Abstract
Translation initiation determines both the quantity and identity of the protein encoded in an mRNA by establishing the reading frame for protein synthesis. In eukaryotic cells, numerous translation initiation factors (eIFs) prepare ribosomes for polypeptide synthesis, yet the underlying dynamics of this process remain enigmatic1,2. A central question is how eukaryotic ribosomes transition from translation initiation to elongation. Here, we applied in vitro single-molecule fluorescence microscopy approaches to monitor directly in real time the pathways of late translation initiation and the transition to elongation using a purified yeast Saccharomyces cerevisiae translation system. This transition was remarkably slower in our eukaryotic system than that reported for Escherichia coli3–5. The slow entry to elongation was defined by a long residence time of eIF5B on the 80S ribosome after joining of individual ribosomal subunits, which is catalyzed by this universally conserved initiation factor. Inhibition of eIF5B GTPase activity following subunit joining prevented eIF5B dissociation from the 80S complex, thereby preventing elongation. Our findings illustrate how eIF5B dissociation serves as a kinetic checkpoint for the transition from initiation to elongation, and its release may be governed by a conformation of the ribosome complex that triggers GTP hydrolysis.
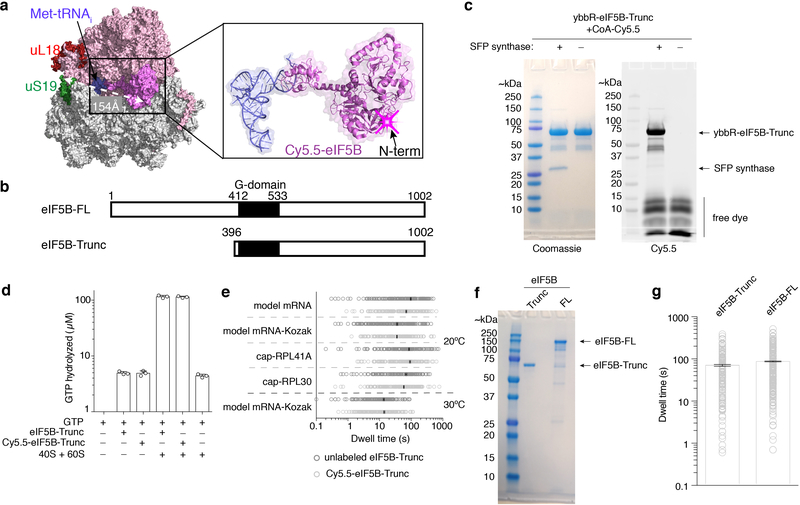
Despite decades of study, the transition from eukaryotic translation initiation to elongation remains poorly understood. During translation initiation, the 40S ribosome subunit in complex with eIFs is recruited to a 7-methylguanosine (m7G)-capped mRNA and scans the 5’ untranslated region until the correct AUG start codon is recognized (termed “scanning”), forming a post-scanning 48S pre-initiation complex (PIC)1,2. The subsequent joining of the 60S subunit to this 48S PIC is catalyzed by the universally conserved factor eIF5B, independent of its GTPase activity6–9. The conformation of yeast eIF5B when bound to GTP is remodeled upon its association with a ribosome, enabling eIF5B to directly contact both Met-tRNAi and the GTPase activation center on the 60S subunit10–12. These contacts are likely essential for triggering GTP hydrolysis and the subsequent release of eIF5B-GDP, allowing the ribosomal A site to accept the first elongator aminoacyl-tRNA (aa-tRNA) and enter the elongation phase12. Bypassing eIF5B GTP hydrolysis with a mutation that weakens its ribosome binding affinity increases the frequency of bypassing the first AUG codon in an mRNA (“leaky scanning”) in vivo8, implying that the timing of eIF5B dissociation from the 80S complex and the subsequent entry into elongation is critical to start site selection. The steps and their timescales that govern this transition have not been elucidated in eukaryotes because its complex, multi-step nature challenges traditional biochemical approaches. Here, we overcome these limitations by directly observing translation initiation using single-molecule fluorescence microscopy to parse out the dynamics and pathways involved in 80S assembly and subsequent acceptance of the first elongator aa-tRNA. We first reconstituted an active yeast translation system from purified ribosomes and core eIFs and eEFs (Extended Data Fig. 1) suitable for single-molecule analyses13. We then applied this system to study real-time 80S assembly with a zero-mode waveguide (ZMW)-based instrumentation platform14, which is capable of multi-color single-molecule fluorescence imaging at high fluorescently-labeled ligand concentrations.
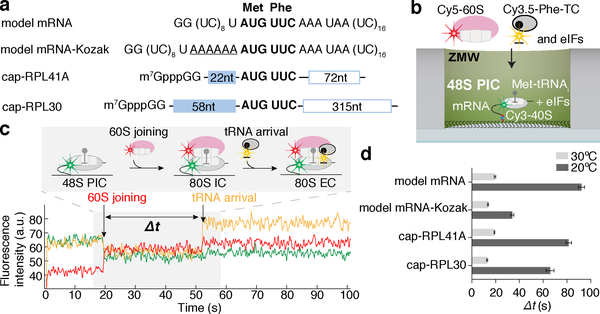
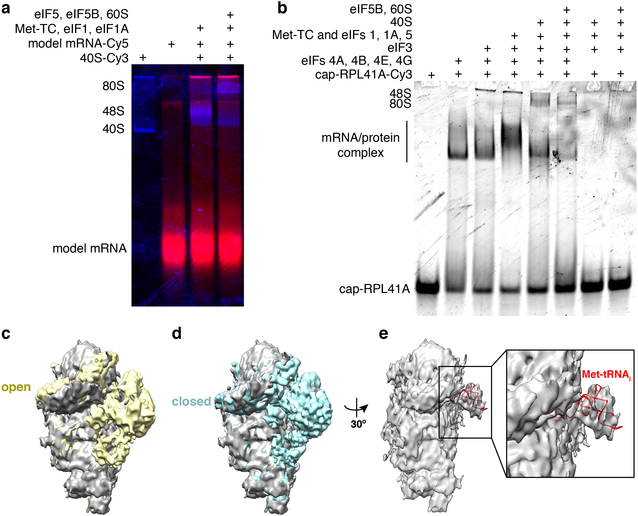
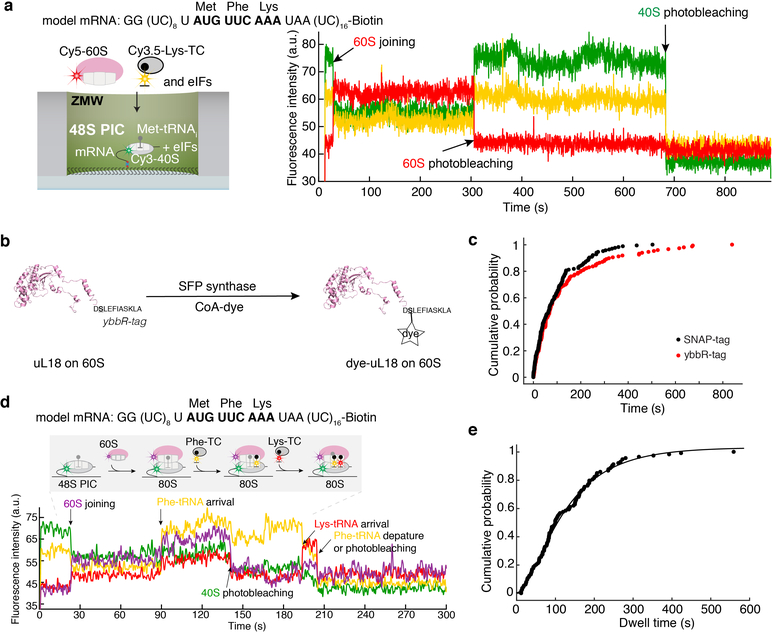
Since we observed efficient and stable 48S PIC formation on a previously well-characterized, uncapped, unstructured model mRNA (Fig. 1a, Extended Data Fig. 2a), we formed and immobilized single 48S PICs (with Cy3–40S) within ZMWs and delivered Cy5–60S along with the other required factors (Extended Data Fig. 3a). We observed efficient 60S subunit joining on single 48S PICs, with ~ 90% (n = 267 out of 300) of the 48S PICs binding to Cy5–60S to form the 80S complex (Extended Data Fig. 3b,c), which required eIF5B; in the absence of eIF5B, no 80S formation was observed in the 15-min imaging time window (n = 500) (see Extended Data Fig. 3d,e for data that may reconcile how yeast lacking eIF5B survive, albeit with severe growth defect8, given the critical role of this factor: raised polyamine concentrations can give functional subunit joining in the absence of eIF5B). The association rate of 60S subunits was comparable to that from prior bulk measurements (Extended Data Fig. 3c)9. When the first elongator Phe-(Cy3.5)-tRNAPhe was included in the delivery mix as a ternary complex with eEF1A and GTP (Cy3.5-Phe-TC) (Fig. 1b), ~80% (n = 164 out of 200) of the 60S joining smFRET events were followed by a subsequent Cy3.5-Phe-TC arrival event (Fig. 1c). This A-site aa-tRNA association was codon/tRNA specific and leads to further elongation (Extended Data Fig. 4). These results demonstrate that real-time 80S assembly occurs in the correct reading frame and that our single-molecule translation initiation system is fully active.
Fig. 1. Real-time observation of eukaryotic translation initiation and the transition to elongation.
a, mRNA constructs used in single-molecule assays (also see Extended Data Fig. 5d). All the mRNAs contain a UUC phenylalanine (Phe) codon after the AUG start codon and are biotinylated at their 3’ ends. b, Experimental setup for single-molecule assays. 48S preinitiation complexes (PICs) containing Cy3–40S, Met-tRNAi, and the 3’-biotyinlated mRNA of interest were immobilized in ZMWs in the presence of required eIFs. Experiments were started by illuminating ZMWs with a green laser and delivering Cy5–60S, Cy3.5-Phe-tRNAPhe:eEF1A:GTP ternary complex (TC) and eIFs. c, Example experimental trace (bottom) and schematic illustration (top) of the molecular events along the reaction coordinate. The dwell times between the 60S joining and the A-site Phe-TC arrival were measured for n = 118, 130, 130, 121, 189, 159, 164 and 136 molecules (d, from bottom to top for each bar), and fit to single-exponential distributions to estimate the average time, “Δt” (with 95% confidence interval), of the transition from initiation to elongation.
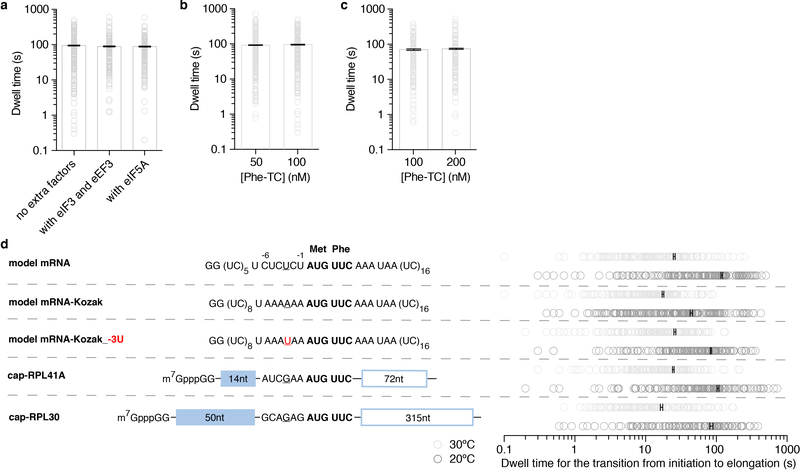
To determine an initial rate for the transition from initiation to elongation, we measured the dwell times between 60S joining and the arrival of the Cy3.5-Phe-TC and determined the average time of this transition (“Δt”) (Fig. 1c). The Δt was 30.2 ± 1.0 s (n = 147) when assayed under the same condition as in prior bulk kinetic analyses of yeast translation initiation (26ºC and buffer containing 3 mM free Mg2+)9. This was significantly longer than the bacterial counterpart, which was reported to be ~1 s at 20ºC3–5. For direct comparisons with the bacterial rates, we also determined the Δt at 20ºC (92.2 ± 2.5 s) (Fig. 1d). The Δt remained similar upon addition of 150 nM eIF3 and 200 nM eEF3 (and 1 mM ATP:Mg2+), or 500 nM hypusine-modified eIF5A15, indicating that the transition from initiation to elongation is independent of these factors (Extended Data Fig. 5a). Changing the concentration of Cy3.5-Phe-TC did not significantly alter the Δt (Extended Data Fig. 5b). Thus, the rate of the transition was limited by a step prior to binding of the first A-site aa-tRNA.
We next tested whether the slow transition was due to the cap-independence of initiation on the model mRNA (i.e., the missing m7G cap on the model mRNA and eIF4F proteins in the reaction). We formed 48S PICs on cap-RPL41A or cap-RPL30 mRNAs (Fig. 1a, mRNAs were completely capped with m7G, Methods) in the presence of eIF4F, eIF4B and eIF3 proteins and performed the same experiments as above. The 48S PICs readily formed in a cap-dependent manner (Extended Data Fig. 2b) and were in the post-scanning, mRNA channel-“closed” state as demonstrated by cryo-electron microscopy (cryo-EM) analyses of our complexes (Extended Data Fig. 2c,d,e)16. Thus, the 48S PIC central to our single-molecule assays was an authentic and on-pathway complex. Yet, the Δt values at 20ºC were 81.4 ± 2.5 s and 65.7 ± 3.4 s for cap-RPL41A and cap-RPL30, respectively, which were similar to the Δt measured on the model mRNA (Fig. 1d). Increasing the Cy3.5-Phe-TC concentration did not significantly change the Δt value (Extended Data Fig. 5c), further demonstrating that the rate-limiting step occurs prior to A-site aa-tRNA binding. Our findings suggest that the slow transition to elongation may be a general feature of yeast translation.
We next tested whether the sequence context near the start codon impacted entry into the elongation phase of translation. When the optimal yeast Kozak sequence17 was inserted upstream of the AUG codon in the model mRNA (designated as model mRNA-Kozak), the Δt was reduced by ~3-fold to 34.1 ± 1.3 s at 20ºC (Fig. 1a,d). This effect was largely abolished by an A to U substitution at the −3 position (Extended Data Fig. 5d), consistent with the critical role of that residue within the Kozak sequence18. Thus, we have uncovered an unexpected role of the Kozak sequence in the control of this transition. However, even the fastest rate we observed at 20ºC was still more than 30-fold slower than that observed in E. coli3–5. To understand the energetics of the rate-limiting step in the eukaryotic transition, we increased the reaction temperature to 30ºC (in the range of the optimal growth temperature of Saccharomyces cerevisiae19), and found a striking dependence of Δt on temperature across all assayed mRNAs (Fig. 1d, Extended Data Fig. 5d): a ~4–5-fold decrease for the model and cap-RPL41A/30 mRNAs, a ~2.5-fold decrease for the model mRNA-Kozak.
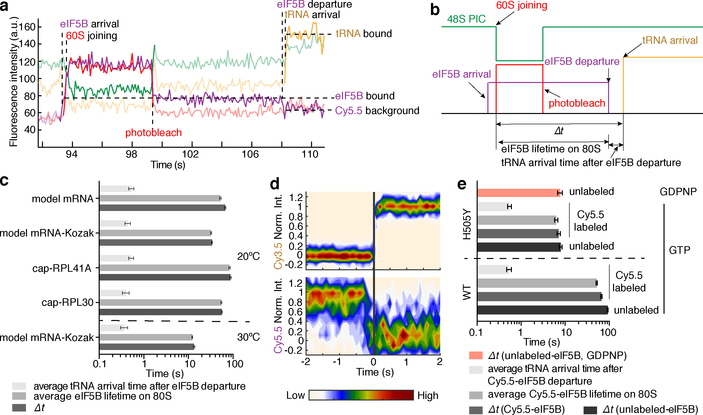
The temperature dependence of Δt suggested that enthalpically-driven slow conformational rearrangements and/or factor dissociation limited the transition. We hypothesized that eIF5B dissociation was the rate-limiting step given that its binding site on the ribosome overlaps with that of the elongator aa-tRNAs12. Early kinetic analyses of in vitro rabbit reticulocyte translation suggest that GTP hydrolysis by an initiation factor limits the rate of the first peptide bond formation after ribosomal subunit joining20, which could plausibly be eIF5B GTP hydrolysis. If eIF5B indeed gates the transition to elongation, we expect that its lifetime on the 80S would be similar to the Δt measured above.
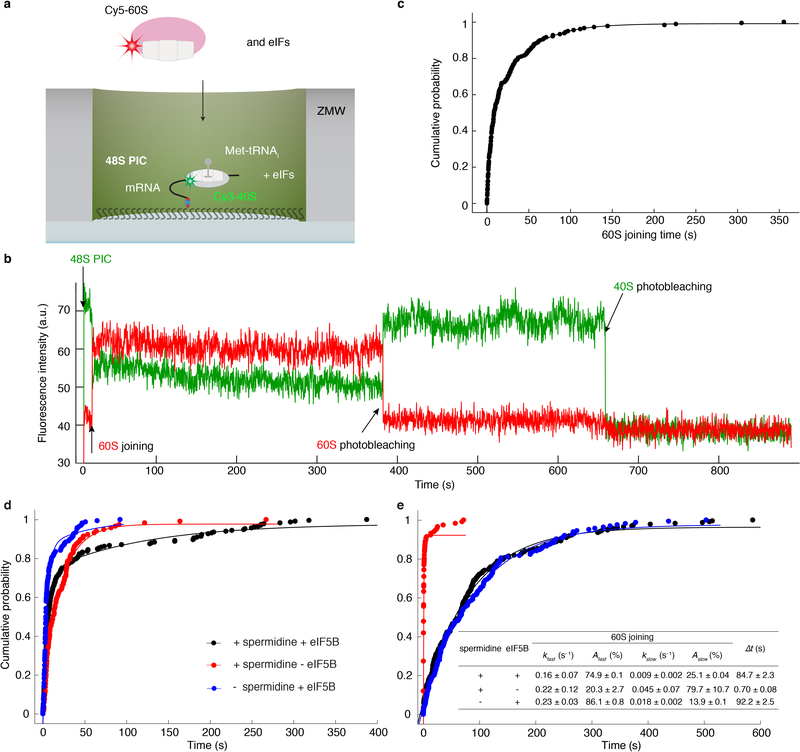
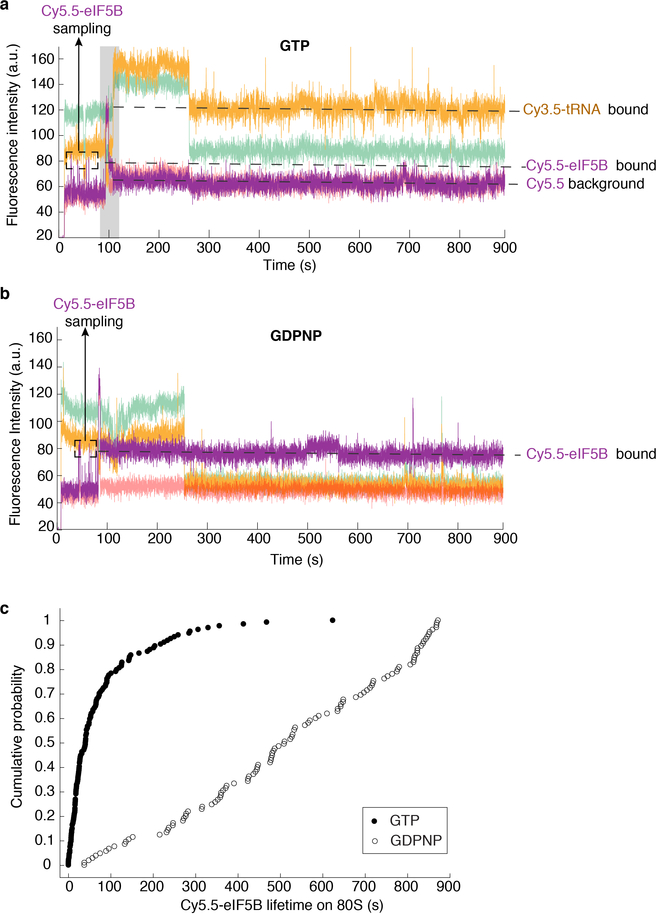
To test this hypothesis, we fluorescently labeled eIF5B (Extended Data Fig. 6a) and tracked the protein throughout the reaction pathways using a similar experimental scheme as above. A Cy5.5 dye was attached to N-terminally truncated eIF5B (remaining residues 396–1002), via a ybbR-tag fused to its N-terminus. This truncated eIF5B is the standard construct in the eukaryotic translation field6,9,21,22 and was used in all our measurements above (Extended Data Fig. 6b), and neither the tag nor dye impacted eIF5B function (Extended Data Fig. 6c,d). After immobilizing pre-formed Cy3–48S PICs in ZMWs, we co-delivered Cy5.5-eIF5B, Cy5–60S and Cy3.5-Phe-TC in the presence of other required factors to the immobilized complex. With the model mRNA and direct illumination of all fluorophores, we observed transient Cy5.5-eIF5B sampling events to the 48S PIC prior to 60S joining (Extended Data Fig. 7a), consistent with its dynamic interaction with that complex as suggested by ensemble fluorescence measurements9. Upon Cy5–60S joining to form the 80S complex, as signaled by smFRET, the average Cy5.5-eIF5B lifetime on the ribosome was prolonged and nearly equal to Δt (Fig. 2a,b,c, Extended Data Fig. 7a). Importantly, the Δt measured here matched that measured with unlabeled eIF5B, reassuring us of the non-perturbed function post labeling (Extended Data Fig. 6e). Furthermore, Cy5.5-eIF5B dissociation always preceded Cy3.5-Phe-TC binding to the A-site of the 80S ribosome, which occurred very rapidly (within 1 s) after eIF5B departure (Fig. 2a,d). Similar results were observed with all mRNAs at both 20ºC and 30ºC (Fig. 2c). Notably, the Δt was not reduced by using full-length eIF5B, indicating that the lack of the non-conserved N-terminal region8,10 of the protein did not interfere with the transition rate (Extended Data Fig. 6b,f,g). Thus, the presence of eIF5B on the de novo assembled 80S ribosome limits the rate of the transition from initiation to elongation.
Fig. 2. eIF5B gates the transition between initiation and elongation.
Sample trace (a) and schematic illustration (b) showing the correlation of the Cy5.5-eIF5B occupancy on the ribosomal complex with Cy5–60S joining and Cy3.5-Phe-TC arrival. c, For all assayed mRNAs, the Δt values, average eIF5B lifetimes on 80S, and average tRNA arrival times after eIF5B departure (with 95% confidence intervals) were estimated by fitting the dwell times (from bottom to top for each group of three bars, n = 141, 164, 133, 131 and 134) to single-exponential distributions (also see Extended Data Fig. 9g). d, Contour plots of Cy5.5-eIF5B departure (bottom, normalized Cy5.5 fluorescence intensity changing from 1 to 0) and Cy3.5-Phe-TC arrival (top, normalized Cy3.5 intensity changing from 0 to 1) in experiments performed with model mRNA in the presence of GTP, generated by superimposing all the analyzed fluorescence traces such that the Cy3.5-Phe-TC arrival was set at time 0. Contours are plotted from tan (lowest population) to red (highest population), n = 134. e, The same types of average dwell times (with 95% confidence intervals from fitting to single-exponential distributions) as in (c) were determined for experiments performed with the model mRNA at 3 mM free Mg2+ and 20ºC, in the presence of GTP with either unlabeled wild-type (WT) eIF5B (n = 164, data taken from Fig. 1d); Cy5.5-labeled WT eIF5B (n = 134, data taken from c); unlabeled eIF5B-H505Y (n = 119); or Cy5.5-labeled eIF5B-H505Y (n = 80); or separately in the presence of GDPNP with unlabeled eIF5B-H505Y (n = 80) (also see Extended Data Fig. 9h).
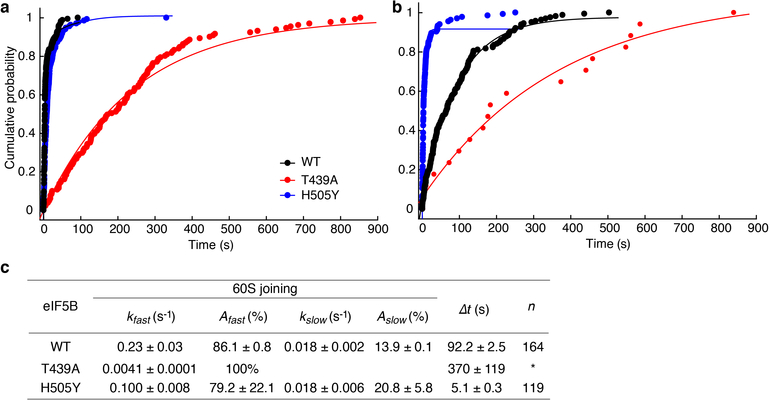
We hypothesize that the rate of GTP hydrolysis by eIF5B defines its residence time on the 80S complex. The Δt measured here on the model mRNA was similar to the mean time of eIF5B GTP hydrolysis during initiation on the same type of model mRNA (~30.2 s vs. ~28 s at 26ºC)21. Importantly, inclusion of a non-hydrolysable GTP analog, GDPNP, trapped eIF5B on the 80S complex (average Cy5.5-eIF5B lifetime on the 80S was ~850 s and photobleaching/imaging-time limited, n = 105), and prevented A-site aa-tRNA association (Extended Data Fig. 7b,c). To further explore this, we leveraged two previously reported eIF5B mutants that disrupt its GTPase activity8. The first (T439A) inhibited A-site tRNA association (only 17 out of 131 assembled 80S ribosomes yielded A-site tRNA association) and increased Δt to ~ 370 s at 20ºC (Extended Data Fig. 8). On the other hand, the second mutant (H505Y), which also has weakened affinity for the ribosome, decreased Δt by ~15-fold (n = 119) compared to wild-type eIF5B (Fig. 2e, Extended Data Fig. 8). This mutant instead reduced the occupancy time of eIF5B on the ribosome independent of GTP hydrolysis (Fig. 2e). Thus, eIF5B departure is indeed controlled by GTP hydrolysis.
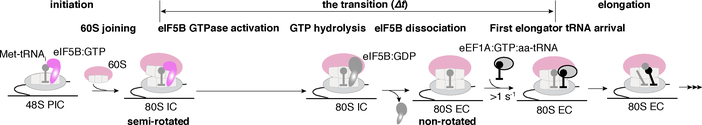
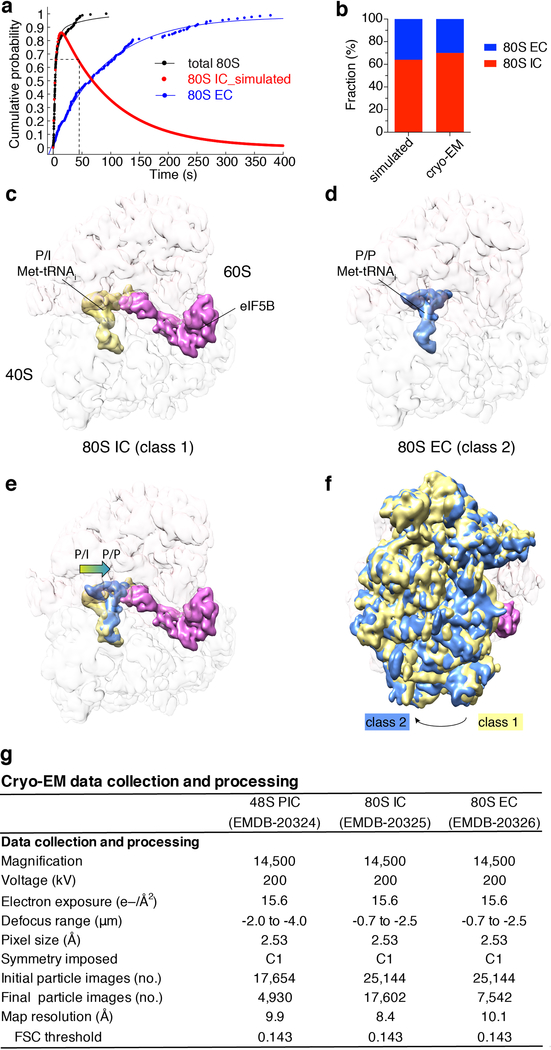
The striking dependence of the eIF5B departure rate on temperature and free Mg2+ concentration (Extended Data Fig. 9) suggests that its GTP hydrolysis activity is sensitive to the conformation of the ribosome. We therefore applied cryo-EM guided by our single-molecule kinetics to analyze the conformations of on-pathway ribosomal complexes assembled in the presence of wild-type eIF5B and GTP. We added eIF5B:GTP and 60S subunits to assembled 48S PICs (on the model mRNA, Fig. 1a), and the reaction was allowed to proceed for 45 s (from mixing to grid plunge-freezing, Methods) at ~20ºC, a pre-steady-state time point according to our single-molecule measurements (Methods and Extended Data Fig. 10a). Two distinct classes of 80S complexes were resolved: (1) the eIF5B bound-80S initiation complex (IC); and (2) the 80S elongation complex (EC) (Extended Data Fig. 10c,d). The two states were populated in agreement with predictions from our single-molecule kinetics (Extended Data Fig. 10a,b). Similar to that of the reported eIF5B:GDPCP bound 80S complex12, the eIF5B bound-80S IC (class 1) adopts a semi-rotated inter-subunit conformation with the Met-tRNAi in the P/I configuration. Moreover, eIF1A was absent in this complex. Its dissociation thus occurs prior to eIF5B departure and is not the rate-limiting step9. Upon eIF5B dissociation, the 80S IC matured to the non-rotated-state 80S EC (class 2) with Met-tRNAi shifted to the P/P configuration, poising the ribosome for rapid A-site tRNA association. Taken together, we propose that the irreversible GTP hydrolysis by eIF5B and its subsequent dissociation serves as a final kinetic quality control checkpoint of the 80S complex assembly prior to entry into the energy-expensive elongation phase.
In summary, we demonstrate that eIF5B gates the transition from eukaryotic translation initiation to elongation (Fig. 3). The lowest transition time we measured in m7G-cap-dependent translation was ~8 s in the presence of 2 mM free Mg2+ at 30ºC (Extended Data Fig. 9b) – a timescale relevant to the estimated rates of yeast translation initiation in vivo (varying from several seconds to minutes at 30ºC)23,24. The transition to elongation is likely gated by eIF5B GTP hydrolysis, as shown by the correspondence of GTP hydrolysis rate21 and eIF5B occupancy time measured here, and our observation that the eIF5B (likely with GTP)-bound 80S complex is in a similar conformation as observed with non-hydrolysable GTP12. We speculate that the long lifetime of eIF5B-bound 80S complexes could partially account for the enrichment of ribosomal protected mRNA fragments at the start codon observed in ribosome profiling experiments in eukaryotic systems25–27. The relatively slow transition between translation initiation and elongation in the eukaryotic system versus the bacterial counterpart provides a mechanism to control the ribosomal density in the early part of the mRNA open reading frame23,28 to prevent ribosome collisions29,30, and a final kinetic checkpoint for proper start codon selection. Consistent with this latter proposal, disrupted eIF5B GTPase activities cause higher leaky scanning propensities in vivo8, and eIF5B residence time on the 80S is strongly dependent on the start codon surrounding sequence context, with non-optimal Kozak sequences yielding longer residence times. Subtle conformational signaling, perhaps through the initiator tRNA, may modulate the final conformation of the 80S-eIF5B complex for rapid GTP hydrolysis, and subsequent formation of an elongation competent 80S complex. Finally, we expect that there are many kinetic checkpoints in the early stages of eukaryotic translation. The application of similar single-molecule approaches will unveil the complicated dynamics underlying mRNA selection, scanning and start codon selection, and their regulation1,2.
Fig. 3. Model of the late eukaryotic translation initiation and its transition to elongation.
eIF5B catalyzes 60S subunit joining to the 48S PIC to form the 80S initiation complex (IC), and its dissociation from the 80S IC requires GTP hydrolysis, plausibly leading to an altered eIF5B conformation thereby lowering its affinity to the 80S. Thus, the dissociation of eIF5B from the 80S IC gates the transition to elongation, marked by the binding of an elongator aa-tRNA to the elongation 80S complex (80S EC). Effects of free Mg2+ concentration, the sequence context surrounding the start codon, and temperature on the rate of the transition indicate that conformational rearrangements may play key roles in governing the rate of eIF5B dissociation, likely by controlling its GTPase activity.
Methods
1. The ybbR-tag labeling strategy
The ybbR-tag labeling strategy was established by Yin et al., wherein the SFP synthase specifically transfers a CoA-conjugate to a serine residue in the ybbR-tag (an 11 amino acids peptide sequence), resulting in a covalently linked CoA-conjugate to the ybbR-tag31. The SFP synthase used in this study was purified as described, and the Cy3-like dye analog CoA 547 was purchased from (New England BioLabs, Cat. S9349). Following the published protocol, we synthesized CoA-Cy5.5 dye by reacting CoA trilithium salt (Sigma-Aldrich, Cat. C3019–25MG; 0.52 mg, 0.66 μmol, 0.75 equiv) with sulfo-Cyanine5.5 maleimide (Lumiprobe, Cat. 17380; 1 mg, 0.88 μmol, 1 equiv) at room temperature for 2 hours in 1 mL volume in 100 mM potassium phosphate pH 7.05 and 10% DMSO. This was followed by quenching the excess amount of maleimide with final 8 mM DTT at room temperature for 10 min, enabling the use of the product without the need for further purification.
2. Unlabeled and labeled tRNAs and yeast ribosomal subunits
Native yeast Met-tRNAi was purchased from tRNA Probes, LLC (Cat. MI-60), and E. coli Phe-tRNAPhe and Lys-tRNALys labeled with Cy3.5 or Cy5 were previously characterized32. Unlabeled yeast ribosomal subunits were purified by sucrose density gradient centrifugation as described13. Cy3–40S was prepared by the ybbR-tag labeling method as follows:
1). Genetically tagging 40S subunit at uS19 with a ybbR-tag
The yeast uS19 knockout strain JD1346 [his3△1; leu2△0; ura3△0; YOL040c::KanMX4; pFL38(Pgal::RPS15; URA3)] and plasmid pJD1055 (pRPS15-HIS3) were kindly provided by J.D. Dinman33. A ybbR-tag sequence (GATTCTCTTGAATTTATTGCTAGTAAGCTTGCG)31 was inserted to at the N-terminal end of the uS19 sequence in the plasmid pJD1055, resulting in the plasmid pJFW1. This was introduced into yeast JD1346 using a standard lithium acetate yeast transformation protocol34 followed by a 5’-FOA plasmid shuffle35, to obtain the yeast strain yJFW11 in which all the 40S ribosomal subunits were tagged. For ybbR-uS19–40S purification, the yeast strain yJFW11 was streaked onto a SC-His selection plate, and one single colony was used to inoculate 5mL of fresh SC-His liquid medium and grown at 30ºC overnight with shaking at 200 RPM. The next morning, the stationary phase yeast culture was used to inoculate a 1.8 liter culture of YPAD medium with a 1/4000 dilution for large scale growth until OD600nm = ~0.6 before harvesting by fast centrifugation at 4 ºC and 6,000 RPM in a Fiberlite™ F9–6×1000 LEX Fixed Angle Rotor (ThermoFisher) for 12 min. The cell pellet was flash frozen in liquid nitrogen before storage at −80ºC for further ribosome purification.
2). Labeling of ybbR-uS19–40S
For labeling of the 40S subunit via the ybbR-tag, a 300 μL labeling reaction was prepared by mixing 1 μM 40S, 2 μM SFP synthase and 10 μM CoA-547 dye in a buffer containing 50 mM HEPES-KOH pH 7.5, 1 mM DTT and 10 mM MgCl2. The reaction was incubated at 30ºC for 30 min. To purify the labeled 40S from free dye, 150 μL of the reaction mixture was loaded on top of a 750 μL sucrose cushion [30 mM HEPES-KOH pH 7.5, 2 mM DTT, 100 mM KOAc, 5 mM Mg(OAc)2 and 0.5 M sucrose] for ultracentrifugation at 90,000 RPM at 4ºC for 90 min in a TLA100.2 rotor. The pellet containing labeled 40S was washed once with and then resuspended in the ribosome storage buffer [50 mM HEPES-KOH pH 7.5, 2 mM DTT, 100 mM KOAc, 3 mM Mg(OAc)2 and 0.25 M sucrose] and stored at −80ºC after flash freezing with liquid nitrogen.
The Cy5-labeled SNAP-tagged 60S was prepared as described and characterized before32. The Cy5.5-labeled ybbR-tagged 60S was prepared in the same way as described above for the Cy3–40S. For yeast genetics, the pRS315 plasmid used for the SNAP-tagged 60S yeast strain was mutated to carry the native uL18 (RPL5) gene sequence with a ybbR-tag sequence fused in frame at the C-terminus before the stop codon. The new plasmid was transformed into JWY3733 strain (MATα; ura3-52; trp1△101; leu2△1; ade1; his3△200; rpl1::TRP1+ pRS316-RPL5) and plated onto SC-Leu plates followed by screening for the loss of the pRS316-RPL5 on 5’-FOA-containing SC-Leu plates. The resulting strain, (MATα; ura3-52; trp1△101; leu2△1; ade1; his3△200; rpl1::TRP1+ pRS315-RPL5-ybbR), expresses uL18-ybbR as the sole source of the protein. The ybbR-ribosome labeling efficiencies were generally around ~75% based on absorbance measurements.
3. Purification of protein factors
Published strains/constructs and protocols were used for the purification of yeast eIF2, eIF3, eIF4E, eIF5, eIF5A, eIF5B (residues 396–1002), eEF1A, eEF1Bα, eEF2 and eEF313,15,32,36.
1). eIF1
The SUI1 gene was cloned into a pET28 vector such that the overexpressed protein had the following sequence: MGSSHHHHHH MENLYFQG-native eIF1, wherein the underlined sequence is a TEV protease cleavage site, which is followed by the native eIF1 sequence, and the sequence in bold is the final purified protein sequence. The plasmid was transformed into Rosetta2(DE3) E. coli cells. Protein overexpression was performed at OD600nm ~0.6 by induction with 1 mM IPTG at 30ºC for 4 hours. Cells were pelleted and stored at −80ºC. The pellet was rapidly thawed and suspended in 50 mM potassium phosphate (pH 7.5), 500 mM KCl, 10 mM imidazole, 5 mM β-mercaptoethanol (βME), 10% (v/v) glycerol and 1 mM phenylmethylsulfonyl fluoride (PMSF). The cells were lysed by sonication and the lysate was clarified by centrifugation at 18,000 RPM for 30 min at 4ºC in a F21–8×50y rotor (ThermoScientific). The clarified lysate was loaded onto a 2 mL Ni-NTA column and washed with 50 mL wash buffer [50 mM potassium phosphate (pH 7.5), 300 mM KCl, 20 mM imidazole, 5 mM βME, 10% (v/v) glycerol]. His-tagged protein was eluted with wash buffer containing 250 mM imidazole. The eluted protein was diluted five times in volume by addition of 20 mM HEPES-KOH pH 7.5, 10% (v/v) glycerol and 2 mM DTT, and loaded to a 5 mL HiTrap Heparin HP column for further purification as described previously13. The eluate was treated with TEV protease overnight on ice to cleave off the His6-tag and eIF1 was recovered by flowing through a Ni-column to remove the free tag and TEV protease. The final eIF1 protein was purified on a HiLoad 26/600 Superdex 200 pg gel filtration column in storage buffer [20 mM HEPES-KOH pH 7.5, 100 mM KOAc, 10% (v/v) glycerol, and 2 mM DTT], flash frozen with liquid nitrogen, and stored at −80ºC.
2). eIF1A
The TIF11 gene was cloned into a pET28 vector such that the overexpressed protein had the following sequence: MGSSHHHHHH MENLYFQG-native eIF1A, wherein the underlined sequence is a TEV protease cleavage site, which is followed by the native eIF1A sequence, and the sequence in bold is the final purified protein sequence. The protein overexpression and purification were performed in exactly the same way as for eIF1.
3). eIF4A
The TIF1 gene was cloned into a pET28 vector such that the overexpressed protein had the following sequence: MGSSHHHHHH MENLYFQG-native eIF4A, wherein the underlined sequence is a TEV protease cleavage site, which is followed by the native eIF4A sequence, and the sequence in bold is the final purified protein sequence. The protein overexpression and Ni-NTA purification were performed in the same way as for eIF1. After Ni-NTA purification, the protein was treated with TEV protease overnight to cleave off the His6-tag while dialyzing at 4ºC against a buffer containing 50 mM potassium phosphate (pH 7.5), 300 mM NaCl and 5 mM βME. The eIF4A protein was recovered by flowing through a Ni-column to remove the cleaved His6-tag and TEV protease. The final protein was purified on a HiLoad 26/600 Superdex 200 pg gel filtration column in storage buffer [20 mM HEPES-KOH pH 7.5, 100 mM KOAc and 2 mM DTT], flash frozen with liquid nitrogen and stored at −80ºC.
4). eIF4B
The TIF3 gene was cloned into a pET22b vector such that the overexpressed protein had the following sequence: native eIF4B-ENLYFQGAAAELHHHHHH, wherein native eIF4B sequence is followed by the underlined TEV protease cleavage site, and the sequence in bold is the final purified protein sequence. The protein overexpression and purification were performed in exactly the same way as for eIF4A.
5). eIF4G
The TIF4631 gene was cloned into the multiple cloning site 1 of pETDuet-1 vector such that the overexpressed protein had the following sequence: native eIF4G-ENLYFQGHHHHHH, wherein native eIF4G sequence is followed by the underlined TEV protease cleavage site, and the sequence in bold is the final purified protein sequence. On the same plasmid, native eIF4E gene was cloned into the multiple cloning site 2, which has been proven to facilitate the overexpression of the full-length eIF4G in E. coli37. The protein overexpression and purification were performed in the same way as for eIF4A, except 500 mM KCl was included in all the buffers used for the Ni-column purification. The final protein contained both eIF4G and wild-type eIF4E and the full-length eIF4G migrates slower than expected on SDS-PAGE for its molecular weight, which has been shown previously by others37–39. The full-length nature of eIF4G was further verified by Edman degradation and mass spectrometry.
6). eIF5B variants
When applicable, a ybbR-tag sequence was introduced into the pMGAHisTev5B plasmid13 after the TEV cleavage site, with/without a T439A or H505Y mutation, by site-directed mutagenesis. The protein was overexpressed and purified following the previously described protocol13. For full-length eIF5B, we started from the plasmid pC100540, deleted the Flag-tag sequence, inserted in a 8xHis tag and a TEV cleavage site before the native eIF5B sequence, and cloned into the multiple cloning site 1 of the pETDuet-1 vector. Proteins were overexpressed in Rosetta2(DE3) cells as described for eIF1 and the purification was performed as described13. To label the ybbR-tagged proteins with Cy5.5, 10 μM protein was incubated with 2 μM SFP synthase and 15 μM CoA-Cy5.5 dye in a buffer containing 50 mM HEPES-KOH pH 7.5, 1 mM DTT and 10 mM MgCl2 at room temperature for 30 min. The reaction mixture was loaded onto a Ni-NTA column equilibrated with the same buffer to remove His-tagged SFP synthase, and the flow-through was passed through a 10-DG desalting column to remove free dye from the protein. The final protein was purified on a Superdex 75 10/300 GL column in storage buffer [20 mM HEPES-KOH pH 7.5, 100 mM KOAc, 10% glycerol and 2 mM DTT] and stored at −80ºC. The ribosome-dependent GTPase activity was determined as previously described using the malachite green assay32.
4. mRNAs
All the mRNAs were prepared by T7 RNA polymerase run-off transcription. The RNA transcript sequences are as follows (with the start and stop codons underlined):
model mRNA: GG(UC)8UAUGUUCAAAUAA(UC)16;
model mRNA-Kozak: GG(UC)8UAAAAAAAUGUUCAAAUAA(UC)16;
model mRNA-Kozak_−3U: GG(UC)8UAAAUAAAUGUUCAAAUAA(UC)16;
RPL41A: GGAGACCACAUCGAUUCAAUCGAAAUGUUCGCCAAGUGGAGAAAGAAGAGAACUAGAAGACUUAAGAGAAAGAGACGGAAGGUGAGAGCCAGAUCCAAAUAA;
RPL30: GGAAACAGACCGGAGUGUUUAAGAACCUACAGCUUAUUCAAUUAAUCAAUAUACGCAGAGAUGUUCCCAGUUAAAUCCCAAGAAUCUAUCAACCAAAAGUUGGCUUUGGUUAUCAAGUCUGGUAAGUACACCUUAGGUUACAAGUCCACUGUCAAGUCUUUGAGACAAGGUAAGUCUAAGUUGAUCAUCAUUGCCGCUAACACUCCAGUUUUGAGAAAGUCCGAAUUGGAAUAUUACGCUAUGUUGUCCAAGACUAAGGUCUACUACUUCCAAGGUGGUAACAACGAAUUGGGUACUGCUGUCGGUAAGUUAUUCAGAGUCGGUGUUGUCUCUAUUUUGGAAGCUGGUGACUCUGAUAUCUUGACCACCUUGGCUUAAAUAAGGUAAGUUCAAACGAUUU.
For the model mRNA, model mRNA-Kozak and model mRNA-Kozak_−3U, 3’end biotinylation or Cy5-labeling was performed using periodate chemistry followed by hydrazide reaction as described41. The RNA was purified from the free hydrazide on a Superdex 75 10/300 GL column in storage buffer (20 mM HEPES-KOH pH 7.5 and 100 mM KOAc) and stored at −80ºC. The hydrazide analogs used were EZ-Link™ Biotin-LC-Hydrazide (ThermoFisher Scientific, Cat. 21340) and Cyanine5 hydrazide (Lumiprobe, Cat. 13070).
To obtain 3’end biotinylated or Cy3-labeled, fully m7G-capped, RPL41A mRNA, the RNA transcript was: 1) ligated to a DNA oligonucleotide 5’pd(CT)12-Biotin-3’ or 5’pd(CT)6-Cy3- d(CT)6-3’ (TriLink Biotechnologies) and gel purified; 2) m7G-capped using the Vaccinia capping system (New England BioLabs, Cat. M2080S); 3) treated with RNA 5’polyphosphatase (Epicentre, Cat. RP8092H) and Terminator™ 5’-Phosphate-Dependent Exonuclease (Epicentre, Cat. TER51020) to remove uncapped-RNA, followed by phenol/chloroform extraction and ethanol precipitation.
The 3’end biotinylated, fully m7G-capped, RPL30 mRNA was prepared by: 1) 3’end biotinylation using the periodate chemistry and hydrazide reaction41; 2) m7G-capping and uncapped-RNA depleting as described above for RPL41A mRNA.
5. Native gel shift analyses (related to Extended Data Fig. 2a,b)
The formation of pre-initiation and initiation complexes was monitored by 4% native PAGE analyses using protocols as described13. Samples were prepared in a 1xRecon buffer [30 mM HEPES-KOH pH 7.5, 100 mM KOAc, 3 mM Mg(OAc)2]. A ternary complex was prepared by incubating 3.9 μM eIF2 with 1 mM GTP:Mg2+ at 30ºC for 10 min, followed by addition of 2.7 μM Met-tRNAi and incubation at 30ºC for another 5 min.
For reactions with the model mRNA (Extended Data Fig. 2a), the 48S PIC was formed by mixing a final concentration of 1 μM eIF1, 1 μM eIF1A, 3-fold diluted ternary complex (final 1.3 μM eIF2 and 0.9 μM Met-tRNAi), 50 nM Cy3–40S, and 2 μM model mRNA-Cy5 and incubating at 30ºC for 15 min; the 80S PIC was formed by addition of a final concentration of 1 μM eIF5, 1 μM eIF5B and 200 nM 60S to the 48S PIC and further incubation at 30ºC for 5 min. Typically, 10 μL of each sample was mixed with 2 μL of native gel dye (50% sucrose, 0.02% bromophenol blue in 1xRecon buffer) and loaded into the 4% native PAGE gel for electrophoresis13 followed by scanning the gel for the Cy3 and Cy5 fluorescence with a Typhoon scanner.
For reactions with the cap-RPL41A mRNA (Extended Data Fig. 2b), the samples were prepared similarly as above, except 1 mM ATP:Mg2+ was supplemented into the reaction buffer, cap-RPL41A-Cy3 mRNA was at 100 nM, 40S was unlabeled, and when applicable, 1 μM eIF4A, 1 μM eIF4B, 400 nM eIF4G, 200 nM eIF4E and 200 nM eIF3 were added.
6. Single-molecule inter-subunit FRET experiments on a TIRF microscope
The ribosomal inter-subunit FRET signal was characterized on a home-built, prism-based TIRF microscope as previously described42,43. The 80S complexes were assembled using Cy3–40S and Cy5–60S on the model mRNA-biotin in the same way as for the native gel shift assays. The 80S complexes were immobilized on a Neutravidin-derivatized quartz slide according to reported protocols32,43. Unbound components were washed from the slide with 1xRecon buffer supplemented with 1 mM GTP:Mg2+ and an oxygen-scavenging system [containing 2.5 mM of PCA (protocatechuic acid), 2.5 mM of TSY, and 2x PCD (protocatechuate-3,4-dioxygenase), purchased from Pacific Bioscience]. Cy3 fluorophores were excited with a diode-pumped solid-state 532-nm laser at 1 kW/cm2. Fluorescence intensity trajectories were recorded at 10 frames per second with a total of 600 frames collected per movie by using the MetaMorph imaging software (Molecular Devices). Analysis of fluorescence intensity trajectories was performed in MATLAB using custom scripts42. Baseline correction of raw fluorescence traces and Cy3 signal bleedthrough into the Cy5 channel were corrected on a trace-by-trace basis. The FRET states were assigned with a hidden Markov model-based approach as previously described and visually corrected42.
7. Real-time single-molecule experiments on the ZMW instrument
All the experiments were performed in the 1xRecon buffer supplemented with 1 mM GTP:Mg2+ (and 1 mM ATP:Mg2+ for cap-mRNAs) unless otherwise specified. A ZMWs-based PacBio RSII instrumentation14 was used for all the real-time measurements at 20ºC or 30ºC. For experiments performed with the model mRNA, model mRNA-Kozak and model mRNA-Kozak_−3U, in general, immediately before each experiment, the 48S PIC was formed as described for the native gel shift assays using final 80 nM Cy3–40S, 500 nM biotinylated mRNA, 1 μM eIF1, 1 μM eIF1A, 300 nM eIF5, 1.3 μM eIF2 and 0.9 μM Met-tRNAi. In parallel, a ZMW chip was treated with 0.2% (w/v) Tween20 at room temperature for 10 minutes followed by the treatment with Neutravidin at room temperature for 5 minutes, which bound to the Biotin-PEG (polyethylene glycol) on the chip. After immobilizing the 48S PIC via the biotin on the mRNA onto the Neutravidin-coated surface, the chip was washed twice using an imaging buffer containing the oxygen-scavenging system and 62.5 μg/mL casein to remove unbound complexes, and loaded onto the RSII instrument in the presence of 20 μL of the imaging buffer. At the same time, a delivery mixture, containing 200 nM Cy5–60S, 1 μM eIF5, eIF5B (2 μM unlabeled or 40 nM Cy5.5-labeled), Cy3.5-Phe-TC [pre-formed by incubating 8 μM eEF1A, 10 μM eEF1Bα with 1 mM GTP:Mg2+ at room temperature for 5 min, followed by the addition of 0.8 μM Phe-(Cy3.5)tRNAPhe and another incubation at room temperature for 5 min; diluted 8-fold in the delivery mixture for final 50 nM TC or 4-fold for final 100 nM TC], oxygen-scavenging system and 62.5 μg/mL casein, was prepared and loaded onto the instrument. For each experiment, the instrument delivered 20 μL of the delivery mixture to the chip at the beginning of the experiment, and recorded a 15-minute movie at 10 frames per second, with 0.32 μW/μm2 of 532 nm laser illumination (for unlabeled eIF5B experiments), or 0.6 μW/μm2 of 532 nm laser and 0.1 μW/μm2 of 642 nm laser dual illumination (for Cy5.5-labeled eIF5B experiments). For the elongation assay with tRNA-tRNA smFRET, both the imaging buffer and the delivery mixture were supplemented with 1 mM ATP, and the delivery mixture contained 115 nM Cy5.5–60S, 1 μM eIF5, 2 μM eIF5A, 2 μM eIF5B, 1 μM eEF2, 400 nM eEF3, 100 nM Cy3.5-Phe-TC and 100 nM Cy5-Lys-TC [pre-formed separately by incubating 8 μM eEF1A, 10 μM eEF1Bα with 1 mM GTP:Mg2+ at room temperature for 5 min, followed by the addition of 0.8 μM Phe-(Cy3.5)tRNAPhe or Lys-(Cy5)tRNALys, and another incubation at room temperature for 5 min], oxygen-scavenging system and 62.5 μg/mL casein. The imaging was performed at 20ºC with 0.32 μW/μm2 of 532 nm laser illumination.
Experiments with the cap-RPL41A and cap-RPL30 mRNAs were performed in a similar manner, except 1 μM eIF4A, 1 μM eIF4B, 0.67 μM eIF4E, 300 nM eIF4G, and 200 nM eIF3 were also included in the second incubation step for the 48S PIC formation, and 300 nM eIF3 was also added to the delivery mixture. When titrating free Mg2+ concentrations, the corresponding amount of Mg(OAc)2 was used in both the imaging buffer and the delivery mixture. When additional protein factors were added to each experiment, they were added to the delivery mixture at the denoted amounts. For experiments performed with GDPNP, the 48S PIC was formed in the presence of GTP as described above. After immobilization, the imaging buffer used contained 0.2 mM GTP, while no GTP was added in the delivery mixture and 6 mM GDPNP:Mg2+ was added instead.
The resulting experimental movies were analyzed using custom MATLAB scripts, as previously described14. Briefly, fluorescence traces from the ZMWs were filtered based on the presence of the fluorophores of interests at different time points (the signal from immobilized 48S PICs was expected to be present at the beginning of the movie, while signals from fluorophores attached to the 60S, eIF5B and tRNA were expected not to be). The different arrival and departure events were manually assigned based on the appearance and disappearance of the corresponding fluorescent signals.
8. Cryo-EM analysis of the 48S PIC formed with cap-RPL30 mRNA
The 48S PIC sample for the cryo-EM analysis was prepared in the same way as that for the real-time single-molecule experiments in the presence of ATP, GTP and 3 mM free Mg2+. After the incubation at 30ºC, 30 μL of the sample was diluted with 100 μL 1xRecon buffer and then concentrated to 30 μL using a Vivaspin 500 concentrator (100, 000 MWCO PES, Sartorius, Cat. VS0141) by centrifugation at 9,000 RPM for 5 min at 4ºC. This step mimics the washing step in our single-molecule experiments for removing unbound components. 3 μL sample was applied to each 25-second glow-discharged (in a PELCO easiGlow glow discharger, Ted Pella, INC) 200-mesh Quantifoil R2/1 gold grid (Electron Microscopy Sciences, Cat. Q250AR1) at 21ºC and 95% humidity. The grid was blotted for 2 seconds before plunging into liquid ethane using a Leica EM GP (Leica Microsystems). Automated data acquisition was performed using SerialEM44 on a Tecnai F20 microscope operated at 200 kV at a dose rate of 10 e−/pixel/s using a defocus range of −2.0 to −4.0 μm. Eighty movie-mode images with 10-second exposure time and 50 frames for each were recorded on a Gatan K2 Summit direct electron detector camera in a counting mode at a nominal magnification of 14,500 (yielding a pixel size of 2.53 Å on the specimen level). The drift correction was performed using MotionCor245 and the contrast transfer function (CTF) estimation were performed using CTFFIND-4.146. The images with bad CTF fitting were excluded and 78 images were selected for further data processing. The initial particle picking was performed using EMAN247 and the saved box files were imported into Relion-3.0-beta-248 for the first 2D classification to generate some good 2D class averages as a new template for automatic particle picking in Relion. The 3D initial model was built from Relion. After the first 6-class 3D classification, 11,478 particles within one class from total 17,654 good particles were selected for the 3D auto-refine to obtain a map at 9.0 Å resolution (not shown). Then a soft mask around the Met-tRNAi region was created to perform the particle subtraction followed by the focused classification (without image alignment) into 2 classes: with Met-tRNAi and without Met-tRNAi. Finally, 4,930 particles were selected for the final 3D auto-refine to reconstruct the map at 9.9-Å resolution. The scanning-competent open (EMD 3049) or scanning-incompetent closed (EMD 3048) 48S PIC structures were low-pass filtered to the same resolution (9.9 Å) and scaled to the same pixel size using Relion’s command relion_image_handler for visual comparisons in Chimera (UCSF) (related to Extended Data Fig. 2c,d,e).
9. Cryo-EM analysis of the on-pathway 80S complexes during initiation and the transition to elongation
To probe the conformational states of the 80S complexes along the native initiation and the transition to elongation pathways, we decided to analyze the sample corresponding to the reaction time point 45 s (under pre-steady-state condition), when 80S IC should account for ~64.5% (and 80S EC ~35.5%, see Supplementary Note) of the total 80S complexes assembled on the model mRNA at 20ºC and 3 mM Mg2+ in the presence of WT-eIF5B and GTP. Also, to ease the post-acquisition particle classification procedure, we decided to use excess 48S PICs to 60S, such that free 60S particles would be minimized. To this end, we assembled 48S PICs in the same way as in the single-molecule experiments with the model mRNA at 3 mM free Mg2+, and the only difference was that the concentration of the unlabeled 40S was at 300 nM (assuming 100% complex assembly, the 48S PIC concentration should be 300 nM). In parallel, an initiation mixture contained 150 nM unlabeled 60S, 1 μM eIF5, and 2 μM eIF5B was prepared. After mixing equal volumes of the 48S PIC with the initiation mixture, the final concentrations were: 150 nM 48S PIC, 75 nM 60S and 1 μM eIF5B. At 20ºC under this condition, we would expect the reaction kinetics to be similar to that of the single-molecule experiments described above.
While the 48S PIC and initiation mixtures were kept on ice, the 200-mesh R2/1 Quantifoil grids were glow-discharged for 25 s. When the Leica EM GP system was ready for grid plunging, 5 μL of the 48S PIC mixture was mixed with 5 μL of the initiation mixture, and incubated at ~20ºC for 30 s before ~3 μL of the sample was applied to each grid. The grid was immediately blotted for 2 s using a filter paper before being plunged into liquid ethane with 95% chamber humidity at ~21ºC (resulting in a reaction time from mixing to grid-freezing at 45 s). Automated data acquisition was done using SerialEM44 on a Tecnai F20 microscope operated at 200 kV at a dose rate of 10 e−/pixel/s using a defocus range of −0.7 to −2.5 μm. Total 226 movie-mode images with 10-second exposure time and 50 frames for each were recorded on a Gatan K2 Summit direct electron detector camera in a counting mode at a nominal magnification of 14,500 (yielding a pixel size of 2.53 Å on the specimen level). The drift correction was performed using MotionCor245 and the contrast transfer function (CTF) estimation were done using CTFFIND-4.146. The images with bad CTF fitting or too much drift or being over-focused were excluded, and eventually 134 images were selected for further 2D and 3D classifications. The initial particle picking was performed using EMAN247 and the saved box files were imported into Relion-3.0.648 for the first 2D classification to generate some good 2D class averages as a new template for better 80S particle picking in Relion. The 3D initial model of the 80S complex was built using Relion. The following 2D and 3D classifications were used to eliminate junk or bad particles or even free 40S or 60S particles. Totally 25,144 80S particles were selected for the 3D auto-refine to obtain the map at 7.8-Å resolution (not shown). The eIF5B PDB model was selected from the pdb file 4V8Z12 and fit into the 7.8-Å 80S map. A zone with radius 6 around the fitted eIF5B PDB was generated using Chimera. Then a soft mask around eIF5B density was made from Relion’s Mask Creation. The particle subtraction was done with this soft mask to perform the focused 3D classification (without image alignment) into 2 classes of 80S: 17,602 particles with eIF5B bound (class 1, 80S IC, Extended Data Fig. 10c) and 7,542 without eIF5B (class 2, 80S EC, Extended Data Fig. 10d). Finally 3D auto-refine reconstructed the 80S IC map at 8.4 Å resolution and the 80S EC map at 10.1 Å resolution.
Extended Data
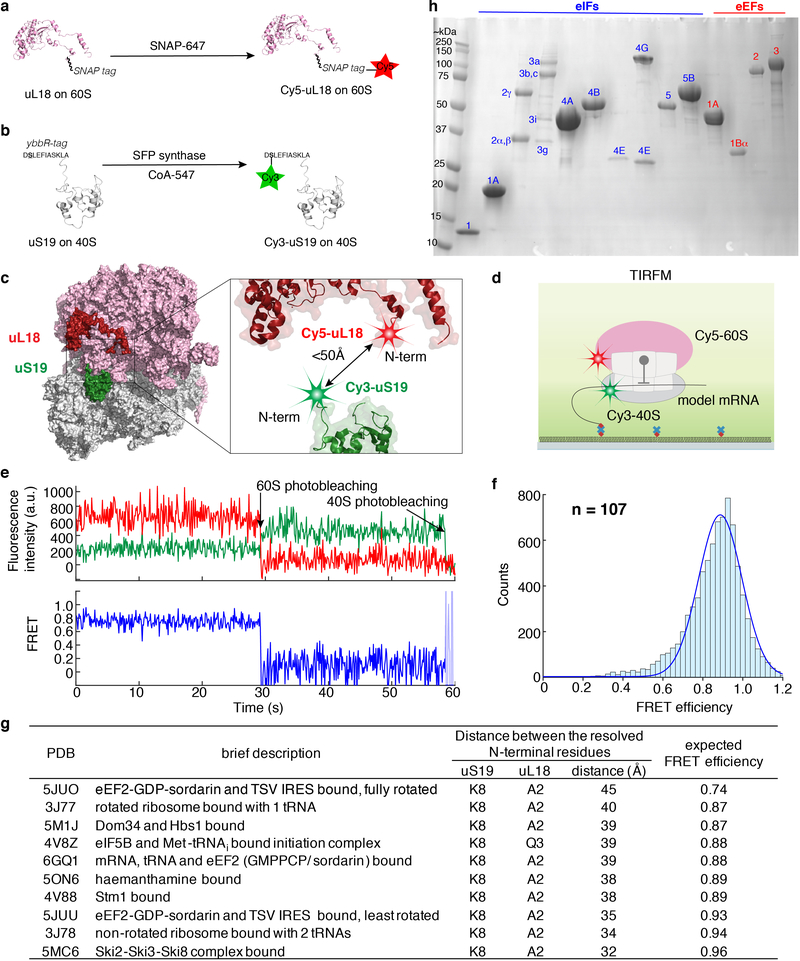
Extended Data Fig. 1 |. A reconstituted yeast translation system with fluorescently labeled ribosomes for inter-subunit smFRET.
a, We previously have established the Cy5 labeling of yeast 60S ribosomal subunit via a SNAP tag fused to the uL18 protein and the reaction with a SNAP-647 dye32. b, In this work, we engineered a yeast strain in which all the 40S subunits carried the N-terminal ybbR-tagged uS19 protein. Upon purification, the 40S was labeled by SFP synthase with CoA-547 at the serine residue (in bold and underlined) of the ybbR tag, resulting in the Cy3–40S. c, The estimated distance between the two ribosomal labeling sites is within 50Å from published yeast 80S 3D structures (g). The ribosome model was created in PyMOL with PDB 4V8Z12. d, TIRFM experimental setup to characterize the inter-subunit FRET signal. 80S complexes were assembled from Cy3–40S and Cy5–60S on the model mRNA (Fig. 1a) in the presence of required factors (Method) and were immobilized on a quartz slide used for TIRFM imaging with green laser illumination. e and f, Sample TIRFM experimental trace (e) and the inter-subunit smFRET efficiency histogram (f, fit with a single-Gaussian distribution, with a mean FRET efficiency at 0.89 ± 0.15 s.e.m.), n = 107 molecules. g, Estimated distances between the two labeling sites on the ribosomal subunits from a few examples of the published yeast 80S structures in different functional states, and the expected FRET efficiencies based on a Förster radius (R0) of 54 Å for the Cy3/Cy5 FRET pair. h, A representative SDS-PAGE analysis of the purified core eIFs (blue numbering) and eEFs (red numbering) used for the reconstitution of the translation system. Each component was analyzed at least three times with similar results.
Extended Data Fig. 2 |. Native gel shift assays and cryo-EM analysis showing active translation initiation with our purified yeast translation system.
a, A representative gel showing initiation on the model mRNA. A merged view of Cy5 (red) and Cy3 (blue) scans of the same gel is shown. For gel source data, see Supplementary Figure 1. The model mRNA was labeled with Cy5 and 40S was labeled with Cy3. Addition of Cy3–40S, Met-tRNAi:eIF2:GTP (Met-TC), eIF1 and eIF1A to the model mRNA-Cy5 resulted in the formation of a distinct 48S PIC band. Further addition of eIF5, eIF5B and 60S led to the formation of the 80S band. The experiment was repeated three times with similar results. b, A representative gel showing initiation on the cap-RPL41A mRNA. For gel source data, see Supplementary Figure 1. The cap-RPL41A mRNA was labeled with Cy3, and other components were unlabeled. The gel was scanned for Cy3 fluorescence. Various mRNA/protein complexes were formed in the absence of the 40S. Upon adding 40S to the mixture, a distinct 48S PIC band was formed and further addition of eIF5B and 60S shifted this band to the 80S band. The apparent electrophoretic mobility of the 48S PIC formed with the capped-RPL41A mRNA differs from that with the model mRNA, likely due to the different charges/hydrodynamic radius of the complex brought about by the capped-mRNA. Both 48S PIC and 80S formation were very inefficient when the cap-binding eIF4F (eIFs 4A, 4E and 4G) and eIF4B proteins were omitted from the reaction, demonstrating the cap-dependence of the initiation when the full set of eIFs were added. The experiment was repeated three times with similar results. c,d and e, With our 48S PIC assembly regime, we would expect that the 48S PIC in the post-scanning state, with the eIFs required during the scanning process potentially dissociated from the complex. A 9.9Å cryo-EM map was obtained for the 48S PIC formed on the cap-RPL30 mRNA (grey), which was compared with the reported scanning-competent, mRNA channel-open (EMD 3049, yellow, c), or scanning-incompetent, mRNA channel-closed (EMD 3048, cyan, d) 48S PIC structures16 (see Methods). The cap-RPL30/48S PIC was assembled in the same way as for our single-molecule experiments, and the comparisons showed that it resembles the post-scanning closed state, with the Met-tRNAi positioned in the P-site (e, in red was the modeled Met-tRNAi in the EMD 3048 structure, PDB 3JAP).
Extended Data Fig. 3 |. Real-time 80S assembly on the model mRNA at the single-molecule level.
a, smFRET assay for subunit joining in ZMWs. 48S PICs were formed by incubating Cy3–40S, Met-TC, model mRNA-biotin, eIF1, eIF1A and eIF5 at 30ºC for 15 min before immobilization in the ZMWs. After washing away free components, the experiment was started with green laser illumination and delivery of Cy5–60S, eIF5, and eIF5B. The reaction was performed in the 1x Recon buffer supplemented with 1 mM GTP:Mg2+ at 20ºC. b, Example experimental trace showing real-time observation of Cy5–60S joining to immobilized Cy3–48S PIC to form the 80S complex, identified by the appearance of smFRET. Single photobleaching events are denoted. Similar results were obtained from three independent experiments. c, The cumulative probability distribution of 60S joining dwell times was fit to a double-exponential equation, resulting in a fast phase rate of ~0.22 s−1 with ~46% amplitude, and a slow phase rate of ~0.03 s−1 with ~54% amplitude. n = 178. The kinetics is comparable to prior bulk measurement of the same reaction under similar condition (~77% fast phase with a rate of ~0.076 s−1; ~23% slow phase with a rate of 0.019 s−1)9. d and e, Spermidine-driven initiation in the absence of eIF5B. The cumulative probability distributions of the dwell times for 60S joining (d) and the transition to elongation (e) from experiments performed in the presence or absence of 3 mM spermidine and/or 1 μM eIF5B at 20ºC with the model mRNA were fit to a double-exponential (d) or a single-exponential (e) equation. The estimated average fast and slow phase rates (kfast and kslow) and amplitudes (Afast and Aslow) of 60S joining (from d) and Δt values (from e) with the 95% confidence intervals were shown in the inset table in e. n = 232 (for + spermidine + eIF5B), 117 (for + spermidine – eIF5B) and 164 (for – spermidine + eIF5B). Notably, the Δt is small in the presence of only spermidine, likely due to the lack of the rate-limiting eIF5B dissociation step. Consistently, this Δt falls in the same range of the average tRNA arrival times after eIF5B departure in those experiments performed with labeled eIF5B (Fig. 2).
Extended Data Fig. 4 |. Single-molecule assays demonstrate that the first A-site aa-tRNA association is codon/tRNA specific and can lead to further elongation.
a, Experimental setup (left) and sample fluorescence trace (right) for the single-molecule assay to assess the A-site tRNA binding specificity. 48S PICs containing Cy3–40S, Met-tRNAi, and the 3’-biotyinlated model mRNA (encoding Met-Phe-Lys-stop) were immobilized in ZMWs in the presence of required eIFs. Experiments were started by illuminating ZMWs with a green laser and delivering Cy5–60S, Cy3.5-Lys-tRNALys:eEF1A:GTP ternary complex (TC) and eIFs. No Cy3.5-Lys-tRNALys binding events were observed in the 15-min imaging window, demonstrating that the A-site aa-tRNA association was codon/tRNA specific. n = 150. b-e, The elongation competence of the 80S complex is scored by (Cy3.5)tRNAPhe-(Cy5)tRNALys smFRET after initiation and elongation to the second elongation codon in the model mRNA. In order to study the tRNA-tRNA smFRET in the context of ribosomal inter-subunit smFRET, we decided to use Cy3 and Cy5.5 for the labeling of the 40S and 60S, respectively. Therefore, we engineered the yeast 60S to carry a ybbR-tag at the C-terminus of uL18 and labeled the 60S with Cy5.5-CoA by SFP synthase (b), and show that the different tag/label did not significantly affect the kinetics of the transition to elongation (red curve, Δt = 95.4 ± 2.6 s, n = 146, model mRNA, 20ºC and 3 mM free Mg2+) compared with that when using the original Cy5-SNAP-tagged 60S (black curve, Δt = 92.2 ± 2.5 s, n = 164) (c, errors represent 95% confidence intervals of the average dwell times from fitting the lifetimes to single-exponential distributions). d shows a sample fluorescence trace from the experiments where Cy5.5–60S, Cy3.5-Phe-TC, Cy5-Lys-TC, eEF2, eEF3:ATP, eIF5A and other required eIFs were delivered to ZMWs immobilized with 48S PICs containing Cy3–40S, Met-tRNAi, and the 3’-biotyinlated model mRNA, and illuminated with a green laser at 20ºC. Out of n = 152 molecules showing the sequential 60S and Cy3.5-Phe-tRNA association events, n = 113 molecules showed the subsequent Cy3.5-tRNAPhe to Cy5-tRNALys FRET signal (d). And the distribution of the dwell times between the appearance of the Cy3.5 and Cy5 signals was fit to a single-exponential equation, with the average time being 142 ± 8 s (e, the error represents the 95% confidence interval, n = 113). Thus 74% of the first A-site aa-tRNA association yielded the elongation to the next codon.
Extended Data Fig. 5 |. Comparisons of the kinetics of the transition from initiation to elongation under various conditions.
The transition dwell times (open circles) were fit to single-exponential distributions to estimate the average dwell times (i.e. Δt values, gray bars, with error bars in black represent the 95% confidence intervals), and compared when no extra factors were added (data taken from Fig. 1d, n = 164), or in presence of eIF3 and eEF3 (n = 130), or with addition of eIF5A (n = 143) (a); or when the concentration of Cy3.5-Phe-TC was at 50 nM (n = 221) or 100 nM (data taken from Fig. 1d, n = 164) (b) in experiments performed with the model mRNA at 20ºC. Similar comparison was shown in (c) for experiments performed with the cap-RPL30 mRNA at 20ºC when the concentration of Cy3.5-Phe-TC was at 100 nM (n = 118) or 200 nM (n = 132). d, The Δt values (the average dwell time estimated from fitting the transition dwell times, open circles, to a single-exponential equation, with 95% confidence intervals in black) compared across all assayed mRNAs at 20ºC and 30ºC (related to Fig. 1a,d). In model mRNA-Kozak_−3U, the −3 position A of the optimal Kozak sequence was mutated to U, which largely abolished the Kozak sequence effect. From bottom to top for each group, n = 118, 130, 130, 121, 149, 161, 189, 159, 164 and 136.
Extended Data Fig. 6 |. Neither truncation nor fluorescent labeling of eIF5B perturbs its function.
a, Fluorescent labeling of eIF5B with Cy5.5 via a ybbR-tag at the N-terminal end, which is distal from the ribosomal subunit labels and hence not expected to interfere with the inter-subunit smFRET. The ribosome model was created in PyMOL with PDB 4V8Z12. b, A N-terminal domain truncated version of eIF5B (eIF5B-Trunc) was used in most of our assays, as in other reported reconstituted, purified yeast translation assays8,9,11–15,21,25. Previous reports failed to purify the full-length protein and have demonstrated that the truncated protein supported initiation in vitro and in vivo8,9. c, We tagged eIF5B-Trunc at the N-terminus with a ybbR tag and labeled the protein by SFP synthase with a CoA-Cy5.5 dye. A representative gel is shown, which was first scanned for Cy5.5 fluorescence (right) and subsequently stained with Coomassie blue (left) following SDS-PAGE analysis of ybbR-eIF5B-Trunc post-labeling with and without SFP synthase. The experiment was repeated three times with similar results. d, The GTPase activity of Cy5.5-eIF5B-Trunc was not perturbed by the labeling. Multiple turnover GTP hydrolysis was performed in 50 mM HEPES-KOH pH 7.5, 10 mM Mg(OAc)2, 100 mM KOAc at 30ºC 30min before quenching with malachite green assay solution. Where applicable, concentrations were: GTP 100 μM; eIF5B-Trunc 2.5 μM; Cy5.5-eIF5B-Trunc 2.5 μM; 40S+60S 0.2 μM each. The GTP only group was used as negative controls and the values were normalized to 0. Bars represent mean, and error bars indicate standard deviations of three biological replicates (individual data points are indicated with open circles). e, The dwell times (open circles) between 60S arrival and A-site Phe-TC arrival were fit to single-exponential distributions (n = 141, 159, 164, 118, 133, 130, 131, 189, 134 and 164 from bottom to top for each group) for experiments performed with Cy5.5-eIF5B (related to Fig. 2c) versus those with unlabeled eIF5B (related to Fig. 1d) and at 20ºC or 30ºC. Error bars (in black) represent the 95% confidence intervals of the average dwell times (Δt values). f, Despite it being reported that recombinant yeast eIF5B-FL purification cannot be achieved49, we were able to recombinantly express and purify it as shown by a 12% SDS-PAGE gel analysis. The experiment was repeated three times with similar results. g, Use of the full-length eIF5B in our assay did not lead to faster transition to elongation in experiments performed with the cap-RPL30 mRNA at 3 mM free Mg2+ 20ºC. Error bars (in black) represent the 95% confidence intervals of the average dwell times (Δt values, gray bars) from fitting of the dwell times (open circles) to single-exponential distributions. From left to right, n = 118 (related to Fig. 1d) and 205.
Extended Data Fig. 7 |. The use of a non-hydrolysable GTP analog, GDPNP, traps eIF5B on the 80S and prevents the transition to elongation.
a, Sample trace from experiments performed with GTP (n = 134). The grey highlighted part of the trace is shown in Fig. 2a as a zoomed-in view. b, Sample trace from experiments performed with non-hydrolysable GDPNP (n = 105), whereupon 60S joining eIF5B is trapped on the 80S and no A-site tRNA binding was observed. Black dash boxed Cy5.5 events are transient eIF5B sampling events to the 48S PIC prior to 60S joining. c, The cumulative probability distributions of the Cy5.5-eIF5B lifetimes on 80S in experiments performed with the model mRNA at 3 mM Mg2+ and 20ºC in the presence of GTP (n = 134, related to Fig. 2d) or GDPNP (n = 105). In the presence of GTP, the mean lifetime was 51.9 s (± 1.8 s, 95% confidence interval from fitting to a single-exponential equation); in the presence of GDPNP, the mean lifetime was 848 s (± 19 s, 95% confidence interval from fitting to a linear equation).
Extended Data Fig. 8 |. The effects of eIF5B mutations on the kinetics of 60S joining and the transition to elongation.
The cumulative probability distributions of the 60S joining (a) and the transition to elongation (b) dwell times from experiments performed with unlabeled wild-type (WT, n = 164), or T439A mutant (n = 131), or H505Y mutant (n = 119) eIF5B and the model mRNA at 20ºC in the presence of 3 mM Mg2+ and 1mM GTP. The estimated average fast and slow phase rates (kfast and kslow) and amplitudes (Afast and Aslow) of 60S joining (from a) and Δt values (from b) with the 95% confidence intervals were shown in (c), from fitting the distributions to a double-exponential (WT and H505Y) or a single-exponential (T439A) equation for the 60S joining; to a single-exponential equation for the transition dwell times. *Note: in case of T439A mutant eIF5B, out of n = 131 molecules showing 60S joining signal, only n = 17 molecules showed the subsequent A-site aa-tRNA association signal. Thus the T439A mutant is defective in catalyzing subunit joining (consistent with bulk measurement9), and inhibits the A-site tRNA association.
Extended Data Fig. 9 |. Free Mg2+ concentration modulates 60S joining and the transition to elongation.
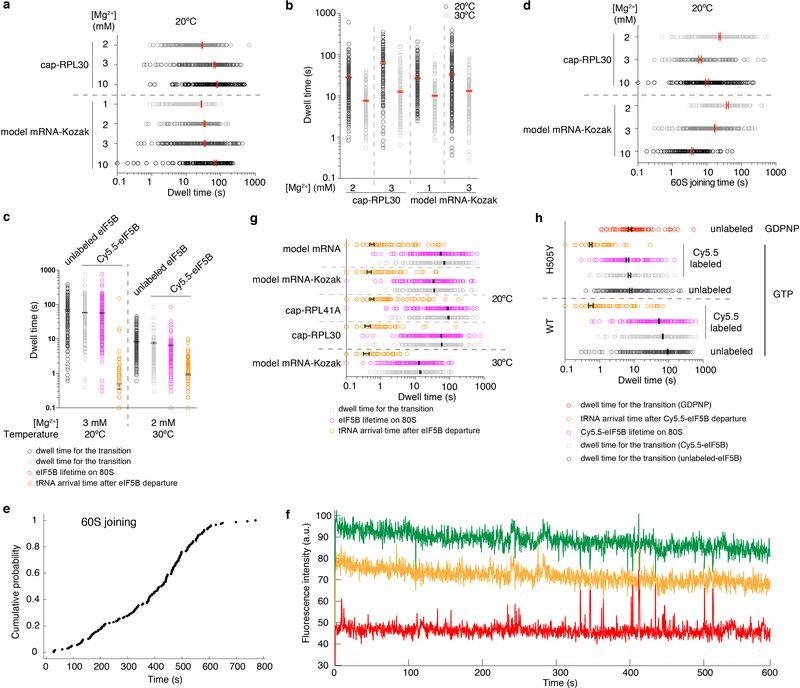
a, The dwell times for the transition from initiation to elongation (open circles) were fit to single-exponential distributions to estimate the average transition dwell times (Δt values, with 95% confidence intervals, in red) from experiments performed with unlabeled eIF5B and cap-RPL30 mRNA or model mRNA-Kozak at 20ºC in the presence of 1 to 10 mM Mg2+ (data for 3 mM Mg2+ were taken from Fig. 1d). For cap-RPL30, unstable 80S formation was observed at 1 mM Mg2+ (see (f) below) and thus no Δt values were obtained. From bottom to top for each group, n = 108, 189, 124, 150, 144, 118 and 195. b, The dwell times for the transition from initiation to elongation (open circles) were fit to single-exponential distributions to estimate the average transition dwell times (Δt values, with 95% confidence intervals, in red) from experiments performed with unlabeled eIF5B and cap-RPL30 or model mRNA-Kozak under denoted conditions. From left to right for each group, n = 195, 152, 118, 130, 150, 132, 189 and 159. c, The dwell times (open circles) for the transition from initiation to elongation, eIF5B lifetimes on 80S, and tRNA arrival times after eIF5B departure were fit to single-exponential distributions to estimate the average dwell times (with 95% confidence intervals, in black). Experiments were performed with cap-RPL30 mRNA at 3 mM free Mg2+ and 20ºC in the presence of unlabeled (n = 195, data taken from Fig. 1d) or Cy5.5-eIF5B (n = 164, data taken from Fig. 2c); or at 2 mM free Mg2+ and 30ºC in the presence of unlabeled (n = 152, data taken from b) or Cy5.5-eIF5B (n = 150). d, The 60S joining dwell times (open circles) from the same experiments as in (a) were fit to single-(for model mRNA-Kozak) or double- (for cap-RPL30, with the fast phase average times were plotted here) exponential distributions to estimate the average dwell time (with 95% confidence intervals, in red). e, The cumulative probability distribution of the 60S joining times showing compromised 60S joining rate in experiments performed with the model mRNA-Kozak at 1 mM Mg2+ and 20ºC as in (a) (n = 150). However, we still observed that A-site tRNA arrival occurred readily after 80S formation. The 60S joining kinetics under this reaction condition were not well fit by a single- nor double-exponential distributions and therefore no average time was deduced for the bar plot in (d). f, Sample trace from experiments performed with cap-RPL30 mRNA at 1 mM Mg2+ and 20ºC as described in (a), n = 200. g and h, Dot plots for Fig. 2c and 2e, showing the single-exponential distributions of the dwell times, with the average dwell times (with 95% confidence interval) in black. Notes: (1) Increasing free Mg2+ concentrations increased the Δt values (i.e. higher Mg2+ concentration favors the eIF5B-bound semi-rotated 80S conformation, Extended Data Fig. 10). This seems opposite to the known effects of free Mg2+ concentrations on the conformation of the bacterial pre-translocation 70S elongation complex50: lower Mg2+ concentration favors the rotated state and higher Mg2+ concentration favors the non-rotated state. The seeming discrepancy might be a result of the different compositions of the complexes: in our case, the semi-rotated state of the 80S contains an acylated Met-tRNAi in the P site and a protein factor eIF5B bound in the A site; while in the pre-translocation 70S complex, the P-site tRNA is deacylated and the A site is bound with a peptidyl-tRNA. The energy landscape differences between these two types of complexes can also be explained by no apparent fluctuations between semi-rotated and non-rotated 80S conformations with eIF5B bound vs. frequent fluctuations between rotated and non-rotated 70S conformations in the pre-translocation complexes. (2) The different magnitudes of temperature (Fig. 1d)/Mg2+ concentration dependence of Δt values for different mRNAs suggest that there are mRNA sequence context differences in the thermodynamics governing the transition from initiation to elongation. Similarly, the minimal Mg2+ concentrations required for stable 80S assembly on different mRNAs are different.
Extended Data Fig. 10 |. Cryo-EM analyses of the on-pathway 80S complexes during initiation and the transition to elongation.
a, Cumulative probability distributions of the dwell times for 60S joining to 48S PIC (i.e. the formation of 80S complex, black, designated as “total 80S”) and the subsequent A-site aa-tRNA association (corresponding to the Δt value, blue, designated as “80S EC”) were from the single-molecule experiment with the model mRNA and wild-type eIF5B at 20ºC and 3 mM free Mg2+ (n = 164). The red curve corresponds to the time-evolution of the fraction of the eIF5B-bound 80S complex (80S IC) simulated with the kinetics of the prior two curves. Dashed lines indicate that at time point 45 s, ~64.5% of the 80S complexes should be in the 80S IC state under this condition. b, The cryo-EM observed population sizes of the two classes of 80S complexes (80S IC, 17,602 particles, 70%; 80S EC, 7,542 particles, 30%) were in agreement with predictions from our single-molecule kinetics at time point 45 s. c-f show the compositional and conformational differences between the 80S IC and EC complexes. g. Summary of cryo-EM data collection and processing for the 48S PIC (Extended Data Fig. 2c–e), 80S IC (c) and 80S EC (d) maps.
Supplementary Material
Acknowledgements
We are grateful to J. Lorsch, T. Dever, J. Dinman, J. Yin and C. Aitken for sharing constructs, strains and protocols; C. Sitron and O. Brandman’s lab for generously sharing equipment and knowledge for use of their Freezer/Mill for yeast lysis; the Stanford PAN facility for protein mass spectrometry analyses; and members of Puglisi laboratory for discussion and input. This work was supported by the US National Institutes of Health (NIH) grants GM113078 and AI047365 to J.D.P.; a Knut and Alice Wallenberg Foundation postdoctoral scholarship (KAW 2015.0406) to J.W.; a National Science Foundation Graduate Research Fellowship (DGE-114747) to A.G.J.; a Damon Runyon Fellowship funded by the Damon Runyon Cancer Research Foundation (DRG-#2321–18) to C.P.L.; a Stanford Bio-X fellowship to J.C.; a Stanford Interdisciplinary Graduate Fellowship and NIH Molecular Biophysics Training Grant T32-GM008294 to A.P..
Footnotes
The authors declare no competing interests. Readers are welcome to comment on the online version of the paper.
Supplementary Information is available in the online version of the paper.
Data Availability Source data for figures are available as accompanying Source Data files. The cryo-EM maps have been deposited in the Electron Microscopy Data Band (EMDB) server under the accession codes EMD-20324 (48S PIC), EMD-20325 (80S IC) and EMD-20326 (80S EC). Single-molecule fluorescence trace data are available upon request.
Code Availability The MATLAB scripts used in this study are available at: https://drive.google.com/drive/u/1/folders/1-yKC0Nui7T7yoJScCuODPhCT4G52emLj
References
- 1.Hinnebusch AG The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem 83, 779–812 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Sokabe M & Fraser CS Toward a kinetic understanding of eukaryotic translation. Cold Spring Harb. Perspect. Biol a032706 (2018). doi: 10.1101/cshperspect.a032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaledhonkar S et al. Real-time structural dynamics of late steps in bacterial translation initiation visualized using time-resolved cryogenic electron microscopy. Nature 570, 400–404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai A et al. Heterogeneous pathways and timing of factor departure during translation initiation. Nature 487, 390–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyal A, Belardinelli R, Maracci C, Milón P & Rodnina MV Directional transition from initiation to elongation in bacterial translation. Nucleic Acids Res. 43, 10700–10712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pestova TV et al. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403, 332–335 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Lee JH et al. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc. Natl. Acad. Sci. U. S. A 99, 16689–16694 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin BS et al. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell 111, 1015–1025 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Acker MG et al. Kinetic analysis of late steps of eukaryotic translation initiation. J. Mol. Biol 385, 491–506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roll-Mecak A, Cao C, Dever TE & Burley SK X-ray structures of the universal translation initiation factor IF2/eIF5B: Conformational changes on GDP and GTP binding. Cell 103, 781–792 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Kuhle B & Ficner R eIF5B employs a novel domain release mechanism to catalyze ribosomal subunit joining. EMBO J. 33, 1177–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernández IS et al. Molecular architecture of a eukaryotic translational initiation complex. Science 342, 1240585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acker MG, Kolitz SE, Mitchell SF, Nanda JS & Lorsch JR Reconstitution of yeast translation initiation. Methods Enzymol. 430, 111–145 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Chen J et al. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc. Natl. Acad. Sci 111, 664–669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez E et al. eIF5A promotes translation of polyproline motifs. Mol. Cell 51, 35–45 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llácer JL et al. Conformational differences between open and closed states of the eukaryotic translation Initiation complex. Mol. Cell 59, 399–412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton R, Watanabe CK & de Boer HA Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 15, 3581–3593 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li JJ, Chew GL & Biggin MD Quantitating translational control: mRNA abundance-dependent and independent contributions and the mRNA sequences that specify them. Nucleic Acids Res. 45, 11821–11836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh RM & Martin PA Growth of Saccharomyces cerevisiae and Saccharomyces uvarum in a temperature gradient incubator. J. Inst. Brew 83, 169–172 (1977). [Google Scholar]
- 20.Lorsch JR & Herschlag D Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. EMBO J. 18, 6705–6717 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Acker MG, Shin BS, Dever TE & Lorsch JR Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J. Biol. Chem 281, 8469–8475 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Pisareva VP & Pisarev AV EIF5 and eIF5B together stimulate 48S initiation complex formation during ribosomal scanning. Nucleic Acids Res. 42, 12052–12069 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah P, Ding Y, Niemczyk M, Kudla G & Plotkin JB Rate-limiting steps in yeast protein translation. Cell 153, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu D et al. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 33, 21–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuller AP, Wu CCC, Dever TE, Buskirk AR & Green R eIF5A functions globally in translation elongation and termination. Mol. Cell 66, 194–205.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingolia NT, Ghaemmaghami S, Newman JRS & Weissman JS Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heyer EE & Moore MJ Redefining the Translational Status of 80S Monosomes. Cell 164, 757–769 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Ingolia NT et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8, 1365–1379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juszkiewicz S et al. ZNF598 Is a Quality Control Sensor of Collided Ribosomes. Mol. Cell 72, 469–481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeuchi K et al. Collided ribosomes form a unique structural interface to induce Hel2‐driven quality control pathways. EMBO J e100276 (2019). doi: 10.15252/embj.2018100276 [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 31.Yin J, Lin AJ, Golan DE & Walsh CT Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc 1, 280–285 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Petrov A, Grosely R, Chen J, O’Leary SE & Puglisi JD Multiple parallel pathways of translation initiation on the CrPV IRES. Mol. Cell 62, 92–103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowen AM et al. Ribosomal protein uS19 mutants reveal its role in coordinating ribosome structure and function. Translation 3, e1117703 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito H, Fukuda Y, Murata K & Kimura A Transformation of intact yeast cells treated with alkali cations. J. Bacteriol 153, 163–168 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maniatis T, Fritsch EF & Sambrook J Molecular cloning: a laboratory manual. (Cold Spring Harbor Laboratory Press, 1982). [Google Scholar]
- 36.O’Leary SE, Petrov A, Chen J & Puglisi JD Dynamic recognition of the mRNA cap by Saccharomyces cerevisiae eIF4E. Structure 21, 2197–2207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell SF et al. The 5’−7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol. Cell 39, 950–962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Algire MA et al. Development and characterization of a reconstituted yeast translation initiation system. RNA 8, 382–397 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goyer C et al. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell. Biol 13, 4860–4874 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi SK et al. Physical and Functional Interaction between the Eukaryotic Orthologs of Prokaryotic Translation Initiation Factors IF1 and IF2. Mol. Cell. Biol 20, 7183–7191 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rinaldi AJ, Suddala KC & Walter NG Native purification and labeling of RNA for single molecule fluorescence studies. Methods Mol. Biol 1240, 63–95 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall RA, Dorywalska M & Puglisi JD Irreversible chemical steps control intersubunit dynamics during translation. Proc. Natl. Acad. Sci 105, 15364–15369 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanchard SC, Kim HD, Gonzalez RL, Puglisi JD & Chu S tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci 101, 12893–12898 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang G et al. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Scheres SHW RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin BS et al. Structural integrity of α-helix H12 in translation initiation factor eIF5B is critical for 80S complex stability. RNA 17, 687–696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Munro JB, Sanbonmatsu KY, Spahn CMT & Blanchard SC Navigating the ribosome’s metastable energy landscape. Trends Biochem. Sci 34, 390–400 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.