Fungi produce a diverse array of secondary metabolites, many of which are of pharmacological importance whereas many others are noted for mycotoxins, such as aflatoxin and citrinin, that can threaten human and animal health. The polyketide-derived compound ascochitine, which is structurally similar to citrinin mycotoxin, has been considered to be important for pathogenicity of legume-associated Ascochyta species. Here, we identified the ascochitine polyketide synthase (PKS) gene in Ascochyta fabae and its neighboring genes that may be involved in ascochitine biosynthesis. Interestingly, the ascochitine PKS genes in other legume-associated Ascochyta species have been mutated, encoding truncated PKSs. This indicated that point mutations may have contributed to genetic diversity for secondary metabolite production in these fungi. We also demonstrated that ascochitine is not a pathogenicity factor in A. fabae. The antifungal activities and production of ascochitine during sporulation suggested that it may play a role in competition with other saprobic fungi in nature.
KEYWORDS: Ascochyta fabae, Didymellaceae, ascochitine, citrinin, nonsense mutation, polyketide, secondary metabolite
ABSTRACT
The polyketide-derived secondary metabolite ascochitine is produced by species in the Didymellaceae family, including but not restricted to Ascochyta species pathogens of cool-season food legumes. Ascochitine is structurally similar to the well-known mycotoxin citrinin and exhibits broad-spectrum phytotoxicity and antimicrobial activities. Here, we identified a polyketide synthase (PKS) gene (denoted pksAC) responsible for ascochitine production in the filamentous fungus Ascochyta fabae. Deletion of the pksAC prevented production of ascochitine and its derivative ascochital in A. fabae. The putative ascochitine biosynthesis gene cluster comprises 11 genes that have undergone rearrangement and gain-and-loss events relative to the citrinin biosynthesis gene cluster in Monascus ruber. Interestingly, we also identified pksAC homologs in two recently diverged species, A. lentis and A. lentis var. lathyri, that are sister taxa closely related to ascochitine producers such as A. fabae and A. viciae-villosae. However, nonsense mutations have been independently introduced in coding sequences of the pksAC homologs of A. lentis and A. lentis var. lathyri that resulted in loss of ascochitine production. Despite its reported phytotoxicity, ascochitine was not a pathogenicity factor in A. fabae infection and colonization of faba bean (Vicia faba L.). Ascochitine was mainly produced from mature hyphae at the site of pycnidial formation, suggesting a possible protective role of the compound against other microbial competitors in nature. This report highlights the evolution of gene clusters harnessing the structural diversity of polyketides and a mechanism with the potential to alter secondary metabolite profiles via single nucleotide polymorphisms in closely related fungal species.
IMPORTANCE Fungi produce a diverse array of secondary metabolites, many of which are of pharmacological importance whereas many others are noted for mycotoxins, such as aflatoxin and citrinin, that can threaten human and animal health. The polyketide-derived compound ascochitine, which is structurally similar to citrinin mycotoxin, has been considered to be important for pathogenicity of legume-associated Ascochyta species. Here, we identified the ascochitine polyketide synthase (PKS) gene in Ascochyta fabae and its neighboring genes that may be involved in ascochitine biosynthesis. Interestingly, the ascochitine PKS genes in other legume-associated Ascochyta species have been mutated, encoding truncated PKSs. This indicated that point mutations may have contributed to genetic diversity for secondary metabolite production in these fungi. We also demonstrated that ascochitine is not a pathogenicity factor in A. fabae. The antifungal activities and production of ascochitine during sporulation suggested that it may play a role in competition with other saprobic fungi in nature.
INTRODUCTION
Polyketides are the most abundant fungal secondary metabolites and have been studied for centuries for either their beneficial bioactivity or their toxicity. The genes underlying the biosynthetic pathways for these and other secondary metabolites are arranged in contiguous clusters on the genome (1). Linking of the biosynthetic pathways to their respective gene clusters has been facilitated by technological advances in metabolomics and genomics.
Ascochitine, also known as ascochytine, is an o-quinone methide produced via a polyketide biosynthetic pathway. Ascochitine was originally reported as a selective antifungal agent, as some tested fungi were more sensitive to ascochitine whereas others were able to detoxify ascochitine (2, 3). In addition, applied externally, ascochitine can cause electrolyte leakage from the leaf disc of Clematis cultivars susceptible to an ascochitine producer, Phoma clematidina. However, Clematis cultivars resistant to P. clematidina were largely insensitive to ascochitine, suggesting that ascochitine is a host-selective phytotoxin (4).
Ascochitine was first discovered in culture extracts of Ascochyta pisi Lib. (5), and later in A. fabae Speg. (6), causal agent of Ascochyta blight of pea (Pisum sativum L.) and faba bean (Vicia faba L.), respectively (7). The ability to produce ascochitine is prevalent among legume-associated Ascochyta but has been lost in some of the lineages (8). Ascochitine production has been also reported in species in other genera in the family Didymellaceae (4, 9) and in the sister family Pleosporaceae (10) in the Dothideomycetes (11–14). However, ascochitine has not been found in A. lentis, which is placed as a sister taxon to the ascochitine producers such as A. fabae, A. pisi, and A. viciae-villosae (8). Instead, new phytotoxic compounds have been discovered in culture extracts from A. lentis and A. lentis var. lathyri (15, 16).
Ascochitine is structurally similar to the well-known mycotoxin citrinin, which is produced by diverse fungal species in the genera Aspergillus, Monascus, and Penicillium in the Eurotiomycetes. The citrinin biosynthesis gene cluster, including a polyketide synthase (PKS) gene that initiates the biosynthetic pathway, as well as a pathway-specific transcription regulator, has been identified previously in Monascus spp. (17–21). Ascochitine biosynthetic pathways of ascochitine have been proposed previously (9, 22). However, identification of the underlying gene cluster is lacking.
All described fungal PKSs belong to the type I iterative PKS group that contains several functional domains, and all synthesize structurally diverse compounds, such as the health-threatening aflatoxin, the cholesterol-lowering drug lovastatin, and melanin pigments (23). The type I iterative PKSs can be broadly classified into nonreducing PKS (NR-PKS) and highly reducing PKS (HR-PKS). Many NR-PKSs have an N-terminal starter unit–ACP transacylase (SAT) domain–which mediates the loading of a starter unit, followed by chain extension components ketosynthase (KS), acyl transferase (AT), and acyl carrier protein (ACP). The product template (PT) domain is often located between the AT and ACP domains and may be involved in polyketide chain length determination (24). The C-terminal processing domains of NR-PKS are variable and often include some of the following domains: methyltransferase (MeT), thioesterase (TE), Claisen-cyclase (CLC), thiolester reductase domain (R), and additional ACP domains (24). HR-PKSs often possess dehydratase (DH), ketoreductase (KR), and enoyl reductase (ER) and terminate with an ACP domain, producing complex highly reduced compounds. To date, there are no identified HR-PKSs that have the N-terminal SAT and PT domains commonly found in NR-PKSs. Among the diverse PKS domains, the KS domain has been used in classification of PKSs in fungi due to the high amino acid sequence similarity (25–27).
Since the first discovery of ascochitine in 1956 (5), the compound has been found in many different fungal taxa and researchers have reported its phytotoxicity and antifungal activities. However, direct evidence is lacking for the roles of ascochitine in pathogenicity and competition with other microbes, primarily due to lack of information about the genetic control of biosynthesis. Thus, the objectives of this study were (i) to identify genes responsible for ascochitine biosynthesis and the associated gene cluster in A. fabae, (ii) to examine the roles of ascochitine in the fungal biology through targeted gene knockout study, and (iii) to investigate genetic diversity in ascochitine biosynthetic genes among legume-associated Ascochyta species.
RESULTS
Homology-based search for putative ascochitine PKS gene.
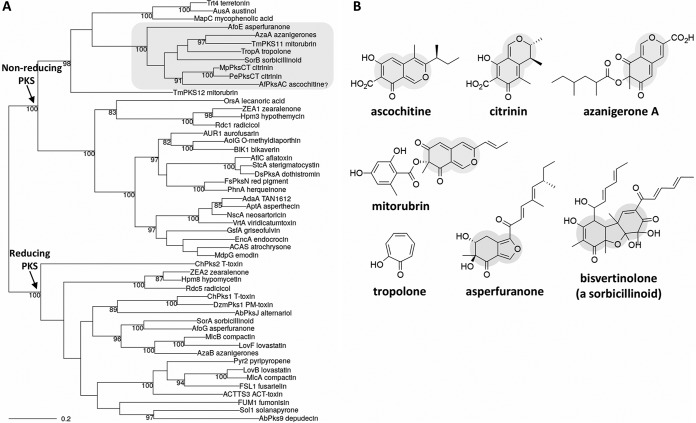
To search for a PKS gene responsible for ascochitine biosynthesis, we identified a total of nine putative type I PKS genes from the genome sequence of A. fabae isolate AF1, using the antiSMASH program (28). Then, the KS domain of the citrinin PKS gene (pksCT) in Monascus purpureus was queried against the putative PKS genes found in A. fabae. We found a PKS gene (here referred to as pksAC) similar to the pksCT gene, showing 67% identity in the KS domains at the amino acid sequence level. The PKS domain architectures of the pksAC and pksCT genes were identical (SAT-KS-AT-PT-ACP-MeT-R). To predict a PKS function in an evolutionary framework, we reconstructed the genealogy of fungal PKSs, using the deduced amino acid sequences of the KS domains of the pksAC gene and 51 PKS genes that have been definitively linked to the biosynthesis of specific compounds (see Text S1 in the supplemental material). It seems reasonable to believe that the resulting polyketides produced by phylogenetic orthologs are likely more similar to each other in their chemical structures. As expected, the pksAC gene was placed next to the pksCT genes in the phylogenetic tree (Fig. 1A), providing additional evidence of the link between the pksAC gene and ascochitine biosynthesis. The polyketide products within the subclade that includes the pksAC and pksCT genes share a common characteristic, the heavily oxygenated heterocyclic core, which is observed most often in pyranoquinones, such as ascochitine, azanigerones, citrinin, and mitorubrin (Fig. 1B).
FIG 1.
Genealogy of fungal PKS. (A) The maximum likelihood phylogeny was estimated from deduced amino acid sequences of the KS domain of PKS genes with known products. The numbers at the internal nodes indicate percentages of bootstrap support greater than 80% from 1,000 bootstrap replications. Branch lengths are proportional to the inferred amount of evolutionary change, and the scale represents 0.2 amino acid sequence substitutions per site. A subclade that includes pksCT and putative ascochitine PKS genes (denoted pksAC) was highlighted. (B) Chemical structures of polyketides in the highlighted subclade within the nonreducing PKS clade. The hallmark oxygenated heterocyclic cores are shaded.
The deduced amino acid sequences of the KS domains of fungal PKS genes. Download Text S1, DOCX file, 0.02 MB (26.6KB, docx) .
Copyright © 2019 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of pksAC in A. fabae.
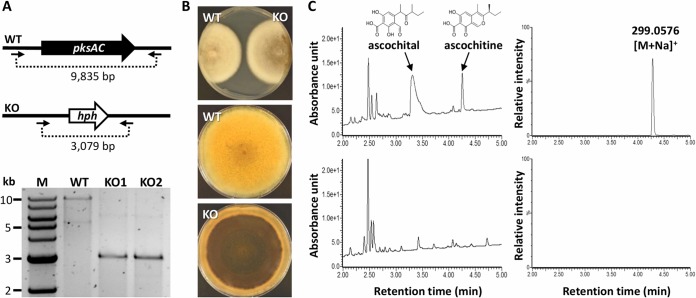
To validate the involvement of the pksAC gene in ascochitine production in A. fabae isolate AF1 (wild type [WT]), we conducted targeted gene replacement of the pksAC gene with a hygromycin phosphotransferase gene cassette. Successful replacement in putative transformants was verified with PCR using a primer pair that annealed to flanking regions of the introduced cassette (Fig. 2A). The resulting knockout mutants (ΔpksAC) were similar to the WT in growth rate (Fig. 2B; top panel). In the aged culture, the colony of the ΔpksAC gene became highly melanized (Fig. 2B), which was likely caused by the excess of acetates that otherwise should have been used in ascochitine biosynthesis. Similar phenomena have been observed in solanapyrone-negative mutants in Ascochyta rabiei (29) and in a mutant defective in a red pigment production in Fusarium neocosmosporiellum (30).
FIG 2.
Generation of ascochitine-negative mutants. (A) The pksAC coding sequence of A. fabae isolate AF1 (WT) was replaced with a hygromycin phosphotransferase gene cassette (hph). Gene replacement was confirmed by PCR; the primer pair is indicated by arrows. (B) A front view of the WT and ΔpksAC (KO) strains after 10 days of hyphal growth on potato dextrose agar is shown in the top panel; reverse views of hyphal growth after 20 days are shown in the middle and bottom panels. (C) UV-visible light (UV-Vis) chromatograms (left panels) and selected ion mass chromatograms (right panels) of WT (upper panels) and KO (bottom panels) culture extract. Peaks characteristic of ascochitine and its derivative ascochital are indicated with arrows where present. Denoted is the ascochitine observed m/z; for reference, the m/z calculated for C15H16NaO5 [M+Na]+ is 299.0895.
The ΔpksAC gene lost the ability to produce ascochitine and its derivatives, as evidenced by the chemical profiles of the culture extracts (Fig. 2C). Liquid chromatography-mass spectrometry (LC-MS) analysis of a culture extract of the WT confirmed the production of ascochitine (eluted at 4.28 min) and ascochital (another end product from the ascochitine biosynthetic pathway; eluted at 3.32 min) (9) (Fig. 2C; top panels). In the ΔpksAC gene, there was no detectable production of ascochitine and ascochital, indicating that the pksAC gene is required for ascochitine biosynthesis (Fig. 2C; bottom panels).
Identification of ascochitine biosynthesis gene cluster.
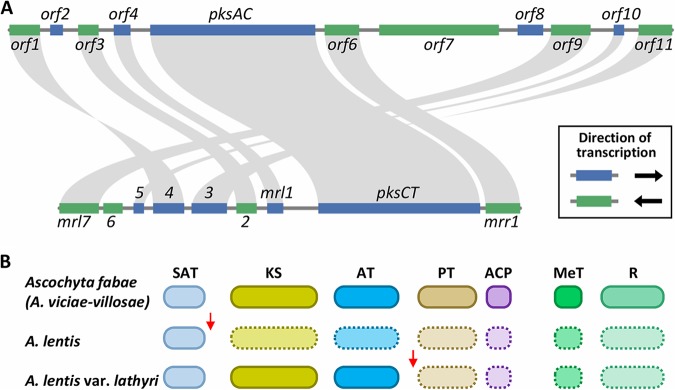
The identity and function of each gene in the citrinin biosynthesis gene cluster in Monascus ruber were recently reported (21). On the basis of the structural similarity between citrinin and ascochitine (Fig. 1B), we hypothesized that the contents of the citrinin and ascochitine gene clusters are conserved. Since the genome assembly of A. fabae isolate AF1 around the pksAC locus was not complete, we used the genome sequences of another A. fabae isolate (isolate AF247/15) and an A. viciae-villosae isolate (isolate AV22) that were retrieved from the NCBI database to search for ascochitine biosynthesis gene cluster. There was one nucleotide difference in the pksAC sequence in the AF247/15 isolate (G to A at position 5437) that caused a nonsynonymous substitution (valine to isoleucine) relative to the pksAC sequences in the AF1 and AF55/01 isolates. By comparing genes neighboring the pksAC gene to the gene members of the citrinin gene cluster, we identified a minimal set of conserved genes (orf1 to orf11; Table 1) that are likely involved in ascochitine biosynthesis in the genomes of the AF247/15 and AV22 isolates. The gene content and order of the ascochitine biosynthesis gene cluster in the two ascochitine producers were identical, and genes further downstream of orf1 or upstream of orf11 did not have significant sequence similarities with the members of the citrinin gene cluster (not shown).
TABLE 1.
Annotation of the ascochitine biosynthesis gene cluster in A. fabae
| Gene | Length (aa)a |
Protein function | Citrinin gene |
% aa |
|---|---|---|---|---|
| orf1 | 494 | NAD(P)+-dependent aldehyde dehydrogenase | mrl4 | 54 |
| orf2 | 186 | Cupin domain-containing protein | ||
| orf3 | 331 | Nonheme Fe(II)-dependent oxygenase | mrl2 | 62 |
| orf4 | 270 | Serine hydrolase | mrl1 | 53 |
| pksAC (orf5) | 2,645 | Nonreducing polyketide synthase (NR-PKS) | pksCT | 52 |
| orf6 | 517 | Transporter | mrr1 | 60 |
| orf7 | 1,886 | Highly reducing polyketide synthase (HR-PKS) | ||
| orf8 | 379 | SAT domain-containing protein | ||
| orf9 | 621 | NAD(P)+-dependent oxidoreductase | mrl7 | 60 |
| orf10 | 128 | Glyoxylase-like domain-containing protein | mrl5 | 46 |
| orf11 | 573 | Transcription factor | mrl3 | 45 |
aa, amino acids.
The orf1, orf3, and orf9 genes were homologous to the citrinin biosynthesis genes encoding NAD(P)+-dependent aldehyde dehydrogenase (mrl4), nonheme Fe(II)-dependent oxygenase (mrl2), and NAD(P)+-dependent oxidoreductase (mrl7), respectively. These enzymes were involved in subsequent oxidations of methyl groups to the carboxylic acid of the heterocyclic ring of citrinin (21). The mrl6 gene, encoding an enzyme involved in the generation of carbonyl groups in the heterocyclic ring of citrinin, was missing in the putative ascochitine gene cluster in both A. fabae and A. viciae-villosae. An additional PKS gene (orf7) was found adjacent to the pksAC gene (Fig. 3A). This PKS gene is classified as a highly reducing PKS gene. As such, it may be involved in the addition of the sec-butyl group in ascochitine via mechanisms analogous to the biosynthesis of asperfuranone and azanigerones in Aspergillus spp. (31). The gene cluster also includes a transporter gene (orf6) and a gene encoding a putative transcription factor (orf11). Overall, despite the observed structural similarity in ascochitine and citrinin, the responsible gene clusters appeared to have diverged through gene order rearrangement and through gene gains and losses (Fig. 3A).
FIG 3.
Divergence of ascochitine biosynthesis gene cluster. (A) Synteny plots of ascochitine (top) and citrinin (bottom) biosynthesis gene clusters. Annotations of genes in the citrinin biosynthesis gene cluster are presented as described previously by He and Cox (21). (B) Schematic diagrams of PKS domain structures of pksAC orthologs in legume-associated Ascochyta species. Arrows indicate lineage-specific SNPs that caused nonsense mutations. Domains surrounded by dashed lines indicate absence of translation due to premature stop codons.
Sequence diversity in pksAC orthologs.
Genetic diversity in secondary metabolite gene clusters can be introduced by different mechanisms in fungi (32). We investigated how ascochitine gene clusters have diverged during the evolution of legume-associated Ascochyta species. First, the presence of pksAC orthologous genes was checked in A. lentis and A. lentis var. lathyri, which are closely related to ascochitine producers such as A. fabae and A. viciae-villosae (33). We surveyed genome sequences of three A. lentis isolates, namely, isolates AL1, AL4, and Kewell, and one A. lentis var. lathyri isolate, namely, isolate ER1415, and found orthologous pksAC genes largely as complete copies in the four isolates. However, many single nucleotide polymorphisms (SNPs) were observed in the pksAC orthologs that included nonsense mutations introducing a premature stop codon that resulted in truncated PKS gene products (Fig. 3B). Interestingly, the nucleotide positions of the first nonsense mutation differed in A. lentis and A. lentis var. lathyri: G to T at position 828 in A. lentis and C to T at position 4116 in A. lentis var. lathyri (Table 2). The A. lentis genome sequences were all from Australian isolates, so we examined additional isolates of different geographical origins, including Asia, Europe, and South America (Table 2). Since A. lentis var. lathyri is an species endemic to Italy, we checked one more isolate collected in a different year (Table 2). The SNPs that caused the nonsense mutation were confirmed in all the tested isolates by Sanger sequencing.
TABLE 2.
Single nucleotide polymorphisms that caused nonsense mutations in orthologous pksAC genesa
| Species (isolate code) | Origin of isolation |
Location (collector, yr) | SNP1/ SNP2 |
|---|---|---|---|
| Ascochyta fabae (AF1) | Vicia faba | Saskatoon, Canada (A. Vandenberg, 1992) (8, 33) | GGA/CAG |
| Ascochyta fabae (AF55/01) | Vicia faba | Cochaleechie, Australia (R. Kimber, 2001) | GGA/CAG |
| Ascochyta fabae (AF247/15) | Vicia faba | Tarlee, Australia (NA) | GGA/CAG |
| Ascochyta viciae-villosae (AV22) | Vicia villosae | Apriltsi, Bulgaria (W. Kaiser, 1996) (33) | GGA/CAG |
| Ascochyta lentis (AL1) | Lens culinaris | Australia (W. Kaiser, NA) (8, 33) | TGA/CAG |
| Ascochyta lentis (AL2) | Lens culinaris | Brazil (W. Kaiser, NA) (8, 33) | TGA/CAG |
| Ascochyta lentis (AL3) | Lens culinaris | Canada (W. Kaiser, NA) (8, 33) | TGA/CAG |
| Ascochyta lentis (AL4) | Lens culinaris | Horsham, Australia (M. Nasir and T. W. Bretag, 1998) (64, 65) | TGA/CAG |
| Ascochyta lentis (AL6) | Lens culinaris | Russia (W. Kaiser, NA) (8, 33) | TGA/CAG |
| Ascochyta lentis (AL11) | Lens culinaris | India (W. Kaiser, NA) (8, 33) | TGA/CAG |
| Ascochyta lentis (Kewell) | Lens culinaris | Kewell, Australia (M. Nasir and T. W. Bretag, 2001) (65) | TGA/CAG |
| Ascochyta lentis var. lathyri (ER1415) | Lathyrus sativus | Salerno, Italy (A. Infantino, 2007) (8, 42) | GGA/TAG |
| Ascochyta lentis var. lathyri (ER1478) | Lathyrus sativus | Salerno, Italy (A. Infantino, 2008) (8, 42) | GGA/TAG |
SNP1 and SNP2 occurred at nucleotide 828 and 4,116 positions from the A of the ATG translation initiation codon of the pksAC gene in A. fabae isolate AF247/15, respectively. NA, not available. Bold letter Ts in column 4 represent nucleotide substitutions to be a stop codon.
To investigate possible selection acting on ascochitine biosynthesis genes, we estimated synonymous and nonsynonymous substitution rates between pksAC orthologs. Although the degrees of sequence divergence between pksAC orthologs were similar at both the DNA and deduced amino acid sequence levels, the ratio of synonymous to nonsynonymous substitution rates between A. fabae and A. viciae-villosae (ascochitine producers) was slightly higher than between A. fabae and the two ascochitine-negative species (Table 3), which could be ascribed to some degree of purifying selection acting on the pksAC genes in the ascochitine producers.
TABLE 3.
Estimation of synonymous and nonsynonymous substitution rates in pksAC orthologsa
| Ascochyta fabae vs.: | No. of Sd |
No. of Sn |
No. of S | No. of N | Ratio | nt% | aa% |
|---|---|---|---|---|---|---|---|
| Ascochyta viciae-villosae | 107 | 50 | 1,787.0 | 5,875.0 | 7.04 | 97.95 | 98.08 |
| Ascochyta lentis | 87 | 54 | 1,786.2 | 5,875.8 | 5.30 | 98.16 | 97.92 |
| Ascochyta lentis var. lathyri | 102 | 57 | 1,787.2 | 5,874.8 | 5.88 | 97.92 | 97.77 |
Sd, observed synonymous substitutions; Sn, observed nonsynonymous substitutions; S, potential synonymous substitutions; N, potential nonsynonymous substitutions; Ratio, the ratio of synonymous to nonsynonymous substitutions = (Sd/S)/(Sn/N); nt%, percentage of nucleotides; aa%, percentage of amino acids.
Possible roles of ascochitine.
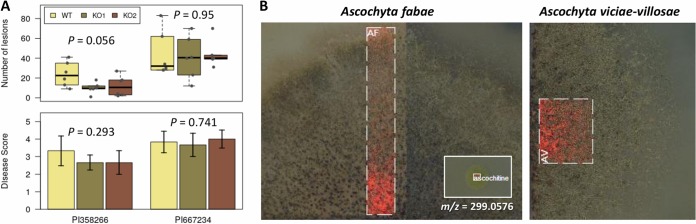
Ascochitine has been postulated to represent a pathogenicity factor (4). Thus, the role of ascochitine in the A. fabae and faba bean pathosystem was evaluated by comparing the WT and the ΔpksAC mutant with respect to their disease-causing abilities. Faba bean strains with accession numbers PI 358266 (moderately resistant) and PI 667234 (susceptible) served as hosts. The ΔpksAC mutants maintained the ability to cause disease on faba bean, and there was no significant difference between the WT and the ΔpksAC mutant with respect to disease incidence or severity (one-way analysis of variance [ANOVA] P > 0.05; Fig. 4A). These results suggested that ascochitine production is not essential for pathogenicity in A. fabae on faba bean.
FIG 4.
Characterization of ascochitine-negative mutants. (A) Pathogenicity tests were performed on faba bean strains (accession no. PI 358266, moderately resistant; accession no. PI 667234, susceptible). P values are for one-way ANOVA for each strain. (B) In situ detection of ascochitine in the cultures of A. fabae (AF) and A. viciae-villosae (AV). A selected area (dashed box) was analyzed with a Fourier-transform ion cyclotron resonance mass spectrometry instrument. Detection of ascochitine (m/z calculated for C15H16NaO5 [M+Na]+, 299.0895; observed, 299.0576). The inset shows an ascochitine standard spotted onto a separate place, and reference mass spectrometry signals were obtained in a simultaneous run. Small black dots visible on the fungal colony represent pycnidia (asexual fruiting bodies). The difference in brightness of the left and right side of the A. fabae colony is due to the background of the embedded microscopic slide on which the fungus grew.
matrix-assisted laser desorption ionization (MALDI) imaging mass spectrometry enabled the detection of ascochitine on the surface of the fungal colony, and ascochitine accumulated in aged hyphae where pycnidia (the asexual fruiting bodies) formed (Fig. 4B), suggesting that ascochitine was associated with sporulation and might play a protective role against competing microbes in nature.
DISCUSSION
With the advent of high-throughput sequencing technologies, fungal genomes are becoming available in exponentially increasing numbers (34). Nevertheless, relatively few secondary metabolite pathways have been linked to their respective biosynthetic gene clusters. Although classes of compounds can be made through completely distinct biochemical pathways, phylogenetic orthologs that encode enzymes with identical or nearly identical biochemical functions are expected to produce similar compounds. Therefore, homology-based prediction within an evolutionary framework can facilitate linking of genes to molecules, and, ultimately, the genetic information can be used for developing heterologous systems for expression of compounds of pharmaceutical and industrial interest in the postgenomic era (31, 35).
To date, ascochitine has been found in many species in legume-associated Ascochyta and other genera in the Dothideomycetes, such as Calophoma, Phoma, Pleospora, and Stagonosporopsis (4, 8–10). Recent efforts to resolve the structure of complex Didymellaceae, including Ascochyta and Phoma taxa in the Dothideomycetes, reclassified P. clematidina into Calophoma, which is a sister genus to the Ascochyta, and A. salicorniae into Stagonosporopsis (11, 13, 14). Ascochyta hyalospora was also renamed Pleospora chenopodii, which belongs to the Pleosporaceae (12). These widespread distributions of ascochitine production in the Dothideomycetes suggested that the trait of producing ascochitine is ancestral to legume-associated Ascochyta species. Despite the phylogenetic distance between ascochitine- and citrinin-producing fungi, the ascochitine and citrinin biosynthetic genes exhibited high degrees of sequence similarities (up to 62%), suggesting a common origin of ascochitine and citrinin gene clusters. The two gene clusters showed gene content polymorphisms and appear to have experienced multiple gene translocation and inversion events during divergence from the gene cluster that shared the most recent common ancestry. Interestingly, the ascochitine gene cluster included a HR-PKS (orf7) with an unusual PKS domain architecture (KS-DH-ER-KR-ACP), missing AT and MeT domains found in many HR-PKSs, such as lovastatin diketide synthase lovF (KS-AT-DH-MeT-ER-KR-ACP) (36). Most PKS require an AT domain to transfer acyl groups to the KS and ACP components. It is currently unknown whether the orf7 without an AT domain is truly functional. Since the orf7 ortholog was also found in A. viciae-villosae, it is less likely that the unusual PKS architecture of the orf7 was due to genome assembly or annotation errors. Thus, the precise role of the HR-PKS in ascochitine biosynthesis needs to be determined.
It was previously hypothesized that a reduced diketide is used as a starter unit for citrinin biosynthesis by the pksCT gene (37). Recently, however, He and Cox demonstrated that acetate is used as a starter unit for subsequent 4-fold extension by the pksCT gene, yielding an unreduced trimethylated pentaketide without the need of another PKS (21). The only structural difference between ascochitine and citrinin is that ascochitine has a sec-butyl group in the heterocyclic ring. As an alternative to the previously proposed biosynthetic scheme (9), the finding of the orf7 in the ascochitine gene cluster in A. fabae suggested that the HR-PKS makes a diketide starter unit which is passed to the pksAC gene for further extension, producing ascochital and ascochitine via a mechanism analogous to that of asperfuranone biosynthesis (38). Notably, in the NR-PKS subclade that includes ascochitine (Fig. 1B), asperfuranone, azanigerone A, and bisvertinolone (a sorbicillinoid) are synthesized by the combination of an NR-PKS (AfoE, AzaA, and SorB, respectively) and a HR-PKS (AfoG, AzaB, and SorA, respectively) (31, 38, 39).
The ascochitine gene cluster also included a gene encoding a short peptide with a cupin domain (orf2) that is often found in secondary metabolite gene clusters (30, 40). The orf11 gene encodes a fungus-specific Zn(II)2Cys6-type transcription factor and exhibited the least sequence similarity with the counterpart gene (mrl3) in the citrinin gene cluster. Unlike the mrl3 gene, the deduced amino acid sequence of the orf11 gene lacked a DNA-binding domain and contained only the conserved domain (termed “middle homology region”) that is commonly found in Zn(II)2Cys6-type transcription factors (41). Interestingly, the sol4 transcription factor with only the middle homology region was shown to regulate the entire solanapyrone gene cluster in A. rabiei (29). This type of transcription factor without an apparent DNA-binding domain may activate genes by interacting with cognate repressor proteins, enabling timely induction of the repressed gene clusters.
When A. lentis and A. lentis var. lathyri are sexually crossed in vitro, they produce viable progeny, and segregation of molecular markers is normal, indicating that no intrinsic mating barrier exists between these taxa (42). However, pathogenic specialization of these two recently diverged species is evident (42). Although molecular phylogenies of protein-coding genes cannot clearly differentiate A. lentis from A. lentis var. lathyri (99.6% to 100% similar) (42), the two SNPs that caused a nonsense mutation in each taxon were consistent within each taxon, suggesting a prezygotic mating barrier and a lack of gene flow between the two probably sympatric species. Despite the presence of the nonsense mutation, transcripts of the pksAC gene in A. lentis isolate AL4 were detected on the basis of transcriptome sequencing (RNA-seq) data (see Fig. S1 in the supplemental material), suggesting that the gene is still active and likely undergoing pseudogenization. Compounds other than ascochitine were identified from cultures of A. lentis and A. lentis var. lathyri (15, 16, 43). Among the diverse mechanisms that produce genetic diversity in fungal secondary metabolism, nonsense mutation would be the fastest and the most effective, as the whole biosynthetic pathway of a compound can be disrupted by a single point mutation and indels in the responsible gene cluster (32, 44–48).
Expression of the pksAC gene in Ascochyta lentis isolate AL4. A schematic diagram of the pksAC gene is shown. Yellow arrows indicate exons, and black blocks represent RNA-seq reads mapped on the pksAC gene. Download FIG S1, PDF file, 0.01 MB (15KB, pdf) .
Copyright © 2019 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Production of ascochitine by P. clematidina was evident during the infection process, and the amount of ascochitine production in axenic culture was positively correlated with the ability to cause the disease among P. clematidina isolates (4). In contrast, there was no such correlation between the aggressiveness of activity on faba bean cultivars and the amount of ascochitine production among A. fabae isolates (49). In this study, we demonstrated that ascochitine was not involved in the pathogenicity of A. fabae. Solanapyrone, another phytotoxic compound produced by A. rabiei, was also proven not to be a pathogenicity factor (29, 50). Therefore, caution should be taken in claiming the involvement of secondary metabolites in pathogenicity based on in vitro tests of phytotoxicity.
It is always a challenging task to determine the role of secondary metabolites in an interaction with hosts or in the environment (51). However, the role of secondary metabolites may be deduced from their biological activity and spatiotemporal production patterns during the life cycle of the producing fungi. As with solanapyrone, ascochitine was produced during sporulation, while another polyketide, pinolidoxin, produced by A. pinodes, was detected mainly in young, advanced hyphae (52). Together with the reported antifungal activities, these observations suggest that ascochitine may play a role in competition with other fungi during saprobic growth stages in nature (2, 52). Genes involved in secondary metabolite production are generally believed to confer a fitness advantage to the producing organism (51, 53). Otherwise, the highly plastic fungal genomes would have purged out costly genes encoding megasize proteins such as PKS by selective pressure over the course of evolution. To end on a speculative note, some lineages of legume-associated Ascochyta may have lost the ability to produce ascochitine, a compound to which some saprobic fungi are insensitive (2). Studying the precise role of ascochitine in fungal ecology will provide an opportunity for tracing the fate of secondary metabolite biosynthesis gene clusters in species divergence.
MATERIALS AND METHODS
KS domain phylogeny.
The predicted KS domains of 51 PKS genes with known polyketide products plus a putative ascochitine PKS gene (pksAC) were aligned using the MUSCLE program (v3.8.31) (54) (see Text S1 in the supplemental material). Subsequently, spurious sequences or poorly aligned regions from the 52 KS domains were automatically removed, using the trimAl program (v1.2) (55) with the following option argument: “-gappyout.” A maximum likelihood phylogeny based on a general empirical model (WAG) (56) was constructed using the Phangorn R package (v2.4.0) (57). Nodal support was evaluated by 1,000 bootstrap replications using nearest-neighbor interchange rearrangements.
Gene replacement.
The pksAC gene in A. fabae isolate AF1 (WT) was deleted, using the split marker method (58). The DNA fragments used to construct the split marker were amplified using double-joint PCR (59). In the first round of PCR, a 756-bp upstream region of the pksAC coding sequence was amplified using primer pair L5 and L3, and a 947-bp downstream region of the pksAC coding sequence was amplified using primer pair R5 and R3. A 1,372-bp hph cassette was amplified from pDWJ5 plasmid (60) using primer pair HYG-F and HYG-R. The L3 and R5 primers carried 27-bp sequence tails that overlapped the 5′ and 3′ ends of the hph cassette, respectively. In the second round of PCR, the flanking DNA fragments were fused to the hph cassette through PCR by overlap extension. In the third round of PCR, the resulting PCR product was used as a template to generate the split-marker constructs that were amplified using primer pair N5 and HY-R and primer pair N3 and YG-F for the upstream and downstream flanking DNA fragments, respectively. Preparation of fungal protoplasts and transformation were conducted as previously described (61) with a minor modification. Volumes of 1 to 2 μg of each upstream and downstream split-marker DNA construct were used for genetic transformation. Putative transformants were subcultured on peptone-dextrose agar (PDA) containing hygromycin B (200 μg/ml). The gene replacement was confirmed by PCR with primer pair L5 and R3. Primers used in this study are listed in Table S1 in the supplemental material.
Primers used in this study. Download Table S1, DOCX file, 0.01 MB (14.8KB, docx) .
Copyright © 2019 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LC-MS analyses.
The WT and ΔpksAC mutant were grown on PDA for 14 days. Whole-agar chunks with fungal colonies were excised, soaked in 10 ml of ethyl acetate (EtOAc) in 50-ml Falcon tubes, and incubated at 4°C overnight. An aliquot of 200 μl of the extracts was transferred to a microcentrifuge tube, evaporated to dryness, reconstituted with 30 μl of MeOH, and subjected to LC-MS analyses. Chromatographic separation was achieved using an Acquity ultraperformance liquid chromatography (UPLC) system (Waters Corp., Milford, MA, USA), as previously described (50). MS analysis was performed on an inline Synapt G2-S high-definition mass spectrometry (HDMS; Waters Corp.) time of flight mass spectrometer using the positive-ion electrospray mode for data collection. The UV data were acquired using a wavelength of 210 to 400 nm, resolution of 1.2 nm, and 20 points/s with an Acquity photodiode array detector (Waters Corp.).
Synonymous-nonsynonymous analysis.
SNAP (v2.1.1) (62) was used to estimate synonymous and nonsynonymous substitution rates and to calculate the ratio of synonymous to nonsynonymous substitutions between pksAC homologs in A. fabae AF247/15, A. viciae-villosae AV22, A. lentis AL4, and A. lentis var. lathyri ER1415 isolates. Since the genome assembly in the pksAC gene locus in A. lentis var. lathyri ER1415 was incomplete (with two N stretches [176 bp and 111 bp] of gap sequence in the coding region), the corresponding 96 codons from the other pksAC genes were excluded in this analysis.
Pathogenicity tests.
Two faba bean strains (accession no. P1 667234 and PI 358266) were planted in Deepots (Stuewe & Sons, Inc., Corvallis, OR, USA) (6 by 25 cm) in a growth chamber. The inoculum was prepared by harvesting conidia (asexual spores) from 2-week-old fungal colonies on V8 agar media and was adjusted to 2 × 105 conidia/ml. Two-week-old plants were sprayed with inoculum until runoff (approximate 2 ml per plant) and were immediately covered with an inverted translucent plastic cup to form a minidome to produce uniformly high levels of relative humidity for 24 h to facilitate infection. Plants were then placed in a growth chamber (Conviron model PGR 15, Winnipeg, MB, Canada) with settings of 12 h day (20°C) and 12 h night (16°C) at 100% relative humidity. Control plants were sprayed with water but were otherwise treated in the same manner as the inoculated plants. At 2 weeks after inoculation, disease scores were determined and numbers of lesions were measured, as previously described (63).
MALDI imaging analysis.
Ascochyta fabae isolate AF1 and A. viciae-villosae isolate AV1 (33) was grown on half-strength PDA in which a microscopic slide was embedded. After 2 weeks of growth, the embedded slide with the fungal colony growing on the thin layer of PDA (about 2 mm in diameter) was removed from the plate and dried for 1 h at 65°C for MALDI imaging analysis. The dried specimen on the slide was sprayed with 20 mg/ml of 2,5-dihydroxybenzoic acid (in a mixture that also contained 0.1% trifluoroacetic acid and 50% methanol) as a matrix, using a TM-sprayer system (HTX Technologies, Carrboro, NC, USA) with the following parameters: 12 passes; track spacing, 1.2 mm; nozzle temperature, 70°C; linear velocity, 110 cm/min; flow rate, 60 μl/min. A selected area of the specimen was analyzed on a 9.4T Fourier transform ion cyclotron resonance (FTICR) MS instrument (Bruker Daltonics, Billerica, MA, USA) in positive-ion mode. Data were acquired using ftmsControl software (Bruker Daltonics). Approximately 100 shots per spot were acquired with a Smartbeam II Nd:YAG laser using repetition rate of 1 kHz. Image acquisition was carried out using flexImaging 4.1 (Bruker Daltonics), and data were normalized to the total ion current. Ascochitine standard compound purified in our previous study (8) was spotted, and reference MS signals were obtained in a simultaneous run.
Data accessibility.
The genome assemblies for A. fabae isolate AF247/15 (GCA_004335285.1), A. lentis isolate AL4 (GCA_004011705.1), and A. viciae-villosae AV22 (deposited as ONG-16-641; GCA_004335205.1) were retrieved from the NCBI database. The ascochitine gene cluster orf1 to orf11 in A. fabae isolate AF247/15 was deposited in NCBI, and the data are accessible through accession no. MN052622 to MN052632.
ACKNOWLEDGMENTS
We thank Alessandro Infantino (CREA, Rome, Italy), Walter Kaiser (USA), Jenny Davidson and Rohan Kimber (SARDI, Australia), and Rebecca Ford (Griffiths University, Australia) for providing Ascochyta isolates. We also thank Yung-Chun Chen for technical assistance in pathogenicity testing and Anna Berim and David Gang (Washington State University, USA) for performing LC-MS analyses. We appreciate the assistance of Julie Lawrence in generating the first genome assembly drafts for Ascochyta isolates AF1, AF55/10, AV22, AL4, AL1, Kewell, and ER1415.
This publication is based on work supported in part by a U.S. Dry Pea and Lentil Council grant (to W.C.) and by Australian Grains Research and Development Corporation grants GRDC CUR00014 and CUR00023/7 (to J.L.). J.L. and R.A.S. were supported by resources provided by the Pawsey Supercomputing Centre with funding from the Government of Australia and the Government of Western Australia. A.H.W. was supported by resources provided by the National Computational Infrastructure Specialised Facility for Bioinformatics (NCI-SF Bioinformatics).
Author contributions were as follows: conceptualization, W.K. and W.C.; investigation, W.K.; resources, J.L. and T.L.P., data acquisition, W.K., J.L., R.A.S., and A.H.W.; data analysis, W.K., J.L., R.A.S., and A.H.W.; original draft preparation, W.K.; review and editing, J.L., R.A.S., T.L.P., and W.C.; funding acquisition, J.L. and W.C.
REFERENCES
- 1.Keller NP, Turner G, Bennett JW. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol 3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 2.Oku H, Nakanishi T. 1964. Reductive detoxification of an antibiotic, ascochitine, by an insensitive fungus, Fusarium lycopersici Sacc. Naturwissenschaften 51:538–538. doi: 10.1007/BF00601453. [DOI] [Google Scholar]
- 3.Nakanishi T, Oku H. 1969. Mechanism of selective toxicity: absorption and detoxication of an antibiotic, ascochitine, by sensitive and insensitive fungi. Phytopathology 59:1563–1565. [PubMed] [Google Scholar]
- 4.Smith GR, Munro MHG, Fineran BA, Cole A. 1994. Evidence for the involvement of ascochitine in phoma leafspot-wilt disease of Clematis. Physiol Mol Plant Pathol 45:333–348. doi: 10.1016/S0885-5765(05)80063-3. [DOI] [Google Scholar]
- 5.Bertini S. 1956. Su di un composto ad antibiotica prodotto da Ascochyta pisi Lib. Annali Sperimentaz Agraria (Roma) 11:545–556. [Google Scholar]
- 6.Oku H, Nakanishi T. 1963. A toxic metabolite from Ascochyta fabae having antibiotic activity. Phytopathology 53:1321–1325. [Google Scholar]
- 7.Hernandez-Bello MA, Chilvers MI, Akamatsu H, Peever TL. 2006. Host specificity of Ascochyta spp. infecting legumes of the Viciae and Cicerae tribes and pathogenicity of an interspecific hybrid. Phytopathology 96:1148–1156. doi: 10.1094/PHYTO-96-1148. [DOI] [PubMed] [Google Scholar]
- 8.Kim W, Peever TL, Park J-J, Park C-M, Gang DR, Xian M, Davidson JA, Infantino A, Kaiser WJ, Chen W. 2016. Use of metabolomics for the chemotaxonomy of legume-associated Ascochyta and allied genera. Sci Rep 6:20192. doi: 10.1038/srep20192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seibert SF, Eguereva E, Krick A, Kehraus S, Voloshina E, Raabe G, Fleischhauer J, Leistner E, Wiese M, Prinz H, Alexandrov K, Janning P, Waldmann H, König GM. 2006. Polyketides from the marine-derived fungus Ascochyta salicorniae and their potential to inhibit protein phosphatases. Org Biomol Chem 4:2233–2240. doi: 10.1039/b601386d. [DOI] [PubMed] [Google Scholar]
- 10.Venkatasubbaiah P, Chilton WS. 1992. Phytotoxins of Ascochyta hyalospora, causal agent of lambsquarters leaf spot. J Nat Prod 55:461–467. doi: 10.1021/np50082a010. [DOI] [Google Scholar]
- 11.Chen Q, Jiang JR, Zhang GZ, Cai L, Crous PW. 2015. Resolving the Phoma enigma. Stud Mycol 82:137–217. doi: 10.1016/j.simyco.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Gruyter J, Woudenberg JHC, Aveskamp MM, Verkley GJM, Groenewald JZ, Crous PW. 2013. Redisposition of phoma-like anamorphs in Pleosporales. Stud Mycol 75:1–36. doi: 10.3114/sim0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aveskamp MM, de Gruyter J, Woudenberg JHC, Verkley GJM, Crous PW. 2010. Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Stud Mycol 65:1–60. doi: 10.3114/sim.2010.65.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Q, Hou LW, Duan WJ, Crous PW, Cai L. 2017. Didymellaceae revisited. Stud Mycol 87:105–159. doi: 10.1016/j.simyco.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andolfi A, Cimmino A, Villegas-Fernández AM, Tuzi A, Santini A, Melck D, Rubiales D, Evidente A. 2013. Lentisone, a new phytotoxic anthraquinone produced by Ascochyta lentis, the causal agent of Ascochyta blight in Lens culinaris. J Agric Food Chem 61:7301–7308. doi: 10.1021/jf4026663. [DOI] [PubMed] [Google Scholar]
- 16.Masi M, Nocera P, Boari A, Cimmino A, Zonno MC, Infantino A, Vurro M, Evidente A. 2018. Lathyroxins A and B, phytotoxic monosubstituted phenols isolated from Ascochyta lentis var. lathyri, a fungal pathogen of grass pea (Lathyrus sativus). J Nat Prod 81:1093–1097. doi: 10.1021/acs.jnatprod.7b01034. [DOI] [PubMed] [Google Scholar]
- 17.Shimizu T, Kinoshita H, Ishihara S, Sakai K, Nagai S, Nihira T. 2005. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Appl Environ Microbiol 71:3453–3457. doi: 10.1128/AEM.71.7.3453-3457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimizu T, Kinoshita H, Nihira T. 2007. Identification and in vivo functional analysis by gene disruption of ctnA, an activator gene involved in citrinin biosynthesis in Monascus purpureus. Appl Environ Microbiol 73:5097–5103. doi: 10.1128/AEM.01979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai K, Kinoshita H, Shimizu T, Nihira T. 2008. Construction of a citrinin gene cluster expression system in heterologous Aspergillus oryzae. J Biosci Bioeng 106:466–472. doi: 10.1263/jbb.106.466. [DOI] [PubMed] [Google Scholar]
- 20.Li Y-P, Xu Y, Huang Z-B. 2012. Isolation and characterization of the citrinin biosynthetic gene cluster from Monascus aurantiacus. Biotechnol Lett 34:131–136. doi: 10.1007/s10529-011-0745-y. [DOI] [PubMed] [Google Scholar]
- 21.He Y, Cox RJ. 2016. The molecular steps of citrinin biosynthesis in fungi. Chem Sci 7:2119–2127. doi: 10.1039/c5sc04027b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colombo L, Gennari C, Scolastico C, Aragozzini F, Merendi C. 1979. Biosynthesis of ascochitine: incorporation studies with advanced precursors. J Chem Soc Chem Commun 1979:492–493. doi: 10.1039/c39790000492. [DOI] [Google Scholar]
- 23.Staunton J, Weissman KJ. 2001. Polyketide biosynthesis: a millennium review. Nat Prod Rep 18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 24.Cox RJ. 2007. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem 5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 25.Hopwood DA. 1997. Genetic contributions to understanding polyketide synthases. Chem Rev 97:2465–2498. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 26.Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci U S A 100:15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallo A, Ferrara M, Perrone G. 2013. Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins. Toxins (Basel) 5:717–742. doi: 10.3390/toxins5040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim W, Park J-J, Gang DR, Peever TL, Chen W. 2015. A novel type pathway-specific regulator and dynamic genome environments of a solanapyrone biosynthesis gene cluster in the fungus Ascochyta rabiei. Eukaryot Cell 14:1102–1113. doi: 10.1128/EC.00084-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim W, Cavinder B, Proctor RH, O'Donnell K, Townsend JP, Trail F. 2019. Comparative genomics and transcriptomics during sexual development gives insight into the life history of the cosmopolitan fungus Fusarium neocosmosporiellum. Front Microbiol 10:1247. doi: 10.3389/fmicb.2019.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zabala AO, Xu W, Chooi Y-H, Tang Y. 2012. Discovery and characterization of a silent gene cluster that produces azaphilones from Aspergillus niger ATCC 1015 reveal a hydroxylation-mediated pyran-ring formation. Chem Biol 19:1049–1059. doi: 10.1016/j.chembiol.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lind AL, Wisecaver JH, Lameiras C, Wiemann P, Palmer JM, Keller NP, Rodrigues F, Goldman GH, Rokas A. 2017. Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol 15:e2003583. doi: 10.1371/journal.pbio.2003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peever TL, Barve MP, Stone LJ. 2007. Evolutionary relationships among Ascochyta species infecting wild and cultivated hosts in the legume tribes Cicereae and Vicieae. Mycologia 99:59–77. doi: 10.1080/15572536.2007.11832601. [DOI] [PubMed] [Google Scholar]
- 34.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tietz JI, Mitchell DA. 2016. Using genomics for natural product structure elucidation. Curr Top Med Chem 16:1645–1694. doi: 10.2174/1568026616666151012111439. [DOI] [PubMed] [Google Scholar]
- 36.Fisch MK. 2013. Biosynthesis of natural products by microbial iterative hybrid PKS–NRPS. RSC Adv 3:18228–18247. doi: 10.1039/c3ra42661k. [DOI] [Google Scholar]
- 37.Carter RH, Garson MJ, Staunton J. 1979. Biosynthesis of citrinin: incorporation studies with advanced precursors. J Chem Soc Chem Commun 1979:1097–1098. doi: 10.1039/c39790001097. [DOI] [Google Scholar]
- 38.Chiang Y-M, Oakley CE, Ahuja M, Entwistle R, Schultz A, Chang S-L, Sung CT, Wang CCC, Oakley BR. 2013. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J Am Chem Soc 135:7720–7731. doi: 10.1021/ja401945a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salo O, Guzmán-Chávez F, Ries MI, Lankhorst PP, Bovenberg RAL, Vreeken RJ, Driessen A. 2016. Identification of a polyketide synthase involved in sorbicillin biosynthesis by Penicillium chrysogenum. Appl Environ Microbiol 82:3971–3978. doi: 10.1128/AEM.00350-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao S-S, Duan A, Xu W, Yu P, Hang L, Houk KN, Tang Y. 2016. Phenalenone polyketide cyclization catalyzed by fungal polyketide synthase and flavin-dependent monooxygenase. J Am Chem Soc 138:4249–4259. doi: 10.1021/jacs.6b01528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schjerling P, Holmberg S. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res 24:4599–4607. doi: 10.1093/nar/24.23.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Infantino A, Zaccardelli M, Costa C, Ozkilinc H, Habibi A, Peever T. 2016. A new disease of grasspea (Lathyrus sativus) caused by Ascochyta lentis var. lathyri. J Plant Pathol 98:541–548. [Google Scholar]
- 43.Masi M, Nocera P, Zonno MC, Tuzi A, Pescitelli G, Cimmino A, Boari A, Infantino A, Vurro M, Evidente A. 2018. Lentiquinones A, B, and C, phytotoxic anthraquinone derivatives isolated from Ascochyta lentis, a pathogen of lentil. J Nat Prod 81:2700–2709. doi: 10.1021/acs.jnatprod.8b00556. [DOI] [PubMed] [Google Scholar]
- 44.O'Hanlon KA, Cairns T, Stack D, Schrettl M, Bignell EM, Kavanagh K, Miggin SM, O'Keeffe G, Larsen TO, Doyle S. 2011. Targeted disruption of nonribosomal peptide synthetase pes3 augments the virulence of Aspergillus fumigatus. Infect Immun 79:3978–3992. doi: 10.1128/IAI.00192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campbell MA, Rokas A, Slot JC. 2012. Horizontal transfer and death of a fungal secondary metabolic gene cluster. Genome Biol Evol 4:289–293. doi: 10.1093/gbe/evs011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Proctor RH, Busman M, Seo J-A, Lee YW, Plattner RD. 2008. A fumonisin biosynthetic gene cluster in Fusarium oxysporum strain O-1890 and the genetic basis for B versus C fumonisin production. Fungal Genet Biol 45:1016–1026. doi: 10.1016/j.fgb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 47.Schumacher J, Gautier A, Morgant G, Studt L, Ducrot P-H, Pêcheur PL, Azeddine S, Fillinger S, Leroux P, Tudzynski B, Viaud M. 2013. A functional bikaverin biosynthesis gene cluster in rare strains of Botrytis cinerea is positively controlled by VELVET. PLoS One 8:e53729. doi: 10.1371/journal.pone.0053729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang P-K, Horn BW, Dorner JW. 2005. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet Biol 42:914–923. doi: 10.1016/j.fgb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 49.Beed FD, Strange RN, Onfroy C, Tivoli B. 1994. Virulence for faba bean and production of ascochitine by Ascochyta fabae. Plant Pathol 43:987–997. doi: 10.1111/j.1365-3059.1994.tb01648.x. [DOI] [Google Scholar]
- 50.Kim W, Park C-M, Park J-J, Akamatsu HO, Peever TL, Xian M, Gang DR, Vandemark G, Chen W. 2015. Functional analyses of the Diels-alderase gene sol5 of Ascochyta rabiei and Alternaria solani indicate that the solanapyrone phytotoxins are not required for pathogenicity. Mol Plant Microbe Interact 28:482–496. doi: 10.1094/MPMI-08-14-0234-R. [DOI] [PubMed] [Google Scholar]
- 51.Raffa N, Keller NP. 2019. A call to arms: mustering secondary metabolites for success and survival of an opportunistic pathogen. PLoS Pathog 15:e1007606. doi: 10.1371/journal.ppat.1007606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim W, Park J-J, Dugan FM, Peever TL, Gang DR, Vandemark G, Chen W. 2017. Production of the antibiotic secondary metabolite solanapyrone A by the fungal plant pathogen Ascochyta rabiei during fruiting body formation in saprobic growth. Environ Microbiol 19:1822–1835. doi: 10.1111/1462-2920.13673. [DOI] [PubMed] [Google Scholar]
- 53.Romero D, Traxler MF, López D, Kolter R. 2011. Antibiotics as signal molecules. Chem Rev 111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 57.Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Catlett NL, Lee B, Yoder OC, Turgeon BC. 2003. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet Newsl 50:9–11. doi: 10.4148/1941-4765.1150. [DOI] [Google Scholar]
- 59.Yu J-H, Hamari Z, Han K-H, Seo J-A, Reyes-Domínguez Y, Scazzocchio C. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 60.White D, Chen W. 2006. Genetic transformation of Ascochyta rabiei using Agrobacterium-mediated transformation. Curr Genet 49:272–280. doi: 10.1007/s00294-005-0048-8. [DOI] [PubMed] [Google Scholar]
- 61.Akamatsu HO, Chilvers MI, Stewart JE, Peever TL. 2010. Identification and function of a polyketide synthase gene responsible for 1,8-dihydroxynaphthalene-melanin pigment biosynthesis in Ascochyta rabiei. Curr Genet 56:349–360. doi: 10.1007/s00294-010-0306-2. [DOI] [PubMed] [Google Scholar]
- 62.Korber B. 2000. HIV signature and sequence variation analysis, p 55–72. In Rodrigo AG, Learn GH (ed), Computational analysis of HIV molecular sequences. Kluwer Academic Publishers, Netherlands. [Google Scholar]
- 63.Chen W, Coyne CJ, Peever TL, Muehlbauer FJ. 2004. Characterization of chickpea differentials for pathogenicity assay of ascochyta blight and identification of chickpea accessions resistant to Didymella rabiei. Plant Pathol 53:759–769. doi: 10.1111/j.1365-3059.2004.01103.x. [DOI] [Google Scholar]
- 64.Nguyen TT, Taylor PWJ, Brouwer JB, Pang ECK, Ford R. 2001. A novel source of resistance in lentil (Lens culinaris ssp. culinaris) to ascochyta blight caused by Ascochyta lentis. Austral Plant Pathol 30:211–215. doi: 10.1071/AP01021. [DOI] [Google Scholar]
- 65.Davidson J, Smetham G, Russ MH, McMurray L, Rodda M, Krysinska-Kaczmarek M, Ford R. 2016. Changes in aggressiveness of the Ascochyta lentis population in Southern Australia. Front Plant Sci 7:393. doi: 10.3389/fpls.2016.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The deduced amino acid sequences of the KS domains of fungal PKS genes. Download Text S1, DOCX file, 0.02 MB (26.6KB, docx) .
Copyright © 2019 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of the pksAC gene in Ascochyta lentis isolate AL4. A schematic diagram of the pksAC gene is shown. Yellow arrows indicate exons, and black blocks represent RNA-seq reads mapped on the pksAC gene. Download FIG S1, PDF file, 0.01 MB (15KB, pdf) .
Copyright © 2019 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S1, DOCX file, 0.01 MB (14.8KB, docx) .
Copyright © 2019 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The genome assemblies for A. fabae isolate AF247/15 (GCA_004335285.1), A. lentis isolate AL4 (GCA_004011705.1), and A. viciae-villosae AV22 (deposited as ONG-16-641; GCA_004335205.1) were retrieved from the NCBI database. The ascochitine gene cluster orf1 to orf11 in A. fabae isolate AF247/15 was deposited in NCBI, and the data are accessible through accession no. MN052622 to MN052632.