Abstract
Genetic susceptibility to type 2 diabetes is primarily due to β cell dysfunction. However, a genetic study to directly interrogate β cell function ex vivo has never been previously performed. We isolated 233,447 islets from 483 Diversity Outbred (DO) mice maintained on a Western-style diet, and measured insulin secretion in response to a variety of secretagogues. Insulin secretion from DO islets ranged greater than 1000-fold even though none of the mice were diabetic. The insulin secretory response to each secretagogue had a unique genetic architecture; some of the loci were specific for one condition, whereas others overlapped. Human loci that are syntenic to many of the insulin secretion quantitative trait loci (QTL) from mice are associated with diabetes-related SNPs in human genome-wide association studies. We report on 3 genes, Ptpn18, Hunk, and Zfp148, where the phenotype predictions from the genetic screen were fulfilled in our studies of transgenic mouse models. These 3 genes encode a nonreceptor type protein tyrosine phosphatase, a serine/threonine protein kinase, and a Krϋppel-type zinc-finger transcription factor, respectively. Our results demonstrate that genetic variation in insulin secretion that can lead to type 2 diabetes is discoverable in nondiabetic individuals.
Keywords: Cell Biology, Genetics
Keywords: Diabetes, Glucose metabolism, Mouse models
Introduction
Type 2 diabetes (T2D) is a syndrome of relative insulin deficiency wherein the demand for insulin exceeds the ability of β cells to meet that demand. The increased demand is usually caused by insulin resistance, often a consequence of obesity. Limited insulin secretion can be due to insufficient β cell mass or an impaired capacity of the β cells to secrete insulin.
The most common cause of insulin resistance is obesity. Over the past several decades, there has been a dramatic increase in the proportion of the population that is obese or overweight. This has led to an unprecedented diabetes epidemic. Although the diabetes epidemic is primarily due to obesity, only approximately 20% of obese people develop diabetes. This is because most obese people can respond to insulin resistance by increasing insulin production. Thus, the inability to boost β cell function in response to obesity-induced insulin resistance is a tipping point that can lead to T2D. The heritability of insulin secretion is higher than that of insulin resistance (1). Consequently, most of the candidate genes associated with risk of T2D directly or indirectly affect β cell function and/or β cell mass (2–4). Most importantly, the diabetes epidemic is primarily due to dietary changes and their effect on obesity, rather than major changes in the gene pool (5).
The β cell is a nutrient sensor (6). In response to specific nutrients, it mobilizes insulin-containing secretory granules and triggers the exocytosis of insulin. The quintessential insulin secretagogue is glucose, which must be transported into the β cell and metabolized in order to stimulate insulin secretion. However, β cells are also responsive to amino acids and fatty acids. In addition, the response to glucose is amplified by a hormone produced in intestinal L cells and pancreatic α cells, glucagon-like peptide-1 (GLP1).
Despite decades of intense study, much remains unknown about the pathways that mediate insulin exocytosis and what features are unique to the response to specific secretagogues. Nutrient sensing in the β cells involves multiple intracellular signaling pathways, followed by changes in key proteins that participate in the trafficking of secretory vesicles. In addition to specific nutrients, β cell insulin secretion can be experimentally evoked by a high concentration of external K+ ions, which depolarizes the membrane potential, leading to elevations in internal Ca2+ ions.
The inaccessibility of human islets makes it impossible to conduct ex vivo functional phenotyping in human subjects in the sample sizes required to carry out genetic association studies. Thus, insulin secretion has been interrogated in vivo by measuring the insulin and C-peptide response to a secretagogue challenge, usually glucose. The variation in serum insulin can reflect variation in β cell mass or β cell function. In addition, in vivo circulating insulin levels are affected by variation in the insulin clearance rate (7). Without direct access to pancreatic islets, it is not possible to distinguish between these possibilities.
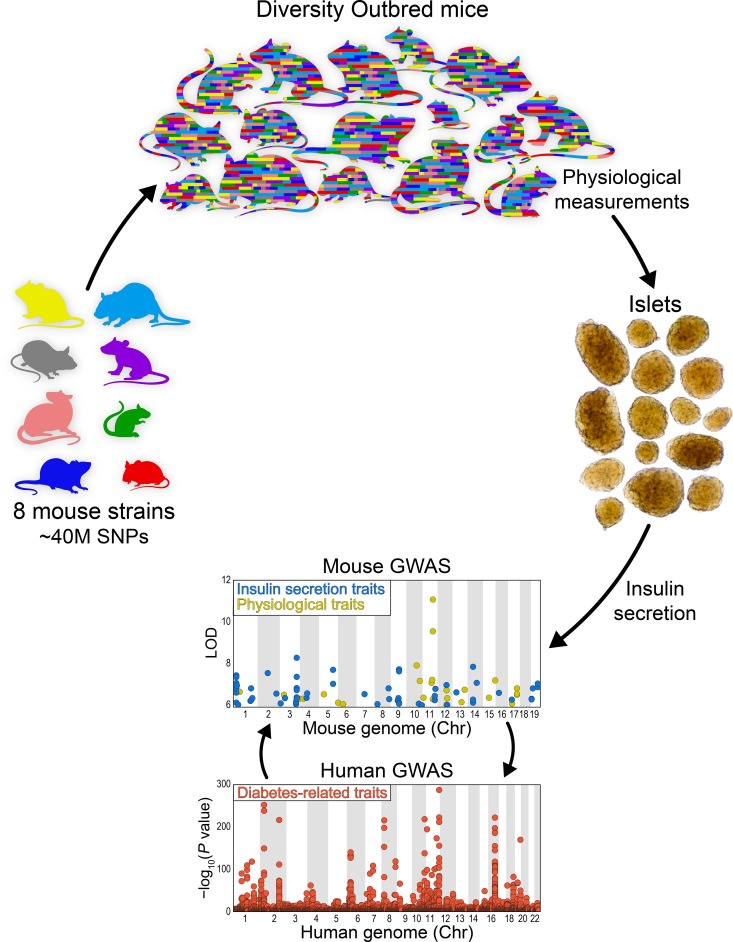
The genetic diversity of the human population can be modeled by the collection of inbred mouse strains available to the research community. Fifteen years ago, the Collaborative Cross project was launched to develop mouse resources that capture this genetic diversity (8). Eight mouse strains that capture most of the genetic diversity of known inbred mouse strains were intercrossed and have been maintained as an outbred stock, called the Diversity Outbred (DO): 129S1/SvImJ, A/J, C57BL/6J, NOD/ShiLtJ, NZO/HILtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ (9). Together, these 8 strains carry about 40 million single nucleotide polymorphisms (SNPs), a level of genetic diversity comparable to that of the entire human population, making the DO mouse population an ideal genetic model resource to link genetic variation with physiological variation in pancreatic islets.
We recently reported on the use of DO mice to describe the genetic architecture of gene expression in pancreatic islets (10). We observed strong heritability for islet mRNA transcripts, yielding approximately 40,000 local and distal expression quantitative trait loci (eQTL), and the identification of genomic hotspots where several hundred distal eQTL co-mapped in response to the same genetic variation. In parallel, we surveyed insulin secretion in pancreatic islets that were isolated from the 8 founder strains used to generate the DO mouse panel (11). In response to several secretagogues, we observed a wide range in insulin secretion among the strains and between male and female mice, suggesting that genetic background and sex have major effects on the insulin secretory capacity of pancreatic islets.
To investigate the pathways that lead from nutrient sensing to insulin secretion, we carried out a genetic screen of insulin secretion ex vivo in DO mice. We isolated pancreatic islets from 483 DO mice and measured insulin secretion in response to 7 different secretagogue challenges: 3 different glucose concentrations, KCl, a mixture of amino acids (leucine, alanine, and glutamine), palmitic acid, and the incretin hormone GLP1. We genotyped the mice at approximately 150,000 SNP loci, allowing us to reconstruct the entire DNA sequence of every mouse. We observed a greater than 1000-fold range in insulin secretion among the mice, enabling us to derive the genetic architecture of insulin secretion corresponding to each individual secretagogue. Finally, we identified 3 candidate genes, derived transgenic mouse models, and validated their role in insulin secretion.
Results
A genetic screen for insulin secretion.
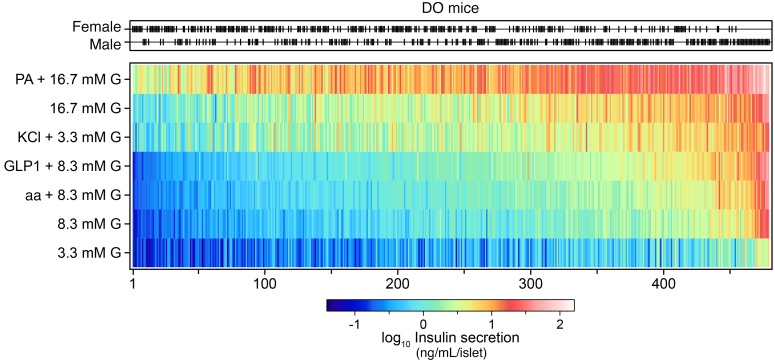
To identify the gene loci that underlie the insulin secretory differences among the DO mice, we surveyed the insulin secretory responses of 233,447 islets collected from 483 DO mice (half for each sex). There was a wide variance in the number of islets harvested per mouse (42–1096 islets). The whole-islet content of insulin (85 ± 2 ng/islet) was not different between the sexes, whereas islet glucagon levels were approximately 2-fold lower in males than females (360 ± 15 pg/islet vs. 678 ± 33 pg/islet, respectively). To interrogate the genetic architecture of insulin secretion, islets from each mouse were challenged with 7 different secretagogue conditions: low (3.3 mM), medium (8.3 mM), or high (16.7 mM) glucose; low glucose plus KCl (40 mM); medium glucose plus a cocktail of 3 amino acids (aa) (leucine, 0.5 mM; alanine, 1.25 mM; glutamine, 2 mM) or the incretin hormone GLP1 (100 nM); and high glucose plus palmitic acid (PA) (0.5 mM). Across these conditions, insulin secretion varied more than 1000-fold (Figure 1).
Figure 1. Ex vivo insulin secretion measurements from 479 DO mice maintained on a Western-style diet.
Distribution of female (n = 240) and male (n = 239) DO mice. Black ticks show sex for each mouse. Insulin secretion was determined from single isolated islets in response to 7 conditions: 3.3, 8.3, and 16.7 mM glucose (G); the amino acids (aa) leucine (0.5 mM), alanine (1.25 mM), and glutamine (2 mM) plus 8.3 mM G; KCl (40 mM) plus 3.3 mM G; GLP1 (100 nM) plus 8.3 mM G; and the fatty acid (PA) palmitate (0.5 mM) plus 16.7 mM G. Heatmap illustrates the amount of insulin secreted into the medium for each condition. Mice are ordered by the median value of their insulin secretory responses to the 7 conditions, highlighting mice that demonstrated low (left side) versus high (right side) secretory capacity. Insulin secretion values are the geometric mean of 6 individual measurements/condition/mouse for 479 DO mice, yielding a total of approximately 20,000 measures.
Ordering the mice by their median response to all 7 insulin secretion conditions revealed a smooth transition from mice that generally showed a poor secretory response, to mice that were highly responsive to all conditions. Male mice tended to belong to the highly responsive group; the 25 mice with the highest secretory response were all male (Figure 1). Under all test conditions, islets from male mice secreted more insulin than did islets from female mice (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI129143DS1). The sex difference was greatest in response to GLP1, where males secreted approximately 3-fold more insulin than females.
In 365 DO mice, we measured glucagon secretion in response to KCl plus 3 mM glucose (Supplemental Figure 1). A greater than 10-fold range in glucagon secretion was observed among the mice, from about 2 pg/islet to about 33 pg/islet, and on average was approximately 50% greater in islets from female than male mice (P < 3 × 10–7). KCl-induced glucagon secretion was only weakly correlated with KCl-induced insulin secretion (r ~ 0.19), despite both measurements deriving from the same islet samples in response to the same stimulus. This suggests that the gene loci and molecular components that mediate the KCl-induced entry of Ca2+ ions and mobilization and exocytosis of glucagon granules in α cells are distinct from those that mediate that for insulin granules in β cells.
Prior to collecting islets for the ex vivo secretion measurements, we measured several whole-body physiological traits in all of the mice (10). These traits included an oral glucose tolerance test (oGTT), homeostatic model assessments (HOMA) for insulin resistance (IR) and pancreatic β cell function (B), measures of plasma glucose, insulin, and triglyceride (TG), and body weight at 6, 10, and 14 weeks of age, number of islets isolated per mouse, and the total islet insulin content. In addition, we measured total islet glucagon content, body weight, plasma glucose, insulin, and TG when the mice were euthanized (Supplemental Figure 2). Fasting plasma glucose at the age the mice were euthanized (22–26 weeks) exceeded 300 mg/dL in only 7 of 483 mice, all male. In summary, a large dynamic range was observed for all phenotypes measured among the mice, most of which were strongly influenced by sex. That the vast majority of the DO mice were not diabetic indicates that these phenotypes are linked to genetic variation among the mice, and are not a consequence of diabetes.
We asked if the whole-body physiological phenotypes were correlated with the insulin and glucagon secretion phenotypes from the isolated islets. Because of the strong sex bias observed in both sets of traits, we computed the pairwise Pearson’s correlations between all traits separately in females and males (Supplemental Figure 3). The insulin secretion responses evoked by each of the 7 conditions were positively intercorrelated. For example, secretion in response to aa or GLP1 was strongly correlated (r ~ 0.9) with secretion in response to 8.3 mM glucose in both male and female mice. Similarly, PA-induced insulin secretion correlated with 16.7 mM glucose (r ~ 0.8) and KCl-induced secretion correlated with 3.3 mM glucose (r ~ 0.6). These results demonstrate that for some of the nutrient secretagogues (e.g., aa, GLP1, and PA), glucose played a dominant role in controlling the amount of insulin secretion among the mice. Consistent with their “amplifying” role (12), the metabolic nutrients (aa, GLP1, and PA) exerted a positive but smaller influence on the secretory response of isolated islets.
Insulin secretion was correlated with several of the physiological phenotypes that were measured in the mice prior to euthanasia. Sex had a strong influence on these relationships. For example, the total amount of insulin/islet, body weight at all ages, AUCinsulin, and HOMA-IR were all positively correlated with insulin secretion. Interestingly, with the exception of insulin/islet, these relationships were generally stronger in males than females. The average correlation between AUCinsulin and the 7 insulin secretion traits was approximately 0.26 and approximately 0.48 in females and males, respectively (P < × 10–4). An opposite pattern was observed for islet insulin content. In male mice, insulin content correlated with the insulin secretion traits with an r ~0.37, whereas in female mice, this correlation was ~0.47 (P = 0.005). These results suggest that in response to a high-fat/high-sucrose (HF/HS) diet, male mice become more insulin resistant and require greater amounts of insulin secretion from islets than female mice to maintain euglycemia. This increased insulin secretion may in turn result in a small decrease in insulin content in islets from male mice.
Both glucagon secretion and islet glucagon content were negatively correlated with several whole-body physiological traits (e.g., body weight, plasma glucose, AUCglucose); these correlations were stronger in male than in female mice. In summary, these results show that the insulin and glucagon secretory responses determined in the ex vivo islet studies correlate with diabetes-related physiological phenotypes of the whole animal, and that these relationships can be strongly influenced by sex.
Genetic architecture of insulin secretion.
The heritability (h2) of the ex vivo insulin secretion traits was consistently high (Supplemental Figure 4). Basal insulin secretion (3.3 mM glucose) had h2 = 0.36, whereas stimulated insulin secretion ranged from h2 = 0.51 (8.3 mM glucose) to 0.62 (PA + 16.7 mM glucose). We observed high heritability for the number of islets per mouse (h2 = 0.53), insulin per islet (h2 = 0.57), and whole pancreas insulin content (h2 = 0.60), computed as the number of islets per mouse multiplied by the insulin content per islet. Despite high heritability, none of the insulin secretion traits resulted in large-effect QTL. Thus, the ex vivo insulin secretion traits were highly heritable and their genetic architecture complex, involving multiple loci. Overall, the ex vivo islet phenotypes displayed higher heritability compared with the in vivo, whole-body physiological phenotypes (e.g., plasma glucose and insulin), where h2 ranged from 0.2–0.47.
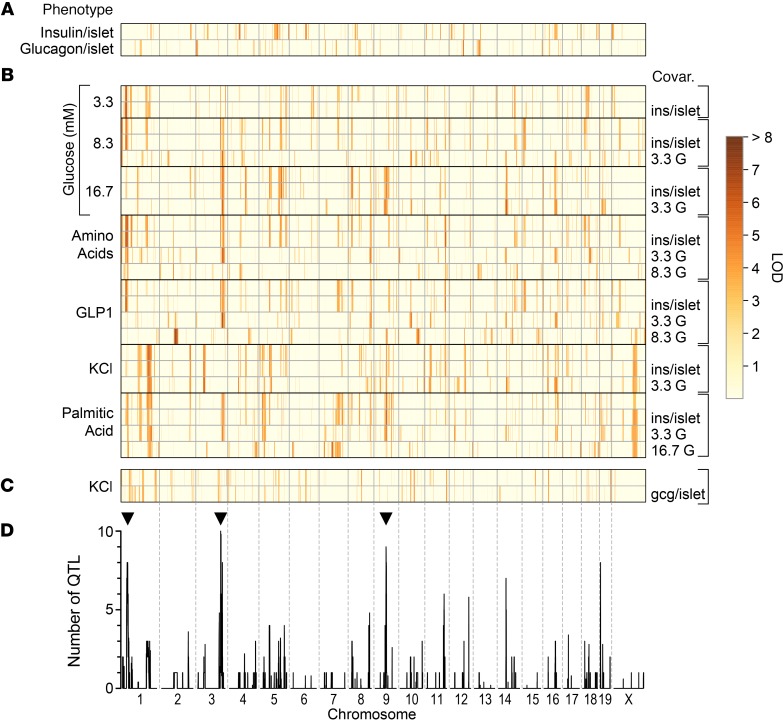
To identify the genetic loci that modulate insulin and glucagon secretion from isolated islets, we performed whole-genome scans for each of the secretagogue conditions, using sex as an additive covariate. Ex vivo insulin secretion is typically represented by 1 of 3 metrics (11): the amount of insulin secreted into the medium (secretion), the proportion of the total islet content that is secreted (fractional secretion), or the fold-stimulation of secretion relative to basal conditions (fold-change relative to 3.3 mM). Using a covariate approach to provide the fractional and fold-stimulatory secretory responses (see Methods), we mapped all 3 metrics for each secretion trait in addition to islet insulin and glucagon content. Application of a suggestive genome-wide threshold (LOD > 6; see Methods) identified 60 QTL across these traits, 5 of which exceeded genome-wide P < 0.05 significance (LOD > 7.4) (Figure 2).
Figure 2. The genetic architecture of insulin and glucagon secretion.
Inferred QTL (LOD ≥ 6) for ex vivo islet phenotypes determined from DO mice maintained on HF/HS diet. (A) Total islet content for insulin and glucagon (n = 482 mice each). (B) Insulin secretion in response to 7 different conditions; low (3.3 mM), medium (8.3 mM) or high (16.7 mM) glucose (G), medium glucose plus amino acids (aa; Leu, Ala and Gln) or the incretin hormone GLP1 (100 nM), high glucose plus palmitic acid (PA; 0.5 mM), or low glucose plus KCl (40 mM) (n = 479 mice). (C) Glucagon secretion in response to low glucose plus KCl (n = 365 mice). Secretion traits were mapped without or with conditioning ( | ) on the islet content for insulin (ins/islet) or glucagon (gcg/islet), or the concentration of glucose used for the condition (e.g., 16.7 mM for PA-induced secretion). Red = high LOD, yellow = low LOD. (D) Profile illustrating the number of QTL (LOD > 5) occurring within a 4 Mb genomic window. QTL hotspots with 7 or more co-mapping traits were identified on chromosomes 1, 3, and 9 (see black arrowheads). Supplemental Table 1 lists all QTL, their LOD scores, genomic position, and allele effect values at the peak.
We discovered several secretion QTL hotspots where 3 or more secretion traits co-mapped. A hotspot on proximal Chr 1 at approximately 30 Mb had QTL for 3.3 mM and 8.3 mM glucose, GLP1, and aa-induced insulin secretion. Mapping to distal Chr 3 at approximately 135 Mb was insulin secretion in response to 8.3 mM or 16.7 mM glucose, aa, and fatty acid (PA). A third secretion hotspot was identified on Chr 9 at approximately 66 Mb for 16.7 mM glucose, GLP1, and PA. Other QTL where 2 secretion traits co-mapped included chromosomes 8 (~115 Mb; aa and GLP1), 11 (~103 Mb; aa and GLP1), 14 (~46 Mb; 16.7 mM glucose and GLP1), and 19 (~5 Mb; 8.3 mM glucose and GLP1). At each quantitative trait locus, distinct combinations of secretion traits co-mapped, suggesting that the genetic variation at the locus selectively affects the insulin exocytotic response, depending on the metabolic or nonmetabolic (e.g., KCl) triggers that were used to stimulate insulin secretion.
The founder strain alleles associated with higher or lower secretion were very similar across traits that co-map to a common hotspot, suggesting the presence of shared genetic driver(s). For example, at Chr 1, insulin secretion was increased when the genotype at the locus was derived from the CAST founder strain (Supplemental Figure 5). The hotspot on Chr 3 showed CAST and PWK as the low alleles and NOD and WSB as the high alleles. We used the similarity of founder allele effects on co-mapping traits to determine whether one quantitative trait locus or multiple linked QTL is affecting these traits. Using this approach, we reduced our 60 QTL to 25 distinct QTL for insulin secretion, and one quantitative trait locus for glucagon secretion. In addition, we identified a quantitative trait locus for islet insulin content and 3 QTL for islet glucagon content (Supplemental Table 1).
In addition to secretion hotspots, we identified QTL that were selective for a single secretagogue. For example, fold-change in insulin secretion in response to GLP1, a measure of the GLP1-dependent secretory response, separate from glucose, mapped to 3 QTL: Chr 2 at approximately 84 Mb, LOD = 7.5; Chr 10 at approximately 102 Mb, LOD = 6.1; and Chr 14 at approximately 79 Mb, LOD = 7.1. No other secretion traits mapped to these QTL, suggesting that these loci are specific for GLP1-dependent insulin secretion. Similarly, insulin secretion in response to KCl mapped exclusively to a QTL on proximal Chr 3 at approximately 41 Mb, LOD = 6.35.
We identified 3 QTL for islet glucagon content (Chrs 3, 12, and 13) and 1 quantitative trait locus for KCl-induced glucagon secretion (Chr 19). The Chr 19 quantitative trait locus was observed whether we mapped glucagon secretion with (LOD = 7.0), or without (LOD = 6.9) conditioning on islet glucagon content, indicating that the amount of glucagon secreted in response to KCl was not determined by the glucagon content within the islets. The NZO allele conferred low glucagon secretion whereas the PWK was the high allele. Interestingly, the glucagon secretion QTL did not overlap with the quantitative trait locus for KCl-induced insulin secretion, consistent with a weak correlation between KCl-induced insulin and glucagon secretion, suggesting that distinct pathways downstream of KCl mediate insulin and glucagon secretion.
Identification of candidate regulators of insulin secretion.
In order to identify the most promising candidate genes in the QTL support intervals, we performed a mediation test for each target phenotype and its associated QTL, based on changes in LOD after including the mRNA expression of candidate mediators as covariates in the genome scan model (13). This approach identified a reduced number of candidate genes that are potential mediators for ex vivo secretion QTL on chromosomes 1, 3, 8, 11, and 19 (Supplemental Table 2). For each candidate mediator, we applied the causal model selection test (CMST) to determine which of the competing models (causal, reactive, independent, or complex) is most consistent with the data (14).
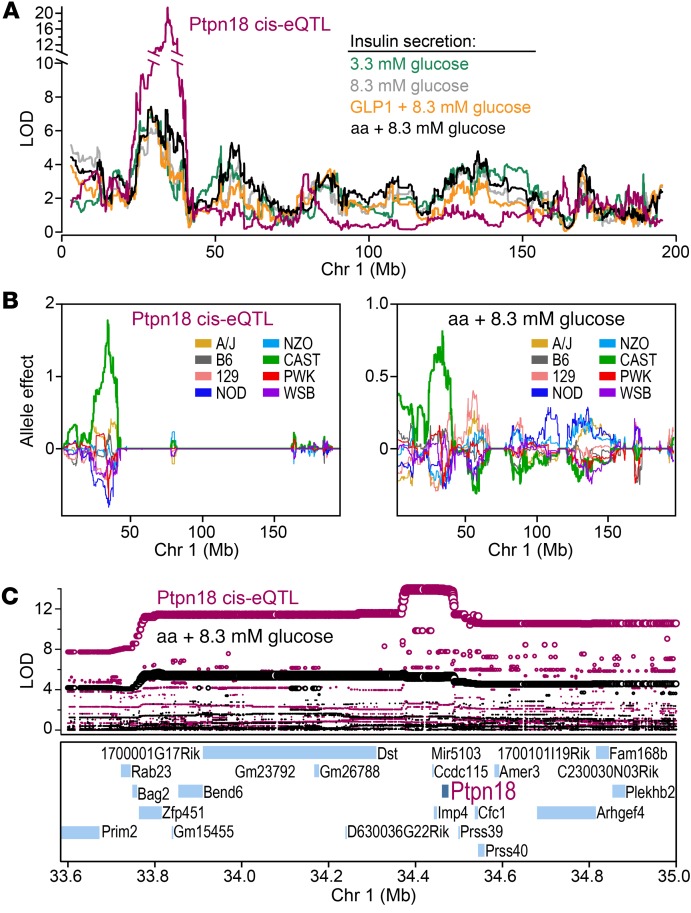
Mediation analysis predicted that Ptpn18, a protein tyrosine phosphatase, is a candidate driver for the insulin secretion QTL hotspot on Chr 1 at approximately 34 Mb (Supplemental Table 2). Several insulin secretion traits mapped to this locus, including secretion in response to 3.3 mM and 8.3 mM glucose, GLP1, and aa (Figure 3A). A local expression quantitative trait locus for Ptpn18 (Figure 3A) shared the same allele effect pattern as the insulin secretion traits; both were associated with CAST alleles (Figure 3B; Supplemental Figure 5). The directionality of the association suggests that CAST alleles lead to an increase in the expression of Ptpn18, and an increase in insulin secretion. SNPs most strongly associated with insulin secretion were the same as those associated with Ptpn18 expression, and spanned a region that contains 30 genes, from approximately 33.7 to approximately 34.7 Mb (Figure 3C). Of these, only Ptpn18 produced a significant drop in the LOD score for the secretion QTL in the mediation analysis. The CMST supported a causal model (P = 1.6 × 10–10), suggesting that CAST-private SNPs are associated with changes in the expression of Ptpn18 that mediate the insulin secretion QTL.
Figure 3. An insulin secretion hotspot on Chr 1 demonstrates shared genetic architecture with islet expression of Ptpn18.
(A) LOD profiles on Chr 1 for the islet expression of Ptpn18, and insulin secretion in response to 3.3 mM or 8.3 mM glucose, GLP1, or amino acids (aa). A quantitative trait locus was identified for all 5 traits at approximately 34 Mb. (B) Allele effect plots for Ptpn18 local eQTL and insulin secretion in response to aa. Both traits were linked to CAST as the high allele at the quantitative trait locus. (C) SNP association plots for Ptpn18 local eQTL and aa-induced insulin secretion. Twenty-four annotated genes are located within the region showing highest SNP association (~33.6 Mb and ~35 Mb), including Ptpn18. Ptpn18 was the only gene to show a local expression quantitative trait locus with a matching allele dependence to the insulin secretion quantitative trait locus. All data derive from QTL analysis of traits measured in 479 DO mice.
A catalytically inactive, substrate-trapping mutant of Ptpn18 results in improved insulin sensitivity and reduced insulin secretion from pancreatic islets.
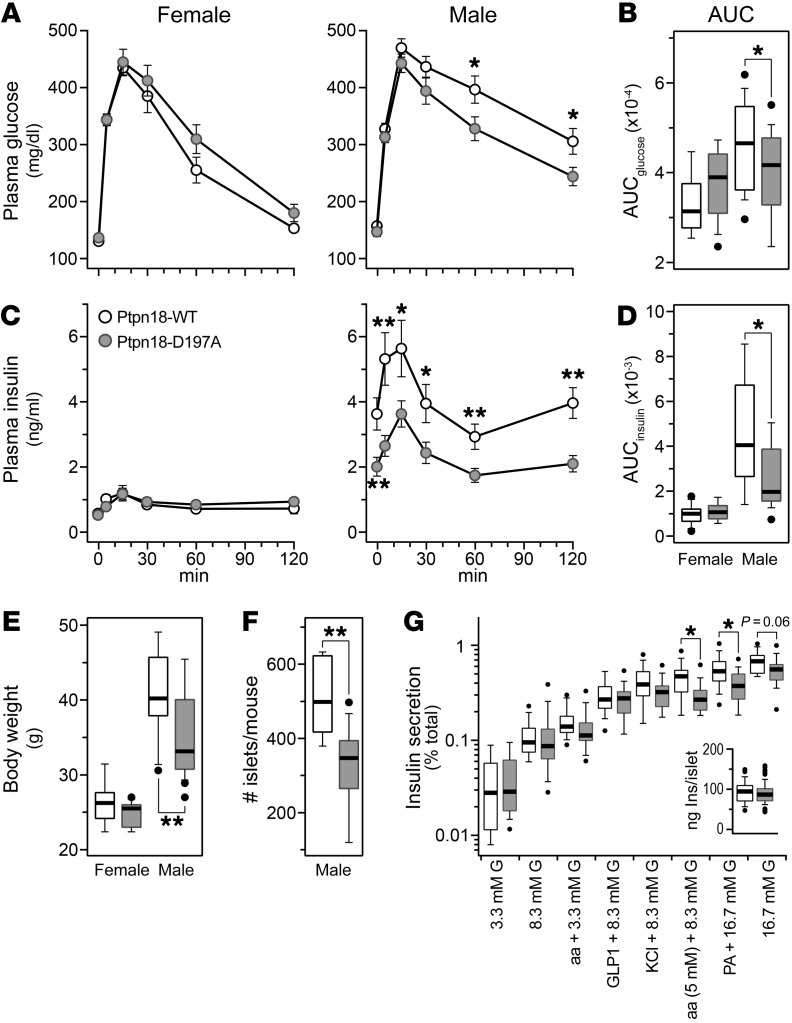
To test the role of Ptpn18 as a gene responsible for the Chr 1 QTL hotspot, we generated a mouse with catalytically inactive Ptpn18. We used CRISPR to substitute a conserved aspartate residue (Asp197) that is within the catalytic domain and required for tyrosine phosphatase activity, with an alanine residue. This strategy was originally used to create a catalytically inactive, substrate-trapping mutant of PTP1B (15), and later for Ptpn18 (16), to identify their direct protein substrates. We asked if the islets from mice carrying the Ptpn18D197A mutation have altered insulin secretion compared with WT mice.
To evaluate islet function in vivo, we measured circulating insulin levels during an oGTT. Glucose tolerance and insulin dynamics were not different between Ptpn18D197A and WT male or female mice maintained on chow diet (Supplemental Figure 6, A–D), or female mice maintained on the HF/HS diet (Figure 4, A and C). However, male Ptpn18D197A mice fed a HF/HS diet for 16 weeks showed improved glucose tolerance during an oGTT (Figure 4, A–D). Circulating insulin was significantly reduced in male Ptpn18D197A mice during the oGTT, including the fasting value at time zero (Figure 4, C and D), suggesting that male Ptpn18D197A mice maintained on a HF/HS diet have enhanced insulin sensitivity. In parallel with improved insulin sensitivity was a reduction in body weight in male mutant mice on the HF/HS diet (Figure 4E). Body weight was not different between chow-fed Ptpn18D197A and WT mice of either sex (Supplemental Figure 6E). These results suggest that HF/HS diet–induced insulin resistance, which is much more pronounced in male than in female B6 mice (Figure 4D), is required to reveal the role played by Ptpn18 in pancreatic islets.
Figure 4. Ptpn18D197A mutant mice show reduced body weight, improved insulin sensitivity, and reduced insulin secretion from pancreatic islets.
Plasma glucose (A) and insulin (C) during an oGTT in female (n = 12, 13) and male (n = 19, 22) Ptpn18-WT and Ptpn18D197A mice, respectively, that were maintained on the HF/HS diet for 4 months; area under the curve (AUC) values for glucose (B) and insulin (D). Body weight at 18 weeks of age in female (n = 12, 13) and male (n = 19, 22) Ptpn18-WT and Ptpn18D197A mice, respectively (E). Number of islets harvested per mouse (F) (n = 8, 12) and insulin secretion (G) (n = 13, 16; G indicates glucose) from male Ptpn18-WT and Ptpn18D197A mice, respectively. *P < 0.05; **P < 0.01 for Student’s 2-tailed t test.
We asked if the protection from diet-induced insulin resistance observed in the male Ptpn18D197A mice translated to differential insulin secretion from isolated islets. Because male but not female mice showed enhanced insulin sensitivity in mice carrying the Ptpn18D197A mutation, we focused on male mice for our ex vivo islet studies. In parallel with reduced body weight, the number of islets isolated from Ptpn18D197A mice was significantly lower than that from WT mice (Figure 4F); 315 versus 511 islets, respectively (P = 0.001). Islets from Ptpn18D197A mice secreted significantly less insulin in response to amino acids and fatty acid (P < 0.05), and showed a strong trend for reduced secretion in response to high glucose (P = 0.07) (Figure 4G). Similarly, islets from chow-fed Ptpn18D197A mice showed reduced insulin secretion in response to 8.3 mM glucose and amino acids (Supplemental Figure 6F). The average number of islets isolated per mouse was not different between chow-fed Ptpn18D197A and WT mice: 302 versus 287 (female) and 311 versus 296 (male), respectively.
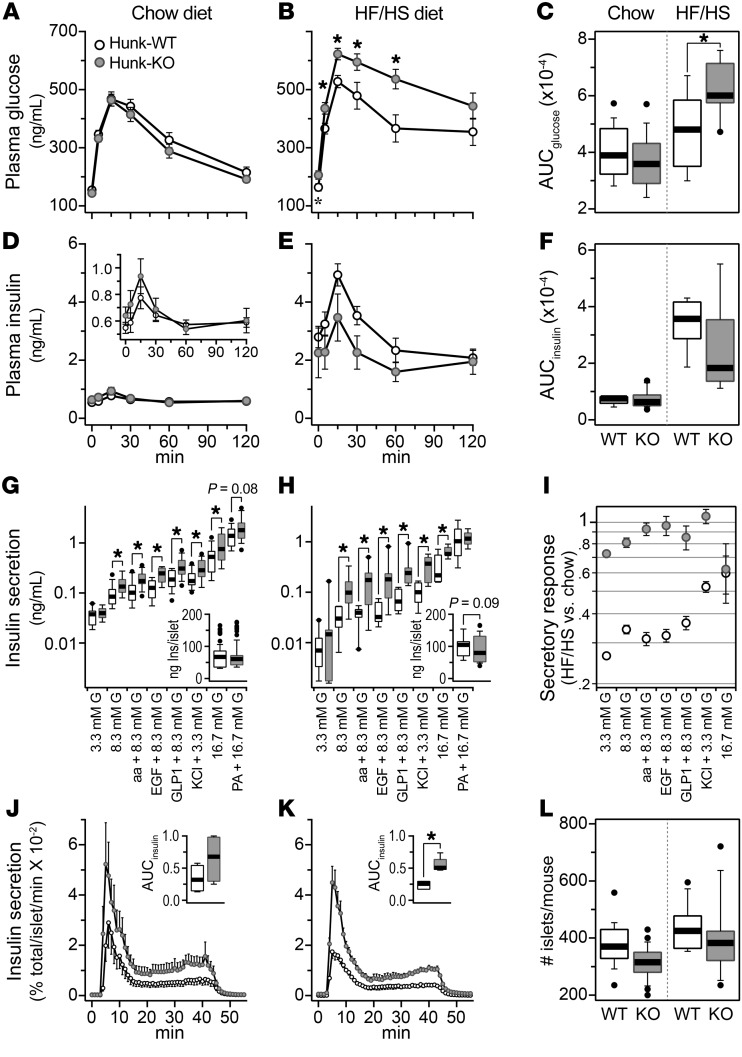
Loss of Hunk results in protection from Western-style diet-induced islet dysfunction, leading to enhanced insulin secretion.
At the midpoint of the study, we carried out a planned interim QTL mapping analysis (see Discussion) and mapped insulin secretion in response to GLP1 to a locus on Chr 16 at approximately 90.7 Mb, LOD = 7.4. Insulin secretion is low when the allele at this locus is derived from CAST mice. The gene for hormonally upregulated Neu-associated kinase (Hunk) is located on Chr 16 at 90.4 Mb. In our eQTL study, we identified an islet local expression quantitative trait locus for Hunk with a dependence on the CAST allele, but in the opposite direction (10). The high allele for the local expression quantitative trait locus was associated with CAST, suggesting that Hunk expression is elevated in response to SNPs present in CAST at the Hunk locus, predicting that Hunk is a negative regulator of insulin secretion.
We derived Hunk-knockout mice Hunk-KO (17) and evaluated insulin secretion in their islets. The Hunk-KO and Hunk-WT mice were maintained on chow diet, or the HF/HS diet used for the insulin secretion screen of the DO mice. Prior to isolating their islets for the ex vivo insulin secretion measurements, an oGTT was performed on each mouse during which we measured glucose and insulin at multiple time points. Similar to our observations with Ptpn18D197A mice (Supplemental Figure 6), no difference in glucose or insulin was observed between Hunk-KO and Hunk-WT male mice maintained on chow diet (Figure 5, A–F). Consistent with diet-induced insulin resistance, plasma insulin levels were significantly higher in mice maintained on the HF/HS diet (Figure 5F). However, like chow-fed mice, there was no difference in the insulin values during the oGTT between Hunk-KO and Hunk-WT mice fed the HF/HS diet. Hunk-KO mice showed delayed recovery of plasma glucose during the oGTT (Figure 5, B and C), suggesting that Hunk-KO mice develop glucose intolerance in response to the HF/HS diet.
Figure 5. Hunk is necessary for Western diet–induced islet dysfunction.
Plasma glucose (A, B) and insulin (D, E) for Hunk-WT and Hunk-KO male mice maintained on either chow diet (n = 15 and 17, respectively) or the HF/HS diet (n = 8 each); AUC values for glucose (C) and insulin (F). Ex vivo insulin secretion measures for Hunk-WT and Hunk-KO male mice maintained on chow diet (G) (n = 14, 15) or HF/HS diet (H) (n = 8 each). Insets in G and H show total insulin content per islet for Hunk-WT and Hunk-KO mice. Ratio (HF/HS vs. chow diet) of insulin secretory responses (I). Insulin secretion during dynamic perifusion assay for Hunk-WT and Hunk-KO male mice maintained on chow diet (J) (n = 4 each) or HF/HS diet (K) (n = 3 and 5, respectively). Glucose was increased from 3.3 mM to 16.7 mM from time 0 to 40 minutes, after which glucose was returned to 3.3 mM. Insets in J and K show AUC for insulin responses during perifusion. Number of islets harvested per mouse (L); n = 23, 21 (chow), and n = 12 each (HF/HS) for Hunk-WT and Hunk-KO male mice, respectively. *P < 0.05 for Hunk-WT versus Hunk-KO, for Student’s 2-tailed t test. In G, H, and I, G indicates glucose.
In contrast to the in vivo observations, we observed a robust difference between Hunk-KO and Hunk-WT mice in the ex vivo insulin secretion measurements. During static insulin secretion measurements, islets from Hunk-KO mice maintained on either chow diet (Figure 5G) or the HF/HS diet (Figure 5H) secreted significantly more insulin than islets from Hunk-WT mice in response to nearly all secretagogue challenges. Insulin secretion in response to 8.3 mM glucose, aa, EGF, or GLP1 was reduced about 70% in islets isolated from WT mice on the HF/HS diet (Figure 5I). This diet-induced reduction in insulin secretion was not observed in islets from Hunk-KO mice, suggesting that Hunk is required for islet dysfunction evoked by a HF/HS diet.
We performed perifusion experiments to evaluate the distinct phases of insulin secretion. Both the first and second phases of insulin secretion in response to 16.7 mM glucose were augmented in Hunk-KO islets. This was observed in mice maintained on either chow diet (Figure 5J) or the HF/HS diet (Figure 5K), suggesting that a general secretory enhancement occurs in the absence of Hunk. Total islet insulin content was significantly reduced in Hunk-KO mice maintained on HF/HS diet (Figure 5H, inset), which may be a consequence of the elevation in insulin secretion. We performed a 2-way ANOVA to evaluate the number of islets isolated for Hunk-KO versus Hunk-WT mice that were maintained on either chow, or the HF/HS diet (Figure 5L). The HF/HS diet resulted in an approximately 20% increase (P = 6.6 × 10–4) in islet number for both mice, suggesting that Hunk is not required for the compensatory increase in β cell mass that results from diet-induced insulin resistance, in contrast to its negative effect on β cell function. After accounting for this diet effect, there was an approximately 15% decrease (P = 3.1 × 10–3) in the number of islets for Hunk-KO versus Hunk-WT mice.
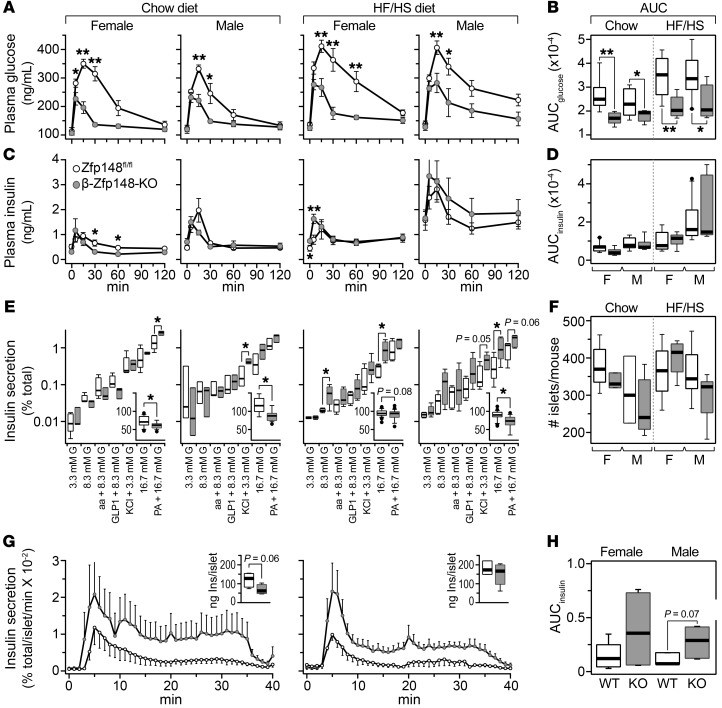
β cell–specific deletion of Zfp148 improves glucose tolerance and enhances insulin secretion from pancreatic islets.
In the interim QTL mapping analysis, we identified a second quantitative trait locus for insulin secretion on Chr 16 at approximately 32 Mb. The Zfp148 gene is located at this locus and its mRNA abundance maps to this site, a local expression quantitative trait locus. Across the DO mice, the expression of Zfp148 was negatively correlated with glucose-stimulated insulin secretion. Thus, we predicted that, like Hunk, Zfp148 is a negative regulator of insulin secretion. Zfp148 is highly expressed in mouse and human (ZNF148) β cells.
We generated mice with a β cell–specific knockout of Zfp148 (β-Zfp148-KO) and performed oGTTs on Zfp148fl/fl and β-Zfp148-KO mice that were maintained on either a chow or HF/HS diet. Male and female β-Zfp148-KO mice showed greatly enhanced glucose recovery during an oGTT, regardless of diet (Figure 6). Glucose values at 15 and 30 minutes during the oGTT were significantly lower in β-Zfp148-KO versus Zfp148fl/fl mice (Figure 6A), resulting in a significant reduction in the AUCglucose (Figure 6B). In parallel with accelerated glucose recovery, the kinetics of the insulin response in β-Zfp148-KO and Zfp148fl/fl mice were distinct. In mice maintained on either diet, insulin peaked at 5 minutes in β-Zfp148-KO mice, whereas in Zfp148fl/fl mice, insulin peaked at 15 minutes (Figure 6C). Due to the greater variability of the dynamic changes in insulin compared with glucose during the oGTT, AUCinsulin was not significantly different between β-Zfp148-KO and Zfp148fl/fl mice (Figure 6D).
Figure 6. β-Zfp148-KO mice show enhanced glucose tolerance during oGTT.
Plasma glucose (A) and insulin (C) responses during an oGTT performed on female and male control (Zfp148fl/fl) and β cell–specific Zfp148 knockout (β-Zfp148-KO) mice maintained on either chow diet (n = 8, 6 Zfp148fl/fl and β-Zfp148-KO female and male mice, respectively) or the HF/HS diet (n = 8 female each and n = 11, 7 for male Zfp148fl/fl and β-Zfp148-KO mice, respectively); AUC for glucose (B) and insulin (D). Ex vivo secretion measurements on Zfp148fl/fl and β-Zfp148-KO female and male mice maintained on either chow diet (n = 5, 3, 3 and 3, respectively), or the HF/HS diet (n = 5, 6, 5 and 5, respectively) (E). Insets show total islet insulin content (ng Ins/islet) for each sex/diet group. Total number of islets harvested per mouse maintained on either chow diet (n = 5, 3, 3, and 5, respectively) or the HF/HS diet (n = 6, 6, 8 and 8, respectively) for Zfp148fl/fl and β-Zfp148-KO female and male mice, respectively (F). Insulin secretion measurements during islet perifusion for HF/HS diet–fed female (n = 5, 4) and male (n = 5, 7) Zfp148fl/fl and β-Zfp148-KO mice, respectively (G). Islets were exposed to 16.7 mM glucose from 0 minutes to 30 minutes. Insets, total insulin content per islet for perifusion studies. AUC for insulin values in response to 16.7 mM glucose determined during perifusion studies (H). *P < 0.05; **P < 0.01 for Zfp148fl/fl versus β-Zfp148-KO mice, for Student’s 2-tailed t test.
Islets from female and male β-Zfp148-KO mice maintained on the HF/HS diet secreted about 3-fold more insulin in response to high glucose than control mice (Figure 6E). In chow-fed mice, female β-Zfp148-KO mice showed enhanced responsiveness to fatty acid-induced insulin secretion, whereas male β-Zfp148-KO mice secreted more insulin in response to KCl. Under all conditions, islets from β-Zfp148-KO mice had a lower total insulin content than Zfp148fl/fl mice (Figure 6E, insets). The number of islets isolated from Zfp148fl/fl or β-Zfp148-KO male or female mice maintained on either chow diet or the HF/HS diet was not significantly different (Figure 6F).
We performed perifusion experiments to evaluate the first and second phases of insulin secretion. In male and female mice maintained on HF/HS diet, 16.7 mM glucose stimulated higher insulin secretion during both the first and second phase in β-Zfp148-KO mice (Figure 6G). The total AUC for glucose-induced insulin secretion during the perifusion measurement was elevated in islets from β-Zfp148-KO mice (Figure 6H).
An interactive QTL viewer.
All data from this study are available on the web-based analysis tool QTL viewer (10) (https://churchilllab.jax.org/qtlviewer/attie/islets). This powerful data resource allows the user to scan the entire genome for evidence of genetic association for any trait, and displays the allele dependence and SNP associations at selected loci (13, 18). Local eQTL that demonstrate an allele dependence similar to that for secretion QTL are strong candidates as mediators of the secretion trait.
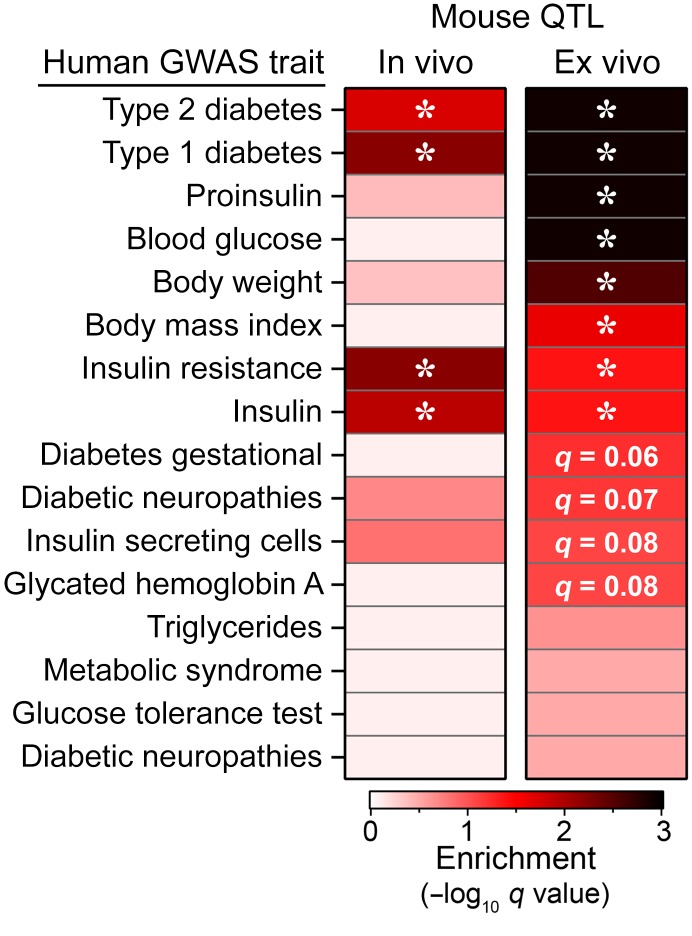
Insulin secretion QTL from mice are enriched with diabetes-related SNPs in human GWAS.
We asked if the ex vivo insulin and glucagon secretion QTL and the QTL associated with the in vivo whole-body physiological traits (e.g., plasma insulin) from the DO mice are associated with diabetes risk alleles identified in human genome-wide association studies (GWAS). We downloaded 16,415 SNP-trait associations from GWAS Central (www.gwascentral.org) that were nominally associated (P < 10–4) with 16 diabetes-related traits (e.g., blood glucose and insulin, glucose tolerance test, proinsulin, and type 1 and type 2 diabetes) that were measured in 216 human genetic studies (Supplemental Table 5).
To determine if the QTL identified in our DO mouse study correspond to loci associated with diabetes risk in humans, we computed a Bayesian support interval based on the local architecture of linkage disequilibrium (LD) at each quantitative trait locus. These regions were mapped onto the human genome using the LiftOver utility (see Methods). We then determined if these syntenic regions were enriched with the diabetes-associated SNPs. We considered ex vivo and in vivo traits separately, and computed a q value to control FDR at 5%, correcting for the 16 GWAS traits evaluated (19–21).
Our results show significant enrichment (q < 0.05) at both the ex vivo and in vivo QTL for SNPs associated with type 1 and type 2 diabetes, as well as insulin resistance and serum insulin (Figure 7). The ex vivo QTL were further enriched for proinsulin, blood glucose, body weight, and body mass index. Ex vivo QTL showed suggestive enrichment (q < 0.1) for gestational diabetes, diabetic nephropathies, insulin-secreting cells, and HbA1c. These results suggest that loci associated with our ex vivo islet studies more closely align with the diabetes risk alleles in human GWAS than the whole-body physiological traits, consistent with the risk variants affecting islet function, as has been previously suggested (2–4).
Figure 7. Insulin secretion QTL enrich for diabetes GWAS SNPs.
SNPs nominally associated (P < 10–4) to 16 diabetes-related traits were obtained from GWAS Central. QTL identified for whole-body physiological phenotypes in live mice (in vivo), or for insulin secretion traits from isolated islets (ex vivo) were mapped onto the human genome to identify syntenic regions. Enrichment for SNPs associated with one or more traits was determined for mapped regions; q values are corrected for number of tests; *q < 0.05.
We asked which GWAS trait(s) were associated with specific in vivo and ex vivo QTL. As we reported previously (18), a strong QTL for plasma insulin is located at the Hnf1b locus on Chr 11 at approximately 82 Mb (see also Supplemental Table 1). This locus contains GWAS SNPs associated with insulin resistance and insulin (Supplemental Figure 7), suggesting that genetic factors controlling circulating insulin are common between mice and humans. The strongest associations were observed for GWAS triglyceride levels at terminal plasma insulin QTL on chromosomes 5 and 10, and for T2D at a terminal body weight quantitative trait locus on Chr 17. Half of the islet-based ex vivo QTL were significantly associated with one or more GWAS traits. A glucagon islet content quantitative trait locus on Chr 12 contained GWAS SNPs for all traits except proinsulin, suggesting that glucagon may play a critical role in diabetes risk, as proposed by Unger and colleagues (22). Several insulin secretion QTL aligned with multiple GWAS traits, including QTL on chromosomes 1, 2, 7, 8, 14, and 17. In summary, our results demonstrate that the QTL we identified in our DO genetic screen correspond to human loci associated with diabetes risk in human GWAS.
Discussion
This is the first genetic screen of insulin secretion on ex vivo pancreatic islets ever performed. To distinguish insulin secretion from insulin clearance or insulin resistance, we carried out direct measurements of insulin secretion from isolated pancreatic islets ex vivo. We asked, to what extent do the pathways that are responsive to each secretagogue overlap? Since each DO mouse inherited a unique collection of alleles that affect islet function, we can determine the degree to which their insulin secretory responses are correlated. We found that in some mice, there was a uniformly strong or weak response to all of the secretagogues, whereas in other mice there was a strong response to some but not all secretagogues. One interpretation of these results is that the earlier stages of the response, those affecting nutrient sensing and signal transduction, are more secretagogue-specific, whereas the later stages of insulin secretion (e.g., vesicle trafficking or docking) are less specific, and therefore show a response to all secretagogues. This would be reflected in the alleles that are segregating among the DO mice. We predict that the loci affecting the response to all the secretagogues affect vesicle trafficking rather than nutrient signaling.
A major goal of our study was to discover specific novel genes that affect insulin secretion, with the premise that these genes would reveal new pathways and mechanisms. To arrive at individual candidate genes within genomic mapping intervals, we used 2 additional sources of data: allele effects and eQTL. Because there are 8 alleles segregating in the DO mice, we were able to trace the contribution of each allele to the physiological trait. In addition, since we carried out RNA-seq on islets from the same DO mice that were used for insulin secretion, we screened for local-acting eQTL whose allele effect matched that of the physiological QTL. This proved to be a stringent filter, typically resulting in fewer than 5 candidate genes.
We mapped insulin secretion, in response to essentially all of the tested secretagogues, to a locus on chromosome 1. The strongest association was with aa-stimulated insulin secretion. At this locus, we identified a small number of genes that, when we mediated on their eQTL, significantly diminished the association with insulin secretion. Only one gene, Ptpn18, met all of the statistical criteria for mediation of the secretion phenotype.
Ptpn18 is a protein tyrosine phosphatase. A known substrate of this enzyme is a human EGF receptor-related protein HER2 (ERBB2 gene product), a receptor that plays a role in breast cancer through its regulation of EGF-induced proliferation and cell migration (reviewed in 23). Ptpn18 increases the ubiquitination and degradation of HER2 (24). Its role in β cells has not been previously studied, and substrates other than HER2 have not yet been discovered.
We derived a model system for studying Ptpn18 consisting of a mouse with a knock-in mutation in an aspartate residue that is required for releasing the covalent intermediate between the enzyme and its substrate. This enabled us to abolish the phosphatase activity while also providing an enzyme that traps its substrates and enables their identification (15, 16). We believe that the phenotype of these mice likely resulted from an increase in the phosphorylation state of one or more substrates of Ptpn18, and/or the depletion of the substrates after being trapped by the mutant phosphatase.
We carried out a planned interim QTL mapping analysis of the data at the N = 300 point of this study. We mapped insulin secretion in response to GLP1 to a locus on Chr 16 at approximately 90.7 Mb, LOD = 7.4. We identified a second significant QTL peak on Chr 16 for KCl-induced insulin secretion at approximately 32 Mb, LOD = 6.5. These findings and other supporting evidence led us to nominate Hunk and Zfp148 as candidate genes at each locus, and we initiated validation experiments. At the completion of our study (N = 500) support for these QTL fell below our suggestive threshold. Although this was disappointing, it was not unexpected. It is an example of the winner’s curse — also known in QTL mapping literature as the Beavis effect (25), after Bill Beavis, who first reported it. When many loci of small effect are impacting a trait, there is a substantial element of chance involved in their detection. The problem is exacerbated by the application of stringent genome-wide selection criteria. Thus, in any given experiment, or even in an interim analysis of a single experiment, only a handful of causal genes are likely to have significant LOD scores. The validation experiments were successful in both cases and we continue to find consistent support for the role of these genes in insulin secretion. Further, both HUNK and ZNF148 are associated with T2D in human GWAS (see below). We conclude that the effects of these genes on insulin secretion are real but weak, and that chance fluctuations in the LOD scores favored their detection in the interim analysis.
To validate a role for Hunk in regulating insulin secretion, we obtained a whole-body Hunk-KO mouse and studied insulin secretion ex vivo. Islets from Hunk-KO mice, maintained on either chow or the HF/HS diet used in the genetic screen, showed increased insulin secretion during a perifusion. The increase occurred in both the first phase and sustained second phase of insulin secretion. Hunk-KO islets showed an enhanced insulin response to all secretagogue conditions, except basal glucose (3 mM). We and others have found that when B6 mice are chronically maintained on a Western-style diet, their insulin secretion level is reduced (26, 27). However, islets from Hunk-KO mice are protected from this diet effect, suggesting that Hunk activity is necessary for diet-induced suppression of insulin secretion.
A SNP near the HUNK gene is associated with T2D in multi-ethnic cohorts from Southeast Asia (28). In that population, the risk allele frequency is 0.57. In the Singapore prospective genome-wide association study (29), there were several 3′-UTR variants associated with T2D with an allele frequency of 0.36 (http://www.type2diabetesgenetics.org/gene/geneInfo/ZNF148).Supplemental Figure 8 illustrates a regional association plot for these diabetes-associated variants.
HUNK is required for HER2-induced breast cancer (30), and has become a potential drug target for patients who are resistant to the HER2 antagonists lapatinib and trastuzumab (31, 32). HUNK mediates HER2-dependent suppression of cellular apoptosis (30, 33) and activation of autophagy (31). Thus, it is intriguing that 2 genes that we discovered to regulate insulin secretion, Ptpn18 and Hunk, are associated with HER2-dependent signaling in breast cancer. Further studies will establish whether Ptpn18 and Hunk also participate in a common signaling pathway in pancreatic β cells.
At a separate locus on chromosome 16, we validated a role for Zfp148 in insulin secretion. Zfp148 is a transcription factor that has been shown to suppress Nr4a1 expression in β cells (34). Palmitate relieves this suppression by dislodging Zfp148 protein, resulting in recruitment of Sp1 protein to a common binding site within the Nr4a1 promoter. Nr4a1 has been implicated in a number of critical signaling pathways in β cells, including Nkx6.1-mediated β cell proliferation (35), and protection from ER stress–induced β cell apoptosis (36). Zfp148 has been shown to regulate Irs2 protein levels in brain (37). β cell Irs2 is a critical regulator of β cell function and survival (38). Zfp148 has also been suggested to regulate myocyte differentiation in culture, specifically controlling the expression of cytochrome c oxidase Vb (Cox5b) (39), a component of complex IV in the mitochondrial electron transport chain also expressed in β cells (40). Additionally, Zfp148 interacts with downstream targets of Stat3 in cell culture models (41, 42). Stat3 can regulate DNA damage responses within the islet and increase β cell survival (43).
Human GWAS have identified several variants within the ZNF148 gene associated with fasting insulin, glucose, and HOMA-IR (http://www.type2diabetesgenetics.org/gene/geneInfo/ZNF148). These include several nonsynonymous coding mutations, near the Zn2+-finger domains (G293S) and nuclear localization signal sequence (T320M) (44), and multiple noncoding variants (45, 46). Further, several SNPs that are within an intron of SLC12A8, which is downstream of ZNF148, are strongly associated with T2D, with P values of approximately 10–10 (see Supplemental Figure 9 for a regional association plot). It is possible these SNPs alter the activity of an enhancer locus for ZNF148.
Islets from β-Zfp148-KO mice showed enhanced glucose-stimulated insulin secretion. Zfp148 can function as either a repressor or an activator (47). Genes induced in the β-Zfp148-KO islets may be repressed by Zfp148, whereas genes downregulated in β-Zfp148-KO islets may be activated by Zfp148. Zfp148 competes with Sp1 for binding to its target genes (37, 48–50). Sp1 has itself been implicated in insulin secretion and peripheral insulin sensitivity (51–54).
Two of the 3 genes we validated in this study, Hunk and Zfp148, are negative regulators of insulin secretion. In a previous screen, we identified an additional negative regulator of insulin secretion, Tomosyn-2 (Stxbp5l) (55). We went on to show that glucose derepresses insulin secretion by stimulating the phosphorylation and degradation of Tomosyn-2 (56). Collectively, our results suggest that negative regulation might be an important regulatory mechanism for insulin secretion. β cells contain sufficient insulin to cause death if secreted in excess or at inappropriate times. Negative regulatory mechanisms safeguard against uncontrolled insulin secretion and likely were subject to positive selection in evolution. Follow-up studies will be required to determine the mechanisms by which β cells lift the brake exerted by Hunk and Zfp148 in response to nutrient signals.
Our studies to validate Ptpn18, Hunk, and Zfp148 as regulators of insulin secretion revealed an interesting contrast between in vivo phenotypes of live mice versus the ex vivo phenotypes measured in their cultured islets. To probe islet function in vivo, we performed an oGTT and measured the resulting change in plasma insulin. For the ex vivo studies, we used glucose (as well as other secretagogues) to stimulate insulin secretion from cultured islets. However, unlike the ex vivo studies, insulin-dependent glucose disposal in mice results in a tightly regulated feedback loop. As glucose is removed from circulation, insulin secretion diminishes. Further, as much as 50% of insulin that is secreted from islets into the portal vein is cleared by the liver (57). Hepatic insulin clearance is regulated by Zn2+ ions that are cosecreted with insulin (58), contributing to the genetic association of the Zn2+ transporter SLC30A8 with diabetes (59).
In our studies of Hunk-KO mice, we did not observe enhanced insulin secretion during the oGTT, despite showing clear evidence for that from cultured islets. Similarly, for the β-Zfp148-KO mice, we measured a more rapid rise in plasma insulin during the oGTT, but not an overall increase in plasma insulin, despite showing that high glucose evoked approximately 3-fold higher insulin secretion in β-Zfp148-KO islets versus control islets. Glucose tolerance was improved in Ptpn18D197A mice, in parallel with reduced in vivo and ex vivo insulin secretion. Finally, in our genetic screen we performed an oGTT on all DO mice and detected several loci associated with plasma insulin, including a strong quantitative trait locus at the Hnf1b gene locus on Chr 11 at approximately 84 Mb, LOD = 11 (10). However, we did not identify an insulin secretion quantitative trait locus at the Hnf1b locus, suggesting that the genetic regulation of plasma insulin during an oGTT and insulin secretion from isolated islets is distinct, which may in part reflect genetic regulation of insulin sensitivity or clearance (7).
The islets we harvested from Western diet–fed DO mice and used to perform the genetic screen for insulin secretion were also used for RNA-sequencing, yielding about 40,000 eQTL (10). We identified transcriptional hotspots, where hundreds of expression traits co-mapped as distal eQTL. At each hotspot, mediation analysis was used to identify novel candidate gene drivers, including Sat2, Il6st, and Fam83e, all of which have nominal association with diabetes in human GWAS. In our current study, we asked whether the QTL we identified for the insulin secretion traits or the whole-body physiological traits (e.g., plasma insulin) are syntenic to loci associated with diabetes risk in humans.
To evaluate enrichment for diabetes GWAS at our insulin secretion QTL, we computed a Bayesian support interval around the mouse QTL and then mapped this region onto the human genome. We then asked if the mapped regions were enriched with diabetes-associated SNPs obtained from GWAS Central (Figure 7). The vast majority of SNPs associated with diabetes occur in noncoding genomic regions, suggesting that they may influence enhancer activity at regulatory loci (60–63), similar to the T2D-associated SNPs proximal to the ZNF148 gene locus.
QTL associated with fasting plasma insulin in our mouse study were enriched with loci associated with insulin resistance in humans (Supplemental Figure 7). This observation demonstrates that similar traits measured in mice and humans have common genetic architecture. It is not feasible to perform a population-based genetic screen of insulin secretion in human islets, thus we cannot determine if this trait would identify common loci in mice and humans. However, numerous studies have suggested that a large proportion of diabetes-associated SNPs affect islet health or function (3, 4, 64). Our results, which show that insulin secretion QTL were much more highly enriched with several GWAS traits than were the whole-body physiological traits, provide further support for the conclusion that many diabetes GWAS SNPs directly influence islets.
Although very few of the DO mice that we screened became diabetic, we still identified loci that had been previously linked to diabetes in human GWAS, as well as several novel genes. Thus, carrying out a screen with granular phenotypes, like our direct measure of β cell function, does not require diseased individuals. Among nondiabetic humans, there is a wide range in insulin secretory capacity. In the absence of insulin resistance, a reduction of insulin secretion does not usually lead to diabetes, yet it still remains a discoverable phenotype that may contribute to diabetes risk in the context of obesity. The results of our study suggest that it would be productive to study the genetics of insulin secretion in nondiabetic human subjects. We predict that those studies would also identify novel diabetes-susceptibility loci.
Methods
Animals.
Animal care and study protocols were approved by the University of Wisconsin-Madison Animal Care and Use Committee. The DO mice used for the study were previously described (10). All DO mice were maintained on a Western-style HF/HS diet (44.6% kcal fat, 34% carbohydrate, and 17.3% protein) from Envigo Teklad (TD.08811) for 16 weeks. Transgenic mice for Hunk, Zfp148, and Ptpn18 were maintained on chow diet (Purina 5008) until sacrifice at approximately 12 weeks of age, or were maintained on the HF/HS diet for 16 weeks to mimic the conditions used in the DO genetic screen. A detailed description of the origins and molecular characterizations of the transgenic mouse models used in the study is provided in the Supplementary Material. The genotypes of the Hunk, Zfp148, Ins1Cre, and Ptpn18 transgenic mice were confirmed by PCR using the primer sequences provided in Supplemental Table 3.
In vivo physiological measurements.
At various ages and following a 4 hour fast (8 am to noon), body weights were recorded, and whole blood was collected by retro-orbital bleed and used to determine plasma glucose, insulin, and triglyceride (TG). Glucose was measured with a commercially available kit (TR15221, Thermo Fisher Scientific). TG was measured by commercially available kit (TR22421, Thermo Fisher Scientific). Insulin was measured by radioimmunoassay (SRI-13K, Millipore). To evaluate insulin dynamics in vivo, an oGTT was performed. Mice were fasted (6 am to 10 am), given an oral glucose bolus (2 gm/kg body weight), and whole blood collected at 0, 5, 15, 30, 60, and 120 minutes. Plasma insulin and glucose was measured as described above.
Ex vivo insulin and glucagon secretion measurements.
We previously described a 96-well assay that we developed to conduct the ex vivo screen of insulin and glucagon secretion from 500 DO mice (see also Supplementary Material) (11). We applied 3 different transformations to each secretion trait for mapping: (i) log-transformed direct measures of secretion, (ii) log fractional secretion relative to the total amount of hormone (insulin or glucagon) present in the islets, and (iii) log fold-change relative to the level of secretion at the corresponding baseline glucose exposure (e.g., secretion in response to amino acids was evaluated relative to secretion at 8.3 mM glucose). Fractional and fold-change measures were computed as residual values from log-scale regressions (65).
Genome scans.
We performed genomes scans using R/qtl2 software (66). Sex and wave were included as additive covariates. The genome scan model included a random effect to account for kinship among the DO animals using the LOCO method (67). Significance thresholds were determined for individual traits by permutation analysis (68). We applied a suggestive threshold (LOD > 6.0) for reporting QTL loci in order to capture a more complete picture the genetic architecture of the ex vivo traits. However, several QTL loci also exceeded the genome-wide 0.05 threshold (LOD > 7.4). We established Bayesian support intervals (69) around each quantitative trait locus peak using the R/qtl2 function bayes_int.
Identification of candidate gene drivers.
Mediation tests for causality were performed using conditional regression of the target phenotype on the gene expression of the candidate mediator and the local quantitative trait locus genotype (13, 14). We evaluated all mRNA expression traits that had significant peaks (LOD > 5.5) within 10 Mb of the LOD peak for the target phenotype as candidate mediators. In addition, we performed CMSTs to compare causal, reactive, independent, and undecided models for the target and mediator (14). CMST evaluates which among the causal, reactive, independent, or complex models is most consistent with the data. We modified the original CMST method to use nonparametric Wilcoxon tests and accounted for covariates as well as kinship relationships.
Integration of mouse QTL with human GWAS.
For each medical subject headings–coded (MeSH-coded) human trait that corresponded to a mouse phenotype (Supplemental Table 4), we downloaded from GWAS Central all SNPs that have an association P value above a threshold of –log10 ≥ 4, resulting in a total of 12,829 unique SNPs and 16,415 SNP-trait associations (Supplemental Table 5).
At each quantitative trait locus, we first computed a Bayesian support interval based on the local architecture of linkage disequilibrium (LD). To identify synteny in humans, these intervals from mice were mapped onto the human genome using USCS’s LiftOver tool (https://genome.ucsc.edu/cgi-bin/hgLiftOver). GWAS SNPs within the syntenic regions were then identified. Syntenic regions separated by less than 1 Kb were combined to reduce the number of human genomic segments. For in vivo whole-body physiological traits, 19 QTL yielded genomic regions ranging in size from between 45,891 bp to 12.69 Mb, with a median size of 4.46 Mb. For ex vivo islet traits, 30 QTL yielded genomic regions ranging in size from between 676,305 bp to 13.36 Mb, with a median size of 4.54 Mb. We asked if the syntenic regions were enriched with the diabetes GWAS SNPs by a permutation test where we randomly selected the same number and size of human genomic regions (excluding the Y chromosome). We repeated this procedure 1000 times, and computed a q value to control the FDR at 5%, adjusting for the 16 diabetes-related GWAS traits.
To identify which specific QTL were enriched with one or more of the 16 diabetes-related GWAS traits, we computed an enrichment score that was based on the P value for each SNP (Supplemental Table 5). The enrichment score is defined as:
(Equation 1)
where n is the number of SNPs that are located within our syntenic human regions, and p is the SNP’s P value for a particular GWAS trait. Higher scores indicate greater enrichment of GWAS traits with highly significant P values.
Statistics.
Statistical computations were carried out using R software package (http://www.R-project.org/). Estimated values are reported as mean ± standard error. For most of the experimental validation studies, we used a Student’s 2-tailed t test (e.g., β-Zfp148-KO vs. β-Zfp148fl/fl, Ptpn18D197A vs. Ptpn18-WT or Hunk-KO vs. Hunk-WT for one set of conditions). For the comparison of the number of islets harvested for Hunk-KO versus Hunk-WT mice (Figure 5L), we performed a 2-way ANOVA (diet × genotype). We first tested for an interaction effect, and if significant, we reported the mean difference between genotype for each diet. Otherwise, we reported the effects of diet as average across genotypes and the effects of genotype as an average across diets. A P value less than 0.05 was considered significant for all comparisons that used a Student’s 2-tailed t test. A P value less than 0.017 was considered significant in the case of the ANOVA for islet number in the Hunk studies, allowing for the multiple test correction (0.05/3 = 0.017).
Study approval.
All experiments involving mice were approved by the Institutional Animal Care and Use Committee, an AAALAC-accredited unit, by the College of Agricultural and Life Sciences (CALS) at the University of Wisconsin-Madison. ADA is the Principal Investigator for the CALS Animal Care and Use Protocol (A005821).
Author contributions
MER, KLS, DSS, KAM, SPS, CHE, and RD conducted the experiments. DMG, MV, ZW, TI, BSY, KWB, CK, TB, MPK, and GAC performed analyses. MPK, GAC, and ADA, designed the experiments and wrote the paper.
Supplementary Material
Acknowledgments
This work was supported by NIH grants DK101573, DK102948, DK066369 (to ADA), GM102756, U54 AI117924 (to CK) and GM070683 (to GAC and KWB). We are grateful for graphical expertise provided by Laura Vanderploeg within the Media Center of the Department of Biochemistry, University of Wisconsin–Madison.
Version 1. 07/25/2019
In-Press Preview
Version 2. 09/09/2019
Electronic publication
Version 3. 10/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors declare that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(10):4419–4432.https://doi.org/10.1172/JCI129143.
See the related Commentary at One small step for mice, one giant leap for GWAS?.
Contributor Information
Mark P. Keller, Email: mkeller3@wisc.edu.
Mary E. Rabaglia, Email: rabaglia@biochem.wisc.edu.
Kathryn L. Schueler, Email: kschuele@wisc.edu.
Donnie S. Stapleton, Email: dsstaple@biochem.wisc.edu.
Daniel M. Gatti, Email: dmgatti@coa.edu.
Matthew Vincent, Email: Matt.Vincent@jax.org.
Kelly A. Mitok, Email: mitok@wisc.edu.
Ziyue Wang, Email: zwang667@wisc.edu.
Takanao Ishimura, Email: takanao@seas.upenn.edu.
Shane P. Simonett, Email: ssimonett@wisc.edu.
Christopher H. Emfinger, Email: emfinger@wisc.edu.
Rahul Das, Email: rdas@medicine.wisc.edu.
Tim Beck, Email: timbeck@leicester.ac.uk.
Christina Kendziorski, Email: kendzior@biostat.wisc.edu.
Karl W. Broman, Email: kbroman@biostat.wisc.edu.
Brian S. Yandell, Email: brian.yandell@wisc.edu.
Gary A. Churchill, Email: gary.churchill@jax.org.
Alan D. Attie, Email: adattie@wisc.edu.
References
- 1.Poulsen P, Levin K, Petersen I, Christensen K, Beck-Nielsen H, Vaag A. Heritability of insulin secretion, peripheral and hepatic insulin action, and intracellular glucose partitioning in young and old Danish twins. Diabetes. 2005;54(1):275–283. doi: 10.2337/diabetes.54.1.275. [DOI] [PubMed] [Google Scholar]
- 2.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci. 2010;1212:59–77. doi: 10.1111/j.1749-6632.2010.05838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohlke KL, Boehnke M. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum Mol Genet. 2015;24(R1):R85–R92. doi: 10.1093/hmg/ddv264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prasad RB, Groop L. Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel) 2015;6(1):87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popkin BM. Nutrition transition and the global diabetes epidemic. Curr Diab Rep. 2015;15(9):64. doi: 10.1007/s11892-015-0631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prentki M, Matschinsky FM, Madiraju SR. Metabolic signaling in fuel-induced insulin secretion. Cell Metab. 2013;18(2):162–185. doi: 10.1016/j.cmet.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 7.Goodarzi MO, et al. Insulin sensitivity and insulin clearance are heritable and have strong genetic correlation in Mexican Americans. Obesity (Silver Spring) 2014;22(4):1157–1164. doi: 10.1002/oby.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Threadgill DW, Miller DR, Churchill GA, de Villena FP. The collaborative cross: a recombinant inbred mouse population for the systems genetic era. ILAR J. 2011;52(1):24–31. doi: 10.1093/ilar.52.1.24. [DOI] [PubMed] [Google Scholar]
- 9.Churchill GA, Gatti DM, Munger SC, Svenson KL. The Diversity Outbred mouse population. Mamm Genome. 2012;23(9-10):713–718. doi: 10.1007/s00335-012-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keller MP, et al. Genetic drivers of pancreatic islet function. Genetics. 2018;209(1):335–356. doi: 10.1534/genetics.118.300864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitok KA, et al. Islet proteomics reveals genetic variation in dopamine production resulting in altered insulin secretion. J Biol Chem. 2018;293(16):5860–5877. doi: 10.1074/jbc.RA117.001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henquin JC. The dual control of insulin secretion by glucose involves triggering and amplifying pathways in β-cells. Diabetes Res Clin Pract. 2011;93 Suppl 1:S27–S31. doi: 10.1016/S0168-8227(11)70010-9. [DOI] [PubMed] [Google Scholar]
- 13.Chick JM, et al. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534(7608):500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neto EC, et al. Modeling causality for pairs of phenotypes in system genetics. Genetics. 2013;193(3):1003–1013. doi: 10.1534/genetics.112.147124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci USA. 1997;94(5):1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gensler M, Buschbeck M, Ullrich A. Negative regulation of HER2 signaling by the PEST-type protein-tyrosine phosphatase BDP1. J Biol Chem. 2004;279(13):12110–12116. doi: 10.1074/jbc.M309527200. [DOI] [PubMed] [Google Scholar]
- 17.Reed KR, et al. Hunk/Mak-v is a negative regulator of intestinal cell proliferation. BMC Cancer. 2015;15:110. doi: 10.1186/s12885-015-1087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller MP, et al. Genetic drivers of pancreatic islet function. Genetics. 2018;209(1):335–356. doi: 10.1534/genetics.118.300864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. The Annals of Statistics. 2003;31(6):2013–2035. doi: 10.1214/aos/1074290335. [DOI] [Google Scholar]
- 20.Storey JD, Taylor JE, Siegmund D. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J R Stat Soc Series B Stat Methodol. 2004;66(1):187–205. doi: 10.1111/j.1467-9868.2004.00439.x. [DOI] [Google Scholar]
- 21.Storey JD. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol. 2002;64(3):479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 22.Lee YH, Wang MY, Yu XX, Unger RH. Glucagon is the key factor in the development of diabetes. Diabetologia. 2016;59(7):1372–1375. doi: 10.1007/s00125-016-3965-9. [DOI] [PubMed] [Google Scholar]
- 23.Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel) 2017;9(5):E52. doi: 10.3390/cancers9050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang HM, et al. The catalytic region and PEST domain of PTPN18 distinctly regulate the HER2 phosphorylation and ubiquitination barcodes. Cell Res. 2014;24(9):1067–1090. doi: 10.1038/cr.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu S. Theoretical basis of the Beavis effect. Genetics. 2003;165(4):2259–2268. doi: 10.1093/genetics/165.4.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta D, et al. Temporal characterization of beta cell-adaptive and -maladaptive mechanisms during chronic high-fat feeding in C57BL/6NTac mice. J Biol Chem. 2017;292(30):12449–12459. doi: 10.1074/jbc.M117.781047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peyot ML, et al. Beta-cell failure in diet-induced obese mice stratified according to body weight gain: secretory dysfunction and altered islet lipid metabolism without steatosis or reduced beta-cell mass. Diabetes. 2010;59(9):2178–2187. doi: 10.2337/db09-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sim X, et al. Transferability of type 2 diabetes implicated loci in multi-ethnic cohorts from Southeast Asia. PLoS Genet. 2011;7(4):e1001363. doi: 10.1371/journal.pgen.1001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan CE1, Emmanuel SC, Tan BY, Jacob E. Prevalence of diabetes and ethnic differences in cardiovascular risk factors. The 1992 Singapore National Health Survey. Diabetes Care. 1999;22(2):241–247. doi: 10.2337/diacare.22.2.241. [DOI] [PubMed] [Google Scholar]
- 30.Yeh ES, Yang TW, Jung JJ, Gardner HP, Cardiff RD, Chodosh LA. Hunk is required for HER2/neu-induced mammary tumorigenesis. J Clin Invest. 2011;121(3):866–879. doi: 10.1172/JCI42928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh ES, Abt MA, Hill EG. Regulation of cell survival by HUNK mediates breast cancer resistance to HER2 inhibitors. Breast Cancer Res Treat. 2015;149(1):91–98. doi: 10.1007/s10549-014-3227-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phelps-Polirer K, Abt MA, Smith D, Yeh ES. Co-Targeting of JNK and HUNK in Resistant HER2-Positive Breast Cancer. PLoS ONE. 2016;11(4):e0153025. doi: 10.1371/journal.pone.0153025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh ES, Belka GK, Vernon AE, Chen CC, Jung JJ, Chodosh LA. Hunk negatively regulates c-myc to promote Akt-mediated cell survival and mammary tumorigenesis induced by loss of Pten. Proc Natl Acad Sci USA. 2013;110(15):6103–6108. doi: 10.1073/pnas.1217415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazuy C, et al. Palmitate increases Nur77 expression by modulating ZBP89 and Sp1 binding to the Nur77 proximal promoter in pancreatic β-cells. FEBS Lett. 2013;587(23):3883–3890. doi: 10.1016/j.febslet.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 35.Tessem JS, et al. Nkx6.1 regulates islet β-cell proliferation via Nr4a1 and Nr4a3 nuclear receptors. Proc Natl Acad Sci USA. 2014;111(14):5242–5247. doi: 10.1073/pnas.1320953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu C, et al. The orphan nuclear receptor NR4A1 protects pancreatic β-cells from endoplasmic reticulum (ER) stress-mediated apoptosis. J Biol Chem. 2015;290(34):20687–20699. doi: 10.1074/jbc.M115.654863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Udelhoven M, Pasieka M, Leeser U, Krone W, Schubert M. Neuronal insulin receptor substrate 2 (IRS2) expression is regulated by ZBP89 and SP1 binding to the IRS2 promoter. J Endocrinol. 2010;204(2):199–208. doi: 10.1677/JOE-09-0266. [DOI] [PubMed] [Google Scholar]
- 38.Rhodes CJ, White MF, Leahy JL, Kahn SE. Direct autocrine action of insulin on β-cells: does it make physiological sense? Diabetes. 2013;62(7):2157–2163. doi: 10.2337/db13-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boopathi E, et al. Regulation of murine cytochrome c oxidase Vb gene expression during myogenesis: YY-1 and heterogeneous nuclear ribonucleoprotein D-like protein (JKTBP1) reciprocally regulate transcription activity by physical interaction with the BERF-1/ZBP-89 factor. J Biol Chem. 2004;279(34):35242–35254. doi: 10.1074/jbc.M403160200. [DOI] [PubMed] [Google Scholar]
- 40.Martens GA, et al. Protein markers for insulin-producing beta cells with higher glucose sensitivity. PLoS ONE. 2010;5(12):e14214. doi: 10.1371/journal.pone.0014214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonawane PJ, et al. Transcriptional regulation of the novel monoamine oxidase renalase: Crucial roles of transcription factors Sp1, STAT3, and ZBP89. Biochemistry. 2014;53(44):6878–6892. doi: 10.1021/bi500798n. [DOI] [PubMed] [Google Scholar]
- 42.Wu Y, Diab I, Zhang X, Izmailova ES, Zehner ZE. Stat3 enhances vimentin gene expression by binding to the antisilencer element and interacting with the repressor protein, ZBP-89. Oncogene. 2004;23(1):168–178. doi: 10.1038/sj.onc.1207003. [DOI] [PubMed] [Google Scholar]
- 43.Linnemann AK, et al. Interleukin 6 protects pancreatic β cells from apoptosis by stimulation of autophagy. FASEB J. 2017;31(9):4140–4152. doi: 10.1096/fj.201700061RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guan M, et al. An exome-wide association study for type 2 diabetes-attributed end-stage kidney disease in African Americans. Kidney Int Rep. 2018;3(4):867–878. doi: 10.1016/j.ekir.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salmon M, Owens GK, Zehner ZE. Over-expression of the transcription factor, ZBP-89, leads to enhancement of the C2C12 myogenic program. Biochim Biophys Acta. 2009;1793(7):1144–1155. doi: 10.1016/j.bbamcr.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Diab IH, Zehner ZE. ZBP-89 represses vimentin gene transcription by interacting with the transcriptional activator, Sp1. Nucleic Acids Res. 2003;31(11):2900–2914. doi: 10.1093/nar/gkg380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merchant JL, et al. ZBP-89, a Krüppel-like zinc finger protein, inhibits epidermal growth factor induction of the gastrin promoter. Mol Cell Biol. 1996;16(12):6644–6653. doi: 10.1128/MCB.16.12.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai L, Merchant JL. Transcription factor ZBP-89 cooperates with histone acetyltransferase p300 during butyrate activation of p21waf1 transcription in human cells. J Biol Chem. 2000;275(39):30725–30733. doi: 10.1074/jbc.M004249200. [DOI] [PubMed] [Google Scholar]
- 51.Moreno-Aliaga MJ, Swarbrick MM, Lorente-Cebrián S, Stanhope KL, Havel PJ, Martínez JA. Sp1-mediated transcription is involved in the induction of leptin by insulin-stimulated glucose metabolism. J Mol Endocrinol. 2007;38(5):537–546. doi: 10.1677/JME-06-0034. [DOI] [PubMed] [Google Scholar]
- 52.Li T, Bai L, Li J, Igarashi S, Ghishan FK. Sp1 is required for glucose-induced transcriptional regulation of mouse vesicular glutamate transporter 2 gene. Gastroenterology. 2008;134(7):1994–2003. doi: 10.1053/j.gastro.2008.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malenczyk K, et al. A TRPV1-to-secretagogin regulatory axis controls pancreatic β-cell survival by modulating protein turnover. EMBO J. 2017;36(14):2107–2125. doi: 10.15252/embj.201695347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samson SL, Wong NC. Role of Sp1 in insulin regulation of gene expression. J Mol Endocrinol. 2002;29(3):265–279. doi: 10.1677/jme.0.0290265. [DOI] [PubMed] [Google Scholar]
- 55.Bhatnagar S, et al. Positional cloning of a type 2 diabetes quantitative trait locus; tomosyn-2, a negative regulator of insulin secretion. PLoS Genet. 2011;7(10):e1002323. doi: 10.1371/journal.pgen.1002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhatnagar S, et al. Phosphorylation and degradation of tomosyn-2 de-represses insulin secretion. J Biol Chem. 2014;289(36):25276–25286. doi: 10.1074/jbc.M114.575985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eaton RP, Allen RC, Schade DS. Hepatic removal of insulin in normal man: dose response to endogenous insulin secretion. J Clin Endocrinol Metab. 1983;56(6):1294–1300. doi: 10.1210/jcem-56-6-1294. [DOI] [PubMed] [Google Scholar]
- 58.Tamaki M, et al. The diabetes-susceptible gene SLC30A8/ZnT8 regulates hepatic insulin clearance. J Clin Invest. 2013;123(10):4513–4524. doi: 10.1172/JCI68807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rutter GA, Chimienti F. SLC30A8 mutations in type 2 diabetes. Diabetologia. 2015;58(1):31–36. doi: 10.1007/s00125-014-3405-7. [DOI] [PubMed] [Google Scholar]
- 60.Parker SC, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci USA. 2013;110(44):17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46(2):136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varshney A, et al. Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc Natl Acad Sci USA. 2017;114(9):2301–2306. doi: 10.1073/pnas.1621192114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaulton KJ, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47(12):1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci. 2010;1212:59–77. doi: 10.1111/j.1749-6632.2010.05838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karp NA, Melvin D, Sanger Mouse Genetics Project, Mott RF. Robust and sensitive analysis of mouse knockout phenotypes. PLoS ONE. 2012;7(12):e52410. doi: 10.1371/journal.pone.0052410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Broman KW, et al. R/qtl2: software for mapping quantitative trait loci with high-dimensional data and multiparent populations. Genetics. 2019;211(2):495–502. doi: 10.1534/genetics.118.301595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138(3):963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159(1):371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahajan A, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.