortex, hippocampus
Neural information is transmitted and processed by synapses. Synaptic plasticity, the bidirectional modification of synaptic strength based on activation history, is thought to play a key role in development, learning, and memory. The induction of long-lasting synaptic changes and memory formation both require tightly regulated, activity-driven protein synthesis (Davis and Squire, 1984; Bailey et al., 1996). The mRNAs encoding these newly synthesized proteins can have two distinct histories: some are transcribed in direct response to neural activity, whereas others are stored in the cytoplasm and are regulated at the translational level (Wells et al., 2000; Aakalu et al., 2001). Characterization of the molecular mechanisms regulating activity-induced transcription and translation is essential for understanding how experience and neural activity can be transformed into discrete, stable synaptic changes. Our knowledge of transcriptionally based mechanisms is relatively advanced. In the best studied case of activity-induced transcriptional activation, the relevant synaptic stimuli [NMDA-type glutamate receptors (NMDARs)], several putative intracellular signaling molecules (cAMP and protein kinase A), the cognatetrans-acting DNA-binding protein (cAMP response element-binding protein), and the structure of the gene promoter region (cAMP response element) have been identified (Shaywitz and Greenberg, 1999). In sharp contrast, little is known about the molecular pathways underlying translational regulation in neurons. In particular, in no case have defined aspects of synaptic activation been functionally linked to specific structural elements in an identified, translationally activated mRNA.
One approach to this problem is to draw on mechanisms that have been elucidated in non-neural systems. Oocyte maturation and early embryonic development require the translational activation of maternal mRNAs that are stored in the cytoplasm. A process known as cytoplasmic polyadenylation regulates one key set of maternal messages. These mRNAs harbor a specific cis-element in their 3′-untranslated regions (UTRs), the cytoplasmic polyadenylation element (CPE) that binds a trans-acting binding protein [CPE-binding protein (CPEB)]. CPEB in turn is part of a protein complex that regulates the translational state of CPE-containing mRNAs (Richter, 2000).
This mechanism not only regulates the activation of mRNA translation but is directly involved in keeping CPE-containing mRNA translationally dormant before progesterone stimulation in the oocyte (de Moor and Richter, 1999). A CPEB-associated protein, maskin, binds to the translation initiation factor 4E (eIF-4E). The maskin complex precludes the activation of translation by prohibiting the binding of eIF-4G to eIF-4E. Progesterone stimulation of the oocyte leads to the phosphorylation of CPEB by the Aurora serine–threonine kinase (also known as Eg2 or IAK1 kinase) (Bischoff and Plowman, 1999; Mendez et al., 2000). CPEB phosphorylation is required for cytoplasmic polyadenylation and the subsequent disassociation of maskin and eIF-4E, which in turn permits translational initiation. In the rodent brain, both CPEB and Aurora have been localized to synapses (Wu et al., 1998) (Y.-S. Huang, M.-Y. Jung, M. Sarkissian, and J. D. Richter, unpublished observations), suggesting that this mechanism could play an important role in local synaptic protein synthesis (Wells et al., 2000).
In a recent study, we showed that CPEB is expressed in the visual cortex and that the CPE-containing the α-subunit of calcium/calmodulin-dependent kinase II (α-CaMKII) mRNA is polyadenylated and translated in this brain region in response to visual experience (Wu et al., 1998). In this report, we demonstrate that NMDAR activation is essential for α-CaMKII protein synthesis in the visual cortex and that this synthesis is also sensitive to inhibitors of cytoplasmic polyadenylation. We also introduce a new cell culture-based assay for studying translational regulation in neurons. Using this system, we show that NMDAR-stimulated translation requires the CPEs in the 3′-UTR of α-CaMKII mRNA. These findings link a specific mechanism of translational regulation to many of the key molecular elements thought to play critical roles in synaptic plasticity, learning, and memory formation.
MATERIALS AND METHODS
Analysis of α-CaMKII in synaptoneurosomes. Long–Evans rats were born in a room specifically designed for rearing of animals in a light-free environment. They were raised in this room between 4 and 6 weeks before use in these experiments. Dark-reared rats were either anesthetized in the dark (DR) or anesthetized after 30 min of light exposure (DR + 30′). Treated DR rats were injected intraperitoneally in the dark and either kept in the dark for 1 hr or brought into the light for 30 min, 0.5 hr after injection. The primary visual cortex was rapidly dissected in cold, sterile PBS and immediately homogenized in ice-cold buffer (10 mm HEPES, 2.0 mm EDTA, 2.0 mm EGTA, 0.5 mm DTT, 0.1 mm PMSF, 10 mg/l leupeptin, 50 mg/l soybean trypsin inhibitor, and 100 nm microcystin). Synaptoneurosome fractions were isolated as by Quinlan et al. (1999). Briefly, the tissues were homogenized and passed through two 100 μm nylon mesh filters, followed by a 5 μm pore filter. The filtrate was then centrifuged at 1000 × g for 10 min.
Equal amounts of total protein (25 μg) from the synaptoneurosome fractions were resolved on a 5–15% polyacrylamide gel, blotted, and probed simultaneously with monoclonal antibodies to α-CaMKII (clone #6G9; Boehringer Mannheim, Indianapolis, IN) and NMDAR1 (PharMingen, San Diego, CA), followed by an alkaline phosphatase-conjugated secondary antibody. Digital images of the α-CaMKII Western blots were obtained using a ScanJet IIcx (Hewlett-Packard, Palo Alto, CA) with DeskScan II (Hewlett-Packard) software, and quantitative densitometry was performed with NIH Image 1.60 software.
Hippocampal neuron cultures. Cultures of rat hippocampal neurons were made as described previously (Goslin and Banker, 1991). Briefly, the hippocampus was removed from embryonic day 18 (E18) rat embryos, trypsinized (0.25%), dissociated by trituration, and plated onto poly-l-lysine (1 mg/ml)-coated glass coverslips (240,000 cells/ml) for 3 hr. The coverslips were then transferred to dishes containing a monolayer of glial cells in growth medium. After 7–10 d in vitro, individual coverslips were transferred to 12 well plates for transfection with 0.5 μg of DNA per coverslip for 1 hr using Effectene (Qiagen, Hilden, Germany) or 1 μg of DNA per coverslip for 5 hr using Lipofectamine 2000 (Invitrogen, San Diego, CA). In preliminary experiments, we attempted transfection with calcium phosphate and an earlier version of Lipofectamine (Lipofectamine Plus), but transfection efficiencies were low and cell viability after transfection was often compromised. The highest efficiency and greatest viability 24 hr after transfection was obtained with Lipofectamine 2000. After transfection, the coverslips were then washed and placed back into the dishes containing the glial feeder layer. Removal from the transfection media was considered time 0 for all experiments. Cultures were stimulated at indicated times after transfection with bath application of either 100 μm glutamate (30 sec) or 35 mm KCl (5 min). The cultures were washed and remained with the glial feeder layer for an additional 1.5 hr before fixation with 4% paraformaldehyde. Ind-aminophosphonovalerate (APV)-treated cultures, the drug was applied immediately after transfection and remained in the media for the duration the stimulation and through the remainder of the experiment. In actinomycin D-, cycloheximide-, and cordycepin-treated cultures, the drugs were applied 30 min before stimulation and remained in the media for the duration of the experiment.
In situ hybridization. At indicated times after transfection, the coverslips were washed once in 1× PBS, fixed in 4% formaldehyde–PBS for 10 min at room temperature, and washed three more times in PBS. The coverslips were then incubated in 1× SSC for 5 min at room temperature and permeabilized by 1% Triton X-100–1× SSC for 30 min at room temperature. The coverslips were placed cell-side down on Parafilm and hybridized overnight at 37°C in 40 μl of hybridization mix (50% formamide, 2× SSC, 10% dextran sulfate, 1 mg/ml tRNA, 0.02% RNase-free BSA, and 2 mmvanadyl–ribonucleoside complex) plus 30 ng of digoxygenin (DIG)-labeled DNA oligonucleotide probe against green fluorescent protein (GFP) coding region having the following sequence: 5′-ATATAGACGXTGTGGCTGTTGTAGTTGTACTCCAGCTTCT-3′ (X indicates the DIG-modified base) (Oligos Etc., Wilsonville, OR). After hybridization, the coverslips were washed twice with 50% formamide–2× SSC for 30 min at 37°C and then incubated for 1 hr at 37°C in blocking solution (2× SSC, 8% formamide, 2 mm vanadyl–ribonucleoside complex, and 0.2% RNase-free BSA) and washed four times for 5 min each in 8% formamide–2× SSC at room temperature. The DIG-labeled oligo probes were detected with monoclonal mouse anti-digoxygenin antibody (1:250; Boehringer Mannheim) overnight at 4°C, followed by biotinylated anti-mouse IgG (1:100; Vector Laboratories, Burlingame, CA) for 45 min and Cy3 streptavidin (1:500; Jackson ImmunoResearch, West Grove, PA) for 30 min. Finally, the coverslips were incubated with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO) for 5 min at room temperature to stain the nuclei.
Scoring of GFP-fluorescent neurons and GFP-in situhybridization (ISH)-positive neurons cell counts was performed on a Nikon (Tokyo, Japan) E800 fluorescent microscope. Cultures derived from E18 rat embryos (as above) consist of two types of neurons. The majority (∼94%) are glutamatergic pyramidal neurons, and the remaining 6% are GABAergic interneurons (Benson et al., 1994). Non-neural cells comprise a small proportion (∼1%) of the total number of cells in these cultures and were distinguished from neurons based on their distinctive cellular morphology. No effort was made to distinguish between neuronal cell types. For each coverslip, the total number of neurons and the total number of GFP-fluorescent neurons was counted. Neurons were scored as GFP-expressing if they exhibited intense fluorescence throughout the entire cell. Intermediate levels of GFP expression and non-uniform distribution of fluorescence were rare. GFP-ISH-positive neurons were counted from 10 random fields per coverslip using a 40× objective. The scoring was performed blind to the stimulation history of the cultures. The absolute numbers of transfected neurons was the same at 24 hr under all conditions, indicating that viability was not influenced by these transfections (mean number of GFP-CPEWT, 169 ± 7.5 per 1800 total neurons; mean number of GFP-CPEMUT, 193 ± 26.9 per 2000 total neurons).
Images were recorded with a PhotoMetrics Inc. (Huntington Beach, CA) CCD camera using IP Lab Systems software and imported into an Adobe Photoshop (Adobe Systems, San Jose, CA) file.
Construction of GFP–α-CaMKII–3′-UTR plasmids. pEGFP-C1 vector (Clontech, Cambridge, UK) was digested with EcoRI and blunt-ended with Klenow to generate the stop codon TAA and self-ligated. The resulting plasmid was digested with SalI and XbaI, and a partial α-CaMKII 3′-UTR (∼170 bp) with wild-type (WT) CPEs or mutant (MUT) CPEs (Wu et al., 1998) were ligated into this vector between these sites.
RESULTS
Experience-induced protein synthesis in the visual cortex is NMDAR dependent
We used the visual cortex of dark-reared rats exposed to light for brief periods as a model for robust, experience-driven synaptic reorganization (Carmignoto and Vicini, 1992; Kirkwood et al., 1996;Quinlan et al., 1999). In this paradigm, α-CaMKII mRNA is polyadenylated after 30 min of visual experience (Wu et al., 1998). This polyadenylation is accompanied by an increase in α-CaMKII protein in the synaptic fraction isolated from the visual cortex (Fig.1A) (Wu et al., 1998). In contrast, the level of NR1 (NMDA receptor subunit-1) protein in the same fractions remains unchanged after light exposure. This elevation in α-CaMKII protein is sensitive to the protein synthesis inhibitor cycloheximide (Wu et al., 1998). The increased production of α-CaMKII protein could be attributable to newly synthesized message or to the enhanced translation of existing mRNA. To distinguish between these possibilities, rats were injected with the transcription inhibitor actinomycin D before light exposure. As shown in Figure 1, this treatment failed to block the increase in α-CaMKII protein in the synaptoneurosome fraction of dark-reared, light-exposed animals. Therefore, the activation or enhancement of mRNA translation is required for the visual experience-induced increase in synaptic α-CaMKII.
Fig. 1.
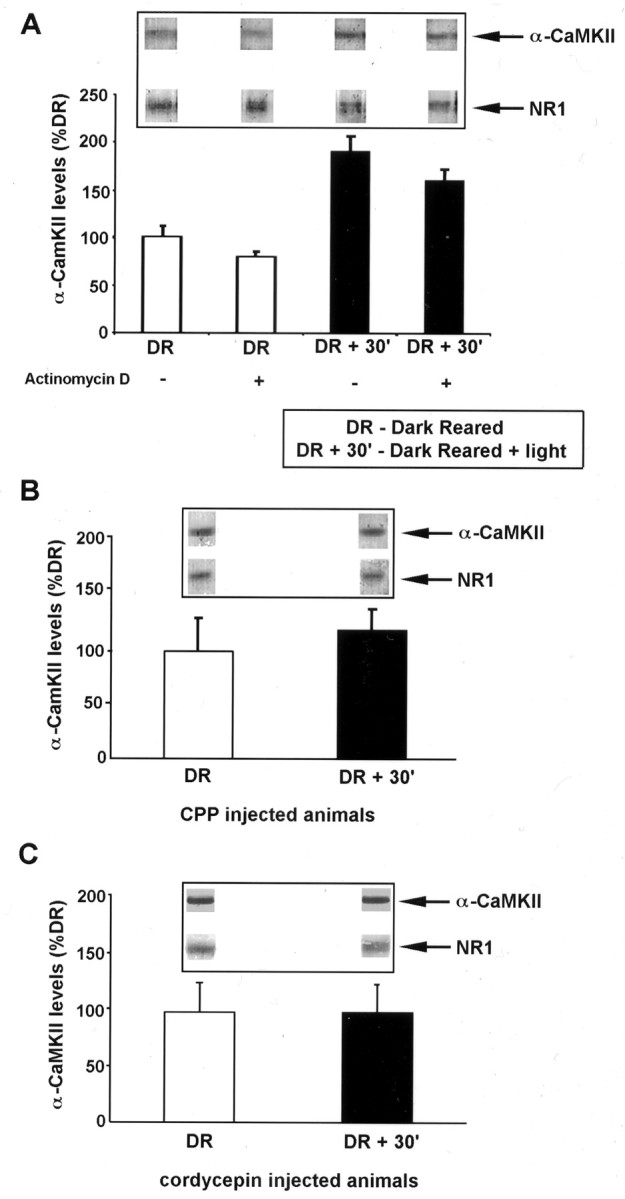
Experience-induced increase in α-CaMKII protein in the visual cortex mediated by NMDAR activation and mRNA polyadenylation. A, Quantification of α-CaMKII levels in synaptoneurosome (SN) fractions isolated from the visual cortex of animals reared in complete darkness (DR) and animals reared in the dark and exposed to light for 30 min (DR + 30′). Western blots for α-CaMKII and NMDAR subunit NR1 were performed from PAGE loaded with equal total protein of SN samples isolated from DR and DR + 30′ visual cortex. Quantitative densitometry was performed on the α-CaMKII bands, and these were normalized to the level of NR1 in the same lane [the amount of NR1 subunit in SN fraction does not change with visual experience (Quinlan et al., 1999)]. Where indicated, actinomycin D (1 mg/kg) was injected (intraperitoneally) 30 min before light exposure. This dose of actinomycin D is effective in blocking protein synthesis in the brain (Jackson, 1972; Pickering and Fink, 1976). Each experiment consisted of two to four rats per treatment group, and results shown are the mean ± SEM of three experiments. Insets show representative bands from one experiment. B, Quantification of α-CaMKII expression as in A, in animals injected with the NMDAR antagonist CPP (10 mg/kg) 30 min before light exposure. Each experiment consisted of two to four rats per treatment group, and results shown are the mean ± SEM of three experiments. C, Quantification of α-CaMKII performed as in A, in animals injected with cordycepin (6 mg/kg) 30 min before light exposure. Each experiment consisted of two to four rats per treatment group, and results shown are the mean ± SEM of three experiments.
The activation of NMDARs is thought to drive experience-induced synaptic plasticity during postnatal development of the visual cortex (Bear et al., 1990; Daw et al., 1999). To examine whether NMDAR activation triggers this new protein synthesis, rats were injected with the NMDAR antagonist 3-(2 carboxypiperazin-4yl) propyl-1-phosphonic acid (CPP) just before visual experience. As shown in Figure1B, NMDAR blockade of dark-reared, light-exposed rats inhibited the increase in α-CaMKII protein in synaptic fractions. Thus, NMDAR activation is essential for experience-induced translation of α-CaMKII mRNA in the visual cortex.
As one test of whether mRNA polyadenylation is required for this new α-CaMKII protein synthesis in the visual cortex, we injected the dark-reared animals with 3′-deoxyadenosine (cordycepin), an adenosine analog that inhibits mRNA polyadenylation (Beach and Ross, 1978;Ulibarri and Yahr, 1987; McGrew et al., 1989; Groisman et al., 2000). Cordycepin treatment blocked the light-induced increase in α-CaMKII protein in the synaptic fraction of visual cortex (Fig. 1C). This finding suggests that visual experience-induced NMDAR activation triggers α-CaMKII protein synthesis via a mechanism that requires mRNA polyadenylation.
A role for cytoplasmic polyadenylation elements in activity-dependent mRNA translation in neurons
To elucidate the mechanistic basis of this process in more detail, we next developed a cell culture model to directly assess the role of the α-CaMKII CPEs in translational regulation. We based this assay on the well defined low-density hippocampal neuron culture system (Bartlett and Banker, 1984; Fletcher et al., 1991, 1994; Goslin and Banker, 1991). These cultures are comprised of ∼99% neurons and ∼1% glial cells. In all of our experiments, we scored only neurons, which were identified based on morphology using phase contrast microscopy and/or fluorescence imaging of GFP-transfected cells. Control experiments with anti-microtubule-associated protein 2, synaptic markers, and anti-GFAP confirmed the reliability of these identification methods (data not shown). Neurons cultured for 7–10 d were transfected with plasmids containing the GFP coding sequence linked to a fragment of the α-CaMKII 3′-UTR that harbored either intact or mutated CPEs (GFP-CPEWT and GFP-CPEMUT, respectively) (Fig.2). We then monitored the expression of the GFP-encoding mRNA and GFP protein in the transfected neurons by ISH and GFP fluorescence, respectively.
Fig. 2.

Transfection of hippocampal cells in culture with reporter GFP constructs. A, Schematic of α-CaMKII mRNA and the GFP constructs used for transfections. GFP constructs were modified to contain the last ∼160 nucleotides of α-CaMKII 3′-UTR with either intact CPE sequences (GFP-CPEWT;top) or mutated CPEs (GFP-CPEMUT;bottom). B, Hippocampal neurons grown in culture for 7 d, transfected with GFP-CPEWT. This culture was processed for GFP fluorescence 8 hr after transfection. GFP-fluorescing neurons are readily distinguished from non-GFP-fluorescing neurons, and GFP is detected throughout the entire neuron (right). Scale bar, 20 μm.
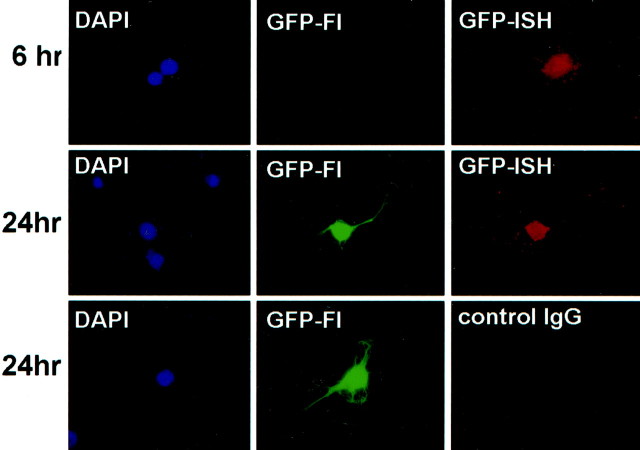
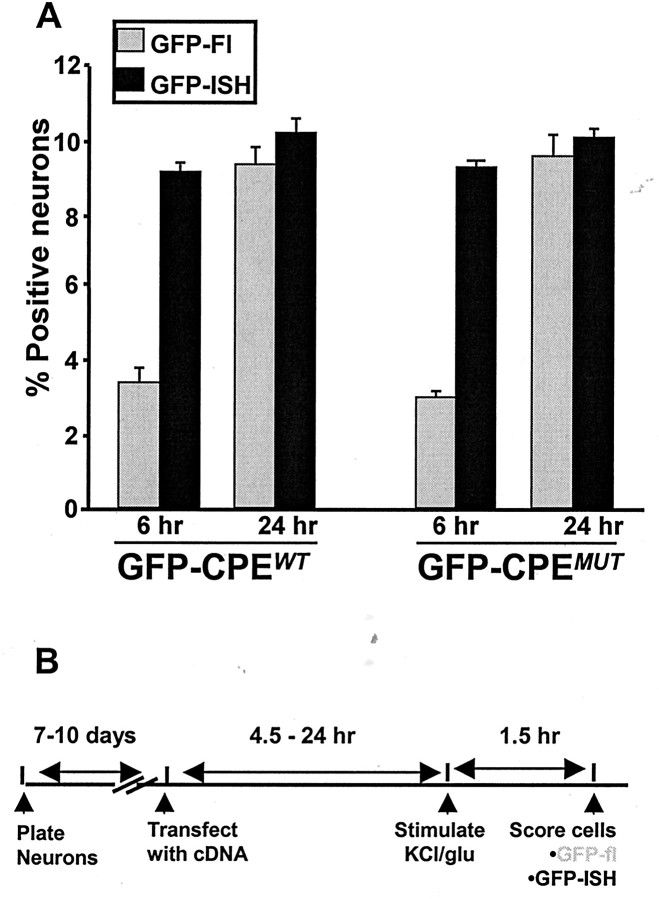
To enable reliable quantification, we established conditions in which ∼10% of the neurons (∼200 per coverslip) were transfected (see Materials and Methods). As shown in Figure3, GFP-encoding mRNA was detected in transfected neurons. We then compared the presence of GFP-encoding mRNA to the expression of GFP protein in neurons as detected by intrinsic GFP fluorescence. At 24 hr, all neurons containing GFP mRNA also expressed GFP protein. In contrast, at 6 hr after transfection, only a fraction of the GFP mRNA-containing neurons expressed detectable GFP protein (Fig. 3). Scoring GFP protein and mRNA expression at these time points showed that the same percentage of neurons were transfected; however, at 6 hr after transfection, only ∼35% of the neurons containing GFP mRNA expressed detectable GFP protein (Fig.4A). Importantly, the transfection efficiencies observed using the unmodified GFP construct, GFP-CPEWT and GFP-CPEMUT were indistinguishable (Fig.4A and data not shown).
Fig. 3.
GFP mRNA and GFP protein expression in transfected neurons. Neurons were processed for GFP fluorescence (GFP-Fl) and fluorescent in situhybridization (GFP-ISH) at either 6 or 24 hr after transfection. Neurons at 6 hr can contain GFP mRNA without expressing detectable GFP fluorescence (top panel). In contrast, at 24 hr after transfection, all GFP mRNA-containing neurons also express the fluorescent GFP protein (middle panel). In the bottom panel, the anti-DIG primary antibody was replaced with a nonspecific normal mouse IgG antibody (control IgG). DAPI staining reveals the nuclei of cells within each field.
Fig. 4.
Quantification of GFP expression in cultured hippocampal neurons and experimental design. A, GFP fluorescence is not correlated with GFP mRNA expression at early times after transfection. In cultures transfected with GFP-CPEWT, 9.23 ± 0.002% of all neurons expressed GFP mRNA at 6 hr. However, significantly fewer neurons (3.4 ± 0.002%) expressed GFP protein at detectable levels (p ≤ 0.005). In contrast, at 24 hr after transfection, 10.27 ± 0.003% of the total neuronal population contained GFP mRNA, and 9.41 ± 0.004% exhibited GFP fluorescence (p = 0.09). Similar results were obtained when cultures were transfected with GFP-CPEMUTconstructs: at 6 hr after transfection, 9.3 ± 0.002% contained GFP mRNA, with only 3.07 ± 0.002% expressing detectable protein (p ≤ 0.01). This difference was not present at 24 hr after transfection (10.2 ± 0.002% contained GFP mRNA, and 9.7 ± 0.005% contained GFP fluorescence;p = 0.4). Data represent mean ± SEM.B, Experimental design. Seven- to 10-d-old cultures were transfected and then stimulated with either glutamate or KCl at time points between 4.5 and 24 hr after transfection. GFP fluorescence and GFP mRNA presence (using fluorescent in situhybridization) was scored 1.5 hr after stimulation.GFP-Fl, GFP fluorescence; GFP-ISH, GFP-fluorescent in situ hybridization.
The presence of neurons containing GFP mRNA without detectable levels of GFP protein suggested a time window in which activity-regulated translation might be readily revealed. Because the number of cells transfected was the same under all conditions tested, we predicted that, at early times after transfection, new translation would manifest as an increase in the number of neurons expressing detectable GFP protein.
To test this hypothesis, we designed experiments in which neurons were stimulated by either direct glutamate application or KCl depolarization at varying times after transfection (Fig.4B). Figure5A shows that glutamate stimulation (100 μm for 30 sec) at an early time after transfection triggered an increase in the number of GFP protein-expressing neurons. This increase was completely blocked by the translation inhibitor cycloheximide, indicating that the GFP is newly synthesized (Fig. 5B). This increase in GFP-expressing neurons could also be induced by KCl depolarization (35 mm for 5 min) (Fig. 5C) and was insensitive to treatment with actinomycin D (Fig. 5C). An examination of the time course showed that the number of GFP protein-expressing neurons in unstimulated cultures increased gradually over the first 10 hr after transfection, reaching a plateau at ∼14 hr (Fig. 5D). KCl treatment at early times after transfection resulted in a significant increase in the number of neurons expressing detectable GFP protein compared with unstimulated control cultures. On the other hand, such increases were not observed when neurons were treated with KCl at later times after transfection. This result is completely consistent with our observations of GFP mRNA and protein expression at 6 and 24 hr (Fig. 4). We thus conclude that activity-induced stimulation of mRNA translation is manifested by an increase in the number of neurons with detectable GFP protein expression.
Fig. 5.
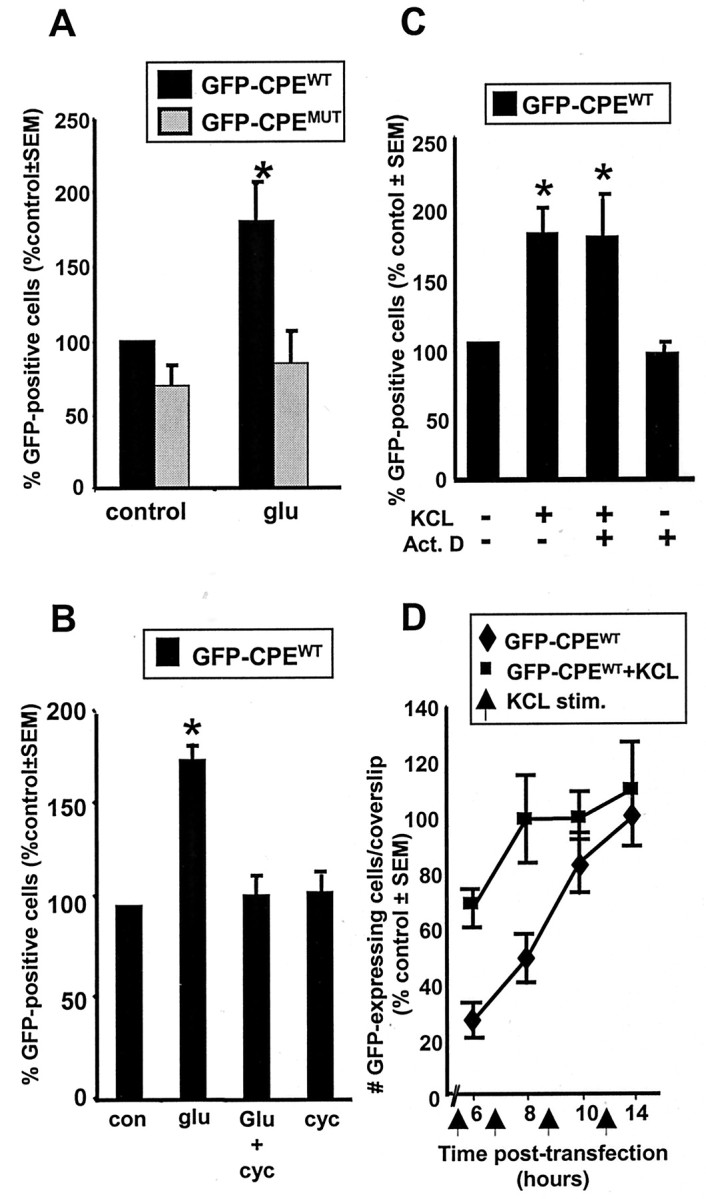
Activity induces an increase in translation of CPE-containing mRNA at early times after transfection.A, Hippocampal neuron cultures transfected with either GFP-CPEWT (black bars) or GFP-CPEMUT (gray bars) were stimulated 6 hr after transfection by a 30 sec application of glutamate (glu; 100 μm) and processed for GFP fluorescence 1.5 hr later. A significant increase in the number of GFP-expressing neurons was detected only in the cultures transfected with the CPE-containing construct (n = 3).B, The glutamate-induced increase in GFP-expressing neurons is dependent on protein synthesis. Cultures transfected with GFP-CPEWT and stimulated with glutamate (glu) as above were treated with cycloheximide 30 min before glutamate stimulation. Cycloheximide (cyc) treatment blocked the increase in the number of GFP-expressing neurons. Cycloheximide treatment alone for the duration of the post-stimulation period (1.5 hr) had no effect on the number of GFP-expressing neurons (n = 3). con, Control.C, Depolarization induces an increase in GFP-expressing neurons that is not dependent on new gene transcription. Where indicated, cultures transfected with GFP-CPEWT were depolarized with KCl (35 mm, 5 min) 1.5 hr before fixation. KCl depolarization induced a significant increase in the number of GFP-expressing neurons. Addition of actinomycin D (Act. D; 25 μm) 30 min before KCl application did not alter the response to KCl depolarization (n = 3).D, Time course of GFP expression in neurons transfected with GFP-CPEWT. Hippocampal neurons were transfected with GFP-CPEWT and then processed for GFP fluorescence at 6, 8, 10, and 14 hr after transfection (♦). In parallel experiments, cultures were stimulated with 35 mmKCl for 5 min (arrows at 4.5, 6.5, 8.5, and 10.5 hr) 1.5 hr before fixation (▪). Three coverslips were counted at each time point in each experiment, and results are the mean ± SEM of two experiments. All coverslips were counted blind to treatment protocol (control is unstimulated; *p ≤ 0.05).
The experiments described above show that glutamate stimulation activates the translation of GFP reporter constructs that contain the wild-type CPE elements from the 3′-UTR of the α-CaMKII mRNA (GFP-CPEWT). To determine whether these CPEs are necessary for the observed translational activation, we tested neurons transfected with a construct in which these elements were mutated (GFP-CPEMUT) (Fig. 2). Glutamate stimulation of these cultures at 6 hr after transfection failed to elicit a significant increase in the number of GFP protein-expressing neurons (p > 0.7) (Fig. 5A). Glutamate stimulation also failed to stimulate translation in cells transfected with an unmodified GFP reporter construct that lacks CPEs (Clontech). We conclude that the CPE is required for an activity-driven increase in translation in hippocampal neurons.
NMDAR-dependent protein synthesis mediated by polyadenylation
Our in vivo experiments indicated that NMDAR activation is necessary for the experience-induced translation of α-CaMKII mRNA (Fig. 1). However, because the entire animal was treated with the antagonist, it was impossible to determine whether the NMDAR activation and the protein synthesis occurred in the same neuron. Therefore, we examined NMDAR-driven protein synthesis in our in vitromodel system using the receptor-specific antagonist APV to determine whether the NMDAR mediates activity-induced translation in neurons. As shown in Figure 6A, APV treatment completely inhibited the glutamate-induced increase in GFP-expressing neurons transfected with GFP-CPEWT. The KCl-induced increase in GFP synthesis was also APV sensitive (Fig. 6B). These results place the NMDAR in the signal transduction pathway leading from synaptic stimulation to the translational activation of CPE-containing mRNA.
Fig. 6.
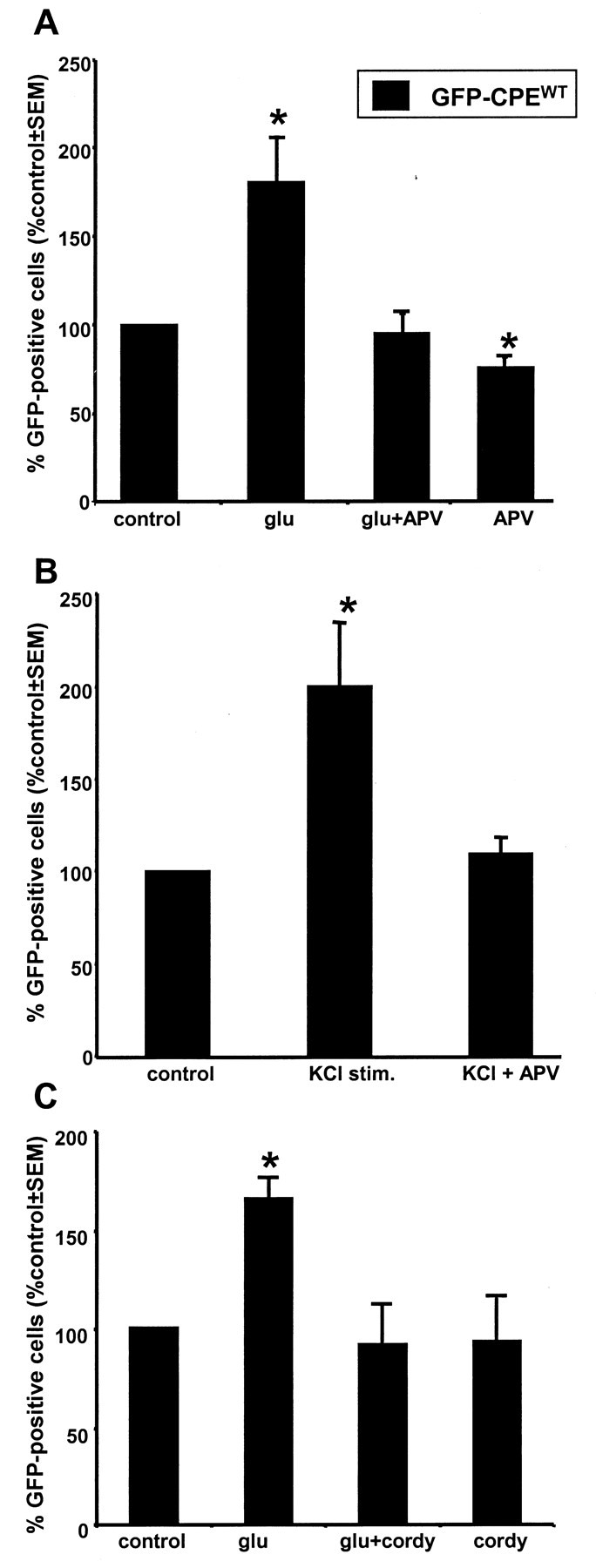
Activity-dependent translation in cultured hippocampal neurons regulated by NMDAR activation and mediated by polyadenylation. A, Neurons cultured and transfected as in Figure 5 were treated with the NMDAR antagonist APV (300 μm) starting immediately after transfection and continuing through the end of the stimulation protocol (total of 7.5 hr). The glutamate (glu)-induced increase in the number of neurons expressing GFP in cultures transfected with GFP-CPEWT was inhibited by APV. APV treatment alone for the entire post-transfection interval (7.5 hr) caused a small but significant (p < 0.05) decrease in GFP expression.B, The KCl depolarization-induced increase in GFP-expressing neurons was similarly inhibited by APV (n = 4). C, The glutamate-induced stimulation of GFP translation is blocked by the treatment of cordycepin (cordy; 200 μm) for 30 min before glutamate stimulation (n = 3). Cordycepin alone did not affect GFP expression in these neurons (control is unstimulated; *p ≤ 0.05).
In the visual cortex, α-CaMKII mRNA was polyadenylated in response to visual experience, a process reminiscent of CPE-dependent translation activation during Xenopus oocyte maturation (Richter, 1996;Wu et al., 1998). To test whether the NMDAR-mediated, CPE-dependent increase in GFP expression in neurons requires polyadenylation, we incubated the cultures in cordycepin for 30 min before and during glutamate stimulation. Cordycepin blocked the increase in GFP expression observed after glutamate stimulation (Fig. 6C). Note that, in these experiments, direct activation of glutamate receptors bypasses the need for synaptic release. Thus, possible presynaptic effects of cordycepin can be ruled out. Together, both thein vivo and in vitro data indicate that NMDAR activation can stimulate cytoplasmic polyadenylation and translation of CPE-containing mRNA.
DISCUSSION
New protein synthesis triggered by neural activity is required for invoking long-lasting changes in synaptic strength and for memory formation. Although some of these polypeptides arise as a consequence of increased transcription, recent evidence suggests that the synthesis of others is regulated at the translational level. Here, we used bothin vivo and cell culture systems to demonstrate a molecular mechanism for the activity-driven translation of a specific mRNA.
We used the rat visual cortex as a model system to examine the changes in protein synthesis during experience-induced synaptic plasticity. Dark-rearing rats from birth results in a relatively immature visual cortex that maintains the high degree of synaptic plasticity characteristic of the critical period (Kirkwood et al., 1995). Exposure of dark-reared rats to light results in a rapid, robust and coordinated burst of experience-driven synaptic plasticity that can be readily monitored at the biochemical and electrophysiological level (Quinlan et al., 1999). In previous work, we showed that visual experience evokes the polyadenylation of α-CaMKII mRNA in visual cortex and the elevation of α-CaMKII protein in synaptic fractions from this brain region. Moreover, this increase was a direct result of new synthesis because it was sensitive to the translation inhibitor cycloheximide (Wu et al., 1998). Here we show that the experience-induced increase of α-CaMKII protein does not require new transcription. Thus, the source of newly synthesized α-CaMKII protein is derived from the translational activation of already existing mRNA. This process of translational activation was blocked by an NMDAR antagonist, indicating that NMDAR signaling is necessary for this experience-evoked increase in synaptic α-CaMKII protein. Finally, the increase in synaptic α-CaMKII protein was blocked by the polyadenylation inhibitor cordycepin. Together, this data suggests that neural activity, transduced by the NMDAR, activates mRNA translation mediated by mRNA polyadenylation.
We developed a novel cell culture assay to elucidate both the cellular signaling mechanisms and the mRNA regulatory sequences that underlie this activity-dependent translation. In this system, hippocampal neurons are transfected with constructs encoding GFP and either wild-type or mutated α-CaMKII 3′-UTR sequences. To quantify the results, we took advantage of two observations. First, the transfection efficiency was the same regardless of which construct was used (Fig. 4A). Second, the number of cells expressing GFP protein at early times after transfection was only ∼35% of the neurons containing GFP mRNA, with detectable GFP fluorescence increasing slowly during the first 14 hr after transfection. Accordingly, we reasoned that stimulating translation of a given GFP-encoding mRNA during these early times after transfection would result in more of the transfected cells exhibiting detectable GFP expression. Our findings with both KCl depolarization and glutamate stimulation at 5–8 hr after transfection supported this interpretation (Fig. 5). Moreover, the increase in the number of GFP-expressing cells was a consequence of mRNA translation because it was sensitive to cycloheximide but resistant to actinomycin D. Furthermore, as predicted by this model, stimulation of cultures >14 hr after transfection did not result in an increase in the number of cells with detectable fluorescence. We exploited the disparity between the presence of transfected mRNA and protein expression at the early times after transfection to investigate the signaling mechanisms and mRNA sequence elements that function in activity-regulated translation in neurons. Guided by our in vivo data, we stimulated cultured neurons with glutamate and demonstrated that translation was only enhanced if the reporter contained the intact CPEs from the 3′-UTR of α-CaMKII. Furthermore, consistent with our observations in the visual cortex, NMDAR activation is required for this increase in translation. It should be noted that, in the current study, the mRNA encoding the GFP reporter constructs are expressed in cell bodies, as well as in dendrites (Fig. 3). Therefore, it is not possible to determine the localization of the translation detected. Studies to address this issue are currently in progress.
Our results in vitro and in vivo suggest that cytoplasmic polyadenylation is a mechanism for regulating activity-induced mRNA translation in neurons. When activity is induced by visual experience, NMDAR activation induces the translation of α-CaMKII, a message that contains two CPE sequences in its 3′-UTR (Wu et al., 1998). In a previous study, we demonstrated that the CPE-binding protein CPEB is localized to synapses in the brain (Wu et al., 1998). CPEB is likely to be a key regulator of CPE-dependent cytoplasmic polyadenylation (Richter, 2000; Wells et al., 2000), suggesting that CPE-mediated translation could be occurring synaptically. Furthermore, inhibiting polyadenylation, by injecting cordycepin, blocks the increase in synaptic α-CaMKII after light exposure. In vitro, GFP-CPEWT-transfected neurons stimulated by direct glutamate receptor activation in the presence of cordycepin produces a complete inhibition of the increase in GFP-expressing neurons. In addition, translation was not induced in hippocampal neurons transfected with either unmodified constructs or constructs that contained mutated CPEs (GFP-CPEMUT).
NMDAR activation and α-CaMKII are critical for synaptic plasticity and memory formation (Morris et al., 1986; Bear et al., 1990; Silva et al., 1992; Tsien et al., 1996; Shimizu et al., 2000). The data presented here suggests that one signal transduction pathway leading from NMDAR activation to an increase in protein by a mechanism that is CPE-dependent and involves cytoplasmic polyadenylation. This proposal is supported by recent results demonstrating NMDAR-dependent cytoplasmic polyadenylation in synaptic fractions and the accompanying activation of an Aurora kinase (Huang, Jung, Sarkissian, and Richter, unpublished observations). Together, these findings link many of the key molecular elements in synaptic plasticity and establish a pathway whereby experience-induced synaptic activation can generate new synaptic protein synthesis. Knowledge of this pathway, combined with other recently described translational mechanisms in dendrites (Martin et al., 1997; Scheetz et al., 2000; Aakalu et al., 2001), opens the way for elucidating the molecular basis of long-term changes in synaptic efficacy. In turn, such understanding could provide the insights and tools for determining the role of how synaptic modification contributes to learning and memory.
Footnotes
This work was supported by National Institutes of Health Grants NS39321 and RR15578 (J.F.) and by National Research Scientist postdoctoral Award NS10919 (D.W.) Y.-S.H. was supported by the Charles A. King trust postdoctoral fellowship.
D.G.W. and X.D. contributed equally to this work.
Correspondence should be addressed to Justin R. Fallon at the above address. E-mail: justin_fallon@brown.edu.
D. G. Wells's present address: Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT 06511.
E. M. Quinlan's present address: Department of Biology, University of Maryland, College Park, MD 20742.
REFERENCES
- 1.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 2.Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci USA. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett WP, Banker GA. An electron microscopic study of the development of axons and dendrites by hippocampal neurons in culture. II. Synaptic relationships. J Neurosci. 1984;4:1954–1965. doi: 10.1523/JNEUROSCI.04-08-01954.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beach LR, Ross J. Cordycepin. An inhibitor of newly synthesized globin messenger RNA. J Biol Chem. 1978;253:2628–2633. [PubMed] [Google Scholar]
- 5.Bear MF, Kleinschmidt A, Gu Q, Singer W. Disruption of experience-dependent synaptic modifications in striate cortex by infusion of NMDA receptor antagonist. J Neurosci. 1990;10:909–925. doi: 10.1523/JNEUROSCI.10-03-00909.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson DL, Watkins FH, Steward O, Banker G. Characterization of GABAergic neurons in hippocampal cell cultures. J Neurocytol. 1994;23:279–295. doi: 10.1007/BF01188497. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff JR, Plowman GD. The aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. doi: 10.1016/s0962-8924(99)01658-x. [DOI] [PubMed] [Google Scholar]
- 8.Carmignoto G, Vicini S. Activity-dependent decrease in NMDA receptor responses during development of the visual cortex. Science. 1992;258:1007–1011. doi: 10.1126/science.1279803. [DOI] [PubMed] [Google Scholar]
- 9.Davis HP, Squire LR. Protein synthesis and memory: a review. Psychol Bull. 1984;96:518–559. [PubMed] [Google Scholar]
- 10.Daw NW, Gordon B, Fox KD, Flavin HJ, Kirsch JD, Beaver CJ, Ji QH, Reid SNM, Czepita D. Injection of MK-801 affects ocular dominance shifts more than visual activity. J Neurophysiol. 1999;81:204–215. doi: 10.1152/jn.1999.81.1.204. [DOI] [PubMed] [Google Scholar]
- 11.de Moor CH, Richter JD. Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J. 1999;18:2294–2303. doi: 10.1093/emboj/18.8.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher TL, Cameron P, De Camilli P, Banker G. The distribution of synapsin I and synaptophysin in hippocampal neurons developing in culture. J Neurosci. 1991;11:1617–1626. doi: 10.1523/JNEUROSCI.11-06-01617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher TL, De Camilli P, Banker G. Synaptogenesis in hippocampal cultures: evidence indicating that axons and dendrites become competent to form synapses at different stages of neuronal development. J Neurosci. 1994;14:6695–6706. doi: 10.1523/JNEUROSCI.14-11-06695.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goslin K, Banker G. Rat hippocampal neurons in low-density culture. In: Banker G, Goslin K, editors. Culturing Nerve Cells. MIT; Cambridge, MA: 1991. pp. 251–282. [Google Scholar]
- 15.Groisman I, Huang Y-S, Mendez R, Cao Q, Theurkauf W, Richter JD. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- 16.Jackson GL. Effect of actinomycin D on estrogen-induced release of luteinizing hormone in ovariectomised rats. Endocrinology. 1972;91:1284–1287. doi: 10.1210/endo-91-5-1284. [DOI] [PubMed] [Google Scholar]
- 17.Kirkwood A, Lee HK, Bear MF. Co-regulation of long-term potentiation and experience-dependent synaptic plasticity in visual cortex by age and experience. Nature. 1995;375:328–331. doi: 10.1038/375328a0. [DOI] [PubMed] [Google Scholar]
- 18.Kirkwood A, Rioult MC, Bear MF. Experience-dependent modification of synaptic plasticity in visual cortex. Nature. 1996;381:526–528. doi: 10.1038/381526a0. [DOI] [PubMed] [Google Scholar]
- 19.Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 20.McGrew LL, Dworkin-Rastl E, Dworkin MB, Richter JD. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989;3:803–815. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- 21.Mendez R, Murthy KGK, Manley JL, Richter JD. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell. 2000;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- 22.Morris RG, Anderson E, Lynch GS, Baudry M. Selective impairment of learning and blockade of long-term potentiation by an N-methyl-d-aspartate receptor antagonist, AP5. Nature. 1986;319:774–776. doi: 10.1038/319774a0. [DOI] [PubMed] [Google Scholar]
- 23.Pickering AJ, Fink G. Priming effect of luteinizing hormone releasing factor: in-vitro and in-vivo evidence consistent with its dependence upon protein and RNA synthesis. J Endocrinol. 1976;69:373–379. doi: 10.1677/joe.0.0690373. [DOI] [PubMed] [Google Scholar]
- 24.Quinlan EM, Philpot BD, Huganir RL, Bear MF. Rapid, experience-dependent expression of synaptic NMDA receptors in visual cortex in vivo. Nat Neurosci. 1999;2:352–357. doi: 10.1038/7263. [DOI] [PubMed] [Google Scholar]
- 25.Richter JD. Dynamics of poly(A) addition and removal during development. In: Hershey J, Mathews M, Sonenburg S, editors. Translational control. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1996. pp. 481–503. [Google Scholar]
- 26.Richter JD. Influence of polyadenylation-induced translation on metazoan development and neuronal synaptic function. In: Sonenberg N, Hershey J, Mathews M, editors. Translational control of gene expression. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 2000. pp. 785–805. [Google Scholar]
- 27.Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3:211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- 28.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu E, Tang YP, Rampon C, Tsien JZ. NMDA receptor-dependent synaptic reinforcement as a crucial process for memory consolidation. Science. 2000;290:1170–1174. doi: 10.1126/science.290.5494.1170. [DOI] [PubMed] [Google Scholar]
- 30.Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium/calmodulin kinase II mutant mice. Science. 1992;257:201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- 31.Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- 32.Ulibarri C, Yahr P. Poly-A+ mRNA and defeminization of sexual behaviour and gonadotripin secretion in rats. Physiol Behav. 1987;39:767–774. doi: 10.1016/0031-9384(87)90264-2. [DOI] [PubMed] [Google Scholar]
- 33.Wells DG, Richter JD, Fallon JR. Molecular mechanisms for activity-regulated protein synthesis in the synapto-dendritic compartment. Curr Opin Neurobiol. 2000;10:132–137. doi: 10.1016/s0959-4388(99)00050-1. [DOI] [PubMed] [Google Scholar]
- 34.Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]