Intracerebral haemorrhage (ICH) and small vessel ischaemic stroke (SVS) are the most severe manifestations of cerebral small vessel disease. In a cross-phenotype genome-wide association analysis, Chung et al. identify two novel associations at 2q33 and 13q34 plus a previously identified locus at 1q22 for non-lobar ICH and SVS risk.
Keywords: genome-wide association studies, cerebral small vessel disease, multi-trait analysis
Abstract
Intracerebral haemorrhage and small vessel ischaemic stroke (SVS) are the most acute manifestations of cerebral small vessel disease, with no established preventive approaches beyond hypertension management. Combined genome-wide association study (GWAS) of these two correlated diseases may improve statistical power to detect novel genetic factors for cerebral small vessel disease, elucidating underlying disease mechanisms that may form the basis for future treatments. Because intracerebral haemorrhage location is an adequate surrogate for distinct histopathological variants of cerebral small vessel disease (lobar for cerebral amyloid angiopathy and non-lobar for arteriolosclerosis), we performed GWAS of intracerebral haemorrhage by location in 1813 subjects (755 lobar and 1005 non-lobar) and 1711 stroke-free control subjects. Intracerebral haemorrhage GWAS results by location were meta-analysed with GWAS results for SVS from MEGASTROKE, using ‘Multi-Trait Analysis of GWAS’ (MTAG) to integrate summary data across traits and generate combined effect estimates. After combining intracerebral haemorrhage and SVS datasets, our sample size included 241 024 participants (6255 intracerebral haemorrhage or SVS cases and 233 058 control subjects). Genome-wide significant associations were observed for non-lobar intracerebral haemorrhage enhanced by SVS with rs2758605 [MTAG P-value (P) = 2.6 × 10−8] at 1q22; rs72932727 (P = 1.7 × 10−8) at 2q33; and rs9515201 (P = 5.3 × 10−10) at 13q34. In the GTEx gene expression library, rs2758605 (1q22), rs72932727 (2q33) and rs9515201 (13q34) are significant cis-eQTLs for PMF1 (P = 1 × 10−4 in tibial nerve), NBEAL1, FAM117B and CARF (P < 2.1 × 10−7 in arteries) and COL4A2 and COL4A1 (P < 0.01 in brain putamen), respectively. Leveraging S-PrediXcan for gene-based association testing with the predicted expression models in tissues related with nerve, artery, and non-lobar brain, we found that experiment-wide significant (P < 8.5 × 10−7) associations at three genes at 2q33 including NBEAL1, FAM117B and WDR12 and genome-wide significant associations at two genes including ICA1L at 2q33 and ZCCHC14 at 16q24. Brain cell-type specific expression profiling libraries reveal that SEMA4A, SLC25A44 and PMF1 at 1q22 and COL4A1 and COL4A2 at 13q34 were mainly expressed in endothelial cells, while the genes at 2q33 (FAM117B, CARF and NBEAL1) were expressed in various cell types including astrocytes, oligodendrocytes and neurons. Our cross-phenotype genetic study of intracerebral haemorrhage and SVS demonstrates novel genome-wide associations for non-lobar intracerebral haemorrhage at 2q33 and 13q34. Our replication of the 1q22 locus previous seen in traditional GWAS of intracerebral haemorrhage, as well as the rediscovery of 13q34, which had previously been reported in candidate gene studies with other cerebral small vessel disease-related traits strengthens the credibility of applying this novel genome-wide approach across intracerebral haemorrhage and SVS.
Introduction
Cerebral small vessel disease (CSVD) is a term used to categorize a variety of pathological and neurological processes that affect the small arteries, arterioles, venules and capillaries of the brain (Pantoni, 2010). The main phenotypes of CSVD include small vessel ischaemic stroke (SVS) and intracerebral haemorrhage (ICH), as well as radiographically-apparent lesions in subcortical regions of the brain. Collectively, the chronic effects of CSVD are major contributors to vascular cognitive impairment, late-life gait disorders and depression (Sacco et al., 2006; Pantoni, 2010). ICH and SVS are the most acute manifestations of CSVD. ICH and SVS may result from acute rupture or occlusion of small perforating arteries and arterioles in white or deep grey matter of the brain (Qureshi et al., 2001; Wardlaw et al., 2013; Ter Telgte et al., 2018). Approximately 238 500 individuals experience ICH or SVS each year worldwide, with about three in four being first-time strokes (Benjamin et al., 2018). Because ICH and SVS incidence rates increase with age, the overall burden of these conditions is expected to grow as the world’s population is ageing. Other than management of hypertension, we lack effective treatments to reduce the risk of CSVD. Pathways involved in ICH and SVS pathogenesis must be elucidated to develop new effective prevention and treatment strategies.
Despite their substantial heritability, estimated to be at least 29.0% for ICH and 16.1% for SVS (Bevan et al., 2012; Devan et al., 2013), relatively few genetic variants have been associated with increased risk for ICH and SVS. One possible explanation for this could be a highly polygenic architecture for these traits. The ability to boost the statistical power of genome-wide association study (GWAS) by increasing the available sample size could uncover novel genetic loci for CSVD, as has been the case for other complex diseases or phenotypes including schizophrenia (Andreassen et al., 2015) and hypertension (Andreassen et al., 2014).
The most recent and largest to-date GWAS of SVS substantially boosted sample sizes for SVS (SVS cases n = 4453, controls n = 233 058) allowing identification of two novel associations (Malik et al., 2018). In comparison, ICH has a limited sample size for genetic studies (current lobar ICH n = 755, non-lobar ICH n = 1005) in part because of its overall lower incidence and higher mortality (Benjamin et al., 2018) despite strong collaborations across the International Stroke Genetics Consortium (ISGC; www.strokegenetics.org). Furthermore, the mechanisms underlying ICH differ based on the haemorrhage location within the brain (lobar versus non-lobar), which has been corroborated by previous studies (Biffi et al., 2010; Falcone et al., 2012, 2013; Martini et al., 2012), in addition to neuropathological series. Lobar ICH, occurring in either the cerebral cortex or cortical-subcortical junction, shows a strong association with cerebral amyloid angiopathy but weak association with hypertension (Vinters, 1987). In contrast, hypertensive arteriopathy is most commonly associated with non-lobar ICH (Fisher, 1971). This heterogeneity in biological mechanisms underlying ICH location necessitates ICH subgroup analysis by the haemorrhagic location in the brain, which also limits the available sample size for genetic studies. While there are plans for genotyping of additional ICH samples for future larger GWAS, a potentially useful near-term solution is to leverage larger sample sizes through cross-phenotype (i.e. pleiotropy) analyses of GWAS, integrating genetic association signals of biologically related traits for novel discovery. These cross-phenotype analyses estimate the common polygenic structure shared by two different diseases (i.e. genetic overlap), and the genetic associations of variants from the two diseases are combined and weighted according to the degree of overlap. Such cross-phenotype analyses increase the statistical power for a main phenotype of interest by utilizing another proxy phenotype obtained in large studies (Lam et al., 2017; Hill et al., 2018). Furthermore, recently developed methods for cross-phenotype analysis, which do not require the two traits to arise from the same dataset, are robust to potential environmental confounds by applying corrections for human genome structure (e.g. minor allele frequency and recombination rate) (Gratten and Visscher, 2016; Evans et al., 2018).
In the present study, we aimed to identify novel genetic risk associations for ICH using cross-phenotype analyses with GWAS summary statistics of SVS. While analyses were performed for non-lobar ICH, lobar ICH, and all ICH, our expectation was that non-lobar ICH was most likely to demonstrate pleiotropic associations with SVS because both are manifestations of hypertensive CSVD.
Materials and methods
Subjects, genotyping and data processing for intracerebral haemorrhage study
Genotype and phenotype data for 1543 well-characterized ICH cases and 1711 stroke-free control subjects were drawn from three ICH GWAS datasets: the North American (USA) multi-centre Genetics of Cerebral Haemorrhage on Anticoagulation (GOCHA) study, the European member sites contributing to the International Stroke Genetics Consortium (ISGC-EUR), and Genetic and Environmental Risk Factors for Haemorrhagic Stroke (GERFHS) I, II and III. Details of subject recruitment, genotyping, quality control and population structure for the included datasets are described in the Supplementary material and have also been reported previously (Woo et al., 2014; Anderson et al., 2016). Demographic information for the ICH GWAS subjects is presented in Table 1.
Table 1.
Sample demography of GWAS for ICH by location and SVS
| ICH GWAS | SVS GWAS | |||||||
|---|---|---|---|---|---|---|---|---|
| GOCHA | ISGC-EUR | GERFHS I-III | MEGASTROKE | |||||
| Cases | Controls | Cases | Controls | Cases | Controls | SVS | Controls | |
| Subjects, n | 386 | 387 | 574 | 530 | 848 | 794 | 4453 | 233 058 |
| Female, n (%) | 178 (46.1) | 179 (46.3) | 258 (45.0) | 258 (48.7) | 420 (49.5) | 420 (52.9) | 45.50 | 49.90 |
| Lobar ICH, n (%) | 210 (54.3) | - | 205 (35.7) | - | 340 (40.1) | - | - | - |
| Non-lobar ICH, n (%) | 173 (44.7) | - | 334 (58.2) | - | 508 (59.9) | - | - | - |
| Age, mean (SD) | 73.8 (10.2) | 72.4 (7.89) | 71.1 (12.2) | 66 (15.8) | 69.2 (14.1) | 68.4 (13.3) | 65.6 (12.4) | 61.0 (9.8) |
GERFHS = Genetic and Environmental Risk Factors for Haemorrhagic Stroke; GOCHA = Genetics of Cerebral Haemorrhage on Anticoagulation; ISGC-EUR = European member sites contributing to the International Stroke Genetics Consortium; SD = standard deviation.
Imputation of intracerebral haemorrhage GWAS datasets
All ICH samples were imputed separately in the three datasets to the Haplotype Reference Consortium (HRC) haplotype reference panel using post-QC genotypes from the participating ICH GWAS studies. Imputation was performed on the Michigan Imputation Server (https://imputationserver.sph.umich.edu/) running MiniMac3.8 (McCarthy et al., 2016). Across all samples, allele dosages of about 39 235 000 single nucleotide polymorphisms (SNPs) were imputed, with the actual number of SNPs imputed for each individual varying based on the density of genotype arrays available. Imputed SNPs with the minor allele frequency ≥1% and an imputation quality estimate (R2) ≥ 0.40, as done in prior analyses (Lambert et al., 2013; Jun et al., 2016; Chung et al., 2018), were included in the final single nucleotide polymorphism (SNP) set for association analyses.
Samples for small vessel ischaemic stroke study
SVS GWAS summary statistics for European-ancestry individuals were derived from MEGASTROKE (Malik et al., 2018) (n = 237 511) through www.cerebrovascularportal.org. Demographic information for the SVS sample is presented in Table 1.
Statistical analysis
Meta-analysis for intracerebral haemorrhage
Within each dataset, primary genome-wide association analyses were performed for all ICH, lobar ICH, and non-lobar ICH using logistic regression including covariates of age, sex and the first four principal components of population structure using the R software environment with the ‘glm’ function. All autosomal SNPs were analysed as predictors using quantitative measures (i.e. continuous dosages) between zero and two of the number of effect alleles, which were estimated from the HRC imputation. The genome-wide association results for ICH from the three included ICH GWAS studies were combined by meta-analysis using inverse variance weighting as implemented in the METAL software (Willer et al., 2010).
Multi-trait analysis of GWAS
Multi-trait analysis of GWAS (MTAG) requires GWAS summary statistics for traits, rather than individual-level data (Turley et al., 2018). MTAG estimates the degree of statistical boost (i.e. newly added sample size) for one trait statistically gained from the other trait, providing summary-level statistics as MTAG results for each trait. There was a known small overlap in control subjects between the ICH and SVS genetic studies. However, MTAG has been demonstrated to be effective in accounting for (even unknown) sample overlap between the GWAS summary data of the traits by conducting a bivariate linkage disequilibrium score regression of the traits. Further information about the MTAG methodology and procedure is described in the Supplementary material.
Because our main goal was to increase statistical power for identifying genetic loci associated with ICH by adding the genetic variance shared with SVS, we applied MTAG directionally to return results for ICH boosted by the large-scale GWAS of SVS.
We tested whether the results from MTAG were consistent with other pleiotropy association methods. We carried out a Bayesian pleiotropy association test as used in the GWAS-PW software tool (Pickrell et al., 2016) to detect genomic regions that influence both diseases. GWAS-PW combines GWAS summary data of two correlated traits and calculated four posterior probabilities of association (PPA) of a genomic region under four models: (i) a region specifically associated with ICH only; (ii) a region specifically associated with SVS; (iii) a region associated with both traits; and (iv) a region with separate alleles independently associated with each trait (Supplementary material). Because GWAS-PW can differentiate the genomic regions that are associated exclusively with one of the traits, not both, we used GWAS-PW to confirm that our findings from MTAG were not solely driven by one of the traits. Genomic regions with PPA ≥ 0.9 of model 3 (PPA3) were considered to significantly influence both traits (i.e. pleiotropy effect), while regions with PPA ≥ 0.6 were considered suggestive.
Gene-based association analysis using S-PrediXcan
Given the results of our SNP-based MTAG analysis, we applied genome-wide gene-based association testing using S-PrediXcan (Barbeira et al., 2018) for the SNP-level results from the MTAG analysis of non-lobar ICH. S-PrediXcan integrates reference transcriptome data [e.g. GTEx eQTL (expression quantitative trait locus) data] with genetic associations for a phenotype. S-PrediXcan first estimates gene expressions by using tissue-dependent prediction models trained in the reference data, and then correlates the estimated gene expressions with genetic associations to identify the genes involved with the phenotype. We used 11 tissue-specific expression models (PredictDB; http://predictdb.org), chosen for their representation of nervous system, blood vessel, and non-lobar brain tissues from the GTEx project v7p: one tibial nerve tissue, three artery relevant tissues in aorta, coronary, and tibia and seven non-lobar related brain regions including cerebellum, hypothalamus, substantia nigra, amygdala, caudate, nucleus accumbens and putamen. Genome-wide significance was set as α = 0.05 with Bonferroni correction for the number of genes tested within the tissue. Experiment-wide significance was set conservatively with Bonferroni correction for the total number of tests performed in the 11 tissues (P < 8.5 × 10−7).
Functional and eQTL analysis
We evaluated potential regulatory functions of genome-wide significant results (MTAG P-value < 5.0 × 10−8) using publicly available databases including HaploReg (Ward and Kellis, 2012) and ENCODE (Consortium, 2012) via UCSC Genome Browser (Kent et al., 2002). For the exonic SNPs among the genome-wide significant SNPs, we determined the likelihood that a non-synonymous amino acid substitution has a deleterious effect on protein function using SIFT (Sim et al., 2012). To identify shared functions by top-ranked genes from our MTAG analysis, pathway analysis based on gene ontology (GO) categories was also conducted using the competitive gene-set analysis of MAGMA, which is used in the FUMA pipeline (de Leeuw et al., 2015; Watanabe et al., 2017). A total of 5935 GO categories were tested. A nominal P-value threshold was set to 8.42 × 10−6 after multiple testing correction (0.05/5938). Default parameter values were applied for filtering and clumping SNPs for the use of FUMA: r2 threshold to define linkage disequilibrium (LD) block of independent SNPs > 0.6; the maximum distance of LD blocks to merge into a locus: 250 kb; P-value cut-off < 0.05; minor allele frequency ≥ 0.01.
We examined the association between genome-wide significant SNPs (allele counts) and transcription-level expression (i.e. eQTL) using the GTEx Portal (Consortium, 2015) and the Brain eQTL Almanac (Braineac) (Trabzuni et al., 2011). In the GTEx Portal database, we selected the same 11 tissues used for S-PrediXcan. In Braineac, we focused on three non-lobar relevant brain regions including putamen, substantia nigra and thalamus. GTEx Portal and Braineac have been used for eQTL mapping in multiple prior studies (Loh et al., 2018; Maguire et al., 2018; Malik et al., 2018; Zhou et al., 2018). GTEx includes a diverse tissue library of eQTLs (53 types of tissues from at least 80 participants each), and Braineac is an independent dataset that includes gene expression data from 10 regions of the human brain in 134 participants.
We determined a conservative significance level of 1.6 × 10−4, which was calculated as the nominal level (α = 0.05) divided by the total number of eQTL testing (n = 322). The total number of testing was counted by the number of genes to test at each locus (eight genes at 1q22; 11 genes at 2q33; four genes at 13q34) with the number of tissues for expression data (11 tissues from the GTEx Portal and three tissues from the Braineac).
Cell type-specific expression analysis in human brain
We investigated cell type-specific expression for the candidate genes located within or near the top-ranked associations using two independent single cell RNA sequencing (scRNA-Seq) datasets from (i) the anterior temporal lobe of adult human brains; and (ii) cerebrovascular cells of adult mouse brains.
The human brain scRNA-Seq data classified each single brain cell into the six major cell types of astrocytes, oligodendrocytes, oligodendrocyte precursor cells (OPCs), neurons, microglia and endothelial cells (Darmanis et al., 2015). Foetal quiescent or hybrid cells were not considered for this study. Mapping results of this human brain scRNA-Seq were downloaded from Gene Expression Omnibus (GEO; Accession number: GSE67835) and the mapped read counts were library normalized into count per million (CPM). Expression levels of the candidates in each cell type were summarized as the log2 of the mean expression plus one.
The scRNA-Seq data in mouse brains catalogued differential gene expressions in various cell types including endothelial cell, smooth muscle and pericytes in whole brain vascular tissue. This scRNA-Seq data were reported by Vanlandewijck and colleagues (2018) and were obtained from http://betsholtzlab.org/VascularSingleCells/database.html.
Data availability
The data that support the findings of this study are openly available in Cerebrovascular Disease Knowledge Portal (CDKP) at http://www.cerebrovascularportal.org (Crawford et al., 2018).
Results
Intracerebral haemorrhage GWAS
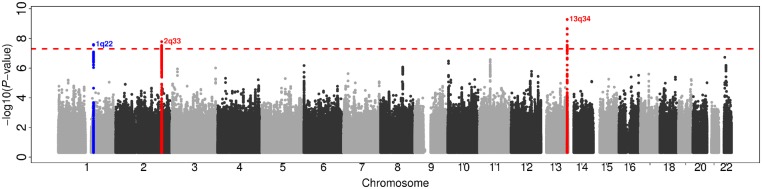
Genome-wide association (i.e. Manhattan) and QQ plots of the meta-analyses for all, lobar and non-lobar ICH are provided in Supplementary Figs 1 and 2. There was little evidence for genomic inflation in genome-wide association results for all ICH [genomic control (λ) = 1.04], for lobar ICH (λ = 1.01), and for non-lobar ICH (λ = 1.01). Genome-wide significant association was found in the APOE region for the lobar ICH [best SNP: rs5117; odds ratio (OR) = 1.65 and P-value = 6.2 × 10−10]. The SNP, rs5117, is in relatively high LD (R2 = 0.58 and D’ = 0.94) with one of the allele-defining SNPs for APOE ɛ4 (rs429358). No other genome-wide significant association was observed for all ICH and non-lobar ICH, but SNPs at 13q34 reached suggestively significant (P < 10−7) association for non-lobar ICH (best SNP: rs9515200; meta-analysis P-value = 2.7 × 10−7; Supplementary Table 1A). Differences between these results and a previously published analysis (notably at 1q22) (Woo et al., 2014) are attributable to differences in sample size as well as imputation to the HRC reference panel in the present study, as opposed to the 1000 Genomes reference panel, which was used for imputation in the previous publication.
Multi-trait analysis of GWAS for intracerebral haemorrhage and small vessel ischaemic stroke
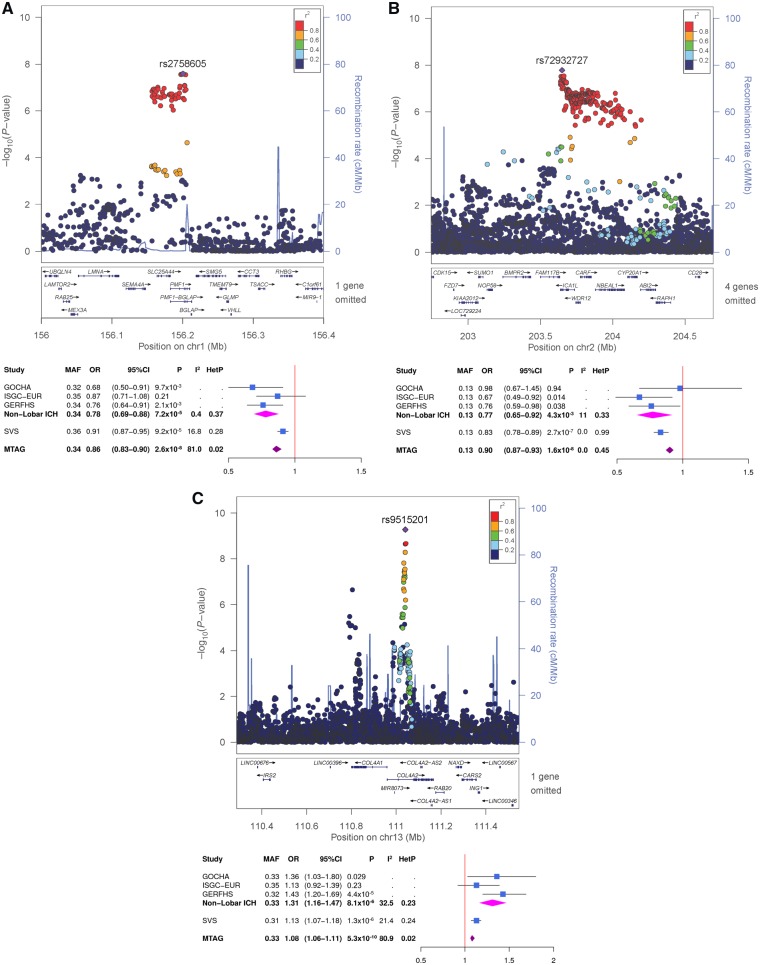
There was limited genomic inflation in the QQ plot of the MTAG results for non-lobar ICH (Supplementary Fig. 3) although its genomic control value remained close to one (λ = 1.02). We did not observe genomic inflation for lobar (λ = 1.02) and all ICH (λ = 0.97) (Supplementary Fig. 3 and 4). No genome-wide significant associations were found in the MTAG analyses for all ICH or lobar ICH with SVS and so neither of these phenotypes were carried into the later stages of analysis. However, the MTAG analysis of non-lobar ICH and SVS revealed genome-wide significant associations with SNPs at the previously identified 1q22 locus, as well as two novel loci at 2q33 and 13q34 (Table 2 and Figs 1 and 2). At 1q22 locus, genome-wide significant association was attained with five SNPs between SLC25A44 and PMF1 (best SNP: rs2758605, MTAG P-value = 2.6 × 10−8). Within 2q33, there were genome-wide significant associations with 10 SNPs within ICA1L (best SNP: rs72932727, MTAG P-value = 1.7 × 10−8). At 13q34, nine SNPs within COL4A2 (best SNP: rs9515201, MTAG P-value = 5.3 × 10−10) surpassed genome-wide significance. Suggestive associations (MTAG P-value < 1.0 × 10−5) are summarized in Supplementary Table 1A.
Table 2.
Genome-wide significant associations with non-lobar ICH and/or SVS in the MTAG analysis
| Non-lobar | SVS | MTAG (non-lobar ICH) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CH | SNP | GENE | EA | NEA | EAF | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P |
| 1 | rs2758605 | PMF1 | C | G | 0.34 | 0.78 (0.69–0.88) | 7.15 x 10−5 | 0.91 (0.88–0.95) | 9.15 x 10−5 | 0.86 (0.83–0.90) | 2.57 x 10−8 |
| 2 | rs72932727 | ICA1L | C | G | 0.13 | 0.77 (0.65–0.92) | 4.32 x 10−3 | 0.84 (0.77–0.90) | 2.66 x 10−7 | 0.90 (0.87–0.94) | 1.65 x 10−8 |
| 13 | rs9515201 | COL4A2 | A | C | 0.33 | 1.31 (1.16–1.47) | 8.13 x 10−6 | 1.13 (1.08–1.17) | 1.34 x 10−6 | 1.08 (1.06–1.10) | 5.27 x 10−10 |
CI = confidence interval; EA = effect allele; EAF = effect allele frequency; NEA = non-effect allele; OR = odds ratio.
Figure 1.
Manhattan plot of the MTAG result for non-lobar ICH GWAS, enhanced by SVS of MEGASTROKE. Red dashed horizontal line denotes genome-wide significance (P = 5.0 × 10−8). Blue and red dots indicate a previously identified ICH locus and novel findings, respectively.
Figure 2.
Regional association plots with forest plots for the top-ranked associations. (A) 1q22, (B) 2q33 and (C) 13q34 from the MTAG analysis for non-lobar ICH, enhanced by SVS.
We observed consistent association patterns from GWAS-PW model 3 (the pleiotropy model) with our MTAG results. All three loci detected by MTAG (1q22, 2q33 and 13q34) attained PPA3 > 0.6 in the GWAS-PW model 3 using non-lobar ICH and SVS (Supplementary Fig. 5): 1q22 PPA3 was 0.67, 2q33 PPA3 was 0.83, and 13q34 PPA3 was 0.97. No other genomic regions were observed with PPA3 > 0.4 for any of the tested trait pairs (Supplementary Fig. 6).
Gene-based associations using multi-trait analysis results of non-lobar intracerebral haemorrhage
Tissue-specific gene-level associations were generated by S-PrediXcan using the MTAG results of non-lobar ICH. Table 3 lists the top-ranked genes with suggestive significance (P < 10−4). Three genes at 2q33 met experiment-wide significance in various types of tissues: WDR12 in brain amygdala, FAM117B in aorta, and NBEAL1 in coronary artery, tibial artery, and tibial nerve. Two genes attained genome-wide significance: ICA1L at 2q33 in tibial nerve and ZCCHC14 at 16q24 in tibial artery. No genes at 1q22 reached genome-wide significance, but we observed suggestive association at PMF1 in tibial nerve tissue. COL4A1 and COL4A2 could not be tested by S-PrediXcan because tissue-specific expression models for these genes were not available in the PredictDB database.
Table 3.
Top-ranked genes with P-value < 10−4 in S-PrediXcan
| Tissue | Genes, n | GWS | Band | Gene | Effect | P-value | Var | R2 | SNPs available, n | SNPs for model, n |
|---|---|---|---|---|---|---|---|---|---|---|
| Nerve – tibial | 9378 | 5.3 x 10−6 | 1q22 | PMF1 | 0.41 | 6.8 x 10−5 | 0.02 | 0.06 | 12 | 13 |
| 2q33 | ICA1L | −0.11 | 1.3 x 10−6 | 0.13 | 0.14 | 37 | 37 | |||
| 2q33 | NBEAL1 | 0.15 | 5.9 x 10−7 | 0.08 | 0.13 | 22 | 23 | |||
| 11p11 | CELF1 | 0.20 | 1.8 x 10−5 | 0.03 | 0.06 | 19 | 19 | |||
| Artery – aorta | 6595 | 7.6 x 10−6 | 2q33 | FAM117B | 0.38 | 5.9 x 10−7 | 0.01 | 0.07 | 10 | 10 |
| Artery – coronary | 3468 | 1.4 x 10−5 | 2q33 | NBEAL1 | 0.12 | 7.6 x 10−7 | 0.11 | 0.17 | 17 | 17 |
| Artery – tibial | 8178 | 6.1 x 10−6 | 2q33 | ICA1L | −0.26 | 9.8 x 10−6 | 0.02 | 0.05 | 16 | 16 |
| 2q33 | NBEAL1 | 0.08 | 1.7 x 10−7 | 0.31 | 0.39 | 17 | 17 | |||
| 16q24 | ZCCHC14 | −0.40 | 2.1 x 10−6 | 0.01 | 0.06 | 2 | 2 | |||
| Brain – amygdala | 2342 | 2.1 x 10−5 | 2q33 | WDR12 | 0.31 | 2.3 x 10−7 | 0.02 | 0.08 | 14 | 14 |
| Brain – cerebellar hemisphere | 4723 | 1.1 x 10−5 | 11p11 | FAM180B | 0.33 | 4.2 x 10−5 | 0.01 | 0.05 | 13 | 13 |
| Brain – spinal cord | 2500 | 2.0 x 10−5 | 17q21 | KAT7 | 0.14 | 3.2 x 10−5 | 0.06 | 0.05 | 22 | 22 |
GWS = genome-wide significance for the tissue; Var = variance of the gene expression.
Experiment-wide significance = 8.5 x 10−7. Results that remained experiment-wide significant after multiple test correction are highlighted in bold.
Functional annotation for 1q22, 2q33 and 13q34 and pathway analysis
Functional annotations from HaploReg and ENCODE for 350 genome-wide or suggestively significant SNPs at the three genome-wide significant loci are summarized in Supplementary Table 1B. There were three missense SNPs in exon 2 of PMF1 (rs1052053; MTAG P-value = 3.3 × 10−10; p.Q75P), exon 3 of WDR12 (rs35212307; MTAG P-value = 6.3 × 10−7; p.I75V), and exon 16 of CARF (rs72932557; MTAG P-value = 4.7 × 10−7; p.Y571F). According to SIFT, these missense SNPs were predicted to be neutral to their secondary protein structures. Of the remaining non-coding SNPs (302 intronic and 45 intergenic), 50 are located in promoter histone marks, 152 in enhancer histone marks, 68 in DNase I marks, and 297 altered the binding sites of regulatory proteins in various types of tissues including blood and brain. We conducted competitive gene set tests for the pathway analysis, but none of the sets (i.e. GO) attained the significance threshold after multiple testing correction (Supplementary Table 2).
eQTL association results for 1q22, 2q33 and 13q34
We examined whether expression levels of the genes near the top-ranked genome-wide significant SNPs for each locus detected from the MTAG analysis of non-lobar ICH with SVS were associated with risk allele counts by SNP (Table 4). We tested five genes at 1q22 (SEMA4A, SLC25A44, PMF1, BGLAP and SMG5), five genes at 2q33 (FAM117B, ICA1L, WDR12, CARF and NBEAL1), and two genes at 13q34 (COL4A1 and COL4A2).
Table 4.
eQTL association summary table
| eQTL (effect allele) | Gene | Database | P-value | Effect dir | Tissue |
|---|---|---|---|---|---|
| rs2758605-C | SEMA4A | GTEx | 1.8 × 10-5 | + | Artery - tibial |
| GTEx | 4.3 × 10-3 | + | Brain - cerebellar hemisphere | ||
| GTEx | 6.7 × 10-3 | + | Artery - coronary | ||
| GTEx | 1.5 × 10-3 | + | Nerve - tibial | ||
| GTEx | 0.01 | + | Brain - cerebellum | ||
| GTEx | 0.03 | + | Brain - caudate | ||
| SLC25A44 | GTEx | 3.9 × 10-4 | − | Nerve - tibial | |
| GTEx | 1.9 × 10-4 | − | Artery - aorta | ||
| GTEx | 2.8 × 10-3 | − | Artery - tibial | ||
| PMF1 | GTEx | 1.1 × 10-4 | − | Nerve - tibial | |
| GTEx | 0.02 | − | Brain - substantia nigra | ||
| Braineac | 2.3 × 10−4 | + | Brain - thalamus | ||
| Braineac | 0.02 | + | Brain - putamen | ||
| SMG5 | GTEx | 1.1 × 10−4 | − | Nerve - tibial | |
| GTEx | 0.02 | − | Brain - substantia nigra | ||
| LMA | GTEx | 0.01 | − | Brain - caudate | |
| GTEx | 0.02 | − | Brain - putamen (basal ganglia) | ||
| rs72932727-C | CARF | GTEx | 1.1 × 10−10 | − | Artery – tibial |
| GTEx | 0.01 | − | Artery - aorta | ||
| FAM117B | GTEx | 2.1 × 10−7 | − | Artery - aorta | |
| GTEx | 0.03 | − | Artery - coronary | ||
| GTEx | 0.03 | − | Artery - tibial | ||
| ICA1L | GTEx | 7.7 × 10−4 | + | Artery - tibial | |
| GTEx | 0.04 | + | Artery - aorta | ||
| GTEx | 0.04 | − | Brain - nucleus accumbens (basal ganglia) | ||
| NBEAL1 | GTEx | 1.2 × 10−34 | − | Artery - tibial | |
| GTEx | 1.3 × 10−19 | − | Artery - aorta | ||
| GTEx | 4.9 × 10−8 | − | Artery - coronary | ||
| GTEx | 0.03 | − | Brain - hypothalamus | ||
| WDR12 | GTEx | 7.6 × 10−4 | − | Brain - putamen (basal ganglia) | |
| GTEx | 5.0 × 10−4 | − | Brain - amygdala | ||
| GTEx | 1.4 × 10−3 | − | Brain - nucleus accumbens (basal ganglia) | ||
| GTEx | 0.04 | − | Brain - substantia nigra | ||
| ABI2 | GTEx | 0.01 | − | Nerve - tibial | |
| rs9515201-A | COL4A1 | GTEx | 0.02 | − | Nerve - tibial |
| Braineac | 2.1 × 10−3 | + | Brain - putamen | ||
| COL4A2 | GTEx | 0.01 | + | Brain - putamen (basal ganglia) |
Significance threshold: 1.6 × 10−4.
The eQTLs that remained significant after multiple test correction are highlighted in bold.
The minor allele C of intronic SNP rs2758605 at 1q22 was significantly (P < 1.6 × 10−4) associated with increased expression of SEMA4A in tibial artery, decreased expression of SLC25A44 in aorta, and decreased expressions of PMF1 and SMG5 in nerve cells of tibia. According to Braineac, the C allele of rs2758605 was also significantly associated with increased expression of PMF1 in the thalamus. According to GTEx, significant cis-eQTL evidence was observed for rs72932727 at 2q33, with the minor C allele significantly associated with decreased expression of CARF in tibial artery, FAM117B in aorta, and NBEAL1 in tibial artery. We did not identify significant cis-eQTL evidence for rs9515201 at 13q34 in either of the expression databases although its minor allele A is positively associated with the expression level of COL4A1 in putamen using Braineac (P = 2.1 × 10−3).
Cell type-specific expression analysis at 1q22, 2q33 and 13q34
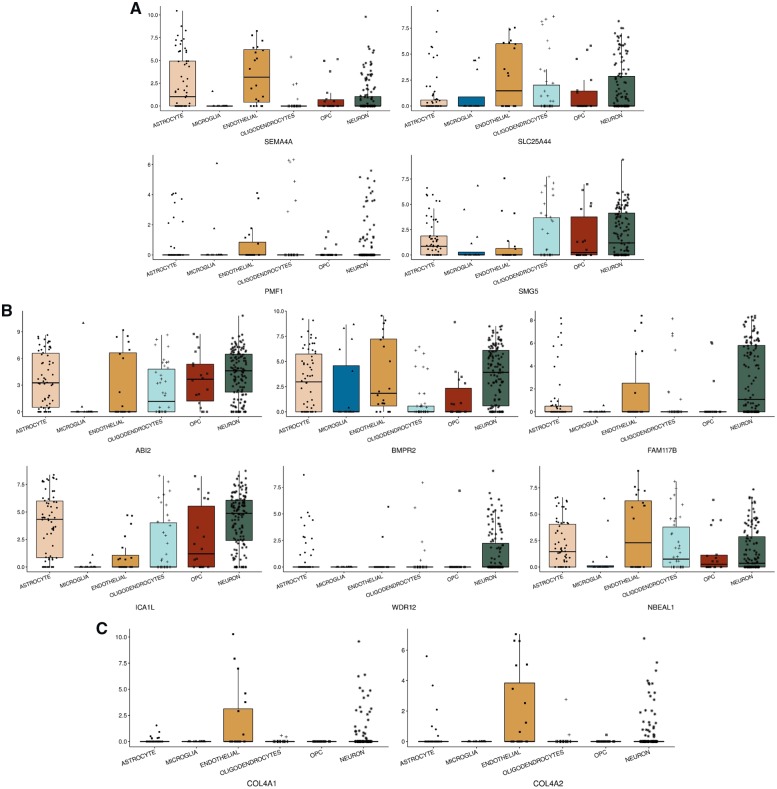
We explored cell type-specific gene expression patterns for the genes at the three loci identified by our MTAG analysis via scRNA-Seq data. According to the human brain scRNA-Seq data (Fig. 3), SLC25A44 and PMF1 at 1q22 were predominantly expressed in endothelial cells, while SEMA4A was observed in other examined cell types except microglia. For 2q33, FAM117B, ICA1L and NBEAL1 were expressed in various cell types except microglia, but WDR12 was observed only in neurons. At 13q34, COL4A1 and COL4A2 were observed solely in endothelial cells. In the mouse cerebrovascular scRNA-Seq data, SLC25A44 and PMF1 at 1q22 and COL4A1 and COL4A2 at 13q34 were expressed in other vessel-related cell types including pericytes, smooth muscle cells, vascular fibroblast-like cells, as well as endothelial cells. For 2q33, we observed ABI2, CARF and NBEAL1 were mostly expressed in smooth muscle cells, and BMPR2, FAM117B and ICA1L were mainly expressed in endothelial cells.
Figure 3.
Human brain cell type-specific expression profiling. X-axes are cell types, and y-axes are log-transformed counts per million (CPM) of expressions for genes at (A) 1q22, (B) 2q33, and (C) 13q34. CARF at 2q33 was not available across any cell types in the RNA-Seq data (Darmanis et al., 2015). OPC = oligodendrocyte precursor cell.
Discussion
The goal of this genetic study was to identify novel genes for ICH risk with statistically increased sample size by combining genetic associations with its proxy phenotype, SVS. Using the cross-phenotype meta-analysis method, MTAG, we identified two novel genetic loci at 2q33 and 13q34, which attained genome-wide significant association with non-lobar ICH, although 13q34 has been observed in a previous candidate-gene association study for COL4A1/COL4A2 in ICH and SVS (Rannikmae et al., 2015). We also confirmed the previously identified loci at 1q22 (Woo et al., 2014) and at APOE (Biffi et al., 2010). This doubles the total of genome-wide significant loci for ICH from two to four.
Annotation data for the genome-wide significant SNPs at the three loci (1q22, 2q33 and 13q24) suggest a potential to act as functional regulatory SNPs for their neighbour genes via modifying regulatory domains or affecting gene expression via a cis-regulatory mechanism. We also attempted to connect our genetic findings for non-lobar ICH to specific brain cell types as defined by a previous scRNA-Seq expression profile of human brain and thereby to better interpret biological roles of the genes detected in this study.
Genome-wide significant association at 1q22 has been identified in our previous GWAS of non-lobar ICH (Woo et al., 2014) as well as in other CSVD-related traits in large scale of genetic studies including white matter hyperintensity burden (Verhaaren et al., 2015; Traylor et al., 2016) and all stroke (Malik et al., 2018). The expression level of PMF1 among the genes at 1q22 was the most significantly associated with the genome-wide significant SNP at 1q22 (rs2758605), and PMF1 showed the strongest association (P = 6.8 × 10−5 in tibial nerve) from expression-based gene-level association test (S-PrediXcan). PMF1, encoding polyamine-modulated factor 1, also regulates polyamine metabolism, which has been repeatedly linked to cerebrovascular disease through blood–brain barrier breakdown and NMDA receptor regulation (Koenig et al., 1989; Georgiev et al., 2008).
The intronic SNPs in the genes FAM117B, NBEAL1 and WDR12 at 2q33 have been detected previously in other genetic studies including coronary artery disease (Consortium et al., 2013; Dichgans et al., 2014), total cholesterol (Willer et al., 2013), early-onset myocardial infarction (Myocardial Infarction Genetics Consortium et al., 2009), and white matter hyperintensity burden (Verhaaren et al., 2015; Traylor et al., 2016; Jian et al., 2018). Among the genes at 2q33, NBEAL1 and FAM117B showed significant associations in both eQTL and S-PrediXcan results. Unfortunately, the biological functions of NBEAL1 and FAM117B have not been studied in detail.
The genome-wide significant SNP, rs9515201, at 13q34 is located within COL4A2 and also in the upstream of COL4A1. This SNP is a cis-acting eQTL and the minor allele A, which is risk for non-lobar ICH, is positively associated with expression levels of COL4A1 and COL4A2 in the putamen region of the brain. However, the strength of these eQTL associations for COL4A1 and COL4A2 is nominal (P < 10−3). This could be because the standard eQTL analysis could not capture the changes of transcription levels in specific tissues or cell types due to high constitutive expression levels or subtle alterations in the expressed ratios between COL4A1 and COL4A2 (Zhang et al., 2018). Alternatively, this may reflect that causal variants tagged by our GWAS finding (rs9515201) could actually be coding variants that influence protein structure rather than expression. Using the human brain cell type-specific expression profiling, we observed that COL4A1 and COL4A2 were expressed solely in endothelial cells in the brain. COL4A2 encodes the alpha-2 chain of type IV collagen, which forms the sheet-like basement membrane that separates epithelium from the connective tissue (Khoshnoodi et al., 2008; Bignon et al., 2011; Bahramsoltani et al., 2014; Loscertales et al., 2016). In mouse studies, COL4A2 has been shown to cause multifocal ICH in the subcortical region of the forebrain and in the cerebellum as well as porencephaly and decreased stability of small vessels. It should be noted that rare non-synonymous mutations in COL4A1 and COL4A2 were observed in familial ICH and SVS patients (Gould et al., 2006; Locatelli et al., 2009; Weng et al., 2012; Jeanne et al., 2015). Moreover, common variants in high LD (r2 > 0.8) with rs9515201 have been associated with sporadic small vessel disease (Rannikmae et al., 2015, 2017) but did not reach genome-wide thresholds. Our genetic study for non-lobar ICH enhanced by SVS provides further evidence that COL4A2 and COL4A1 contribute to the risk of sporadic non-lobar ICH.
We also observed one genome-wide significant association with the gene ZCCHC14 by S-PrediXcan (P = 2.2 × 10−6 in tibial artery) although this association did not attain our conservative experiment-wide threshold (P < 8.5 × 10−7). This region of ZCCHC14 has been associated with SVS (Traylor et al., 2017; Malik et al., 2018). The most significant SNP at 16q24 in the MTAG result was rs12445022 (MTAG P-value = 3.1 × 10−6), which is located between ZCCHC14 and JPH3.
Another notable finding in our study is the gene CELF1 by S-PrediXcan (P = 1.8 × 10−5 in tibial nerve; best SNP from the MTAG result: rs61895112 with OR = 0.93 and P-value = 2.7 × 10−7; Supplementary Table 1). An intronic SNP rs1083728, which is in high LD with rs61895112 (r2 > 0.9), in CELF1 (CUGBP, elav-like family member 1 gene) was identified in the genetic studies of late onset Alzheimer’s disease in 74 046 subjects by International Genomics of Alzheimer’s Project (IGAP) (Lambert et al., 2013; Jun et al., 2017; Kunkle et al., 2019). The effect direction of the minor allele C of the SNP rs61895112 for ICH and SVS (OR = 0.93) was opposite to its effect direction for Alzheimer’s disease in the latest IGAP study (OR = 1.07 and P = 2.9 × 10−7) (Kunkle et al., 2019) and in the Jansen et al. (2019) study (OR = 1.01 and P = 8.9 × 10−4).
Our study has limitations. First, there was an imbalance of sample sizes between ICH and SVS datasets in our analyses, which may lead to a potential increase of type I error rate. The main reason for this imbalance was because the control population used in the SVS GWAS in MEGASTROKE was almost five times larger than the SVS case population. However, according to a recent simulation study about the type I error rates by the ratios between cases and controls in GWAS (Cook et al., 2017), the type I error rate was not affected in a scenario where the number of controls was more than five times larger than the cases (300 cases versus 1700 controls), compared with the type I error rate in the scenario where the sample sizes of cases and controls were balanced (1000 cases versus 1000 controls). Therefore, the potential type I error rates rooted from the imbalanced sample size in the SVS dataset are unlikely to have resulted in an increase of type I error rate in our study. Furthermore, the genome-wide significant associations at 1q22 and 13q34 from our MTAG analysis, except for 2q33, were equally contributed by the associations with the single traits (non-lobar ICH and SVS). This suggests that our MTAG result was not driven solely by the GWAS of SVS. In addition, our MTAG results were consistent with those from another method, GWAS-PW, suggesting that the loci detected by MTAG are less likely to arise from artefacts in the statistical methodology. Moreover, our findings include a replication of a previous non-lobar ICH GWAS result (1q22) and two additional loci, which have been previously associated with other CSVD phenotypes sharing biological substrates with ICH and SVS (2q33 and 13q34). Second, because MTAG improves the statistical power by aggregating the shared genetic components between the traits, we may have missed associations for ICH that are not shared with SVS. Therefore, the methodology employed in the present study should not be considered a substitute for improving the power of dedicated ICH GWAS through increasing available sample size. Third, while more shared genetic associations between two diseases detected from cross-phenotype analysis suggest that more genes are involved in the pathogenesis of both diseases, it does not necessarily follow that the two diseases share biological pathways or disease aetiology in toto (Gratten and Visscher, 2016; Pickrell et al., 2016) because a gene could have effects on the diseases via different pathways, in different tissues, or in response to different metabolic perturbations. Fourth, we did not detect significantly enriched gene sets in our pathway analysis, but this could be because we used Bonferroni adjustment for multiple testing correction, which is more conservative than the other approaches (e.g. false discovery rate) (Noble, 2009). Finally, as mentioned above, some of the controls in the ICH datasets were used in the MEGASTROKE study for SVS genetic studies. However, MTAG permits sample overlap between the GWAS summary data of the traits by conducting bivariate LD score regression of the traits.
Future work in large whole genome sequencing datasets will be required to replicate, fine map and annotate the candidate loci identified in this study, which will narrow the pool of putative causal variants at each locus. These putative causal variants could be examined in massively parallel reporter assays or gene-editing in relevant isogenic cell lines (e.g. endothelia) to further delineate the functional consequences of these variants and uncover related molecular mechanisms that could be used for defining novel therapeutic targets. Furthermore, other CSVD-related traits such as cerebral microbleeds, white matter hyperintensities, dilated perivascular spaces, and cortical superficial siderosis could be added into the next wave of cross-phenotype analyses of ICH and SVS, which may identify novel genetic susceptibility loci for ICH or across the CSVD spectrum.
In summary, we identified novel loci at 2q33 and 13q34 associated with non-lobar ICH by using cross-phenotype analysis with SVS. The genes at these loci have been implicated in other common diseases or traits related to CSVD, and are now implicated in ICH. We selected the genes at the loci by eQTL association and tissue-specific expression analyses. Cell type-specific expression profiling suggests these genes play diverse roles in vascular pathology and inflammation.
Funding
C.D.A. was supported in this work by NIH R01NS103924 and K23NS086873. A.L. was funded by Region Skåne, Lund University, the Swedish Heart and Lung Foundation, the Freemasons Lodge of Instruction Eos Lund, Skåne University Hospital, the Foundation of Färs&Frosta—one of Sparbanken Skåne’s ownership Foundations, and the Swedish Stroke Association. J.R.G was funded by Instituto de Salud Carlos III FEDER, RD12/0042/0020 INVICTUS-PLUS.
Competing interests
A.L. reports honoraria for speech and seminar participation for Bayer and BMS Pfizer, and advisory board consulting for Bayer, Astra Zeneca, Boehringer Ingelheim, and BMS Pfizer. B.N. has received honoraria for DMC work from Astra Zeneca and Bayer. C.D.A reports funding from the NIH, the American Heart Association, the Massachusetts General Hospital Center for Genomic Medicine, and has consulted for ApoPharma, Inc.
Supplementary Material
Glossary
Abbreviations
- CSVD
cerebral small vessel disease
- eQTL
expression quantitative trait locus
- GWAS
genome-wide association study
- ICH
intracerebral haemorrhage
- MTAG
multi-trait analysis of GWAS
- PPA
posterior probabilities of association
- SNP
single nucleotide polymorphism
- SVS
cerebral small vessel ischaemic stroke
References
- Anderson CD, Falcone GJ, Phuah CL, Radmanesh F, Brouwers HB, Battey TW et al. . Genetic variants in CETP increase risk of intracerebral hemorrhage. Ann Neurol 2016; 80: 730–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen OA, Harbo HF, Wang Y, Thompson WK, Schork AJ, Mattingsdal M et al. . Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Mol Psychiatry 2015; 20: 207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen OA, McEvoy LK, Thompson WK, Wang Y, Reppe S, Schork AJ et al. . Identifying common genetic variants in blood pressure due to polygenic pleiotropy with associated phenotypes. Hypertension 2014; 63: 819–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahramsoltani M, Slosarek I, De Spiegelaere W, Plendl J. Angiogenesis and collagen type IV expression in different endothelial cell culture systems. Anat Histol Embryol 2014; 43: 103–15. [DOI] [PubMed] [Google Scholar]
- Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM et al. . Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 2018; 9: 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S et al. . Heart disease and stroke statistics-2018 update: a report from the American Heart Association. Circulation 2018; 137: e67–e492. [DOI] [PubMed] [Google Scholar]
- Bevan S, Traylor M, Adib-Samii P, Malik R, Paul NL, Jackson C et al. . Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke 2012; 43: 3161–7. [DOI] [PubMed] [Google Scholar]
- Biffi A, Sonni A, Anderson CD, Kissela B, Jagiella JM, Schmidt H et al. . Variants at APOE influence risk of deep and lobar intracerebral hemorrhage. Ann Neurol 2010; 68: 934–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignon M, Pichol-Thievend C, Hardouin J, Malbouyres M, Brechot N, Nasciutti L et al. . Lysyl oxidase-like protein-2 regulates sprouting angiogenesis and type IV collagen assembly in the endothelial basement membrane. Blood 2011; 118: 3979–89. [DOI] [PubMed] [Google Scholar]
- Chung J, Wang X, Maruyama T, Ma Y, Zhang X, Mez J et al. . Genome-wide association study of Alzheimer’s disease endophenotypes at prediagnosis stages. Alzheimers Dement 2018; 14: 623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium CAD, Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL et al. . Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 2013; 45: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015; 348: 648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JP, Mahajan A, Morris AP. Guidance for the utility of linear models in meta-analysis of genetic association studies of binary phenotypes. Eur J Hum Genet 2017; 25: 240–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford KM, Gallego-Fabrega C, Kourkoulis C, Miyares L, Marini S, Flannick J et al. . Cerebrovascular disease knowledge portal: an open-access data resource to accelerate genomic discoveries in stroke. Stroke 2018; 49: 470–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM et al. . A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA 2015; 112: 7285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 11: e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devan WJ, Falcone GJ, Anderson CD, Jagiella JM, Schmidt H, Hansen BM et al. . Heritability estimates identify a substantial genetic contribution to risk and outcome of intracerebral hemorrhage. Stroke 2013; 44: 1578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichgans M, Malik R, Konig IR, Rosand J, Clarke R, Gretarsdottir S et al. . Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke 2014; 45: 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LM, Tahmasbi R, Vrieze SI, Abecasis GR, Das S, Gazal S et al. . Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat Genet 2018; 50: 737–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone GJ, Biffi A, Brouwers HB, Anderson CD, Battey TW, Ayres AM et al. . Predictors of hematoma volume in deep and lobar supratentorial intracerebral hemorrhage. JAMA Neurol 2013; 70: 988–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone GJ, Biffi A, Devan WJ, Jagiella JM, Schmidt H, Kissela B et al. . Burden of risk alleles for hypertension increases risk of intracerebral hemorrhage. Stroke 2012; 43: 2877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CM. Pathological observations in hypertensive cerebral hemorrhage. J Neuropathol Exp Neurol 1971; 30: 536–50. [DOI] [PubMed] [Google Scholar]
- Georgiev D, Taniura H, Kambe Y, Takarada T, Yoneda Y. A critical importance of polyamine site in NMDA receptors for neurite outgrowth and fasciculation at early stages of P19 neuronal differentiation. Exp Cell Res 2008; 314: 2603–17. [DOI] [PubMed] [Google Scholar]
- Gould DB, Phalan FC, van Mil SE, Sundberg JP, Vahedi K, Massin P et al. . Role of COL4A1 in small-vessel disease and hemorrhagic stroke. N Engl J Med 2006; 354: 1489–96. [DOI] [PubMed] [Google Scholar]
- Gratten J, Visscher PM. Genetic pleiotropy in complex traits and diseases: implications for genomic medicine. Genome Med 2016; 8: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WD, Marioni RE, Maghzian O, Ritchie SJ, Hagenaars SP, McIntosh AM et al. . A combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Mol Psychiatry 2018; 24: 169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S et al. . Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet 2019; 51: 404–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanne M, Jorgensen J, Gould DB. Molecular and genetic analyses of collagen type IV mutant mouse models of spontaneous intracerebral hemorrhage identify mechanisms for stroke prevention. Circulation 2015; 131: 1555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X, Satizabal CL, Smith AV, Wittfeld K, Bis JC, Smith JA et al. . Exome chip analysis identifies low-frequency and rare variants in MRPL38 for white matter hyperintensities on brain magnetic resonance imaging. Stroke 2018; 49: 1812–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC et al. . A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry 2016; 21: 108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun GR, Chung J, Mez J, Barber R, Beecham GW, Bennett DA et al. . Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement 2017; 13: 727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM et al. . The human genome browser at UCSC. Genome Res 2002; 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnoodi J, Pedchenko V, Hudson BG. Mammalian collagen IV. Microsc Res Tech 2008; 71: 357–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig H, Goldstone AD, Lu CY. Blood-brain barrier breakdown in cold-injured brain is linked to a biphasic stimulation of ornithine decarboxylase activity and polyamine synthesis: both are coordinately inhibited by verapamil, dexamethasone, and aspirin. J Neurochem 1989; 52: 101–9. [DOI] [PubMed] [Google Scholar]
- Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC et al. . Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet 2019; 51: 414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M, Trampush JW, Yu J, Knowles E, Davies G, Liewald DC et al. . Large-scale cognitive GWAS meta-analysis reveals tissue-specific neural expression and potential nootropic drug targets. Cell Rep 2017; 21: 2597–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al. . Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45: 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli F, Bersano A, Ballabio E, Lanfranconi S, Papadimitriou D, Strazzer S et al. . Stem cell therapy in stroke. Cell Mol Life Sci 2009; 66: 757–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat Genet 2018; 50(7): 906–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscertales M, Nicolaou F, Jeanne M, Longoni M, Gould DB, Sun Y et al. . Type IV collagen drives alveolar epithelial-endothelial association and the morphogenetic movements of septation. BMC Biol 2016; 14: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire LH, Handelman SK, Du X, Chen Y, Pers TH, Speliotes EK. Genome-wide association analyses identify 39 new susceptibility loci for diverticular disease. Nat Genet 2018; 50: 1359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A et al. . Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 2018; 50: 524–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini SR, Flaherty ML, Brown WM, Haverbusch M, Comeau ME, Sauerbeck LR et al. . Risk factors for intracerebral hemorrhage differ according to hemorrhage location. Neurology 2012; 79: 2275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A et al. . A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016; 48: 1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myocardial Infarction Genetics Consortium, Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D et al. . Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 2009; 41: 334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble WS. How does multiple testing correction work? Nat Biotechnol 2009; 27: 1135–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoni L. Cerebral small vessel disease: from pathogenesis and clinical characteristics to therapeutic challenges. Lancet Neurol 2010; 9: 689–701. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet 2016; 48: 709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi AI, Tuhrim S, Broderick JP, Batjer HH, Hondo H, Hanley DF. Spontaneous intracerebral hemorrhage. N Engl J Med 2001; 344: 1450–60. [DOI] [PubMed] [Google Scholar]
- Rannikmae K, Davies G, Thomson PA, Bevan S, Devan WJ, Falcone GJ et al. . Common variation in COL4A1/COL4A2 is associated with sporadic cerebral small vessel disease. Neurology 2015; 84: 918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannikmae K, Sivakumaran V, Millar H, Malik R, Anderson CD, Chong M et al. . COL4A2 is associated with lacunar ischemic stroke and deep ICH: Meta-analyses among 21,500 cases and 40,600 controls. Neurology 2017; 89: 1829–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco RL, Adams R, Albers G, Alberts MJ, Benavente O, Furie K et al. . Guidelines for prevention of stroke in patients with ischemic stroke or transient ischemic attack: a statement for healthcare professionals from the American Heart Association/American Stroke Association Council on Stroke: co-sponsored by the Council on Cardiovascular Radiology and Intervention: the American Academy of Neurology affirms the value of this guideline. Stroke 2006; 37: 577–617. [DOI] [PubMed] [Google Scholar]
- Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012; 40 (Web Server issue): W452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Telgte A, van Leijsen EMC, Wiegertjes K, Klijn CJM, Tuladhar AM, de Leeuw FE. Cerebral small vessel disease: from a focal to a global perspective. Nat Rev Neurol 2018; 14: 387–98. [DOI] [PubMed] [Google Scholar]
- Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A et al. . Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem 2011; 119: 275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traylor M, Malik R, Nalls MA, Cotlarciuc I, Radmanesh F, Thorleifsson G et al. . Genetic variation at 16q24.2 is associated with small vessel stroke. Ann Neurol 2017; 81: 383–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traylor M, Zhang CR, Adib-Samii P, Devan WJ, Parsons OE, Lanfranconi S et al. . Genome-wide meta-analysis of cerebral white matter hyperintensities in patients with stroke. Neurology 2016; 86: 146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA et al. . Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet 2018; 50: 229–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandewijck M, He L, Mae MA, Andrae J, Ando K, Del Gaudio F et al. . A molecular atlas of cell types and zonation in the brain vasculature. Nature 2018; 554: 475–80. [DOI] [PubMed] [Google Scholar]
- Verhaaren BF, Debette S, Bis JC, Smith JA, Ikram MK, Adams HH et al. . Multiethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ Cardiovasc Genet 2015; 8: 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinters HV. Cerebral amyloid angiopathy. A critical review. Stroke 1987; 18: 311–24. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012; 40 (Database issue): D930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw JM, Smith EE, Biessels GJ, Cordonnier C, Fazekas F, Frayne R et al. . Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol 2013; 12: 822–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8: 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng YC, Sonni A, Labelle-Dumais C, de Leau M, Kauffman WB, Jeanne M et al. . COL4A1 mutations in patients with sporadic late-onset intracerebral hemorrhage. Ann Neurol 2012; 71: 470–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S et al. . Discovery and refinement of loci associated with lipid levels. Nat Genet 2013; 45: 1274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD et al. . Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet 2014; 94: 511–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Samocha KE, Rivas MA, Karczewski KJ, Daly E, Schmandt B et al. . Base-specific mutational intolerance near splice sites clarifies the role of nonessential splice nucleotides. Genome Res 2018; 28: 968–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Dinh HQ, Ramjan Z, Weisenberger DJ, Nicolet CM, Shen H et al. . DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat Genet 2018; 50: 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in Cerebrovascular Disease Knowledge Portal (CDKP) at http://www.cerebrovascularportal.org (Crawford et al., 2018).