Summary
As modern humans dispersed from Africa throughout the world, they encountered and interbred with archaic hominins, including Neandertals and Densiovans [1,2]. Although genome-scale maps of introgressed sequences have been constructed [3–6], considerable gaps in knowledge remain about the functional, phenotypic, and evolutionary significance of archaic hominin DNA that persists in present day individuals. Here, we describe a comprehensive set of analyses that identified 126 high frequency archaic haplotypes as putative targets of adaptive introgression in geographically diverse populations. These loci are enriched for immune related genes (such as OAS1/2/3, TLR1/6/10, and TNFAIP3) and also encompass genes (including OCA2 and BNC2) that influence skin pigmentation phenotypes. Furthermore, we leveraged existing and novel large-scale gene expression datasets to show many positively selected archaic haplotypes act as expression QTL (eQTL), suggesting modulation of transcript abundance was a common mechanism facilitating adaptive introgression. Our results demonstrate that hybridization between modern and archaic hominins provided an important reservoir of advantageous alleles that enabled adaptation to Out-of-Africa environments.
Results and Discussion
Identifying adaptively introgressed loci in geographically diverse populations
Non-African individuals inherited ~2% of their genomes from Neandertal ancestors [1,7], and individuals of Melanesian ancestry have an additional 2%-4% of their genomes inherited from Denisovan ancestors [2,8]. Considerable progress has been made in cataloging Neandertal and Denisovan sequences that persist in modern individuals [3–6,9], but the consequences of hybridization remain poorly understood. Recent studies suggest that introgressed sequences experienced widespread purifying selection [3–6] and influence susceptibility to a broad spectrum of diseases [4,10]. Conversely, archaic admixture may have also resulted in the acquisition of advantageous alleles that allowed modern humans to adapt to emergent selective pressures as they dispersed into new environments. Indeed, several examples of adaptive introgression have been hypothesized [11,12], including a Denisovan like haplotype of the EPAS1 gene that confers adaptation to high-altitude in Tibetans [13]. Nonetheless, many important questions remain including the number of loci subjected to adaptive introgression, the population genetics characteristics of such loci, and what the functional and phenotypic consequences of adaptive Neandertal and Denisovan sequences are in modern humans.
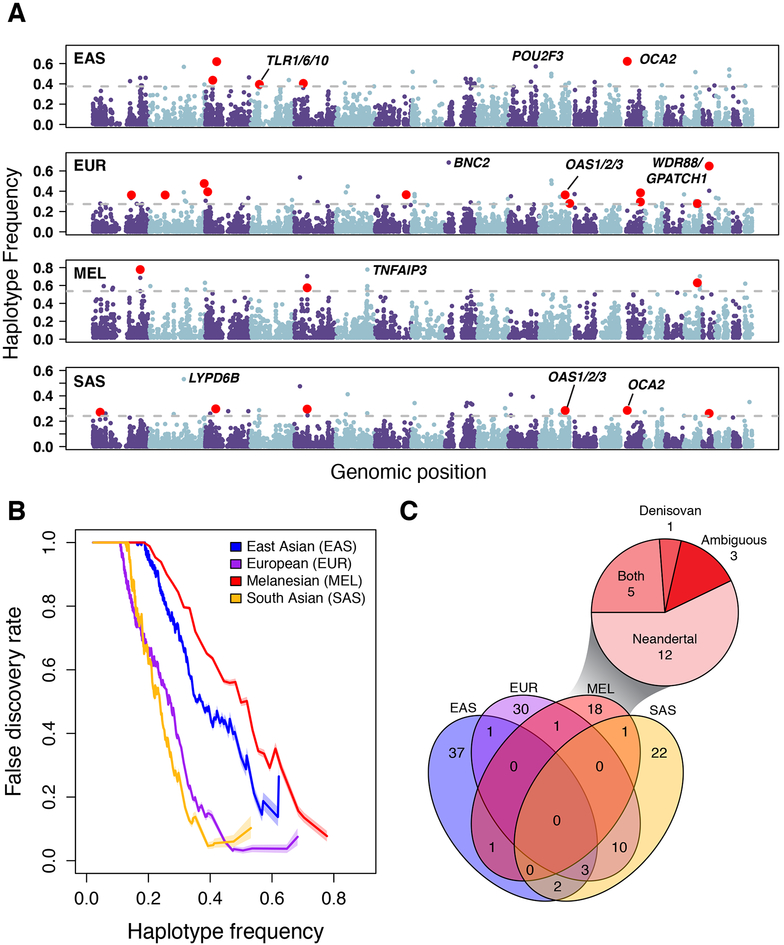
To more comprehensively understand how adaptive introgression has shaped patterns of human genomic variation, we leveraged recently constructed genome-scale maps of Neandertal and Denisovan sequences identified in 1,523 geographically diverse individuals [5] (Fig. 1a). Collectively, we analyzed 1.34Gb and 303Mb of Neandertal and Denisovan sequences, respectively, that segregates in 504 East Asian (EAS), 503 European (EUR), 489 South Asian (SAS), and 27 individuals from Island Melanesia (MEL). We first carefully identified SNPs that “tag” archaic haplotypes (Experimental Procedures), and performed extensive coalescent simulations to compare the number of observed high frequency haplotypes to neutral expectations across a range of demographic models (Experimental Procedures; Figure S1). Consistent with previous studies [14,15], our simulations suggest that simple outlier approaches are effective in enriching for positively selected loci, although the false discovery rate (FDR) can be high (Fig. 1b). For example, at a FDR ≤50% (corresponding to extreme outliers in the >99th percentile of the empirical frequency distribution) we identify 126 candidate adaptively introgressed loci (44, 45, 22, and 38 in EAS, EUR, MEL, and SAS, respectively; Table S1). Thus, we estimate that there are on the order of 10–20 true cases of adaptive introgression per population, and this estimate is robust to different FDR thresholds (Supplemental Experimental Procedures). Note, recent work [16,17] suggests Neandertal sequences were on average deleterious in modern human backgrounds, which is reflected in the positive correlation of B-values [18] and introgressed haplotype frequency (P = 3.×10−5, 1.9×10−05, 3.4×10−05, 1.5×10−05 for East Asians, Europeans, Melanesians, and South Asians, respectively), we therefore included mild purifying selection against introgressed sequences in our simulation framework. However, our inferences are robust if Neandertal sequences are instead assumed to be on average neutral (Supplementary Experimental Procedures). Unless otherwise noted, we focus subsequent analyses on this set of 126 loci (and provide locus-specific estimates of FDR Table S1).
Figure 1. Genomic distribution and characteristics of high-frequency archaic haplotypes in geographically diverse populations.
A) Each dot represents the frequency and genomic position of an introgressed archaic haplotype. Loci above the grey lines correspond to putative targets of adaptive introgression (outliers in the ≥99th percentile; FDR ≤ 50%). Outlier loci that had a significant phenotypic association (GWAS or eQTL) are highlighted in red. B) Relationship between the archaic haplotype frequency threshold for identifying adaptively introgressed loci and FDR for each population. Shaded regions delimit 95% confidence intervals. C) Venn diagram showing overlap of high frequency archaic haplotypes between populations. The inset pie chart shows how many of the Melanesian high frequency haplotypes are Neandertal, Denisovan, or Ambiguous. See also Figure S1 and Tables S1 and S2.
Of the 126 distinct archaic haplotypes that are found at unusually high frequencies, seven have previously been highlighted as putative targets of adaptive introgression [5,11,12]. High frequency archaic haplotypes either span or are proximal to 7 genes involved in skin traits and 31 genes involved in immunity, with significant GO enrichments for defense response (Benjamini and Hochberg corrected P = 8×10−4) and cytokine receptor activity (Benjamini and Hochberg corrected P = 3.64×10−6) among others (Table S2). We estimate the strength of selection acting on these loci to be ~10−3, which is an order of magnitude lower than selection coefficients associated with strong recent selective sweeps, such as loci that confer lactase persistence and malaria resistance [19,20] (Figure S1C).
Population characteristics of high frequency introgressed loci
Interestingly, 107 of the 126 distinct regions are at high frequency in only one population (Fig. 1c). For instance, 66% and 58% of archaic haplotypes are population specific in Europeans and South Asians, respectively, whereas 84% and 86% of haplotypes are population specific in East Asians and Melanesians. These data are consistent with additional distinct pulses of introgression into East Asians and Melanesians [5] (Fig. 1c).
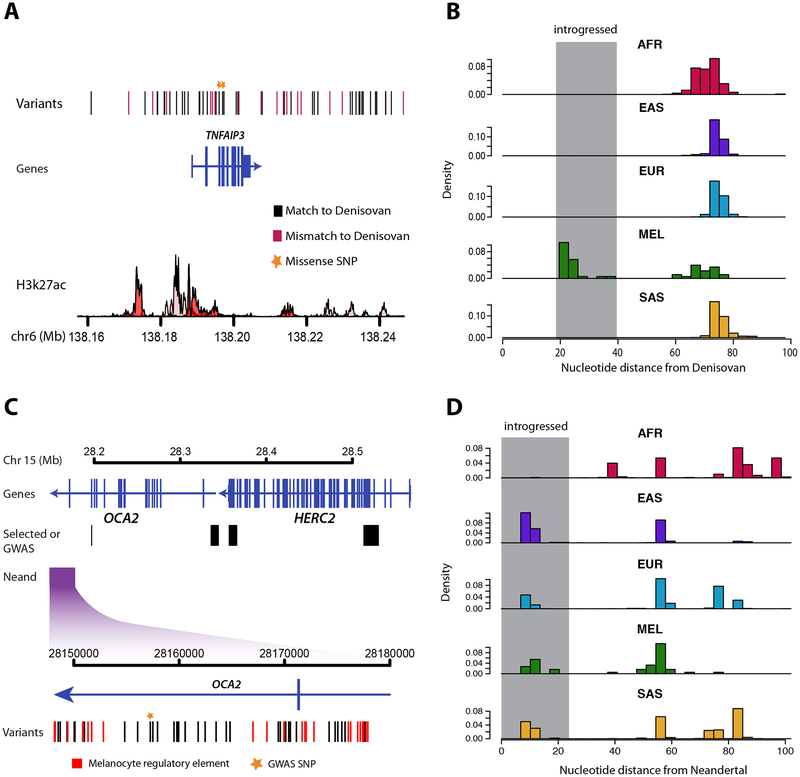
We next analyzed the number of high frequency archaic haplotypes that were inherited from Neandertals or Denisovans. As described in Vernot et al5, some archaic haplotypes exhibit high sequence similarity to both the Neandertal and Denisovan reference genomes and thus cannot be confidently labeled; we refer to these haplotypes as ambiguous. As expected, all high-frequency archaic haplotypes in Europeans, East Asians, and South Asians are of Neandertal origin. Strikingly, however, 59% of high frequency haplotypes in Melanesians are inherited solely from Neandertal, despite the fact that these individuals have considerably more Denisovan compared to Neandertal ancestry [5] (Fig. 1c). We also identified five regions segregating both Neandertal and Denisovan sequence (Figure S2; Table S1), including archaic haplotypes that are mostly Denisovan and span the TNFAIP3 (Fig. 2a–b), a ubiquitin-editing enzyme involved in the attenuation of cytokine-induced innate immune responses [21].
Figure 2. Adaptive introgression of archaic sequence at the TNFAIP3 and OCA2 loci.
A. Schematic of the TNFAIP3 region is shown with vertical bars indicating introgressed SNPs. Black and red denote matches and mismatches to the Denisovan reference genome, respectively. Two missense SNPs are highlighted with stars. The track along the bottom depicts H3K27ac signal from seven ENCODE cell types [41]. B. Distributions of absolute genetic distance to the Denisovan reference genome for all haplotypes within the four populations studied, as well as Africans. The grey box indicates the portion of the distribution comprised of introgressed sequence. C. Schematic of the OCA2/HERC2 region. The purple box indicates the introgressed region, and the black boxes indicate previously identified positively selected and pigment associated regions in East Asians and Europeans. Below, a zoomed view of the introgressed region is shown with vertical bars indicating introgressed variants. Variants that overlap melanocyte regulatory elements are shown in red and GWAS study variants are indicated with a yellow star. D. Distributions of absolute genetic distance to Neandertal for all haplotypes within the four populations studied as well as Africans. The grey box indicates the portion of the distribution comprised of introgressed sequence. See also Figure S2.
Inferring the functional effects of high frequency loci
To better understand the phenotypic consequences of high frequency archaic haplotypes, we analyzed previously published GWAS results [22]. We found that 10 haplotypes are associated with 17 traits, including breast and nasopharyngeal carcinoma, bone abnormalities (Paget’s disease), Celiac Disease, rheumatoid arthritis, optic disk size, and atopic dermatitis (Table S1).
The median length of putative adaptively introgressed haplotypes is 81kb. Thus, compared to analyses of recent selective sweeps that typically identify large genomic regions, adaptively introgressed sequences often result in single gene resolution [9]. 59% of loci overlap protein-coding genes, and there are 49 protein-coding variants in 36 distinct genes. However, 80% of high frequency haplotypes contain no protein-coding variants, indicating that regulatory evolution is the prominent mechanism through which adaptively introgressed sequences act. Given the likelihood that many high frequency archaic haplotypes influence patterns of gene expression, we leveraged extensive RNA-seq data from the GTEx [23] and Geuvadis [24] projects to identify expression quantitative trait loci (eQTL; see Supplemental Experimental Procedures). Of the 48 high-frequency introgressed haplotypes that could be tested, 13 act as eQTLs to 34 different genes across multiple tissues (Permutation FDR = 0.05; Fig. 1a).
Adaptive introgression of archaic OCA2 haplotypes is associated with pigmentation traits
Notably, the highest frequency introgressed haplotype in East Asians (62%) encompasses a 29.7kb region of the OCA2 gene (Fig. 2c), and is also found at appreciable frequencies in South Asians (29%), Europeans (20%), and Melanesians (35%). OCA2 encodes a transmembrane protein involved in iris, skin, and hair pigmentation [25], and both coding and non-coding variants are under strong selection in Europeans and East Asians [26]. The introgressed haplotype contains an average of 10.6 differences with Neandertal (Fig. 2d). In contrast, African haplotypes contain an average of 74.3 differences, with the exception of four haplotypes that appear introgressed, likely a result of recent gene flow [27] (Fig. 2d). We used a coalescent framework to calculate the probability that a haplotype of this length and divergence from Neandertal is caused by incomplete lineage sorting as opposed to introgression (Supplemental Experimental Procedures), and across a wide range of parameters can robustly reject the hypothesis of ILS (P < 0.003). The introgressed haplotype does not overlap, nor is it in LD (maximum r2 < 0.03), with variants that have previously been inferred to be targets of positive selection or are most strongly associated with pigmentation traits (Fig. 2c). However, it does contain a variant that is associated with blue versus brown eyes in Europeans (P = 4×10−10) [28] (Fig. 2c). Finally, although there are no coding variants on the Neandertal haplotype, 25 introgressed variants overlap regulatory elements active in melanocytes (Fig. 2c). Thus, these data are consistent with the hypothesis of recurrent positive selection acting on multiple variants of OCA2, some of which arose in modern humans and some that were inherited through hybridization with Neandertals.
An archaic haplotype encompassing three OAS genes is associated with gene expression and isoform usage
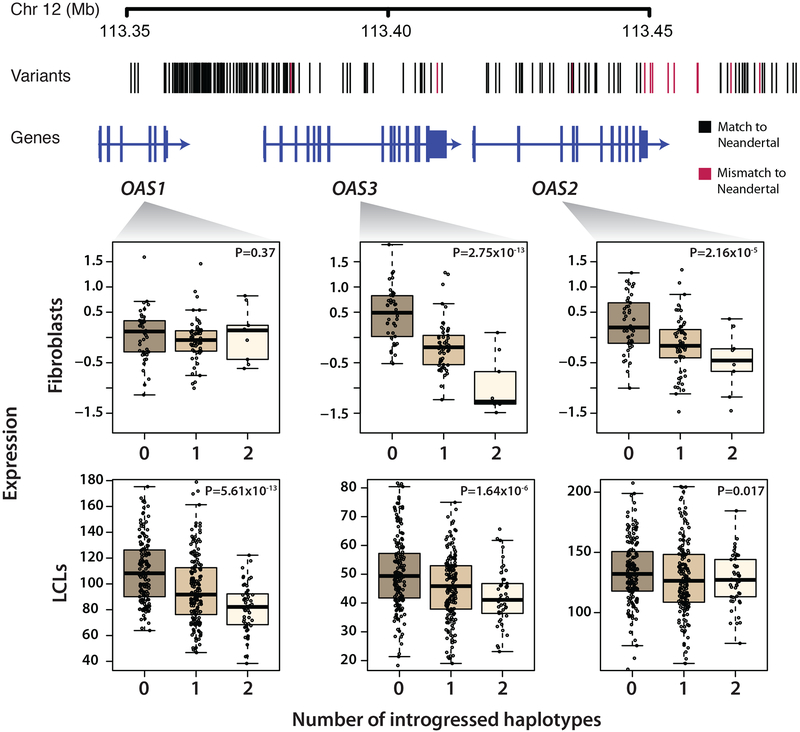
Our eQTL analyses provide significant new insights into both novel and previously hypothesized targets of adaptive introgression. For instance, Mendez et al. [29] proposed a Neandertal haplotype encompassing the genes OAS1, OAS2, and OAS3, which encode for antiviral proteins [30], was driven to high frequency by positive selection. Although this haplotype is only an outlier in Europeans, it is also found at appreciable frequencies in other populations (17%, 13%, and 13% in East Asians, Melanesians, and South Asians, respectively). This introgressed haplotype is strongly associated with expression of all three OAS genes across various tissues (Fig. 3). When looking at expression aggregated across isoforms, the eQTL is specific to OAS2 and OAS3 in transformed fibroblasts and OAS1 and OAS3 in transformed lymphoblast cell lines (LCLs), with individuals containing the introgressed allele show lower expression in all cases (Fig. 3). Further analyses of exon-level expression suggests the Neandertal haplotype results in differential splicing of OAS1 and OAS2 (Figure S3). In particular, the introgressed haplotype contains a 3’ splice variant between exons 6 and 7 of OAS1, leading to the production of a higher activity isoform of OAS1 [31]. It is important to note that the introgressed haplotype also harbors protein-coding variants that could be targets of selection (Table S3).
Figure 3. Introgression at the OAS locus and its impact on gene expression.
A schematic of the OAS1/2/3 region is shown with vertical bars indicate introgressed variants. Black and red denote matches and mismatches to the Neandertal reference genome, respectively. Below, gene expression for OAS1, OAS2, and OAS3 is shown stratified by the number of Neandertal haplotypes an individual has in fibroblasts and LCLs. See also Figure S3 and Table S3.
Tissue specific eQTL effects of an introgressed TLR1/6/10 haplotype
Our eQTL analyses reveal novel insights into the recently discovered TLR1/6/10 haplotype [11,32] inherited from Neandertals, which is at high frequency (39%) in East Asians and intermediate frequency in other populations (22%, 6%, and 17% in Europeans, Melanesians, and South Asians, respectively; Fig. 4a). Toll-like receptors play a key role in the innate immune system, and TLR1 and TLR6 are well-characterized non-viral receptors [33], whereas TLR10 has only recently been identified as a possible receptor for influenza [34] and other pathogens [35]. The introgressed haplotype contains 25 variants that have been associated with several immune traits, including helicobacter pylori status [36], allergy burden [37], and cellular response to Pam3CSK4 [38] (a TLR1 agonist). Consistent with previous results [11], the introgressed haplotype results in significantly higher expression (P < 2×10−5) of all three genes in transformed lymphoblast cell lines (LCLs; Fig. 4b, Figure S4). However, the Neandertal haplotype is associated with significantly lower expression of TLR6 (P < 0.019) in transformed fibroblasts and primary B cells from healthy volunteers31 (Fig. 4b). The differential effect in B cells is particularly interesting because they are closely related to the LCLs, differing only in their lack of viral transformation. Finally, TLR6 expression in other GTEx cell types does not show significant association with the Neandertal haplotype (data not shown).
Figure 4. Patterns of eQTL effects for the introgressed TLR1/6/10 haplotype across multiple cell types.
A. Schematic of the TLR1/6/10 region is shown with vertical bars indicating introgressed variants. Black and red denote matches and mismatches to the Neandertal reference genome, respectively. B. Gene expression for TLR6 in three different cell types is shown stratified by the number of Neandertal alleles each sample has. P values are indicated in each plot. The table below corresponds to each cell type, and check marks indicate characteristics of each cell type. C. Gene expression for TLR6 is shown for whole blood samples before and after stimulation with LPS. Expression is stratified by the number of Neandertal alleles each sample has. See also Figure S4.
We hypothesized that the tissue specific patterns of eQTL observed for the Neandertal haplotype reflects differential states of innate immune activation. To test this hypothesis, we measured expression levels in whole blood samples from healthy volunteers before and after stimulation with LPS (lipopolysaccharide), a TLR4 agonist (see Supplemental Experimental Procedures). Before stimulation, the Neandertal haplotype showed no association with levels of TLR1, TLR6, or TLR10 expression (P > 0.05; Fig. 4c, Figure S4A). However, after stimulation both TLR10 and TLR6 show a significant positive association between expression and number of Neandertal alleles (P = 0.003; Fig. 4c, Figure S4A). The effect is modest, but is likely attenuated due to the low proportion of immune cells in whole blood. These data are consistent with the hypothesis that the introgressed Neandertal haplotype influences TLR expression in a context-dependent manner, increasing TLR6 expression specifically in stimulated immune cells (Fig. 4b). Fine-scale mapping suggests that the causal regulatory variant, or variants, may fall within the promoter of TLR1 or TLR10 (Figure S4B).
Conclusions
In summary, hybridization with Neandertals and Denisovans provided an important reservoir of advantageous mutations for modern humans that enabled adaptation to emergent selective pressures as they dispersed out-of-Africa. Our results show that immune and pigmentation traits were frequent substrates of adaptive introgression, and that in many cases adaptive archaic haplotypes also contribute to the disease susceptibility in contemporary individuals (Fig. 1a; Table S1). Finally, our ability to interpret adaptively introgressed loci was facilitated by large-scale functional genomics and GWAS data [22–24]. However, many of these data sets were generated in individuals of European ancestry, and thus better geographic representation would accelerate efforts to understand adaptively introgressed loci, and more generally, human genomic diversity.
Experimental Procedures
Description of samples
We analyzed recent genome-scale maps of Neandertal and Denisovan sequences constructed with high coverage whole genome sequence data from 1,523 geographically diverse individuals [5]. These individuals included 504 East Asian (EAS), 503 European (EUR), 489 South Asian (SAS) samples sequenced as part of phase 3 release of the 1000 Genomes Project [39], as well as 27 additional individuals from 11 sampling locations in the Bismark Archipelago of Northern Island Melanesia, Papua New Guinea. In total we analyzed 1.34Gb and 303Mb of Neandertal and Denisovan sequences, respectively.
Estimating introgressed haplotype frequencies
We began with the complete set of phased SNPs that tag archaic haplotypes identified in any individual, analyzing each population separately. We filtered these SNPs to include only those that match Neandertal (or the correct archaic sequence in the case of Melanesians), and required that each of these tag SNPs belong to a 50kb window with at least two other tag SNPs. Then, in order to aggregate SNPs into cohesive haplotypes, we calculated LD among tag SNPs using phased 1000 Genomes data [39] for each population and clustered SNPs with r2 ≥ 0.3. We used VCFtools [40] to calculate r2 statistics. After obtaining these initial haplotypes, we extended each haplotype by identifying all additional SNPs at r2 ≥ 0.8 with any tag SNPs on the haplotype. Finally, we filtered any haplotypes that had less than 5 tag SNPs, less than 10 total SNPs, or were shorter than 10kb. Unless otherwise specified, when referring to “variants on a haplotype”, we are referring to this final set of tag SNPs and variants in LD with tag SNPs. To estimate the frequency of each haplotype, we calculated the median frequency of all tag SNPs on a haplotype in each population separately.
Coalescent Simulations
We developed a two-part approximate likelihood coalescent simulation framework to a) estimate the false discovery rate (FDR) for adaptive introgression across haplotype frequencies, and b) estimate selection coefficients for adaptively introgressed haplotypes. Briefly, in the first step we simulate introgression across a wide variety of demographic models in order to identify the demographic model that best matches the observed data in each population, and thus allowed us to calculate a FDR for the observed data. In the second step, we use this demographic model to simulate introgression under a range of selection coefficients to estimate the maximum likelihood estimate of the selection coefficient across all frequencies. See the Supplemental Experimental Procedures for detail.
Supplementary Material
Acknowledgements
We thank members of the Akey laboratory for helpful feedback and comments on the manuscript. This work is supported by NIH grant R01GM110068 to JMA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. (2010) A draft sequence of the Neandertal genome. Science. 328, 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, et al. (2010) Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 468, 1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Vernot B, Akey JM (2014) Resurrecting surviving Neandertal lineages from modern human genomes. Science. 343, 1017–1021. [DOI] [PubMed] [Google Scholar]
- [4].Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D (2014) The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 507, 354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, Dannemann M, Grote S, McCoy RC, Norton H, et al. (2016) Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 352, 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sankararaman S, Mallick S, Patterson N, Reich D (2016) The Combined Landscape of Denisovan and Neanderthal Ancestry in Present-Day Humans. Curr Biol. 26, 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. (2014) The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 505, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay, Prüfer K, de Filippo C, et al. (2012) A high-coverage genome sequence from an archaic Denisovan individual. Science. 338, 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vattathil S, Akey JM (2015) Small Amounts of Archaic Admixture Provide Big Insights into Human History. Cell. 163, 281–284. [DOI] [PubMed] [Google Scholar]
- [10].Simonti CN, Vernot B, Bastarache L, Bottinger E, Carrell DS, Chisholm RL, Crosslin DR, Hebbring SJ, Jarvik GP, Kullo IJ, et al. (2016) The phenotypic legacy of admixture between modern humans and Neandertals. Science. 351, 737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dannemann M, Andrés AM, Kelso J (2016) Introgression of Neandertal- and Denisovan-like Haplotypes Contributes to Adaptive Variation in Human Toll-like Receptors. Am J Hum Genet. 98, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Racimo F, Sankararaman S, Nielsen R, Huerta-Sánchez E (2015) Evidence for archaic adaptive introgression in humans. Nat Rev Genet. 16, 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Huerta-Sánchez E, Jin X, Asan, Bianba Z, Peter BM, Vinckenbosch N, Liang Y, Yi X, He M, Somel M, et al. (2014) Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 512, 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Teshima KM, Coop G, Przeworski M (2006) How reliable are empirical genomic scans for selective sweeps? Genome Res. 16, 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM (2006) Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 16, 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fu Q, Posth C, Hajdinjak M, Petr M, Mallick S, Fernandes D, Furtwängler A, Haak W, Meyer M, Mittnik A, et al. (2016) The genetic history of Ice Age Europe. Nature. 534, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Juric I, Aeschbacher S, Coop G The Strength of Selection Against Neanderthal Introgression. bioRxiv 2015:030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McVicker G, Gordon D, Davis C, Green P (2009) Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 5, e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vitti JJ, Grossman SR, Sabeti PC (2013) Detecting natural selection in genomic data. Annu Rev Genet. 47, 97–120. [DOI] [PubMed] [Google Scholar]
- [20].Fu W, Akey JM (2013) Selection and adaptation in the human genome. Annu Rev Genomics Hum Genet. 14, 467–489. [DOI] [PubMed] [Google Scholar]
- [21].Heyninck K, De Valck D, Vanden Berghe W, Van Criekinge W, Contreras R, Fiers W, Haegeman G, Beyaert R (1999) The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol. 145, 1471–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li MJ, Liu Z, Wang P, Wong MP, Nelson MR, Kocher J-PA, Yeager M, Sham PC, Chanock SJ, Xia Z, et al. (2016) GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 44, D869–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Consortium GTEx. (2015) The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lappalainen T, Sammeth M, Friedländer MR, ‘t Hoen PAC, Monlong J, Rivas MA, Gonzàlez-Porta M, Kurbatova N, Griebel T, Ferreira PG, et al. (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 501, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, Manolescu A, Karason A, Palsson A, Thorleifsson G, et al. (2007) Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 39, 1443–1452. [DOI] [PubMed] [Google Scholar]
- [26].Donnelly MP, Paschou P, Grigorenko E, Gurwitz D, Barta C, Lu R-B, Zhukova OV, Kim JJ, Siniscalco M, New M, et al. (2012) A global view of the OCA2-HERC2 region and pigmentation. Hum Genet. 131, 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sikora M, Carpenter ML, Moreno-Estrada A, Henn BM, Underhill PA, Sánchez-Quinto F, Zara I, Pitzalis M, Sidore C, Busonero F, et al. (2014) Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe. PLoS Genet. 10, e1004353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Jakobsdottir M, Steinberg S, Gudjonsson SA, Palsson A, Thorleifsson G, et al. (2008) Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 40, 835–837. [DOI] [PubMed] [Google Scholar]
- [29].Mendez FL, Watkins JC, Hammer MF (2013) Neandertal origin of genetic variation at the cluster of OAS immunity genes. Mol Biol Evol. 30, 798–801. [DOI] [PubMed] [Google Scholar]
- [30].Hornung V, Hartmann R, Ablasser A, Hopfner K-P. (2014) OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 14, 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bonnevie-Nielsen V, Field LL, Lu S, Zheng D-J, Li M, Martensen PM, Nielsen TB, Beck-Nielsen H, Lau YL, Pociot F (2005) Variation in antiviral 2’,5’-oligoadenylate synthetase (2”5”AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am J Hum Genet. 76, 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Deschamps M, Laval G, Fagny M, Itan Y, Abel L, Casanova JL, Patin E, Quintana-Murci L (2016) Genomic Signatures of Selective Pressures and Introgression from Archaic Hominins at Human Innate Immunity Genes. Am J Hum Genet. 98, 5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kawai T, Akira S (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 11, 373–84. [DOI] [PubMed] [Google Scholar]
- [34].Lee SMY, Kok K-H, Jaume M, Cheung TKW, Yip T-F, Lai JCC, Guan Y, Webster RG, Jin DY, Peiris JS (2014) Toll-like receptor 10 is involved in induction of innate immune responses to influenza virus infection. Proc Natl Acad Sci USA. 111, 3793–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nagashima H, Iwatani S, Cruz M, Jiménez Abreu JA, Uchida T, Mahachai V, Vilaichone RK, Graham DY, Yamaoka Y (2015) Toll-like Receptor 10 in Helicobacter pylori Infection. J Infect Dis. 212, 1666–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mayerle J, Hoed den CM, Schurmann C, Stolk L, Homuth G, Peters MJ, Capelle LG, Zimmermann K, Rivadeneira F, Gruska S, et al. (2013) Identification of genetic loci associated with Helicobacter pylori serologic status. JAMA. 309, 1912–1920. [DOI] [PubMed] [Google Scholar]
- [37].Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM, St Pourcain B, Ring SM, Mountain JL, Francke U, et al. (2013) A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 45, 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mikacenic C, Reiner AP, Holden TD, Nickerson DA, Wurfel MM (2013) Variation in the TLR10/TLR1/TLR6 locus is the major genetic determinant of interindividual difference in TLR1/2-mediated responses. Genes Immun. 14, 52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. (2015) A global reference for human genetic variation. Nature. 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. (2011) The variant call format and VCFtools. Bioinformatics. 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].ENCODE Project Consortium. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature. 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.